Abstract
Background & Aims
Colorectal cancer is an epigenetically heterogeneous disease, however, the extent and spectrum of the CpG island methylator phenotype (CIMP) is not clear.
Methods
Genome-scale methylation and transcript expression were measured by DNA Methylation and RNA expression microarray in 216 unselected colorectal cancers, and findings were validated using The Cancer Genome Atlas 450K and RNA sequencing data. Mutations in epigenetic regulators were assessed using CIMP-subtyped Cancer Genome Atlas exomes.
Results
CIMP-high cancers dichotomized into CIMP-H1 and CIMP-H2 based on methylation profile. KRAS mutation was associated significantly with CIMP-H2 cancers, but not CIMP-H1 cancers. Congruent with increasing methylation, there was a stepwise increase in patient age from 62 years in the CIMP-negative subgroup to 75 years in the CIMP-H1 subgroup (P < .0001). CIMP-H1 predominantly comprised consensus molecular subtype 1 cancers (70%) whereas consensus molecular subtype 3 was over-represented in the CIMP-H2 subgroup (55%). Polycomb Repressive Complex-2 (PRC2)-marked loci were subjected to significant gene body methylation in CIMP cancers (P < 1.6 × 10-78). We identified oncogenes susceptible to gene body methylation and Wnt pathway antagonists resistant to gene body methylation. CIMP cluster–specific mutations were observed in chromatin remodeling genes, such as in the SWItch/Sucrose Non-Fermentable and Chromodomain Helicase DNA-Binding gene families.
Conclusions
There are 5 clinically and molecularly distinct subgroups of colorectal cancer. We show a striking association between CIMP and age, sex, and tumor location, and identify a role for gene body methylation in the progression of serrated neoplasia. These data support our recent findings that CIMP is uncommon in young patients and that BRAF mutant polyps in young patients may have limited potential for malignant progression.
Keywords: DNA Methylation, CIMP, Colorectal Cancer, Epigenetics, BRAF, KRAS
Abbreviations used in this paper: CGI, CpG Island; CHD, Chromodomain Helicase DNA-Binding; CIMP, CpG island methylator phenotype; CMS, consensus molecular subtyping; DMP, differentially methylated probes; FDR, false-discovery rate; hES, human embryonic stem; mRNA, messenger RNA; MSI, microsatellite instability; NCG, Network of Cancer Genes; PRC2, Polycomb Repressive Complex-2; RBWH, Royal Brisbane and Women’s Hospital; RPMM, recursively partitioned mixed model; SUZ12, Supressor Of Zeste 12; SWI/SNF, SWItch/Sucrose Non-Fermentable; TCGA, The Cancer Genome Atlas
Graphical abstract
See editorial on page 293.
Summary.
We have identified 5 molecularly and clinically relevant subtypes of the CpG island methylator phenotype (CIMP) in colorectal cancer. We show that CIMP-high cancers segregate into distinct subgroups, which display different frequencies of BRAF and KRAS mutation. These CIMP subtypes are associated with important clinical and molecular features, are correlated with mutations in different epigenetic regulator genes, and show a marked relationship with patient age.
Colorectal cancer is a heterogeneous disease characterized by distinct genetic and epigenetic changes that drive proliferative activity and inhibit apoptosis. The conventional pathway to colorectal cancer is distinguished by APC mutation and chromosomal instability, and accounts for approximately 75% of sporadic cancers.1, 2 The remaining colorectal cancers arise from serrated polyps and have activating mutations in the BRAF proto-oncogene, frequent microsatellite instability (MSI), and the CpG island methylator phenotype (CIMP).2, 3
The development of CIMP is critical in the progression of serrated neoplasia.3 It is well established that CIMP can result in the silencing of key genes important for tumor progression, including the tumor-suppressor gene CDKN2A and the DNA mismatch repair gene MLH1.4, 5 Gene silencing mediated by MLH1 promoter hypermethylation impairs mismatch repair function, which leads to MSI.5 CIMP can be detected using a standardized marker panel to stratify tumors as CIMP-high, CIMP-low, or CIMP-negative.3 Activation of the mitogen-activated protein kinase signaling pathway as a result of the BRAF mutation is associated highly with CIMP-high. CIMP-high cancers frequently arise proximal to the splenic flexure and are more common in elderly female patients,2, 3 whereas CIMP-low cancers have been associated with KRAS mutation.6, 7
More recently, consensus molecular subtyping (CMS) was proposed for classifying colorectal cancers based on transcriptional signatures. Guinney et al8 identified 4 major molecular subtypes (CMS1–CMS4). CMS1, or MSI immune subtype, is characterized by MSI, BRAF mutation, and enhanced immunogenicity. CMS2 can be distinguished by chromosomal instability and WNT pathway perturbations. CMS3, or metabolic subtype, is characterized by KRAS mutation, CIMP-low status, and infrequent copy number alterations. CMS4, or mesenchymal subtype, shows high copy number aberrations, activation of the transforming growth factor-β signaling cascade, stromal infiltration, and the worst overall survival. The relationship between CIMP and CMS subtypes is currently unclear.
Methylation is not a phenomenon distinct to neoplasia. Changes in the epigenome also occur with age and in response to environmental factors.9, 10 We previously showed that the promoter region of certain genes becomes increasingly methylated in normal colonic mucosa with age.9 CIMP-high cancers are identified primarily in older patients,2 hence, age-related hypermethylation might prime the intestinal epigenome for serrated neoplasia-type colorectal cancers. Methylation also is critical in the progression of serrated pathway precursors to invasive cancer, primarily through methylation of MLH1 at the transition to dysplasia.11, 12 Thus, the natural history of the cancer within the colorectum may dictate the methylation profile of the cancer once malignancy develops.
DNA methylation alone can be insufficient to induce transcriptional repression.13 Gene repression also is associated with repressive histone marks such as the H3K27me3 mark,14 which is catalyzed by the polycomb-repressor-complex 2. Modification of histone tails is catalyzed by a series of enzymes including epigenetic readers, which scan for histone modifications; writers, which effect the addition of a modification; and erasers, which are responsible for the removal of histone marks. Mutations in genes encoding epigenetic enzymes have been shown to occur frequently in cancer.15 Although DNA methylation is associated classically with gene silencing, the relationship between DNA methylation and histone modifications has not been fully elucidated, and the role of somatic mutations in enzymes that catalyze these epigenetic processes has not been examined comprehensively.
In this study, we define the extent and spectrum of DNA methylation changes occurring in colorectal cancers and relate this to key clinical and molecular events characteristic of defined pathways of tumor progression. We investigate the role of DNA methylation in the modulation of gene transcription, and assess mutation of genes encoding epigenetic regulatory proteins.
Results
Clinical and Molecular Features of the Consecutive Cohort in Comparison With the Cancer Genome Atlas Cohort
Genome-wide DNA methylation levels were assessed in 216 unselected colorectal cancers (Table 1). The mean age of patients at surgery was 67.9 years. Twenty-nine of 216 (13.4%) cancers had a BRAF V600E mutation, and 75 of 216 (34.7%) cancers were mutated at KRAS codons 12 or 13. Mutation of BRAF and KRAS were mutually exclusive. Patients with BRAF mutated cancers were significantly older than patients with BRAF wild-type cancers (mean age, 74.9 vs 66.9 y; P = .01). TP53 was mutated in 78 of 185 (42.2%) cancers. MSI was associated significantly with BRAF mutation (18 of 29 BRAF mutant vs 9 of 187 BRAF wild-type cancers; P < .0001). By using the Weisenberger et al3 panel to determine CIMP status, 24 of 216 (11.1%) were CIMP-high, 44 of 216 (20.4%) were CIMP-low, and 148 of 216 (68.5%) were CIMP-negative. CIMP-high was associated significantly with BRAF mutation compared with BRAF wild-type cancers (19 of 29 vs 5 of 186; P < .0001). CIMP-low was associated significantly with KRAS mutation compared with KRAS wild-type cancers (26 of 75 [34.6%] vs 18 of 141 [12.8%]; P < .001).
Table 1.
Clinicopathologic Details of the 216 Colorectal Adenocarcinomas as Stratified for Methylation-Based CIMP Clustering, Measured on Illumina HM450 Arrays, Using the 5000 Most Variable CpG Sites That Were Not Hypermethylated in Normal Mucosal Tissue
| n | CIMP-H1 | CIMP-H2 | CIMP-L1 | CIMP-L2 | CIMP-Neg | P value | |
|---|---|---|---|---|---|---|---|
| Total, n | 216 | 23 | 22 | 52 | 66 | 53 | |
| Mean age, y | 67.9 | 75.2 | 73.4 | 70.1 | 66.8 | 61.9 | <.0001 |
| Sex | |||||||
| Male | 100 (46.4%) | 5 (21.7%) | 9 (40.9%) | 24 (46.2%) | 35 (53.0%) | 27 (50.9%) | .11 |
| Female | 116 (53.7%) | 18 (78.3%) | 13 (59.1%) | 28 (53.8%) | 31 (47.0%) | 26 (49.1%) | |
| Site | |||||||
| Proximal | 75/213 (35.2%) | 19 (82.6%) | 13 (59.1%) | 20 (39.2%) | 15 (23.4%) | 8 (15.1%) | <.0001 |
| Distal | 96/213 (45.1%) | 4 (17.4%) | 6 (27.3%) | 21 (41.2%) | 32 (50.0%) | 33 (62.3%) | |
| Rectal | 42/213 (19.7%) | 0 | 3 (13.6%) | 10 (19.6%) | 17 (26.6%) | 12 (22.6%) | |
| CIMP status | |||||||
| CIMP-high | 24 (11.1%) | 16 (69.6%) | 3 (13.6%) | 3 (5.8%) | 2 (3.0%) | 0 | <.0001 |
| CIMP-low | 44 (20.4%) | 6 (26.1%) | 13 (59.1%) | 16 (30.8%) | 8 (12.1%) | 1 (1.9%) | |
| CIMP-neg | 148 (68.5%) | 1 (4.3%) | 6 (27.3%) | 33 (63.5%) | 56 (84.8%) | 52 (98.1%) | |
| Mutation | |||||||
| KRAS mutant | 75 (34.7%) | 4 (17.4%) | 12 (54.5%) | 34 (65.4%) | 19 (28.8%) | 7 (13.2%) | <.0001 |
| BRAF mutant | 29 (13.4%) | 17 (73.9%) | 2 (9.1%) | 6 (11.5%) | 4 (6.0%) | 0 (0%) | <.0001 |
| TP53 mutant | 77/185 (41.6%) | 12/21 (57.1%) | 6/21 (28.6%) | 18/45 (40.0%) | 22/54 (40.7%) | 19/44 (43.2%) | .45 |
| Microsatellite instability | |||||||
| MSI | 26 (12.0%) | 11 (47.8%) | 1 (4.8%) | 8 (15.4%) | 6 (9.1%) | 0 | <.0001 |
| MSS | 190 (88.0%) | 12 (52.2%) | 21 (95.2%) | 44 (84.6%) | 60 (90.9%) | 0 | |
| CMS | |||||||
| CMS1 | 35 (16.2%) | 16 (69.6%) | 4 (18.2%) | 5 (9.6%) | 9 (13.6%) | 1 (1.9%) | <.0001 |
| CMS2 | 68 (31.5%) | 0 | 4 (18.2%) | 10 (19.2%) | 30 (45.5%) | 24 (45.3%) | |
| CMS3 | 53 (24.5%) | 3 (13.0%) | 12 (54.5%) | 21 (40.4%) | 10 (15.2%) | 7 (13.2%) | |
| CMS4 | 60 (27.8%) | 4 (17.4%) | 2 (9.1%) | 16 (30.8%) | 17 (25.8%) | 21 (39.6%) | |
| Stage | |||||||
| I | 30/111 | 0/15 | 5/11 (45.5%) | 8/30 (26.7%) | 13/35 (37.1%) | 4/20 (20.0%) | .15 |
| II | 33/111 | 7/15 (46.7%) | 1/11 (9.1%) | 10/30 (33.3%) | 10/35 (28.6%) | 5/20 (25.0%) | |
| III | 34/111 | 6/15 (40.0%) | 4/11 (36.4%) | 7/30 (23.3%) | 11/35 (31.4%) | 6/20 (30.0%) | |
| IV | 14/111 | 2/15 (13.3%) | 1/11 (9.1%) | 5/30 (16.7%) | 1/35 (2.9%) | 5/20 (25.0%) | |
| LINE1 | 70.3 | 68.75 | 68.96 | 72.05 | 70.45 | 69.67 | .38 |
NOTE. P values reported were obtained using analysis of variance for continuous variables and chi-squared analysis for categoric variables.
MSS, microsatellite stable.
We collected a subset of 32 matched noncancerous mucosal samples from patients in the consecutive cohort. The mean age of patients within the cohort of matched normal samples was 68.9, and was not significantly different than the mean age of patients in the wider cohort (P = .71).
Methylation-Based Clustering Shows 5 Subtypes of Colorectal Cancer With Distinct Clinical and Molecular Features
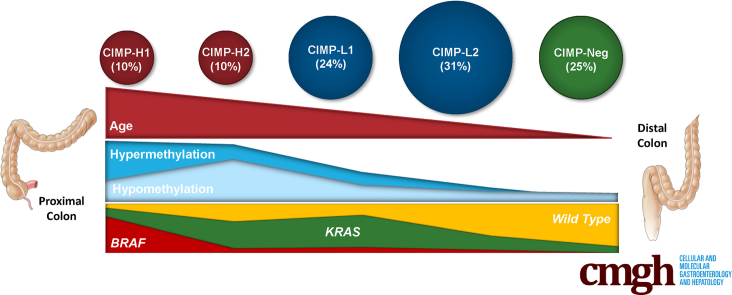
We examined the extent and spectrum of DNA methylation changes in these 216 colorectal cancers using Illumina HumanMethylation450 BeadChip arrays (Illumina Inc, San Diego, CA). Five clusters were identified by recursively partitioned mixed model (RPMM) clustering (Figure 1). These included 2 clusters with high levels of methylation that we have designated CIMP-H1 and CIMP-H2; 2 clusters with intermediate levels of methylation, CIMP-L1 and CIMP-L2; and a single cluster with low levels of methylation, CIMP-neg. There was a significant stepwise increase in age between clusters concordant with increasing genomic methylation (CIMP-neg, 61.9 y; CIMP-L2, 66.8 y; CIMP-L1, 70.1 y; CIMP-H2, 73.4 y; and CIMP-H1, 75.2 y; P < .0001) (Table 1).
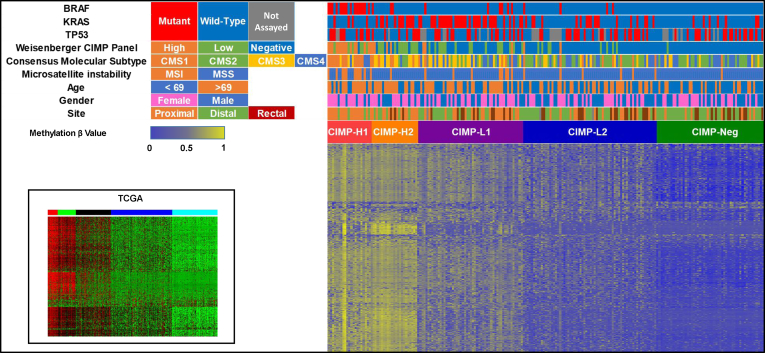
Figure 1.
Methylation heatmap of unselected 216 colorectal cancers using the 5000 most variable β values in CpG sites that were not hypermethylated in normal mucosal tissue. Clustering was performed using the RPMM R package. Clustering showed 5 distinct clusters, termed CIMP-H1, CIMP-H2, CIMP-L1, CIMP-L2, and CIMP-Neg. This was faithfully recapitulated in TCGA.
The CIMP-H1 subgroup comprised 23 of all 216 (10.6%) cancers and was enriched for female patients (18 of 23, 78.3%; P < .0001) and for tumors located proximal to the splenic flexure (19 of 23, 82.6%; P < .0001). We observed no differences in cancer stage at diagnosis and methylation cluster. The CIMP-H1 cluster was strikingly enriched for cancers with features characteristic of serrated neoplasia, including BRAF mutation (17 of 23, 73.9%; P < .0001), CIMP-H status was determined using the Weisenberger et al3 marker panel (16 of 23, 69.6%; P < .0001), MSI (11 of 23, 47.8%; P < .0001), and consensus molecular subtype CMS1 (16 of 23, 69.6%; P < .0001) (Table 1, Figure 1). TP53 was mutated in 12 of 21 (57.1%) CIMP-H1 cluster cancers.
CIMP-H2 cluster cancers also frequently arose in the proximal colon (consecutive cohort, 13 of 22; 59.1%). CIMP-H2 cancers were KRAS mutant more often than CIMP-H1 cancers (54.5% vs 17.4%), and were less often TP53 mutant when compared with the rest of the cohort (28.6%). The incidence of MSI within these cancers was low (4.8%). The frequency of the metabolic CMS3 subtype was higher than in the other CIMP subtypes (54.5%). CIMP-H2 cancers were significantly less likely to be identified as CIMP-high using the Weisenberger et al3 MethyLight panel when compared with CIMP-H1 cancers (13.6% vs 69.6%; P < .001).
CIMP-L1 cancers were significantly enriched for KRAS mutation (65.4%; P < .0001), and were identified equally in the distal and proximal colon. These cancers were rarely MSI (15.4%), and were often the CMS3 (40.4%) or CMS4 (30.8%) subtype. CIMP-L2 cancers mutate KRAS with relative infrequency when compared with CIMP-H2 and CIMP-L1 cancers (28.8%), and are significantly enriched for distal colonic and rectal locations (50% and 26.6%, for distal and rectal locations, respectively; P < .0001). The proportion of CMS2 cancers was significantly higher in CIMP-L2 cancers when compared with CIMP-H1, CIMP-H2, and CIMP-L1 cancers (P < .001). The frequency of distal colonic location was the highest among CIMP-neg cancers (62.3%) and were identified in patients with the youngest mean age (61.9 y). We did not identify a BRAF mutation in any CIMP-neg cancers. CMS2 and CMS4 were the most frequent CMS subtypes in CIMP-neg cancers (45.3% and 39.6%, respectively). The proportion of CMS4 was highest in CIMP-neg cancers when compared with other subtypes (P < .001).
We sequenced hotspots on exons 11 and 15 of BRAF, codon 61 in KRAS, and exon 18 in EGFR in CIMP-H1/H2 cancers that were wild-type at BRAF V600E and KRAS codons 12 and 13, however, we did not identify any mutations in these regions.
Validation of the Association Between CIMP Subtype and Clinical and Molecular Features in The Cancer Genome Atlas
DNA methylation was previously measured using the HumanMethylation 450 array in 392 colorectal cancers from The Cancer Genome Atlas (TCGA) project.16 We observed several differences in the TCGA cohort when compared with the consecutive Royal Brisbane and Women's Hospital (RBWH) cohort. The mean age of patients at the time of diagnosis was significantly lower in the TCGA cohort when compared with the consecutive cohort (64.5 vs 67.9; P < .01). Male sex was slightly over-represented (199 of 373; 53.4%). The distribution of cancers throughout the colon was significantly different in the TCGA cohort. Cancers in the TCGA were significantly enriched for proximal location in comparison with the RBWH cohort (47.0% vs 35.2%; P < .01), and less likely to be located in the distal colon (40.3% vs 45.1%; P < .01) or rectum (12.7% vs 19.7%; P < .01).
There were many similarities between the TCGA and RBWH cohorts. The frequency of BRAF mutations was 9.4%, and was not significantly different from the proportion observed in the RBWH cohort. Likewise, there was no significant difference in the frequency of KRAS mutations between the cohorts (40.1% vs 34.7%, for TCGA and RBWH cohorts, respectively). The proportion of microsatellite unstable cancers was not significantly different between the 2 cohorts (15.9% vs 12%; P = .1).
Despite underlying differences in the clinical and molecular features of the cohorts, unsupervised clustering using the same methods as was used in the RBWH cohorts also resulted in the 5 distinct CIMP clusters identified in the TCGA series (Table 2, Figure 1). There was a similar, striking association between CIMP subtype and biological age (P < .0001). In keeping with the RBWH cohort, increasing CIMP in the TCGA cohort was associated with proximal colonic location (P < .0001), and was correlated inversely with distal and rectal locations (P < .0001 and P < .05, for distal and rectal locations, respectively). The distribution of KRAS mutations in CIMP subtypes followed a similar bell-shaped distribution, and were most common in CIMP-L1 cancers (48 of 81; 59.3%), and least common in CIMP-H1 (5 of 22; 26.3%) and CIMP-negative cancers (21 of 102; 20.6%). Notably, KRAS mutation was more common in CIMP-H2 cancers when compared with CIMP-H1 cancers in the TCGA cohort (43.6% vs 26.3%).
Table 2.
Clinicopathologic and Molecular Details of 374 Colorectal Adenocarcinomas From TCGA Stratified for CIMP Subtype
| n | CIMP-H1 | CIMP-H2 | CIMP-L1 | CIMP-L2 | CIMP-neg | P value | |
|---|---|---|---|---|---|---|---|
| Total, n | 374 | 19 (5.1%) | 39(10.4%) | 81 (21.7%) | 133 (35.6%) | 102 (27.3%) | |
| Mean age, y | 64.5 | 72.2 | 67.8 | 66.5 | 64.5 | 57.1 | <.0001 |
| Sex | |||||||
| Male | 199 | 7 (36.8%) | 21 (53.8%) | 47 (58.0%) | 74 (55.6%) | 50 (49.5%) | NS |
| Female | 174 | 12 (63.2%) | 18 (46.2%) | 34 (42.0%) | 59 (44.4%) | 51 (50.5%) | |
| Site | |||||||
| Proximal | 167 | 17 (100%) | 28 (84.8%) | 53 (67.9%) | 53 (40.8%) | 16 (16.5%) | <.0001 |
| Distal | 143 | f0 | 4 (12.1%) | 18 (23.1%) | 57 (43.8%) | 64 (65.9%) | |
| Rectal | 45 | 0 | 1 (3.0%) | 7 (9.0%) | 20 (15.4%) | 17 (17.5%) | |
| Mutation | |||||||
| BRAF | 35 | 10 (52.6%) | 19 (48.7%) | 5 (6.2%) | 1 (0.8%) | 0 | <.0001 |
| KRAS | 150 | 5 (26.3%) | 17 (43.6%) | 48 (59.3%) | 59 (44.4%) | 21 (20.6%) | <.0001 |
| TP53 | 234 | 10 (52.6%) | 19 (48.7%) | 44 (54.3%) | 85 (63.9%) | 76 (74.5%) | .01 |
| Microsatellite instability | |||||||
| MSI | 51 | 10 (52.6%) | 17 (50%) | 11 (16.7%) | 7 (6.2%) | 6 (6.7%) | <.0001 |
| MSS | 269 | 9 (47.4%) | 17 (50%) | 55 (83.3%) | 105 (93.8%) | 83 (93.3%) | |
| CMS | |||||||
| CMS1 | 42 | 10 (58.8%) | 20 (69%) | 9 (14.3%) | 3 (2.8%) | 0 (0%) | <.0001 |
| CMS2 | 121 | 2 (11.8%) | 1 (3.4%) | 25 (39.7%) | 48 (45.3%) | 45 (51.1%) | |
| CMS3 | 45 | 4 (23.5%) | 4 (13.8%) | 16 (25.4%) | 14 (13.2%) | 7 (8%) | |
| CMS4 | 95 | 1 (5.9%) | 4 (13.8%) | 13 (20.6%) | 41 (38.7%) | 36 (40.9%) | |
| Stage | |||||||
| I | 54 | 3 (15%) | 9 (23.7%) | 16 (20.8%) | 11 (8.7%) | 15 (16%) | <.01 |
| II | 133 | 9 (45%) | 18 (47.4%) | 32 (41.6%) | 50 (39.4%) | 24 (25.5%) | |
| III | 119 | 5 (25%) | 11 (28.9%) | 20 (26%) | 46 (36.2%) | 37 (39.4%) | |
| IV | 50 | 3 (15%) | 0 (0%) | 9 (11.7%) | 20 (15.7%) | 18 (19.1%) |
NOTE. P values reported were obtained using analysis of variance for continuous variables and chi-squared for categoric variables and represent the P value for an association between all subtypes and the feature in question.
MSS, microsatellite stable.
In both cohorts, CMS2 cancers were most frequent in CIMP-L2 (TCGA, 45.3%; RBWH, 45.5%) and CIMP-negative (TCGA, 51.1%; RBWH, 45.3%). Likewise, CIMP-neg cancers were strongly enriched for the CMS4 subtype in both cohorts (TCGA, 40.9%; RBWH, 39.6%)
In contrast to the RBWH cohort, CIMP-H1 cancers were less frequent overall (TCGA, 5.1%; RBWH, 10.6%) and BRAF mutation was associated with CIMP-H1 and CIMP-H2 (CIMP-H1: TCGA, 52.6%; RBWH, 73.9%; CIMP-H2: TCGA, 48.7%; RBWH, 9.1%). Perhaps as a consequence of the increased frequency of BRAF mutations in TCGA CIMP-H2 cancers, MSI was significantly more enriched in CIMP-H2 cancers in the TCGA cohort (50%). Although we did not identify any association between stage and CIMP subtype in the RBWH cohort, late-stage disease was associated significantly with decreasing CIMP in the TCGA cohort (stage IV: CIMP-H1, 15%; CIMP-H2, 0%; CIMP-L1, 11.7%; CIMP-L2, 15.7%; and CIMP-neg, 19.1%; P < .01).
The Colorectal Cancer Methylome Is Altered in Comparison With Normal Mucosa
We identified differentially methylated probes in each cluster compared with 32 normal mucosal samples that matched a subset of cancers in the unselected series (Table 3). In all 4 CIMP clusters (CIMP-H1, -H2, -L1, and -L2), the number of differentially hypermethylated CpG sites greatly exceeded those that were hypomethylated (Table 3). By contrast, in the single CIMP-negative cluster, hypomethylation was more common than hypermethylation. Probe hypermethylation was most frequent in the CIMP-H1 cluster, including 21,168 hypermethylated probes occurring within 5165 unique CpG islands. Of these, 4333 also were hypermethylated in CIMP-H2, whereas 832 were uniquely hypermethylated in CIMP-H1. An additional 523 CpG islands were uniquely hypermethylated in the CIMP-H2 cluster relative to CIMP-H1. The highest number of hypomethylation events was seen in the CIMP-H2 cluster compared with all other clusters (P < .0001), with the majority occurring in open sea regions of the genome.
Table 3.
Distribution of Differentially Hypermethylated Probes in Reference to CpG Islands Vs Normal Mucosal Tissue
| CpG location | CIMP-H1 |
CIMP-H2 |
CIMP-L1 |
CIMP-L2 |
CIMP-neg |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| + | - | + | - | + | - | + | - | + | - | |
| Island | 21,011 | 204 | 19,651 | 426 | 11,297 | 118 | 5685 | 127 | 754 | 162 |
| South Shore | 3196 | 586 | 3003 | 1359 | 1253 | 426 | 513 | 284 | 78 | 242 |
| North Shore | 4745 | 890 | 4641 | 1885 | 2095 | 617 | 911 | 420 | 184 | 346 |
| South Shelf | 229 | 743 | 181 | 1620 | 83 | 574 | 49 | 331 | 19 | 238 |
| North Shelf | 280 | 738 | 259 | 1660 | 92 | 591 | 58 | 342 | 35 | 246 |
| Sea | 2056 | 8396 | 1721 | 15,575 | 647 | 6812 | 297 | 4189 | 104 | 3428 |
| Total | 31,517 | 11,557 | 29,453 | 22,525 | 15,467 | 9138 | 7513 | 5693 | 1174 | 4662 |
NOTE. Cancers were stratified for CIMP clustering. Differential methylation was deemed as an absolute β value change of more than 0.2 and an FDR corrected P value less than .01 compared with 32 normal colorectal mucosal samples.
+, differential hypermethylation; -, differential hypomethylation.
Next, we examined the impact of our chosen β value change threshold on the number of differential methylation events we were able to detect. Shifting the β value change threshold to 0.3 substantially reduced the number of differentially methylated probes identified (to 47.1%, 47.8%, 24.9%, 13.4%, and 5.8% of the probes identified at 0.2 for CIMP-H1 to CIMP-neg, respectively). When we increased the threshold to 0.4 we saw a similar, and more drastic, reduction in our ability to identify differentially methylated probes (DMPs) (18.9%, 19.5%, 4.1%, 1.2%, 0.3% of probes identified at 0.2 for CIMP-H1 to CIMP-Neg, respectively). There was a significant relationship between CIMP subtype and the magnitude of the DMPs identified (P < .0001).
We compared the probes that were differentially hypermethylated (vs normal mucosa) in the RBWH cohort with those differentially hypermethylated in the TCGA cohort. There was a remarkable degree of overlap in differentially methylated loci. In CIMP-H1, 80.2% of differentially hypermethylated loci were detected in both the RBWH and TCGA cohorts. Of the remaining 7481 probes, 6009 were detected solely in the TCGA and 1472 in the RBWH cohorts. We hypothesized that the β cut-off value (>0.2 mean β value difference vs normal) may have resulted in the filtering of many of the probes that were detected in 1 cohort only. Indeed, of the 7481 DMPs detected in 1 cohort only, the methylation level of 98.5% was statistically significantly different from normal colonic mucosa in the other cohort, but were filtered as a result of the difference in the β cut-off value. This was consistent across all CIMP subtypes.
The events that were recognized in 2 independent cohorts are likely to be bona fide differential methylation events. These data indicated that the selection of an appropriate difference in the β cut-off value is critical and that applying stringent cut-off values may significantly increase the type II error rate when reporting differentially methylated events.
CIMP Subtypes Are Associated With Different Stromal Immune Cell Composition
We hypothesized that CIMP subtypes may differ in their stromal cell type composition. We used CIBERSORT to deconvolute the relative composition of immune cells in the tumor microenvironment.17 CIMP-H1 cancers were enriched for M1 macrophages in comparison with all other CIMP subtypes, with the exception of CIMP-L2 cancers (P < .01 vs CIMP-H2, P = .02 vs CIMP-L1, and P = .01 vs CIMP-neg). CIMP-H2 cancers were enriched for resting CD4 T memory cells (P < .01), and were depleted for M1 macrophages (P = .01). Mast cells were associated inversely with DNA methylation subtype, with mast cells contributing least to the immune microenvironment in CIMP-H1 cancers and increasing in a stepwise manner from CIMP-H1 to CIMP-neg (P = .01). Conversely, natural killer cells were associated with CIMP-H cancers (analysis of variance, P < .05), but did not differ between CIMP-H1 and CIMP-H2.
CIMP-H1 and CIMP-H2 Cancers Can Be Delineated by Expression Profiles
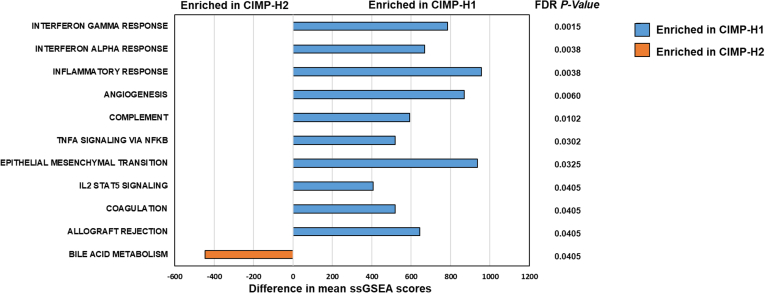
To examine the extent to which CIMP-H1 and CIMP-H2 are transcriptionally distinct, we analyzed differential expression for each cluster with respect to normal mucosa using Illumina HT-12 expression arrays. We then performed single-sample gene set enrichment analysis18 to evaluate enrichments in the Hallmark gene set19 in individual samples (false-discovery rate [FDR] corrected, P < .05). We identified 10 gene sets significantly enriched in CIMP-H1 cancers, 7 of which were related to the immune response (Figure 2). The bile acid metabolism gene set was significantly enriched in CIMP-H2 cancers. In TCGA we did not identify any significant differences in immune response or bile acid metabolism. This may be owing to the increased frequency of BRAF mutant MSI cancers in CIMP-H2 cancers in TCGA.
Figure 2.
Differentially regulated hallmark gene sets between CIMP-H1 and CIMP-H2 cancers as assessed by single-sample gene set enrichment analysis. IL, interleukin; ssGSEA, single sample gene set enrichment analysis.
Relationship Between Promoter Hypermethylation and Gene Transcriptional Activity
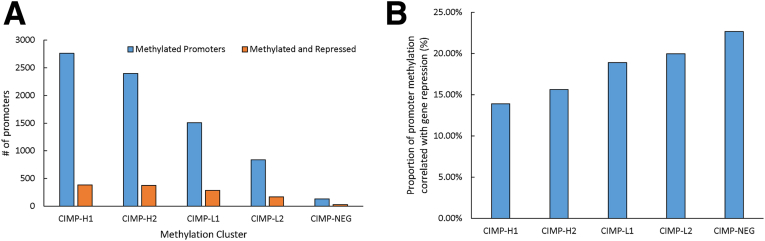
To determine the frequency of which DNA hypermethylation in promoter regions controls transcription of downstream genes, we examined the transcript levels for genes where the promoter was hypermethylated relative normal mucosa. Although promoter methylation was most common in CIMP-H1 and CIMP-H2 clusters (Figure 3A), these subgroups had the lowest proportion of genes in which hypermethylation correlated with reduced transcript expression (13.9% and 15.6%, respectively). This inverse relationship continued for CIMP-L1 (18.9%), CIMP-L2 (19.9%), and with the CIMP-negative cancers, with reduced transcription in 22.7% of hypermethylated promoters (P < .0001) (Figure 3B). We observed a similar relationship between gene transcription and promoter methylation in cancers in TCGA. In TCGA, the proportion of methylated genes that resulted in gene transcription repression did not differ between CIMP subtypes.
Figure 3.
(A) Number of differentially methylated promoters in each CIMP cluster vs the cohort of normal mucosal samples. (B) The proportion of methylation events within each cluster that resulted in gene repression at the transcript level.
We considered that loci that were methylated and repressed in multiple CIMP clusters may be genes that are important for cancer development. Strikingly, of the 1273 genes that were methylated and repressed in at least 1 CIMP cluster, 82.3% were methylated and repressed in 2 or more CIMP clusters, 16.9% silenced in 3 or more CIMP subtypes, and 8.0% in all 4 CIMP subtypes (excluding CIMP-negative). We identified 21 tumor-suppressor genes, as per the Network of Cancer Genes (NCG)6.0 database, that were recurrently methylated and silenced in 3 or more CIMP subtypes (Table 4).
Table 4.
Tumor-Suppressor Genes That Were Recurrently Methylated and Repressed in More Than 3 CIMP Subtypes
| Gene name | Description |
|---|---|
| PCDH9 | Protocadherin 9 (source: HGNC symbol; Acc: HGNC: 8661) |
| CDO1 | Cysteine dioxygenase type 1 (source: HGNC symbol; Acc: HGNC: 1795) |
| MAL | Mal, T-cell differentiation protein (source: HGNC symbol; Acc: HGNC: 6817) |
| EPB41L3 | Erythrocyte membrane protein band 4.1-like 3 (source: HGNC symbol; Acc: HGNC: 3380) |
| AKAP12 | A-kinase anchoring protein 12 (source: HGNC symbol; Acc: HGNC: 370) |
| NDRG4 | NDRG family member 4 (source: HGNC symbol; Acc: HGNC: 14466) |
| LIFR | LIF-receptor α (source: HGNC symbol; Acc: HGNC: 6597) |
| SCUBE2 | Signal peptide, CUB domain, and EGF-like domain containing 2 (source: HGNC symbol; Acc: HGNC: 30425) |
| TMEFF2 | Transmembrane protein with EGF-like and 2 follistatin-like domains 2 (source: HGNC symbol; Acc: HGNC: 11867) |
| DUSP26 | Dual-specificity phosphatase 26 (source: HGNC symbol; Acc: HGNC: 28161) |
| C2orf40 | Chromosome 2 open reading frame 40 (source: HGNC symbol; Acc: HGNC: 24642) |
| SFRP1 | Secreted frizzled-related protein 1 (source: HGNC symbol; Acc: HGNC: 10776) |
| UCHL1 | Ubiquitin C-terminal hydrolase L1 (source: HGNC symbol; Acc: HGNC: 12513) |
| IKZF1 | IKAROS family zinc finger 1 (source: HGNC symbol; Acc: HGNC: 13176) |
| CADM2 | Cell adhesion molecule 2 (source: HGNC symbol; Acc: HGNC: 29849) |
| CXCL12 | C-X-C motif chemokine ligand 12 (source: HGNC symbol; Acc: HGNC: 10672) |
| IRF4 | Interferon regulatory factor 4 (source: HGNC symbol; Acc: HGNC: 6119) |
| ZBTB16 | Zinc finger and BTB domain containing 16 (source: HGNC symbol; Acc: HGNC: 12930) |
| CHFR | Checkpoint with forkhead and ring finger domains (source: HGNC symbol; Acc: HGNC: 20455) |
| SLIT2 | Slit guidance ligand 2 (source: HGNC symbol; Acc: HGNC: 11086) |
| ZFP82 | ZFP82 zinc finger protein (source: HGNC symbol; Acc: HGNC: 28682) |
Acc, accession number; BTB, Broad-Complex, Tramtrack and Bric a brac; EGF, epidermal growth factor; HGNC, Human Genome Organisation Gene Nomenclature Committee; LIF, leukocyte inhibitory factor; NDRG, N-Myc downregulated gene.
Polycomb-Repressive Complex 2 Occupancy at Hypermethylated CpGs Is Correlated Inversely With Global Hypermethylation
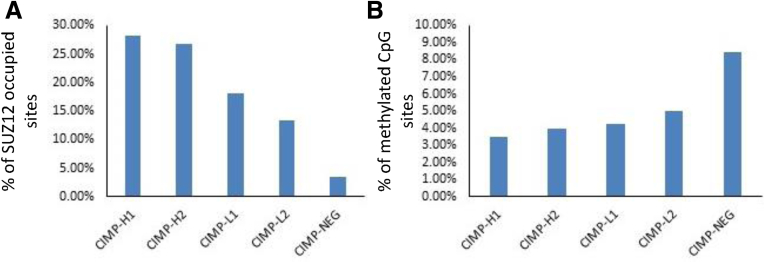
Supressor Of Zeste 12 (SUZ12) occupancy is a surrogate for polycomb-repressor complex 2 occupancy and in embryonic stem cells this has been shown to associate with transcriptional repression of hypermethylated loci.6, 20 Consistent with this, we observed an increase in the number of methylated CpG sites that overlap with SUZ12-occupied regions with increasing CIMP cluster (P < .0001) (Figure 4A). Conversely, and in keeping with our findings with promoter methylation, an inverse association between the proportion of hypermethylated loci genes that overlapped with SUZ12-occupied sites with increasing CIMP cluster was observed (P < .0001) (Figure 4B). This further supports our finding that although DNA hypermethylation occurs more frequently with increasing CIMP cluster, these methylation events are more likely to result in gene silencing in CIMP-negative cancers.
Figure 4.
(A) Proportion of SUZ12-occupied regions in hESC1 cells that contained hypermethylated probes in respective CIMP clusters. (B) Proportion of differential hypermethylation events that overlapped with Polycomb Repressive Complex-2 (PRC2)-occupied regions.
CIMP-H1 and CIMP-H2 Promoter Methylation Is Defined by the Enrichment of Distinct Transcription Factor Binding Sites
Transcription factor binding sites often contain CpG sequences and therefore are a target of DNA methylation, which may explain some of the effects of methylation on transcription. To explore whether DNA methylation is targeted to specific transcription factor binding sites we performed an enrichment analysis using the CentriMo21 tool to examine the 2-kb region immediately upstream of hypermethylated genes. There were 128 significantly enriched binding sites that overlapped in CIMP-H1 and CIMP-H2 cancers. An additional 323 sites were uniquely enriched in CIMP-H1 cancers and an additional 330 sites in CIMP-H2 cancers. SMAD4 and FOXP3 (adjusted P values = 1.2 × 10-24 and 4.1 × 10-23, respectively) were the most significantly enriched motifs in CIMP-H1 cancers. SPDEF, FLI1, and NKX6 (adjusted P values = 7.2 × 10-30, 1.1 × 10-16, and 3.5 × 10-16, respectively) were most significantly enriched in CIMP-H2 cancers. Table 5 presents the top 10 enriched consensus binding sites that were exclusive to CIMP-H1 and CIMP-H2.
Table 5.
Motifs That Were Most Significantly and Exclusively Enriched at Methylated Promoters in CIMP-H1 and CIMP-H2
| CIMP-H1 |
CIMP-H2 |
||||||
|---|---|---|---|---|---|---|---|
| Motif name | Motif | Raw P value | Adjusted P value | Motif name | Motif | Raw P value | Adjusted P value |
| Smad4 | TGTCTRGM | 1.2E-21 | 1.2E-24 | SPDEF_DBD_2 | GTGGTCCCGGATTAT | 7.2E-33 | 7.2E-30 |
| FOXP3_DBD | RTAAACA | 4.1E-20 | 4.1E-23 | UP00142_1 | VNTAATTAATTAABGSG | 2.4E-20 | 2.4E-17 |
| FOXP3 | RTAAACA | 4.1E-20 | 4.1E-23 | FLI1_full_2 | ACCGGAAATCCGGT | 1.1E-19 | 1.1E-16 |
| POU2F2_DBD_2 | HWTRMATATKCAWA | 4.5E-19 | 4.5E-22 | UP00200_1 | GVWAATTAATTAMYBBG | 3.5E-19 | 3.5E-16 |
| Zscan4_primary | DHNATGTGCACAYAHWN | 1.2E-18 | 1.3E-21 | NHLH1_DBD | CGCAGCTGCS | 2.1E-18 | 2.1E-15 |
| HOXC10 | GTCRTAAAAH | 1.3E-18 | 1.3E-21 | ERG_full_2 | ACCGGAWATCCGGT | 4.8E-18 | 4.8E-15 |
| Bbx_secondary | HVWNNGTTAACASHNRV | 3.1E-16 | 3.1E-19 | MA0680.1 | TAATCGATTA | 8.7E-18 | 8.6E-15 |
| Foxc1_DBD_1 | GTAAAYAAACA | 1.3E-15 | 1.3E-18 | PAX7_DBD | TAATYRATTA | 1.4E-16 | 1.4E-13 |
Gene Bodies of Wnt Pathway Antagonists Are Resistant to Methylation
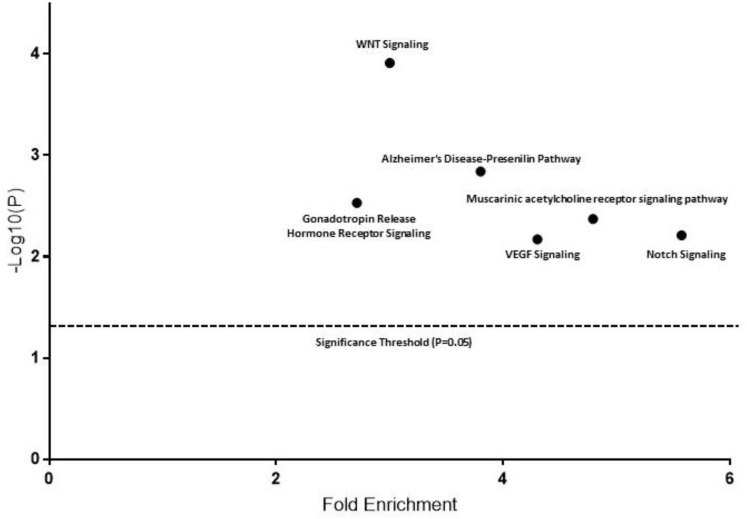
We further explored gene bodies that were unmethylated but had more than 10 CpG island probes, and performed pathway analysis to identify pathways that were devoid of gene body methylation. There were 6 pathways that were significantly enriched among these genes, including the WNT signaling pathway (Figure 5). The WNT signaling pathway was most heavily enriched. PCDHA6, PCDHGA2, PCDHA7, and PCDHA2 contained 36, 15, 10, and 20 gene body CpG island probes, respectively, which were all unmethylated. These protocadherins have been implicated in the regulation of the WNT signal and may act as a tumor-suppressor gene. Likewise, AXIN1, a gene critical to the β-catenin destruction complex, contained 11 unmethylated intragenic CpG Island (CGI) probes. TCF3, a WNT pathway repressor, contained 19 unmethylated intragenic CGI probes. We considered whether gene body methylation within WNT antagonists could alter gene transcription, however, we did not observe any differences in expression profiles of these genes vs normal mucosa tissue, and they were not expressed in normal mucosa tissue. In the remaining WNT genes we did not identify any consistent expression changes.
Figure 5.
Pathways significantly enriched for genes that contained CpG islands that were devoid of methylation in both CIMP-H clusters. VEGF, vascular endothelial growth factor.
Oncogenes Are Significantly More Likely Than Tumor-Suppressor Genes to Undergo Gene Body Methylation in CIMP-H1 and CIMP-H2 Cancers
Gene body methylation is correlated positively with gene expression.22 We examined hypermethylation in gene body CpG islands, defined as a minimum of 2 probes in the CpG island as hypermethylated relative to normal (P < .01) and there was a mean absolute difference in β values vs normal of greater than 0.2 to evaluate whether gene body methylation was a phenomena enriched in oncogenes of CIMP-H–type cancers, or was driven more nonspecifically by CIMP itself. In total, 239 genes were annotated as known oncogenes, and 239 as known tumor-suppressor genes in the NCG6.0 cancer gene database.23 Of these, 121 tumor suppressors and 116 oncogenes had a CpG island within the gene body that was probed on the array. In CIMP-H1 cancers, 21.5% (20.2% in TCGA) of oncogenes had significant gene body methylation in reference to normal, by comparison, significantly fewer tumor-suppressor genes underwent gene body methylation (12.4% in the RBWH cohort, P < .05; 8.1% in TCGA; P < .001). Likewise, gene body methylation was significantly more likely to occur in oncogenes than tumor-suppressor genes in CIMP-H2 cancers (23.3% vs 11.6%; P = .01). The gene expression of 5 oncogenes in CIMP-H1 and CIMP-H2 differed significantly from normal mucosa (FEV, BCL2, and KIT were down-regulated and PAX3 and SND1 were up-regulated in CIMP-H1; LMO2 and CTNND2 were down-regulated and SND1, CNTTA2, and TLX1 were up-regulated in CIMP-H2). Table 6 presents the oncogenes that had significantly higher gene body methylation in CIMP-H1 and CIMP-H2 cancers compared with normal colonic mucosa.
Table 6.
Oncogenes With Significantly Higher Methylation Within the Body of the Gene
| CIMP-H1 |
CIMP-H2 |
||||
|---|---|---|---|---|---|
| Gene | Expression | Description | Gene | Expression | Description |
| FEV | Down-regulated | FEV, ETS transcription factor | LMO2 | Down-regulated | LIM domain only 2 |
| BCL2 | Down-regulated | BCL2, apoptosis regulator | CTNND2 | Down-regulated | Catenin Δ 2 |
| KIT | Down-regulated | KIT proto-oncogene receptor tyrosine kinase | SND1 | Up-regulated | Staphylococcal nuclease and tudor domain containing 1 |
| PAX3 | Up-regulated | Paired box 3 | CTNNA2 | Up-regulated | Catenin α2 |
| SND1 | Up-regulated | Staphylococcal nuclease and tudor domain containing 1 | TLX1 | Up-regulated | T-cell leukemia homeobox 1 |
| LMO2 | No difference | LIM domain only 2 | PREX2 | No difference | PI-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| RSPO3 | No difference | R-spondin 3 | RSPO3 | No difference | R-spondin 3 |
| CTNND2 | No difference | Catenin delta 2 | RET | No difference | Ret proto-oncogene |
| TLX3 | No difference | T-cell leukemia homeobox 3 | LMO1 | No difference | LIM domain only 1 |
| SIX1 | No difference | SIX homeobox 1 | FLT3 | No difference | Fms-related tyrosine kinase 3 |
| HOXC13 | No difference | Homeobox C13 | CACNA1D | No difference | Calcium voltage-gated channel subunit α1 D |
| LMO1 | No difference | LIM domain only 1 | WWTR1 | No difference | WW domain containing transcription regulator 1 |
| ZNF521 | No difference | Zinc finger protein 521 | CHST11 | No difference | Carbohydrate sulfotransferase 11 |
| SALL4 | No difference | Spalt like transcription factor 4 | PAX3 | No difference | Paired box 3 |
| ZEB1 | No difference | Zinc finger E-box binding homeobox 1 | FLT4 | No difference | Fms-related tyrosine kinase 4 |
| PREX2 | No difference | PI-3,4,5-trisphosphate dependent Rac exchange factor 2 | CXCR4 | No difference | C-X-C motif chemokine receptor 4 |
| OLIG2 | No difference | Oligodendrocyte transcription factor 2 | TLX3 | No difference | T-cell leukemia homeobox 3 |
| SMO | No difference | Smoothened, frizzled class receptor | TAL1 | No difference | TAL bHLH transcription factor 1, erythroid differentiation factor |
| FLT3 | No difference | Fms related tyrosine kinase 3 | SIX1 | No difference | SIX homeobox 1 |
| GATA2 | No difference | GATA binding protein 2 | HOXC11 | No difference | Homeobox C11 |
| TLX1 | No difference | T-cell leukemia homeobox 1 | OLIG2 | No difference | Oligodendrocyte transcription factor 2 |
| TAL1 | No difference | TAL bHLH transcription factor 1, erythroid differentiation factor | MYOD1 | No difference | Myogenic differentiation 1 |
| CACNA1D | No difference | Calcium voltage-gated channel subunit a1 D | ZEB1 | No difference | Zinc finger E-box binding homeobox 1 |
| MYOD1 | No difference | Myogenic differentiation 1 | HOXC13 | No difference | Homeobox C13 |
| CTNNA2 | No difference | Catenin a2 | ZNF521 | No difference | Zinc finger protein 521 |
| CHST11 | No difference | Carbohydrate sulfotransferase 11 | SMO | No difference | Smoothened, frizzled class receptor |
| NR4A3 | No difference | Nuclear receptor subfamily 4 group A member 3 | GATA2 | No difference | GATA binding protein 2 |
| NR4A3 | No difference | Nuclear receptor subfamily 4 group A member 3 | |||
BCL, B-cell lymphoma; bHLH, basic helix-loop-helix; ETS, E26 transformation specific; FEV, fifth ewing variant; PI, phosphatidylinositol; SIX, Sineoculis homeobox homolog; TAL, T-cell acute lymphocyctic.
Loci Marked by the PRC2 Complex in Human Embryonic Stem Cells Are Prone to Gene Body Methylation During Cancer Development
Polycomb Repressive Complex-2 (PRC2) marking in human embryonic stem cells has been shown previously to overlap significantly with promoter hypermethylation in colorectal cancers.6 We hypothesized that a similar phenomenon would occur with regard to gene body hypermethylation. In CIMP-H1 and CIMP-H2 cancers, 30.59% and 31.04%, respectively, of loci marked with H3K27me3 in human embryonic stem cells developed significant gene body hypermethylation (Table 7) (P = 1.34 × 10-280 for CIMP-H1 and P = 2.5 × 10-300 for CIMP-H2 overlap). We observed a lesser, but still highly significant, overlap between H3K27me3 marked loci and gene body methylation in CIMP-L1 (13.1%; P = 6.11 × 10-122) and CIMP-L2 (8.5%; P = 1.6 × 10-78) cancers, but did not observe any correlation in CIMP-neg cancers, which likely is owing to the scarcity of which gene body methylation occurs in these cancers. We observed similar overlaps for embryonic ectoderm development (EED) targets, SUZ12 targets, and PRC2 targets.
Table 7.
Overlap Between Genes Marked by the PRC2 Complex and H3K27Me3 in hEScells and Genes That Undergo Significant Gene Body Methylation in Colorectal Cancer Development
| Gene set name | CIMP-H1 |
CIMP-H2 |
CIMP-L1 |
CIMP-L2 |
||||
|---|---|---|---|---|---|---|---|---|
| Overlap fraction | FDR P value | Overlap fraction | FDR P value | Overlap fraction | FDR P value | Overlap fraction | FDR P value | |
| BENPORATH_ES_WITH_H3K27ME3 | 30.59% | 1.34E-280 | 31.04% | 2.50E-300 | 13.06% | 6.11E-122 | 8.50% | 1.60E-78 |
| BENPORATH_EED_TARGETS | 30.70% | 3.91E-267 | 31.07% | 1.12E-284 | 12.81% | 8.75E-112 | 8.66% | 8.47E-77 |
| BENPORATH_SUZ12_TARGETS | 30.92% | 5.05E-264 | 30.73% | 9.67E-273 | 12.91% | 1.29E-110 | 8.48% | 2.02E-72 |
| BENPORATH_PRC2_TARGETS | 37.27% | 1.04E-218 | 38.04% | 8.59E-235 | 16.41% | 4.56E-98 | 11.04% | 2.05E-66 |
NOTE. The overlap fraction represents the gene bodies that are methylated (k) divided by the number of genes marked by each respective mark in hES cells (K) (k/K). The FDR corrected P value was obtained through modeling a hypergeometric distribution (k-1, K, N-K, n; where k is the number of genes methylated in each cluster; K is the number of genes in the gene set; N is the number of genes in the human genome; and n is the number of genes in the query set) using the compute overlaps tool on the Gene Set Enrichment Analysis (GSEA) web portal using the Benporath gene sets, which were obtained though ChIP-on a Chip analysis of human embryonic stem cells.
Epigenetic Regulator Gene Mutations Are Common in TCGA Cancers
Mutations in epigenetic modifier genes have been shown previously to modulate transcriptional profiles in cancer.15 We assessed the mutational frequency of 719 epigenetic regulator genes in cancers from the TCGA colon adenocarcinoma and rectal adenocarcinoma projects using the CIMP subtypes identified earlier. For these analyses we included only mutations that were truncating in nature (nonsense or indels), were predicted to alter splicing, or were predicted to have a deleterious effect by PolyPhen.24
Overall, 92.8% of cancers had a deleterious mutation in an epigenetic regulator gene (347 of 374). There were 94.7% and 100% of CIMP-H1 and CIMP-H2 cancers that had at least 1 mutation in an epigenetic regulator. The proportion of CIMP-L1, CIMP-L2, and CIMP-negative cancers with deleterious mutations in these genes was slightly lower (93.8%, 89.5%, and 93.1%, respectively), however, these proportions were not significantly different from CIMP-H1 or CIMP-H2. Of the 719 genes we investigated, 95.7% were mutated in at least 1 cancer (688 of 719).
Figure 6 shows the most commonly mutated epigenetic regulators in each cluster. Mutations were least common in cancers classified as CIMP-neg, with increasing global methylation being associated with a concordant increase in epigenetic mutational load. However, when we examined epigenetic mutation frequency in relation to microsatellite instability, there was no significant relationship between CIMP cluster and epigenetic mutation frequency, indicating that the differences observed between CIMP clusters may be driven by the increasing frequency of microsatellite instability in CIMP clusters with higher genomic methylation.
Figure 6.
High-impact mutations in epigenetic regulator genes are frequent in cancers with higher genomic methylation. Del, deletion; Ins, insertion.
CIMP-H1 and H2 Subtypes Have Similar Mutational Patterns in Epigenetic Regulator Genes
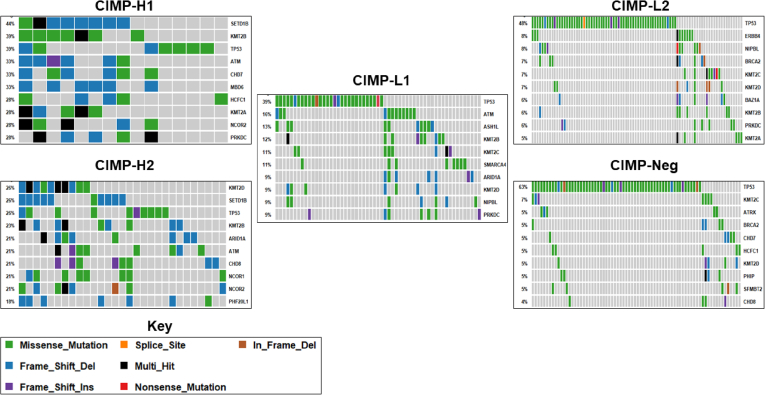
We examined the top 25 mutated epigenetic regulator genes in CIMP-H1 and CIMP-H2 to identify mutational targets that are common to CIMP-H and those that are exclusive to either the CIMP-H1 or CIMP-H2 subtypes. This was not influenced by MSI, which was equally represented in these cancer subtypes (53% CIMP-H1, 50% CIMP-H2). A total of 31.6% of these genes were identifiable in the top 25 epigenetic mutational targets in both CIMP-H1 and CIMP-H2. Such genes included 4 histone lysine methyltransferases (SETD1B, KMT2A, KMT2B, and KMT2D), the SWItch/Sucrose Non-Fermentable (SWI/SNF) complex member ARID1A, and the chromohelicase domain gene CHD7. Thirteen genes were identified in the top 25 mutated epigenetic regulators in CIMP-H1, but not CIMP-H2, these included the DNA demethylases TET1 (mutated in 15.8% of CIMP-H1 cancers vs 10.3% of CIMP-H2 cancers) and TET3 (mutated in 26.3% of CIMP-H1 cancers vs 10.3% of CIMP-H2 cancers). Mutations in histone lysine demethylase KDM2B were enriched in CIMP-H1 cancers (mutated in 36.8% of CIMP-H1 cancers vs 7.7% of CIMP-H2 cancers; P = .01).
In contrast, 13 genes were found in the top 25 mutated epigenetic regulators of CIMP-H2 but not CIMP-H1. The NCOR1 transcription factor was mutated in 20.5% of CIMP-H2 cancers compared with 5.3% of CIMP-H1 cancers, and the cohesin complex subunit NIPBL in 15.4% of CIMP-H2 cancers, despite not being identified as mutated in any CIMP-H1 cancer.
Epigenetic Regulator Gene Mutation Exclusivity Supports the Dichotomization of CIMP-L Clusters
We used a similar approach (top 25 epigenetic gene mutations) to investigate whether CIMP-L1 and CIMP-L2 subtype cancers also target similar epigenetic regulators for somatic mutation. Here, 11 epigenetic regulator genes were commonly mutated in both CIMP-L1 and CIMP-L2. The histone lysine methyltransferases KMT2B and KMT2C were among the top 25 mutated epigenetic regulators in both CIMP-L1 and CIMP-L2, however, the frequency of mutation in both KMT2B and KMT2C was lower in CIMP-L2 cancers (KMT2B CIMP-L1, 11.8%; CIMP-L2, 5.7%; KMT2C CIMP-L1, 10.5%; and CIMP-L2, 6.5%), but this was not statistically significant. There was a nonsignificant trend (P = .06) for increased ASH1L mutation in CIMP-L1 cancers (13.2%) vs CIMP-L2 cancers (4.9%). Fourteen genes were in the top 25 mutated epigenetic regulators of CIMP-L1 or CIMP-L2 alone. SETD1B, a histone lysine methyltransferase identified as a commonly mutated gene in CIMP-H cancers was mutated in 6 CIMP-L1 cancers, but was only mutated in a single CIMP-L2 cancer (P < .01). Likewise, we identified recurrent ARID1A mutations in CIMP-L1 (9.2%), however, we identified significantly fewer in CIMP-L2 cancers (1.6%; P < .01).
The SWI/SNF Complex Is a Commonly Aberrantly Mutated Chromatin Remodeling Complex in CIMP-H1, CIMP-H2, and CIMP-L1 Cancers
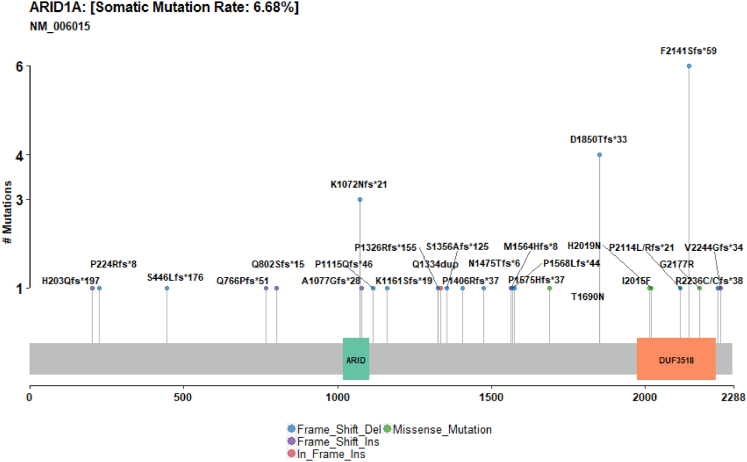
Next, we examined the SWI/SNF complex (MARCA2, ARID1A, ARID1B, ARID2, PBRM1, SMARCB1, and SMARCA4) for high-impact somatic mutations. Mutations in any of the SWI/SNF subunits occurred in 19.06% of cancers. An ARID1A mutation was the most frequent genetic alteration of the complex (6.7%). We observed a number of recurrently mutated positions in ARID1A, including 6 frameshift deletions at codon 2141, 4 deletions at codon 1850, and 3 deletions at codon 1072 (Figure 7). ARID2 was mutated in 6% of cancers, but unlike ARID1A we did not identify any recurrently mutated positions. The distribution of the mutations between CIMP subtypes was significantly skewed toward subtypes with higher overall methylation (P < .0001). SWI/SNF mutations were observed in 50% of CIMP-H1 cancers, and 38.5% of CIMP-H2 cancers. A total of 26.3% of CIMP-L1 samples mutated a SWI/SNF member, and in contrast to CIMP-H1 and CIMP-H2, the most frequently mutated member of the complex was SMARCA4 (11%). The R885C mutation was observed in 3 cancers in CIMP-L1. Mutations in SWI/SNF subunits were similarly infrequent and significantly less prevalent in CIMP-L1 and CIMP-neg (10.6% and 11.6%, respectively; P < .0001).
Figure 7.
High impact mutations in ARID1A are common in colorectal adenocarcinomas. Del, deletion; Ins, insertion.
Synthetic lethality in the SWI/SNF complex was established previously.25 CIMP-H1, CIMP-H2, and CIMP-L1 cancers may be more vulnerable to treatments targeting the other element of the SWI/SNF complex. To test whether 1 SWI/SNF mutation confers dependency on other SWI/SNF subunits in vitro, we correlated exome capture data from 15 cell lines26 with cell line–dependency data from Meyers et al.27 Five cell lines had an ARID1A truncating mutation and these were significantly more dependent on ARID1B expression for survival (0.31 vs 0.06; P < .05).
The Frequency of Genetic Perturbation of Chromodomain Helicase DNA Binding Genes Is Associated With DNA Methylation
CHD genes are members of another chromatin remodeling family. High-impact CHD family gene mutations were present in 22.4% of colorectal cancers in the TCGA. CHD mutations were markedly more common in CIMP-H1 and CIMP-H2 cancers. Family members were mutated in 50% and 51.3% of CIMP-H1 and CIMP-H2 cancers, respectively. CHD7 was the most frequently altered gene in CIMP-H1 (33% of cancers), and CHD8 in CIMP-L2 (22%). CHD mutations were less common, but still frequent, in CIMP-L1 cancers (19.7%). In these cancers, CHD4 was the most commonly mutated gene (8%). The frequency of CHD mutations continued to decline as concordant with DNA methylation. The frequency of CHD mutations in CIMP-L2 was 11.7%, and was lower than the frequency observed in CIMP-neg cancers (15%).
We examined the CHD genes for recurrently mutated positions. At the CHD7 locus, which was mutated in 5.5% of cancers, we observed 5 frameshift deletions (D2988fs del 3) at the 3’ end of the gene. This mutation has been observed in a number of colorectal cancer cell lines. For CHD3, CHD4, and CHD9 we observed 3 recurrently mutated positions at R540fs del 16, R975H, and F760fs del 16.
Discussion
Remodeling of the epigenome is fundamental to colorectal cancer progression. One of the most common epigenetic phenomena altered throughout carcinogenesis is the DNA methylation landscape. Here, we aimed to better understand the extent and heterogeneity of aberrant DNA methylation in colorectal cancers, and characterize the interplay between DNA methylation, somatic variation in epigenetic regulator genes, and gene transcription. Through the genome-scale interrogation of the largest unselected and consecutive series of colorectal cancers to date, we identified 5 clinically and molecularly distinct DNA methylation subtypes. The 5 subtypes identified in this study are highly correlated with key clinical and molecular features, including patient age, tumor location, microsatellite instability, and oncogenic mitogen-activated protein kinase mutations. We show that cancers with high DNA methylation show an increased preponderance for mutating genes involved in epigenetic regulation, and namely those that are implicated in the chromatin remodeling process.
Hinoue et al6 previously reported the presence of 4 colorectal cancer methylation subgroups by assessing 125 colorectal cancers using Illumina 27K DNA methylation arrays. In the present study, we have considerably increased the power to assess subgroups based on differential methylation by studying 216 unselected cancers using the Illumina 450K DNA methylation platform. The Illumina 450K DNA methylation platform is capable of assessing more than 10 times more CpG sites and thus can identify methylation subtypes more robustly. A major difference of our study was the identification of 2 discrete CIMP-high subtypes: CIMP-H1 and CIMP-H2. The dichotomization of these CIMP-H cancers identified a homogeneous subgroup of CIMP-H1 cancers with an average age of 75 years, striking over-representation of female sex, and BRAF mutant cancers arising in the proximal colon. The newly identified CIMP-H2 subtype encompasses more KRAS mutant cancers than CIMP-H1, and the majority of cancers in this subtype would be CIMP-low using the 5-marker CIMP panel proposed by Weisenberger et al.3 Our genome-scale analyses of both our cohort and the TCGA indicate this is not the case. Together, our CIMP-H1/H2 clusters represent 21% of our unselected cohort, and 16.3% of the TCGA cohort. Collectively, the current findings indicate that CIMP is more prevalent than previously thought, and classification of cancers using existing panels may not identify all CIMP-high colorectal cancers.
We observed a consistent increase in patient age with CIMP cluster, from 62 years in CIMP-neg cancers to 75 years in CIMP-H1 cancers. This is in contrast to the Hinoue et al6 study. The variance in our assay was mostly contained in uniquely mapping probes that were not present in the Illumina HumanMethylation27 BeadChip array used by Hinoue et al.6 Numerous studies have shown age-related methylation in different tissues9, 28, 29 and we previously identified hypermethylated loci in the colons of patients even with no history of colonic disease.9 In the present study, we detected a significant correlation between methylation and patient age. After removal of all probes that were significantly hypermethylated in normal mucosal tissue, we still observed distinct, age-linked clustering. This association was faithfully reproduced in cancers from TCGA.
The subtype with the highest degree of methylation (CIMP-H1) was strongly associated with mutations in the BRAF oncogene. BRAF mutations are a hallmark of the serrated neoplasia pathway, and indicate that these cancers probably arose in serrated precursor lesions. We previously showed that the colonoscopic incidence of sessile serrated adenomas does not differ between patients aged in their 30s and patients who are much older, whereas BRAF mutant cancers were restricted to older individuals,30 suggesting these BRAF mutant polyps may have limited malignant potential in young patients. We also reported a striking association between patient age and CIMP in sessile serrated adenomas.31 Here, we report that the vast majority of BRAF mutant cancers in both the RBWH and TCGA cohorts are CIMP-H and arise in older individuals. Collectively, these findings suggest that sessile serrated adenomas may be relatively benign in young patients. In older patients with more advanced DNA methylation changes in the colon, the risk of progression to cancer will be significantly greater. Recently, we recapitulated this process in a murine model for serrated neoplasia and showed that early onset Braf mutation leads to the temporal accumulation of DNA methylation and ultimately to malignancy.32 Additional studies are necessary to fully determine the natural history of BRAF mutant cancers, and elucidate the determinants of malignant potential to inform the development of patient-centric surveillance for young and older patients who present with sessile serrated adenomas.
Differential CpG island and shore hypermethylation were the most frequently observed methylation events in the study. Probes on the north and south CpG shelves, as well as those in the open seas, frequently were hypomethylated across most cancers. The implications of hypomethylated CpG dinucleotides outside of CpG islands are unclear. We did not observe any relationship between hypomethylation and gene transcription, however, it is possible that hypomethylation of specific regions of the genome may affect chromatin accessibility elsewhere and hence may modulate transcription in a trans-acting manner. Open sea hypomethylation was also the most frequent methylation event in CIMP-neg cancers. These are predominately conventional pathway cancers with a high degree of chromosomal instability. One hypothesis that may explain this association is that hypomethylation outside of CpG islands may predispose to copy number changes in these cancers.33, 34 Functional studies are necessary to explore the implications of shelf and open sea hypomethylation and whether this is relevant to the cancer development process for these cancers.
There were marked differences in transcriptional deregulation of key cancer-related pathways between methylation clusters. CIMP-H1 cancers activated several immune pathways, including those involved in the interferon response, inflammatory response, and complement signaling, consistent with the over-representation of CMS1 cancers in this group. This likely is owing to the higher mutational burden in these cancers, largely driven by the increased incidence of epigenetically induced microsatellite instability. MSI cancers have been associated with greater immune infiltrate and hence some of this signaling may originate in the stromal immune cells, rather than from within the tumor itself.35 In the RBWH cohort, CIMP-H2 cancers were uniquely enriched for altered bile acid metabolism, consistent with the previously described relationship between silencing of the farnesoid X bile acid receptor in KRAS mutant cancers.36 Bile acids are more concentrated in the proximal colon and metabolism is influenced by the gut microbiome.37 The increased bile acid metabolism signaling in this group of cancers may identify a subset of cancers that have arisen owing to aberrant bile acid accumulation. We did not observe such an effect in the TCGA cohort. This may be owing to the increased frequency of BRAF mutant MSI cancers in CIMP-H2 in TCGA. A better understanding of the role of bile acid signaling in KRAS mutant cancers of the proximal colon may have therapeutic implications for this cancer subgroup.
Paradoxically, despite observing less differential methylation, we observed an increase in gene silencing that correlated with promoter hypermethylation in the least methylated cancer clusters. This may indicate that promoter hypermethylation in CIMP-L1/2 and CIMP-neg cancers is more specifically selected based on a functional advantage in these cancers. Alternatively, the increased frequency of mutations in epigenetic regulators of CIMP-H1/2 cancers may result in a reduced capacity to induce gene repression at certain loci. This may be owing to the loss of a repressive histone-modifying enzyme, or mutation of locus-specific repressive transcription factors. Methylation alone may be insufficient to induce gene repression in certain instances. Instead, relevant chromatin remodeling and histone modifications, such as the addition of the repressive PRC2 mark, may be required in tandem with methylation changes to reduce gene expression. Indeed, we showed that PRC2 occupancy was most frequently related to transcriptionally repressed and methylated genes in the CIMP-neg subgroup. We also observed instances of promoter methylation that correlated with increased gene transcription. It is possible that some transcription factors preferentially bind methylated DNA,38 and that binding sites for these transcription factors become available after promoter methylation. These data may indicate that the genomic context of methylation is important for determining whether gene expression changes will occur. In TCGA, however, we were unable to discern any significant differences in the proportion of methylated and repressed genes vs all methylated genes between CIMP subtypes. This may be owing to technological differences between the array-based methods used to evaluate gene transcription in the current study and the RNA sequencing-based methods used in TCGA. Direct comparisons between the expression values derived from each of these studies is difficult and should be approached with caution.
A major novel finding of the current study was the discovery that gene body methylation may be a major driver of serrated tumorigenesis, and that this may be mediated by H3K27me3 histone marks. Gene body hypermethylation recently was correlated with increased oncogene expression.22 Here, we identified many well-characterized oncogenes, such as BCL2 and ZEB1, with methylation of their gene bodies in CIMP-H1/2 cancers, and noted a significant preference for the methylation of gene bodies of oncogenes compared with tumor-suppressor genes. We also identified Wnt pathway antagonists that are resistant to gene body methylation. In the present study, we did not identify distinct transcriptional differences in these Wnt pathway antagonists. It is possible that gene body methylation affects other aspects of the transcriptional process that were not assessed in this study, such as splicing and isoform switching. Alternatively, this gene body methylation may be a stochastic result of the overall increase in aberrant DNA methylation in these cancers.
The epigenome is regulated by proteins that interact with histones or DNA. We assessed the coding sequence of 719 epigenetic regulator genes in the TCGA data set. The chromodomain-helicase-DNA (CHD) binding protein family was a frequent mutational target in CIMP-H1 cancers. Recently, Fang et al39 showed that CHD8 operates in a transcriptional repression complex to direct methylation in the setting of BRAF mutation. In the current study we showed BRAF and CHD8 mutations were associated with CIMP-H1. Thus, these data suggest that CHD8 mutation may enhance repression complex activity in the setting of BRAF mutation, resulting in hypermethylation. Moreover, CHD8 has been associated with the CCCTC-binding factor (CTCF) protein, which is essential for promoter-enhancer looping and regional insulation. CHD8 mutations may influence CIMP by decreasing the ability of CTCF to insulate regions of the genome, and could encourage methylation spreading throughout the genome.40 Similarly, we report frequent mutations in different members of the CHD family. CHD7 was the most mutated CHD gene, and some positions in the CHD7 locus were recurrently mutated. Tahara et al41 identified mutations in CHD7 and CHD8 in 42% of CIMP1 colorectal cancers. The functional consequences of CHD7 mutations are unclear. In pancreatic duct adenocarcinoma, CHD7 expression has been shown to correlate with gemcitabine sensitivity.42 The most commonly mutated CHD gene in CIMP-L1 cancers was CHD4. Recently, Xia et al43 in 2017 proposed an oncogenic role for CHD4 through facilitating the hypermethylation of tumor-suppressor genes. In contrast, Li et al44 in 2018 showed that CHD4 mutations that promote protein degradation enhance stemness and contribute to the progression of endometrial cancers via the transforming growth factor-β signaling cascade. Indeed, we identified 3 mutations at the R975H hotspot of CHD4 that were studied by Li et al44 and a number of other mutations that were predicted to be damaging. It is not possible to conclude from our data whether these mutations promote the hypermethylation proposed by Xia et al,43 and therefore support the oncogenic role of the protein or whether the enhanced protein degradation and increased stemness proposed by Li et al44 is the predominant purpose of these mutations.
Chromatin remodeling is an essential process whereby condensed euchromatin is modified in a context-specific manner to give rise to regions of heterochromatin that can be actively transcribed. Chromatin remodeling is driven by a series of complexes that are able to enzymatically catalyze reactions that modify histone tails and, in turn, modulate the accessibility of the chromatin. In mammalian cells, 5 key chromatin-modifying complexes predominate, the CHD binding complex, the INO80 complex, the SWI/SNF complex, Imitation SWItch (ISWI) complex, and the NuRD complex.45 Here, we have examined the frequency of mutations in the SWI/SNF complex, which has been shown previously to be perturbed in various cancers. Interestingly, half of CIMP-H1 and more than 25% of CIMP-H2 and CIMP-L1 cancers harbored somatic mutations in SWI/SNF members that were predicted to be deleterious. We hypothesized that mutation of 1 member of the subunit would increase the reliance of the cancer on other otherwise redundant subunits. To test this hypothesis we used public colorectal cancer cell line dependency data in conjunction with mutational data, and identified a strong dependency conferred upon ARID1B after genetic perturbation of ARID1A. These data support the investigation of SWI/SNF inhibitors to exploit synthetic lethality presented by SWI/SNF mutations in CIMP-L1 cancers. Although we have shown associations between genomic methylation and SWI/SNF mutations, and between mutations of SWI/SNF members and synthetic lethality, functional causation is difficult to infer from our study. Collectively, these data indicate a need for further functional experiments to elucidate the role of these mutations in the carcinogenic process of CIMP-H1, CIMP-H2, and CIMP-L1 cancers, and to determine whether the potential synthetic lethalities they create can be exploited.
We leveraged the publicly available DNA methylation data from the TCGA project to validate findings in our consecutive cohort. Key findings, including relationships between CIMP subtype and age, proximal location, BRAF mutation, and KRAS mutation also were identified in an analysis of the TCGA data. In our unselected and consecutively collected series we observed a strong relationship between the BRAF mutation and CIMP-H1 and the KRAS mutation and CIMP-H2. Although BRAF was still enriched in the TCGA CIMP-H1 cancers, and KRAS among the CIMP-H2 cancers, we observed a higher proportion of BRAF mutant CIMP-H2 cancers in the TCGA cohort. The increased proportion of BRAF mutant/CIMP-H2 cancers skewed these cancers toward a preference for microsatellite instability, and the CMS1 subtype. It is notable that more than 40% of CIMP-H2 cancers in the validation cohort are KRAS mutant, and, of these, the majority are microsatellite stable and follow similar CMS patterns to that observed in our consecutive series. The discrepancies observed between the 2 cohorts may be owing to structural differences in each cohort. The mean age of patients in our study was 3.4 years older than those in the TCGA cohort. Cancers were identified most often in the distal colon of the patient, as is typical for colorectal cancers,46 however, in contrast, the TCGA consisted of a marked over-representation of proximal cancers (47.7%).
It is important to recognize the limitations of our study. First, our samples were collected in a consecutive manner in which there was sufficient sample available for DNA and RNA analyses. This excluded very small cancers and those in patients in whom surgery was not possible. This presents a slight bias, however, this is standard practice and unavoidable in studies of this nature. As technologies improve and analyses are possible on smaller amounts of tissue it will be important to replicate the key findings of this study. Moreover, because we collected fresh tissue we were not able to make any assessments of tumor purity. One alternative would have been to perform analyses on formalin-fixed, paraffin-embedded samples, in which we could perform accurate histologic assessments of the purity of the samples. Although the Illumina HM450 platform and newer platforms such as the EPIC arrays are amenable to formalin-fixed, paraffin-embedded–derived DNA, co-extraction of high-quality RNA from formalin-fixed, paraffin-embedded remains challenging. We note that the findings of this study are largely correlative and as such we cannot draw causation from our data. In depth, mechanistic follow-up evaluation is necessary to fully examine many of the key associations we have identified in the present study.
Another limitation of our study was the use of normal mucosal samples from patients with cancer. Field DNA methylation defects have been reported in colorectal cancer.47 Thus, we cannot exclude the possibility that field DNA defects impacted our analysis. In the current study, we performed all analyses on bulk tissue samples. As such, we have collected the DNA methylome and transcript profile of an aggregate of cells that includes epithelial cells, immune cells, and stromal cells. The interplay between these cell types is crucial and it is important to note that some of the expression and methylation differences observed here may be driven by any of the cells in the bulk cell sample.
Conclusions
The past decade has heralded an era in which the importance of the cancer epigenome increasingly is recognized, in which treatments targeting different epigenetic modifications are entering the clinic and improving patient outcomes. Although early strategies targeting epigenetic modifications in colorectal cancers largely have proved ineffective, it has become apparent that a comprehensive understanding of the epigenetic drivers of cancer will be crucial in the rational design of clinical trials and the development of precision medicine strategies. Here, we have identified 5 clinically and molecularly distinct subgroups based on a comprehensive assessment of a large, unselected series of colorectal cancer methylomes. We have validated these subtypes in an additional cohort of 374 cancers from TCGA. In contrast to earlier studies, we identified 2 clinically and molecularly distinct CIMP-H clusters. We observed a striking association between genomic methylation and age, which further supports the investigation of the epigenetic clock in serrated neoplasia risk. We identified an association between gene body methylation CIMP-H cancers, which may be mediated by H3K27me3 histone marks. Our interrogation of the coding regions of epigenetic regulatory genes shows that they frequently are mutated in colorectal cancers and this may be partially influenced by the degree of genomic methylation. Our analyses have identified potentially druggable vulnerabilities in cancers of different methylation subtypes. Inhibitors targeting synthetic lethalities, such as SWI/SNF component inhibitors for those with ARID mutations, should be evaluated because these agents may be clinically beneficial to certain patient subsets.
Methods
Patient Samples
Colorectal cancer (N = 216) and matched normal (N = 32) samples were obtained from patients undergoing surgery at the Royal Brisbane and Women’s Hospital in Brisbane, Australia, in a consecutive manner between 2009 and 2012. Tissue was snap-frozen in liquid nitrogen to preserve sample integrity. Written informed consent was obtained from each patient. The study protocol was approved by the Royal Brisbane and Women’s Hospital and QIMR Berghofer Medical Research Institute Research Ethics Committees. TCGA colon adenocarcinoma exome and methylation data (N = 278) were used for independent validation.16
DNA and Messenger RNA Extractions
DNA and messenger RNA (mRNA) were extracted simultaneously from approximately 30 mg of homogenized tissue using the AllPrep DNA/RNA Kit (QIAGEN, Hilden, Germany) in accordance with the manufacturer’s protocols. Double-stranded DNA concentration was assessed using the PicoGreen quantitation assay (Thermofisher Scientific, Waltham, MA). mRNA quality was measured using the Bioanalyzer 2100 platform (Agilent, Santa Clara, CA). Microarray analysis was performed on samples with a RNA integrity number greater than 7.
Molecular Characterization of Cancer Samples
Cancer sample DNA was analyzed for the BRAF V600E mutation using allelic discrimination as previously reported.48 In addition, we assayed mutations in KRAS codons 12 and 13, and TP53 exons 4 to 8 using previously reported methods.49, 50 We assessed CIMP status by methylation-specific polymerase chain reaction using the 5-marker panel (CACNA1G, IGF2, NEUROG1, RUNX1, and SOCS1) proposed by Weisenberger et al.3 Samples were considered CIMP-high if 3 or more markers were methylated, CIMP-low if 1 or 2 markers were methylated, and CIMP-negative if no markers were methylated. MSI was assessed using the criteria of Nagasaka et al51 in which instability in 1 or more mononucleotide markers, and 1 or more additional nonmononucleotide markers, using the marker set reported by Boland et al,52 was indicative of MSI, the remainder being microsatellite stable. LINE1 methylation was assessed using pyrosequencing as per Irahara et al.53 CIMP-high cancers that were both KRAS and BRAF wild-type at hotspot codons were Sanger sequenced for BRAF exons 11 and 15 (exon 11, forward: 5’-TTCCTGTATCCCTCTCAGGCA-3’, reverse: 5’-AAAGGGGAATTCCTCCAGGTT-3’; exon 15, forward 5’-GGAAAGCATCTCACCTCATCCT-3’, reverse 5’-TAGAAAGTCATTGAAGGTCTCAACT-3’), KRAS codon 61 (forward: 5’-TCCAGACTGTGTTTCTCCCTTC-3’, reverse: 5’-TGAGATGGTGTCACTTTAACAGT-3’), and EGFR exon 18 (forward: 5’-ATGTCTGGCACTGCTTTCCA-3’, reverse: 5’-ATTGACCTTGCCATGGGGTG-3’).
DNA Methylation Microarray
Genome-scale DNA methylation was measured using the HumanMethylation450 BeadChip array (Illumina). The BeadChip array interrogates cytosine methylation at more than 480,000 CpG sites. A total of 500 ng DNA was bisulfite-converted using the EZ-96 DNA Methylation Kit (Zymo Research, Irvine, CA) per the manufacturer’s protocol. Whole-genome amplification and enzymatic fragmentation was performed on post-treatment DNA, which subsequently was hybridized to the array at 48°C for 16 hours. Arrays were scanned using the iScan System (Illumina).
Gene Expression Microarray
Gene expression levels for more than 47,000 transcripts were measured for all samples using the HumanHT-12 v3 Expression BeadChip array (Illumina). Total mRNA (500 ng) was reverse-transcribed, amplified, and biotinylated using the TotalPrep-96 RNA Amplification Kit (Illumina). The labeled complementary RNA (750 ng) was hybridized to the array followed by washing, blocking, and staining with streptavidin-Cy3. Arrays were scanned on the iScan System and the data were extracted using GenomeStudio Software (Illumina).
Data Analysis
Methylation microarray data were checked for quality against parameters provided by Illumina using the GenomeStudio Software package. IDAT files were read into the R environment using Limma.54 We used subset-within-array normalization to correct for biases resulting from type 1 and type 2 probes on the array. We used the BEclear R package to assess for probe-level batch effects and excluded probes that were significantly batch-affected (n = 1072) from downstream analysis. We filtered probes that had a detection of P > .05 in more than 50% of samples, as well as probes that were on the X or Y chromosome, where the CpG site was within 10 bp of a single-nucleotide polymorphism, or where a probe mapped to the genome ambiguously. At the conclusion of filtering, 377,612 probes remained and were used for subsequent analyses.
The RPMM clustering method55 was used for unsupervised clustering. To capture cancer-specific methylation we followed the methods used based on TCGA.56 DNA methylation drift with age has been characterized in a number of different normal and cancerous tissues.10 To limit confounding from methylation that occurs through age, probes with a mean β value greater than 0.3 in normal samples were excluded from clustering analysis. A total of 144,542 probes were unmethylated (mean β value, <0.3) in normal mucosa, of these the 5000 probes with the greatest variance in tumor samples were selected for clustering. The RPMM clustering method is particularly suited to analysis of methylation data generated from the HumanMethylation450 array because output β values are between 0 and 1, and can be modeled using a β-like distribution.55 We accessed level 1 DNA methylation data from the TCGA project and performed an identical analysis as mentioned earlier for validation.
For motif analysis, the CentriMo tool was used.21 CentriMo identifies over-represented motifs within sequences, correlating these with known DNA protein-binding motifs.21 β values were transformed to M values using the following formula: M = log2 (β/[1 - β]). For differential methylation analysis vs the subset of normal mucosal samples, a probe was considered to be differentially methylated in a comparison if the Benjamini–Hochberg57 adjusted P value for the comparison was less than 0.05 and had an average absolute Δβ ≥ 0.2 vs normal mucosal samples. For examination of methylation in oncogenes and tumor-suppressor genes we consulted the NCG6.0 cancer gene database.23 For these analyses we included only cancer genes that were annotated in NCG6.0 without ambiguity (were not annotated as both tumor-suppressor genes and oncogenes) and those that we probed on the array.
Expression data were preprocessed and normalized using quantile normalization with the Limma R package. For between-group comparisons the empiric Bayes function was used, and adjusted for multiple testing using the Benjamini–Hochberg57 method to control for FDR and avoid type 1 errors. We examined gene expression in the TCGA by accessing level 3 expression data in Fragments Per Kilobase of transcript per Million reads (FPKM) format from Genome Data Commons.58 We used Limma to perform a voom transformation to correct for heteroscedasticity and examine differential expression against normal colonic mucosal samples using the same methods as used in the consecutive series. We considered 0.05 to be the FDR threshold for significance. For integrated expression and methylation data analysis, genes were considered to be methylated if 1 probe within 2 kb upstream of the gene transcription start site was methylated differentially by FDR and had an average Δβ ≥ 0.2 at that site. If a gene met this criterion, and had a significant FDR corrected P value for the cancer vs normal expression value, it was predicted to be influenced by methylation. Single-sample gene-set enrichment analysis was used for between-groups comparisons of transcriptomes.18 We used the CIBERSORT algorithm to compute the relative proportion of stromal cells within each subtype.17
The CMS classifier package was used to classify cancers into CMS as previously reported.8
To examine the mutational frequency of epigenetic regulators, level 3 somatic variant data were downloaded from the Genome Data Commons portal. Silent variants were discarded and variants in epigenetic regulator genes present in the EpiFactors database extracted for further analyses. We assessed the potential pathogenicity of missense mutations using the PolyPhen2 tool.59 PolyPhen2 predicts functional effects of missense mutations by examining how evolutionarily conserved the affected residue is, and computes the likelihood that the event will induce a structural change. Only variants that were predicted to be probably or possibly damaging were retained. Variants predicted to be benign were not included as part of these analyses
PRC2 and Methylation Overlap Analysis
Polycomb occupancy was inferred from SUZ12 Chromatin Immunoprecipitation (ChIP) sequencing data from hESC1 cells analyzed as part of the Encyclopedia of DNA Elements (ENCODE) ENCODE consortium.60 SUZ12 was chosen as a surrogate for PRC2 occupancy because previous studies have indicated that it is an essential subunit of the PRC2 complex.20, 61 The overlap function within BedTools62 was used to overlap differentially methylated probes within each cluster vs normal with regions where SUZ12 was bound in hESC1 cells, producing a list of regions where methylation and PRC2 occupancy co-occurred.
Synthetic Lethality Analysis
Cell line dependency data from Meyers et al27 was correlated with colorectal cancer cell line mutation data.26 Synthetic lethal relationships were inferred if a high-impact mutation (truncating mutations or those in splice sites) occurred in 1 subunit of a molecular complex, and the cell line had relatively higher dependence values on other subunits when compared with cell lines that lacked a mutation. Cell lines were grouped as having a mutation in a specific gene and those not having a mutation, and a Student t test was performed on dependence values in every other subunit within the complex.
Statistical Analysis
For statistical analyses a combination of different types of software were used, including R and GraphPad Prism 7 (GraphPad Software, San Diego, CA). The Fisher exact test was used for hypothesis testing on 2 × 2 contingencies. The Pearson chi-squared test was used to compare contingencies larger than 2 × 2. The Student t test or the Wilcoxon rank-sum test was used to compare continuous variables where appropriate. One-way analysis of variance was used for continuous variable comparisons with more than 2 groups.
Acknowledgments
The authors are thankful for the insightful comments offered by the reviewers and for their contribution in greatly improving the manuscript.
The microarray data have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-8148 (expression) and E-MTAB-7036 (methylation).
Footnotes
Author contributions Lochlan Fennell performed bioinformatic and statistical analyses on the data, was involved in the conceptualization aspects of the study, and prepared the manuscript; Troy Dumenil performed molecular and bioinformatic analyses and revised the manuscript for content; Gunter Hartel was involved in the statistical and bioinformatic analysis of the data; Katia Nones was involved in the bioinformatic analysis of methylation data and revised the manuscript for content; Catherine Bond performed molecular analyses and revised the manuscript for content; Diane McKeone was involved in molecular analyses; Lisa Bowdler processed the microarrays; Grant Montgomery processed the microarrays; Leesa Wockner was involved in bioinformatic analyses; Kerenaftali Klein was involved in bioinformatic analyses; Ann-Marie Patch was involved in bioinformatic analyses of The Cancer Genome Atlas exome data; Stephen Kazakoff was involved in bioinformatic analyses of The Cancer Genome Atlas exome data; John Pearson was involved in bioinformatic analyses of The Cancer Genome Atlas exome data; Nicola Waddell was involved in bioinformatic analyses of The Cancer Genome Atlas exome data; Pratyaksha Wirapati performed consensus molecular subtype analysis; Paul Lochhead performed Long Interspersed Nuclear Element-1 methylation assays and analysis; Yu Imamura performed Long Interspersed Nuclear Element-1 methylation assays and analysis; Shuji Ogino performed Long Interspersed Nuclear Element-1 methylation assays and analysis and provided supervision for this aspect of the study; Renfu Shao supervised the study and was involved in the conceptualization aspects of the study; Sabine Tejpar performed Consensus Molecular Subtype analysis and provided supervision for this aspect of the study; Barbara Leggett supervised the study, was involved in conceptualization aspects of the study, revised the manuscript for content, and secured funding for the study; Cheng Liu performed molecular analyses and revised the manuscript for content; Jennifer Borowsky performed molecular analyses and revised the manuscript for content; Isabell Hoffmann performed bioinformatic analysis and revised the manuscript for content; and Vicki Whitehall conceptualized the study, performed statistical analyses, revised the manuscript for content, secured funding for the study, and provided overarching supervision of the study.
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported through funding from the National Health and Medical Research Council (1050455 and 1063105), the US National Institutes of Health (R01 CA151933 and R35 CA197735), and Pathology Queensland. Also supported by a Senior Research Fellowship from the Gastroenterological Society of Australia (V.W.); and by a Research Training Program Living Scholarship from the Australia Government and a Top-Up award from QIMR Berghofer and Australian Rotary Health (L.F.).
Supplementary Material
References
- 1.Fearon E.R., Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 2.Leggett B., Whitehall V. Role of the serrated pathway in colorectal cancer pathogenesis. Gastroenterology. 2010;138:2088–2100. doi: 10.1053/j.gastro.2009.12.066. [DOI] [PubMed] [Google Scholar]
- 3.Weisenberger D.J., Siegmund K.D., Campan M., Young J., Long T.I., Faasse M.A., Kang G.H., Widschwendter M., Weener D., Buchanan D., Koh H., Simms L., Barker M., Leggett B., Levine J., Kim M., French A.J., Thibodeau S.N., Jass J., Haile R., Laird P.W. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 4.Guan R.J., Fu Y., Holt P.R., Pardee A.B. Association of K-ras mutations with p16 methylation in human colon cancer. Gastroenterology. 1999;116:1063–1071. doi: 10.1016/s0016-5085(99)70009-0. [DOI] [PubMed] [Google Scholar]
- 5.Herman J.G., Umar A., Polyak K., Graff J.R., Ahuja N., Issa J.-P.J., Markowitz S., Willson J.K.V., Hamilton S.R., Kinzler K.W., Kane M.F., Kolodner R.D., Vogelstein B., Kunkel T.A., Baylin S.B. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinoue T., Weisenberger D.J., Lange C.P.E., Shen H., Byun H.-M., Van Den Berg D., Malik S., Pan F., Noushmehr H., van Dijk C.M., Tollenaar R.A.E.M., Laird P.W. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2012;22:271–282. doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogino S., Kawasaki T., Kirkner G.J., Loda M., Fuchs C.S. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8:582–588. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guinney J., Dienstmann R., Wang X., de Reyniès A., Schlicker A., Soneson C., Marisa L., Roepman P., Nyamundanda G., Angelino P., Bot B.M., Morris J.S., Simon I.M., Gerster S., Fessler E., De Sousa E Melo F., Missiaglia E., Ramay H., Barras D., Homicsko K., Maru D., Manyam G.C., Broom B., Boige V., Perez-Villamil B., Laderas T., Salazar R., Gray J.W., Hanahan D., Tabernero J., Bernards R., Friend S.H., Laurent-Puig P., Medema J.P., Sadanandam A., Wessels L., Delorenzi M., Kopetz S., Vermeulen L., Tejpar S. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Worthley D.L., Whitehall V.L.J., Buttenshaw R.L., Irahara N., Greco S.A., Ramsnes I., Mallitt K.A., Le Leu R.K., Winter J., Hu Y., Ogino S., Young G.P., Leggett B.A. DNA methylation within the normal colorectal mucosa is associated with pathway-specific predisposition to cancer. Oncogene. 2010;29:1653–1662. doi: 10.1038/onc.2009.449. [DOI] [PubMed] [Google Scholar]
- 10.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bettington M., Walker N., Clouston A., Brown I., Leggett B., Whitehall V. The serrated pathway to colorectal carcinoma: current concepts and challenges. Histopathology. 2013;62:367–386. doi: 10.1111/his.12055. [DOI] [PubMed] [Google Scholar]
- 12.Bettington M., Walker N., Rosty C., Brown I., Clouston A., McKeone D., Pearson S.-A., Leggett B., Whitehall V. Clinicopathological and molecular features of sessile serrated adenomas with dysplasia or carcinoma. Gut. 2017;66:97–106. doi: 10.1136/gutjnl-2015-310456. [DOI] [PubMed] [Google Scholar]
- 13.Ford E.E., Grimmer M.R., Stolzenburg S., Bogdanovic O., de Mendoza A., Farnham P.J., Blancafort P., Lister R. Frequent lack of repressive capacity of promoter DNA methylation identified through genome-wide epigenomic manipulation. bioRxiv. 2017 Available at: https://www.biorxiv.org/content/10.1101/170506v3. [Google Scholar]
- 14.Allis C.D., Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y., Thomas A., Lau C., Rajan A., Zhu Y., Killian J.K., Petrini I., Pham T., Morrow B., Zhong X., Meltzer P.S., Giaccone G. Mutations of epigenetic regulatory genes are common in thymic carcinomas. Sci Rep. 2014;4:7336. doi: 10.1038/srep07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., Hoang C.D., Diehn M., Alizadeh A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov Jill P., Tamayo P. The molecular signatures database hallmark gene set collection. Cell Systems. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nayak V., Xu C., Min J. Composition, recruitment and regulation of the PRC2 complex. Nucleus. 2011;2:277–282. doi: 10.4161/nucl.2.4.16266. [DOI] [PubMed] [Google Scholar]
- 21.Bailey T.L., Machanick P. Inferring direct DNA binding from ChIP-seq. Nucleic Acid Res. 2012;40 doi: 10.1093/nar/gks433. e128-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X., Han H., De Carvalho D.D., Lay Fides D., Jones P.A., Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Repana D., Nulsen J., Dressler L., Bortolomeazzi M., Kuppili Venkata S., Tourna A., Yakovleva A., Palmieri T., Ciccarelli F.D. The Network of Cancer Genes (NCG): a comprehensive catalogue of known and candidate cancer genes from cancer sequencing screens. bioRxiv. 2018:389858. doi: 10.1186/s13059-018-1612-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Software tool. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4480630/.
- 25.St. Pierre R., Kadoch C. Mammalian SWI/SNF complexes in cancer: emerging therapeutic opportunities. Curr Opin Genet Dev. 2017;42:56–67. doi: 10.1016/j.gde.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mouradov D., Sloggett C., Jorissen R.N., Love C.G., Li S., Burgess A.W., Arango D., Strausberg R.L., Buchanan D., Wormald S., Connor L., Wilding J.L., Bicknell D., Tomlinson I.P.M., Bodmer W.F., Mariadason J.M., Sieber O.M. Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res. 2014;74:3238. doi: 10.1158/0008-5472.CAN-14-0013. [DOI] [PubMed] [Google Scholar]
- 27.Meyers R.M., Bryan J.G., McFarland J.M., Weir B.A., Sizemore A.E., Xu H., Dharia N.V., Montgomery P.G., Cowley G.S., Pantel S., Goodale A., Lee Y., Ali L.D., Jiang G., Lubonja R., Harrington W.F., Strickland M., Wu T., Hawes D.C., Zhivich V.A., Wyatt M.R., Kalani Z., Chang J.J., Okamoto M., Stegmaier K., Golub T.R., Boehm J.S., Vazquez F., Root D.E., Hahn W.C., Tsherniak A. Computational correction of copy number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nat Genet. 2017;49:1779. doi: 10.1038/ng.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson A.A., Akman K., Calimport S.R.G., Wuttke D., Stolzing A., de Magalhães J.P. The role of DNA methylation in aging, rejuvenation, and age-related disease. Rejuvenation Res. 2012;15:483–494. doi: 10.1089/rej.2012.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steegenga W.T., Boekschoten M.V., Lute C., Hooiveld G.J., de Groot P.J., Morris T.J., Teschendorff A.E., Butcher L.M., Beck S., Müller M. Genome-wide age-related changes in DNA methylation and gene expression in human PBMCs. Age. 2014;36:9648. doi: 10.1007/s11357-014-9648-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bettington M., Brown I., Rosty C., Walker N., Liu C., Croese J., Rahman T., Pearson S.-A., McKeone D., Leggett B., Whitehall V. Sessile serrated adenomas in young patients may have limited risk of malignant progression. J Clin Gastroenterol. 2019;53:e113–e116. doi: 10.1097/MCG.0000000000001014. [DOI] [PubMed] [Google Scholar]
- 31.Liu C., Bettington M.L., Walker N.I., Dwine J., Hartel G.F., Leggett B.A., Whitehall V.L.J. CpG island methylation in sessile serrated adenomas increases with age, indicating lower risk of malignancy in young patients. Gastroenterology. 2018;155:1362–1365.e2. doi: 10.1053/j.gastro.2018.07.012. [DOI] [PubMed] [Google Scholar]
- 32.Bond C.E., Liu C., Kawamata F., McKeone D.M., Fernando W., Jamieson S., Pearson S.-A., Kane A., Woods S.L., Lannagan T.R.M., Somashekar R., Lee Y., Dumenil T., Hartel G., Spring K.J., Borowsky J., Fennell L., Bettington M., Lee J., Worthley D.L., Leggett B.A., Whitehall V.L.J. Oncogenic BRAF mutation induces DNA methylation changes in a murine model for human serrated colorectal neoplasia. Epigenetics. 2018;13:40–48. doi: 10.1080/15592294.2017.1411446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez J., Frigola J., Vendrell E., Risques R.A., Fraga M.F., Morales C., Moreno V., Esteller M., Capella G., Ribas M., Peinado M.A. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res. 2006;66:8462–9468. doi: 10.1158/0008-5472.CAN-06-0293. [DOI] [PubMed] [Google Scholar]
- 34.Sheaffer K.L., Elliott E.N., Kaestner K.H. DNA hypomethylation contributes to genomic instability and intestinal cancer initiation. Cancer Prev Res (Phila) 2016;9:534–546. doi: 10.1158/1940-6207.CAPR-15-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao Y., Freeman G.J. The microsatellite instable (MSI) subset of colorectal cancer is a particularly good candidate for checkpoint blockade immunotherapy. Cancer Discov. 2015;5:16–18. doi: 10.1158/2159-8290.CD-14-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bailey A.M., Zhan L., Maru D., Shureiqi I., Pickering C.R., Kiriakova G., Izzo J., He N., Wei C., Baladandayuthapani V., Liang H., Kopetz S., Powis G., Guo G.L. FXR silencing in human colon cancer by DNA methylation and KRAS signaling. Am J Physiol Gastrointest Liver Physiol. 2014;306:G48–G58. doi: 10.1152/ajpgi.00234.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee M.S., Menter D.G., Kopetz S. Right versus left colon cancer biology: integrating the consensus molecular subtypes. J Natl Compr Canc Netw. 2017;15:411–419. doi: 10.6004/jnccn.2017.0038. [DOI] [PubMed] [Google Scholar]
- 38.Chatterjee R., Vinson C. CpG methylation recruits sequence specific transcription factors essential for tissue specific gene expression. Biochim Biophys Acta. 2012;1819:763–770. doi: 10.1016/j.bbagrm.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fang M., Ou J., Hutchinson L., Green M.R. The BRAF oncoprotein functions through the transcriptional repressor MAFG to mediate the CpG island methylator phenotype. Mol Cell. 2014;55:904–915. doi: 10.1016/j.molcel.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kemp C.J., Moore J.M., Moser R., Bernard B., Teater M., Smith L.E., Rabaia N., Gurley K.E., Guinney J., Busch S.E., Shaknovich R., Lobanenkov V.V., Liggitt D., Shmulevich I., Melnick A., Filippova G.N. CTCF haploinsufficiency destabilizes DNA methylation and predisposes to cancer. Cell Rep. 2014;7:1020–1029. doi: 10.1016/j.celrep.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tahara T., Yamamoto E., Madireddi P., Suzuki H., Maruyama R., Chung W., Garriga J., Jelinek J., Yamano H.-O., Sugai T., Kondo Y., Toyota M., Issa J.-P.J., Estécio M.R.H. Colorectal carcinomas with CpG island methylator phenotype 1 frequently contain mutations in chromatin regulators. Gastroenterology. 2014;146:530–538.e5. doi: 10.1053/j.gastro.2013.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colbert L.E., Petrova A.V., Fisher S.B., Pantazides B.G., Madden M.Z., Hardy C.W., Warren M.D., Pan Y., Nagaraju G.P., Liu E.A., Saka B., Hall W.A., Shelton J.W., Gandhi K., Pauly R., Kowalski J., Kooby D.A., El-Rayes B.F., Staley C.A., 3rd, Adsay N.V., Curran W.J., Jr., Landry J.C., Maithel S.K., Yu D.S. CHD7 expression predicts survival outcomes in patients with resected pancreatic cancer. Cancer Res. 2014;74:2677–2687. doi: 10.1158/0008-5472.CAN-13-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xia L., Huang W., Bellani M., Seidman M.M., Wu K., Fan D., Nie Y., Cai Y., Zhang Y.W., Yu L.-R., Li H., Zahnow C.A., Xie W., Chiu Yen R.-W., Rassool F.V., Baylin S.B. CHD4 has oncogenic functions in initiating and maintaining epigenetic suppression of multiple tumor suppressor genes. Cancer Cell. 2017;31:653–668.e7. doi: 10.1016/j.ccell.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y., Liu Q., McGrail D.J., Dai H., Li K., Lin S.-Y. CHD4 mutations promote endometrial cancer stemness by activating TGF-beta signaling. Am J Cancer Res. 2018;8:903–914. [PMC free article] [PubMed] [Google Scholar]
- 45.Langst G., Manelyte L. Chromatin remodelers: from function to dysfunction. Genes (Basel) 2015;6:299–324. doi: 10.3390/genes6020299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomez D., Dalal Z., Raw E., Roberts C., Lyndon P.J. Anatomical distribution of colorectal cancer over a 10 year period in a district general hospital: is there a true “rightward shift”? Postgrad Med J. 2004;80:667. doi: 10.1136/pgmj.2004.020198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bernstein C., Nfonsam V., Prasad A.R., Bernstein H. Epigenetic field defects in progression to cancer. World J Gastrointest Oncol. 2013;5:43–49. doi: 10.4251/wjgo.v5.i3.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benlloch S., Payá A., Alenda C., Bessa X., Andreu M., Jover R., Castells A., Llor X., Aranda F.I., Massutí B. Detection of BRAF V600E mutation in colorectal cancer: comparison of automatic sequencing and real-time chemistry methodology. J Mol Diagn. 2006;8:540–543. doi: 10.2353/jmoldx.2006.060070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whitehall V.L.J., Rickman C., Bond C.E., Ramsnes I., Greco S.A., Umapathy A., McKeone D., Faleiro R.J., Buttenshaw R.L., Worthley D.L., Nayler S., Zhao Z.Z., Montgomery G.W., Mallitt K.-A., Jass J.R., Matsubara N., Notohara K., Ishii T., Leggett B.A. Oncogenic PIK3CA mutations in colorectal cancers and polyps. Int J Cancer. 2012;131:813–820. doi: 10.1002/ijc.26440. [DOI] [PubMed] [Google Scholar]
- 50.Bond C.E., Umapathy A., Ramsnes I., Greco S.A., Zhen Zhao Z., Mallitt K.-A., Buttenshaw R.L., Montgomery G.W., Leggett B.A., Whitehall V.L.J. p53 mutation is common in microsatellite stable, BRAF mutant colorectal cancers. Int J Cancer. 2012;130:1567–1576. doi: 10.1002/ijc.26175. [DOI] [PubMed] [Google Scholar]
- 51.Nagasaka T., Koi M., Kloor M., Gebert J., Vilkin A., Nishida N., Shin S.K., Sasamoto H., Tanaka N., Matsubara N., Boland C.R., Goel A. Mutations in both KRAS and BRAF may contribute to the methylator phenotype in colon cancer. Gastroenterology. 2008;134:1950–1960.e1. doi: 10.1053/j.gastro.2008.02.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boland C.R., Thibodeau S.N., Hamilton S.R., Sidransky D., Eshleman J.R., Burt R.W., Meltzer S.J., Rodriguez-Bigas M.A., Fodde R., Ranzani G.N., Srivastava S. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 53.Irahara N., Nosho K., Baba Y., Shima K., Lindeman N.I., Hazra A., Schernhammer E.S., Hunter D.J., Fuchs C.S., Ogino S. Precision of pyrosequencing assay to measure LINE-1 methylation in colon cancer, normal colonic mucosa, and peripheral blood cells. J Mol Diagn. 2010;12:177–183. doi: 10.2353/jmoldx.2010.090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acid Res. 2015;43 doi: 10.1093/nar/gkv007. e47-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Houseman E.A., Christensen B.C., Yeh R.-F., Marsit C.J., Karagas M.R., Wrensch M., Nelson H.H., Wiemels J., Zheng S., Wiencke J.K., Kelsey K.T. Model-based clustering of DNA methylation array data: a recursive-partitioning algorithm for high-dimensional data arising as a mixture of beta distributions. BMC Bioinformatics. 2008;9:365. doi: 10.1186/1471-2105-9-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.The Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B (Method) 1995;57:289–300. [Google Scholar]
- 58.Jensen M.A., Ferretti V., Grossman R.L., Staudt L.M. The NCI Genomic Data Commons as an engine for precision medicine. Blood. 2017;130:453–459. doi: 10.1182/blood-2017-03-735654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adzhubei I., Jordan D.M., Sunyaev S.R. Predicting functional effect of human missense mutations using polyphen-2. Curr Protoc Hum Genet. 2014;76:7.20–7.41. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.The Encode Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Kruijsbergen I., Hontelez S., Veenstra G.J.C. Recruiting polycomb to chromatin. Int J Biochem Cell Biol. 2015;67:177–187. doi: 10.1016/j.biocel.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.