Abstract
Background
Antibiotic resistance is a global challenge in the public health sector and also a major challenge in Ethiopia. It is truly difficult to report bacterial antibiotic resistance pattern in Ethiopia due to the absence of a review which is done comprehensively. The aim of this systematic review was to provide an overview of the works of literature on the antibiotic resistance pattern of the specific bacterial isolates that can be obtained from different clinical samples in the context of Ethiopia.
Materials and Methods
A web-based search using PubMed, Google Scholar, Hinari, Sci Hub, Scopus and the Directory of Open Access Journals was conducted from April to May 2018 for published studies without restriction in the year of publication. Works of literature potentially relevant to the study were identified by Boolean search technique using various keywords: Bacterial infection, antimicrobial resistance, antibiotic resistance, drug resistance, drug susceptibility, anti-bacterial resistance, Ethiopia. Study that perform susceptibility test from animal or healthy source using <10 isolates and methods other than prospective cross-sectional were excluded.
Results
The database search delivered a total of 3459 studies. After amendment for duplicates and inclusion and exclusion criteria, 39 articles were found suitable for the systematic review. All studies were prospective cross-sectional in nature. The review encompasses 12 gram-positive and 15 gram-negative bacteria with their resistance pattern for around 12 antibiotics. It covers most of the regions which are found in Ethiopia. The resistance pattern of the isolates ranged from 0% up to 100%. The overall resistance of M. tuberculosis for antituberculosis drugs ranges from 0% up to 32.6%. The percentage of resistance increases among previously treated tuberculosis cases. Neisseria gonorrhea, S. typhimurium, S. Virchow, Group A Streptococci (GAS), and Group B Streptococci (GBS) were highly susceptible for most of the tested antibiotics. Methicillin-Resistant Staphylococcus aureus was highly resistant to most of the antibiotics with a slightly increased susceptibility to gentamycin.
Conclusions
Total bacterial isolates obtained from a different source of sample and geographic areas were 28, including M. tuberculosis. Majority of the bacterial isolates were resistant to commonly used antibiotics. A continuous monitoring and studies on the multidrug-resistant bacterial isolates are important measures.
1. Introduction
Human beings have been living unfriendly with a lot of microorganisms that can be a potential cause of infections and diseases. In the case of bacterial infections, due to the introduction of Penicillin for treatment in the early 1940s, there was an improvement [1]. Majority of naturally derived antibiotics are produced from Actinomycetes [2, 3]. In this day, even though the struggle to defeat bacterial pathogens continues, bacteria are evolving ever more clever by manifesting different forms of resistance [4].
The current antimicrobial profile studies have been proved that, bacteria that can cause nosocomial as well as community acquired infections become pan resistant for different groups of antibiotics. Hence, this situation becomes a clinical threat to the human beings [5–13]. Most of the bacterial antibiotic resistance mechanisms are acquired by altering of target genes or acquisition of plasmid encoding resistance genes. These encoded genes may lead to the production of lytic enzymes, change of membrane permeability, efflux action, and hiding from the action of antibiotics [14].
Centers for Disease Control and Prevention (CDC) stated that antibiotic resistance is responsible for around 2 million infections, more than twenty thousand deaths and, costs $55 billion each year in the United States [15]. The national pharmaceutical sales data on global antibiotic consumption (2000-2010) reveals that total antibiotic consumption grew by more than 30%. The greatest increase in antibiotics use was recorded in Low and Middle Income Countries (LMIC) [16].
As long as Ethiopia is one of the LMICs, antibiotic resistance is a major challenge. Yet, there is no antibiotic stewardship that helps to establish surveillance system for tracking current antibiotic use and its resistance in Ethiopia. Therefore, it is truly difficult to report bacterial antibiotic resistance pattern in Ethiopia. So, the aim of this systematic review emphasizes on the antibiotic resistance pattern of the specific bacterial isolates that can be obtained from different clinical samples in the context of Ethiopia.
2. Materials and Methods
2.1. Eligibility Criteria
All available studies and data were incorporated based on the following predefined eligibility criteria:
should be published and written in English,
had to describe the microbial isolation, identification, and antimicrobial sensitivity test methods according to the criteria of the Clinical Laboratory Standards Institute (CLSI) and defined antimicrobial resistance range according to CLSI manual,
studies which used human infection sample (isolated from a diseased individual),
had to report the number of tested isolates (>10) and the number of isolates resistant or sensitive,
should be a prospective cross-sectional study.
2.2. Study Selection Procedure and Search
The search and selection of eligible studies were shown in Figure 1. To make it more elaborative; a web-based search using PubMed, Google Scholar, Hinari, Sci Hub, Scopus, and the Directory of Open Access Journals (DOAJ) was conducted from April to May 2018 and pieces of literature potentially relevant to the study were identified. The search was performed using various keywords: Bacterial infection, antimicrobial resistance, antibiotic resistance, drug resistance, drug susceptibility, anti-bacterial resistance, Ethiopia. These key terms were used in various combinations using Boolean search technique. The search was not limited to the year of publication.
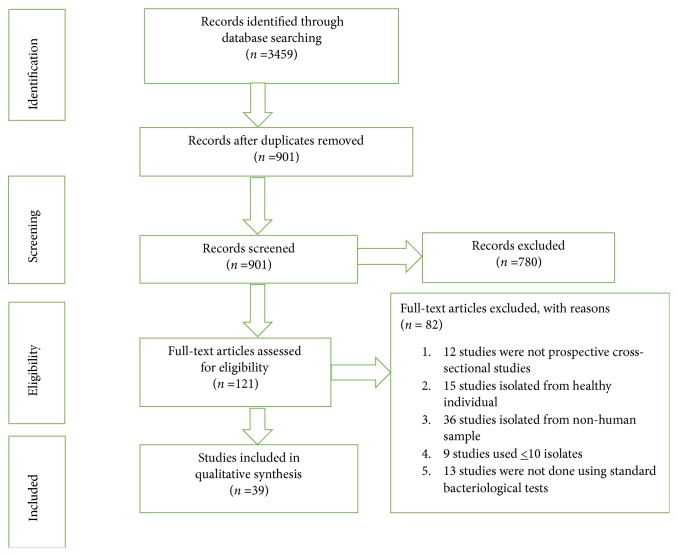
Figure 1.
A flow diagram of the selection of eligible studies. The flow diagram shows the procedure of selecting an eligible study to undergo the systematic review. To perform this we start by identifying 3459 studies using a web-based search and goes to a screening of 901 studies after removing duplicates. Using eligibility criteria only 121 studies went to eligibility testing. Finally, 39 studies were included.
Relevant search results from the above site were individually downloaded and the reference lists of the identified studies were used to scrutinize to identify extra articles.
2.3. Data Extraction
Essential data were extracted from eligible studies using Excel spreadsheet format prepared for this purpose and any discrepancies were resolved by the author. The data extracted from eligible studies include the name of regions, study area/city, the name of the author(s), year of the study, study design, types of specimens, numbers of patients/study participants, number of resistant isolates, resistance pattern of the isolates, and references.
3. Results
3.1. Literature Search Results
The search from PubMed, Google Scholar, Hinari, Sci Hub, Scopus, and DOAJ delivered a total of 3459 studies. After amendment for duplicates, 901 endured. Of these, 780 studies were castoff, since, after a review of their titles and abstracts, they did not meet the criteria. The full texts of the remaining 121 studies were reviewed in detail. Of these, 85 studies were discarded after the full text had been reviewed for appropriate study method, sample source, number of isolates, and standard bacteriological test. Finally, 39 studies were included in the review (Figure 1).
3.2. Antibiotic Resistance for Gram-Positive Bacteria
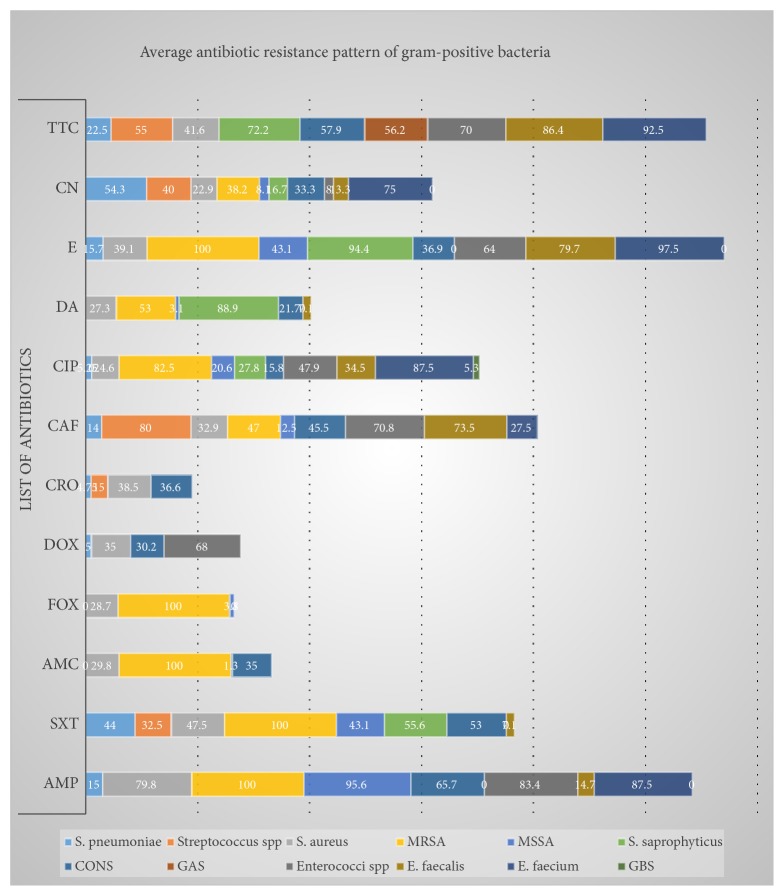
The review tries to encompass 12 gram-positive bacteria and their resistance pattern for around 12 antibiotics. It covers most of the regions which are found in Ethiopia. The resistance pattern of the isolates ranged from 0% up to 100%. GAS and GBS were highly susceptible for most of the tested antibiotics, but they have a relatively increased resistance to tetracycline. In contrast with these, MRSA was highly resistant for most of the antibiotics with a slightly increased susceptibility to gentamycin. The rest bacterial isolates have a different resistance pattern for different antibiotics and also a variable pattern from sample to sample (Table 1). The average resistance pattern of gram-positive bacteria shows a cumulative antibiotic resistance pattern of bacterial isolates from different clinical cases, geographic location, and source of sample for a similar antibiotic. Still, the average resistance range of the gram-positive bacteria is similar with the detailed resistance pattern (Figure 2).
Table 1.
Antibiotic resistance for gram-positive bacteria.
| Microorganisms | Type of sample | Study area | No of isolates | Antibiotics and resistance (%) | Reference | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | SXT | AMC | FOX | DOX | CRO | CAF | CIP | DA | E | CN | TTC | |||||
| S. pneumoniae | Nasal swab | GUH | 96 | 10 | 22 | - | 0 | - | 0 | 14 | 2 | - | 32 | - | 32 | [20] |
| Conjunctival swabs | West Gojjam | 25 | - | 44 | - | 4 | 32 | 4 | 8 | 8 | [21] | |||||
| Ocular swab | JUSH | 20 | 65 | 5 | 15 | 0 | 5 | 10 | 85 | 20 | [22] | |||||
| Ocular swab | HUH | 20 | 20 | 45 | 0 | 0 | 10 | 10 | 5 | 70 | 30 | [23] | ||||
|
| ||||||||||||||||
| Streptococcus spp | Conjunctival swabs | West Gojjam | 40 | 32.5 | 15 | 80 | 0 | 40 | 55 | [21] | ||||||
|
| ||||||||||||||||
| S. aureus | Urine | Bahir Dar | 10 | 100 | 87.5 | 33.3 | 50 | 60 | [24] | |||||||
| Blood | Mekelle | 54 | 66.7 | 13 | 53.7 | 57.4 | 38.9 | 44.4 | 33.3 | [25] | ||||||
| Nasal swab | Adigrat, Wukro | 29 | 48.3 | 51.7 | 48.3 | 17.2 | 37.9 | 17.2 | 62.1 | 37.9 | 55.2 | [26] | ||||
| Ocular swab | JUSH | 42 | 21.4 | 19.1 | 23.8 | 57.1 | 4.8 | 35.7 | 4.8 | 21.4 | [22] | |||||
| Urine | GUH | 28 | 100 | 100 | 100 | 50 | 100 | 50 | 0 | [27] | ||||||
| Wound | JUSH | 73 | 94.5 | 60.3 | 28.8 | 82.2 | 20.5 | 69.9 | 13.7 | 85 | 79.4 | 16.5 | [28] | |||
| Ear discharge | HUCSH | 48.5 | 18.2 | [29] | ||||||||||||
| Wound | JUSH | 47 | 95.7 | 6 | 27.6 | 14.9 | 14.9 | 4 | 14.9 | 4 | 51 | [30] | ||||
| Pus∗ | GGH | 15 | 80 | 26.7 | 26.7 | 20 | 0 | 40 | 20 | 20 | 20 | 26.7 | 0 | 13.3 | [31] | |
| Blood | GUH | 17 | 47 | 58.3 | 35.3 | 23.5 | 29.4 | 58.8 | 29.4 | 35 | [32] | |||||
| Eye discharge | GUH | 13 | 76.9 | 69.2 | 53.8 | 23.1 | 30.8 | 15.4 | 46.2 | 46.2 | 61.6 | [33] | ||||
| Wound | Mekelle | 40 | 90 | 50 | 90 | 22.5 | 22.2 | 90 | [34] | |||||||
| Ocular swab | HUH | 30 | 70 | 23.3 | 10 | 6.7 | 6.7 | 6.7 | 16.7 | 6.7 | 60 | [23] | ||||
| Ocular specimen | QOH | 40 | 42.5 | 27.5 | 27.5 | 12.5 | 22.5 | 20 | 12.5 | 45 | [35] | |||||
| Ocular specimen | GUH | 96 | 92.7 | 7.3 | 17.7 | 20.8 | 11.5 | 7.3 | 7.3 | 28.1 | 28.1 | 27.1 | [36] | |||
| Conjunctival swabs | West Gojjam | 120 | 66 | 38 | 30 | 80 | 13 | 16.6 | 21 | [21] | ||||||
| Specimen∗ | YHMC | 194 | 96.4 | 53.1 | 18.5 | 17.5 | 18.6 | 31.4 | 11.9 | 53.1 | 13.4 | [37] | ||||
|
| ||||||||||||||||
| MRSA | Specimen∗ | YHMC | 34 | 100 | 100 | 100 | 100 | 47 | 82.5 | 53 | 100 | 38.2 | [37] | |||
|
| ||||||||||||||||
| MSSA | Specimen∗ | YHMC | 160 | 95.6 | 43.1 | 1.3 | 3.8 | 12.5 | 20.6 | 3.1 | 43.1 | 8.1 | [37] | |||
|
| ||||||||||||||||
| S. saprophyticus | Urine | AAML | 18 | 55.6 | 27.8 | 88.9 | 94.4 | 16.7 | 72.2 | [38] | ||||||
|
| ||||||||||||||||
| CONS | Blood | GUH | 30 | 40 | 53.3 | 36.6 | 40 | 23.3 | 30 | 30 | 73.3 | [32] | ||||
| Ocular swab | JUSH | 15 | 46.6 | 6.7 | 26.7 | 73.4 | 0 | 26.7 | 53.3 | 33.3 | [22] | |||||
| Eye discharge | GUH | 17 | 76.5 | 70.6 | 64.7 | 23.5 | 52.9 | 23.5 | 64.7 | 58.8 | 64.7 | [33] | ||||
| Blood | Mekelle | 44 | 81.8 | 11.4 | 61.4 | 50 | 25 | 40.9 | 18.1 | [25] | ||||||
| Ocular specimen | GUH | 64 | 89.1 | 23.4 | 17.2 | 17.2 | 15.6 | 10.9 | 4.7 | 42.2 | 26.6 | 32.8 | [36] | |||
| Wound | Mekelle | 18 | 77.8 | 77.8 | 72.3 | 50 | 50 | 77.8 | [34] | |||||||
| Ocular swab | HUH | 26 | 30.8 | 38.5 | 3.8 | 26.7 | 26.9 | 18.2 | 11.5 | 23.1 | 65.4 | [23] | ||||
| Conjunctival swabs | West Gojjam | 110 | 80 | 41.8 | 40 | 81 | [21] | |||||||||
| Ocular specimen | QOH | 31 | 67.7 | 22.6 | 29 | 9.7 | 38.7 | 29 | 6.5 | 58.1 | [35] | |||||
|
| ||||||||||||||||
| GAS | Throat swab | Addis Ababa | 52 | 0 | 0 | 0 | 0 | 0 | 34.6 | [39] | ||||||
| Dire Dawa | 16 | 0 | 0 | 0 | 0 | 0 | 75 | [39] | ||||||||
| Gondar | 22 | 0 | 0 | 0 | 0 | 0 | 59.1 | [39] | ||||||||
|
| ||||||||||||||||
| Enterococci spp | Conjunctival swabs | West Gojjam | 25 | 100 | 25 | 8 | 72 | [21] | ||||||||
| Urine, wound, blood | GUH | 24 | 66.7 | 68 | 70.8 | 70.8 | 64 | 68 | [40] | |||||||
|
| ||||||||||||||||
| E. faecalis | Urine | AAML | 14 | 7.1 | 73.5 | 7.1 | 7.1 | 85.8 | 0 | 78.6 | [38] | |||||
| Stool, rectal swab | Jimma | 34 | 14.7 | 73.5 | 61.8 | 73.5 | 26.5 | 94.1 | [41] | |||||||
|
| ||||||||||||||||
| E. faecium | Stool, rectal swab | Jimma | 40 | 87.5 | 27.5 | 87.5 | 97.5 | 75 | 92.5 | [41] | ||||||
|
| ||||||||||||||||
| GBS | Vaginal swab | Mekelle | 19 | 0 | 5.3 | 0 | 0 | [42] | ||||||||
Specimen∗: Nasal swab, pus from wound, ear discharge, blood, throat swab, eye swab, vaginal discharge, urethral discharge, urine, stool, sputum, CSF, and body fluids; Pus∗: collected from leprosy ulcer; AMP: Ampicillin; SXT: Trimethoprim-Sulfamethoxazole; AMC: Amoxicillin-Clavulanic acid; Dox: Doxycycline; CRO: Ceftriaxone; CAF: Chloramphenicol; CIP: Ciprofloxacin; DA: Clindamycin; E: Erythromycin; CN: Gentamicin; TTC: Tetracycline; FOX: Cefoxitin; GUH: Gonder University Hospital; JUSH: Jimma University Specialized Hospital; CONS: Coagulase-negative Staphylococcus; HUH: Hawassa University Hospital; GAS: Group A Streptococcus; GBS: Group B Streptococcus YHMC: Yekatit 12 Hospital Medical College; QOH: Quiha Ophthalmic Hospital; AAML: Arsho Advanced Medical laboratory; HUCSH: Hawassa University Comprehensive Specialized Hospital MRSA: Methicillin-Resistant Staphylococcus aureus; MSSA: Methicillin-Sensitive Staphylococcus aureus
Figure 2.
Average resistance pattern of gram-positive bacteria for different antibiotics.
3.3. Antibiotic Resistance for Gram-Negative Bacteria
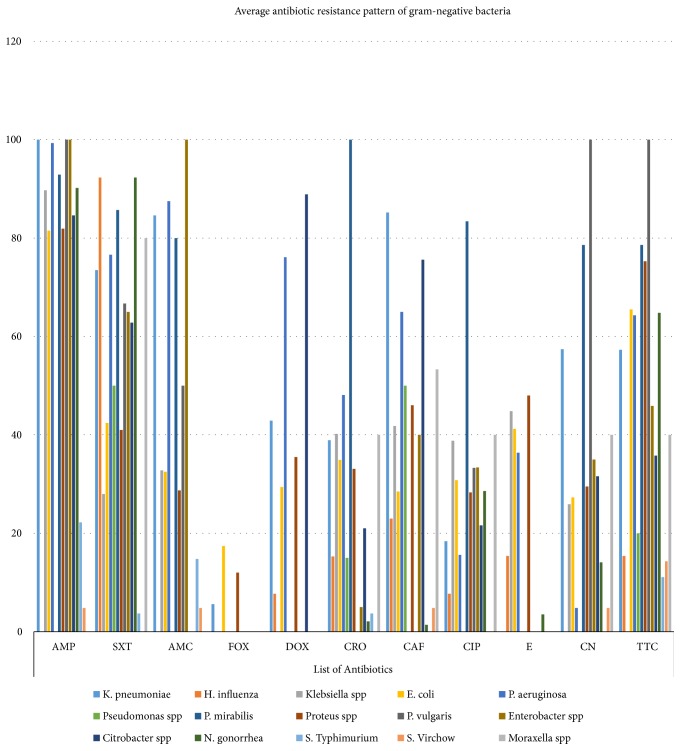
Fifteen gram-negative bacteria were recovered from various specimens. Like the gram-positive bacteria, the resistance pattern ranges from 0% up to 100%. Almost all bacterial isolates were highly resistant for ampicillin. Relatively, isolates obtained from conjunctival swab were highly susceptible to different antibiotics. Neisseria gonorrhea, S. typhimurium, and S. Virchow were susceptible for many antibiotics. Moraxella spp. and S. Virchow had a similar resistance pattern for different antibiotics, even if the number of isolates varies between the two (Table 2). The average resistance pattern of gram-negative bacteria shows a cumulative antibiotic resistance pattern of bacterial isolates from different clinical cases, geographic location, and source of sample for a similar antibiotic. Still, the average resistance range of the gram-negative bacteria is similar with the detailed resistance pattern (Figure 3).
Table 2.
Antibiotic resistance for gram-negative bacteria.
| Microorganisms | Type of sample | Study area | No of isolates | Antibiotics and resistance (%) | Reference | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | SXT | AMC | FOX | DOX | CRO | CAF | CIP | E | CN | TTC | |||||
| K. pneumoniae | sputum, urine, pus | Harar | 57 | 65 | 40 | 70 | 61 | [43] | |||||||
| Urine | Bahir Dar | 17 | 100 | 50 | 84.6 | 21 | 40 | 27.8 | [24] | ||||||
| Urine | AAML | 18 | 100 | 66.7 | 5.6 | 44.4 | 16.7 | 22.2 | 44.4 | [38] | |||||
| Urine | GUH | 28 | 100 | 100 | 0 | 100 | 0 | 100 | 100 | [27] | |||||
| Wound | JUSH | 14 | 100 | 85.7 | 42.9 | 71 | 85.7 | 35.7 | 64 | 57 | [30] | ||||
|
| |||||||||||||||
| H. influenza | Ocular swab | JUSH | 92.3 | 7.7 | 15.3 | 23 | 7.7 | 15.4 | 0 | 15.4 | [22] | ||||
|
| |||||||||||||||
| Klebsiella spp | Wound | Mekelle | 29 | 89.7 | 65.5 | 86.2 | 37.9 | 44.8 | 27.8 | [34] | |||||
| Conjunctival swab | West Gojjam | 20 | 28 | 20 | 55 | 0 | 0 | 0 | [21] | ||||||
| Urine | AAHFH | 14 | 0 | 14.3 | 28.6 | 78.6 | 50 | [44] | |||||||
|
| |||||||||||||||
| E. coli | Urine | Bahir Dar | 64 | 89.1 | 64.5 | 78.6 | 64.4 | 27.6 | 66.1 | [24] | |||||
| Blood | Mekelle | 16 | 6.7 | 6.7 | 40 | 60 | 6.7 | 13.3 | [25] | ||||||
| Urine | GUH | 28 | 66.7 | 66.7 | 50 | 16.7 | 50 | 66.7 | 100 | [27] | |||||
| Vaginal swab | FRH | 15 | 73.3 | 60 | 20 | 26.7 | 73.3 | [45] | |||||||
| Wound | JUSH | 30 | 76.7 | 20 | 23.3 | 66.7 | 13.3 | 3.3 | 0 | [28] | |||||
| Wound | JUSH | 29 | 100 | 55 | 44.8 | 62 | 65.5 | 34 | 51.7 | 79 | [30] | ||||
| Urine | AAML | 135 | 77.8 | 70.4 | 45.2 | 22.9 | 34.8 | 50.4 | 28.1 | 69.6 | [38] | ||||
| Urine | GUH | 19 | 100 | 26.3 | 36.8 | 0 | 0 | 0 | 5.3 | 52.6 | [18] | ||||
| Urine | StHMC | 53 | 79.2 | 22.6 | 71.7 | 45.3 | 30.2 | 54.7 | 22.6 | 83 | [46] | ||||
| Conjunctival swab | West Gojjam | 20 | 15 | 0 | 35 | 0 | 20 | 25 | [21] | ||||||
| Pus∗ | GGH | 17 | 70.6 | 58.8 | 5.9 | 11.8 | 29.4 | 5.9 | 35.3 | 29.4 | 41.2 | 11.8 | 41.2 | [31] | |
| Urine | AAHFH | 65 | 21.6 | 24.6 | 32.2 | 56.9 | 53.8 | [44] | |||||||
|
| |||||||||||||||
| P. aeruginosa | Urine | Bahir Dar | 8 | 100 | 71.4 | 75 | 0 | 0 | 40 | [24] | |||||
| Wound | JUSH | 74 | 97.3 | 87.9 | 83.3 | 9.5 | 74.3 | 5.4 | 10.8 | [28] | |||||
| Ocular swab | JUSH | 31 | 74.2 | 45.1 | 19.4 | 38.7 | 6.4 | 0 | 71 | [22] | |||||
| Wound | JUSH | 11 | 100 | 73 | 100 | 63.6 | 82 | 0 | 18 | 82 | [30] | ||||
| Urine | JUSH | 36 | 0 | 0 | [47] | ||||||||||
| Wound | Mekelle | 11 | 100 | 100 | 100 | 81.8 | 36.4 | 0 | [34] | ||||||
|
| |||||||||||||||
| Pseudomonas spp | Conjunctival swabs | West Gojjam | 20 | 50 | 15 | 50 | 0 | 0 | 20 | [21] | |||||
|
| |||||||||||||||
| P. mirabilis | Urine | Bahir Dar | 7 | 85.7 | 71.4 | 80 | 66.7 | 57.1 | 57.1 | [24] | |||||
| Urine | GUH | 28 | 100 | 100 | 100 | 0 | 100 | 100 | 100 | [27] | |||||
|
| |||||||||||||||
| Proteus spp | Wound | JUSH | 23 | 91 | 39 | 43 | 65 | 30 | 17 | 26 | 74 | [30] | |||
| Wound | Mekelle | 15 | 86.7 | 46.7 | 73.3 | 46.7 | 20 | [34] | |||||||
| Pus∗ | GGH | 25 | 68 | 56 | 20 | 12 | 28 | 8 | 36 | 32 | 48 | 32 | 96 | [31] | |
| Conjunctival swabs | West Gojjam | 25 | 28 | 0 | 76 | 0 | 8 | 56 | [21] | ||||||
| Urine | AAHFH | 31 | 19.4 | 19.4 | 41.9 | 45.9 | 61.3 | [44] | |||||||
|
| |||||||||||||||
| P. vulgaris | Urine | Bahir Dar | 3 | 100 | 66.7 | 50 | 33.3 | 100 | 100 | [24] | |||||
|
| |||||||||||||||
| Enterobacter spp | Urine | Bahir Dar | 3 | 100 | 100 | 100 | 66.7 | 50 | 66.7 | [24] | |||||
| Conjunctival swabs | West Gojjam | 20 | 30 | 5 | 40 | 0 | 20 | 25 | [21] | ||||||
|
| |||||||||||||||
| Citrobacter spp | Urine | GUH | 28 | 69.2 | 38.5 | 46.2 | 76.9 | 53.8 | 61.5 | 61.5 | [27] | ||||
| Wound | JUSH | 18 | 100 | 100 | 88.9 | 16.7 | 100 | 11.1 | 33.3 | [28] | |||||
| Conjunctival swabs | West Gojjam | 20 | 50 | 0 | 50 | 0 | 0 | 10 | [21] | ||||||
|
| |||||||||||||||
| Neisseria gonorrhea | Urethral discharge | Gondar Health Center | 142 | 80.3 | 92.3 | 4.2 | 1.4 | 3.5 | 14.1 | 29.6 | [48] | ||||
| Urethral or endo-cervical swabs | Gambella hospital | 21 | 100 | 0 | 28.6 | 100 | [49] | ||||||||
|
| |||||||||||||||
| S. Typhimurium | Stool | Addis Ababa | 27 | 22.2 | 3.7 | 14.8 | 0 | 3.7 | 0 | 0 | 0 | 11.1 | [50] | ||
|
| |||||||||||||||
| S. Virchow | Stool | Addis Ababa | 21 | 4.8 | 0 | 4.8 | 0 | 0 | 4.8 | 0 | 4.8 | 14.3 | [50] | ||
|
| |||||||||||||||
| Moraxella spp | Conjunctival swabs | West Gojjam | 15 | 80 | 40 | 53.3 | 40 | 40 | 40 | [21] | |||||
GUH: Gonder University Hospital; StHMC: St. Paul's Hospital Millennium Medical College; AAHFH: Addis Ababa Hamlin fistula hospital; FRH: Felegehiwot Referral Hospital; GGH: Gambo General Hospital Pus∗ = collected from leprosy ulcer
Figure 3.
Average resistance pattern of gram-negative bacteria for different antibiotics.
3.4. Drug Resistance of M. tuberculosis
The overall resistance of M. tuberculosis for antituberculosis drugs ranges from 0% up to 32.6%. The percentage of resistance increases among previously treated tuberculosis cases. New pulmonary and extra-pulmonary cases relatively had decreased resistance for antituberculosis drugs (Table 3).
Table 3.
Resistance pattern of M. tuberculosis for anti-tuberculosis drugs.
| Type of tuberculosis case | Type of sample | Study area | No of isolates | Antibiotics and resistance (%) | Reference | ||||
|---|---|---|---|---|---|---|---|---|---|
| H | R | S | E | P | |||||
| New case | Sputum | BLUH | 103 | 8.7 | 1.9 | 7.8 | 0.9 | - | [51] |
| Previously treated | Sputum | BLUH | 18 | 5.6 | 5.6 | 5.6 | 5.6 | - | [51] |
| New case | Sputum | JUSH | 136 | 13.2 | 2.2 | 8.1 | 5.2 | - | [52] |
| New case | Sputum | Amhara region | 93 | 3.2 | 0 | 20.4 | 0 | - | [53] |
| New case | Sputum | Amhara region | 214 | 9.8 | 3.7 | 6.5 | 5.6 | 3.7 | [54] |
| Previously treated | Sputum | Amhara region | 46 | 32.6 | 15.2 | 26.1 | 15.2 | 8.7 | [54] |
| EPTB | pleural, peritoneal and synovial fluids | Addis Ababa | 58 | 8 | 21.6 | 5.4 | 2.7 | - | [56] |
H: isoniazid; R: rifampin; S: streptomycin; E: ethambutol; P: pyrazinamide; EPTB: Extra Pulmonary Tuberculosis; JUSH: Jimma University Specialized Hospital; BLUH: Black Lion University Hospital
4. Discussion
Our results indicate that the antibiotic resistance pattern of both gram-negative and gram-positive bacteria varied across the studies reviewed, ranging from 0% to 100%. This variation was found depending on the type of isolate, the source of the sample, type of infection, type of antibiotics, and geographical difference used in each study.
Even though it is difficult to discuss average resistance pattern of gram positive and gram negative bacteria with a single study for various antibiotics, a study done in Nigeria revealed that gram-negative and gram-positive bacteria had a resistance pattern of 19.8%-92.3% and 10%-87%, respectively [17]. And also a study in Gondar, Northwest Ethiopia, showed 20%–100% and 23.5%–58.8% for gram-negative and gram-positive bacteria, respectively [18]. If we look at the overall resistance pattern of the above studies, it ranges from 10% to 100%. This was relatively comparable to the current review result.
A systematic review done on antimicrobial resistance in Sub-Saharan Africa (1990–2013) has been reported that Gram-positive pathogens show high prevalence of resistance to chloramphenicol, trimethoprim/sulfamethoxazole, and tetracycline and gram negative bacteria specifically the enterobacteriaceae groups show resistance to chloramphenicol, isolated from patients with a febrile illness, ranged between 31.0% and 94.2%, whereas resistance to third-generation cephalosporins ranged between 0.0% and 46.5% [13].
The current review revealed that majority of the bacterial isolates are resistant to commonly used antibiotics in Ethiopia [19–54]. The possible reason might be related to scientific justifications like the following: numerous antibacterial agents, effective previously, are no longer used today because of the rise of resistance genes in the bacterial genome [55]. The emergence of resistance genes can be through natural selection in the environment over a long period of time or by a spontaneous mutation in the bacterial DNA. The resistant pattern has been reported by almost all antibiotics that have been developed [19].
5. Limitations of the Study
Most of the studies done in Ethiopia do not document the antibiogram; due to this we were unable to review and extract data related to multidrug resistance.
6. Conclusions
The main result of this study is to obtain an internationally valid reference to know the antibiotic resistance pattern in Ethiopia. The review encompasses 12 gram-positive bacteria and their resistance pattern for around 12 antibiotics. It covers most of the regions which are found in Ethiopia. The resistance pattern of the isolates ranged from 0% up to 100%. Fifteen gram-negative bacteria were recovered from various specimens. Like the gram-positive bacteria, the resistance pattern ranges from 0% up to 100%. Almost all bacterial isolates were highly resistant for ampicillin. Relatively, isolates obtained from conjunctival swab were highly susceptible to different antibiotics. Neisseria gonorrhea, S. typhimurium, and S. Virchow were susceptible for many antibiotics. The overall resistance of M. tuberculosis for antituberculosis drugs ranges from 0% up to 32.6%. Given the limitations of the current study, these findings should be interpreted carefully but warrant further evaluation in consequent studies.
7. Recommendations
Our study provides evidence that majority of the bacterial isolates were resistant to commonly used antibiotics. Antibiotic resistance should be a substantial concern for Ethiopia as well as for all countries around the globe. So to alleviate such problems and to advance the effectiveness of antibiotics in Ethiopia, the government of Ethiopia as well as the international community should do the following:
Prepare the guidelines for proper use of antibiotics in the health institutions.
Establish antimicrobial resistance stewardship.
Increase proper immunization coverage that may reduce the use of antibiotics.
Implement one health policy (reduce antimicrobial use for agricultural practice and animals).
Create community awareness on rational use of drugs.
Make strong policy for antibiotic dispensing by drug venders.
Control Hospital and community acquired infections.
Ensure political commitment to meet the threat of antibiotic resistance.
Acknowledgments
We would like to acknowledge Debre Markos University Digital Library staffs for their contribution during article searching.
Conflicts of Interest
The authors declare that they have no competing interests.
Authors' Contributions
Alemayehu Reta carried out interpretation of data, drafted the manuscript, revised it critically for important intellectual content, and gave final approval of the version to be published. Abebaw Bitew Kifilie revised it critically and searched literatures. Abeba Mengist revised it critically and searched literatures. All authors have read and approved the final manuscript.
References
- 1.Tenover F. C. Mechanisms of antimicrobial resistance in bacteria. American Journal of Infection Control. 2006;34(5):S3–S10. doi: 10.1016/j.ajic.2006.05.219. [DOI] [PubMed] [Google Scholar]
- 2.Pimentel-Elardo S. M., Kozytska S., Bugni T. S., Ireland C. M., Moll H., Hentschel U. Anti-parasitic compounds from streptomyces sp. strains isolated from mediterranean sponges. Marine Drugs. 2010;8(2):373–380. doi: 10.3390/md8020373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dahal R. H., Shim D. S., Kim J. Development of actinobacterial resources for functional cosmetics. Journal of Cosmetic Dermatology. 2017;16(2):243–252. doi: 10.1111/jocd.12304. [DOI] [PubMed] [Google Scholar]
- 4.Fair R. J., Tor Y. Antibiotics and bacterial resistance in the 21st century . Perspectives in Medicinal Chemistry. 2014;28(6):25–64. doi: 10.4137/PMC.S14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rattan A., Kalia A., Ahmad N. Multidrug-resistant Mycobacterium tuberculosis: molecular perspectives. Emerging Infectious Diseases. 1998;4(2):195–209. doi: 10.3201/eid0402.980207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillespie S. H. Evolution of drug resistance in mycobacterium tuberculosis: clinical and molecular perspective. Antimicrobial Agents and Chemotherapy. 2002;46(2):267–274. doi: 10.1128/AAC.46.2.267-274.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Otto M. Staphylococcus epidermidisthe accidental pathogenInt. Journal of Antimicrobial Agents. 2009;29:S23–S32. [Google Scholar]
- 8.Schaefler S. Staphylococcus epidermidis BV: antibiotic resistance patterns, physiological characteristics, and bacteriophage susceptibility. Journal of Applied Microbiology. 1971;22(4):693–699. doi: 10.1128/am.22.4.693-699.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eady E. A., Gloor M., Leyden J. J. Propionibacterium acnes resistance: a worldwide problem. Dermatology. 2003;206(1):54–56. doi: 10.1159/000067822. [DOI] [PubMed] [Google Scholar]
- 10.Boucher H. W., Talbot G. H., Bradley J. S., et al. Bad bugs, no drugs: no ESKAPE! an update from the infectious diseases society of america. Clinical Infectious Diseases. 2009;48(1):1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 11.Saravanan M., Niguse S., Abdulkader M., et al. Review on emergence of drug-resistant tuberculosis (MDR & XDR-TB) and its molecular diagnosis in Ethiopia. Microbial Pathogenesis. 2018;117:237–242. doi: 10.1016/j.micpath.2018.02.047. [DOI] [PubMed] [Google Scholar]
- 12.Muthupandian S., Balajee R., Barabadi H. The prevalence and drug resistance pattern of extended spectrum β–lactamases (ESBLs) producing enterobacteriaceae in Africa. Microbial Pathogenesis. 2017 doi: 10.1016/j.micpath.2017.11.061. [DOI] [PubMed] [Google Scholar]
- 13.Leopold S. J., van Leth F., Tarekegn H., Schultsz C. Antimicrobial drug resistance among clinically relevant bacterial isolates in sub-Saharan Africa: a systematic review. Journal of Antimicrobial Chemotherapy. 2014;69(9):2337–2353. doi: 10.1093/jac/dku176.dku176 [DOI] [PubMed] [Google Scholar]
- 14.Dever L. A., Dermody T. S. Mechanisms of bacterial resistance to antibiotics. Archives of internal medicine. 1991;151(5):886–895. [PubMed] [Google Scholar]
- 15.Health UDo, Services H. Antibiotic Resistance Threats in The United States, 2013. Georgia, Ga, USA: Centers for Disease Control and Prevention; 2013. [Google Scholar]
- 16.Van Boeckel T. P., Gandra S., Ashok A., et al. Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. The Lancet Infectious Diseases. 2014;14(8):742–750. doi: 10.1016/S1473-3099(14)70780-7. [DOI] [PubMed] [Google Scholar]
- 17.Komolafe A. O., Adegoke A. A. Incidence of bacterial septicaemia in Ile-Ife metropolis, Nigeria. Malaysian Journal of Microbiology. 2008;4(2):51–61. [Google Scholar]
- 18.Alemu A., Moges F., Shiferaw Y., et al. Bacterial profile and drug susceptibility pattern of urinary tract infection in pregnant women at university of gondar teaching hospital, Northwest Ethiopia. BMC Research Notes. 2012;5, article 197(1) doi: 10.1186/1756-0500-5-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martínez J. L., Coque T. M., Baquero F. What is a resistance gene? ranking risk in resistomes. Nature Reviews Microbiology. 2014:1–8. doi: 10.1038/nrmicro3399. [DOI] [PubMed] [Google Scholar]
- 20.Assefa A., Gelaw B., Shiferaw Y., Tigabu Z. Nasopharyngeal carriage and antimicrobial susceptibility pattern of streptococcus pneumoniae among pediatric outpatients at gondar university hospital, north west ethiopia. Pediatrics and Neonatology. 2013;54(5):315–321. doi: 10.1016/j.pedneo.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 21.Abera B., Kibret M. Azithromycin, fluoroquinolone and chloramphenicol resistance of non-chlamydia conjunctival bacteria in rural community of Ethiopia. Indian Journal of Ophthalmology. 2014;62(2):236–239. doi: 10.4103/0301-4738.99974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tesfaye T., Beyene G., Gelaw Y., Bekele S., Saravanan M. Bacterial profile and antimicrobial susceptibility pattern of external ocular infections in jimma university specialized hospital, Southwest Ethiopia. American Journal of Infectious Diseases and Microbiology. 2013;1(1):13–20. doi: 10.12691/ajidm-1-1-3. [DOI] [Google Scholar]
- 23.Aweke T., Dibaba G., Ashenafi K., Kebede M. Bacterial pathogens of exterior ocular infections and their antibiotic vulnerability pattern in southern Ethiopia. African Journal of Immunology Research. 2014;1(1):019–025. [Google Scholar]
- 24.Derbie A., Hailu D., Mekonnen D., Abera B., Yitayew G. Antibiogram profile of uropathogens isolated at bahir dar regional health research laboratory centre, northwest ethiopia. Pan African Medical Journal. 2017;26, article no. 134 doi: 10.11604/pamj.2017.26.134.7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wasihun A. G., Wlekidan L. N., Gebremariam S. A., et al. Bacteriological profile and antimicrobial susceptibility patterns of blood culture isolates among febrile patients in mekelle hospital, Northern Ethiopia. SpringerPlus. 2015;4, article 314(1) doi: 10.1186/s40064-015-1056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Legese H., Kahsay A. G., Kahsay A., et al. Nasal carriage, risk factors and antimicrobial susceptibility pattern of methicillin resistant Staphylococcus aureus among healthcare workers in Adigrat and Wukro hospitals, Tigray, Northern Ethiopia. BMC Research Notes. 2018;11(1, article 250) doi: 10.1186/s13104-018-3353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wondimeneh Y., Muluye D., Alemu A., et al. Urinary tract infection among obstetric fistula patients at Gondar University Hospital, Northwest Ethiopia. BMC Women's Health. 2014;14(1, article 12) doi: 10.1186/1472-6874-14-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Godebo G., Kibru G., Tassew H. Multidrug-resistant bacterial isolates in infected wounds at jimma university specialized hospital, Ethiopia. Annals of Clinical Microbiology and Antimicrobials. 2013;12(1, article 17) doi: 10.1186/1476-0711-12-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deyno S., Toma A., Worku M., Bekele M. Antimicrobial resistance profile of Staphylococcus aureus isolates isolated from ear discharges of patients at University of Hawassa comprehensive specialized hospital. BMC Pharmacology & Toxicology. 2017;18, article 35 doi: 10.1186/s40360-017-0141-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mama M., Abdissa A., Sewunet T. Antimicrobial susceptibility pattern of bacterial isolates from wound infection and their sensitivity to alternative topical agents at jimma university specialized hospital, South-West Ethiopia. Annals of Clinical Microbiology and Antimicrobials. 2014;13, article 14(1) doi: 10.1186/1476-0711-13-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramos J. M., Pérez-Tanoira R., García-García C., et al. Leprosy ulcers in a rural hospital of Ethiopia: Pattern of aerobic bacterial isolates and drug sensitivities. Annals of Clinical Microbiology and Antimicrobials. 2014;13(1, article no. 47) doi: 10.1186/s12941-014-0047-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dagnew M., Yismaw G., Gizachew M., et al. Bacterial profile and antimicrobial susceptibility pattern in septicemia suspected patients attending gondar university hospital, Northwest Ethiopia. BMC Research Notes. 2013;6(1, article 283) doi: 10.1186/1756-0500-6-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muluye D., Wondimeneh Y., Moges F., Nega T., Ferede G. Types and drug susceptibility patterns of bacterial isolates from eye discharge samples at gondar university hospital, Northwest Ethiopia. BMC Research Notes. 2014;7, article 292(1) doi: 10.1186/1756-0500-7-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mengesha R. E., Kasa B. G.-S., Saravanan M., Berhe D. F., Wasihun A. G. Aerobic bacteria in post surgical wound infections and pattern of their antimicrobial susceptibility in Ayder Teaching and Referral Hospital, Mekelle, Ethiopia. BMC Research Notes. 2014;7(1, article 575) doi: 10.1186/1756-0500-7-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teweldemedhin M., Saravanan M., Gebreyesus A., Gebreegziabiher D. Ocular bacterial infections at quiha ophthalmic hospital, Northern Ethiopia: an evaluation according to the risk factors and the antimicrobial susceptibility of bacterial isolates. BMC Infectious Diseases. 2017;17, article 207 doi: 10.1186/s12879-017-2304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Getahun E., Gelaw B., Assefa A., Assefa Y., Amsalu A. Bacterial pathogens associated with external ocular infections alongside eminent proportion of multidrug resistant isolates at the University of Gondar Hospital, northwest Ethiopia. BMC Ophthalmology. 2017;17, article 151 doi: 10.1186/s12886-017-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dilnessa T., Bitew A. Prevalence and antimicrobial susceptibility pattern of methicillin resistant Staphylococcus aureus isolated from clinical samples at yekatit 12 hospital medical college, Addis Ababa, Ethiopia. BMC Infectious Diseases. 2016;16, article 398(1) doi: 10.1186/s12879-016-1742-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bitew A., Tamirat M., Chanie M. Species distribution and antibiotic susceptibility profile of bacterial uropathogens among patients complaining urinary tract infections. BMC Infectious Diseases. 2017;17, article 654 doi: 10.1186/s12879-017-2743-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abdissa A., Asrat D., Kronvall G., et al. Throat carriage rate and antimicrobial susceptibility pattern of group a streptococci (gas) in healthy Ethiopian school children. Ethiopian Medical Journal. 2011;49(2):125–130. [PubMed] [Google Scholar]
- 40.Yilema A., Moges F., Tadele S., et al. Isolation of enterococci, their antimicrobial susceptibility patterns and associated factors among patients attending at the university of gondar teaching hospital. BMC Infectious Diseases. 2017;17, article 276 doi: 10.1186/s12879-017-2363-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abamecha A., Wondafrash B., Abdissa A. Antimicrobial resistance profile of Enterococcus species isolated from intestinal tracts of hospitalized patients in Jimma, ethiopia microbiology. BMC Research Notes. 2015;8, articlec 213 doi: 10.1186/s13104-015-1200-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alemseged G., Niguse S., Hailekiros H., Abdulkadir M., Saravanan M., Asmelash T. Isolation and anti-microbial susceptibility pattern of group B Streptococcus among pregnant women attending antenatal clinics in ayder referral hospital and mekelle health center, Mekelle, Northern Ethiopia Health Services Research. BMC Research Notes. 2015;8, article 518 doi: 10.1186/s13104-015-1475-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seid J., Asrat D. Occurrence of extended spectrum β-lactamase enzymes in clinical isolates of Klebsiella species from Harar region, eastern Ethiopia. Acta Tropica. 2005;95(2):143–148. doi: 10.1016/j.actatropica.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 44.Dereje M., Woldeamanuel Y., Asrat D., Ayenachew F. Urinary tract infection among fistula patients admitted at Hamlin fistula hospital, Addis Ababa, Ethiopia. BMC Infectious Diseases. 2017;17, article 150 doi: 10.1186/s12879-017-2265-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mulu W., Yimer M., Zenebe Y., Abera B. Common causes of vaginal infections and antibiotic susceptibility of aerobic bacterial isolates in women of reproductive age attending at Felegehiwot referral Hospital, Ethiopia: a cross sectional study. BMC Women's Health. 2015;15, article 42 doi: 10.1186/s12905-015-0197-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mamuye Y. Antibiotic resistance patterns of common gram-negative uropathogens in st. paul's hospital millennium medical college. Ethiopian Journal of Health Sciences. 2016;26(2):93–100. doi: 10.4314/ejhs.v26i2.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bekele T., Tesfaye A., Sewunet T., Waktola H. D. Pseudomonas aeruginosa isolates and their antimicrobial susceptibility pattern among catheterized patients at Jimma University Teaching Hospital, Jimma, Ethiopia. BMC Research Notes. 2015;8, article 488 doi: 10.1186/s13104-015-1497-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tadesse A., Mekonnen A., Kassu A., Asmelash T. Antimicrobial sensitivity of Neisseria gonorrhoea in Gondar, Ethiopia. East African Medical Journal. 2001;78(5):259–261. doi: 10.4314/eamj.v78i5.9050. [DOI] [PubMed] [Google Scholar]
- 49.Ali S., Sewunet T., Sahlemariam Z., Kibru G. Neisseria gonorrhoeae among suspects of sexually transmitted infection in Gambella hospital, Ethiopia: risk factors and drug resistance. BMC Research Notes. 2016;9, article 439 doi: 10.1186/s13104-016-2247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eguale T., Gebreyes W. A., Asrat D., Alemayehu H., Gunn J. S., Engidawork E. Non-typhoidal Salmonella serotypes, antimicrobial resistance and co-infection with parasites among patients with diarrhea and other gastrointestinal complaints in Addis Ababa, Ethiopia. BMC Infectious Diseases. 2015;15, article 497(1) doi: 10.1186/s12879-015-1235-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bruchfeld J., Aderaye G., Palme I. B., et al. Molecular epidemiology and drug resistance of Mycobacterium tuberculosis isolates from ethiopian pulmonary tuberculosis patients with and without human immunodeficiency virus infection. Journal of Clinical Microbiology. 2002;40(5):1636–1643. doi: 10.1128/jcm.40.5.1636-1643.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abebe G., Abdissa K., Abdissa A., et al. Relatively low primary drug resistant tuberculosis in southwestern Ethiopia. BMC Research Notes. 2012;5, article 225(1) doi: 10.1186/1756-0500-5-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yimer S. A., Agonafir M., Derese Y., Sani Y., Bjune G. A., Holm-Hansen C. Primary drug resistance to anti-tuberculosis drugs in major towns of Amhara region, Ethiopia. APMIS-Acta Pathologica, Microbiologica et Immunologica Scandinavica. 2012;120(6):503–509. doi: 10.1111/j.1600-0463.2011.02861.x. [DOI] [PubMed] [Google Scholar]
- 54.Tessema B., Beer J., Emmrich F., Sack U., Rodloff A. C. First- and second-line anti-tuberculosis drug resistance in Northwest Ethiopia. The International Journal of Tuberculosis and Lung Disease. 2012;16(6):805–811. doi: 10.5588/ijtld.11.0522. [DOI] [PubMed] [Google Scholar]
- 55.Davies J., Davies D. Origins and evolution of antibiotic resistance. Microbiology and Molecular Biology Reviews. 2010;74(3):417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Korma W., Mihret A., Hussien J., Anthony R., Lakew M., Aseffa A. Clinical, molecular and drug sensitivity pattern of mycobacterial isolates from extra-pulmonary tuberculosis cases in Addis Ababa, Ethiopia. BMC Infectious Diseases. 2015;15(1, article 456) doi: 10.1186/s12879-015-1177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]