Abstract
The discovery and engineering of the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) in the past several years have revolutionized biomedical research. The CRISPR technology showed great potential to advance detection, prevention, and treatment of human diseases in the near future. Compared to previous developed genome editing approaches, such as zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), the CRISPR-based systems have numerous advantages. One example is that the CRISPR systems can be easily adopted to efficiently target multiple genes simultaneously. Several strategies and toolboxes have been developed to achieve multiplexed targeting using the CRISPR systems. In this short review, we will discuss the principle, approach, and application of these strategies.
Introduction
The CRISPR-associated systems were discovered in prokaryotes and shown later as adaptive immune systems to defend against viruses and plasmids [1,2]. In the infected cells, short DNA sequences originating from invading pathogens are stored in the form of “spacers” in the CRISPR array in the host genome [3]. CRISPR array is transcribed and processed into small RNAs containing a single spacer known as CRISPR RNAs (crRNAs) [4]. These crRNAs bind to CRISPR-associated (Cas) proteins and another small RNA molecule called trans-activating CRISPR RNA (tracrRNA), to form the effector complexes, which recognize foreign DNA or RNA sequences by base-pairing with spacer sequences in crRNAs [4,5] (Figure 1A). Cas proteins, such as Cas9, then cleave the invaded target nucleic acid to achieve adaptive immunity response [2]. Since this discovery, extensive CRISPR-based tools for genome editing have been developed because the CRISPR systems provide a simple, quick, and efficient solution to manipulate gene expression. In fact, CRISPR engineering was one of the fastest developing fields in biology and medicine in the past five years. As one of the most successful stories, the powerful CRISPR tools have been applied to almost all model species, from bacteria to mammals, and have significantly accelerated the biological and biomedical research.
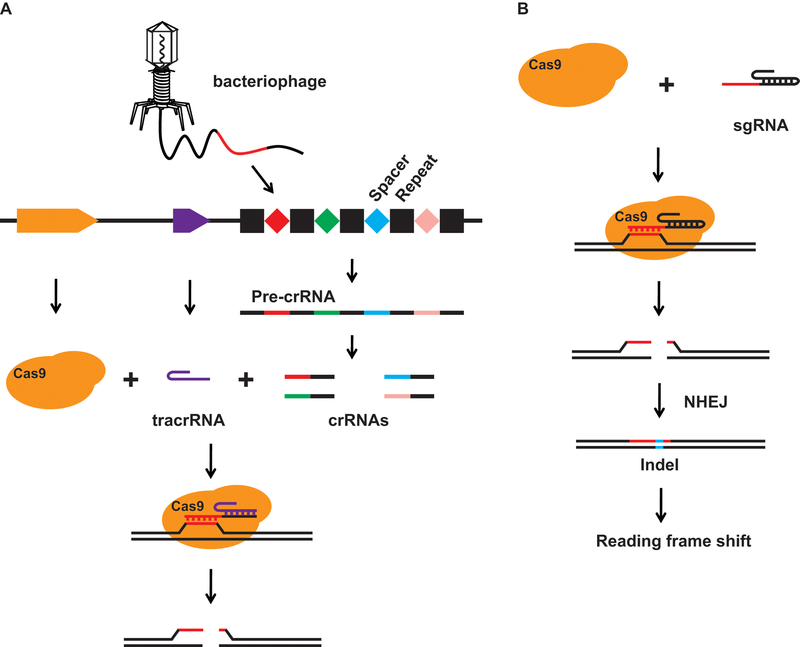
Figure 1. Natural and engineered CRISPR systems.
(A) Schematic of a natural CRISPR pathway. Foreign DNA is captured and inserted between repeats in a CRISPR locus in a bacteria genome. CRISPR array is transcribed and then processed into multiple crRNAs, each carrying a single spacer sequence and a repeat sequence. The crRNA forms a complex with Cas9 and crRNA and directs the complex to the foreign DNA carrying the same sequence as the spacer. Cas9 then generates a double strand break on the foreign DNA. (B) Schematic of an engineered Cas9 genome editing tool. It comprises a Cas9 endonuclease and a single guide RNA (sgRNA). Cas9 and sgRNA form a complex. The guide sequence on sgRNA directs the complex to the target site. Cas9 generates a double strand break, which is repaired via the error-prone non-homologous end-joining (NHEJ), leaving a small insertion or deletion (Indel) at the target site. Indels in protein coding region cause reading frame shift and early termination of protein translation, resulting in loss of protein expression.
The Engineered CRISPR platforms
Streptococcus pyogenes
Cas9 (SpCas9) was the first Cas system engineered to genome editing tools [6–8]. They include a Cas9 endonuclease and a single guide RNA (sgRNA), an artificial chimeric RNA molecule to comprise the function of crRNA and tracrRNA. sgRNA carries a direct sequence of ~20 nucleotides (nt) that pair with ssDNA target, and Cas9 is in charge of generating a double-strand break (DSB) in a targeted DNA locus [6] (Figure 1B). Through the error-prone repairing, DSB introduced by CRISPR/Cas9 abolishes the expression of selected proteins by shifting reading frames [6]. Because the SpCas9-based gene knockout tools are easy to use and highly efficient, they were quickly developed for applications in a wide range of species, including bacteria [9], yeast [10], plants [11,12], worms [13], flies [14], frogs [15], and mammals [16]. Since then, several distinct CRISPR-based tools have been developed based on natural CRISPR effectors. These enzymes include Cas9 from other species, such as FnCas9 [17], SaCas9 [18], St1Cas9 [19], St3Cas9 [20], NmCas9 [21], and other Cas proteins, such as Cas12a (Cpf1) [22] and Cas13a (C2c2) [23]. Additional Cas effectors are still under development for biotechnological applications [24].
Moreover, Cas proteins have been further developed to sequence-specific targeting platforms for a wide range of purposes in addition to generating DSB (Figure 2). These purposes are achieved by fusing nuclease dead mutants of Cas (dCas) proteins with different functional domains [25]. For example, fusing dCas to transcriptional activation domains, such as VP64, or transcriptional repressors, such as KRAB, were used to alter the expression of genes [26–29]. Fusing dCas proteins to modulators of epigenetic markers can manipulate DNA methylation or histone modifications at the selected loci [25]. Additionally, fluorescent proteins fused to dCas9 provided a tool for visualizing native genomic loci in living cells [30].
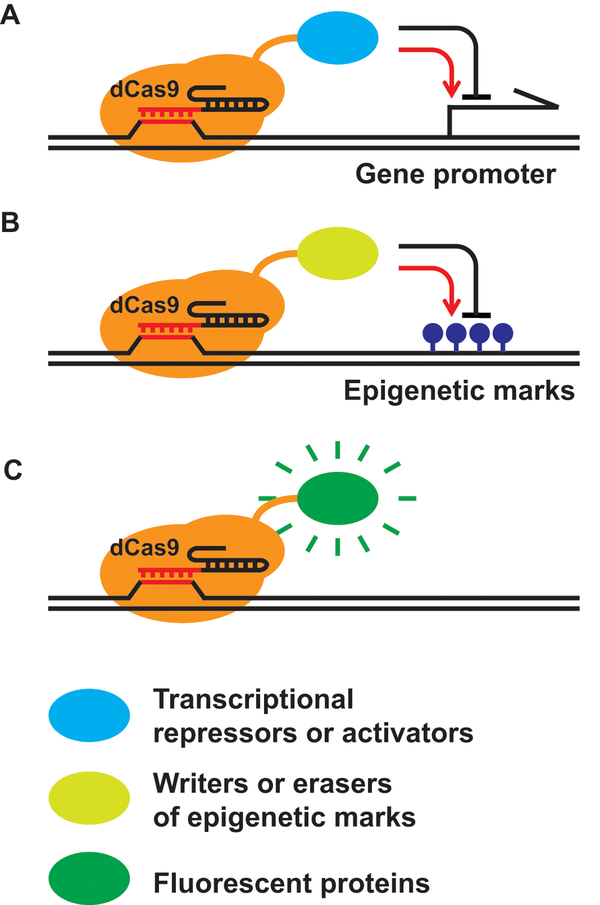
Figure 2. CRISPR-based applications with nuclease-deficient Cas9 (dCas9).
dCas9 serves as a sequence-directed genome target platform. Fusing dCas9 to different functional domains allows for a wide range of applications, including transcriptional up-regulation (with transcriptional activators) and down-regulation (with transcriptional repressors) (A), adding (with epigenetic writers) or removing (with epigenetic erasers) epigenetic marks (B), and visualization of specific genomic regions (C).
Most of these studies have been performed using SpCas9, but it can be replaced with other Cas proteins. Furthermore, sgRNA has also been engineered with additional scaffolds to extend its function [31].
The Multiplexed CRISPR systems
The CRISPR systems can be easily adapted to target multiple genes because CRISPR-introduced targeting relies on base-pairing between guide and target nucleotide sequences. Therefore, multiplexing can be achieved by introducing several sgRNAs simultaneously. In comparison, ZFNs and TALENs, two genome editing techniques developed before CRISPR, recognize targets via protein-DNA interaction. Targeting a new location using either system requires quite extensive engineering, including designing, constituting and introducing a new nuclease.
Multiplexing the single targeting systems
CRISPR-introduced single gene targeting can be achieved by delivering in vitro prepared Cas protein-sgRNA ribonucleoproteins (RNPs), Cas protein mRNA and sgRNA, or plasmid(s) encoding Cas protein and sgRNA. In these approaches, adding additional sgRNAs allows targeting multiple genes at the same time. For examples, multiple in vitro transcribed sgRNAs can be preloaded to purified Cas protein to form pooled RNPs, before transfection into cells [32], animals [33], or plant protoplasts [34] (Figure 3A). An alternative way is to introduce mRNA of Cas protein and several sgRNAs simultaneously into cells [16] (Figure 3B). Similarly, co-transfection of constructs encoding individual gRNAs separately, together with Cas protein, is another option [7] (Figure 3C). However, these approaches are limited by the available ways of delivery RNPs, RNAs or multiple plasmids, respectively. The lack of appropriate approaches to select for cells incorporated all components also restricts their application. On the other hand, vectors derived from different viruses, including lentivirus [35], adenovirus (AdVs) [36], and adeno-associated virus (AAV) [18], and vectors for Agrobacterium-mediated transformation [11], provide the alternative ways to efficiently deliver the CRISPR systems while allowing for incorporation of the selection markers. Several strategies have been developed to generate multiple functional sgRNAs from a single plasmid to achieve multiplexed targeting.
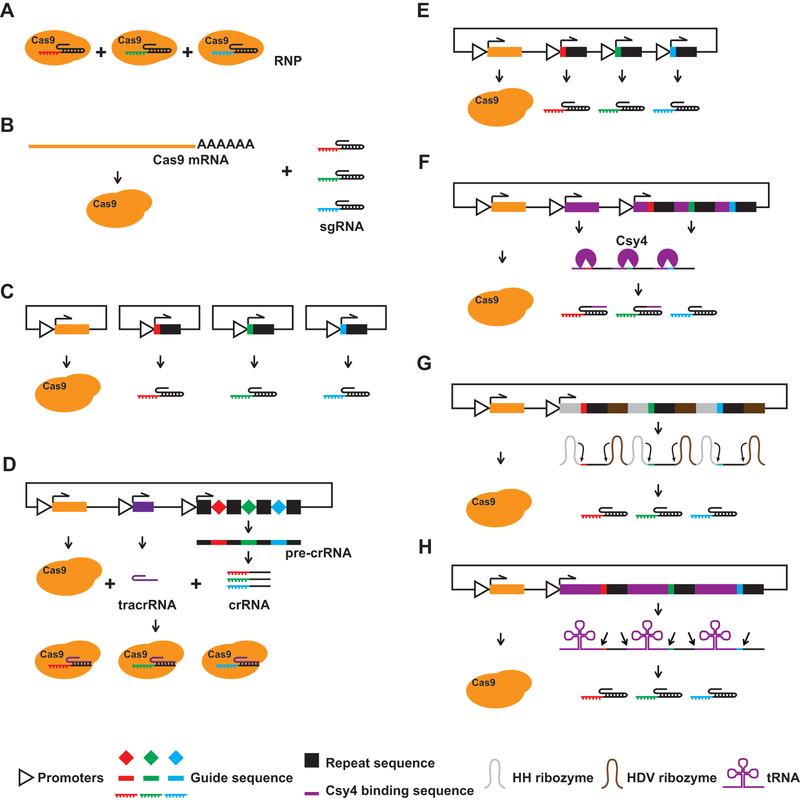
Figure 3. Strategies for multiplexed CRISPR/Cas9-based genome editing.
(A) In vivo pre-assembled RNP of Cas9 and multiple sgRNAs. (B) Introduction of Cas9 mRNA and multiple sgRNAs simultaneously. (C) Introduction of multiple single-sgRNA delivery plasmids simultaneously. (D) Delivery of Cas9, tracrRNA, and a crRNA array with a single plasmid. (E) Delivery of Cas9 and multiple sgRNA expression cassettes with a single plasmid. (F) Delivery of an artificial multi-sgRNA precursor, Cas9, and Csy4 with a single plasmid. sgRNAs are flanked by Csy4 binding sequences. Csy4 digests the precursor to release individual sgRNAs. (G) Delivery of an artificial multi-sgRNA precursor and Cas9 with a single plasmid. sgRNAs are flanked by HH and HDV ribozymes. The ribozymes digest the precursor to release individual sgRNAs. (H) Delivery of an artificial multi-sgRNA precursor and Cas9 with a single plasmid. sgRNAs are flanked by tRNAs. The endogenous tRNA processing machinery digests the precursor to release individual sgRNAs.
The CRISPR RNA array
The natural architecture of CRISPR locus (Figure 1A) provides a solution for multiplexed targeting. In the CRISPR/Cas9 systems, the CRISPR precursor (pre-crRNA) is transcribed from crRNA array, which contains many units of a unique protospacer sequences and a short palindromic repeat sequence. With the presence of tracrRNA, RNase III processes pre-crRNA into individual crRNAs [5]. The released crRNAs form distinct complexes with tracrRNA and Cas9 and direct the complexes to different targets with complementary sequence (Figure 1A).
Similarly, assembly of an artificial crRNA array will allow the genome editing at several foci at the same time when co-introduced with Cas9 and tracrRNA (Figure 3D). Indeed, introducing two guide sequences into a single CRISPR array led to simultaneous editing of both targeted sites in the mammalian genome [6]. However, applying crRNA array in the Cas9 system will require two RNA molecules (crRNA and tracrRNA), instead of a single, engineered, and high efficient sgRNA [7]. The additional requirement of tracrRNA for crRNA maturation and bridging the interaction between crRNA and Cas9 may decrease the efficiency, especially when the number of crRNAs increases. Therefore, crRNA array was not an ideal solution for multiplexed targeting, until the recently developed Cas12a (Cpf1)-based CRISPR tools [37], which does not use tracrRNA. Cas12a itself processes the maturation of crRNA from pre-crRNA and crRNA binds to Cas12a directly [22]. Indeed, a crRNA array and Cas12a can effectively target up to four genes with higher knockout efficiency than pooled individual crRNAs [37].
Expression of multiple sgRNAs using a single plasmid
Production of sgRNAs from separated expression cassettes is the most common approach for multiplexed CRISPR systems. Each cassette has its own promoter, guide sequence, sgRNA scaffold, and transcription terminator (Figure 3E). As sgRNA is a small RNA molecule, RNA polymerase III (Pol III) promoters are used to drive the transcription of sgRNAs in most cases. The human U6 promoter was selected in the earliest studies [6,7] and is still the most popular choice. Other commonly used promoters include the mouse U6 promoter, the human U3 promoter, the human H1 RNA promoter, and the human 7SK RNA promoter [38–42]. For species other than the mammals, the species specific Pol III promoters, e.g. the Arabidopsis U6 promoter [11] and the C. elegans U6 promoters [13], may be required to achieve high level of sgRNA expression in Arabidopsis and C. elegans, respectively. Employing different promoters for each cassette may help avoid potential recombination and maximize the expression efficiency of each sgRNA [38,40]. The length of these promoters is between 200 to 400 base pairs (bp). Guide sequence is around 20 nucleotides (nt) for most of Cas systems [43]. A typical sgRNA is shorter than 100 nt [6–8]. The Pol III transcription stops at a poly-T sequence [44], therefore, six Ts can serve as a terminator for sgRNA transcription [35]. An intergenic sequence of several dozen base pairs is usually cloned to separate sgRNA expression cassettes to avoid potential transcription interference. Adding together, each additional sgRNA expression cassette will add no more than 600 bp in length into the plasmids. Thus, incorporation of 4–6 sgRNA cassettes into a single vector can be achieved without significantly affecting the efficiency of viral packaging in the widely used lentiviral systems [45].
Artificial multi-sgRNA precursors
Another strategy to express multiple sgRNAs is to build up a single artificial precursor that can be processed to several individual sgRNAs. The benefits of using a single long transcript, instead of multiple short transcripts, include: (1) it avoids additional open reading frames (ORFs) in a plasmid, which may affect transcription efficiency; (2) it allows for the usage of Pol II promoters to drive transcription. Compared to Pol III promoters, Pol II promoters allow for tissue-specific or inducible expression. Several endogenous pathways that can cut RNA precisely at sequence- or secondary structure-dependent manners were exploited to catalyze the release of sgRNAs from a poly-sgRNA precursor.
Csy4-introduced cleavage
Csy4 is an endoribonuclease responsible for pre-crRNA processing in Pseudomonas aeruginosa [46]. Csy4 binds to pre-crRNA, recognizes a 28-nt sequence in the CRISPR repeats, and cleaves the immediate downstream nucleotides [46]. Constructing a single transcript with multiple sgRNA separated by Csy4 recognition sites allows for cleavage of this transcript into individual sgRNAs, when co-introducing a Csy4 endoribonuclease [47,48] (Figure 3F). Because Csy4 only cuts at the 3’-end of its recognition sequence, this sequence will remain in the 3’-end of the previous sgRNAs. However, this extra sequence does not seem to block the function of CRISPR/Cas9 in human cells and plants [47,49].
Self-cleaving ribozymes-introduced cleavage
Ribozymes are RNA molecules with enzymatic activities. Some ribozymes, such as Hammerhead (HH) and Hepatitis delta virus (HDV) ribozymes, can catalyze the cleavage of themselves at precise positions [50]. Specifically, the cutting site of HH ribozyme localizes at its 3’-end and HDV ribozyme cuts its 5’-end [51,52]. Therefore, constructing an HH-sgRNA-HDV sequence will ensure the release of an intact sgRNA molecule from the precursor and thus efficient genome editing in mammalian cells, mice, and plants [48,53,54] (Figure 3G). However, a reduction in sgRNA processing efficiency was observed with increased numbers of sgRNAs [53].
tRNA-dependent cleavage
Another intrinsic mechanism engineered for simultaneously producing sgRNAs from one primary transcript is the tRNA processing system. tRNAs are usually transcribed in the forms of polycistronic units, alone or combined with other small RNA molecules [55]. The tRNA precursors (pre-tRNAs) are cleaved by special RNases to remove extra sequences at the 5’- and 3’-end. The precise processing is dependent on the tRNA sequence but not the flanking sequences. Therefore, a tRNA-sgRNA-tRNA structure allows for production of sgRNA singletons. Indeed, polycistronic tRNA-sgRNA simultaneously produced numerous sgRNAs and led to multiplexed targeting in mammalian cells, Drosophila, and plants [37,56,57] (Figure 3H). This strategy has several advantages compared to Csy4 or ribozyme-dependent strategies. Firstly, it uses endogenous enzymes. Secondly, the tRNA processing system is very conservative, providing applications across a wide range of species. Thirdly, tRNAs are abundant, and thus its processing system is capable of cleaving a large number of sgRNAs without being overloaded. Fourthly, tRNAs carry transcriptional enhancers, leading to elevated expression of sgRNAs. However, the extra tRNAs produced may cause un-intended consequences in some situations.
Cloning solutions for constructing multiplexed CRISPR plasmids
It is complicated to generate plasmids to deliver a CRISPR RNA array, a multiplesgRNA expression cassette, or an artificial multi-sgRNA precursor. All three strategies require complicated cloning efforts to add more than one guide sequences into different locations in a plasmid. One basic solution is to conduct sequential cloning, which introduce one guide sequence in each round [58,59]. However, it is time- and effort-consuming, especially with increased number of sgRNAs. Several cloning strategies have been developed to simplify the process and increase the efficiency. Although most of the methods were developed to generate plasmids with separated sgRNA expression cassettes, these methods can be easily adopted to generate crRNA arrays or artificial multi-sgRNA precursors.
Golden Gate assembly
Golden Gate assembly is a cloning approach that enables researchers to simultaneously assemble multiple DNA fragments into one piece through Type-IIs restriction enzymatic digestion and one step ligation [60]. Type-IIs enzymes are a subgroup of restriction enzymes that have separated recognition and cutting sites [61]. Therefore, they can generate overhangs with designed sequences [61]. In contrast, other Type-II enzymes, such as EcoRI, always generate the same overhangs, and usually with palindromic sequences. Type-IIs enzymes make it possible to generate a series of fragments with different overhangs that can be joined in a designed order (Figure 4A). At the same time, non-palindromic overhangs prevent the self-ligation, which blocks the assembly and dramatically decreases the efficiency. Several Golden Gate assembly-based toolboxes have been developed to generate multiplexed CRISPR plasmids [38,45,62–65].
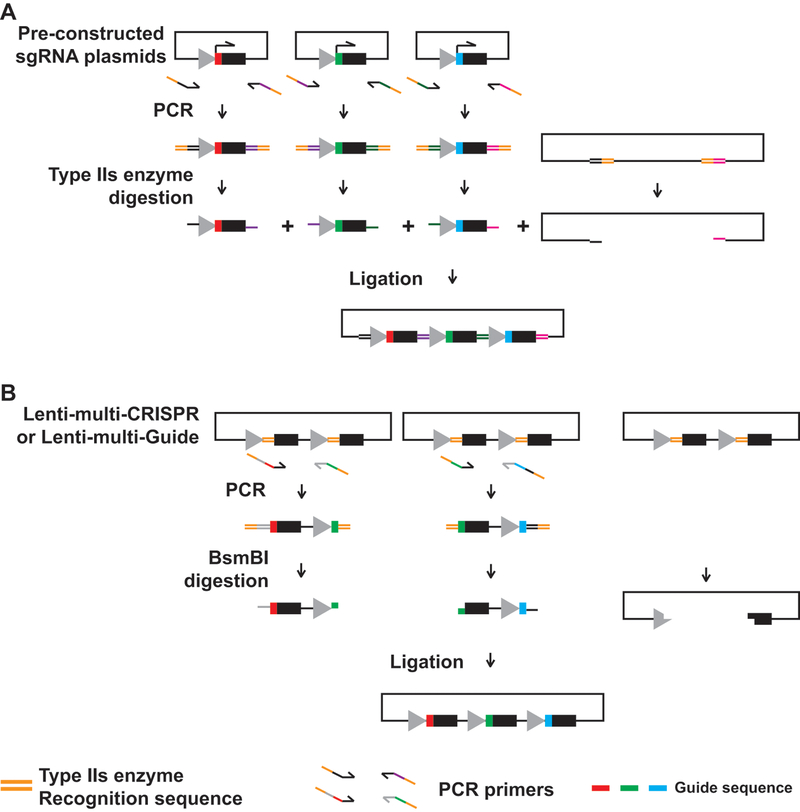
Figure 4. Two Golden Gate assembly-based cloning strategies.
(A) Schematic of a two-step strategy. Guide sequences are first cloned into single sgRNA plasmids. Each sgRNA expression cassette is amplified by primers carrying type IIs enzyme binding and a series of digesting sequences. Type IIs enzyme digestion produces overhangs that allow for assembly of multiple fragments in a designed order. (B) Schematic of a single-step strategy. Primers carrying guide sequences are used to amplify RNA scaffold and promoter sequences from specially designed vectors (Lenti-multi-Guide or Lenti-multi-CRISPR, Addgene #85401 or 85402). BsmBI digestion produces a series of overhangs that allow for assembly of multiple fragments in a designed order.
Gateway assembly
Gateway assembly is a digestion- and ligation-free approach that allows efficiently shuttling fragments between two DNA molecules through site-specific recombination [66]. This method can be further multiplexed by introducing a series of recombination site pairs [42]. Multiple-sgRNA carrying plasmids were generated by this method for genome editing [42,67].
Gibson Assembly
Gibson assembly is another recently developed single-reaction method for assembling multiple DNA fragments [68]. It is dependent on the overlapped sequences at the end of each fragment. 3’-exonuclease is applied to generate long overhangs. The complementary overhangs are then annealed and the fragments are covalently joined [68]. This method is highly efficient, therefore, is capable of assembling DNA fragments of up to several hundred kilobases [69]. It has been applied to clone multiple sgRNA expression cassettes into single plasmids [63,64].
Challenges for the multiplexed CRISPR approaches
With all three cloning strategies, a typical approach needs two steps of plasmid construction: (1). To generate single-sgRNA expression plasmids; (2). To perform assembly assays based on single-sgRNA expression plasmids, or PCR amplicons from these plasmids (Figure 4A). We have recently developed a method to achieve this in a single step of cloning. In short, we used guide sequence containing primers to amplify the elements between guide sequences and assemble them in one step [45] (Figure 4B).
Off-targeting is one of the major challenges for the CRISPR-based platforms. The specificity of CRISPR systems is based on Watson-Crick pairing between guide sequence and its target. Just like other sequence-based targeting approaches, e.g. RNAi, CRISPR can tolerate some mismatches between guide RNAs and targets, leading to off-target effects [70–72]. These effects are more significant when multiple guide sequences are applied. Different approaches have been developed to decrease the binding affinity between Cas9 and off-target sites, including predicting and excluding guide sequences with other binding sites [71], pairing double nicking [47], and engineering Cas proteins [73,74]. For irreversible genome editing, Cas endonucleases is only required for a short period of time. Alternatively, shortening the exposure of the genome to Cas proteins will prevent the accumulation of off-target cutting without affecting the efficiency of on-target editing [45]. Doxycycline-induced Cas9 expression [45] or chemical-controlled Cas9 activity [75,76] allows temporal control of Cas9/sgRNA and makes it possible to shut down Cas9 after achieving on-target editing. Inducible CRISPR also allows for identifying the direct consequences of manipulating the selected gene(s) before the long-term effects of gene manipulation are detectable.
Conclusion
Diverse strategies have been developed to achieve multiplexed CRISPR targeting. Most of the platforms were first designed and tested in mammalian cells. All these strategies we discussed here can be broadly applied to other species with only minor modifications, such as codon optimization for Cas proteins and choosing suitable promoters. For the current systems, the efficiency of cloning and manipulating drops significantly with increased number of targets. Further development of these systems will improve the efficiency for even wider application of the multiplexed CRISPR systems. Combined with the diverse CRISPR platforms, these systems will allow for precise control of genome, epigenome, and transcriptome, and therefore have broad implications in biomedicine research and clinical practices.
Funding support
This work was supported by Lion Heart Fund for Breast Cancer Research (to J. C.), by American Cancer Society Research Scholar Grant RSG-13-384-01-DMC, Department of Defense Breast Cancer Research Program Awards W81XWH-14-1-0308 and W81XWH-15-1-0117, and National Institutes of Health grant R21 CA187862 (to Q. Y.).
Footnotes
Declaration of interest
None.
References
- 1.Koonin EV et al. (2017) Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol 37, 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garneau JE et al. (2010) The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468 (7320), 67–71 [DOI] [PubMed] [Google Scholar]
- 3.Mojica FJ et al. (2005) Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol 60 (2), 174–182 [DOI] [PubMed] [Google Scholar]
- 4.Brouns SJ et al. (2008) Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321 (5891), 960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deltcheva E et al. (2011) CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471 (7340), 602–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cong L et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339 (6121), 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mali P et al. (2013) RNA-guided human genome engineering via Cas9. Science 339 (6121), 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jinek M et al. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337 (6096), 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang W et al. (2013) RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31 (3), 233–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiCarlo JE et al. (2013) Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res 41 (7), 4336–4343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nekrasov V et al. (2013) Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31 (8), 691–693 [DOI] [PubMed] [Google Scholar]
- 12.Li JF et al. (2013) Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31 (8), 688–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedland AE et al. (2013) Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods 10 (8), 741–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gratz SJ et al. (2013) Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194 (4), 1029–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo X et al. (2014) Efficient RNA/Cas9-mediated genome editing in Xenopus tropicalis. Development 141 (3), 707–714 [DOI] [PubMed] [Google Scholar]
- 16.Wang H et al. (2013) One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153 (4), 910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirano H et al. (2016) Structure and Engineering of Francisella novicida Cas9. Cell 164 (5), 950–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ran FA et al. (2015) In vivo genome editing using Staphylococcus aureus Cas9. Nature 520 (7546), 186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleinstiver BP et al. (2015) Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523 (7561), 481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fonfara I et al. (2014) Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Research 42 (4), 2577–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou Z et al. (2013) Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci U S A 110 (39), 15644–15649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zetsche B et al. (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163 (3), 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gootenberg JS et al. (2017) Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356 (6336), 438–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murugan K et al. (2017) The Revolution Continues: Newly Discovered Systems Expand the CRISPR-Cas Toolkit. Mol Cell 68 (1), 15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pulecio J et al. (2017) CRISPR/Cas9-Based Engineering of the Epigenome. Cell Stem Cell 21 (4), 431–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilbert LA et al. (2013) CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154 (2), 442–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maeder ML et al. (2013) CRISPR RNA-guided activation of endogenous human genes. Nat Methods 10 (10), 977–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi LS et al. (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152 (5), 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez-Pinera P et al. (2013) RNA-guided gene activation by CRISPR-Cas9–based transcription factors. Nature Methods 10, 973–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen B et al. (2013) Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155 (7), 1479–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konermann S et al. (2015) Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517 (7536), 583–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bak RO et al. (2017) Multiplexed genetic engineering of human hematopoietic stem and progenitor cells using CRISPR/Cas9 and AAV6. eLife 6, e27873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho SW et al. (2013) Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9-sgRNA ribonucleoproteins. Genetics 195 (3), 1177–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woo JW et al. (2015) DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol 33 (11), 1162–1164 [DOI] [PubMed] [Google Scholar]
- 35.Sanjana NE et al. (2014) Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11 (8), 783–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maggio I et al. (2014) Adenoviral vector delivery of RNA-guided CRISPR/Cas9 nuclease complexes induces targeted mutagenesis in a diverse array of human cells. Sci Rep 4, 5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zetsche B et al. (2017) Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat Biotechnol 35 (1), 31–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kabadi AM et al. (2014) Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Research 42 (19), e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie K et al. (2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proceedings of the National Academy of Sciences of the United States of America 112 (11), 3570–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han K et al. (2017) Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nature biotechnology 35 (5), 463–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aubrey BJ et al. (2015) An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep 10 (8), 1422–1432 [DOI] [PubMed] [Google Scholar]
- 42.Albers J et al. (2015) A versatile modular vector system for rapid combinatorial mammalian genetics. J Clin Invest 125 (4), 1603–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Komor AC et al. (2017) CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell 168 (1–2), 20–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nielsen S et al. (2013) Mechanism of eukaryotic RNA polymerase III transcription termination. Science 340 (6140), 1577–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao J et al. (2016) An easy and efficient inducible CRISPR/Cas9 platform with improved specificity for multiple gene targeting. Nucleic Acids Res 44 (19), e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haurwitz RE et al. (2010) Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329 (5997), 1355–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsai SQ et al. (2014) Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nature biotechnology 32 (6), 569–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nissim L et al. (2014) Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Molecular cell 54 (4), 698–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Čermák T et al. (2017) A Multipurpose Toolkit to Enable Advanced Genome Engineering in Plants. The Plant Cell 29 (6), 1196–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kruger K et al. (1982) Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 31 (1), 147–157 [DOI] [PubMed] [Google Scholar]
- 51.Pley HW et al. (1994) Three-dimensional structure of a hammerhead ribozyme. Nature 372 (6501), 68–74 [DOI] [PubMed] [Google Scholar]
- 52.Ferre-D’Amare AR et al. (1998) Crystal structure of a hepatitis delta virus ribozyme. Nature 395 (6702), 567–574 [DOI] [PubMed] [Google Scholar]
- 53.Xu L et al. (2017) Empower multiplex cell and tissue-specific CRISPR-mediated gene manipulation with self-cleaving ribozymes and tRNA. Nucleic Acids Research 45 (5), e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshioka S et al. (2015) Development of a mono-promoter-driven CRISPR/Cas9 system in mammalian cells. Sci Rep 5, 18341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakajima N et al. (1981) Organization and structure of an E. coli tRNA operon containing seven tRNA genes. Cell 23 (1), 239–249 [DOI] [PubMed] [Google Scholar]
- 56.Dong F et al. (2017) Polycistronic tRNA and CRISPR guide-RNA enables highly efficient multiplexed genome engineering in human cells. Biochem Biophys Res Commun 482 (4), 889–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Port F and Bullock SL (2016) Augmenting CRISPR applications in Drosophila with tRNA-flanked Cas9 and Cpf1 sgRNAs. Nature methods 13 (10), 852–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dow LE et al. (2015) Inducible in vivo genome editing with CRISPR-Cas9. Nat Biotechnol 33 (4), 390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peterson BA et al. (2016) Genome-Wide Assessment of Efficiency and Specificity in CRISPR/Cas9 Mediated Multiple Site Targeting in Arabidopsis. PLoS ONE 11 (9), e0162169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Engler C et al. (2008) A one pot, one step, precision cloning method with high throughput capability. PLoS One 3 (11), e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pingoud A et al. (2005) Type II restriction endonucleases: structure and mechanism. Cell Mol Life Sci 62 (6), 685–707 [DOI] [PubMed] [Google Scholar]
- 62.Sakuma T et al. (2014) Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Sci Rep 4, 5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xing HL et al. (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ma X et al. (2015) A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol Plant 8 (8), 1274–1284 [DOI] [PubMed] [Google Scholar]
- 65.Jao LE et al. (2013) Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci U S A 110 (34), 13904–13909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hartley JL et al. (2000) DNA Cloning Using In Vitro Site-Specific Recombination. Genome Research 10 (11), 1788–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lowder L et al. (2017) Rapid Construction of Multiplexed CRISPR-Cas9 Systems for Plant Genome Editing. Methods Mol Biol 1578, 291–307 [DOI] [PubMed] [Google Scholar]
- 68.Gibson DG et al. (2008) Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science 319 (5867), 1215–1220 [DOI] [PubMed] [Google Scholar]
- 69.Gibson DG et al. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6 (5), 343–345 [DOI] [PubMed] [Google Scholar]
- 70.Fu Y et al. (2013) High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31 (9), 822–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hsu PD et al. (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31 (9), 827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pattanayak V et al. (2013) High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol 31 (9), 839–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kleinstiver BP et al. (2016) High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529 (7587), 490–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Slaymaker IM et al. (2016) Rationally engineered Cas9 nucleases with improved specificity. Science 351 (6268), 84–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zetche B et al. (2015) A Split Cas9 Architecture for Inducible Genome Editing and Transcription Modulation. Nature biotechnology 33 (2), 139–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu KI et al. (2016) A chemical-inducible CRISPR–Cas9 system for rapid control of genome editing. Nature Chemical Biology 12, 980–987 [DOI] [PubMed] [Google Scholar]