Abstract
Aloe vera is a multifunctional plant that has gained acceptance as an excellent home remedy source in Asia and the world. The present study was intended to evaluate the phytochemical contents and in vitro antioxidant, antimicrobial, antileishmanial, and protein kinase inhibition activities in different fractions of A. vera leaf. Methanolic extract of A. vera leaves was fractionated using column chromatography and ten fractions (AV1-AV10) were obtained. Phenolics composition, antioxidant, antimicrobial, antileishmanial, and protein kinase inhibition activities were evaluated using standard protocols. Well-known compounds of A. vera were used for in silico study against enzymes involved in brine shrimp and antileishmanial and hyphae formation inhibition assay on the basis of results. Five fractions (AV3 to AV7) possess potential total phenolics and flavonoids contents along with significant biological activities. AV4 fraction exhibited the highest total phenolics content 332.4 ± 32.6μg GAE/mg and total antioxidant activity 150.4 ± 25.815μg AAE/mg determined by phosphomolybdenum complex assay. Fraction AV6 showed 95% antileishmanial effect as well as the lowest LD50 value of 0.5305μg/mL in brine shrimp lethality assay. The Protein Kinase inhibition potential in A. vera leaves was determined for the first time and three fractions AV1, AV6, and AV7 depicted activity with the highest zone of inhibition up to 21±0.5mm (AV7). Docking analysis showed that A. vera contains anthraquinones, anthrones, chromones, and polysaccharides responsible for synergistic cytotoxic, antileishmanial, antibacterial, and antioxidant potential of this plant. Therefore, with more studies, A. vera could probably have the potential to be used for drug development against leishmaniasis.
1. Introduction
The history of development of medicinal components is based on the fact that over the centuries the natural products such as taxol, artemisinin, and morphine were used to cure a number of diseases [1]. The natural products are more acceptable than purely synthetic products because they match biological intermediates and endogenous substances and have suitability in active transport mechanisms [2]. Nowadays, plants based secondary metabolites extracted from crude extract or fractions are good source of diverse chemical structures that show potent biological profile and pharmacological activities [3, 4].
Thousands of polyphenolics compounds such as phenolics and flavonoids have been discovered so far. These compounds retain different antioxidant, antibacterial, anticancerous, antiviral, and anti-inflammatory activities and antiatherosclerotic properties [5, 6]. The pharmacological and physiological potentials of phenolic compounds depend on their free radical scavenging and antioxidant activities and properties to maintain the activity of enzymes responsible for detoxification [7]. A very unique health effect of flavonoids is that they are cardioprotective due to inhibition of LDL oxidation [8, 9]. The ability of flavonoids to transfer stable free radicals and metal catalyst chelation provides protection to the biological systems [10], antioxidant enzymes activation [11], alpha-tocopherol radical reduction [12], and inhibition of enzyme oxidases [13].
Globally, infectious diseases are considered major cause of mortality and morbidity each year. The infectious disorders are caused by organisms such as bacteria, viruses, fungi, or parasites [14, 15]. Among bacterial infections certain bacteria are well known such as S. aureus, E. coli, and Pseudomonas. Recently most of bacterial strains become resistant to existing antibacterial compounds and pose major problem for treatment [16]. Candida albicans is considered the most commonly occurring fungus responsible for infections [17]. In case of protozole infections leishmaniasis and malaria are main health problems around the world particularly in the developing countries [18].
Therefore, it is essential to screen various plants for their antimicrobial, antiprotozoal potential [14, 19]. Antimicrobial assays include agar well diffusion, disc diffusion, and broth dilution methods. Infections related to cancer also need attention due to emergence of resistance. Cytotoxicity and enzyme inhibitory studies can play an important role in drug development against cancer [20]. The brine shrimp lethality bioassay is important and effective assay to screen cytotoxic potential while various enzymes inhibition assays, for example, protein kinase inhibition, aromatase inhibition, and iNOS inhibition assays, are used for anticancer studies [21]. Protein kinases show a huge class of enzymes that play an important role in regulation of complex molecular mechanisms that control various functions of the cells, including survival, proliferation, and apoptosis. Many pathologic states appear due to abnormal protein kinase functions including cancer, inflammatory and autoimmune disorders, and cardiac diseases. Now the protein kinases are one of the major therapeutic targets [22]. For the treatment of cancer Protein kinase inhibitors are a well-known family of clinically effective drugs, especially in treating cancer [23].
The genus Aloe belonging to family Liliaceae exists almost all over the world and is extensively disseminated in the African and the eastern European continents. The genus Aloe has more than 400 species including globally traded A. vera, Aloe ferox, and Aloe arborescens [24]. Aloe genus is reported for many medicinal uses such as its use in treating constipation, gastrointestinal disorders, and immune system deficiencies. Aloe vera also showed pharmacological activities including antioxidant, antimicrobial, antitumor, hypoglycemic, hypolipidemic, and antidiabetic ones [25]. These properties are mainly contributed by inner gel of the leaves and presence of more than 200 different biologically active substances [24]. The medicinal and pharmacological potential of A. vera revealed that it is quite auspicious as a versatile therapeutic plant and should be further investigated. In this context, the present study was designed to evaluate total phenolics content, in vitro antioxidant and antimicrobial properties, cytotoxicity, and protein kinase inhibition activity in various fractions of Aloe vera leaf extract. Some of the well-known secondary metabolites of A. vera plant belonging to anthraquinone, resins, anthracenes, and polysaccharide class were used for in silico study against enzymes involved in hyphae formation of Streptomyces, hatching of Artemia salina larvae, and growth of leishmanial parasite.
2. Materials and Methods
2.1. Sampling
The plant samples were collected from Haripur, which lies between 34°-34′ and 34°–16′ North latitude and 77°-33′ and 73°-21′ East longitude, Khyber Pakhtunkhwa, Pakistan, September 2014, and is identified by taxonomist at Quaid-i-Azam University Islamabad. Plant samples were collected from private land by the verbal permission of the land owner to conduct this study. About 13 kg, fresh leaves were cleaned with tap water followed by distilled water and then chopped in the grinder and shade-dried with continuous agitation for 2 weeks.
2.2. Extraction
Approximately, 388g dried plant material was macerated in 16 liters of methanol and kept for ten days at room temperature with continuous stirring and shaking at least 4 to 5 times daily. The primary separation was done with muslin cloth followed by filtration using Whatman filter paper. Filtrate was dried and evaporated under reduced pressure at 40°C in a rotary evaporator, and crude extract (20g) was stored for further analysis.
2.3. Fractionation by Column Chromatography
The crude extract was fractionated by using column chromatography [26]. After TLC optimization methanol and chloroform were selected for column chromatography. Concentrated solution of crude extract was prepared in selected solvents and loaded on blank silica gel (1g x 1.5g) to prepare sample for column chromatography.
Silica gel (60, 70-230 mesh, MERCK) was used as stationary phase and the variation of solvent combinations of increasing polarity was used as mobile phase. The bottom of the glass column was stocked with cotton pad and slurry (600 g of silica gel and 600 mL of chloroform) was carefully poured into the column with continuous tapping. Then previously prepared dry power of extract and silica gel was poured on the top of the column and column packing was completed by putting blank silica on the top. Solvent was poured from the top and opened the tap of the column. Elution of the extract was done with 400 mL of each solvent combination (methanol : Chloroform), that is, 0:100, 2:98, 3:97, 5:95, 7:93, 10:90, 13:87, 16:84, 19:81, 23:77, 29:71, 35:65, 40:60, 50:50, 60:40, and 70:30 v/v. In total 55 elution fractions were collected in 100 mL glass bottles. The fractions present in each bottle were studied by TLC method to mix the closely related fractions on the basis of spots observed in UV light. Fractions that showed the same spots were pooled together and total ten fractions were obtained, that is, AV1, AV2, AV3, AV4, AV5, AV6, AV7, AV8, AV9, and AV10.
2.4. Estimation of Total Phenolics
Total phenolics content (TPC) in different fractions of A. vera leaf (AV1-AV10) was determined following the method of [27] with slight modification. In brief, 20 μL of each fraction was taken in 96-well plate followed by addition of 90 μL diluted Folin-Ciocalteu reagent in each well. The mixture was incubated for 5 min at room temperature and 90 μL of 6% sodium carbonate solution was added and again incubated for 1 hour at room temperature. The absorbance was measured at 630 nm wavelength on microplate reader. Final value of total phenolics content was expressed as Gallic acid equivalent (GAE) and data were expressed as ± SD for triplicate analysis.
2.5. Estimation of Total Flavonoids
Total flavonoids content (TFC) was estimated by ammonium chloride calorimetric method as described by [28]. Briefly, 20 μL of each fraction in triplicate was taken in 96-well plate followed by addition of 10 μL of potassium acetate. Then, 10 μL of aluminum chloride (10%) was added to the reaction mixture and constituted up to 200 μL using distilled water. Then, mixture was incubated for 30 minutes at 25°C. Quercetin was used as positive control and DMSO was used as negative control or blank. Absorbance was measured at 405 nm on microplate reader. The TFC value was expressed as Quercetin equivalent and data were presented as ± SD for triplicate analysis.
2.6. Assessment of Antioxidant Activity
Antioxidant activity in different fractions of A. vera leaf was determined using different assays, such as 2, 2-diphenyl-1-picrylhydrazyl (DPPH), ferric ion reducing power assay, and phosphomolybdenum complex assay.
2.6.1. DPPH Assay
The DPPH free radical scavenging assay is established to evaluate the scavenging potentials of antioxidants against the stable 2, 2-diphenyl-1-picrylhydrazyl (DPPH) radical. For the estimation of antioxidant, scavenging activity, DPPH assay is thought to be an authentic and simple assay [29]. The DPPH free radical scavenging activity of A. vera leaf fractions was measured by following the method of [27]. Briefly, 10 μL of the analyte solution was added in 96-well plates, followed by addition of 190 μL of DPPH testing agent. Ascorbic acid (1mg/ml) was positive control while DMSO was used as negative control. Absorbance was measured at 517 nm on microplate reader after one hour of incubation (37°C). The percentage DPPH radical scavenging activity was calculated using the formula
| (1) |
where As is the absorbance of sample and Ac is the absorbance of control. Data were expressed as ± SD for triplicate analysis.
2.6.2. Ferric Ion Reducing Power Assay
Reducing power is powerful impression of antioxidant activity of a compound [30]. Compounds that have to reduce may interact with potassium ferricyanide (Fe3+) and reduce it to potassium ferrocyanide (Fe2+), which subsequently interacts with ferrous chloride in order to generate complex of ferric ferrous which at 700nm wavelength shows maximum absorption [31]. The ferric ion reducing power was estimated by following the method as described earlier [32]. Briefly, 200 μL of each fraction was taken in Eppendorf tube, followed by the addition of 400 μL of 0.2M Phosphate buffer (pH 6.6) and 500 μL of 1% Potassium ferricyanide. Then mixture was incubated for 20 min at 50°C. Then 400 μL of 10% Trichloroacetic acid solution was mixed and centrifuged at 3000 rpm for 10 min. Then 150 μL of supernatant was added in 96-well plates. After that 50 μL of 0.1% of ferric chloride was mixed to each well containing sample mixture. The 200 μL of ascorbic acid (1 mg/mL) was used as positive control and equal volume of DMSO was used as negative control. The absorbance was taken at 630 nm on microplate reader and results of ferric ion reducing power were presented as AA equivalent μg/mL. Data were expressed as ± SD for triplicate analysis.
2.6.3. Phosphomolybdenum Complex Assay
The total antioxidant potential is also accessed through generation of complex compound called phosphomolybdenum. In this assay the Mo (VI) is reduced to Mo (V) through the test sample and further generation of Mo(V) green phosphate complex at a pH which is acidic [33]. Total antioxidant potential of AV1-AV10 fractions was determined by phosphomolybdenum complex assay as explained previously [32]. In short, 100 μL of each fraction was taken in 96-well plate and 900 μL of reagent solution [4 mM ammonium molybdate, 28 mM sodium phosphate, 0.6 M sulfuric acid, 1.63 mL conc. sulfuric acid, 1.6795g NaH2SO4, and 0.247g ammonium molybdate and 50 mL distilled water] was added. The test blend was incubated at 95°C for 90 min. Ascorbic acid (1 mg/mL) solution was positive control, while DMSO was used as negative control. The reading was taken at 630 nm wavelength on microplate reader and results of total antioxidant activity were presented as AA equivalent μg/mL. Data were expressed as ± SD for triplicate analysis.
2.7. Antibacterial Assay
Antibacterial activity of A. vera leaf fractions against gram positive Staphylococcus aureus (ATCC 6538) and Micrococcus luteus (ATCC 10240) and gram negative Escherichia coli (ATCC 15224) and Pseudomonas aeruginosa was determined by microtiter plate method [34]. The bacterial culture was formulated in nutrient broth and incubated for 12 hrs and then stored in refrigerator at 4°C. Before testing, culture was transferred to incubator for 14 hrs and diluted 10 folds with nutrient broth (1:10). 5 μL of the sample solution (4 mg/mL sample solution + 20% DMSO and nutrient broth) was taken in 96-well plate. Then 195 μL of freshly diluted inoculums were added to each well and the final test concentration was 100μg/mL. The active sample was tested at lower concentration by 3 times' serial dilution, that is, 33.3, 11.1, and 3.7μg/mL. The Cefixime monohydrate was used as positive control at 10, 3.33, 1.11, and 0.37 μg/mL, while 5μL of 20% DMSO in nutrient broth was taken as negative control. Absorbance was measured at 630 nm after 24 hrs incubation through microplate reader and percentage inhibition was calculated.
2.8. Antifungal Assay
Four fungal strains, Mucor species, Aspergillus flavus, Fusarium solani, and Aspergillus niger, were used to test the antifungal potential in studied samples using disc diffusion as explained earlier [35]. The fungal strains were stored on SDA at 4°C. Terbinafine (4mg/mL in DMSO) and pure DMSO were used as positive and negative control, respectively. Terbinafine disc concentration was 20 μg. Four growing of fungal strains were cultured in Sabouraud dextrose agar (SDA) having 6.5 pH. 5 μL of each crude extract solution was spew on the surface of filter paper discs and were implanted on the surface of the media in the Petri plate. The concentration of the sample and control was 100μg/disc. The prepared Petri dishes were incubated for 72 hours at 28°C and after incubation the inhibition zones were measured.
2.9. Antileishmanial Activity
The in vitro antileishmanial activity of Aloe fractions was determined by using the method as described before [36] with slight adjustments. The leishmanial strain (Leishmania tropica KWH23) was cultured in medium 199 containing 10% heat-inactivated Fetal Bovine Serum maintained at 24 ± 1°C for 6-7 days. The amphotericin B was used as positive control drug and DMSO was used as negative control. In 96-well plates the stock solutions (4 mg of crude fraction + 1000 μL of DMSO) were diluted serially. The DMSO was used as negative control and Amphotericin B was used as positive control. The 96-well plate was incubated for 72 hours at 24°C. After the incubation, 15 μL of assay culture was transferred to counting chamber (Neubauer) for live promastigotes counting using light microscope. Table curve 2D version 4 was used for LC50 calculation. Data were expressed as ± SD for triplicate analysis.
2.10. Cytotoxicity Assay
The cytotoxic potential in A. vera leaf fractions was estimated through brine shrimp lethality test as discussed earlier [37] with some modifications. At first, stock solution of samples was formulated (100mg crude fraction/mL DMSO); then 500, 250, 100, and 10 μg/mL dilutions were formulated. Different dilutions of Doxorubicin (1mg of doxorubicin + 1 mL DMSO), that is, 10, 1, and 0.1 μg/mL, were used as standard drug. A shallow rectangular tray (22x32 cm) already was filled with simulated sea water (38g sea salt/L of distilled water) in which brine shrimp eggs (Artemia salina L. purchased from Sara, Heidelberg, Germany) hatched. The tray was consisting of two portions, one of which was large and the other was small. Each portion was separated from the other through a wall consisting of many holes. In the large portion of the tray brine shrimp's eggs were spread on the surface of simulated sea water. After spreading the brine shrimp's eggs, the large portion of the tray was concealed with aluminum foil. The surface of smaller portion was lighted up with a lamp and hatching started after 24-26 hours. The newly brine shrimp larvae (nauplii) traveled towards the light portion. These nauplii were taken from the tank and shifted to the beaker by Pasteur pipette.
50 μL of sample solution with four concentrations (500, 250, 100, and 10μg) was poured into respective well of 96-well plate, followed by addition of 200 μL of the simulated sea water and mixed carefully. Ten shrimps were counted under a 3x magnifying glass and then transferred to each well with the help of Pasteur pipette and 300 μL volume was maintained for each well to accomplish the required concentration of crude sample. The microplates containing shrimps were incubated at room temperature for 24 hrs. Then shrimps were taken out from the well with Pasteur pipette and survivor rate was counted under a 3x magnifying glass with 3x resolution against a background that is lighted. The values of LD50 (sample concentration required to kill 50% of shrimps) were calculated by using Table curve 2D version 4.
2.11. Protein Kinase Inhibition Assay
The protein kinase inhibition potential was studied through a method established on the restriction of hyphae formulation in Streptomyces as explained before [38, 39]. The sample solution was formulated by mixing 4 mg fractions/1000μL DMSO. In 20 mL of tryptic soya, broth culture was grown for 3 to 4 days with shaking at 30°C. The production of hyphae inhibition test on Streptomyces was carried out. The Streptomyces mycelia fragments were dispersed on the surface of ISP4 agar media plates. Then 5 μL of the test sample was applied to each disk and placed carefully on the agar surface previously seeded with Streptomyces. Two types of zones were appeared, that is, clear and bald type around the area of paper disk, which were measured after 30 to 48 hrs of incubation. The inhibition zone greater than 9 mm revealed that the sample is active. The Surfactin and DMSO were used as positive control and negative control, respectively.
2.12. Molecular Docking
Chemical structures of compounds 1-10 as mentioned in Table 1 were selected from NCBI database and drawn by ChemBioDraw. The legends were converted into MOL format for further use. The 3D crystal structure of serine protease (UniProt ID: A8D853) (PDB ID: 2HLC), trypanothione reductase (PDB ID: 2W0H), and tyrosinase (PDB ID: 3NM8) enzymes involved in hatching of Artemia larvae, growth of Leishmania, and formation of Streptomyces, respectively, was accessed and downloaded from Protein Data Bank (PDB) database [http://www.rcsb.org/pdb/home/home.do]. The active sites of target proteins were analyzed using the Molecular Operating Environment (MOE) software [https://www.chemcomp.com/MOE-Molecular_Operating_Environment.htm]. An active site was defined from the coordinates of the ligand in the original target protein sites.
Table 1.
Details of compounds used for in silico analysis.
| S # | Code | Name of compound |
|---|---|---|
| 1 | Compound 1 | Aloin (anthraquinone glycoside) |
| 2 | Compound 2 | Aloe emodin (anthraquinone) |
| 3 | Compound 3 | Mannan (polysaccharide) |
| 4 | Compound 4 | Isoaloresin D (chromones) |
| 5 | Compound 5 | Emodin (hydroxyl anthraquinone) |
| 6 | Compound 6 | Aloe barbendol |
| 7 | Compound 7 | Physcion (anthraquinone derivative) |
| 8 | Compound 8 | Aloetic acid |
| 9 | Compound 9 | Cinnamic acid |
| 10 | Compound 10 | Galactan |
A computational ligand-target docking approach was used to determine structural complexes of receptor targets with ligands molecules in order to understand the structural basis of these proteins targets specificity. Finally, docking was carried out by Molecular Operating Environment (MOE) software. The energy of interaction of these compounds with the protein targets is assigned “grid point” [39].
3. Statistical Analysis
All tests relevant to composition and properties assessment of A. vera were performed in triplicate. Data were presented as mean ± SD from at least three replicates.
4. Results and Discussion
4.1. Total Phenolics and Flavonoids Contents
The measured values of TPC and TFC in different fractions of A. vera (AV1 to AV10) are given in Table 2. AV4 fraction exhibited the highest level of total phenolics content (332.4 ± 32.6 μg GAE/mg), followed by AV5, AV3, and AV6 (304.6 ± 29.6, 180.3 ± 21.9, and 157.8 ± 15.4 μg GAE/mg, resp.). The lowest value of total phenolics content was showed by AV10 (97.95 ± 21.5 μg GAE eq./mg). These findings are in agreement with each other as reported previously [40, 41]. It has been reported that plants with high phenolic contents have wound healing ability and reduce inflammation. However, method of extraction has a great impact on the value of total phenolic content, so more phenolics were determined in ethanol and chloroform extracts compared to water extract [42, 43].
Table 2.
TPC∗ and TFC∗ contents in different fractions of A. vera.
| Fractions | TPC (μg GAE. eq/mg) | TFC (μg QE. eq/mg) |
|---|---|---|
| AV1 | 139.7 ± 15.1 | 51.87 ± 12.0 |
| AV2 | 136.2 ± 13.3 | 51.28 ± 7.89 |
| AV3 | 180.3 ± 21.9 | 87.54 ± 15.5 |
| AV4 | 332.4 ± 32.6 | 60.47 ± 13.7 |
| AV5 | 304.6 ± 29.6 | 51.03 ± 9.87 |
| AV6 | 157.8 ± 15.4 | 32.47 ± 6.57 |
| AV7 | 119.7 ± 13.3 | 18.46 ± 7.68 |
| AV8 | 104.4 ± 12.0 | 19.49 ± 5.79 |
| AV9 | 113.3 ± 19.5 | 22.67 ± 4.60 |
| AV10 | 97.95 ± 21.5 | 24.87 ± 3.12 |
TPC:∗ total phenolics; TFC:∗ total flavonoids.
The anti-inflammatory and antimicrobial potential of plants is attributed to flavonoids content. In the studied fractions, TFC content ranged from 18.46 ± 7.68 to 87.54 ± 15.5μg QE/mg. AV3 fraction exhibited the highest TFC, whereas the lowest value was calculated for AV7.In A. vera fractions, measured levels of TFC were comparatively lower than those reported previously [42]. Furthermore, type of solvent may also contribute in the extract amount so more TFC content was reported in methanol-chloroform extract than aqueous extract [35, 41].
4.2. Antioxidant Potential
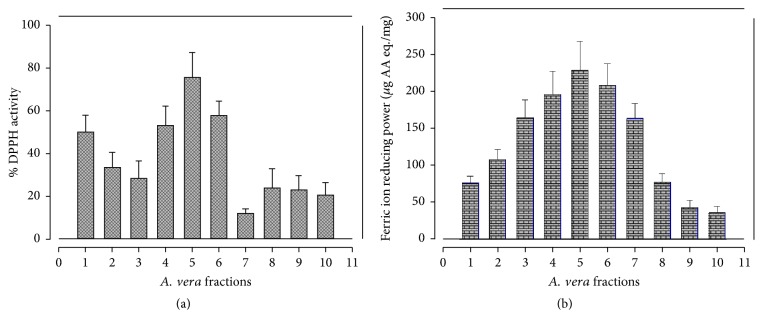
DPPH radical scavenging, ferric ion reducing potential and total antioxidant activity assays were conducted to determine the antioxidant potential in methanolic fractions of A. vera leaf. The percentage of DPPH radical scavenging capacity of various fractions as shown in Figure 1(a) ranged from 11.82 to 75.54%. AV5 fraction has the highest DPPH scavenging activity (75.54 ± 11.6%), followed by AV6 and AV4 (57.87 ± 6.53% and 52.99 ± 9.01%, resp.). These values were comparable to previously reported levels in the ethanolic extract of A. vera leaf [42]; however, they were a little lower than those reported earlier [41]. However, more DPPH radical scavenging activity has been reported in the methanolic and ethanolic extracts of A. vera leaf and pulp than n-hexane and aqueous extracts [44, 45].
Figure 1.
Free radicals scavenging potential in different fractions of A. vera leaf. (a) DPPH activity. (b) Ferric ion reducing antioxidant potential.
Results of ferric ion reducing antioxidant power (FRAP) are shown in Figure 1(b). All tested fractions showed significant ferric ion reducing ability; however AV5 fractions exhibited the highest potential 228.9 ± 39.1μg AA eq./mg to reduce ferric ion, followed by AV6 (208.6 ± 29.3 μg AA eq./mg) and AV4 (195.9 ± 31.2 μg AA eq./mg). However, AV10 fraction exhibited the lowest FRAP value 36.18 ± 7.96μg AA eq./mg. In a study on leaf skin of A. vera ethanol-chloroform extract depicted the highest reducing power activity compared to ethyl acetate, butanol, and n-hexane extract [46]. Therefore, variation in FRAP values might be due to difference in plant parts used, harvesting time, geoclimatic conditions, and solvent system used for the extraction as reported [47]. The results of total antioxidant capacity (TAC) determined by phosphomolybdenum complex assay are given in Figure 2. The total antioxidant activity in AV1 to AV10 A. vera leaf fractions ranged from 28.77 ± 9.36 to 150.4 ± 25.8μg AA eq./mg, with the highest TAC value in AV4 and the lowest in AV8 fraction. Three fractions AV4, AV3, and AV5 exhibited the highest TAC. These findings are in compliance to [46].
Figure 2.
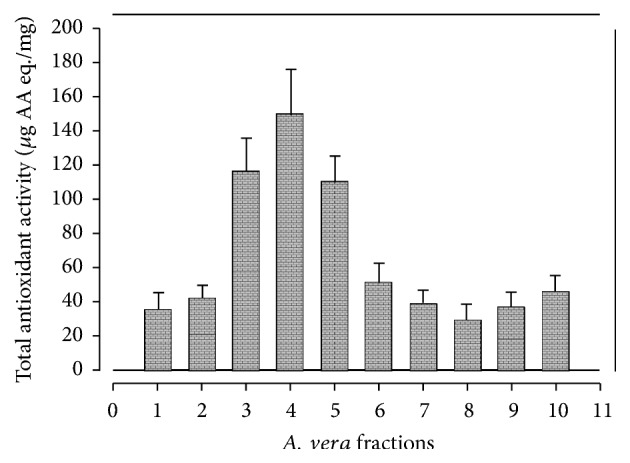
Total antioxidant activity of A. vera leaf fractions.
4.3. Antimicrobial Potential
The antibacterial potential of studied samples given in Table 3 revealed that crude fractions were not much effective against E. coli, but relatively good results were obtained for P. aeruginosa. The most significant inhibition was shown by AV8, AV6, AV10, and AV9 fractions against P. aeruginosa with MIC values of 0.70, 0.71, 7.72, and 8.68μg/mL, respectively. The AV4, AV7, and AV9 fractions exhibited good response against M. luteus with MIC values of 103, 161, and 250μg/mL, respectively. In the previous studies regarding A. vera DMSO gel extract, ethanolic extract was found active against E. coli, P. aeruginosa, and S. aureus [48–50]. However, antibacterial potential of A. vera fractions in the present evaluation against E. coli was not in support with reported data. However, Lawrence et al. [50] reported significant potential in methanolic extract of A. vera against E. coli which is partially in agreement with the current study.
Table 3.
Antibacterial activity of A. vera leaf's fractions.
| Fractions | Minimum inhibitory concentration (MIC) | |||||||
|---|---|---|---|---|---|---|---|---|
| M. luteus | S. aureus | E. coli | P. aeruginosa | |||||
| % inhibition | MIC (μg/mL) | % inhibition | MIC (μg/mL) | % inhibition | MIC (μg/mL) | % inhibition | MIC (μg/mL) | |
| AV1 | 57.93 | 325 | 69.15 | 250 | 55.95 | 673 | 76.00 | 60.20 |
| AV2 | 49.61 | 1000 | 60.26 | 500 | 57.02 | 874 | 70.26 | 18.60 |
| AV3 | 63.14 | 500 | 73.76 | 250 | 48.43 | 1000 | 80.25 | 10.02 |
| AV4 | 63.40 | 103 | 67.44 | <125 | 43.80 | >1000 | 77.10 | 64.66 |
| AV5 | 49.61 | 1000 | 66.75 | <125 | 51.19 | 971 | 77.10 | 123.9 |
| AV6 | 48.07 | 1000 | 69.15 | <125 | 44.49 | >1000 | 69.24 | 0.710 |
| AV7 | 54.57 | 161 | 67.69 | 250 | 45.55 | 1000 | 70.42 | 125.0 |
| AV8 | 48.78 | 1000 | 65.04 | 125 | 42.17 | >1000 | 76.71 | 0.700 |
| AV9 | 52.26 | 250 | 62.74 | 125 | 47.24 | 1000 | 74.82 | 8.670 |
| AV10 | 53.35 | 1000 | 70.60 | 250 | 41.54 | 100 | 74.04 | 7.720 |
| Cefixime | 86.93 | 0.58 | 100.2 | 1.01 | 51.19 | 0.58 | 86.93 | 0.200 |
The antifungal activity of A. vera fractions against four strains, that is, Aspergillus niger, Mucor specie, Aspergillus flavus, and Fusarium solani is mentioned in Table 4. All the tested fractions exhibited minor effects against fungal strains which is not in complete agreement with previous reports [51, 52]. This variation of biological potential may be due to solvent use, harvesting time, seasonal variation, and ecological factors.
Table 4.
Antifungal activity of A. vera leaf's fractions.
| Fractions | Zone of inhibition (mm) | |||
|---|---|---|---|---|
| A. niger | Mucor spp. | A. flavus | F. solani | |
| AV1 | --- | 8.319 ± 1.2 | 7.198 ± 0.7 | --- |
| AV2 | --- | --- | 6.264 ± 1.3 | 7.245 ± 1.9 |
| AV3 | --- | 9.017± 1.5 | 8.451 ± 1.6 | 9.364 ± 2.0 |
| AV4 | --- | 10.25 ± 1.3 | 6.137 ± 0.7 | 8.170 ± 1.3 |
| AV5 | --- | 7.352 ± 0.9 | 9.326 ± 1.5 | 10.03 ± 0.6 |
| AV6 | --- | 8.036 ± 0.7 | 7.367 ± 0.8 | 9.217 ± 1.5 |
| AV7 | --- | 8.247 ± 1.3 | - - - | 8.612 ± 0.8 |
| AV8 | --- | 9.364 ± 1.7 | 8.353 ± 1.7 | --- |
| AV9 | --- | --- | 8.632 ± 2.0 | 8.219 ± 0.5 |
| AV10 | --- | 11.15 ± 2.0 | --- | --- |
| -ve. control (DMSO) | --- | --- | --- | --- |
| +ve. control (Terbinafine ) | 22.15 ± 1.9 | 24.37 ± 2.5 | 27.47 ± 1.9 | 25.19 ± 2.7 |
The antileishmanial activity carried out at 50 μg/mL concentration in different fractions of A. vera leaf extract and results are given in Figure 3. The highest value of mortality was shown by AV6 fraction (95%), followed by AV7, AV9, and AV4 (92, 84, and 83%, resp.), whereas the lowest value was shown by AV10 (18%). The results were comparable with previous studies [53–55].
Figure 3.
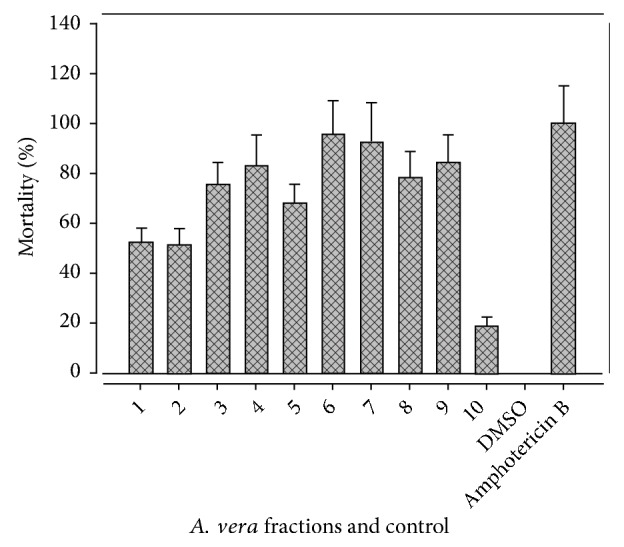
Antileishmanial activity of A. vera leaf fractions.
4.4. Cytotoxicity of A. vera Fractions
The brine shrimp lethality assay is considered as the most appropriate assay for the pharmacological active crude plant extract which may exhibit lethality against newly hatched nauplii [56]. In the present study, cytotoxicity of A. vera leaf fractions was determined by Brine Shrimp Lethality assay and results in LD50 (μg/mL) value are mentioned in Table 5. The fractions with low LD50 value are more active and have high cytotoxic potential. Therefore, AV6 fraction depicted the highest cytotoxicity with LD50 value of 0.530μg/mL, followed by AV4, AV5, and AV3 (4.710, 4.710, and 9.070 μg/mL, resp.). A study carried out on cytotoxicity of A. vera was reported against brine shrimp and LD50 was estimated over 500μg/mL [20], which is partially correlated with the current study. However, brine shrimp lethality activity in methanolic extract of Melia azedarach stem bark with3.27μg/mL LD50 value [57] is in agreement with our findings.
Table 5.
Cytotoxic activity of Aloe vera leaf fractions.
| Fractions | Mortality (%) | ||||
|---|---|---|---|---|---|
| 500 μg/mL | 250 μg/mL | 100 μg/mL | 10 μg/mL | LD50 (μg/mL) | |
| AV1 | 67 | 62 | 58 | 38 | 35.05 |
| AV2 | 69 | 60 | 56 | 41 | 49.72 |
| AV3 | 80 | 67 | 60 | 50 | 9.070 |
| AV4 | 80 | 70 | 62 | 54 | 4.710 |
| AV5 | 80 | 72 | 63 | 52 | 4.710 |
| AV6 | 75 | 69 | 64 | 62 | 0.530 |
| AV7 | 73 | 55 | 54 | 47 | 64.13 |
| AV8 | 50 | 50 | 42 | 37 | 412.8 |
| AV9 | 40 | 30 | 0 | 0 | > 500 |
| AV10 | 60 | 55 | 50 | 35 | 107.6 |
| Doxorubicin | 100 | 94 | 89 | 70 | 0.270 |
4.5. Protein Kinase Inhibitions
Figure 4 depicted protein kinase inhibition (PKI) activity in the methanolic fractions of A. vera leaf. At 4mg/mL concentration, only three fractions AV7, AV6, and AV2 showed PKI activity. The AV7 fraction exhibited the highest zone of inhibition (21 ± 0.58mm), followed by AV2 and AV6 (12 ± 0.45 and 9.5 ± 0.37mm, resp.). The results are strongly supported by a study of Yao and coworkers, which was carried out on evaluation of hyphae formation inhibition in Streptomyces 85E, and isolated compounds showed 21mm zone of inhibition at 80μg/disk and it was hypothesized that the compounds prevent the formation of hyphae in Streptomyces 85E, which may inhibit cancer proliferation [38].
Figure 4.
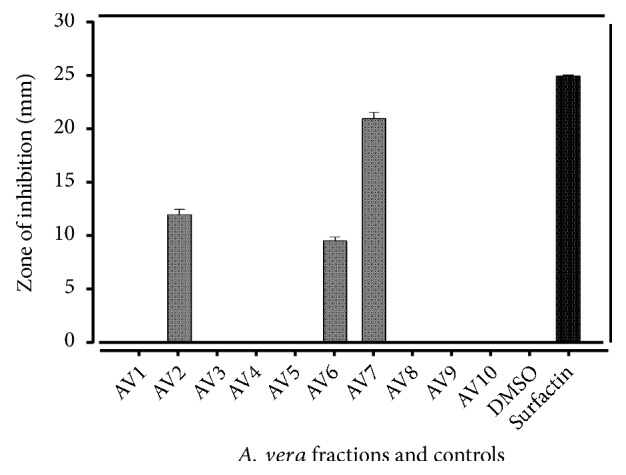
Protein Kinase inhibitions in A. vera leaf fraction.
Comparatively, measured levels of secondary metabolites and biological activities particularly antioxidant potential in different fractions of A. vera were less than those in previous reports. Such variations may be attributed to diverse factors including genetic variation in A. vera reported from various regions, harvesting time and storage conditions, difference in geoclimatic conditions, and growing environment such as temperature, precipitation, humidity, altitude, salinity, and drought stress along with the analytical techniques used.
4.6. Molecular Docking Study
4.6.1. Isolation of Binding Sites
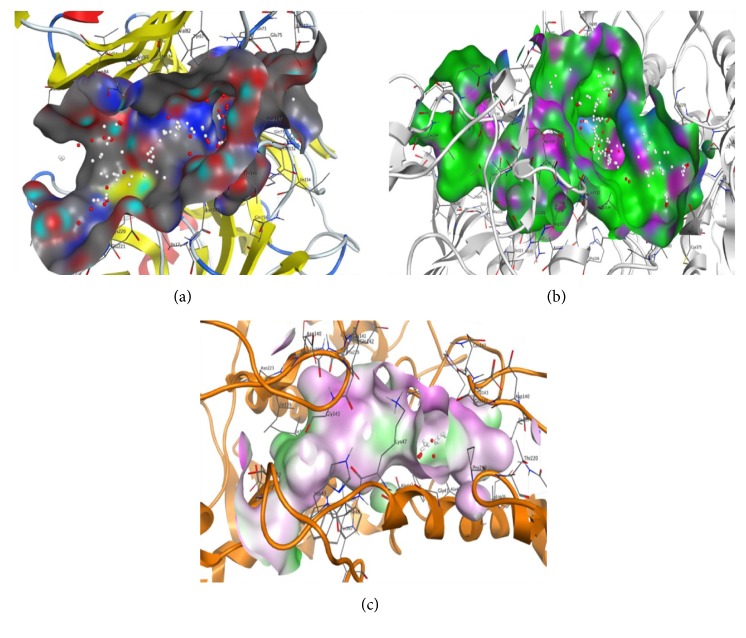
Potential isolated binding sites of each target protein were analyzed by Molecular operator Environment (MOE) software. The possible binding sites residues of serine protease (UniProt ID: A8D853) (PDB ID: 2HLC) were as follows: ILE17, TRP41, GLN73, TYR74, TRP141, GLY142, GLN143, SER144, ASN145, THR146, ASP147, THR152, VAL153, ILE154, GLN156, CYS191, PHE192, GLY193, GLY217A, ALA218, GLY219, CYS220, GLU221, and SER222 (Figure 5(a)). Similarly in case of trypanothione reductase (PDB ID: 2W0H), binding site residues were LEU10, GLY11, ALA12, GLY13, SER14, GLY15, GLY16, VAL34, ASP35, VAL34, ASP35, VAL36, ALA46, ALA47, GLY49, GLY50, THR51, CYS52, VAL55, GLY56, CYS57, LYS60, GLY125, PHE126, GLY127, GLU141, ALA159, THR160, GLY161, SER162, TRP165, ALA200, GLU202, PHE203, GLY229, PHE230, ASP231, LEU283, ALA284, ILE285, GLY286, ARG287, VAL288, PRO289, ARG290, SER291, GLN292, GLN292, ALA293, LEU294, ASN306, ILE325, GLY326, ASP327, VAL328, ASN330, ARG331, VAL332, MET233, LEU334, THR335, PRO336, and ALA338 (Figure 5(b)). Binding site residues of tyrosinase (PDB ID: 3NM8) were ASP36, ILE39, ALA40, TRP41, GLY43, ALA44, LYS47, PHE48, HIS49, ILE139, ASP140, GLU141, GLN142, GLY143, PRO219, THR220, ASN223, and TYR267 (Figure 5(c)). Protein geometric arrangement of amino acid residues in allowed and disallowed regions indicates the quality of target proteins.
Figure 5.
Potential isolated binding sites of each target protein analyzed by Molecular operator Environment (MOE) software. (a) Serine protease. (b) Trypanothione reductase. (c) Tyrosinase.
4.6.2. Docking Analysis
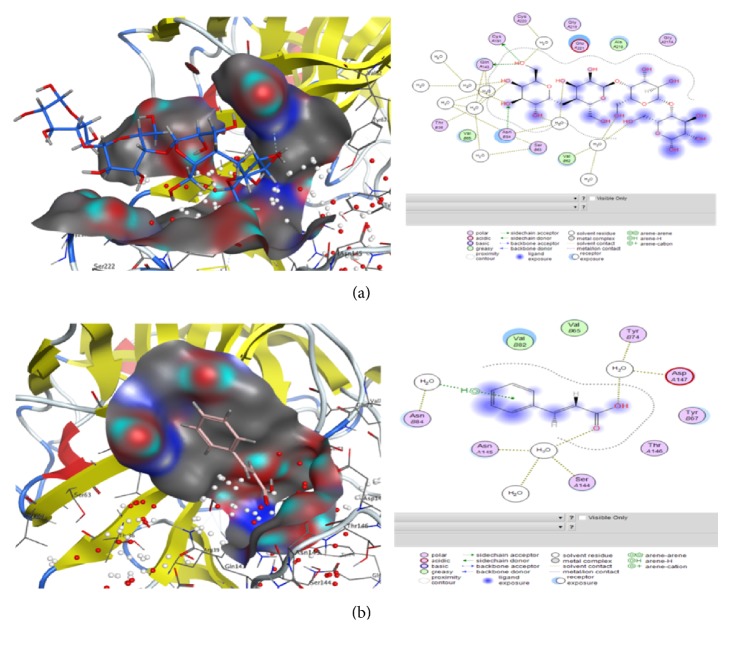
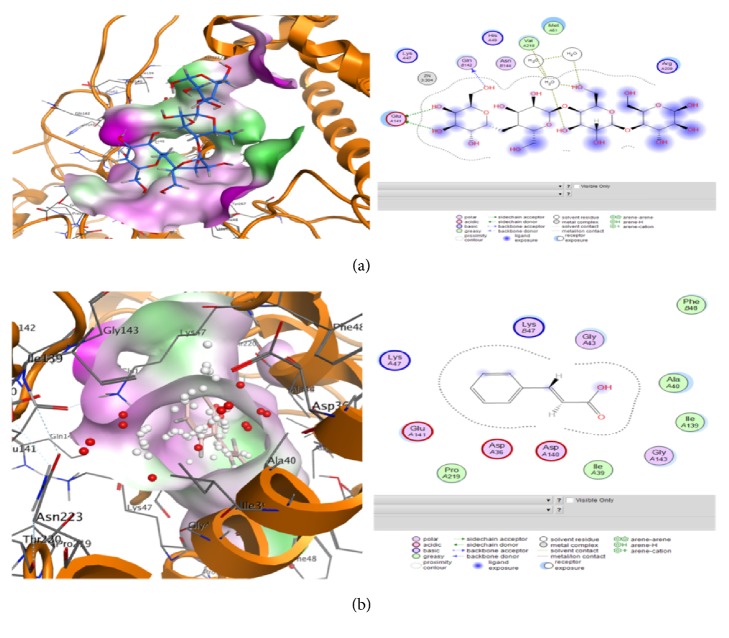
Molecular docking was performed to find ideal ligand orientation with protein for stable complex formation. Strength of association between ligand and protein is evaluated on the basis of scoring function and minimum binding energy. Computational ligand-target binding approach was used in analyzing structural complexes of compounds with three selected enzymes in order to interpret structural basis of target protein specificity. The interaction energy of compounds with target enzyme is assigned, “grid point.” Finally, these ligands were docked with the potential active sites of target molecules. Compounds 3 (mannan), 4 (isoaloresin D), and 9 (cinnamic acid) showed good results against serine protease with binding energy above -5.0 (Table 6/Figures 6(a) and 6(b)) while rest of the compounds also showed binding potential. These finding are also correlated with the fact that A. vera extract showed cytotoxic activities against cell lines [58]. In case of effects of these compounds when analyzed against trypanothione reductase enzyme responsible for leishmanial parasite's growth it was observed that compounds 3 (mannan), 5 (emodin), and 6 (aloe barbendol) showed the lowest binding energy (Table 7) and against tyrosinase compound 8 (aloetic acid) showed better results while compounds 2 (aloe emodin), 3(mannan), 4 (isoaloresin D), and 9 (cinnamic acid) also showed some activity indicating their potential for Streptomyces inhibition (Table 8/Figures 7(a) and 7(b)). These findings are correlated with previous reports that Aloe vera plant contains anthraquinones, anthrones, chromones, and polysaccharides responsible for anticancer, antileishmanial, antibacterial, and antioxidant potential of plant [53, 59]. Docking results indicating the lowest binding energy cluster were considered as representative binding states. The minimum binding energies showed that target proteins were docked successfully with ligand molecules.
Table 6.
Energy values obtained during docking analysis of compounds as ligand molecules serine protease (PDB ID: 2HLC) enzyme as target molecules.
| S. no. | mol | rseq | mseq | S | Rmsd-refine | E-conf | E-place | E-score1 | E-refine | E-score2 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | COMPD1_2HLC | 1 | 1 | -4.522 | 4.681 | 168.3 | -110.5 | -20.26 | -12.83 | -4.522 |
| 2 | COMPD2_2HLC | 1 | 2 | -3.696 | 4.430 | 33.88 | -126.1 | -18.64 | -9.617 | -3.696 |
| 3 | COMPD3_2HLC | 1 | 3 | -5.279 | 4.460 | 338.4 | -126.0 | -26.34 | -13.28 | -5.279 |
| 4 | COMPD4_2HLC | 1 | 4 | -5.249 | 3.720 | 146.2 | -121.2 | -16.25 | -9.643 | -5.249 |
| 5 | COMPD5_2HLC | 1 | 5 | -3.651 | 3.198 | 32.87 | -47.26 | -17.16 | -6.049 | -3.651 |
| 6 | COMPD6_2HLC | 1 | 6 | -4.149 | 3.672 | 40.29 | -97.50 | -16.75 | -11.55 | -4.149 |
| 7 | COMPD7_2HLC | 1 | 7 | -4.204 | 3.778 | 47.10 | -105.1 | -19.84 | -11.38 | -4.204 |
| 8 | COMPD8_2HLC | 1 | 8 | -3.785 | 2.240 | -17.62 | -53.66 | -11.26 | -8.136 | -3.785 |
| 9 | COMPD9_2HLC | 1 | 9 | -5.034 | 4.061 | 281.6 | -116.6 | -22.48 | -16.40 | -5.034 |
| 10 | COMPD10_2HLC | 1 | 10 | -3.932 | 3.282 | 22.61 | -45.39 | -18.20 | -7.129 | -3.932 |
Figure 6.
Views of molecular docking and information of ligands interaction with atoms of serine protease target performed by Molecular Operating Environment (MOE) software comp-4 (a) and comp-9 (b) as mentioned in Table 1.
Table 7.
Energy values obtained during docking analysis of compounds as ligand molecules trypanothione reductase (PDB ID: 2W0H) enzyme as target molecules.
| S. no. | mol | rseq | mseq | S | Rmsd-refine | E-conf | E-place | E-score1 | E-refine | E-score2 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | COMPD1_2W0H | 1 | 1 | -5.950 | 6.927 | 154.3 | -140.6 | -15.74 | -3.376 | -5.950 |
| 2 | COMPD2_2W0H | 1 | 2 | -5.943 | 4.344 | 33.24 | -128.4 | -15.06 | -17.51 | -5.943 |
| 3 | COMPD3_2W0H | 1 | 3 | -7.136 | 3.591 | 369.9 | -192.9 | -17.05 | 13.33 | -7.136 |
| 4 | COMPD4_2W0H | 1 | 4 | -1.220 | 2.907 | 195.1 | -149.7 | -13.59 | 70.64 | -1.220 |
| 5 | COMPD5_2W0H | 1 | 5 | -6.221 | 2.175 | 39.06 | -103.4 | -13.25 | -8.982 | -6.221 |
| 6 | COMPD6_2W0H | 1 | 6 | -6.025 | 3.343 | 38.80 | -115.4 | -15.22 | -15.99 | -6.025 |
| 7 | COMPD7_2W0H | 1 | 7 | -5.581 | 2.251 | 62.32 | -133.5 | -14.86 | 2.153 | -5.581 |
| 8 | COMPD8_2W0H | 1 | 8 | -5.148 | 3.238 | -16.71 | -72.16 | -9.94 | -19.81 | -5.148 |
| 9 | COMPD9_2W0H | 1 | 9 | -2.591 | 3.356 | 343.8 | -182.9 | -15.86 | 51.47 | -2.591 |
| 10 | COMPD10_2W0H | 1 | 10 | -5.958 | 2.460 | 25.19 | -111.8 | -14.74 | -10.42 | -5.958 |
Table 8.
Energy values obtained during docking analysis of compounds as ligand molecules tyrosinase (PDB ID: 3NM8) enzyme as target molecules.
| S. no. | mol | rseq | mseq | S | Rmsd-refine | E-conf | E-place | E-score1 | E-refine | E-score2 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | COMPD1_3NM8 | 1 | 1 | 7.123 | 2.412 | 318.7 | -131.1 | -19.68 | 126.5 | 7.123 |
| 2 | COMPD2_3NM8 | 1 | 2 | -4.240 | 3.577 | 49.18 | -131.2 | -15.90 | 13.81 | -4.241 |
| 3 | COMPD3_3NM8 | 1 | 3 | -4.964 | 6.082 | 338.2 | -153.2 | -23.94 | -14.51 | -4.965 |
| 4 | COMPD4_3NM8 | 1 | 4 | -4.963 | 5.343 | 124.6 | -85.28 | -14.32 | -15.57 | -4.963 |
| 5 | COMPD5_3NM8 | 1 | 5 | -2.949 | 3.719 | 56.49 | -96.54 | -15.63 | 25.46 | -2.949 |
| 6 | COMPD6_3NM8 | 1 | 6 | -2.244 | 1.677 | 56.00 | -94.59 | -16.35 | 36.25 | -2.245 |
| 7 | COMPD7_3NM8 | 1 | 7 | 1.947 | 2.255 | 160.5 | -135.3 | -18.75 | 82.87 | 1.948 |
| 8 | COMPD8_3NM8 | 1 | 8 | -5.016 | 1.550 | -16.19 | -63.69 | -10.49 | -10.49 | -5.017 |
| 9 | COMPD9_3NM8 | 1 | 9 | -4.328 | 3.061 | 306.2 | -138.3 | -19.44 | -9.012 | -4.328 |
| 10 | COMPD10_3NM8 | 1 | 10 | -3.495 | 2.725 | 27.07 | -98.46 | -17.34 | 23.28 | -3.495 |
Figure 7.
Views of molecular docking and information of ligands interaction with atoms of tyrosinase target performed by Molecular Operating Environment (MOE) software comp-3 (a) and comp-8 (b) as mentioned in Table 2.
5. Conclusion
The present study was focused to assess in vitro protein kinase inhibition, brine shrimp lethality assay, and antioxidant activity of different factions (AV1 to AV10) of A. vera leaf extract. The AV4, AV5, AV6, and AV7 methanolic fractions depicted significant TPC and TFC contents and antioxidant and antimicrobial activities along with cytotoxicity and protein kinase inhibition. The Protein Kinase Inhibitory assay was performed first time and AV7 depicted the highest value of 21 ± 0.50mm. Our findings revealed that A. vera is a versatile medicinal plant with significant biological activities and could be used for future drug against cancer and leishmaniasis.
Acknowledgments
Research facilities provided by COMSATS University Islamabad (Abbottabad Campus) are gratefully acknowledged, whereas financial and technical aid provided by Dr. Mingxing Zhang to complete this project is thankfully acknowledged.
Contributor Information
Abdul Mannan, Email: amannan@ciit.net.pk.
Arshad Mehmood Abbasi, Email: arshad799@yahoo.com.
Mingxing Zhang, Email: zmingxing@126.com.
Data Availability
The data used to support the findings of this study are included within the article.
Conflicts of Interest
All authors declared that they have no conflicts of interest.
Authors' Contributions
Huma Tariq performed research experiments; Muhammad Zia and Ihsan-ul-Haq were involved in data analysis; Syed Aun Muhammad and Nighat Fatima contributed in reagents/materials and data interpretation; Abdul Mannan supervised the project; Arshad Mehmood Abbasi contributed in data compilation, analysis, and write-up; Mingxing Zhang contributed in final draft, editing, and funding.
References
- 1.Rishton G. M. Natural products as a robust source of new drugs and drug leads: past successes and present day issues. American Journal of Cardiology. 2008;101(10):S43–S49. doi: 10.1016/j.amjcard.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Ganesan A. The impact of natural products upon modern drug discovery. Current Opinion in Chemical Biology. 2008;12(3):306–317. doi: 10.1016/j.cbpa.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 3.Han X., Shen T., Lou H. Dietary polyphenols and their biological significance. International Journal of Molecular Sciences. 2007;8(9):950–988. doi: 10.3390/i8090950. [DOI] [Google Scholar]
- 4.Owen R. W., Giacosa A., Hull W. E., Haubner R., Spiegelhalder B., Bartsch H. The antioxidant/anticancer potential of phenolic compounds isolated from olive oil. European Journal of Cancer. 2000;36(10):1235–1247. doi: 10.1016/s0959-8049(00)00103-9. [DOI] [PubMed] [Google Scholar]
- 5.Newman D. J., Cragg G. M., Snader K. M. Natural products as sources of new drugs over the period 1981–2002. Journal of Natural Products. 2003;66(7):1022–1037. doi: 10.1021/np030096l. [DOI] [PubMed] [Google Scholar]
- 6.Krishnaraju A. V., Rao T. V., Sundararaju D., Vanisree M., Tsay H. S., Subbaraju G. V. Assessment of bioactivity of Indian medicinal plants using brine shrimp (Artemia salina) lethality assay. International Journal of Applied Science and Engineering. 2005;3(2):125–134. [Google Scholar]
- 7.Báidez A. G., Gómez P., Del Río J. A., Ortuño A. Dysfunctionality of the Xylem in Olea europaea L. Plants Associated with the Infection Process by Verticillium dahliae Kleb. Role of Phenolic Compounds in Plant Defense Mechanism. Journal of Agricultural and Food Chemistry. 2007;55(9):3373–3377. doi: 10.1021/jf063166d. [DOI] [PubMed] [Google Scholar]
- 8.Harborne J. B., Williams C. A. Advances in flavonoid research since 1992. Phytochemistry. 2000;55(6):481–504. doi: 10.1016/S0031-9422(00)00235-1. [DOI] [PubMed] [Google Scholar]
- 9.Mazur A., Bayle D., Lab C., Rock E., Rayssiguier Y. Inhibitory effect of procyanidin-rich extracts on LDL oxidation in vitro. Atherosclerosis. 1999;145(2):421–422. doi: 10.1016/S0021-9150(99)00115-X. [DOI] [PubMed] [Google Scholar]
- 10.Ferrali M., Signorini C., Caciotti B., et al. Protection against oxidative damage of erythrocyte membrane by the flavonoid quercetin and its relation to iron chelating activity. FEBS Letters. 1997;416(2):123–129. doi: 10.1016/s0014-5793(97)01182-4. [DOI] [PubMed] [Google Scholar]
- 11.Elliott A. J., Scheiber S. A., Thomas C., Pardini R. S. Inhibition of glutathione reductase by flavonoids. A structure-activity study. Biochemical Pharmacology. 1992;44(8):1603–1608. doi: 10.1016/0006-2952(92)90478-2. [DOI] [PubMed] [Google Scholar]
- 12.Hirano R., Sasamoto W., Matsumoto A., Itakura H., Igarashi O., Kondo K. Antioxidant ability of various flavonoids against DPPH radicals and LDL oxidation. Journal of Nutritional Science and Vitaminology. 2001;47(5):357–362. doi: 10.3177/jnsv.47.357. [DOI] [PubMed] [Google Scholar]
- 13.Cos P., Ying L., Calomme M., et al. Structure-activity relationship and classification of flavonoids as inhibitors of xanthine oxidase and superoxide scavengers. Journal of Natural Products. 1998;61(1):71–76. doi: 10.1021/np970237h. [DOI] [PubMed] [Google Scholar]
- 14.Colombo M. L., Bosisio E. Pharmacological activities of Chelidonium majus L. (papaveraceae) Pharmacological Research. 1996;33(2):127–134. doi: 10.1006/phrs.1996.0019. [DOI] [PubMed] [Google Scholar]
- 15.Morens D. M., Folkers G. K., Fauci A. S. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430(6996):242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bandow J. E., Brotz H., Leichert L. I., Labischinski H., Hecker M. Proteomic approach to understanding antibiotic action. Antimicrobial Agents and Chemotherapy. 2003;47(3):948–955. doi: 10.1128/AAC.47.3.948-955.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kabir M. A., Hussain M. A., Ahmad Z. Candida albicans: a model organism for studying fungal pathogens. International Scholarly Research Notices. 2012;12:1–15. doi: 10.5402/2012/538694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rocha L., Almedia J., Macedo R., Barbosa-Folho J. A review of natural products with antileishmanial activity. Phytomed. 2005;12:514–535. doi: 10.1016/j.phymed.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 19.Iwu M., Duncan A. R., Okunji C. O. New Antimicrobials of Plant Origin. Alexandria, Egypt: Perspectives on new crops and new uses ASHS Press; 1999. [Google Scholar]
- 20.Thu K., Mon Y. Y., Khaing T. A., Tun O. M. Study on phytochemical properties, antibacterial activity and cytotoxicity of aloe vera l. International Journal of Biotechnology and Bioengineering. 2013;77:102–106. [Google Scholar]
- 21.Fatima N., Kondratyuk T. P., Park E.-J., et al. Endophytic fungi associated with Taxus fauna (West Himalayan Yew) of Pakistan: potential bioresources for cancer chemopreventive agents. Pharmaceutical Biology. 2016;54(11):2547–2554. doi: 10.3109/13880209.2016.1170154. [DOI] [PubMed] [Google Scholar]
- 22.Scapin G. Protein Kinase Inhibition: Different Approaches to Selective Inhibitor Design. Current Drug Targets. 2006;7(11):1443–1454. doi: 10.2174/1389450110607011443. [DOI] [PubMed] [Google Scholar]
- 23.Smyth L. A., Collins I. Measuring and interpreting the selectivity of protein kinase inhibitors. Journal of Chemical Biology. 2009;2(3):131–151. doi: 10.1007/s12154-009-0023-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radha M. H., Laxmipriya N. P. Evaluation of biological properties and clinical effectiveness of aloe vera: a systematic review. Journal of Traditiola and Complementry Medicines. 2014;5(1):21–26. doi: 10.1016/j.jtcme.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boudreau M. D., Mellick P. W., Olson G. R., Felton R. P., Thorn B. T., Beland F. A. Clear evidence of carcinogenic activity by a whole-leaf extract of Aloe barbadensis Miller (Aloe vera) in F344/N rats. Toxicological Sciences. 2013;131(1):26–39. doi: 10.1093/toxsci/kfs275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ode O., Asuza I., Ajayi I. Bioassay-guided fractionation of the crude methanol extract of cassia singueana leaves. Journal of Advanced Scientific Research. 2011;2 [Google Scholar]
- 27.Clarke G., Ting K., Wiart C., Fry J. High correlation of 2,2-diphenyl-1-picrylhydrazyl (DPPH) radical scavenging, ferric reducing activity potential and total phenolics content indicates redundancy in use of all three assays to screen for antioxidant activity of extracts of plants from the malaysian rainforest. Antioxidants. 2013;2(1):1–10. doi: 10.3390/antiox2010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang C. C., Yang M. H., Wen H. M., Chern J. C. Estimation of total flavonoid content in propolis by two complementary colorimetric methods. J Food Drug Anal. 2002;10 [Google Scholar]
- 29.McCune L. M., Johns T. Antioxidant activity in medicinal plants associated with the symptoms of diabetes mellitus used by the Indigenous Peoples of the North American boreal forest. Journal of Ethnopharmacology. 2002;82(2-3):197–205. doi: 10.1016/s0378-8741(02)00180-0. [DOI] [PubMed] [Google Scholar]
- 30.Oktay M., Gülçin I., Küfrevioglu Ö. I. Determination of in vitro antioxidant activity of fennel (Foeniculum vulgare) seed extracts. LWT - Food Science and Technology. 2003;36(2):263–271. doi: 10.1016/S0023-6438(02)00226-8. [DOI] [Google Scholar]
- 31.Jayanthi P., Lalitha P. Reducing power of the solvent extracts of Eichhornia crassipes (Mart.) Solms. International Journal of Pharmacy and Pharmaceutical Sciences. 2011;3(3):126–128. [Google Scholar]
- 32.Ullah R., Hussain I., Ahmad S. Phytochemical and Biological Evaluation of Phlomis bracteosa: A Review. Life Science Journal. 2013;10 [Google Scholar]
- 33.Prieto P., Pineda M., Aguilar M. Spectrophotometric quantitation of antioxidant capacity through the formation of a phosphomolybdenum complex: specific application to the determination of vitamin E. Analytical Biochemistry. 1999;269(2):337–341. doi: 10.1006/abio.1999.4019. [DOI] [PubMed] [Google Scholar]
- 34.Wiegand I., Hilpert K., Hancock R. E. W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nature Protocols. 2008;3(2):163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 35.Akhtar N., Mirza B. Phytochemical analysis and comprehensive evaluation of antimicrobial and antioxidant properties of 61 medicinal plant species. Arabian Journal of Chemistry. 2015 [Google Scholar]
- 36.Khan I., Yasinzai M. M., Mehmood Z., Ilahi I., Khan J. Comparative study of green fruit extract of melia azedarach linn. with its ripe fruit extract for antileishmanial, larvicidal, antioxidant and cytotoxic activity. American Journal of Phytomedicine and Clinical Therapeutics. 2014;2:442–454. [Google Scholar]
- 37.Apu A. S., Bhuyan S. H., Khatun F., Liza M. S., Matin M. Assessment of cytotoxic activity of two medicinal plants using brine shrimp (Artemia salina) as an experimental tool. International Journal of Pharmaceutical Sciences and Research. 2013;4:1125–1130. [Google Scholar]
- 38.Yao G., Sebisubi F. M., Voo L. Y. C., Ho C. C., Tan G. T., Chang L. C. Citrinin derivatives from the soil filamentous fungus Penicillium sp. H9318. Journal of the Brazilian Chemical Society. 2011;22(6):1125–1129. doi: 10.1590/S0103-50532011000600018. [DOI] [Google Scholar]
- 39.Muhammad S. A., Ahmed S., Ali A., et al. Prioritizing drug targets in Clostridium botulinum with a computational systems biology approach. Genomics. 2014;104(1):24–35. doi: 10.1016/j.ygeno.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 40.Kammoun M., Miladi S., Ali Y. B., Damak M., Gargouri Y., Bezzine S. In vitro study of the PLA2 inhibition and antioxidant activities of Aloe vera leaf skin extracts. Lipids in Health and Disease. 2011;10, article no. 30 doi: 10.1186/1476-511X-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siahpoosh A., Dehdari S. Polyphenolic contents and antioxidant activities of leaves of phoenix dactylifera and flowers of aloe vera. Intetnational Journal of Biolosciences. 2014;5:294–304. [Google Scholar]
- 42.Moniruzzaman M., Rokeya B., Ahmed S., Bhowmik A., Khalil M. I., Gan S. H. In vitro antioxidant effects of aloe barbadensis miller extracts and the potential role of these extracts as antidiabetic and antilipidemic agents on streptozotocin-induced type 2 diabetic model rats. Molecules. 2012;17(11):12851–12867. doi: 10.3390/molecules171112851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petti S., Scully C. Polyphenols, oral health and disease: a review. Journal of Dentistry. 2009;37(6):413–423. doi: 10.1016/j.jdent.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 44.Hu Q., Hu Y., Xu J. Free radical-scavenging activity of Aloe vera (Aloe barbadensis Miller) extracts by supercritical carbon dioxide extraction. Food Chemistry. 2005;91(1):85–90. doi: 10.1016/j.foodchem.2004.05.052. [DOI] [Google Scholar]
- 45.Saritha V. Antioxidant and antibacterial activity of Aloe vera gel extracts. International Journal of Pharmaceutical and Biological Archive. 2010;1 [Google Scholar]
- 46.Miladi S., Damak M. In vitro antioxidant activities of aloe vera leaf skin extracts. J de la Société Chimique de Tunisie. 2008;10:101–109. doi: 10.1186/1476-511X-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qader S. W., Abdulla M. A., Chua L. S., Najim N., Zain M. M., Hamdan S. Antioxidant, total phenolic content and cytotoxicity evaluation of selected Malaysian plants. Molecules. 2011;16(4):3433–3443. doi: 10.3390/molecules16043433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Antonisamy J., Beaulah N., Laju R., Anupriya G. Anti-Bacterial and Antifungal activity of Aloe Vera gel extract. International Journal of Biomedical and Advance Research. 2012;3(3):184–187. [Google Scholar]
- 49.Lawrence R., Tripathi P., Jeyakumar E. Isolation, Purification and Evaluation of Antibacterial Agents from Aloe vera. Brazilian Journal of Microbiology. 2009;40(4):906–915. doi: 10.1590/S1517-83822009000400023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stanley M., Ifenyi O., Eziokwu O. Antimicrobial effects of Aloe vera on some human pathogens. International Journal of Current Microbiology and Applied Sciences. 2014;3:1022–1028. [Google Scholar]
- 51.Khaing T. A. Evaluation of the antifungal and antioxidant activities of the leaf extract of aloe vera (aloe barbadensis miller. Proceedings of World Academy of Science, Engineering and Technology. 2011;75:610–612. [Google Scholar]
- 52.Sitara U., Hassan N., Naseem J. Antifungal activity of Aloe vera gel against plant pathogenic fungi. Pakistan Journal of Botany. 2011;43(4):2231–2233. [Google Scholar]
- 53.Dutta A., Mandal G., Mandal C., Chatterjee M. In vitro antileishmanial activity of Aloe vera leaf exudate: a potential herbal therapy in leishmaniasis. Glycoconjugate Journal. 2007;24(1):81–86. doi: 10.1007/s10719-006-9014-z. [DOI] [PubMed] [Google Scholar]
- 54.Dalimi A., Delavari M., Ghaffarifar F., Sadraei J. In vitro and in vivo antileishmanial effects of aloe-emodin on Leishmania major. Journal of Traditional and Complementary Medicine. 2015;5(2):96–99. doi: 10.1016/j.jtcme.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Queiroz A. C. D., Dias T. D. L. F., Matta C. B. B. D., Silva L. H. A. C., Araújo-Júnior J. X. D. Antileishmanial activity of medicinal plants used in endemic areas in northeastern brazil. Evidence-Based Complementary and Alternative Medicine. 2014;2014:9. doi: 10.1155/2014/478290.478290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meyer B. N., Ferrigni N. R., Putnam J. E., Jacobsen L. B., Nichols D. E., McLaughlin J. L. Brine shrimp: a convenient general bioassay for active plant constituents. Planta Medica. 1982;45(5):31–34. doi: 10.1055/s-2007-971236. [DOI] [PubMed] [Google Scholar]
- 57.Shalabi M., Khilo K., Zakaria M. M., Elsebaei M. G., Abdo W., Awadin W. Anticancer activity of Aloe vera and Calligonum comosum extracts separetely on hepatocellular carcinoma cells. Asian Pacific Journal of Tropical Biomedicine. 2015;5(5):375–381. doi: 10.1016/S2221-1691(15)30372-5. [DOI] [Google Scholar]
- 58.Chen S., Lin K., Chang C., Fang C., Lin C. Aloe-emodin-induced apoptosis in human gastric carcinoma cells. Food and Chemical Toxicology. 2007;45(11):2296–2303. doi: 10.1016/j.fct.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 59.Husain S., Alam MA., Jahan N., Ahmed S., KauserSibr H. S. Aloe vera and its therapeutic efficacy described in Unani Medicine: a review. Journal of Scientific and Innovative Research. 2014;3(5):545–551. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are included within the article.