Abstract
The development of CRISPR/Cas9-mediated base editors (BEs) provided a versatile tool for precise genome editing. The recently developed xCas9-derived base editors (xBEs) that recognize the NG PAM substantially expand the targeting scope in the genome, while their editing efficiency needs to be improved. Here, we described an improved version of xBEs by fusing the BPNLS and Gam to the N terminus of xBEs (BPNLS-Gam-xBE3 and BPNLS-xABE), and this version of base editor displayed higher targeting efficiency for the majority of detected sites. By using this improved version of xBEs, we successfully created and corrected pathogenic mutations at genomic sites with the NGN protospacer-adjacent motif in human cells. Lastly, we used BPNLS-Gam-xBE3 to model pathogenic mutations in discarded human tripronuclear (3PN) zygotes, and no obvious off-targets and indels were detected. Taken together, the data in our study offer an efficient tool for precise genome editing and, thus, an enriched base editing toolkit.
Keywords: CRISPR/Cas, base editor, xCas9, base editing efficiency, pathogenic mutation, Wilson disease
Introduction
Base editing is a recently developed approach that can efficiently introduce point mutations in a programmable way without inducing double-strand breaks (DSBs). Two classes of DNA base editor—cytosine base editor (CBE) and adenine base editor1—have been described.2, 3 Both base editors were constructed by fusing a nucleotide deaminase to a catalytically impaired Cas9 nickase, and both possess the ability for single base substitutions.2, 3 The CBE can convert a C⋅G base pair into a T⋅A base pair, and the adenine base editor (ABE) converts an A⋅T base pair into a G⋅C base pair.4 DNA base editors have been widely used in plants, animals, and human embryos to mimic or correct pathogenic mutations.5, 6, 7
One of the limitations for the base editors based on Streptococcus pyogenes Cas9 (spCas9, which recognizes the NGG protospacer-adjacent motif [PAM]) is the lack of suitable PAM sequences for some cases. Therefore, several other base editors recognizing modified PAM were also developed.8, 9, 10 These newly developed base editors have been applied for animal modeling and pathogenic mutation correction.7, 11 Among these variants, xCas9-derived base editors (xBE3 and xABE) substantially broaden the targeting scope with their expanded NG PAM.9 However, the editing efficiency of xBE3 and xABE is relatively low when compared with that of spCas9-derived base editors.10
Previous studies have demonstrated that biparticle nucleus localization signal (BPNLS) efficiently localizes fusion elements to nuclear and, thus, enhances gene editing.1, 12, 13 The Gam protein of bacteriophage Mu binds DSBs to protect them from degradation, thus decreasing the formation of insertions or deletions (indels).14 Recently published studies have also demonstrated that the addition of BPNLS and Gam to the N terminus of base editors can improve editing efficiency and product purity, respectively.15, 16 Therefore, we attempted to optimize xBE3 with the fusion of BPNLS and Gam to the N terminus of xBE3 (BPNLS-Gam-xBE3). Similarly, BPNLS was also fused to the N terminus of xABE (BPNLS-xABE) for further improvement. Our results demonstrated that the editing efficiency of the optimized version of xBE3 and xABE was significantly increased. Furthermore, we applied BPNLS-Gam-xBE3 and BPNLS-xABE for pathogenic mutation creation and correction in human cells and discarded human tripronuclear zygotes. Taken together, the data in our study provide a new tool for precise genome editing and, thus, an enriched base editing toolkit.
Results
BPNLS and Gam Improve the Editing Efficiency and Purity in Human Cells
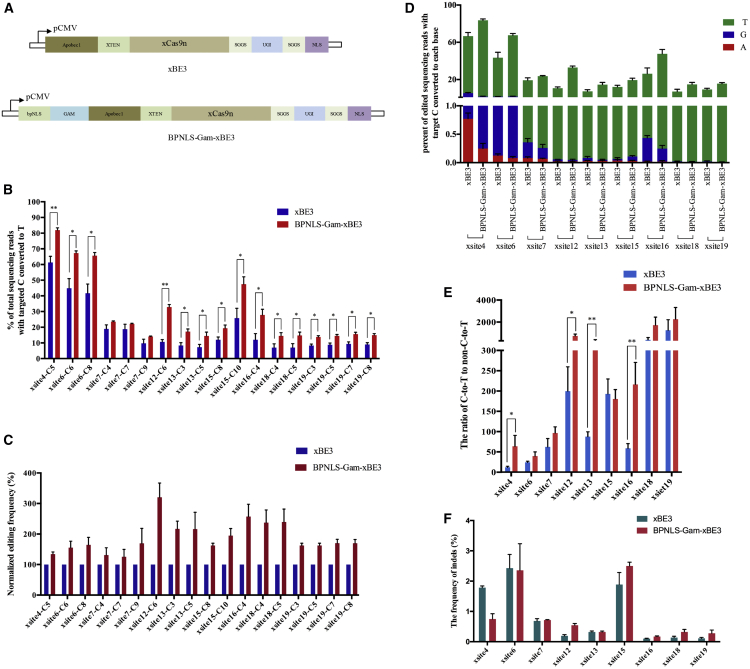
First, we fused Gam and BPNLS to the N terminus of xBE3 individually or simultaneously to generate Gam-xBE3, BPNLS-xBE3, and BPNLS-Gam-xBE3, respectively (Figure 1A; Figure S1A). Then the editing efficiency was compared between xBE3, BPNLS-xBE3, Gam-xBE3, and BPNLS-Gam-xBE3; the results showed that the fusion of BPNLS or Gam increased editing efficiency modestly, while the fusion of BPNLS and Gam simultaneously rendered a large increase in editing efficiency, which suggests that BPNLS and Gam may act synergistically (Figure S1B). Subsequently, the editing efficiency and product purity of xBE3 and BPNLS-Gam-xBE3 were further investigated. HEK293T cells were transfected with plasmids expressing xBE3 or BPNLS-Gam-xBE3 and single guided RNAs (sgRNAs) used in the previous study.9 Editing frequency was estimated using Sanger sequencing and calculated by using the online software EditR (https://moriaritylab.shinyapps.io/editr_v10/).17 The sequence chromatogram showed that, at sites with NG PAM (Figure S1C, xsite4, xsite6, and xsite12 for NGG, NGT, and NGC, respectively, and xsite15 and xsite16 for NGA) and GAT PAM (Figure S1C, xsite19), both xBE3 and BPNLS-Gam-xBE3 induced C-to-T base conversions. This is consistent with the previous study that xCas9 recognizes a broad range of PAM sequences.9 The quantification and normalization of editing efficiency demonstrated that BPNLS-Gam-xBE3 can mediate C-to-T conversion more efficiently than xBE3 at the indicated loci (Figures S1D and S1E). Subsequently, we applied deep sequencing to determine the base substitution frequencies and product purity for several sgRNAs. As confirmed by deep sequencing, BPNLS-Gam-xBE3 mediated higher C-to-T conversion frequencies at the indicated sites (Figures 1B and 1C). In terms of product purity, fewer non-C-to-T substitutions and higher C-to-T editing ratios were induced by BPNLS-Gam-xBE3 when compared to xBE3 (Figures 1D and 1E). The indels were also analyzed by deep sequencing, and the results showed that no significant difference was detected between the two systems (Figure 1F). Taken together, BPNLS-Gam-xBE3 exhibited a higher C-to-T conversion frequency and increased product purity at the indicated loci.
Figure 1.
BPNLS-Gam-xBE3 Has Higher Editing Efficiency and Purity than xBE3 in Human Cells
(A) Schematic diagram illustrating the design of expression vectors of xBE3 and BPNLS-Gam-xBE3. (B) Histogram of editing efficiency between xBE3 and BPNLS-Gam-xBE3. The editing efficiency was indicated as percentage of total sequencing reads with targeted C-to-T conversion at indicated sites. Data from three independent experiments are indicated as means ± SEM. Asterisks indicate statictical significance. *p < 0.05; **p < 0.01. (C) Normalized editing frequency of xBE3 and BPNLS-Gam-xBE3. Set the editing frequency of xBE3 as 100 percent. (D) Bar graph of editing purity of xBE3 and BPNLS-Gam-xBE3. The editing purity is determined by non-C-to-T base substitution at indicated sites. C-to-A, C-to-G, and C-to-T conversions are indicated as bars in red, blue, and green, respectively. Data from three independent experiments are indicated as means ± SEM. (E) The editing purity between xBE3 and BPNLS-Gam-xBE3 was indicated as the ratio of wanted editing (C-to-T) to unwanted editing (non-C-to-T) at indicated sites. Data from three independent experiments are indicated as means ± SEM. Asterisks indicate statistical significance. *p < 0.05; **p < 0.01. (F) The indel frequencies were determined at indicated loci in cells treated with xBE3 or BPNLS-Gam-xBE3.
Similarly, we also generated BPNLS-xABE by fusing BPNLS to the N terminus of xABE (Figure S2A). As expected, both xABE and BPNLS-xABE induced A-to-G base substitutions at the indicated loci (Figure S2B). The quantification and normalization results illustrated that the editing efficiency of BPNLS-xABE is significantly increased when compared to xABE and that the editing frequency of BPNLS-xABE could achieve more than 60% (Figures S2C and S2D).
Modeling Pathogenic Mutations Using the Improved Base Editors in Human Cells
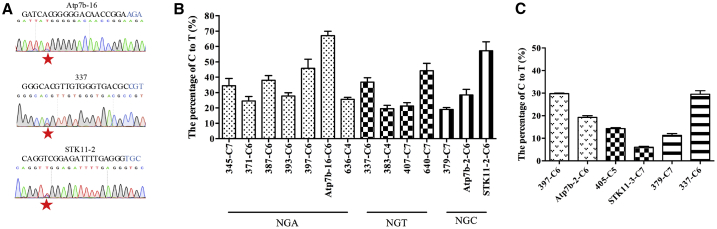
More than half of human genetic diseases are caused by point mutations, and base editors provide useful tools to mimic or correct most of these point mutations.4 Therefore, we chose 14 sgRNAs owing to the NGH PAM based on the UniProt human diseases database (https://www.uniprot.org/docs/humsavar). (The 14 pathogenic sites used here are listed in Table S2, and the pathogenic bases are highlighted in red.) Then, we attempted to mimic the pathogenic mutations using BPNLS-Gam-xBE3 in human cells. As expected, BPNLS-Gam-xBE3 generated the pathogenic mutations at all 14 pathogenic sites in HEK293T cells (Figure 2A; Figure S3C). Quantification of the editing efficiency for each target site showed that all these target sites with NGH PAM could be successfully edited with high targeting efficiencies (Figure 2B). It is noticeable that, at some target sites, BPNLS-Gam-xBE3 induces not only desired base substitutions but also undesired by-stand base editing (e.g., Atp7b-16 and 383 sites), and this was also observed in cells transfected with xBE3 and corresponding sgRNAs (Figures S3A and S3B). We also generated pathogenic mutations using BPNLS-xABE for other 14 target sites owing to the NGH PAM in HEK293T cells (Figures S4 and S5). (The pathogenic sites used here are listed in Table S2, and the target bases are highlighted in red.) The editing frequency demonstrated that BPNLS-xABE could efficiently induce A-to-G conversion at the pathogenic sites with NGH PAM in human cells (Figures S4A, S4B, and S5).
Figure 2.
Modeling of Pathogenic Mutations Using BPNLS-Gam-xBE3 in Human Cells
(A) Sequence chromatogram of representative pathogenic sites with NGA, NGT, or NGC PAM edited by BPNLS-Gam-xBE3. The red stars indicate the desired pathogenic mutations introduced by BPNLS-Gam-xBE3, and the PAM sequences are highlighted in blue. (B) The editing efficiency of BPNLS-Gam-xBE3 on different pathogenic sites with NGH PAM. The editing frequencies were determined by the percentage of C-to-T conversion. The target loci with same PAM were underlined and indicated. Data from at least three independent experiments are shown as means ± SEM. (C) The editing efficiency of BPNLS-Gam-xBE3 at multiple loci in human cells. Cells were transfected with BPNLS-Gam-xBE3 and two sgRNAs simultaneously, and the sgRNAs transfected simultaneously were filled in same pattern. Data from three independent experiments are indicated as means ± SEM.
In some cases, it is necessary to model multiple pathogenic mutations simultaneously. Thus, we evaluated whether BPNLS-Gam-xBE3 and BPNLS-xABE could edit multiple sites simultaneously. We tested two sites simultaneously for three different combinations. As shown in Figures 2C and S4C, both BPNLS-Gam-xBE3 and BPNLS-xABE were able to model two pathogenic mutations simultaneously in human cells. Taken together, BPNLS-Gam-xBE3 and BPNLS-xABE can successfully model pathogenic mutations at single or multiple sites in human cells.
Modeling and Correcting the Pathogenic Mutations in Human Cells
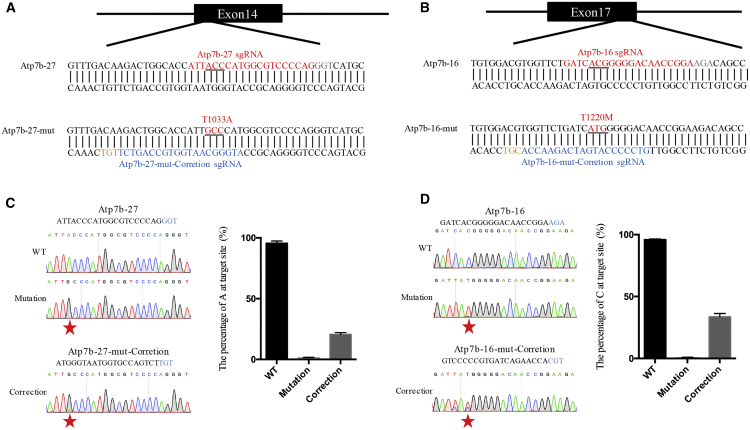
Then we attempted to create the pathogenic mutations of the human Atp7b gene and then correct them using BPNLS-Gam-xBE3 and BPNLS-xABE in human cells. Impaired function of the human copper transport protein Atp7b is associated with Wilson disease,18, 19 and point mutation in the Atp7b gene is the most common manner for pathogenic variants.20 In Wilson disease (WD) patients, copper accumulated in tissues, including liver, brain, muscle, and so on, resulting in neuropsychiatric, hepatic, and systemic disabilities.21 First, we constructed two sgRNAs targeting pathogenic mutations T1033A and T1220M, respectively (hereinafter, Atp7b-27 and Atp7b-16). T1033A is caused by A-to-G substitution and can be modeled by BPNLS-xABE, while T1220M is caused by C-to-T substitution and can be modeled by BPNLS-Gam-xBE3 (Figures 3A and 3B). Then, we transfected BPNLS-Gam-xBE3 or BPNLS-xABE with corresponding sgRNAs into HEK293T cells to generate Atp7b mutant cell lines. A single cell was sorted and plated into 96-well plates using fluorescence-activated cell sorting (FACS), and a monoclone was picked and verified by Sanger sequencing. We chose cell clones with target base conversion for subsequent experiments. The sequence chromatogram demonstrated that the pathogenic mutations were successfully created by using BPNLS-Gam-xBE3 or BPNLS-xABE in human cells (Figures 3C and 3D).
Figure 3.
Modeling and Correction of Pathogenic Mutations in Human Cells
(A) Schematic illustration indicates the sequence and position of the pathogenic site Atp7b-27. The sequence before editing is indicated as Atp7b-27, and the sequence after editing is indicated as Atp7b-27-mut. In the Atp7b-27 sequence, the sgRNA sequence for modeling Atp7b-27 pathogenic mutation is highlighted in red, and the PAM sequence is shown in light gray. In the Atp7b-27-mut sequence, the sgRNA sequence for correcting Atp7b-27 pathogenic mutation is highlighted in blue, and the PAM sequence is indicated in orange. Mutated amino acid codon is underlined and indicated in red in the Atp7b-27-mut sequence. (B) Schematic diagram illustrating the position and sequence of pathogenic site Atp7b-16. The wild-type sequence is shown as Atp7b-16, and the edited sequence is shown as Atp7b-16-mut. In the Atp7b-16 sequence, the sgRNA sequence for modeling Atp7b-16 mutation is highlighted in red, and the PAM sequence is indicated in light gray. In the Atp7b-16-mut sequence, the sgRNA sequence for correcting Atp7b-16 mutation is highlighted in blue, and the PAM sequence is indicated in orange. The mutated amino acid codon is underlined and indicated in red in the Atp7b-16-mut sequence. (C) Modeling and correcting of pathogenic site Atp7b-27. Left: sequence chromatogram of Atp7b-27 in wild-type (WT), mutation, and correction cells. Right: the percentage of target base in wild-type, mutation, and correction cells. Red stars indicate the desired editing. The percentage of each base was calculated using EditR. Data from three independent experiments are indicated as means ± SEM. (D) Modeling and correcting of pathogenic site Atp7b-16. Left: sequence chromatogram of Atp7b-16 in wild-type, mutation, and correction cells. Right: the percentage of target base in wild-type, mutation, and correction cells. Red stars indicate the desired editing. The percentage of each base was calculated using EditR. Data from three independent experiments are indicated as means ± SEM.
With this success, we constructed sgRNAs for correcting the generated mutation sites in the mutant cells. The results showed that both mutations in Atp7b can be successfully corrected with high efficiencies (20% for Atp7b-27 and 40% for Atp7b-16) (Figures 3C and 3D). Taken together, BPNLS-Gam-xBE3 and BPNLS-xABE can efficiently create and correct pathogenic mutations in human cells.
Pathogenic Modeling in Human Tripronuclear Zygotes Using BPNLS-Gam-XBE3
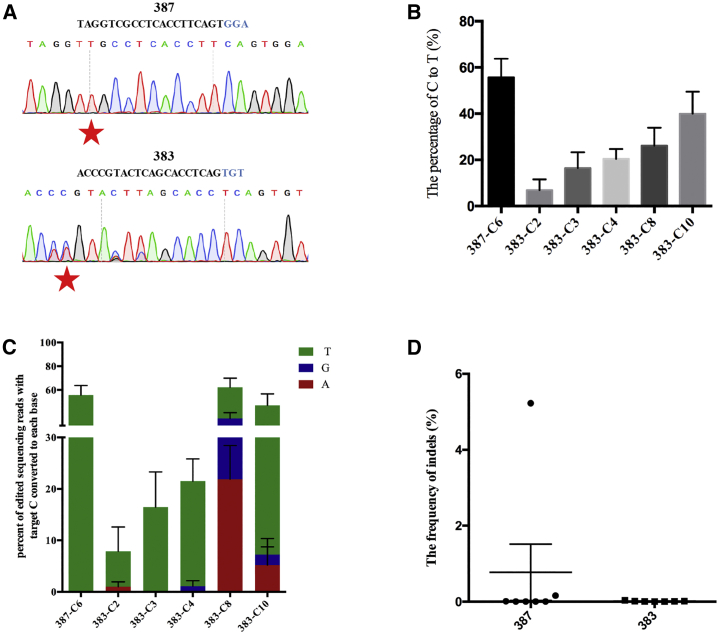
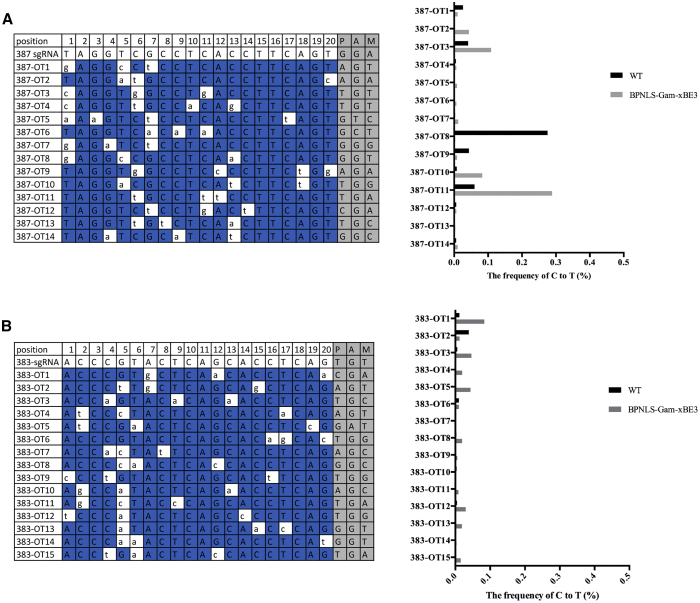
We then attempted to mimic the pathogenic mutations in vivo using discarded human tripronuclear (3PN) zygotes. We carried out base editing in discarded human 3PN zygotes by microinjection of BPNLS-Gam-xBE3 and sgRNAs. The injected 3PN zygotes were cultured for 2 days and then collected for targeted analysis. A sequence chromatogram indicated that targeted base substitutions were successfully achieved in both targeting sites (Figure 4A). Then, targeted deep sequencing was performed to determine the editing frequencies and product purity. The results illustrated that BPNLS-Gam-xBE3 can mediate C-to-T conversion at site 387 at a mean efficiency of about 60% (Figure 4B). For site 383, except for the desired base C4, base substitutions were also observed at undesired bases C2, C3, C8, and C10 in a varied frequency (Figure 4B). Additionally, the product purity and the indels in human 3PN zygotes were also analyzed. The results demonstrated that the product purity of BPNLS-Gam-xBE3 at site 387 was desirable. For site 383, non-C-to-T substitutions were noticeable at C8 and C10 sites (Figure 4C). This editing mode was also observed in human cells at the same site and seems to be site specific (Figure S3). Notably, no obvious indels were detected for the two targeted sites (Figure 4D).
Figure 4.
BPNLS-Gam-xBE3 Can Model Pathogenic Mutations in Discarded Human 3PN Zygotes
(A) The sequence chromatogram of 387 and 383 sites edited using BPNLS-Gam-xBE3 in human 3PN zygotes. The PAM sequences are highlighted in blue. Red stars indicate the desired pathogenic mutation introduced by BPNLS-Gam-xBE3. (B) Histogram of editing efficiency of 387 and 383 sites by BPNLS-Gam-xBE3 in human 3PN zygotes. The editing frequencies were determined by the percentage of C-to-T conversion. Data from seven independent experiments are shown as means ± SEM. (C) Bar graph of editing purity of BPNLS-Gam-xBE3 in human 3PN embryos. The editing purity is indicated as the percentage of edited sequencing reads with targeted C converted to T or A or G. C-to-A, C-to-G, and C-to-T conversions are indicated as bars with red, blue, and green, respectively. (D) The indel frequencies were determined at indicated loci in human 3PN zygotes microinjected with BPNLS-Gam-xBE3 and sgRNAs.
Off-Target Effect Was Analyzed in 3PN Zygotes
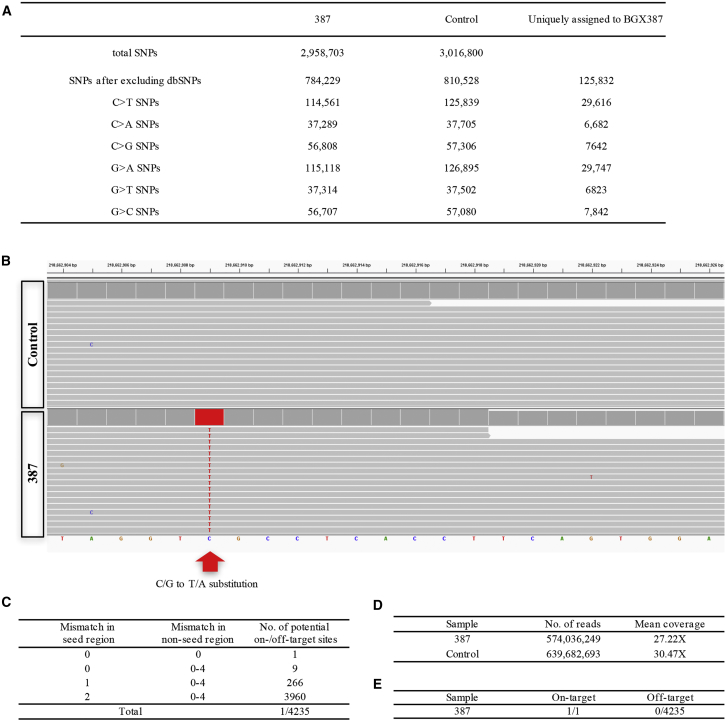
The off-target mutagenesis is a major concern for all genome editing approaches, including the CRISPR/Cas system.22 To assess the off-target effects of BPNLS-Gam-xBE3 in vivo, we performed deep sequencing for the potential off-target sites of 387 and 383, respectively. The potential off-target sites were predicted by an online tool Cas-OFFinder;23 14 and 15 potential off-target sites in the genome with up to 3-nt mismatches for 387 and 383 sites, respectively, were chosen in this study. The PCR products using the genomic DNA of each sample were mixed with an equal amount and subjected to deep sequencing. No obvious base substitution was detected at these potential off-target sites in both the BPNLS-Gam-xBE3 and wild-type (WT) groups (Figures 5A and 5B). To better characterize the off-target effect of BPNLS-Gam-xBE3, whole-genome sequencing was performed at mean depths of 27× and 30× for the BPNLS-Gam-xBE3 and wild-type groups, respectively (Figure 6D). After filtering out human database of SNPs (dbSNPs), a total of 784,229 and 810,528 SNPs were identified over the genome of human 3PN zygotes with the 387 site edited and the control, respectively (Figure 6A). The on-target editing was also confirmed by whole-genome sequencing (WGS) (Figure 6B). The potential off-target sites were explored using the criteria that up to 2-bp mismatch in the seed region and 4-bp mismatch in the non-seed region with NG PAM (Figure 6C). Among 4,234 potential off-target sites, none was detected in the human 3PN zygotes edited by BPNLS-Gam-xBE3 (Figure 6E). Taken together, these results demonstrated that BPNLS-Gam-xBE3 can generate precise C-to-T base conversion for target sites with NG PAM in human 3PN zygotes. These data indicate that our improved xBEs are reliable tools for pathogenic modeling in an ex vivo model.
Figure 5.
Off-Target Detection of the Edited Human 3PN Zygotes at Predicted Sites
(A) The off-target analysis of the 387 site in human 3PN zygotes. Left: the sequence information of 387 sgRNA and predicted off-target sites; mismatches are highlighted in white, and PAM sequences are highlighted in gray. Right: the frequency of C-to-T conversion at predicted off-target sites. (B) The off-target analysis of the 383 site in human 3PN zygotes. Left: the sequence information of 383 sgRNA and predicted off-target sites; mismatches are highlighted in white, and PAM sequences are highlighted in gray. Right: the frequency of C-to-T conversion at predicted off-target sites.
Figure 6.
Whole-Genome Analysis of Human 3PN Zygotes with or without BPNLS-Gam-xBE3 Editing
(A) Summary of SNP analysis. A total of 2,958,703 and 3,016,800 SNPs were identified for the 387 site and control, respectively. After filtering out naturally occurring variants in the human SNP database, 784,229 SNPs were obtained over the edited 3PN zygote genome. (B) Confirmation of the on-target base editing by analyzing the whole-genome sequencing. Red arrow indicates the C/G to T/A substitution within 23 bp of the on-target sequence for the 387 site. (C) Summary of on- and/or off-target site information. A total of 4,235 sites, including 1 on-target site and 4,234 potential off-target sites were analyzed. (D) Summary of the whole-genome sequencing. (E) Summary of on- and off-target analysis.
Discussion
The CRISPR/Cas9 genome editing technology is driving a revolution in biotechnology and biomedicine research.24 The low efficiency of homologous recombination induced by Cas9 nuclease has been a major obstacle for precise editing. Base editors are developed by the fusion of deaminase to catalytically impaired Cas9 nickase and can mediate precise base substitutions, which enable a wide variety of research and medical applications. The targeting scope of base editors are largely expanded by recently developed xBE3 and xABE, which could target the sequences with NG, GAA, or GAT PAM, while the editing efficiency is a limitation for these two base editors.9
In this study, we performed fusion of BPNLS and Gam to the N terminus of xBE3 as well as fusion of BPNLS to xABE to generate BPNLS-gam-xBE3 and BPNLS-xABE, respectively. By assessing the editing efficiency and product purity, we illustrated that BPNLS-Gam-xBE3 could mediate C-to-T base conversion at a higher efficiency and that it induces fewer unwanted base substitutions (Figure 1; Figure S1). Meanwhile, BPNLS-xABE also achieved an editing frequency as high as 3-fold that mediated by xABE (Figure S2). Thus, we succeeded in the optimization of xCas9-derived base editors to improve their editing ability, though the editing efficiency varies. Subsequently, we evaluated the ability of BPNLS-Gam-xBE3 and BPNLS-xABE to model the pathogenic mutations in human cells and demonstrated that both of them can model pathogenic mutations efficiently in human cells (Figure 2; Figures S3, S4, and S5). It is noticeable that the overall editing frequency of BPNLS-xABE is lower than that of BPNLS-Gam-xBE3, and this may be due to the lack of DNA binding ability of TadA.25 Modification of nucleotide deaminase may improve this phenomenon in the future. Furthermore, we attempted to create and correct two pathogenic mutations for human Atp7b gene, which is associated with Wilson disease.18 By transfecting BPNLS-Gam-xBE3 and BPNLS-xABE with corresponding sgRNAs (including mutation modeling sgRNAs and correction sgRNAs), we possessed two cell lines containing A-to-G and C-to-T base substitutions at Atp7b-27 and Atp7b-16, respectively, and achieved correction of the mutant bases (Figure 3). These results suggest that BPNLS-Gam-xBE3 and BPNLS-xABE have the potential for modeling and correcting human pathogenic point mutations in a broad range of genomes. In addition, we performed pathogenic mutation modeling using BPNLS-Gam-xBE3 in discarded human 3PN zygotes; the results illustrated that BPNLS-Gam-xBE3 had the ability to model pathogenic mutation in human 3PN zygotes with no obvious detectable off-target. Another SpCas9 variant SpCas9-NG, which also recognizes NG PAM, was recently reported to potentially have higher editing efficiency than xCas9.10 Since xCas9 has been suggested to possess higher fidelity and also recognize sequences with GAT and GAA PAM,9 these two variants together have thus broadened the genome editing toolkit and could work synergistically.
Two most recently published studies described that, in both plant and animal, spCas9-derived BE3 can mediate genome-wide off-target single-nucleotide variants (SNVs) in vivo while the adenine base editor performs in a better way,26, 27 and this seems mainly due to the high DNA binding affinity of Apobec1. In this study, we performed in vitro off-target detection at predicted sites, and no obvious off-target was detected; this is consistent with the two studies that found that the sgRNA-dependent off-target is very limited.26, 27 In addition, whole-genome sequencing was performed in this study to further analyze the potential off-target effect of BPNLS-Gam-xBE3, and the results indicated that, among 4,234 potential off-target sites, no obvious editing was detected in this study. For BE3-induced off-target SNVs, the use of functional optimized cytidine deaminase may help to reduce off-target mutations; thus, future work in the modification of cytidine deaminases to decrease its DNA binding ability while maintaining cytidine deaminase activity is necessary.
To summarize, we generated BPNLS-Gam-xBE3 and BPNLS-xABE to optimize xCas9-derived base editors as more efficient base editing tools. Indeed, BPNLS-Gam-xBE3 and BPNLS-xABE possess higher editing efficiency and product purity for sequences with NG PAM. These two base editors have the capability to mediate base substitution in a precise manner and can be used for modeling and correcting genetic defects in the future.
Materials and Methods
Discarded Human 3PN Zygotes
All of the human 3PN zygotes used in this study were collected from the Center for Reproductive Medicine of the Third Affiliated Hospital of Guangzhou Medical University, and the study was approved by the Ethics Committee of the hospital. All of the patients have signed the informed consent form before the sample collection. All of the operations on the embryos were conducted at the Center for Reproductive Medicine.
Plasmid Construction
For construction of the pCMV-BPNLS-Gam-xBE3 plasmid, BPNLS and Gam sequence were amplified, verified, and then cloned into the N terminus of the pCMV-xBE3 expression vector. For the pCMV-BPNLS-xABE plasmid, the same strategy was used as that in construction of the pCMV-BPNLS-Gam-xBE3 plasmid. The pGL3-U6-sgRNA-PGK-GFP plasmid was generated from pGL3-U6-sgRNA-PGK-puromycin (Addgene, 51133) by replacing the puromycin element with GFP sequence and was used as the sgRNA mammalian cell expression vector. The pUC57-sgRNA expression vector (Addgene, 51132) was used to generate the template for in vitro transcription. The sgRNA oligos were synthesized, annealed, and cloned into the pGL3-U6-sgRNA-PGK-GFP and pUC57-sgRNA vectors.
Cell Culture and Transfection
HEK293T cells were cultured in DMEM (GIBCO), supplemented with 10% fetal bovine serum (FBS; v/v) (Gemini, 900-108), and incubated at 37°C with 5% CO2 under humid conditions. For plasmid transfection, cells were seeded on poly-D-lysine-coated 12-well plates (JET BIOFIL) the day before and transfected at approximately 70% confluence using Lipofectamine 2000 reagent (Life Technologies) according to the manufacturer’s protocols. 1,000 ng base editor (including BPNLS-Gam-xBE3, xBE3, BPNLS-xABE, and xABE) and 600 ng corresponding sgRNAs were transfected into cells per well. All sgRNAs used in this study are listed in Tables S1 and S2. Three days after transfection, GFP-positive cells were collected from FACS.
Genomic DNA Extraction and Amplification
The genomic DNA of cells harvested from FACS was extracted using QuickExtract DNA Extraction Solution (Lucigen) according to the manufacturer’s protocols. The genomic DNA of zygotes was amplified using the Discover-sc Single Cell Kit (Vazyme, N601-01) according to the manufacturer’s protocols. The target sequences were amplified and sequenced using the primers listed in Table S3.
In Vitro Transcription
In vitro transcription was performed according to the protocols reported previously.28 In brief, the pCMV-BPNLS-Gam-xBE3 vector was linearized by BbsI enzyme (New England Biolabs) and performed to in vitro transcription using the T7 Ultra Kit (Ambion) according to the manufacturer’s protocols. All the sgRNAs used for in vitro transcription were cloned into a pUC57-sgRNA expression vector with T7 promoter. Then, the sgRNAs were amplified and transcribed in vitro using the MEGAshortscript Kit (Ambion) and purified using the MEGAclear Kit (Ambion) according to the manufacturer’s protocols.
Targeted Deep Sequencing
The potential off-target sites of B-383 and B-387 were predicted by Cas-OFFinder (http://www.rgenome.net/cas-offinder). The on-target and off-target sites were amplified from human genomic DNA using Phanta Max Super-Fidelity DNA Polymerase (Vazyme, P505). The paired-end sequencing of purified PCR products was performed using the HiSeq X-10 (2 × 150 bp) platform at the CAS-MPG Partner Institute for Computational Biology Omics Core, Shanghai, China. The BWA-MEM algorithm was used for processing the deep-sequencing data. Primers used for on-target and off-target sequencing are listed in Table S4 and Table S5, respectively.
Whole-Genome Sequencing for the Off-Target Detection
Whole-genome sequencing was performed to analyze the off-target effect induced by BPNLS-Gam-xBE3 in human 3PN zygotes. Genomic DNA of human 3PN zygotes was sequenced using an Illumina HiSeq X Ten system (2 × 150 pair-end [PE]) at the Novogene Bioinformatics Institute, Beijing, China. To map the sequencing data with a human reference genome (GRCh38/hg38), BWA v0.7.16 was used. Sequencing reads were marked for duplicates using Sambamba v0.6.7 and realigned using the Genome Analysis Toolkit (GATK v3.7) IndelRealigner. Variants and the quality were identified and evaluated using GATK HaplotypeCaller and GATK VariantFiltration, respectively. The common SNPs between the edited and control embryos were removed.
Statistical Analysis
Statistical analysis was performed by two-tailed Student’s t test. Data are shown as mean ± SEM, unless stated otherwise. Quantification of editing efficiency with Sanger sequencing data was performed with online software EditR. Statistical analysis was performed using Graph Pad PRISM 7.
Data Availability
All the deep-sequencing data can be accessed at the National Omics Data Encyclopedia (NODE) of CAS-MPG Partner Institute for Computational Biology (PICB), Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (https://www.biosino.org/node) (NODE: OEP000253).
Author Contributions
L.W., J.L., and X.H. conceived, designed, and supervised the project. X.L. and G.L. performed most experiments. X.Z. provided expert technical assistance. X.L. and G.L. wrote the manuscript. R.W. provided expert help in processing the experiments. Y.Q. and S.T. provided advice on the project and edited the manuscript. L.W and X.H. managed the project. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was approved by the Ethics Committee of the Third Affiliated Hospital of Guangzhou Medical University. We thank members of the Wang, Liu, and Huang labs for helpful discussions. This work was supported by the Innovation of Science and Technology Commission of Shenzhen, China (jcyj20170307100911479), the National Key R&D Program (2016YFC0905901), and the China Postdoctoral Science Foundation (2019M650230).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.06.024.
Contributor Information
Jianqiao Liu, Email: liujqssz@gzhmu.edu.cn.
Lisheng Wang, Email: wangls168@163.com.
Xingxu Huang, Email: huangxx@shanghaitech.edu.cn.
Supplemental Information
References
- 1.Zafra M.P., Schatoff E.M., Katti A., Foronda M., Breinig M., Schweitzer A.Y., Simon A., Han T., Goswami S., Montgomery E. Optimized base editors enable efficient editing in cells, organoids and mice. Nat. Biotechnol. 2018;36:888–893. doi: 10.1038/nbt.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R. Programmable base editing of A⋅T to G⋅C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rees H.A., Liu D.R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018;19:770–788. doi: 10.1038/s41576-018-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryu S.-M., Koo T., Kim K., Lim K., Baek G., Kim S.-T., Kim H.S., Kim D.E., Lee H., Chung E., Kim J.S. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat. Biotechnol. 2018;36:536–539. doi: 10.1038/nbt.4148. [DOI] [PubMed] [Google Scholar]
- 6.Li X., Wang Y., Liu Y., Yang B., Wang X., Wei J., Lu Z., Zhang Y., Wu J., Huang X. Base editing with a Cpf1-cytidine deaminase fusion. Nat. Biotechnol. 2018;36:324–327. doi: 10.1038/nbt.4102. [DOI] [PubMed] [Google Scholar]
- 7.Zeng Y., Li J., Li G., Huang S., Yu W., Zhang Y., Chen D., Chen J., Liu J., Huang X. Correction of the Marfan syndrome pathogenic FBN1 mutation by base editing in human cells and heterozygous embryos. Mol. Ther. 2018;26:2631–2637. doi: 10.1016/j.ymthe.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim Y.B., Komor A.C., Levy J.M., Packer M.S., Zhao K.T., Liu D.R. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol. 2017;35:371–376. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu J.H., Miller S.M., Geurts M.H., Tang W., Chen L., Sun N., Zeina C.M., Gao X., Rees H.A., Lin Z., Liu D.R. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556:57–63. doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishimasu H., Shi X., Ishiguro S., Gao L., Hirano S., Okazaki S., Noda T., Abudayyeh O.O., Gootenberg J.S., Mori H. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science. 2018;361:1259–1262. doi: 10.1126/science.aas9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Z., Lu Z., Yang G., Huang S., Li G., Feng S., Liu Y., Li J., Yu W., Zhang Y. Efficient generation of mouse models of human diseases via ABE- and BE-mediated base editing. Nat. Commun. 2018;9:2338. doi: 10.1038/s41467-018-04768-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu J., Corbett A.H., Berland K.M. The intracellular mobility of nuclear import receptors and NLS cargoes. Biophys. J. 2009;96:3840–3849. doi: 10.1016/j.bpj.2009.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki K., Tsunekawa Y., Hernandez-Benitez R., Wu J., Zhu J., Kim E.J., Hatanaka F., Yamamoto M., Araoka T., Li Z. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shee C., Cox B.D., Gu F., Luengas E.M., Joshi M.C., Chiu L.Y., Magnan D., Halliday J.A., Frisch R.L., Gibson J.L. Engineered proteins detect spontaneous DNA breakage in human and bacterial cells. eLife. 2013;2:e01222. doi: 10.7554/eLife.01222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koblan L.W., Doman J.L., Wilson C., Levy J.M., Tay T., Newby G.A., Maianti J.P., Raguram A., Liu D.R. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 2018;36:843–846. doi: 10.1038/nbt.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komor A.C., Zhao K.T., Packer M.S., Gaudelli N.M., Waterbury A.L., Koblan L.W., Kim Y.B., Badran A.H., Liu D.R. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci. Adv. 2017;3:eaao4774. doi: 10.1126/sciadv.aao4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kluesner M.G., Nedveck D.A., Lahr W.S., Garbe J.R., Abrahante J.E., Webber B.R., Moriarity B.S. EditR: a method to quantify base editing from Sanger sequencing. CRISPR J. 2018;1:239–250. doi: 10.1089/crispr.2018.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parnas O., Jovanovic M., Eisenhaure T.M., Herbst R.H., Dixit A., Ye C.J., Przybylski D., Platt R.J., Tirosh I., Sanjana N.E. A genome-wide CRISPR screen in primary immune cells to dissect regulatory networks. Cell. 2015;162:675–686. doi: 10.1016/j.cell.2015.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jayakanthan S., Braiterman L.T., Hasan N.M., Unger V.M., Lutsenko S. Human copper transporter ATP7B (Wilson disease protein) forms stable dimers in vitro and in cells. J. Biol. Chem. 2017;292:18760–18774. doi: 10.1074/jbc.M117.807263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang I.J., Hahn S.H. The genetics of Wilson disease. Handb. Clin. Neurol. 2017;142:19–34. doi: 10.1016/B978-0-444-63625-6.00003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheiber I.F., Brůha R., Dušek P. Pathogenesis of Wilson disease. Handb. Clin. Neurol. 2017;142:43–55. doi: 10.1016/B978-0-444-63625-6.00005-7. [DOI] [PubMed] [Google Scholar]
- 22.Karvelis T., Gasiunas G., Siksnys V. Harnessing the natural diversity and in vitro evolution of Cas9 to expand the genome editing toolbox. Curr. Opin. Microbiol. 2017;37:88–94. doi: 10.1016/j.mib.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Bae S., Park J., Kim J.S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brooks A.K., Gaj T. Innovations in CRISPR technology. Curr. Opin. Biotechnol. 2018;52:95–101. doi: 10.1016/j.copbio.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 25.Losey H.C., Ruthenburg A.J., Verdine G.L. Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nat. Struct. Mol. Biol. 2006;13:153–159. doi: 10.1038/nsmb1047. [DOI] [PubMed] [Google Scholar]
- 26.Jin S., Zong Y., Gao Q., Zhu Z., Wang Y., Qin P., Liang C., Wang D., Qiu J.L., Zhang F. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. 2019;364:292–295. doi: 10.1126/science.aaw7166. [DOI] [PubMed] [Google Scholar]
- 27.Zuo E., Sun Y., Wei W., Yuan T., Ying W., Sun H., Yuan L., Steinmetz L.M., Li Y., Yang H. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. 2019;364:289–292. doi: 10.1126/science.aav9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou C., Zhang M., Wei Y., Sun Y., Sun Y., Pan H., Yao N., Zhong W., Li Y., Li W. Highly efficient base editing in human tripronuclear zygotes. Protein Cell. 2017;8:772–775. doi: 10.1007/s13238-017-0459-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the deep-sequencing data can be accessed at the National Omics Data Encyclopedia (NODE) of CAS-MPG Partner Institute for Computational Biology (PICB), Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (https://www.biosino.org/node) (NODE: OEP000253).