Abstract
The UL4 gene is conserved within the genome of defective interfering particles of equine herpesvirus type 1 (EHV-1) that mediate persistent infection. Here, we show that the UL4 protein inhibits EHV-1 reporter gene expression by decreasing the level of transcribed mRNA. The UL4 protein did not bind any gene class of EHV-1 promoters in electromobility or chromatin immunoprecipitation assays, but directly interacted with the TATA box-binding protein (TBP) and the carboxy-terminal domain of RNA polymerase II both in vitro (GST-pulldown assays) and in infected cells (coimmunoprecipitation analyses). Microarray analyses of the expression of the 78 EHV-1 genes revealed that viral late genes important for virion assembly displayed enhanced expression in cells infected with UL4-null virus as compared to wild-type or UL4-restored EHV-1. Quantitative PCR analyses showed that viral DNA replication was not retarded in cells infected with the UL4-null virus as compared to wild-type EHV-1.
INTRODUCTION
The Alphaherpesvirinae subfamily member equine herpesvirus 1 (EHV-1) is a significant etiologic agent of severe respiratory, neurological, and abortigenic disease in equines worldwide (Allen and Bryans, 1986; O’Callaghan and Osterrieder, 2008). The viral gene program is expressed in a coordinated and temporal fashion, such that the 78 EHV-1 genes (Telford et al., 1992) are expressed at immediate-early (IE), early (E), and late (L) stages of infection (Caughman et al., 1985; Gray et al., 1987). Extensive work has been completed to describe the EHV-1 proteins responsible for controlling the expression of viral genes. The majority of the regulatory proteins are activators of viral gene expression. The essential IE gene encodes the sole IE protein (IEP) that is the major trans-activator of early and some late genes (Buczynski et al., 1999; Caughman et al., 1988; Garko-Buczynski et al., 1998; Grundy et al., 1989; Smith et al., 1992, 1994). The IEP also functions to trans-repress its own gene expression (Smith et al., 1992). Early regulatory proteins IR4P and UL5P function in a synergistic manner with the IEP to mediate the full activation of early and late EHV-1 gene promoters (Albrecht et al., 2004; Holden et al., 1995; Kim et al., 1997; Zhao et al., 1995). The powerful and promiscuous EICP0P can independently activate expression of all three gene classes and is capable of antagonizing the trans-activation potential of the IEP (Bowles et al., 1997, 2000). The late equine α-trans-inducing factor (ETIF) is a tegument protein required for secondary envelopment and virus egress as well as the activation of expression of the IE gene promoter (Kim and O’Callaghan, 2001; Lewis et al., 1993; Purewal et al., 1994; von Einem et al., 2006). The early IR2 protein and the IR3 transcript serve to inhibit EHV-1 gene expression (Ahn et al., 2010; Kim et al., 2006, 2011). The IR2P is a major inhibitory protein and is a truncated version of the IEP (Harty and O’Callaghan, 1991; Kim et al., 2006, 2011). Finally, the IR3 gene is unique to EHV-1 (Holden et., 1992), lies antisense to the IE transcript, does not produce a translated protein, and plays a role in down-regulating IE gene expression (Ahn et al., 2007, 2010).
Serial, high multiplicity passage of EHV-1 in cell culture or Syrian hamsters results in the production of defective interfering particles (DIP) that are capable of interfering with standard viral replication and establishing a state of persistent infection (Campbell et al., 1976; Chen et al., 1996, 1999; Dauenhauer et al., 1982; Ebner et al., 2008; Ebner and O’Callaghan, 2006; Henry et al., 1979). The DIP genome (~7.5 kbp) is a severely truncated and rearranged form of the standard viral genome (~155 kbp) and consists of only three genes: the perfectly conserved UL3 and UL4 genes and a hybrid gene that is comprised of portions of the IR4 and UL5 regulatory genes, which is important for the interference with standard viral replication (Chen et al., 1996, 1999; Ebner et al., 2008; Ebner and O’Callaghan, 2006). Until recently, no functional role for the UL3 and UL4 proteins had been described. It was reported that the UL4 protein was capable of inhibiting gene expression in transient transfection assays, and cells infected with EHV-1 lacking expression of the UL4 protein exhibited increased levels of viral gene transcripts during lytic infection (Charvat et al., 2011). Additionally, an EHV-1 lacking the complete UL4 open-reading frame (ORF) was incapable of producing the DIP genome after serial, undiluted passage, while a mutant EHV-1 that possessed the UL4 ORF but did not express the UL4 protein was still capable of generating the DIP genome (Charvat et al., 2012). In the present study, we elaborate on the properties of the UL4 protein and begin to characterize its mechanism of inhibition. Expression of the UL4 protein decreased reporter gene transcript levels, possibly through direct interactions with the TATA box-binding protein (TBP) and the carboxy-terminal domain of RNA polymerase II. The UL4 protein is not a DNA-binding protein as it fails to interact with EHV-1 promoters in gel shift assays and does not associate with EHV-1 promoters in chromatin immunoprecipitation assays. Microarray analysis of the expression of all 78 EHV-1 genes in cells infected with wild-type or ΔUL4 EHV-1 revealed that late gene expression is enhanced in the absence of the UL4 gene. Quantitative PCR analyses showed that viral DNA synthesis was not retarded in cells infected with ΔUL4 EHV-1 as compared to cells infected with wild type EHV-1.
RESULTS
Expression of the UL4P decreases messenger RNA levels
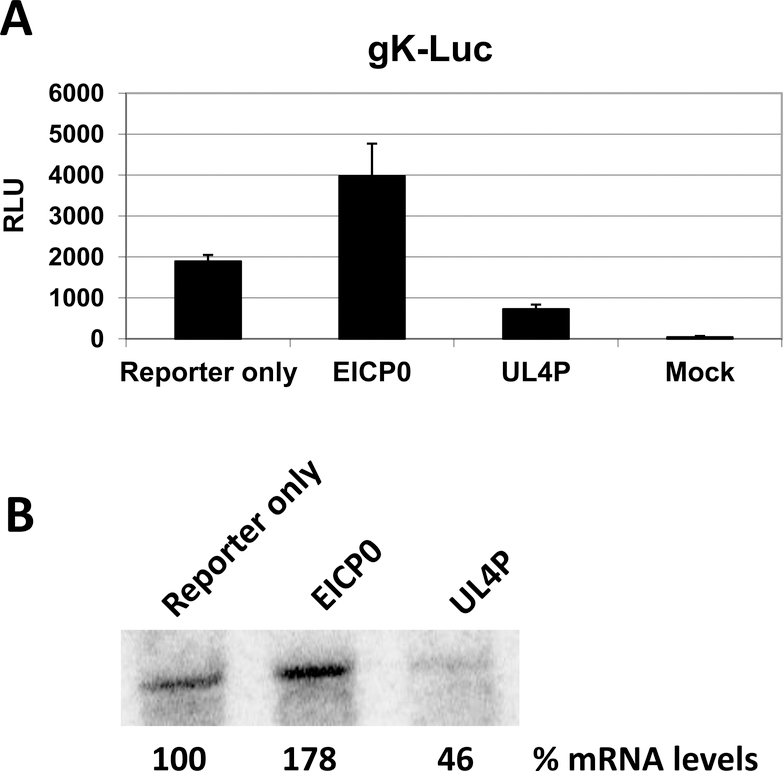
In our previous publication (Charvat et al., 2011), we demonstrated that the UL4 protein was capable of inhibiting luciferase gene expression driven by various EHV-1 promoters, and experiments using the chloramphenicol acetyltransferase (CAT) reporter system validated these original findings (data not shown). Previous data indicated that the absence of UL4 protein synthesis during infection with a mutant UL4 EHV-1 resulted in elevated levels of viral transcripts (Charvat et al., 2011). These data suggested that the UL4P may be responsible for affecting mRNA levels; specifically, it may play a role in reducing transcripts. To assess whether expression of the UL4 protein reduces transcript levels, two groups of RK13 cells were transiently transfected with the gK-Luc reporter plasmid, along with expression plasmids for the EICP0 and UL4 proteins. The EICP0 protein has been shown to activate the gK promoter (Bowles et al., 1997; Kim et al., 1999); thus, it serves as a positive control for a protein that increases gene expression. One group of transfected cells was used to determine mRNA levels by harvesting RNA at 4 h post-transfection (hpt) and performing a northern blot for the luciferase transcript (Fig. 1). The other group of cells was used to perform a luciferase assay (48 hpt) to correlate mRNA levels with luciferase activity (Fig. 1A). As expected, expression of the EICP0 protein increased the luciferase activity driven by the gK promoter, which was approximately two-fold greater than gK-Luc activity alone. Additionally, the increase in luciferase activity corresponded to a 78% increase in mRNA levels (Fig. 1B). Conversely, expression of the UL4 protein decreased the amount of luciferase activity by 50%, which coincided with diminished amounts of the luciferase transcript which were approximately 46% of the levels of the gK-Luc transcript alone. Thus, the decreased luciferase activity correlated to decreased mRNA levels. These findings indicate that inhibition of gene expression by the UL4 protein is mediated at the level of transcription.
Fig. 1.
Luciferase assay and northern blot analysis correlated the activity of the reporter gene to the levels of luciferase gene transcripts. RK13 cells were transfected with the gK-Luc reporter plasmid alone or in conjunction with either the EICP0 protein or UL4 protein expression plasmid. Transfection efficiency was routinely 80%. (A) Luciferase activity was measured at 48 h post-transfection while (B) luciferase transcript levels were examined by northern blot analysis 4 h post-transfection. Densitometry was used to determine the percent mRNA levels. Results were reproducible in three independent experiments. RLU, Relative Luminescence Units
Decreased transcript levels are not a result of mRNA instability
After observing that the gK-Luc transcript was decreased in the presence of the UL4 protein, we examined mRNA stability as an explanation for reduced transcript levels. It is possible that the UL4 protein plays a role in increasing mRNA turnover or decreasing the stability of transcripts. To assess whether the UL4 protein was involved in mRNA stability, we utilized quantitative real-time PCR (qRT-PCR) to examine the half-life of mRNA from cells infected with either wild-type or ΔUL4 EHV-1. Rabbit kidney RK13 cells were infected at a multiplicity of 5 with each virus and incubated for 12 h in normal medium. After 12 h of infection, the medium was replaced with medium supplemented with 1 μM Actinomycin D (Act D) to prevent any further transcription. RNA samples were collected every three hours for a total of 18 h and the levels of the late glycoprotein 2 (gp2) transcript were determined by qRT-PCR. The results are summarized in Table 1 and revealed that the overall levels of the gp2 transcript are not greatly altered during the course of the Act D treatment, suggesting that the gp2 mRNA is inherently stable. Furthermore, the presence of the UL4 protein had no effect on the stability of the gp2 mRNA as indicated by similar levels of message at both 3 and 18 h post-Act D treatment. These observations were reproducible in multiple experiments and revealed that the UL4 protein is not responsible for increasing mRNA instability and turnover, but rather it is likely involved in the process of gene transcription.
Table 1.
EHV-1 glycoprotein 2 transcript levelsa
| Wild-type infected RK13 cells | ||||||
| Act Db | 3 | 6 | 9 | 12 | 15 | 18 |
| Exp 1 | 0.74 | 0.73 | 0.64 | 0.73 | 0.66 | 0.76 |
| Exp 2 | 0.99 | 0.98 | 0.98 | 0.99 | 0.99 | 0.94 |
| Exp 3 | 1.00 | 1.00 | 0.99 | 1.00 | 1.00 | 0.97 |
| Avg | 0.91 | 0.90 | 0.87 | 0.90 | 0.88 | 0.89 |
| ΔUL4 infected RK13 cells | ||||||
| Act Db | 3 | 6 | 9 | 12 | 15 | 18 |
| Exp 1 | 0.89 | 0.84 | 0.71 | 0.63 | 0.71 | 0.21 |
| Exp 2 | 0.99 | 0.99 | 0.99 | 0.99 | 0.98 | 0.99 |
| Exp 3 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 |
| Avg | 0.96 | 0.94 | 0.90 | 0.87 | 0.90 | 0.73 |
Transcript levels normalized to 28S rRNA
Times post-Actinomycin D treatment (hours) added 12 hours post-infection
Exp, experiment; Avg, average
The UL4 protein is not a DNA-binding protein
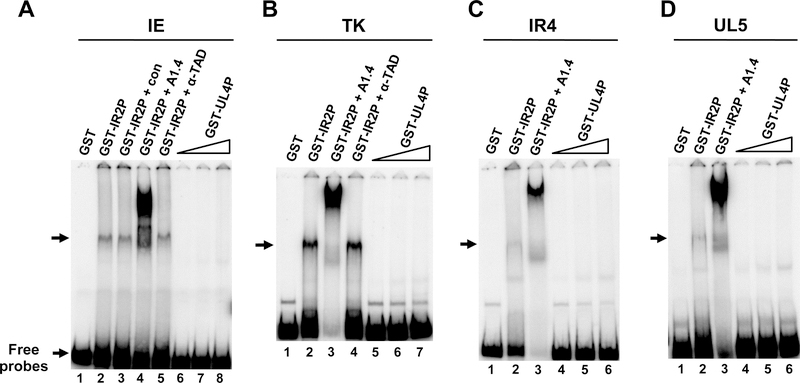
After observing that the expression of the UL4 protein inhibited transient gene expression in reporter assays as well as decreased the levels of viral transcripts, a mechanism to explain this inhibitory activity was investigated. One possible mechanism is that the UL4P binds to DNA in such a way to block transcription from the targeted genes. This potential DNA-binding activity could affect the assembly of the pre-initiation complex (PIC) involved in the transcription of viral genes, as is observed with the EHV-1 IR2 protein (Kim et al., 2006, 2011) and the IE2 (IE86) protein of human cytomegalovirus (Lee et al., 1996; Wu et al., 1993). Inhibition of viral transcription through DNA binding is also documented for human papillomaviruses, where the E1M^E2C fusion protein inhibits viral gene expression as an E2-binding site-specific repressor (Chiang et al., 1991). Therefore, the ability of the UL4 protein to bind DNA was explored. Electromobility shift assays (EMSA) were utilized to determine whether a recombinant GST-UL4 fusion protein possessed DNA-binding activity for the IE, TK, IR4, and UL5 gene promoters, all of which were shown to be inhibited by the UL4P in reporter assays (Charvat et al., 2011; unpublished observation). The GST protein alone was used as a negative control; while the GST-IR2 fusion protein that has documented DNA-binding properties (Kim et al., 1995) was used as a positive control. As expected, the GST protein was unable to bind any of the four radiolabeled promoters (Fig. 2, lanes A1, B1, C1, and D1). In contrast, the GST-IR2 fusion protein readily bound to the four EHV-1 promoters (Fig. 2, lanes A2, B2, C2, and D2), which was confirmed by a shift in the mobility of the radiolabeled DNA-protein complexes using anti-IR2P monoclonal antibody A1.4 (Fig. 2, lanes A4, B3, C3, and D3). As expected, pre-immune serum (Fig. 2 lane3) or antibody to the EHV-1 TAD (Fig. 2, lanes A5 and B4) failed to cause a supershift in the DNA-protein complexes. However, no DNA-binding activity was observed for the GST-UL4 fusion protein with the IE, TK, IR4, or UL5 promoters over a range of protein concentrations (Fig. 2, lanes A6–8, B5–7, C 4–6, and D4–6). Very minor bands were not reproducibly detected and were considered as background.
Fig. 2.
Electromobility shift assays examining whether the UL4 protein possesses the ability to bind EHV-1 promoters. A. Immediate-early (IE) promoter DNA (−120/+73); B. thymidine kinase (TK) promoter DNA (−193/+133); C. IR4 promoter DNA (−267/+17); and D. UL5 promoter DNA (199/+20) were radiolabeled and incubated with various amounts of the protein under standard conditions described in Materials and Methods. The amount of GST and GST-IR2P used in this experiment was 100ng. Triangles: increasing amounts of the GST-UL4P added at 1X (100ng), 3X (300ng), 6X (600ng). Control serum (con), IR2P-specific monoclonal antibody (A1.4; Caughman et al., 1995), and IEP TAD-specific polyclonal antibody (a-TAD; Kim et al., 2011) were used. The position of complexes formed by the IR2P is indicated with arrows.
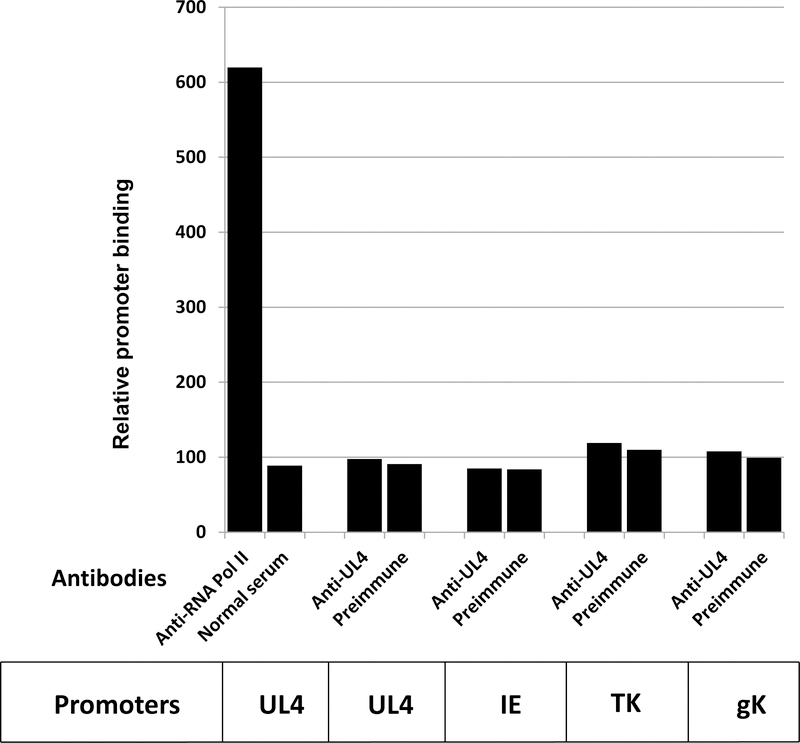
To confirm that the UL4 protein is not a DNA-binding protein, chromatin immunoprecipitation (ChIP) assays were carried out. EHV-1-infected HeLa cells were crosslinked with 1% formaldehyde, cell pellets were lyzed, and lysates were treated with micrococcal nuclease. Immunoprecipitation was performed overnight in spin columns using pre-immune serum, anti-RNA polymerase II antibody, or anti-UL4 antibody. DNAs were eluted and recovered, and PCR assays were carried out to amplify precipitated target sequences. As shown in Fig. 3, RNA polymerase II as the positive control was readily detected and associated with the EHV-1 UL4 promoter. In contrast, antibody to the UL4 protein failed to reveal an association of this EHV-1 protein with the UL4 promoter or with the immediate-early, early TK, or late gK promoters (Fig. 3). Similar findings were obtained at both early (6h) and late times (10h and 16h) after infection and indicate that the UL4 protein is not directly associated with viral DNA sequences and suggest that the inhibition of EHV-1 gene expression by UL4P is not mediated through its binding to promoter DNA sequences.
Fig. 3.
ChIP assays to assess if the UL4 protein was bound to EHV-1 promoters in infected HeLa cells at 16 h post infection. Assays were carried out as described in the Materials and Methods. Similar results were obtained at early (6h) and 10h post infection. PCR results were analyzed for the UL4, IE, TK, and gK promoters. In all assays, PCR bands obtained from DNA samples immunoprecipitated with anti-UL4 antibody did not differ significantly from those obtained with the pre-immune serum.
The UL4P directly interacts with TBP and RNA polymerase II
Whether the UL4 protein interacts with cellular transcription factors to inhibit gene expression was examined as a possible mechanism for decreased transcription. Initial experiments were carried out with general transcription factors (GTFs) involved in the assembly of the pre-initiation complex (PIC) at gene promoters (Buratowski et al., 1989; Davison et al., 1983; Kornberg, 2007; Van Dyke et al., 1988). The TFIID complex, which includes the TATA box-binding protein (TBP), plays a critical role in initiating PIC assembly and transcription and is a target for a number of viral proteins including the E1A protein of adenovirus (Geisberg et al., 1995), the ICP4 protein of herpes simplex virus 1 (Lester and DeLuca, 2011), as well as the EHV-1 IE, IR2, EICP0, and UL5 proteins (Albrecht et al., 2004; Kim et al., 2003, 2006, 2011).
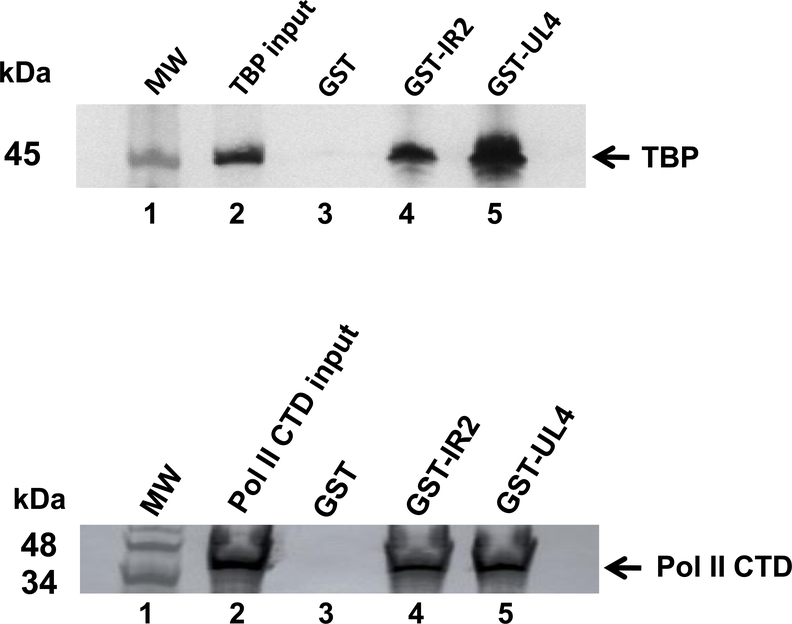
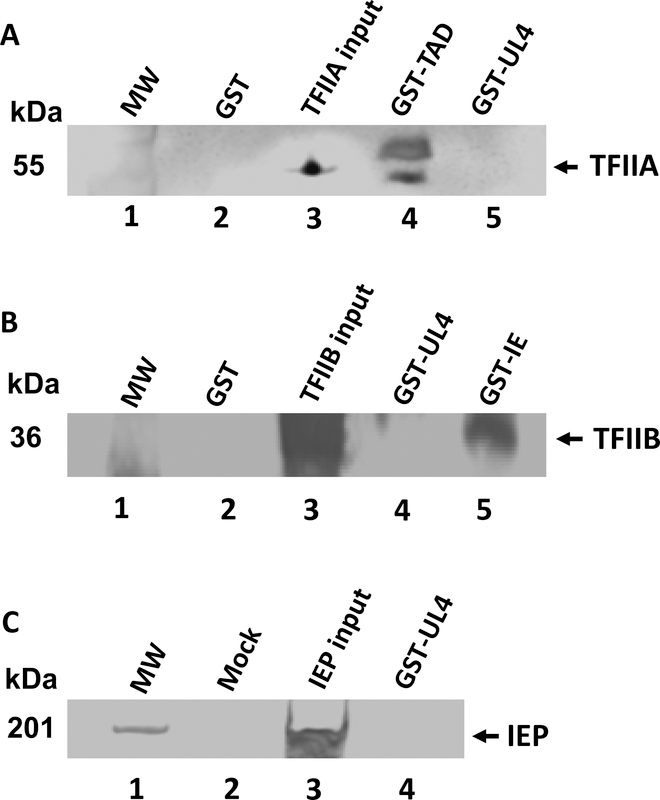
Direct protein-protein interactions were assessed using in vitro GST-pulldown assays with the GST-UL4 fusion protein and purified cellular transcription factors TFIIA, TFIIB, TBP and RNA polymerase II (Pol II). Again, the GST protein and GST-IR2 fusion protein were used as negative and positive controls, respectively. Expectedly, the GST protein alone did not interact with any of the cellular GTFs (Fig. 4, lanes A3 and B3). When purified TBP was incubated with GST-IR2, a direct interaction was observed (Fig. 4A, lane 4). TBP was also found to bind to the GST-UL4 fusion protein (Fig. 4A, lane 5). Similar results were obtained when the GST-IR2 and GST-UL4 fusion proteins were incubated with purified samples of the carboxy-terminal domain (CTD) of Pol II, as shown in Fig. 4B, lanes 4 and 5. However, no interaction was observed between GST-UL4 and TFIIA, TFIIB, or the EHV-1 IEP (Fig. 5). These findings indicate that the UL4 protein directly interacts with TBP and the CTD of Pol II, observations that suggest that the UL4 protein may prevent the assembly of a pre-initiation complex at viral promoters to inhibit gene expression.
Fig. 4.
GST-pulldown assays were completed to determine whether the UL4 protein directly interacts with general transcription factors. Purified GST, GST-IR2, or GST-UL4 fusion proteins were incubated with GST-Bind resin beads for 1.5 h before being combined with (A) TATA box-binding protein (TBP) or (B) RNA polymerase II carboxy-terminal domain (Pol II CTD) for an additional 1.5 h incubation. Captured proteins were eluted and resolved by SDS-PAGE and western blot using TBP or Pol II CTD specific antibodies. MW, molecular weight marker
Fig. 5.
GST-pulldown assays were completed to determine whether the UL4 protein directly interacts with general transcription factor TFIIA or TFIIB or the EHV-1 IE protein. Purified GST (negative control), GST-TAD (positive control), GST-IE (positive control) or GST-UL4 fusion proteins were incubated with GST-Bind resin beads for 1.5 h before being combined with A. TFIIA, B. TFIIB, or C. EHV-1 IE. Captured proteins were eluted and resolved by SDS-PAGE and western blot using anti-TFIIA antibody, anti-TFIIB antibody, or anti-IE specific antibody. MW, molecular weight marker. Details are in the Materials and Methods.
The UL4 protein partners with TBP and Pol II during EHV-1 infection
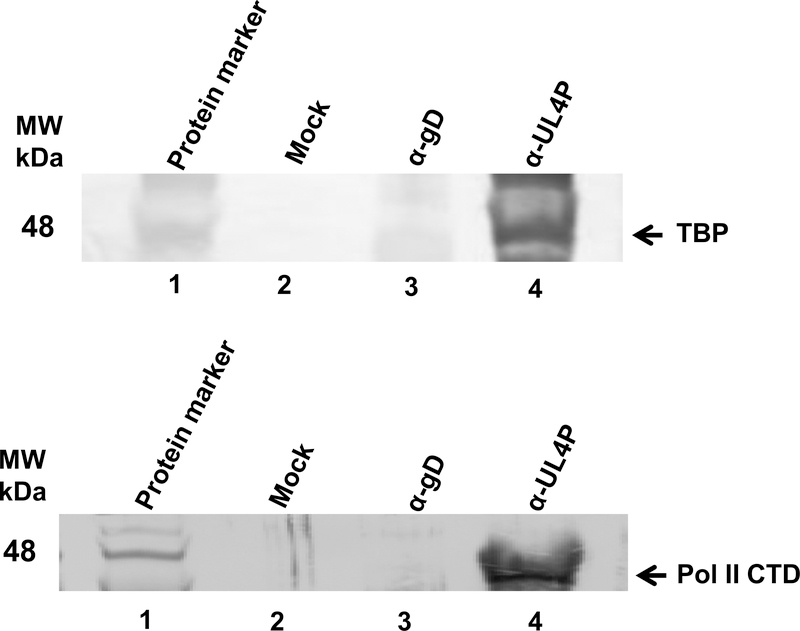
It was demonstrated that the UL4P directly interacts with TBP and RNA polymerase II (Pol II) in vitro through GST-pulldown assays (Fig. 4). However, as this is an artificial system to examine protein-protein interactions, we next addressed whether this interaction occurred during viral infection. HeLa cells were mock infected or infected with the wild-type RacL11 strain of EHV-1. Co-immunoprecipitation assays were performed using the UL4-specific OC95 antibody (Charvat et al., 2011) or a non-related antibody to EHV-1 glycoprotein D (gD). The immunoprecipitated complexes were subjected to SDS-PAGE and western blot analysis using antibodies specific for TBP or Pol II (Fig. 6). Immunoprecipitation with the anti-UL4P antibody of mock infected cell extracts yielded no interaction with either TBP or Pol II (Fig. 6A, lane 2; Fig. 6B, lane 2, respectively). Additionally, assays that included the anti-gD antibody produced no interactions of the UL4 protein with TBP or Pol II from HeLa cells infected with EHV-1 (Fig. 6A, lane 3; Fig. 6B, lane 3, respectively). However, when the anti-UL4P antibody was used to immunoprecipitate protein complexes from EHV-1 infected cells, interactions of the UL4 protein were observed with both TBP and Pol II (Fig. 6A, lane 4; Fig 6B, lane 4, respectively).
Fig. 6.
Co-immunoprecipitation assays with anti-UL4 protein antibody demonstrating that the UL4 protein interacts with cellular transcription factors during EHV-1 lytic infection. HeLa cells were infected with wild-type EHV-1 (MOI=10), and cell lysates were collected at 10 h post-infection. UL4 protein specific antibody and non-specific anti-glycoprotein D (gD) antibody were conjugated to Protein-A agarose beads and were then incubated with cellular lysates from mock-infected or EHV-1-infected cells. Immunoprecipitated proteins were resolved by SDS-PAGE and immunoblotted for either A. TBP or B. Pol II CTD. MW, molecular weight marker
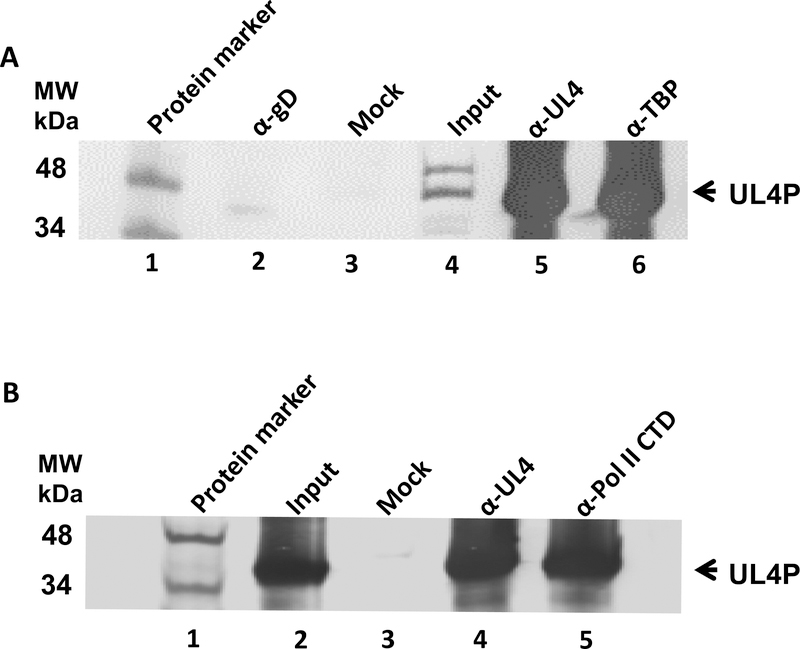
To confirm these interactions of the UL4 protein with these two cell factors essential for transcription, reverse immunoprecipitation assays using antibody to TBP and antibody to Pol II CTD were carried out. Control reactions employed pre-immune serum or anti EHV-1 gD antibody. Precipitates were then probed with pre-immune serum or anti-UL4P antibody. As shown in Fig. 7A, precipitates obtained from EHV-1 infected cells with anti-TBP antibody revealed the presence of the UL4 protein. In contrast negative control precipitates of mock-infected cells or precipitates obtained by use of anti-gD antibody as the primary antibody failed to reveal the UL4 protein. Similarly, reverse precipitation assays using anti-Pol II CTD antibody as the primary antibody confirmed the interaction of the UL4 protein with Pol II CTD during EHV-1 infection (Fig. 7B). The interaction of the UL4 protein with these cellular factors was demonstrated at both early (6h) and late times (10h and 16h) after infection. Overall, findings from GST-pulldown assays and both series of immunoprecipitation analyses indicate that the interaction of the UL4 protein with the TBP and Pol II occurs during viral infection.
Fig. 7.
Co-immunoprecipitation assays with antibodies specific for TBP and Pol II CTD demonstrating that the UL4 protein interacts with cellular transcription factors during EHV-1 lytic infection. HeLa cells were infected with wild-type EHV-1 (MOI = 10), and cell lysates were collected at 6, 10, and 16 h post-infection. A. Anti-TBP antibody or B. anti-Pol II CTD antibody and non-specific anti-glycoprotein D (gD) antibody as a negative control were conjugated to Protein-A agarose beads and were incubated with cellular lysates from mock-infected or EHV-1 infected cells. Immunoprecipitated proteins were resolved by SDS-PAGE and immunoblotted with anti-UL4 protein antibody. MW, molecular weight marker. Results from 16 hpi are show; similar results were obtained at 6 and 10 h post-infection
Late gene expression is augmented in the absence of the UL4 protein
Our previous work (Charvat et al 2011) and findings in Fig. 1 and Table 1 indicate that promoters representative of all three EHV-1 gene classes were inhibited by the expression of the UL4 protein. However, the effect of the UL4 protein on global viral gene expression was unknown. Therefore, the effect of the UL4 protein on the expression of all 78 EHV-1 genes was assessed via microarray analysis using cells that were either mock infected or infected with wild-type EHV-1 or ΔUL4 EHV-1. It was hypothesized that the greatest effect of UL4 protein expression would be evident for the regulatory genes of EHV-1. Unexpectedly, it was observed that the genes with the most enhanced expression in the absence of the UL4 protein were those of the late class responsible for mature virus particle assembly and maturation (Table 2A). The included genes encode tegument proteins, capsid proteins, proteins involved in cleavage and packaging of nascent genomes, and glycoproteins. The only regulatory gene with significantly increased expression in the absence of the UL4 protein was the late ETIF gene that encodes a protein that localizes within the tegument and is necessary for secondary envelopment and egress of viral particles (von Einem et al., 2006). The remaining regulatory genes of EHV-1 were considered to be unaffected in the absence of the UL4 protein (Table 2B). As expected, no UL4 gene expression was observed in the ΔUL4 EHV-1 infected cells. These data suggest that during lytic infection, one role of the UL4 protein is to regulate the expression of late genes.
Table 2A.
EHV-1 genes display enhanced expression in the absence of the UL4 gene
| Fold Increasea | Avg Fold Increased | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Exp 1b | Exp 2c | Exp 3c | SEMe | Name | Functionf | HSV | |
| ORF16 | 0.92 | 3.01 | 2.63 | 2.82 | 0.190 | UL16 | gC | UL44 |
| ORF71 | 1.02 | 2.23 | 2.59 | 2.41 | 0.180 | US4 | gp2 | none |
| ORF57 | 1.17 | 2.41 | 2.40 | 2.40 | 0.005 | UL57 | Helicase/primase | UL5 |
| ORF72 | 1.25 | 2.33 | 2.28 | 2.31 | 0.025 | US5 | gD | US6 |
| ORF54 | 1.15 | 2.14 | 2.34 | 2.24 | 0.100 | UL54 | Helicase/primase | UL8 |
| ORF53 | 1.05 | 2.33 | 2.09 | 2.21 | 0.120 | UL53 | Ori-binding protein | UL9 |
| ORF73 | 1.38 | 1.98 | 2.37 | 2.17 | 0.195 | US6 | gI | US7 |
| ORF40 | 1.46 | 1.86 | 2.14 | 2.00 | 0.140 | UL40 | Tegument | UL21 |
| ORF3 | 0.75 | 1.75 | 2.23 | 1.99 | 0.240 | UL3 | Tegument, conserved in DIP | none |
| ORF15 | 1.47 | 1.86 | 2.10 | 1.98 | 0.120 | UL15 | Membrane | UL45 |
| ORF19 | 1.23 | 1.78 | 2.16 | 1.97 | 0.190 | UL19 | VHS | UL41 |
| ORF58 | 1.40 | 1.75 | 2.02 | 1.88 | 0.135 | UL58 | Virion | UL4 |
| ORF33 | 1.65 | 1.73 | 1.98 | 1.86 | 0.125 | UL33 | gB | UL27 |
| ORF36 | 1.62 | 1.57 | 2.12 | 1.84 | 0.275 | UL36 | Capsid | UL25 |
| ORF41 | 1.50 | 1.57 | 2.10 | 1.83 | 0.265 | UL41 | Virion | UL20 |
| ORF52 | 1.31 | 1.73 | 1.93 | 1.83 | 0.100 | UL52 | gM | UL10 |
| ORF60 | 1.70 | 1.72 | 1.93 | 1.83 | 0.105 | UL60 | Colocalizes w/ IC22 in nuclear bodies | UL3 |
| ORF13 | 1.38 | 1.78 | 1.86 | 1.82 | 0.040 | UL13 | VP13/14 tegument | UL47 |
| ORF6 | 1.58 | 1.51 | 1.97 | 1.74 | 0.230 | UL6 | gK | UL53 |
| ORF56 | 1.29 | 1.61 | 1.87 | 1.74 | 0.130 | UL56 | Portal | UL6 |
| ORF62 | 1.44 | 1.68 | 1.76 | 1.72 | 0.040 | UL62 | gL | UL1 |
| ORF61 | 1.59 | 1.75 | 1.68 | 1.71 | 0.035 | UL61 | Uracil-DNA glycosylase | UL2 |
| ORF12 | 1.39 | 1.60 | 1.83 | 1.71 | 0.115 | UL12 | ET1F | UL48 |
| ORF9 | 1.28 | 1.54 | 1.85 | 1.70 | 0.155 | UL9 | dUTPase | UL50 |
| ORF35 | 1.64 | 1.58 | 1.82 | 1.70 | 0.120 | UL35 | Protease | UL26 |
| ORF11 | 1.72 | 1.73 | 1.66 | 1.70 | 0.035 | UL11 | VP22 tegument | UL49 |
| ORF42 | 1.88 | 1.62 | 1.75 | 1.68 | 0.065 | UL42 | VP5 capsid | UL19 |
| ORF10 | 1.39 | 1.93 | 1.44 | 1.68 | 0.245 | UL10 | Membrane | UL49a |
| ORF55 | 1.38 | 1.65 | 1.70 | 1.67 | 0.025 | UL55 | Tegument/Virion | UL7 |
| ORF14 | 1.68 | 1.68 | 1.66 | 1.67 | 0.010 | UL14 | Tegument | UL46 |
| ORF43 | 1.41 | 1.60 | 1.74 | 1.67 | 0.070 | UL43 | VP23 capsid | UL18 |
| ORF70 | 1.69 | 1.61 | 1.72 | 1.67 | 0.055 | US3 | gG | US4 |
| ORF68 | 1.70 | 1.65 | 1.67 | 1.66 | 0.010 | US1 | Virion | US2 |
| ORF37 | 1.27 | 1.54 | 1.74 | 1.64 | 0.100 | UL37 | Membrane | UL24 |
| ORF47_44 | 1.27 | 1.46 | 1.80 | 1.63 | 0.170 | Packaging | UL15 | |
| ORF23 | 1.46 | 1.49 | 1.77 | 1.63 | 0.140 | UL23 | ICP32 | UL37 |
| ORF22 | 1.39 | 1.48 | 1.77 | 1.63 | 0.145 | UL22 | VP19C capsid | UL38 |
| ORF35.5 | 1.68 | 1.52 | 1.71 | 1.62 | 0.095 | UL35.5 | ICP35 capsid assembly | UL26.5 |
| ORF59 | 1.77 | 1.49 | 1.74 | 1.61 | 0.125 | UL59 | ?? (VZV ORF 57) | none |
| ORF30 | 1.31 | 1.39 | 1.81 | 1.60 | 0.210 | UL30 | DNA pol | UL30 |
Fold increase ΔUL4 EHV-1 vs. wild-type EHV-1. Data are from three independent experiments (Exp)
Infection MOI 5
Infection MOI 10
Average fold increase for Exps 2 & 3
Standard error from mean Exps 2 & 3
Function or location is assigned based on sequence analysis (Telford et al., 1992)
Table 2B.
EHV-1 genes display enhanced expression in the absence of the UL4 gene
| Fold Increasea | Avg Fold Increased | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Exp 1b | Exp 2c | Exp 3c | SEMe | Name | Functionf | HSV | |
| ORF38 | 1.54 | 1.45 | 1.76 | 1.60 | 0.155 | UL38 | Thymidine kinase | UL23 |
| IR3 | 1.52 | 1.17 | 2.03 | 1.60 | 0.430 | IR3 | microRNA | none |
| ORF39 | 1.67 | 1.41 | 1.78 | 1.60 | 0.185 | UL39 | gH | UL22 |
| ORF18 | 1.89 | 1.54 | 1.61 | 1.58 | 0.035 | UL18 | DNA process | UL42 |
| ORF45 | 1.59 | 1.42 | 1.73 | 1.57 | 0.155 | UL45 | Tegument req. for cleavage/packaging | UL17 |
| ORF74 | 1.33 | 1.45 | 1.59 | 1.52 | 0.070 | US7 | gE | US8 |
| ORF17 | 1.10 | 1.61 | 1.43 | 1.52 | 0.090 | UL17 | Membrane | UL43 |
| ORF46 | 1.93 | 1.38 | 1.63 | 1.50 | 0.125 | UL46 | Capsid-assoc. req. for cleavage/packaging | UL16 |
| ORF49 | 1.77 | 1.35 | 1.66 | 1.50 | 0.155 | UL49 | Tyr protein kinase | UL13 |
| ORF75 | 1.96 | 1.40 | 1.60 | 1.50 | 0.100 | US8 | “10K” | none |
| ORF34 | 1.96 | 1.45 | 1.54 | 1.49 | 0.045 | UL34 | ?? | none |
| ORF29 | 1.61 | 1.37 | 1.61 | 1.49 | 0.120 | UL29 | Req. for envelopment | UL31 |
| ORF50 | 1.69 | 1.38 | 1.59 | 1.49 | 0.105 | UL50 | Alkaline Dnase | UL12 |
| ORF28 | 1.56 | 1.35 | 1.59 | 1.47 | 0.120 | UL28 | Cleavage/packaging | UL32 |
| ORF8 | 1.34 | 1.32 | 1.61 | 1.47 | 0.145 | UL8 | Tegument | UL51 |
| ORF27 | 1.29 | 1.27 | 1.64 | 1.46 | 0.185 | UL27 | Packaging | UL33 |
| ORF24 | 1.53 | 1.37 | 1.54 | 1.46 | 0.085 | UL24 | ICP1/2 tegument | UL36 |
| ORF69 | 1.23 | 1.44 | 1.45 | 1.45 | 0.005 | US2 | Protein kinase | US3 |
| ORF25 | 2.18 | 1.27 | 1.61 | 1.44 | 0.170 | UL25 | Capsid | UL35 |
| ORF20 | 1.93 | 1.24 | 1.64 | 1.44 | 0.200 | UL20 | RR2 | UL40 |
| ORF7 | 1.19 | 1.27 | 1.56 | 1.42 | 0.145 | UL7 | Helicase/primase | UL52 |
| ORF26 | 1.66 | 1.30 | 1.52 | 1.41 | 0.110 | UL26 | Membrane | UL34 |
| ORF5 | 1.32 | 1.31 | 1.46 | 1.39 | 0.075 | UL5 | EICP27 | UL54 |
| ORF51 | 2.13 | 1.21 | 1.56 | 1.38 | 0.175 | UL51 | Virion and envelopment | UL11 |
| ORF31 | 1.57 | 1.36 | 1.40 | 1.38 | 0.020 | UL31 | ICP8 ssDNA-binding protein | UL29 |
| ORF48 | 1.22 | 1.17 | 1.58 | 1.38 | 0.205 | UL48 | Tegument | UL14 |
| ORF21 | 1.31 | 1.24 | 1.47 | 1.36 | 0.115 | UL21 | RR1 | UL39 |
| ORF67 | 1.91 | 1.35 | 1.36 | 1.36 | 0.005 | IR6 | Virion forms filaments | none |
| IR4/UL3 | 1.31 | 1.18 | 1.45 | 1.32 | 0.135 | Hybrid | none | |
| ORF32 | 1.09 | 1.19 | 1.30 | 1.24 | 0.055 | UL32 | Cleavage/packaging | UL28 |
| ORF64.1 | 0.74 | 0.95 | 1.50 | 1.23 | 0.275 | IR1 | IE | ICP4 |
| ORF76 | 0.94 | 1.14 | 1.29 | 1.22 | 0.075 | US9 | Tegument | US9 |
| ORF63 | 1.35 | 1.00 | 1.36 | 1.18 | 0.180 | UL63 | EICP0 | IE110 |
| ORF65 | 1.13 | 1.00 | 1.25 | 1.13 | 0.125 | IR4 | EICP22 | US1 |
| ORF1g | −1.06 | 0.69 | 1.50 | 1.10 | 0.405 | UL1 | MHC-I downregulation, immune evasion | UL56 |
| Actin | 0.80 | 1.01 | 1.00 | 1.00 | 0.005 | |||
| GAPDH | 1.00 | 1.00 | 1.00 | 1.00 | 0.000 | |||
| ORF4h | −0.10 | 0.17 | 0.38 | 0.27 | 0.105 | UL4 | Inhibitory protein req. for DIP generation | UL55 |
Fold increase ΔUL4 EHV-1 vs. wild-type EHV-1. Data are from three independent experiments (Exp)
Infection MOI 5
Infection MOI 10
Average fold increase for Exps 2 & 3
Standard error from mean Exps 2 & 3
Function or location is assigned based on sequence analysis (Telford et al., 1992)
ORF1 deleted in both wild-type EHV-1 and ΔUL4 EHV-1
ORF4 deleted in ΔUL4 EHV-1 but present in wild-type EHV-1
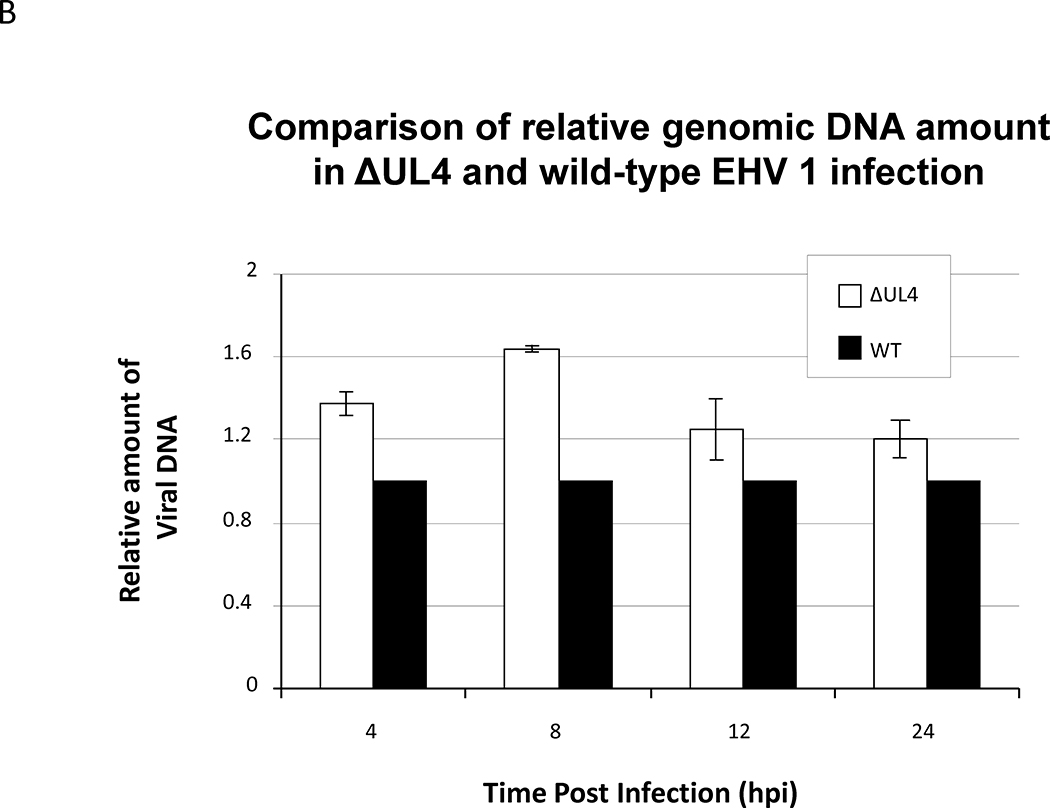
Viral DNA synthesis is not retarded in cells infected with ΔUL4 EHV-1
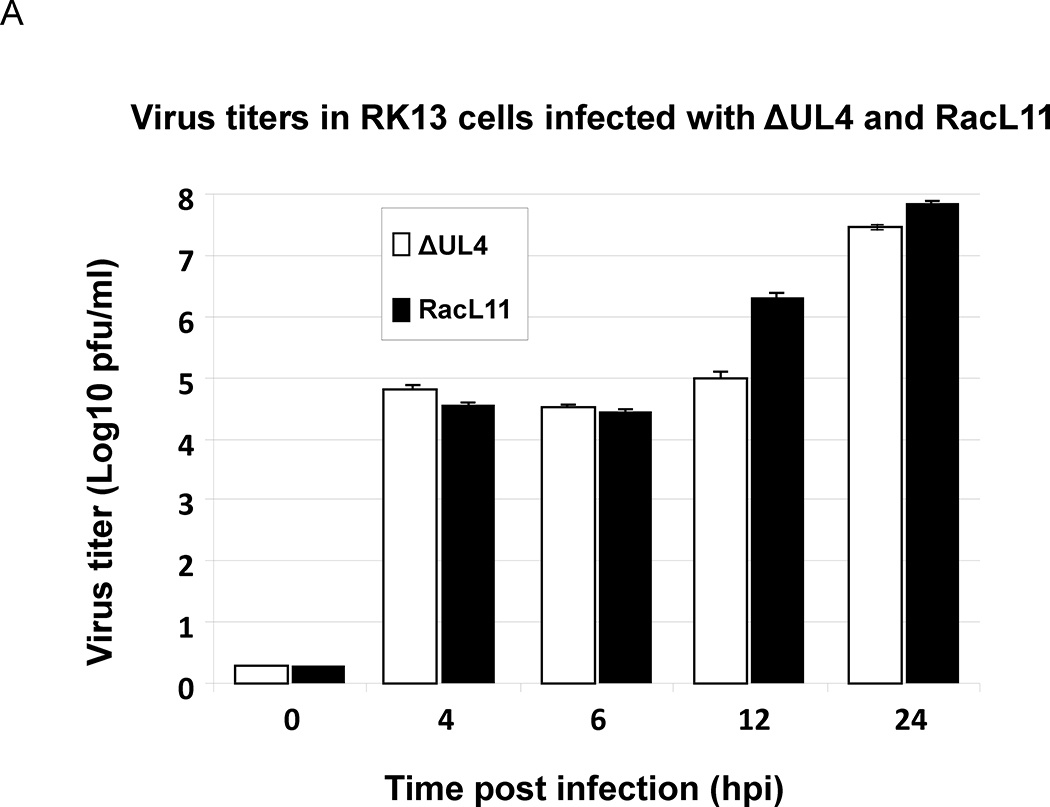
The microarray data indicated that the genes most affected in the absence of UL4 were those of the late class responsible for assembly and maturation of viral particles. However, these changes in viral gene expression did not significantly affect virus replication as the ΔUL4 EHV-1 and wild type EHV-1 replicated with similar kinetics during a 24-hour time course (Fig. 8A). Extensive quantitative PCR analyses revealed that viral DNA synthesis was not retarded in the absence of the UL4 protein, and cells infected with wild type and ΔUL4 EHV-1 produced similar amounts of viral DNA by 24 hours post infection (Fig. 8B).
Fig. 8.
Quantitative PCR and virus titer were employed to examine the growth of UL4-deleted EHV-1 as compared to the wild type EHV-1 RacL11. RK13 cells were infected at MOI of 10, and the infected cells were harvested at 4, 6, 12 and 24 hours post infection. (A) Infectious virus titers were determined by plaque assays as described in the Materials and Methods. (B). DNA was isolated using DNA STAT-60 reagent or DNeasy® Blood &Tissue Kit. Viral DNA was measured by quantitative PCR analyses by detecting the gp2 DNA sequence and normalizing to the levels of cellular GAPDH. Data shown are from four independent assays, and details are in the Material and Methods.
DISCUSSION
It was reported previously that transient expression of the EHV-1 UL4 protein inhibited expression from reporter genes that were under the control of a variety of promoters (Charvat et al., 2011). Furthermore, in the absence of the UL4 protein during EHV-1 lytic infection, expression of representative transcripts of all three gene classes was increased. These observations suggested that the UL4 protein plays a role in regulating gene expression, but no insight into the mechanism of inhibition was provided. In this report, we demonstrate that expression of the UL4 protein is associated with a decrease in the level of reporter gene transcripts (Fig. 1). However, the reduced transcript levels were not a result of increased mRNA instability or turnover (Table 1). Taken together, these observations indicate that the UL4 protein-mediated inhibition of gene expression occurs at the level of transcription. However, the UL4 protein does not inhibit gene expression by binding to viral promoters as demonstrated by gel retardation and ChIP assays, which is in contrast to that demonstrated for the EHV-1 IR2 negative regulatory protein (Kim et al., 2006, 2011).
Protein-protein interactions with cellular transcription factors play a role in inhibiting viral gene expression in a number of viral systems. The human T-cell leukemia virus encodes a bZIP factor that interacts with transcription factor CREB-2 to regulate viral gene expression (Gaudray et al., 2002). Protein interactions between the simian virus 40 (SV40) T antigen and AP-2 prevent AP-2 from binding to DNA to activate viral gene transcription (Mitchell et al., 1987). During infection with high-risk human papillomaviruses, the viral E8^E2C protein mediates repression of viral gene expression by interacting with cellular corepressor molecules, nuclear receptor corepressor 1 (NCoR1), and the chromodomain helicase DNA binding domain 6 protein (CHD6) (Ammermann et al., 2008; Fertey et al., 2010; Powell et al., 2010). Therefore, it was hypothesized that the UL4 protein mediated its inhibitory activity during the process of transcription, presumably at the stage of pre-initiation complex (PIC) assembly and recruitment of the RNA polymerase. Indeed, protein-protein interaction assays revealed that the UL4 protein could directly bind to purified TATA box-binding protein (TBP) and the carboxy-terminal domain (CTD) of Pol II (Fig. 4). Additionally, a series of co-immunoprecipitation analyses confirmed that the UL4 protein interacted with these two cellular factors in EHV-1 infected cells (Figs. 6 and 7). Additional experiments showed that the UL4 protein did not directly associate with general transcription factors TFIIA or TFIIB or with the sole EHV-1 immediate early regulatory protein (Fig. 5). The possibility that the UL4 protein could prevent TBP translocation into the nucleus, thus blocking the critical initiating step in PIC assembly was not substantiated by ongoing immunofluorescence assays to monitor TBP localization in the presence and absence of UL4 protein expression (data not shown). It remains unclear whether the direct interaction of the UL4 protein with TBP and Pol II diminishes their ability to interact with DNA. This question and the Identification of domains within the UL4 protein responsible for its interaction with general transcription factors are the focus of future studies.
It has been well documented that the phosphorylation state of the CTD of Pol II is intimately involved in the progression of transcription from initiation and elongation to termination (Buratowski et al., 1989; Hirose and Ohkuma, 2007; Kobor and Greenblatt, 2002). A potential mechanism for the UL4 protein-mediated inhibition of gene expression may involve dysregulation of the dynamic process of CTD phosphorylation. The direct interaction of the UL4 protein with the CTD may prevent the association between the kinases and/or phosphatases that maintain the appropriate levels of phosphorylation to switch between initiation and elongation or elongation and termination. Likewise, the inhibitory activity may involve binding of the UL4 protein such that it directly eclipses the serines in the heptapeptide repeats that are the targets of phosphorylation (Hengartner et al., 1998; Komarnitsky et al., 2000; Ni et al., 2004). Whether the UL4 protein alters the state of CTD phosphorylation is an interesting avenue of investigation and would represent a novel mechanism for EHV-1 gene regulation.
That the majority of the EHV-1 genes with enhanced expression in the absence of the UL4 protein were those of the late gene class responsible for assembly and maturation of infectious viral particles was unexpected (Tables 2A and B). Thus, during lytic infection, expression of the UL4 protein may prevent the late genes from being transcribed before DNA replication has completed. This could limit the exhaustion of the nucleotide pool and prevent translation of structural proteins that might assemble capsids before significant numbers of daughter genomes can be replicated, which would diminish the assembly of non-infectious empty particles. Additionally, the reduction of late gene expression may facilitate efficient DNA replication, due to the fact that the incoming viral genome would serve as the template for DNA replication as opposed to a template for late gene transcription. Despite these considerations, extensive quantitative PCR analyses clearly revealed that viral DNA synthesis is not retarded in the absence of the UL4 protein (Fig 8B). It is possible that the UL4 protein preferentially regulates late genes by preventing the association of the IE and UL5 proteins with general transcription factors, such as TBP, that were demonstrated to be critical for maximal EHV-1 late gene expression (Albrecht et al., 2004; Zhao et al., 1995). Additional studies are focused on determining if the UL4 protein competes with the IE and UL5 proteins for binding to TBP.
Overall, these findings further demonstrate the inhibitory activity of the UL4 protein and suggest a mechanism by which the UL4 protein affects viral gene expression, namely its direct interactions with two cellular proteins essential for transcription. These interactions may be important for the inhibition of late gene expression as well as for efficient viral DNA replication. Future studies will address how the interactions between the UL4 protein and the transcription machinery contribute to the inhibition of EHV-1 gene expression and efficient viral replication.
MATERIALS AND METHODS
Cell culture and viruses
Mouse fibroblast L-M, rabbit kidney RK13, and human HeLa cells were grown in Dulbecco’s minimum essential medium (DMEM) supplemented with 5% fetal bovine serum at 37°C in a 5% CO2 incubator. Wild-type, ΔUL4, and ΔUL4Res EHV-1 of the pathogenic RacL11 strain background were used for these studies (Charvat et al., 2012).
Plasmids and transfection procedure
The luciferase reporter plasmids, the effector gene expression plasmids, and the GST fusion protein plasmids used in the transient transfection assays were generated elsewhere (Bowles et al., 2000; Charvat et al., 2011; Smith et al., 1992). RK13 cells were transfected using lipofectin (Invitrogen, Carlsbad, CA) and Opti-MEM medium (Gibco, BRL, Carlsbad, CA) as described previously (Ahn et al., 2007). One pmol of reporter plasmid and 0.5 pmol of effector plasmid were employed in most assays. Transfection efficiencies of 70% or greater were routinely obtained.
Luciferase and northern blot analysis
Two groups of RK13 cells were co-transfected with gK-Luc and either the EICP0 or UL4 protein expression plasmids. For one group of cells, luciferase activity was determined 48 h post-transfection utilizing the luciferase activity kit (Promega, Madison, WI) and the POLARstar OPTIMA plate reader (BMG LABTECH Inc., Cary, NC) per manufacturer’s instructions (Ahn et al., 2007). For the other group of cells, total RNA was isolated at 4 h post-transfection using the RNA-Bee RNA isolation reagent (AMS Biotechnology (Europe) Ltd., Abingdon, UK). RNA samples were separated on a 6% denaturing urea-polyacrylamide gel and transferred onto a positively-charged nylon membrane (Ambion, Austin, TX) using a semi-dry electroblotter (Bio-Rad Laboratories). Immobilized RNA was hybridized with a probe specific for the luciferase transcript (5’-GGTGTTGGAGCAAGATGGAT-3’). RNA levels were determined by densitometric measurement of the radiolabeled bands after exposure to a phosphor screen and scanning with the molecular imager FX system.
mRNA half-life
Rabbit kidney cells were infected with either wild-type EHV-1 or ΔUL4 EHV-1 at an MOI of 5. Twelve hours post-infection, the normal growth medium was replaced with growth medium supplemented with 1 μM Actinomycin D. Every three hours, total RNA samples were collected for a total of 18 hours. cDNA was synthesized from the RNA samples using the iScript™ cDNA Synthesis Kit per the manufacturer’s specifications (Bio-Rad Laboratories). Transcript levels were determined by quantitative real-time PCR analysis using the iQ™ SYBR® Green Supermix (Bio-Rad Laboratories). The EHV-1 glycoprotein 2 (gp2, gene EUs4) transcript was detected with forward primer 5’-TACAACAACTGAGACTAC-3’ and reverse primer 5’-GGAGAACTGCTACTATTAG-3’, and the total transcript levels were normalized to cellular 28S rRNA using forward primer 5’-TATCATTGTGAAGCAGAA-3’ and reverse primer 5’-AACAACACATCATCAGTA.
Electromobility shift assays
DNA binding assays were completed using IE, TK, IR4, and UL5 promoter DNA sequences along with GST, GST-IR2, or GST-UL4 fusion proteins. Promoter DNAs were radiolabeled with [α−32P]-dATP and diluted in DNA-binding buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 10 mM β-mercaptoethanol, 0.1% CHAPS, 100 mM NaCl). Radiolabeled promoter DNAs were incubated with equivalent amounts of purified GST fusion proteins for 20 min at room temperature (Kim et al., 1995). DNA-protein complexes were resolved on a 3.2% polyacrylamide gel and then dried onto Whatman chromatography paper (Whatman International Ltd., Maidstone, England). The chromatography paper was exposed to a phosphor screen and scanned with the molecular imager FX system.
Chromatin immunoprecipitation assays (ChIP)
HeLa cells were infected with EHV 1 RacL11 at a MOI of 10 and harvested at 6, 10, 12, and 16 h after infection. The cells were crosslinked with 1% formaldehyde for 10 min followed by adding 1X glycine for 5 min. After adding the Halt protease and phosphatase inhibitor cocktail (Pierce Biotechnology, Rockford, IL), the cells were scrapped and collected. The chromatin preparation was then processed according to the manufacturer’s protocol (Pierce Biotechnology). Lysis buffer was added and micrococcal nuclease digestion was carried out for 15 min at 37°C followed by centrifugation at 9000xg for 5 min. Pre-immune serum, anti-UL4 antibody (Charvat et al., 2011) or anti CTD of Pol II antibody (Protein One, Bethesda, MD) was added in a spin column and incubated overnight at 4°C. After washing, the elution process and DNA purification were carried out according to the manufacturer’s protocol. PCR was performed to amplify the target DNA using appropriate primers : EHV 1 UL4 gene was amplified by the forward primers: 5’-CAT GGT ACC CCA ACG CAA ACA GTT GGC ACC GTG-3’, and reverse primers 5’-CAT AGA TCT CAG GCT GGG AAT TTG CTC GAC TGA AG-3’. The EHV 1 IE gene was amplified by the forward primers: 5’-ACG ACG ATG AGA TGG AGA TG −3’, and reverse primers 5’-ACA GCG ATA CCG AGA CCT G-3’. The EHV 1 TK gene was amplified by the forward primers: 5’-GAG CAC GAC TGG ACG AGT TA −3’, and reverse primers 5’-GTC CGC TTC AAA GAG AGT CC-3’. The EHV 1 gK gene was amplified by the forward primers: 5’-AAA GGT CCT GCT TAG AGC CA −3’, and reverse primers 5’-ACG AGT TCT TAT CGC CGA CT-3’.. PCR products were analyzed in a BioRad XR imaging system. Pre-immune serum was employed as the negative control.
GST-pulldown assays
In vitro protein-protein interaction assays were described previously (Albrecht et al., 2004). Briefly, 2 μg of purified GST, GST-IR2, or GST-UL4 protein was combined with 40 μL of a 50% mix of GST-Bind resin beads (Novagen, Madison, WI) in 650 μL NETN buffer (100 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl [pH 8.0], 0.5% NP-40) and incubated at room temperature for 1.5 h. 1 μg of purified transcription factors TATA box-binding protein (TBP), Pol II CTD, TFIIA, or TFIIB (Protein One, Bethesda, MD) were added to the samples and incubated for an additional 1.5 h. The samples were centrifuged and washed five times with NETN buffer. Precipitated proteins were resolved by SDS-PAGE on 10% polyacrylamide gels and transferred to nitrocellulose membranes. Transcription factor protein complexes were detected with specific antibodies to TBP, Pol II CTD, TFIIA or TFIIB (Protein One). Antibody to the EHV-1 IE protein was described previously (Kim et al 2003).
Co-immunoprecipitation assays
Human HeLa cells were infected with wild-type RacL11 EHV-1 (MOI = 10) and cellular lysates were prepared at 6, 10 and 16 h post-infection. The samples were diluted with a Tris-saline solution (10 mM Tris-HCl [pH 8.0] and 14 mM NaCl) and pre-cleared by mixing with 30 μL of a 50% mix of Protein-A agarose beads (Sigma, St. Louis, MO) for 2 h at room temperature. At the same time, 10μL of the UL4 protein-specific antibody (Charvat et al., 2011) or a non-related anti-glycoprotein D (gD) specific antibody were complexed with a separate 30 μL of a 50% mix of Protein-A agarose beads. The beads were pelleted by centrifugation, and the pre-cleared samples were added to the antibody-bound beads and incubated for 2 h at room temperature. The beads were washed five times with 500 μL of the Tris-saline solution, and after the final wash, the beads were pelleted. SDS sample buffer was added, and the samples were boiled for 5 min before being resolved on 10% polyacrylamide gels. The precipitated protein complexes were detected with either the TBP or Pol II CTD specific antibodies (Protein One). Reverse immunoprecipitation assays using antibody to TBP and antibody to Pol II CTD were carried out by a similar procedure. Control reactions employed pre-immune serum or anti EHV-1 gD antibody. Precipitates were then probed with pre-immune serum or anti-UL4P antibody. The input proteins, TBP and Pol II CTD, were obtained from Protein One.
Microarray analysis
RK13 cells were infected with wild-type or ΔUL4 EHV-1 (MOI = 10) for 18 h before total RNA was isolated using the RNA-Bee reagent. Double-stranded cDNA was synthesized using the Superscript® Double-Stranded cDNA Synthesis Kit and Oligo(dT)12–18 Primer (Invitrogen) following the manufacturer’s protocol. The cDNA was fluorescently labeled with the Label IT® μArray™ Cy™3/Cy™5 Labeling Kit (Mirus, Madison, WI) as per manufacturer’s instructions and purified using the MinElute® PCR Purification Kit (QIAGEN, Germantown, MD). Labeled cDNA was hybridized to the EHV-1 CustomArray 4×2K microarray following the manufacturer’s procedure (CustomArray Inc., Bothell, WA). All 78 EHV-1 genes are represented on the microarray. Each gene is represented by 5 to 10 unique oligonucleotide sequences, each of which is present in triplicate. The microarray also contains host cellular genes GAPDH, GAPDG, and actin. The microarray was scanned using the GenePix 4000B Microarray Scanner (Molecular Devices, LLC, Sunnyvale, CA).
Quantitation of viral genomic DNA and infectious titers
Rabbit kidney cells (RK13) were infected with wild-type (RacL11) or ΔUL4 EHV-1 at a MOI of 10. At 4, 6, 8, 12, and 24 hours post infection, infected cells were pelleted and freeze and thaw for three times for virus titration by plaque assay on RK13 monolayers (Perdue et al., 1974). For quantitative real-time PCR to measure viral DNA, DNA was extracted from infected cell pellets using the DNeasy® Blood &Tissue Kit (QIAGEN, Valencia, CA) following the manufacture’s protocol. Quantitative PCR assays were performed using the iScript™ One-Step RT-PCR Kit SYBR® Green and iQ™ SYBR® Green supermix (Bio-Rad Laboratories, Hercules CA) and primers for cellular GAPDH (forward: 5’-TGCCCCCATGTTTGTGATG-3’ reverse: 5’-TGTGGTCATGAGCCCTTC-3’) and the EHV-1 glycoprotein K (gK) gene (forward: 5’-AAAGGTCCTGCTTAGAGCCA-3’ reverse: 5’-ACGAGTTCTTATCGCCGACT-3’), and the EHV-1 gp2 gene primers (forward:5’-TACAACAACTGAGACTAC-3’ reverse: 5’-GGAGAACTGCTACTATTAG-3’). Statistical analysis for the infectious titer results was completed using the mean of log-transformed data and a two way Student T test, and one way T test for the quantitation of viral genomic DNA.
Highlights.
The UL4 gene is conserved in the genome of DI particles of EHV-1.
The UL4 gene is not essential for EHV-1 lytic replication.
The UL4 protein binds to cellular transcription factors TBP and Pol II.
Late viral gene expression is enhanced in UL4 null virus infection.
Viral DNA synthesis is not retarded in cells infected with the UL4-null virus.
ACKNOWLEDGMENTS
We thank Mrs. Suzanne Zavecz for excellent technical assistance and the members of our laboratory for their helpful suggestions. These investigations were supported by research grant AI-22001 from the National Institute of Allergy and Infectious Diseases, grant 8P20GM103433 from the National Institute of General Medical Sciences, and Agriculture and Food Research Initiative Competitive Grant 2008-35204-04438 from the USDA National Institute of Food and Agriculture.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahn BC, Breitenbach JE, Kim SK, O’Callaghan DJ, 2007. The equine herpesvirus-1 IR3 gene that lies antisense to the sole immediate-early (IE) gene is trans-activated by the IE protein, and is poorly expressed to a protein. Virology 363, 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn BC, Zhang Y, O’Callaghan DJ, 2010. The equine herpesvirus-1 (EHV-1) IR3 transcript downregulates expression of the IE gene and the absence of IR3 gene expression alters EHV-1 biological properties and virulence. Virology 402, 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht RA, Kim SK, Zhang Y, Zhao Y, O’Callaghan DJ, 2004. The equine herpesvirus 1 EICP27 protein enhances gene expression via an interaction with TATA box-binding protein. Virology 324, 311–326. [DOI] [PubMed] [Google Scholar]
- Allen GP, Bryans JT, 1986. Molecular epizootiology, pathogenesis, and prophylaxis of equine herpesvirus-1 infections. Prog. Vet. Microbiol. Immunol 2, 78–144. [PubMed] [Google Scholar]
- Ammermann I, Bruckner M, Matthes F, Iftner T, Stubenrauch F, 2008. Inhibition of transcription and DNA replication by the papillomavirus E8-E2C protein is mediated by interaction with corepressor molecules. J. Virol 82, 5127–5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles DE, Holden VR, Zhao Y, O’Callaghan DJ, 1997. The ICP0 protein of equine herpesvirus 1 is an early protein that independently transactivates expression of all classes of viral promoters. J. Virol 71, 4904–4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles DE, Kim SK, O’Callaghan DJ, 2000. Characterization of the trans-activation properties of equine herpesvirus 1 EICP0 protein. J. Virol 74, 1200–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buczynski KA, Kim SK, O’Callaghan DJ, 1999. Characterization of the transactivation domain of the equine herpesvirus type 1 immediate-early protein. Virus Res. 65, 131–140. [DOI] [PubMed] [Google Scholar]
- Buratowski S, Hahn S, Guarente L, Sharp PA, 1989. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 56, 549–561. [DOI] [PubMed] [Google Scholar]
- Campbell DE, Kemp MC, Perdue ML, Randall CC, Gentry GA, 1976. Equine herpesvirus in vivo: cyclic production of a DNA density variant with repetitive sequences. Virology 69, 737–750. [DOI] [PubMed] [Google Scholar]
- Caughman GB, Lewis JB, Smith RH, Harty RN, and O’Callaghan DJ 1995. Detection and intracellular localization of equine herpesvirus 1 IR1 and IR2 gene products by using monoclonal antibodies. J. Virol 69: 3024–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caughman GB, Robertson AT, Gray WL, Sullivan DC, O’Callaghan DJ, 1988. Characterization of equine herpesvirus type 1 immediate early proteins. Virology 163, 563–571. [DOI] [PubMed] [Google Scholar]
- Caughman GB, Staczek J, O’Callaghan DJ, 1985. Equine herpesvirus type 1 infected cell polypeptides: evidence for immediate early/early/late regulation of viral gene expression. Virology 145, 49–61. [DOI] [PubMed] [Google Scholar]
- Charvat RA, Breitenbach JE, Ahn B, Zhang Y, O’Callaghan DJ, 2011. The UL4 protein of equine herpesvirus 1 is not essential for replication or pathogenesis and inhibits gene expression controlled by viral and heterologous promoters. Virology 412, 366–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvat RA, Zhang Y, O’Callaghan DJ, 2012. Deletion of the UL4 gene sequence of equine herpesvirus 1 precludes the generation of defective interfering particles. Virus Genes 45, 295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Garko-Buczynski KA, Zhang Y, O’Callaghan DJ, 1999. The defective interfering particles of equine herpesvirus 1 encode an ICP22/ICP27 hybrid protein that alters viral gene regulation. Virus Res. 59, 149–164. [DOI] [PubMed] [Google Scholar]
- Chen M, Harty RN, Zhao Y, Holden VR, O’Callaghan DJ, 1996. Expression of an equine herpesvirus 1 ICP22/ICP27 hybrid protein encoded by defective interfering particles associated with persistent infection. J. Virol 70, 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang CM, Broker TR, Chow LT, 1991. An E1M--E2C fusion protein encoded by human papillomavirus type 11 is asequence-specific transcription repressor. J. Virol 65, 3317–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauenhauer SA, Robinson RA, O’Callaghan DJ, 1982. Chronic production of defective-interfering particles by hamster embryo cultures of herpesvirus persistently infected and oncogenically transformed cells. J. Gen. Virol 60, 1–14. [DOI] [PubMed] [Google Scholar]
- Davison BL, Egly JM, Mulvihill ER, Chambon P, 1983. Formation of stable preinitiation complexes between eukaryotic class B transcription factors and promoter sequences. Nature 301, 680–686. [DOI] [PubMed] [Google Scholar]
- Ebner PD, Kim SK, O’Callaghan DJ, 2008. Biological and genotypic properties of defective interfering particles of equine herpesvirus 1 that mediate persistent infection. Virology 381, 98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebner PD, O’Callaghan DJ, 2006. Genetic complexity of EHV-1 defective interfering particles and identification of novel IR4/UL5 hybrid proteins produced during persistent infection. Virus Genes 32, 313–320. [DOI] [PubMed] [Google Scholar]
- Fertey J, Ammermann I, Winkler M, Stoger R, Iftner T, Stubenrauch F, 2010. Interaction of the papillomavirus E8--E2C protein with the cellular CHD6 protein contributes to transcriptional repression. J. Virol 84, 9505–9515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garko-Buczynski KA, Smith RH, Kim SK, O’Callaghan DJ, 1998. Complementation of a replication-defective mutant of equine herpesvirus type 1 by a cell line expressing the immediate-early protein. Virology 248, 83–94. [DOI] [PubMed] [Google Scholar]
- Gaudray G, Gachon F, Basbous J, Biard-Piechaczyk M, Devaux C, Mesnard JM, 2002. The complementary strand of the human T-cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down-regulates viral transcription. J. Virol 76, 12813–12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisberg JV, Chen JL, Ricciardi RP, 1995. Subregions of the adenovirus E1A transactivation domain target multiple components of the TFIID complex. Mol. Cell. Biol 15, 6283–6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WL, Baumann RP, Robertson AT, Caughman GB, O’Callaghan DJ, Staczek J, 1987. Regulation of equine herpesvirus type 1 gene expression: characterization of immediate early, early, and late transcription. Virology 158, 79–87. [DOI] [PubMed] [Google Scholar]
- Grundy FJ, Baumann RP, O’Callaghan DJ, 1989. DNA sequence and comparative analyses of the equine herpesvirus type 1 immediate early gene. Virology 172, 223–236. [DOI] [PubMed] [Google Scholar]
- Harty RN, O’Callaghan DJ, 1991. An early gene maps within and is 3’ coterminal with the immediate-early gene of equine herpesvirus 1. J. Virol 65, 3829–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengartner CJ, Myer VE, Liao SM, Wilson CJ, Koh SS, Young RA, 1998. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Molecular Cell 2, 43–53. [DOI] [PubMed] [Google Scholar]
- Henry BE, Newcomb WW, O’Callaghan DJ, 1979. Biological and biochemical properties of defective interfering particles of equine herpesvirus type 1. Virology 92, 495–506. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Ohkuma Y, 2007. Phosphorylation of the C-terminal domain of RNA polymerase II plays central roles in the integrated events of eucaryotic gene expression. Journal of biochemistry 141, 601–608. [DOI] [PubMed] [Google Scholar]
- Holden VR, Harty RN, Yalamanchili RR, O’Callaghan DJ, 1992. The IR3 gene of equine herpesvirus type 1: a unique gene regulated by sequences within the intron of the immediate-early gene. DNA Seq. 3, 143–152. [DOI] [PubMed] [Google Scholar]
- Holden VR, Zhao Y, Thompson Y, Caughman GB, Smith RH, O’Callaghan DJ, 1995. Characterization of the regulatory function of the ICP22 protein of equine herpesvirus type 1. Virology 210, 273–282. [DOI] [PubMed] [Google Scholar]
- Kim SK, Ahn BC, Albrecht RA, O’Callaghan DJ, 2006. The unique IR2 protein of equine herpesvirus 1 negatively regulates viral gene expression. J. Virol. 80, 5041–5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Bowles DE, O’Callaghan D,J, 1999. The gamma2 late glycoprotein K promoter of equine herpesvirus 1 is differentially regulated by the IE and EICP0 proteins. Virology 256, 173–179. [DOI] [PubMed] [Google Scholar]
- Kim SK, Holden VR, O’Callaghan DJ, 1997. The ICP22 protein of equine herpesvirus 1 cooperates with the IE protein to regulate viral gene expression. J. Virol 71, 1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Jang HK, Albrecht RA, Derbigny WA, Zhang Y, O’Callaghan DJ, 2003. Interaction of the equine herpesvirus 1 EICP0 protein with the immediate-early (IE) protein, TFIIB, and TBP may mediate the antagonism between the IE and EICP0 proteins. J. Virol 77, 2675–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Kim S, Dai G, Zhang Y, Ahn BC, O’Callaghan DJ, 2011. Identification of functional domains of the IR2 protein of equine herpesvirus 1 required for inhibition of viral gene expression and replication. Virology 417, 430–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, O’Callaghan DJ, 2001. Molecular characterizations of the equine herpesvirus 1 ETIF promoter region and translation initiation site. Virology 286, 237–247. [DOI] [PubMed] [Google Scholar]
- Kim SK, Smith RH, O’Callaghan DJ, 1995. Characterization of DNA binding properties of the immediate-early gene product of equine herpesvirus type 1. Virology 213, 46–56. [DOI] [PubMed] [Google Scholar]
- Kobor MS, Greenblatt J, 2002. Regulation of transcription elongation by phosphorylation. Biochimica et biophysica acta 1577, 261–275. [DOI] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S, 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev 14, 2452–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD, 2007. The molecular basis of eukaryotic transcription. Proc. Natl. Acad. Sci. USA 104, 12955–12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Wu J, Luu P, Ghazal P, Flores O, 1996. Inhibition of the association of RNA polymerase II with the preinitiation complex by a viral transcriptional repressor. Proc. Natl. Acad. Sci. USA 93, 2570–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester JT, DeLuca NA, 2011. Herpes simplex virus 1 ICP4 forms complexes with TFIID and mediator in virus-infected cells. J. Virol 85, 5733–5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JB, Thompson YG, Caughman GB, 1993. Transcriptional control of the equine herpesvirus 1 immediate early gene. Virology 197, 788–792. [DOI] [PubMed] [Google Scholar]
- Mitchell PJ, Wang C, Tjian R, 1987. Positive and negative regulation of transcription in vitro: enhancer-binding protein AP-2 is inhibited by SV40 T antigen. Cell 50, 847–861. [DOI] [PubMed] [Google Scholar]
- Ni Z, Schwartz BE, Werner J, Suarez JR, Lis JT, 2004. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Molecular cell 13, 55–65. [DOI] [PubMed] [Google Scholar]
- O’Callaghan DJ, Osterrieder N, 2008. Herpesviruses of Horses, in: Mahy BWJ, VanRegenmortel MHV (Eds.), Encyclopedia of Virology, Third ed. Elsevier; Ltd., Oxford, pp. 411–420. [Google Scholar]
- Perdue ML, Kemp MC, Randall CC, O’Callaghan DJ, 1974. Studies of the molecular anatomy of the L-M cell strain of equine herpes virus type 1: proteins of the nucleocapsid and intact virion. Virology 59, 201–216. [DOI] [PubMed] [Google Scholar]
- Powell ML, Smith JA, Sowa ME, Harper JW, Iftner T, Stubenrauch F, Howley PM, 2010. NCoR1 mediates papillomavirus E8;E2C transcriptional repression. J. Virol 84, 4451–4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purewal AS, Allsopp R, Riggio M, Telford EA, Azam S, Davison AJ, Edington N, 1994. Equid herpesviruses 1 and 4 encode functional homologs of the herpes simplex virus type 1 virion transactivator protein, VP16. Virology 198, 385–389. [DOI] [PubMed] [Google Scholar]
- Smith RH, Caughman GB, O’Callaghan DJ, 1992. Characterization of the regulatory functions of the equine herpesvirus 1 immediate-early gene product. J. Virol 66, 936–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RH, Zhao Y, O’Callaghan DJ, 1994. The equine herpesvirus type 1 immediate-early gene product contains an acidic transcriptional activation domain. Virology 202, 760–770. [DOI] [PubMed] [Google Scholar]
- Telford EA, Watson MS, McBride K, Davison AJ, 1992. The DNA sequence of equine herpesvirus-1. Virology 189, 304–316. [DOI] [PubMed] [Google Scholar]
- Van Dyke MW, Roeder RG, Sawadogo M, 1988. Physical analysis of transcription preinitiation complex assembly on a class II gene promoter. Science 241, 1335–1338. [DOI] [PubMed] [Google Scholar]
- von Einem J, Schumacher D, O’Callaghan DJ, Osterrieder N, 2006. The alpha-TIF (VP16) homologue (ETIF) of equine herpesvirus 1 is essential for secondary envelopment and virus egress. J. Virol 80, 2609–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Jupp R, Stenberg RM, Nelson JA, Ghazal P, 1993. Site-specific inhibition of RNA polymerase II preinitiation complex assembly by human cytomegalovirus IE86 protein. J. Virol 67, 7547–7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Holden VR, Smith RH, O’Callaghan DJ, 1995. Regulatory function of the equine herpesvirus 1 ICP27 gene product. J. Virol 69, 2786–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]