Abstract
Numerous age‐related human diseases have been associated with deficiencies in cellular energy production. Moreover, genetic alterations resulting in mitochondrial dysfunction are the cause of inheritable disorders commonly known as mitochondrial diseases. Many of these deficiencies have been directly or indirectly linked to deficits in mitochondrial gene expression. Transcription is an essential step in gene expression and elucidating the molecular mechanisms involved in this process is critical for understanding defects in energy production. For the past five decades, substantial efforts have been invested in the field of mitochondrial transcription. These efforts have led to the discovery of the main protein factors responsible for transcription as well as to a basic mechanistic understanding of the transcription process. They have also revealed various mechanisms of transcriptional regulation as well as the links that exist between the transcription process and downstream processes of RNA maturation. Here, we review the knowledge gathered in early mitochondrial transcription studies and focus on recent findings that shape our current understanding of mitochondrial transcription, posttranscriptional processing, as well as transcriptional regulation in mammalian systems.
Keywords: mitochondrial transcription, mitochondrial gene expression, POLRMT, TEFM, MTERF1, TFAM, TFB2M, mitochondrial RNA processing
1. INTRODUCTION
Mammalian mitochondria perform numerous essential cellular functions including heme and phospholipid biogenesis,1 the regulation of intracellular calcium signaling, apoptosis,2 reactive oxygen species production,3 and most importantly, energy production through oxidative phosphorylation. These processes are directly or indirectly linked to mitochondrial gene expression, the process by which mitochondrial DNA (mtDNA) is transcribed into RNA and subsequently translated into protein. The mammalian mitochondrial double stranded circular genome is ~16.5 Kilobases (Kb), encoding 13 polypeptide genes, 22 tRNAs for translation, and 2 ribosomal RNAs (rRNAs; Figure 1a). The rRNAs are key constituents of the mitochondrial ribosomes, exclusively dedicated to the translation of the 13 mitochondrial gene products.4, 5 Mitochondria need their own transcription and translation machineries because these two processes are mechanistically distinct from nuclear transcription or translation.6
Figure 1.
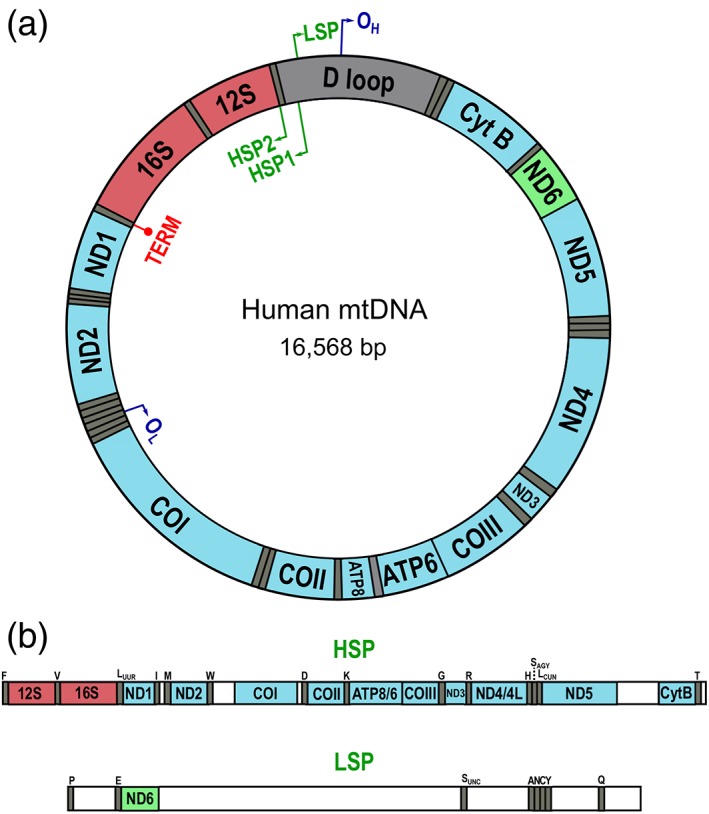
Organization of mitochondrial transcription. (a) Scheme of the mitochondrial genome indicating key functional loci as well as the different gene products. rRNAs (red) and most mRNAs (blue) are coded in the H‐strand, while a single mRNA (ND6; green) is coded by the L‐strand. tRNA genes are show in gray. (b) Scheme of the full‐length polycistronic mitochondrial transcripts indicating the gene products encoded by each transcript
This review will focus on mammalian mitochondrial transcription, covering findings spanning the last five decades and emphasizing recent observations not covered in previous reviews. We review the genomic organization of mitochondrial transcription, the current models for the layout of mitochondrial promoters, our molecular understanding of the transcription machinery, and the process of transcription, posttranscriptional processing as well as transcriptional regulation.
2. MITOCHONDRIAL GENOMIC STRUCTURE: HEAVY VERSUS LIGHT STRANDS
The human mitochondrial genome (mtDNA) is circular and composed of two strands, which were labeled heavy and light strands (H‐strand and L‐strand, respectively). The H‐strand denotes its high content in purine nucleotides compared to the L‐strand (owing mostly to a strong imbalance in the guanine/cytosine ratio), making the strands separable by using density centrifugation in alkaline CsCl2 gradients.7 Early mitochondrial transcription studies using DNA–RNA hybridization experiments showed that most mtRNAs hybridized with one of the DNA strands, suggesting an asymmetry in the utilization of both strands.8, 9 The mtDNA was first sequenced in 1981 by Anderson and colleagues.4 This made it clear that both strands encode genes that are transcribed, although very few genes are encoded in the L‐strand compared to the H‐strand. The H‐strand encodes 12 of the 13 polypeptides, the 2 ribosomal RNAs (12S and 16S) and 14 tRNAs, while the L‐strand encodes one polypeptide, eight tRNAs, and is the source of primer synthesis for mtDNA replication10 (Figure 1b).
It is important to note that numerous other mammalian genomes have been sequenced, and that their genomic organization appears highly conserved with the exception of minor rearrangements in tRNA genes.11 However, in vivo and in vitro functional studies have predominantly been carried out using the human or mouse systems. Thus, although the mechanisms of gene expression are expected to be conserved in all mammals, it is not possible to rule out that differences might exist across species.
3. mtDNA PROMOTERS
Early mitochondrial gene expression studies supported the presence of a single promoter on each strand leading to the transcription of two polycistronic transcripts that are later processed into single RNAs.12 The two promoters are found in the displacement loop (D‐loop) region of mtDNA.4 Termed heavy strand promoter (HSP) and light strand promoter (LSP), HSP and LSP initiate transcription of H‐strand and L‐strand, respectively. This model of mitochondrial transcription was supported by the work of Aloni and Attardi, who used pulse labeling as well as RNA–DNA hybridization followed by electron microscopy to show that the mtDNA is symmetrically transcribed from both strands.13 These early findings were strengthened by the research from three independent groups. These groups showed that the region of mtDNA around the origin of replication contains an initiation site for L‐strand transcription and an initiation site for H‐strand transcription.14, 15, 16
Subsequently, Montoya et al. carried out mapping experiments on the human mtDNA to identify the precise sites of transcription initiation on the H‐strand and L‐strand. They radioactively labeled the 5′ termini of mitochondrial poly(A) containing RNAs using a capping enzyme and mapped those sites by DNA–RNA hybridization. They identified two mapping sites for the capped 5′ ends: one on the H‐strand upstream of the tRNA‐Phe gene (HSP, now known as HSP1; Figure 1a) and another on the L‐strand near the 5′ terminus of the 7S RNA coding sequence (LSP; Fig. 1a). They then confirmed these mapping sites using the 5′ ends of newly synthesized transcripts from in vitro mtDNA transcription experiments.17 Surprisingly, their mapping experiments identified another initiation site on the H‐strand near the 5′ terminus of the 12S rRNA gene (now known as HSP2; Figure 1a). Thus, they proposed a three‐promoter model, consisting of LSP on the L‐strand and HSP1/HSP2 on the H‐strand. The two sites on the H‐strand were interpreted to be responsible for two distinct transcription events: HSP1 primary transcripts would originate at position 561, 16 bp upstream of the tRNA‐Phe gene18, 19 and terminate at the 3′ end of the 16S rRNA,19 consisting of the tRNA‐Phe, the 12S rRNA, the tRNA‐Val, and the 16S rRNA20; HSP2 transcripts would originate at position 646, 2 bp upstream of the 12S rRNA gene19 and proceed to generate near full‐length transcripts, although the precise 3′ end of this transcript was not determined.17, 18 This interpretation is however not consistent with later evidence (see Section 7). Research from Clayton's lab also supported the existence of two HSPs.21, 22 However, it is worth noting that the existence of HSP2 has been contested for various reasons. Transcription from HSP2 was demonstrated in cell‐based studies and remained primarily unreproducible in vitro using recombinant proteins, raising the question of whether it is a functional promoter in vivo.23 In vitro HSP2 transcription was later independently achieved by two research groups, and shown to be highly dependent on the reaction conditions: POLRMT and TFB2M were necessary and sufficient to initiate transcription from HSP2 and the presence of TFAM inhibited transcription from this site,24, 25 suggesting that TFAM could participate in a feedback mechanism to regulate transcription from the two strands of mtDNA. However, the experimental conditions used to observe initiation from HSP2 in vitro have been suggested to be conducive to unspecific initiation.26 Additional support for the existence of HSP2 comes from recent genomic analysis of mitochondrial RNAs.27 However, the existence and functional significance of HSP2 initiation remain open questions in the field.
4. TRANSCRIPTION MACHINERY
The mammalian mitochondrial transcription machinery is mechanistically unique. Although it is not possible to rule out the existence of additional yet unidentified factors, the core transcription machinery consists of an RNA polymerase (RNAP; POLRMT), an initiation factor (TFAM), an activator (TFB2M), an elongation factor (TEFM), and a termination factor (MTERF1). All these proteins are highly conserved in mammals (although TFB2M displays a larger degree of divergence), suggesting that they play similar roles across species.
Our understanding of the mechanism of transcription mostly derives from biochemical and structural observations, but the models are also influenced by the phenotypes observed upon genetic manipulation of the relevant genes. Transcription initiation from all three promoters, LSP, HSP1, and HSP2 can be recapitulated in vitro using only two protein components of the transcription machineries: POLRMT and TFB2M.24, 25, 28, 29, 30 However, substantial evidence suggests that TFAM is a core component of mitochondrial transcription initiation in vivo (see below). Both in vitro and in vivo evidence is consistent with a role for TEFM supporting transcriptional elongation. Finally, strong evidence supports a role for MTERF1 as a transcription termination factor.
5. TRANSCRIPTION INITIATION
Efficient initiation in vitro depends on the presence of POLRMT and the two initiation factors, TFAM and TFB2M. Our mechanistic understanding of the process has been drastically affected by two crystal structures of the initiation complex on the two mitochondrial promoters (LSP and HSP).31 We will begin by reviewing the roles and properties of each of the proteins currently known to be involved in the initiation process: POLRMT, TFAM, and TFB2M.
5.1. POLRMT
Perhaps the most widely studied of the mitochondrial transcription related enzymes, POLRMT is at the heart of the mitochondrial RNA transcription apparatus. The human form of POLRMT was initially discovered by screening the expressed sequence tags database. The protein (1230 residues) was found to be similar to mitochondrial RNAPs from other eukaryotes and to RNAPs from several bacteriophages.32 The single subunit POLRMT does not transcribe nuclear genes and has an exclusively mitochondrial role.33 It is solely responsible for transcription of the circular mitochondrial genome. Furthermore, POLRMT is believed to have another essential role, generating replication primers for DNA replication.34
The catalytic domain of POLRMT is highly similar to the phage T7 RNAP.32 Like T7 RNAP, POLRMT has DNA‐dependent RNAP activity in vitro on synthetic substrates.35, 36 It is also able to promote RNA synthesis on a premelted substrate.37 However, unlike T7 RNAP, it is incapable of initiating promoter‐dependent in vitro transcription.35 A crystal structure of POLRMT (residues 105–1230) identified three structural features that explain why it is by itself unable to facilitate promoter‐dependent transcription.38 Although, like T7 RNAP, the catalytic domain of POLRMT adopts a right‐hand fold with palm, fingers and thumb subdomains (Figure 2a), it is missing promoter‐recognition elements and displays key structural differences in regions that facilitate promoter melting in T7 RNAP.
Figure 2.
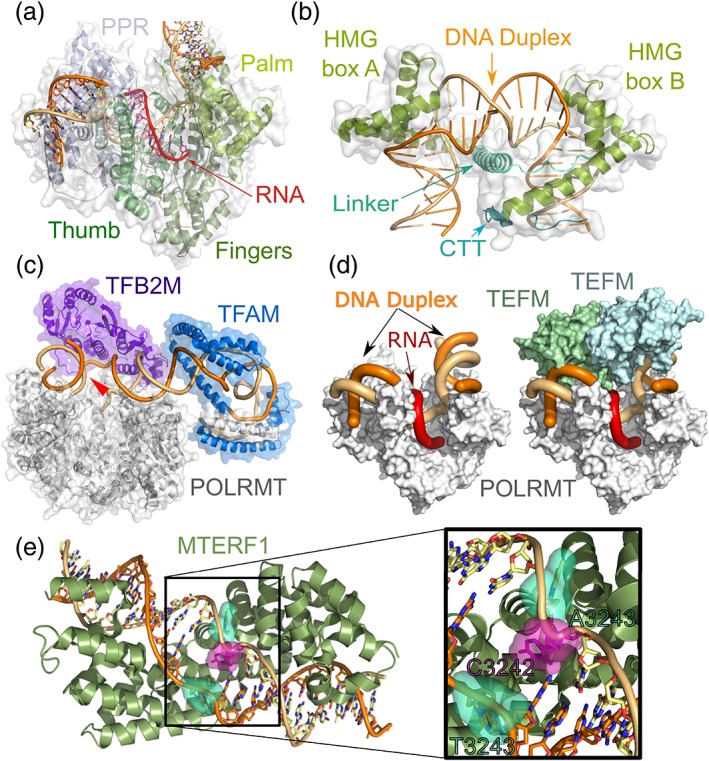
Structures of the mitochondrial transcription machinery. (a) POLRMT (PDBID: 3SPA) adopts a canonical right‐hand fold with fingers, palm, and thumb subdomains. The POLRMT molecular surface is in transparent gray. The fingers are green, the thumb is dark green, the palm is light green, and the N‐terminal and PPR domains are blue. The DNA duplex is show in red‐orange. (b) TFAM (PDBID: 3TMM) is composed of two HMG box domains and is able to induce a 180° bend in the DNA. The TFAM molecular surface is transparent, the two HMG boxes (a and b) are shown in green, the linker is light blue and the C‐terminal tail (CTT) is blue. The DNA duplex backbone is shown in orange ribbon and the bases are represented in sticks. (c) The transcription initiation complex is formed by TFAM (blue), TFB2M (magenta), and POLRMT (gray). TFAM bends the DNA upstream of the transcription start site (red arrow) and recruits POLRMT. TFB2M binds the nontemplate strand and stabilizes the open promoter complex. (d) A TEFM dimer binds to the elongating POLRMT and acts as a sliding clamp, enhancing its processivity. The structure of an elongation complex (PDBID: 5OLA) is shown without (left) and with TEFM (right), highlighting how the TEFM dimer covers the upstream and downstream DNA duplex (orange) as well as the nascent RNA strand (red). (e) MTERF1 (PDBID: 3MVA) makes extensive contacts with the termination sequence and promotes unwinding of the DNA duplex and base‐flipping of three nucleotides, both bases of the A3243 base pair (mutated in MELAS39) as well as C3242
In addition to the T7 RNAP core, POLRMT contains a flexible N‐terminal extension, which shows no homology with T7 RNAP. This N‐terminal extension is dispensable for promoter‐independent transcription. However, it is essential for transcriptional initiation, likely because it mediates an interaction between POLRMT and the initiation factor TFAM.40 Although the structure of the entire N‐terminal domain has not been solved, the POLRMT crystal structure shows that it contains a PPR domain Figure 2a) that makes some contacts with the DNA backbone in the promoter region. This is the only interaction between POLRMT and the DNA template, thus helping to explain why TFAM is required for POLRMT recruitment.41
Thus, although POLRMT is fully active as an RNAP, initiation activity depends on the presence of two protein cofactors: TFAM and TFB2M. Furthermore, although POLRMT is a processive enzyme, able to synthesize long RNA fragments in vitro, the production of longer RNA transcripts seems to require its association with the elongation factor TEFM42 (see below).
5.2. TFAM
TFAM is a multifunctional high mobility group (HMG) box protein that plays essential roles in mitochondria. First discovered in 1985 by separation of fractional pools of protein,43 TFAM was found to facilitate specific transcription, primarily on the L‐strand. The role of TFAM as a critical factor for promoter selection during initiation of mitochondrial transcription and efficient promoter recognition by POLRMT has since been confirmed.44, 45, 46, 47 TFAM activates transcription by specifically binding immediately upstream of transcriptional start sites.31 TFAM plays a second essential role in mitochondria. Owing to its ability to bind mtDNA with no sequence specificity, it coats the entire genome48 and facilitates its compaction into nucleoids.49 It has been proposed that TFAM covers the entire surface of a single copy of the mtDNA to form a nucleoid.50
Consistent with its key roles in transcription and mtDNA maintenance, TFAM deletion is embryogenically lethal, accompanied by a reduction in mtRNA levels and a severe respiratory chain deficiency.51, 52 Its expression level is also known to regulate mammalian mtDNA copy number.52 However, the mechanism by which it modulates mtDNA copy number is unknown: on the one hand transcription is necessary for replication primer formation, but TFAM coating is also thought to protect mtDNA from damage.
Crystal structures of TFAM in complex with different mtDNA substrates show that it is comprised of two HMG boxes and a C‐terminal tail.53, 54 TFAM has the remarkable ability to induce mtDNA into forming a U‐shaped turn (Figure 2b), where each of the singular HMG boxes introduces a 90° turn. These induced turns of the DNA are crucial for interactions with the remainder of the transcription apparatus (see below). Although it is not clear whether this conformation is also relevant to its DNA coating activity, it is tempting to speculate that it may play a role in organizing and compacting the mitochondrial genome.
5.3. TFB2M
TFB2M was discovered in 2002 as a factor key to assist POLRMT with the process of initiation of mitochondrial RNA transcription.35 Interestingly, TFB2M is highly similar to a group of highly conserved rRNA methyltransferases.55 In fact, TFB2M appears to conserve an independent methyltransferase activity of unclear function,56 but this activity is not necessary for mitochondrial transcription.57 TFB2M is required for promoter‐dependent POLRMT transcription in vitro. A crystal structure of TFB2M shows that it adopts a methyltransferase fold and is highly similar to the TFB1M mitochondrial rRNA methyltransferase.41, 58
5.4. Mechanism of initiation
TFAM appears to be the only protein in the initiation complex that can specifically bind to the promoter59 and thus TFAM binding has been proposed as the initial step in initiation.31, 60, 61 Because TFAM and POLRMT can form a direct interaction, TFAM is then believed to recruit POLRMT to the transcription start site. As TFAM is known to coat the mitochondrial genome and its binding mode appears identical at nonpromoter sites,62 it is not clear why this interaction is restricted to the mitochondrial promoters. In this respect, although the POLRMT–TFAM interaction is of low affinity, it can take place in the absence of DNA,60 suggesting that perhaps the initial binding event might involve a preformed TFAM–POLRMT complex. This is consistent with the low affinity of POLRMT–TFAM complexes to DNA observed in vitro61 and might explain the importance of POLRMT for promoter specificity.59
The preinitiation complex formed by TFAM and POLRMT is unable to initiate transcription in the absence of TFB2M. TFB2M is recruited to the preinitiation complex and plays a critical role in facilitating DNA opening, binding the promoter and specifically the nontemplate strand, as well as inducing conformational changes in POLRMT that stabilize the melted promoter complex31 (Figure 2c). The order of events during initiation cannot be established with certainty, and it is important to note that an alternative model for transcription initiation has suggested that a POLRMT–TFAM preinitiation complex might not exist in vivo and that instead TFAM might recruit a POLRMT–TFB2M complex.61
It has been suggested that the mitochondrial promoters are differentially regulated, and this seems to be the case at least in vitro.63 The structures of the initiation complex on both the HSP and LSP promoters however suggest a similar organization of the initiation complex31 and thus whether differential regulation takes place in vivo is still an open question.
6. TRANSCRIPTIONAL ELONGATION
Once transcription initiation takes place and POLRMT switches to elongation mode, TFB2M likely dissociates,37 and TEFM is believed to associate with POLRMT to drive transcription elongation.64 Knockdown of TEFM by RNA interference show reduced levels of H‐ and L‐strand promoter‐distal mitochondrial transcripts, and is accompanied by respiratory deficiencies in human cells.64 Knockdown of TEFM in mouse cells confirms that it is essential for transcriptional elongation and generation of full‐length transcripts.65
TEFM forms a dimer in solution and potently enhances the processivity of POLRMT in vitro by substantially increasing its affinity to DNA.42 The TEFM crystal structure confirms that it dimerizes and reveals that each monomer contains two tandem helix–hairpin–helix domains that are dispensable to enhance POLRMT processivity as well as an RNase H‐like fold that mediates the activities relevant to transcriptional elongation. Moreover, dimerization is essential for its association with POLRMT.66 A crystal structure of a transcription elongation complex showing TEFM complexed with POLRMT shows that TEFM binds POLRMT near the RNA exit channel, forming a sliding clamp that presumably increases the processivity of POLRMT (Figure 2d).66 Importantly, binding of TEFM is incompatible with TFB2M binding, and consistently TEFM is unable to associate with POLRMT during initiation.66 Furthermore, the transition to elongation appears to involve substantial rearrangements in the topology of the DNA upstream of the POLRMT active site.31
The increase in processivity resulting from association of TEFM and POLRMT can overcome transcriptional termination occurring at various sites in the mtDNA and facilitate bypass of oxidative lesions.40 Particularly relevant is termination taking place at the conserved sequence block II region because of the generation of RNA G‐quadruplexes,40, 66 which has been linked to the generation of replication primers.67 This led to the suggestion that TEFM might be key to regulate a switch between mitochondrial transcription and replication, whereby POLRMT elongation in the absence of TEFM would result in the formation of RNA replication primers.67 However, TEFM knockouts do not display an increase in mtDNA replication.65 Instead, the knockout results in an increase in premature termination as well as a subsequent decrease in formation of replication primers. Further processing by RNase H of longer, TEFM‐mediated transcripts is required for their use as replication primers.68 Moreover, the loss of TEFM can negatively affect mitochondrial RNA processing.65 Additional work is required to explain the potential connection between mitochondrial transcription elongation and RNA processing.
7. TRANSCRIPTION TERMINATION
The first studies of mitochondrial transcription termination were motivated by the early conclusion that the rRNA genes situated in the H‐strand proximal region of HSP are transcribed at 15–60 times higher rates than the more distal H‐strand protein coding genes.69 Although the higher steady‐state levels of mitochondrial rRNAs are now understood to be a consequence of higher stability rather than transcription rates,28 subsequent studies of the 3′ ends of the 12S and 16S rRNAs led to the suggestion that their synthesis was mediated by a transcription termination event.70, 71 This led to the discovery of mTERF (now known as MTERF1), a protein factor capable of protecting a 28 base pair segment of mtDNA at the 3′ end of the 16S rRNA and mediate transcription termination72 (TERM in Figure 1a). Subsequent cloning of this factor73 led to the realization that it is a member of a family of nucleic‐acid binding proteins (MTERF proteins) that play various roles related to gene expression in mitochondria and chloroplasts74, 75 and display extreme flexibility in their ability to accommodate a wide range of interactions with nucleic acids.
MTERF1 has been shown to terminate transcription in vitro with very high efficiency.76 Although original models of transcription termination by MTERF1 proposed that it is responsible for termination of HSP2‐driven transcription to facilitate synthesis of rRNAs,19 these models are not supported by any in vivo evidence, and are inconsistent with in vitro biochemical studies.76, 77 Later knockdown78 and knockout28 experiments strongly support the notion that MTERF1 does not play a role in termination of HSP‐driven transcription and is instead responsible for termination of transcription originating at LSP. This is also consistent with structural studies supporting termination polarity. The crystal structure of MTERF1 bound to the DNA termination sequence shows that MTERF1 binds DNA through a unique mechanism that involves a repeated module, the MTERF motif, but also partial unwinding of the DNA double helix, the melting of two base pairs, and eversion of three nucleotides77 (Figure 2e). Sequence recognition by MTERF1 is based on specific recognition of guanine residues in the target sequence by the MTERF modules, and these interactions are prevented by multiple pathogenic mutations.79
MTERF1 appears to play roles beyond LSP termination: it has been shown to mediate more general pausing of RNA transcription by POLRMT80 as well as pausing of mtDNA replication.80, 81 Finally, although MTERF1 might account for LSP termination, termination of HSP transcription occurs through a mechanism that is still poorly understood, appears to be MTERF1‐independent, and might involve additional proteins.82
8. RNA PROCESSING
Mitochondrial primary transcripts must undergo processing for the release of mature and functional mtRNAs. This processing encompasses cleavage, polyadenylation, addition of the cytosine, cytosine, adenine (CCA) tail to tRNAs as well as other base posttranscriptional modifications. It has been proposed that all these processes take place in a compartmentalized area of the mitochondrial matrix known as the mitochondrial RNA granule.83, 84 Ojala et al., first proposed the tRNA punctuation model, whereby cleavage of primary full‐length transcripts occurs at junctions between mRNAs or rRNAs and tRNAs.20, 85 This model has been widely accepted and proven to generally agree with deep sequencing analysis.86 However, deep sequencing also revealed unexpected complexity in RNA processing, including noncanonical processing events87 as well as multiple types of aberrant processing including polyadenylation of some tRNAs.86
Cleavage of polycistronic transcripts is carried out by specific enzymes. Cleavage of tRNAs at their 5′ and 3′ ends is performed by RNase P and RNase Z, respectively, releasing individual or bicistronic RNA species to undergo further maturation (Figure 3). RNase P is composed of three subunits (MRPP1, MRPP2, and MRRP3) and is surprisingly devoid of an RNA component.88 Cleavage at tRNA 3′ ends is performed by RNase Z (ELAC2).89, 90 RNase Z was shown to preferentially process precursors already processed by RNase P.89, 90 Four genes have been identified that are not flanked on both sides by tRNAs and therefore must undergo noncanonical processing. For instance, the mitochondrial cytochrome c oxidase 1 (COX1) is flanked at its 5′ end by noncoding RNA (ncRNA) and at its 3′ end by tRNA‐Cysteine.90 The ncRNA upstream of COX1 is thought to adopt a tRNA‐like structure providing a basis for processing by RNase P at its 5′ end and processing by RNase Z at its 3′ end.90, 91 Unlike COX1, three other transcripts junctions (ND6‐ncRNA, ND5‐CYB, and RNA14‐COX3) do not depend on RNase P or Z for processing and the specific mechanisms by which they are processed are still not clear. Moreover, not all mitochondrial transcripts are cleaved into individual RNA molecules. Some bicistronic transcripts have been observed including ATP8‐ATP6, ATP8/6‐COX3, and ND4L‐ND4.90, 92, 93 Knockdown of any of the previously described mtRNA processing proteins did not affect these junctions.92
Figure 3.
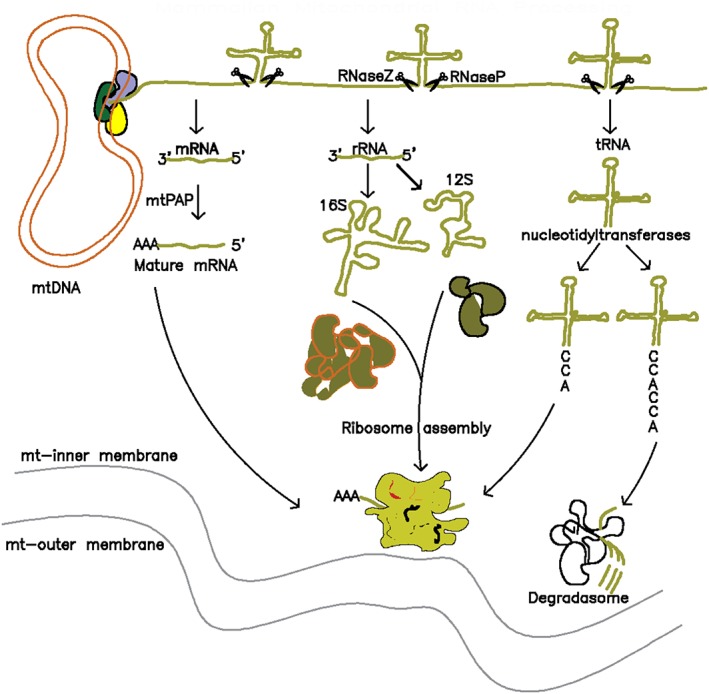
Overview of mammalian mitochondrial RNA processing. RNA processing is believed to take place co‐transcriptionally. Cleavage by RNases P and Z liberates individual mRNAs, tRNAs and rRNAs. These RNAs then undergo different modifications (see text) resulting in maturation and utilization in downstream processes (i.e., ribosome assembly and translation) or, alternatively, in degradation of unstable RNAs
After cleavage, individual or bicistronic transcripts undergo further modification before becoming mature and functionally active. Similar to nuclear mRNAs, most mtRNAs are polyadenylated at their 3′ ends. The human mitochondrial polyA polymerase is responsible for catalyzing polyadenylation of mtRNA.94 The mammalian mitochondrial rRNAs contain very short polyA tails, while mRNAs contain ~40–50 adenine bases.94 The role of mtRNA polyadenylation is controversial. For example, polyadenylation seems to stabilize subunits of complex IV (COX1, COX2, and COX3),94 destabilize subunits of complex I (ND1 and ND2),95 but does not affect ND3.94
Finally, a common tRNA modification is the addition of a CCA tail at their 3′ termini to facilitate attachment of the amino acid group used for translation. CCA tail addition is catalyzed by enzymes belonging to the nucleotidyltransferase superfamily.96 Unlike polyadenylation, CCA addition is associated with clear specific functions: in addition to being central to tRNA maturation, a double CCA tail can be added to unstable tRNA species to target them for degradation97 (Figure 3). In addition to the processes of RNA cleavage, RNA polyadenylation and CCA tail addition, numerous specific and nonspecific posttranscriptional modification must take place during RNA maturation. These are not discussed in this review.
9. TRANSCRIPTIONAL REGULATION
Mitochondria depend on nuclear‐encoded genes for all processes of gene expression. Although there is limited understanding on how mitochondrial transcription is regulated, there is significant evidence from tissue studies that the process is regulated directly and indirectly by nuclear transcription factors.98 Nuclear transcription factors have been localized in mammalian mitochondria where they either bind the mtDNA D‐loop or mtDNA genes to modulate mtDNA transcription. Thyroid hormone/receptor has been observed in vivo in the mitochondrial matrix99, 100 and shown in vitro to bind to the D‐loop and 12S rRNA gene where it regulates mtRNA synthesis.101, 102 Similarly, D‐loop binding by cAMP response element binding protein has been observed and thought to promote mtDNA gene expression.103, 104, 105, 106 In vivo, mouse mtDNA D‐loop binding was also shown for the signal transducer and activator of Transcription 3,107 the nuclear DNA chromatin modulator MOF acetyl transferase,108 and the osteogenic cell line‐specific transcription modulator NFATc1 in human mesenchymal stem cells.109 D‐loop binding has additionally been reported for the DNA methyltransferase Dnmt1, believed to play a role in reducing mtDNA transcript levels.110 Other non‐D‐loop binding nuclear transcription regulators were reported to influence mitochondrial transcription by directly binding on mitochondrial gene coding sequences, which included the myocyte enhancer factor 2D111 and the transcription factors c‐Jun, JunD, and CEBP.112
In addition to regulating transcription by binding directly to the mtDNA, nuclear transcription regulators can modulate expression or activity of the mitochondrial transcriptional machinery. Nuclear respiratory factors, NRF‐1 and NRF‐2 control expression of TFB2M and POLRMT.113, 114 PGC1 family factors, including PGC1‐α, PGC1‐β, and PRC in turn regulate the activity of a variety of transcription factors including NRF‐1,115, 116 NRF2,117, 118 and Yin Yang 1 (YY1),119 all believed to influence mitochondrial gene expression. Surprisingly, PGC1‐α also forms a complex with TFAM at the mouse mtDNA D‐loop to increase mitochondrial gene expression during high energy demands.120
Importantly, additional direct mechanisms appear to modulate mitochondrial transcription. The mitochondrial ribosomal protein L12 (MRPL12) seems to be a positive regulator of the mitochondrial transcription process by controlling POLRMT stability.121 MRPL12 expression correlates with the steady‐state levels of mitochondrial transcripts.122, 123 There is growing evidence suggesting that mitochondrial transcription is also regulated in response to changing mitochondrial ATP concentrations: high ATP levels have been shown to inhibit the activity of POLRMT both in isolated mitochondria and in vitro,124 while intermediate ATP levels activate transcription and very low ATP levels prevent transcription initiation.125 Finally, mtDNA methylation has been proposed to be associated with mitochondrial transcription regulation.126, 127, 128 Recently, a mechanism has been proposed where mtDNA methylation leads to posttranslational modification of TFAM, resulting in increased nucleoid compaction, limiting mtDNA accessibility to POLRMT and TFB2M.129 Despite these observations, we do not have a comprehensive picture of the mechanisms that control mitochondrial transcription, and additional research is needed to increase our understanding of mitochondrial transcription regulation.
10. CONCLUDING REMARKS
Mammalian mitochondrial transcription studies are gaining momentum, especially given the wealth of structural information acquired in recent years. These studies have solidified our mechanistic understanding of the transcription process, although numerous mechanistic questions remain unanswered. Furthermore, recent advances in next‐generation sequencing technology have facilitated a new approach to study mitochondrial RNA biology in an unprecedented way. This has already facilitated strong advances in our understanding of posttranscriptional processing, and it is likely that RNA sequencing approaches will continue to provide insight into mitochondria RNA biology. A large number of observations point to numerous mechanisms that regulate mitochondrial transcription. Understanding how this process is regulated is the next frontier in our understanding of the transcription process and will be the key to help us appreciate how alterations in transcription contribute to human disease.
ACKNOWLEDGMENTS
This work was supported by R01GM100021 (MGD), T32GM007518 (EB and AS), and NSF graduate research fellowships (EB and AS).
Bouda E, Stapon A, Garcia‐Diaz M. Mechanisms of mammalian mitochondrial transcription. Protein Science. 2019;28:1594–1605. 10.1002/pro.3688
Funding information National Institute of General Medical Sciences, Grant/Award Numbers: T32GM007518, R01GM100021; NSF graduate research fellowships
REFERENCES
- 1. Shah R, Schwach J, Frankenberg‐Dinkel N, Gartner W. Complex formation between heme oxygenase and phytochrome during biosynthesis in pseudomonas syringae pv. Tomato. Photochem Photobiol Sci. 2012;11:1026–1031. [DOI] [PubMed] [Google Scholar]
- 2. McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr Biol. 2006;16:R551–R560. [DOI] [PubMed] [Google Scholar]
- 3. Adam‐Vizi V, Chinopoulos C. Bioenergetics and the formation of mitochondrial reactive oxygen species. Trends Pharmacol Sci. 2006;27:639–645. [DOI] [PubMed] [Google Scholar]
- 4. Anderson S, Bankier AT, Barrell BG, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. [DOI] [PubMed] [Google Scholar]
- 5. Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26:167–180. [DOI] [PubMed] [Google Scholar]
- 6. Barrell BG, Bankier AT, Drouin J. A different genetic code in human mitochondria. Nature. 1979;282:189–194. [DOI] [PubMed] [Google Scholar]
- 7. Berk AJ, Clayton DA. Mechanism of mitochondrial DNA replication in mouse L‐cells: asynchronous replication of strands, segregation of circular daughter molecules, aspects of topology and turnover of an initiation sequence. J Mol Biol. 1974;86:801–824. [DOI] [PubMed] [Google Scholar]
- 8. Saccone C, Aaij C, Borst P, Gadaleta MN. Hybridization studies with RNA synthesized by isolated rat‐liver mitochondria. Biochim Biophys Acta. 1970;199:373–381. [DOI] [PubMed] [Google Scholar]
- 9. Tabak HF, Borst P. Erroneous transcription of mitochondrial DNA by RNA polymerase from Escherichia coli . Biochim Biophys Acta. 1970;217:356–364. [DOI] [PubMed] [Google Scholar]
- 10. Gustafsson CM, Falkenberg M, Larsson NG. Maintenance and expression of mammalian mitochondrial DNA. Annu Rev Biochem. 2016;85:133–160. [DOI] [PubMed] [Google Scholar]
- 11. Prada CF, Boore JL. Gene annotation errors are common in the mammalian mitochondrial genomes database. BMC Genomics. 2019;20:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aloni Y, Attardi G. Expression of the mitochondrial genome in HeLa cells. II evidence for complete transcription of mitochondrial DNA. J Mol Biol. 1971;55:251–267. [DOI] [PubMed] [Google Scholar]
- 13. Aloni Y, Attardi G. Symmetrical in vivo transcription of mitochondrial DNA in HeLa cells. Proc Natl Acad Sci USA. 1971;68:1757–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cantatore P, Attardi G. Mapping of nascent light and heavy strand transcripts on the physical map of HeLa cell mitochondrial DNA. Nucleic Acids Res. 1980;8:2605–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hixson JE, Clayton DA. Initiation of transcription from each of the two human mitochondrial promoters requires unique nucleotides at the transcriptional start sites. Proc Natl Acad Sci USA. 1985;82:2660–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bogenhagen DF, Applegate EF, Yoza BK. Identification of a promoter for transcription of the heavy strand of human mtDNA: in vitro transcription and deletion mutagenesis. Cell. 1984;36:1105–1113. [DOI] [PubMed] [Google Scholar]
- 17. Montoya J, Christianson T, Levens D, Rabinowitz M, Attardi G. Identification of initiation sites for heavy‐strand and light‐strand transcription in human mitochondrial DNA. Proc Natl Acad Sci USA. 1982;79:7195–7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chang DD, Clayton DA. Precise identification of individual promoters for transcription of each strand of human mitochondrial DNA. Cell. 1984;36:635–643. [DOI] [PubMed] [Google Scholar]
- 19. Martin M, Cho J, Cesare AJ, Griffith JD, Attardi G. Termination factor‐mediated DNA loop between termination and initiation sites drives mitochondrial rRNA synthesis. Cell. 2005;123:1227–1240. [DOI] [PubMed] [Google Scholar]
- 20. Ojala D, Merkel C, Gelfand R, Attardi G. The tRNA genes punctuate the reading of genetic information in human mitochondrial DNA. Cell. 1980;22:393–403. [DOI] [PubMed] [Google Scholar]
- 21. Chang DD, Clayton DA. Precise assignment of the light‐strand promoter of mouse mitochondrial DNA: a functional promoter consists of multiple upstream domains. Mol Cell Biol. 1986;6:3253–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chang DD, Clayton DA. Precise assignment of the heavy‐strand promoter of mouse mitochondrial DNA: cognate start sites are not required for transcriptional initiation. Mol Cell Biol. 1986;6:3262–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Litonin D, Sologub M, Shi Y, et al. Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. J Biol Chem. 2010;285:18129–18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lodeiro MF, Uchida A, Bestwick M, et al. Transcription from the second heavy‐strand promoter of human mtDNA is repressed by transcription factor a in vitro. Proc Natl Acad Sci USA. 2012;109:6513–6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zollo O, Tiranti V, Sondheimer N. Transcriptional requirements of the distal heavy‐strand promoter of mtDNA. Proc Natl Acad Sci USA. 2012;109:6508–6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shi Y, Dierckx A, Wanrooij PH, et al. Mammalian transcription factor a is a core component of the mitochondrial transcription machinery. Proc Natl Acad Sci USA. 2012;109:16510–16515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Blumberg A, Rice EJ, Kundaje A, Danko CG, Mishmar D. Initiation of mtDNA transcription is followed by pausing, and diverges across human cell types and during evolution. Genome Res. 2017;27:362–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Terzioglu M, Ruzzenente B, Harmel J, et al. MTERF1 binds mtDNA to prevent transcriptional interference at the light‐strand promoter but is dispensable for rRNA gene transcription regulation. Cell Metab. 2013;17:618–626. [DOI] [PubMed] [Google Scholar]
- 29. Shutt TE, Bestwick M, Shadel GS. The core human mitochondrial transcription initiation complex: it only takes two to tango. Transcription. 2011;2:55–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shutt TE, Lodeiro MF, Cotney J, Cameron CE, Shadel GS. Core human mitochondrial transcription apparatus is a regulated two‐component system in vitro. Proc Natl Acad Sci USA. 2010;107:12133–12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hillen HS, Morozov YI, Sarfallah A, Temiakov D, Cramer P. Structural basis of mitochondrial transcription initiation. Cell. 2017;171:1072–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tiranti V, Savoia A, Forti F, et al. Identification of the gene encoding the human mitochondrial RNA polymerase (h‐mtRPOL) by cyberscreening of the expressed sequence tags database. Human Mol Genet. 1997;6:615–625. [DOI] [PubMed] [Google Scholar]
- 33. Kühl I, Kukat C, Ruzzenente B, et al. POLRMT does not transcribe nuclear genes. Nature. 2014;514:E7–E11. [DOI] [PubMed] [Google Scholar]
- 34. Wanrooij S, Fusté JM, Farge G, Shi Y, Gustafsson CM, Falkenberg M. Human mitochondrial RNA polymerase primes lagging‐strand DNA synthesis in vitro. Proc Natl Acad Sci USA. 2008;105:11122–11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat Genet. 2002;31:289–294. [DOI] [PubMed] [Google Scholar]
- 36. Smidansky ED, Arnold JJ, Reynolds SL, Cameron CE. Human mitochondrial RNA polymerase: evaluation of the single‐nucleotide‐addition cycle on synthetic RNA/DNA scaffolds. Biochemistry. 2011;50:5016–5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sologub M, Litonin D, Anikin M, Mustaev A, Temiakov D. TFB2 is a transient component of the catalytic site of the human mitochondrial RNA polymerase. Cell. 2009;139:934–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ringel R, Marina Sologub M, Morozov YI, Litonin D, Cramer P, Temiakov D. Structure of human mitochondrial RNA polymerase. Nature. 2011;478:267–273. [DOI] [PubMed] [Google Scholar]
- 39. Hess JF, Parisi MA, Bennett JL, Clayton DA. Impairment of mitochondrial transcription termination by a point mutation associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1991;351:236–239. [DOI] [PubMed] [Google Scholar]
- 40. Posse V, Hoberg E, Dierckx A, et al. The amino terminal extension of mammalian mitochondrial RNA polymerase ensures promoter specific transcription initiation. Nucleic Acids Res. 2014;42:3638–3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hillen HS, Morozov YI, Sarfallah A, Temiakov D, Cramer P. Structural basis of mitochondrial. Cell. 2017;171:1072–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Posse V, Shahzad S, Falkenberg M, Hallberg BM, Gustafsson C. TEFM is a potent stimulator of mitochondrial transcription elongation in vitro. Nucleic Acids Res. 2015;43:2615–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fisher RP, Clayton DA. A transcription factor required for promoter recognition by human mitochondrial RNA polymerase. J Biol Chem. 1985;260:11330–11338. [PubMed] [Google Scholar]
- 44. Dairaghi DJ, Shadel GS, Clayton DA. Addition of a 29 residue carboxyl‐terminal tail converts a simple HMG box‐containing protein into a transcriptional activator. J Mol Biol. 1995;249:11–28. [DOI] [PubMed] [Google Scholar]
- 45. Fisher RP, Clayton DA. Purification and characterization of human mitochondrial transcription factor 1. Mol Cell Biol. 1988;8:3496–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fisher RP, Lisowsky T, Parisi MA, Clayton DA. DNA wrapping and bending by a mitochondrial high mobility group‐like transcriptional activator protein. J Biol Chem. 1992;267:3358–3367. [PubMed] [Google Scholar]
- 47. Fisher RP, Topper JN, Clayton DA. Promoter selection in human mitochondria involves binding of a transcription factor to orientation‐independent upstream regulatory elements. Cell. 1987;50:247–258. [DOI] [PubMed] [Google Scholar]
- 48. Wang YE, Marinov GK, Wold BJ, Chan DC. Genome‐wide analysis reveals coating of the mitochondrial genome by TFAM. PLoS One. 2013;8:e74513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kaufman BA, Durisic N, Mativetsky JM, et al. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid‐like structures. Mol Biol Cell. 2007;18:3225–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kukata C, Daviesc KM, Wurmd CA, et al. Cross‐strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proc Natl Acad Sci USA. 2015;112:11288–11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Larsson NG, Wang J, Wilhelmsson H, et al. Mitochondrial transcription factor a is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. [DOI] [PubMed] [Google Scholar]
- 52. Ekstrand MI, Falkenberg M, Rantanen A, et al. Mitochondrial transcription factor a regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13:935–944. [DOI] [PubMed] [Google Scholar]
- 53. Rubio‐Cosials A, Sydow JF, Jiménez‐Menéndez N, et al. Human mitochondrial transcription factor a induces a U‐turn structure in the light strand promoter. Nat Struct Mol Biol. 2011;18:1281–1290. [DOI] [PubMed] [Google Scholar]
- 54. Ngo HB, Kaiser JT, Chan DC. The mitochondrial transcription and packaging factor Tfam imposes a U‐turn on mitochondrial DNA. Nat Struct Mol Biol. 2011;18:1290–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. McCulloch V, Seidel‐Rogol BL, Shadel GS. A human mitochondrial transcription factor is related to RNA adenine methyltransferases and binds S‐adenosylmethionine. Mol Cell Biol. 2002;22:1116–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cotney J, Shadel GS. Evidence for an early gene duplication event in the evolution of the mitochondrial transcription factor B family and maintenance of rRNA methyltransferase activity in human mtTFB1 and mtTFB2. J Mol Evol. 2006;63:707–717. [DOI] [PubMed] [Google Scholar]
- 57. Cotney J, McKay SE, Shadel GS. Elucidation of separate, but collaborative functions of the rRNA methyltransferase‐related human mitochondrial transcription factors B1 and B2 in mitochondrial biogenesis reveals new insight into maternally inherited deafness. Hum Mol Genet. 2009;18:2670–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Guja KE, Venkataraman K, Yakubovskaya E, et al. Structural basis for S‐adenosylmethionine binding and methyltransferase activity by mitochondrial transcription factor B1. Nucleic Acids Res. 2013;41:7947–7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gaspari M, Falkenberg M, Larsson NG, Gustafsson CM. The mitochondrial RNA polymerase contributes critically to promoter specificity in mammalian cells. EMBO J. 2004;23:4606–4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yakubovskaya E, Guja KE, Eng ET, et al. Organization of the human mitochondrial transcription initiation complex. Nucleic Acids Res. 2014;42:4100–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ramachandran A, Basu U, Sultana S, Nandakumar D, Patel SS. Human mitochondrial transcription factors TFAM and TFB2M work synergistically in promoter melting during transcription initiation. Nucleic Acids Res. 2017;45:861–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ngo HB, Lovely GA, Phillips R, Chan DC. Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation. Nat Commun. 2014;5:3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Uchida A, Murugesapillai D, Kastner M, et al. Unexpected sequences and structures of mtDNA required for efficient transcription from the first heavy‐strand promoter. Elife. 2017;6:e27283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Minczuk M, He J, Duch AM, et al. TEFM (c17orf42) is necessary for transcription of human mtDNA. Nucleic Acids Res. 2011;39:4284–4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jiang S, Koolmeister C, Misic J, et al. TEFM regulates both transcription elongation and RNA processing in mitochondria. EMBO Rep. 2019;20:e48101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hillen HS, Parshin AV, Agaronyan K, et al. Mechanism of transcription anti‐termination in human mitochondria. Cell. 2017;171:1082–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Agaronyan K, Morozov YI, Anikin M, Temiakov D. Mitochondrial biology. Replication‐transcription switch in human mitochondria. Science. 2015;347:548–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Posse V, Al‐Behadili A, Uhler JP, et al. RNase H1 directs origin‐specific initiation of DNA replication in human mitochondria. PLoS Genet. 2019;15:e1007781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gelfand R, Attardi G. Synthesis and turnover of mitochondrial ribonucleic acid in HeLa cells: the mature ribosomal and messenger ribonucleic acid species are metabolically unstable. Mol Cell Biol. 1981;1:497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dubin DT, Montoya J, Timko KD, Attardi G. Sequence analysis and precise mapping of the 3′ ends of HeLa cell mitochondrial ribosomal RNAs. J Mol Biol. 1982;157:1–19. [DOI] [PubMed] [Google Scholar]
- 71. Van Etten RA, Bird JW, Clayton DA. Identification of the 3′‐ends of the two mouse mitochondrial ribosomal RNAs. The 3′‐end of 16 S ribosomal RNA contains nucleotides encoded by the gene for transfer RNALeuUUR. J Biol Chem. 1983;258:10104–10110. [PubMed] [Google Scholar]
- 72. Kruse B, Narasimhan N, Attardi G. Termination of transcription in human mitochondria: identification and purification of a DNA binding protein factor that promotes termination. Cell. 1989;58:391–397. [DOI] [PubMed] [Google Scholar]
- 73. Fernandez‐Silva P, Martinez‐Azorin F, Micol V, Attardi G. The human mitochondrial transcription termination factor (mTERF) is a multizipper protein but binds to DNA as a monomer, with evidence pointing to intramolecular leucine zipper interactions. EMBO J. 1997;16:1066–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Linder T, Park CB, Asin‐Cayuela J, et al. A family of putative transcription termination factors shared amongst metazoans and plants. Curr Genet. 2005;48:265–269. [DOI] [PubMed] [Google Scholar]
- 75. Roberti M, Polosa PL, Bruni F, et al. The MTERF family proteins: mitochondrial transcription regulators and beyond. Biochim Biophys Acta. 2009;1787:303–311. [DOI] [PubMed] [Google Scholar]
- 76. Asin‐Cayuela J, Schwend T, Farge G, Gustafsson CM. The human mitochondrial transcription termination factor (mTERF) is fully active in vitro in the non‐phosphorylated form. J Biol Chem. 2005;280:25499–25505. [DOI] [PubMed] [Google Scholar]
- 77. Yakubovskaya E, Mejia E, Byrnes J, Hambardjieva E, Garcia‐Diaz M. Helix unwinding and base flipping enable human MTERF1 to terminate mitochondrial transcription. Cell. 2010;141:982–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hyvarinen AK, Kumanto MK, Marjavaara SK, Jacobs HT. Effects on mitochondrial transcription of manipulating mTERF protein levels in cultured human HEK293 cells. BMC Mol Biol. 2010;11:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Guja KE. Garcia‐Diaz M (2012) hitting the brakes: termination of mitochondrial transcription. Biochim Biophys Acta. 2012;1819:939–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hyvärinen AK, Pohjoismäki JLO, Reyes A, et al. The mitochondrial transcription termination factor mTERF modulates replication pausing in human mitochondrial DNA. Nucleic Acids Res. 2007;35(19):6458–6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Shi Y, Posse V, Zhu X, et al. Mitochondrial transcription termination factor 1 directs polar replication fork pausing. Nucleic Acids Res. 2016;44:5732–5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Camasamudram V, Fang JK, Avadhani NG. Transcription termination at the mouse mitochondrial H‐strand promoter distal site requires an a/T rich sequence motif and sequence specific DNA binding proteins. Eur J Biochem. 2003;270:1128–1140. [DOI] [PubMed] [Google Scholar]
- 83. Antonicka H, Shoubridge EA. Mitochondrial RNA granules are centers for posttranscriptional RNA processing and ribosome biogenesis. Cell Rep. 2015;10:920–932. [DOI] [PubMed] [Google Scholar]
- 84. Jourdain AA, Boehm E, Maundrell K, Martinou JC. Mitochondrial RNA granules: compartmentalizing mitochondrial gene expression. J Cell Biol. 2016;212:611–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. [DOI] [PubMed] [Google Scholar]
- 86. Mercer TR, Neph S, Dinger ME, et al. The human mitochondrial transcriptome. Cell. 2011;146:645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Temperley RJ, Wydro M, Lightowlers RN, Chrzanowska‐Lightowlers ZM. Human mitochondrial mRNAs–like members of all families, similar but different. Biochim Biophys Acta. 2010;1797:1081–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Holzmann J, Frank P, Loffler E, Bennett KL, Gerner C, Rossmanith W. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–474. [DOI] [PubMed] [Google Scholar]
- 89. Brzezniak LK, Bijata M, Szczesny RJ, Stepien PP. Involvement of human ELAC2 gene product in 3′ end processing of mitochondrial tRNAs. RNA Biol. 2011;8:616–626. [DOI] [PubMed] [Google Scholar]
- 90. Sanchez MI, Mercer TR, Davies SM, et al. RNA processing in human mitochondria. Cell Cycle. 2011;10:2904–2916. [DOI] [PubMed] [Google Scholar]
- 91. Mercer TR, Gerhardt DJ, Dinger ME, et al. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol. 2011;30:99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wolf AR, Mootha VK. Functional genomic analysis of human mitochondrial RNA processing. Cell Rep. 2014;7:918–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Shimpi GG, Vargas S, Poliseno A, Worheide G. Mitochondrial RNA processing in absence of tRNA punctuations in octocorals. BMC Mol Biol. 2017;18:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Nagaike T, Suzuki T, Katoh T, Ueda T. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria‐specific poly(a) polymerase and polynucleotide phosphorylase. J Biol Chem. 2005;280:19721–19727. [DOI] [PubMed] [Google Scholar]
- 95. Tomecki R, Dmochowska A, Gewartowski K, Dziembowski A, Stepien PP. Identification of a novel human nuclear‐encoded mitochondrial poly(a) polymerase. Nucleic Acids Res. 2004;32:6001–6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yue D, Maizels N, Weiner AM. CCA‐adding enzymes and poly(a) polymerases are all members of the same nucleotidyltransferase superfamily: characterization of the CCA‐adding enzyme from the archaeal hyperthermophile Sulfolobus shibatae. RNA. 1996;2:895–908. [PMC free article] [PubMed] [Google Scholar]
- 97. Betat H, Morl M. The CCA‐adding enzyme: a central scrutinizer in tRNA quality control. Bioessays. 2015;37:975–982. [DOI] [PubMed] [Google Scholar]
- 98. Leigh‐Brown S, Enriquez JA, Odom DT. Nuclear transcription factors in mammalian mitochondria. Genome Biol. 2010;11:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Morrish F, Buroker NE, Ge M, et al. Thyroid hormone receptor isoforms localize to cardiac mitochondrial matrix with potential for binding to receptor elements on mtDNA. Mitochondrion. 2006;6:143–148. [DOI] [PubMed] [Google Scholar]
- 100. Wrutniak C, Cassar‐Malek I, Marchal S, et al. A 43‐kDa protein related to c‐Erb a alpha 1 is located in the mitochondrial matrix of rat liver. J Biol Chem. 1995;270:16347–16354. [DOI] [PubMed] [Google Scholar]
- 101. Casas F, Rochard P, Rodier A, et al. A variant form of the nuclear triiodothyronine receptor c‐ErbAalpha1 plays a direct role in regulation of mitochondrial RNA synthesis. Mol Cell Biol. 1999;19:7913–7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Enriquez JA, Fernandez‐Silva P, Garrido‐Perez N, Lopez‐Perez MJ, Perez‐Martos A, Montoya J. Direct regulation of mitochondrial RNA synthesis by thyroid hormone. Mol Cell Biol. 1999;19:657–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Cammarota M, Paratcha G, Bevilaqua LR, et al. Cyclic AMP‐responsive element binding protein in brain mitochondria. J Neurochem. 1999;72:2272–2277. [DOI] [PubMed] [Google Scholar]
- 104. Lee J, Kim CH, Simon DK, et al. Mitochondrial cyclic AMP response element‐binding protein (CREB) mediates mitochondrial gene expression and neuronal survival. J Biol Chem. 2005;280:40398–40401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ryu H, Lee J, Impey S, Ratan RR, Ferrante RJ. Antioxidants modulate mitochondrial PKA and increase CREB binding to D‐loop DNA of the mitochondrial genome in neurons. Proc Natl Acad Sci USA. 2005;102:13915–13920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. De Rasmo D, Signorile A, Roca E, Papa S. cAMP response element‐binding protein (CREB) is imported into mitochondria and promotes protein synthesis. FEBS J. 2009;276:4325–4333. [DOI] [PubMed] [Google Scholar]
- 107. Macias E, Rao D, Carbajal S, Kiguchi K, DiGiovanni J. Stat3 binds to mtDNA and regulates mitochondrial gene expression in keratinocytes. J Invest Dermatol. 2014;134:1971–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Chatterjee A, Seyfferth J, Lucci J, et al. MOF acetyl transferase regulates transcription and respiration in mitochondria. Cell. 2016;167:722–738. [DOI] [PubMed] [Google Scholar]
- 109. Lambertini E, Penolazzi L, Morganti C, et al. Osteogenic differentiation of human MSCs: specific occupancy of the mitochondrial DNA by NFATc1 transcription factor. Int J Biochem Cell Biol. 2015;64:212–219. [DOI] [PubMed] [Google Scholar]
- 110. Mishra M, Kowluru RA. Epigenetic modification of mitochondrial DNA in the development of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2015;56:5133–5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. She H, Yang Q, Shepherd K, et al. Direct regulation of complex I by mitochondrial MEF2D is disrupted in a mouse model of Parkinson disease and in human patients. J Clin Invest. 2011;121:930–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Blumberg A, Sri Sailaja B, Kundaje A, et al. Transcription factors bind negatively selected sites within human mtDNA genes. Genome Biol Evol. 2014;6:2634–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Bruni F, Polosa PL, Gadaleta MN, Cantatore P, Roberti M. Nuclear respiratory factor 2 induces the expression of many but not all human proteins acting in mitochondrial DNA transcription and replication. J Biol Chem. 2010;28:3939–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Gleyzer N, Vercauteren K, Scarpulla RC. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF‐1 and NRF‐2) and PGC‐1 family coactivators. Mol Cell Biol. 2005;25:1354–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Vercauteren K, Pasko RA, Gleyzer N, Marino VM, Scarpulla RC. PGC‐1‐related coactivator: immediate early expression and characterization of a CREB/NRF‐1 binding domain associated with cytochrome c promoter occupancy and respiratory growth. Mol Cell Biol. 2006;26:7409–7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Shao D, Liu Y, Liu X, et al. PGC‐1 beta‐regulated mitochondrial biogenesis and function in myotubes is mediated by NRF‐1 and ERR alpha. Mitochondrion. 2010;10:516–527. [DOI] [PubMed] [Google Scholar]
- 117. Vercauteren K, Gleyzer N, Scarpulla RC. PGC‐1‐related coactivator complexes with HCF‐1 and NRF‐2beta in mediating NRF‐2(GABP)‐dependent respiratory gene expression. J Biol Chem. 2008;283:12102–12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Vercauteren K, Gleyzer N, Scarpulla RC. Short hairpin RNA‐mediated silencing of PRC (PGC‐1‐related coactivator) results in a severe respiratory chain deficiency associated with the proliferation of aberrant mitochondria. J Biol Chem. 2009;284:2307–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1‐PGC‐1alpha transcriptional complex. Nature. 2007;450:736–740. [DOI] [PubMed] [Google Scholar]
- 120. Safdar A, Little JP, Stokl AJ, Hettinga BP, Akhtar M, Tarnopolsky MA. Exercise increases mitochondrial PGC‐1alpha content and promotes nuclear‐mitochondrial cross‐talk to coordinate mitochondrial biogenesis. J Biol Chem. 2011;286:10605–10617. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 121. Nouws J, Goswami AV, Bestwick M, McCann BJ, Surovtseva YV, Shadel GS. Mitochondrial ribosomal protein L12 is required for POLRMT stability and exists as two forms generated by alternative proteolysis during import. J Biol Chem. 2016;291:989–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Surovtseva YV, Shutt TE, Cotney J, et al. Mitochondrial ribosomal protein L12 selectively associates with human mitochondrial RNA polymerase to activate transcription. Proc Natl Acad Sci USA. 2011;108:17921–17926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Wang Z, Cotney J, Shadel GS. Human mitochondrial ribosomal protein MRPL12 interacts directly with mitochondrial RNA polymerase to modulate mitochondrial gene expression. J Biol Chem. 2007;282:12610–12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Enriquez JA, Fernandez‐Silva P, Perez‐Martos A, Lopez‐Perez MJ, Montoya J. The synthesis of mRNA in isolated mitochondria can be maintained for several hours and is inhibited by high levels of ATP. Eur J Biochem. 1996;237:601–610. [DOI] [PubMed] [Google Scholar]
- 125. Lodeiro MF, Uchida AU, Arnold JJ, Reynolds SL, Moustafa IM, Cameron CE. Identification of multiple rate‐limiting steps during the human mitochondrial transcription cycle in vitro. J Biol Chem. 2010;285:16387–16402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Infantino V, Castegna A, Iacobazzi F, et al. Impairment of methyl cycle affects mitochondrial methyl availability and glutathione level in Down's syndrome. Mol Genet Metab. 2011;102:378–382. [DOI] [PubMed] [Google Scholar]
- 127. Bellizzi D, D'Aquila P, Scafone T, et al. The control region of mitochondrial DNA shows an unusual CpG and non‐CpG methylation pattern. DNA Res. 2013;20:537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Ghosh S, Sengupta S, Scaria V. Comparative analysis of human mitochondrial methylomes shows distinct patterns of epigenetic regulation in mitochondria. Mitochondrion. 2014;18:58–62. [DOI] [PubMed] [Google Scholar]
- 129. van der Wijst MG, Rots MG. Mitochondrial epigenetics: an overlooked layer of regulation? Trends Genet. 2015;31:353–356. [DOI] [PubMed] [Google Scholar]


