Abstract
Q192R and L55M polymorphism were considered to be associated with the development of multiple cancers. Nevertheless, the results of these researches were inconclusive and controversial. Therefore, we conducted a meta-analysis of all eligible case-control studies to assess the association between PON1 (Q192R and L55M) gene polymorphisms and risk of cancer. With the STATA 14.0 software, we evaluated the strength of the association by using the odds ratios (ORs) and 95% confidence intervals (CIs). A total of 43 case-control publications 19887 cases and 23842 controls were employed in our study. In all genetic models, a significant association between PON1-L55M polymorphisms and overall cancer risk was observed. Moreover, in the stratified analyses by cancer type, polymorphism of PON1-L55M played a risk factor in the occurrence of breast cancer, hematologic cancer, and prostate cancer. Similarly, an increased risk was observed in the Caucasian and Asian population as well as hospital-based group and population-based group. For PON1-Q192R polymorphisms, in the stratified analyses by cancer type, PON1-Q192R allele was associated with reduced cancer risks in breast cancer. Furthermore, for racial stratification, there was a reduced risk of cancer in recession model in Caucasian population. Similarly, in the stratification analysis of control source, the overall risk of cancer was reduced in the heterozygote comparison and dominant model in the population-based group. In conclusion, PON1-Q192R allele decreased the cancer risk especially breast cancer; there was an association between PON1-L55M allele and increased overall cancer risk. However, we need a larger sample size, well-designed in future and at protein levels to confirm these findings.
1. Introduction
Cancer is one of the diseases caused by a combination of genetic and environmental factors [1]. The PON1 gene, located on the long arm of chromosome 7q21.3, is an antioxidant enzyme that has strong lipophilic antioxidant properties, which can maintain the balance of antioxidant-oxidant [2, 3]. Simultaneously, PON1 is also an esterase involved in scavenging reactive oxygen species by binding to high-density lipoprotein (HDL). Studies have shown that oxidative stress may participate in the process of cell proliferation and malignant transformation and damage DNA as well as other biological molecules, resulting in the occurrence of tumors [4]. The ability of PON1 detoxification of carcinogenic oxidative stress products makes it possible for researchers to predict PON1 gene polymorphism in cancer susceptibility [5].
At present, with the deep development of genetic studies of PON1, studies have found that PON1-Q192R and PON1-L55M, the two most common functional genetic polymorphisms in PON1, were identified at positions 192 and 55 [6]. PON1-Q192R polymorphism (rs662A > G) was caused by the glutamine (Q genotype) substituted for the arginine (R genotype) 192 of the gene 6 exon of the PON 1 gene [7]. PON1-L55M (rs854560) was originated from the replacement of 55 leucines (L genotype) by methionine (M genotype) at third exon 55[8]. In addition, it has been shown that the two functional SNP, Q192R and L55M, were associated with the risk of multiple tumors [9, 10], such as oral cancer [11], lung cancer [12], and embryonal tumors [13].
According to the important role of PON1 in the development of tumor and the correlation between genotype and phenotype, we speculate that the variation of PON1 gene Q192 R and L55M may be related to tumor susceptibility. However, the data of many studies are contradictory and uncertain at present. Therefore, a comprehensive meta-analysis should be conducted to determine the relationship between Q192R and L55M polymorphism and cancer risk.
2. Materials and Methods
2.1. Search Strategy
We conducted a systematic literature search in the PubMed, Embase, and Web of Science for all related studies before June 10, 2019 via utilizing the following terms: “polymorphism OR paraoxonase 1 OR PON1” AND “tumor OR malignancy OR cancer OR carcinoma OR neoplasm”. In addition, we extracted the reference of the original articles on this issue to carry out a hand search for extra studies. The results deduced from these articles were limited to humans. When the publication referred to more than one cancer type or ethnicity, we deled with data respectively. Besides, if different authors published articles based on the same population or one author used similar data in an article, we picked out the report with the latest study and largest sample size.
2.1.1. Inclusion Criteria and Exclusion Criteria
The enrolled studies must contain the following inclusion criteria: (1) publication that evaluated the association between PON1-L55M, or PON1-Q192R polymorphism and the risk of cancer. (2) The genotype frequency may be obtainable from cases and controls, or we could gain it via computing. In addition, studies were excluded when they would meet these exclusion criteria: (1) reviews, case reports, or case-only studies; (2) studies with deficient genotype frequency date; (3) animals reports; and (4) replicate studies.
2.2. Data Extraction
The authors were able to excerpt relevant data from these qualified studies independently, and the following information would be seized: first author's last name, publishing year, the ethnicity of each population, the genotyping methods, the control of source, cancer types, number of cases and controls, and P value of Hardy–Weinberg equilibrium. When encountering divergences, we analyzed the report and reached a consistent agreement lastly.
2.3. Statistical Analysis
95% confidence interval (CI) and odds ratio (OR) were utilized to estimate the relation between PON1-Q192R, or PON1-L55M polymorphism and the risk of cancer with five genetic models: heterozygote comparison (ML versus LL; RQ versus QQ), allele contrast (M versus L; R versus Q), homozygote (MM versus LL; RR versus QQ), recessive (MM versus ML+LL; RR versus RQ+QQ), and dominant (ML+MM versus LL; RR+RQ versus QQ). Besides, stratified analyses were conducted via ethnicity, cancer type, control source, and genotyping method. However, when any cancer type is less than two studies, we would segment it into the “other cancers” group. In addition, χ2-test-based Q-statistic test [14] was taken to assess the research heterogeneity while I2 values and P values [15] were used for quantifying. When I2< 50% and P>0.10, it indicates that there was no significant heterogeneity, and ORs could be pooled by a fixed-effects model. Otherwise, the random effects model would be adopted [16]. Furthermore, sensitivity analysis, from the qualified removing a single research study and revealing the individual data set to merge OR influence, was applied to estimate the stability of these data. (P<0.05 was regarded as statistically significant [17].) Finally, potential publication bias was estimated by symmetry of funnel plot of Begg's test as well as Egger's test [15, 18], and being statistically significant was considered when P<0.05. All statistical tests were performed with STATA Software (version 14.0, state Corp), and P<0.05 for any genetic models or tests was identified as statistically significant.
3. Result
3.1. Publication Characteristics
According to the inclusion criteria after detailed examination, a total of 43 case-control publications including 19977 cases and 23932 controls were employed in our study [11–13, 19–59]. The flow chart of the study screening process was summarized in Figure 1. Moreover, there were 43 studies with 11412 cases and 13936 controls for PON1-Q192R polymorphism (Table 1), and, for PON1 L55M polymorphism, 28 studies involved a total of 8565 cases and 9996 controls (Table 2). For PON1 Q192R polymorphism, a total of 8 cancer types were processed, including breast cancer [21, 27, 31, 32, 37, 39, 50], prostate cancer [22, 23, 40, 41], gastrointestinal cancer [19, 20, 48, 59, 60], hematologic tumor [25, 29, 33, 44], lung cancer [11, 12, 54], brain tumors [30, 35, 38, 45, 56, 57], ovarian cancer [34, 43] and other cancers [13, 26, 28, 42, 53, 58] (uterine leiomyoma, childhood embryonal tumors, metastatic gastric cancer, bladder cancer, and renal cell cancer). Besides, we disposed a total of 7 cancer types when dealing with PON1-L55M polymorphism nearly like PON1 Q192R polymorphism. In addition, For PON1 Q192R polymorphism, 9 publications were conducted in Asians, 9 in mixed group, and 25 publications in Caucasians. Besides, there were 15 studies divided by TaqMan assay, while 28 studies conducted by PCR- RFLP. Moreover, the majority of control groups in the case group are gender and age matching, including 23 hospital based and 20 population based. For PON1 L55M polymorphism, we also conducted 6, 6, and 16 studies in Asian, mixed group, Caucasians, respectively. Moreover, 10 studies were divided by TaqMan assay as well as 18 studies conducted by PCR- RFLP.
Figure 1.
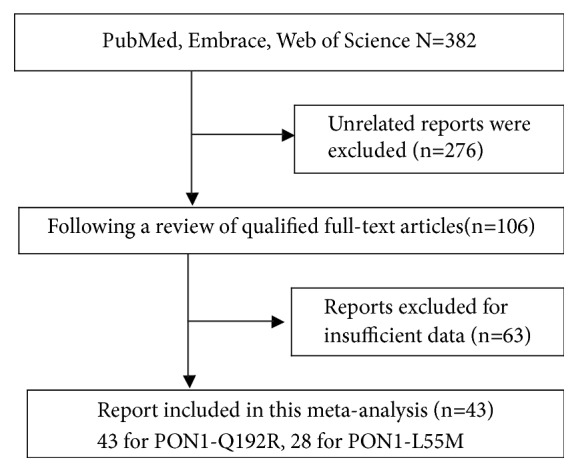
Flow chart of the report selection process.
Table 1.
Characteristics of qualified case-control studies included in the meta-analysis of PON1-Q192R.
| Author | Year | Ethnicity | Genotyping Method | Control of source | Cancer Type | Case | Control | HWE | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| QR | RR | QR | RR | χ 2 | p | p(HWE) | ||||||||
| Stevens et al. | 2006 | Caucasian | PCR-RFLP | P-B | Breast Cancer | 259 | 182 | 42 | 238 | 198 | 47 | 0.38 | 0.54 | Y |
| Gallicchio et al. | 2007 | Caucasian | PCR-RFLP | P-B | Breast Cancer | 38 | 15 | 5 | 469 | 353 | 82 | 1.93 | 0.19 | Y |
| Antognelli et al. | 2009 | Caucasian | PCR-RFLP | P-B | Breast Cancer | 484 | 50 | 13 | 340 | 152 | 52 | 27.19 | 0.00 | N |
| Hussein et al. | 2011 | Caucasian | PCR-RFLP | P-B | Breast Cancer | 51 | 41 | 8 | 46 | 42 | 12 | 0.25 | 0.62 | Y |
| Naidu et al. | 2010 | Asian | PCR-RFLP | H-B | Breast Cancer | 200 | 158 | 29 | 115 | 115 | 22 | 0.81 | 0.37 | Y |
| Tang et al. | 2017 | Asian | TaqMan | P-B | Esophagogastric | 408 | 501 | 132 | 691 | 776 | 207 | 0.23 | 0.63 | Y |
| Uluocak et al. | 2017 | Caucasian | PCR-RFLP | H-B | Prostate Cancer | 24 | 17 | 8 | 45 | 42 | 11 | 0.06 | 0.80 | Y |
| Wu et al. | 2017 | Asian | TaqMan | H-B | Breast Cancer | 155 | 156 | 54 | 167 | 156 | 55 | 3.42 | 0.06 | Y |
| Kaya et al. | 2016 | Caucasian | TaqMan | H-B | Breast Cancer | 10 | 11 | 11 | 5 | 13 | 17 | 0.88 | 0.35 | Y |
| Tomatir et al. | 2015 | Caucasian | PCR-RFLP | H-B | Hematologic Cancer | 36 | 20 | 4 | 58 | 24 | 2 | 0.07 | 0.79 | Y |
| Tomatir et al. | 2015 | Caucasian | PCR-RFLP | H-B | Hematologic Cancer | 33 | 21 | 6 | 58 | 24 | 2 | 0.07 | 0.08 | Y |
| Attar et al. | 2015 | Caucasian | PCR-RFLP | H-B | Uterine Leiomyoma | 60 | 8 | 8 | 50 | 47 | 6 | 1.39 | 0.24 | Y |
| Eom et al. | 2015 | Asian | PCR-RFLP | H-B | Lung Cancer | 37 | 170 | 209 | 48 | 188 | 180 | 0.01 | 0.92 | Y |
| Ahmed et al. | 2015 | Asian | PCR-RFLP | P-B | Colorectal Cancer | 30 | 16 | 4 | 20 | 36 | 24 | 0.76 | 0.38 | Y |
| Akkız et al. | 2013 | Caucasian | PCR-RFLP | P-B | Hepatocellular Carcinoma | 109 | 95 | 13 | 115 | 88 | 14 | 0.27 | 0.60 | Y |
| Vasconcelos et al. | 2014 | Mixed | TaqMan | H-B | Embryonal Tumors | 36 | 85 | 41 | 104 | 160 | 72 | 0.51 | 0.48 | Y |
| Conesa-Zamora et al. | 2013 | Caucasian | TaqMan | H-B | Lymphomas | 83 | 99 | 33 | 100 | 104 | 10 | 7.00 | 0.01 | N |
| Zhao et al. | 2012 | Asian | TaqMan | H-B | Glioma | 161 | 158 | 52 | 159 | 167 | 52 | 0.59 | 0.44 | Y |
| De Aguiar Goncalves et al. | 2012 | Mixed | TaqMan | H-B | Hematologic Tumor | 96 | 102 | 40 | 74 | 106 | 54 | 1.790 | 0.180 | Y |
| Kokouva et al. | 2012 | Caucasian | PCR-RFLP | H-B | Hematologic Cancer | 213 | 88 | 15 | 181 | 141 | 29 | 0.04 | 0.83 | Y |
| Aksoy-Sagirli et al. | 2011 | Caucasian | PCR-RFLP | H-B | Lung Cancer | 93 | 111 | 19 | 121 | 93 | 20 | 0.13 | 0.72 | Y |
| Uyar et al. | 2011 | Caucasian | PCR-RFLP | P-B | Renal Cell Cancer | 38 | 21 | 1 | 27 | 27 | 6 | 0.04 | 0.84 | Y |
| Lurie et al. | 2008 | Mixed | TaqMAN | P-B | Ovarian Cancer | 66 | 120 | 86 | 122 | 211 | 111 | 1.07 | 0.30 | Y |
| Ergen et al. | 2010 | Caucasian | PCR-RFLP | H-B | Osteosarcoma | 27 | 21 | 2 | 15 | 33 | 2 | 0.06 | 0.80 | Y |
| Martinez et al. | 2010 | Caucasian | TaqMan | H-B | Brain Tumor | 31 | 33 | 9 | 22 | 89 | 109 | 0.37 | 0.54 | Y |
| Ozturk et al. | 2009 | Caucasian | PCR-RFLP | H-B | Bladder Cancer | 8 | 53 | 15 | 37 | 84 | 14 | 10.71 | <0.001 | N |
| Gold et al. | 2009 | Mixed | PCR-RFLP | P-B | Multiple Myeloma | 10 | 19 | 13 | 9 | 27 | 19 | 0.01 | 0.91 | Y |
| Arpaci et al. | 2009 | Caucasian | PCR-RFLP | H-B | Ovarian Cancer | 38 | 6 | 6 | 17 | 29 | 6 | 1.46 | 0.23 | Y |
| Rajaraman et al. | 2008 | Mixed | TaqMan | H-B | Brain Tumor | 266 | 207 | 39 | 244 | 165 | 44 | 4.10 | 0.04 | N |
| Searles Nielsen et al. | 2005 | Mixed | TaqMan | P-B | Brain Tumor | 32 | 28 | 6 | 100 | 105 | 31 | 0.17 | 0.68 | Y |
| Van der Logt et al. | 2005 | Caucasian | PCR-RFLP | P-B | Colorectal Cancer | 180 | 150 | 24 | 158 | 120 | 17 | 0.87 | 0.35 | Y |
| Lincz et al. | 2004 | Caucasian | PCR-RFLP | P-B | Multiple Myeloma | 33 | 41 | 16 | 103 | 74 | 22 | 2.35 | 0.13 | Y |
| Kerridge et al. | 2002 | Caucasian | PCR-RFLP | P-B | Lymphoma | 73 | 50 | 39 | 103 | 74 | 22 | 2.35 | 0.13 | Y |
| Antognelli et al. | 2005 | Caucasian | PCR-RFLP | H-B | Prostate Cancer | 197 | 168 | 20 | 212 | 85 | 64 | 67.85 | <0.001 | N |
| Herrera et al. | 2015 | Mixed | TaqMan | H-B | Brain Tumor | 15 | 32 | 20 | 12 | 32 | 14 | 0.64 | 0.42 | Y |
| Kafadar et al. | 2006 | Caucasian | PCR-RFLP | H-B | Brain Tumor | 43 | 26 | 15 | 24 | 18 | 8 | 1.96 | 0.16 | Y |
| J. De Roos et al. | 2006 | Mixed | TaqMan | P-B | Hematologic Cancer | 540 | 453 | 127 | 415 | 403 | 117 | 1.53 | 0.22 | Y |
| Stevens et al. | 2008 | Mixed | TaqMan | P-B | Prostate Cancer | 624 | 537 | 95 | 656 | 487 | 121 | 4.74 | 0.03 | N |
| Antognelli et al. | 2013 | Caucasian | PCR-RFLP | H-B | Prostate Cancer | 291 | 250 | 30 | 707 | 258 | 203 | 244.08 | <0.001 | N |
| Wang et al. | 2012 | Asian | PCR-RFLP | P-B | Lung Cancer | 36 | 177 | 143 | 38 | 84 | 62 | 0.93 | 0.33 | Y |
| Lee et al. | 2005 | Asian | TaqMan | P-B | Lung Cancer | 24 | 80 | 73 | 11 | 89 | 77 | 4.999 | 0.025 | N |
| Agachan et al. | 2006 | Caucasian | PCR-RFLP | P-B | Breast Cancer | 17 | 4 | 12 | 6 | 29 | 17 | 1.461 | 0.230 | Y |
| Hemati et al. | 2019 | Asian | PCR-RFLP | H-B | Gastric Cancer | 39 | 41 | 10 | 62 | 26 | 2 | 0.03 | 0.87 | Y |
Abbreviations: PCR-RFlP, polymerase chain reaction-restriction fragment length polymorphism; HWE, Hardy–Weinberg equilibrium; Y, polymorphisms conforming to HWE in the control group; N, polymorphisms not conforming to HWE in the control group; H-B, hospital based; P-B, population based.
Table 2.
Characteristics of qualified case-control studies included in the meta-analysis of PON1- L55M.
| Author | Year | Ethnicity | Genotyping Method | Control of source | Cancer Type | Case | Control | HWE | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LL | LM | MM | LL | LM | MM | χ 2 | p | p(HWE) | ||||||
| Stevens et al. | 2006 | Caucasian | PCR-RFLP | P-B | Breast Cancer | 176 | 230 | 77 | 202 | 233 | 58 | 0.88 | 0.77 | Y |
| Antognelli et al. | 2009 | Caucasian | PCR-RFLP | P-B | Breast Cancer | 107 | 115 | 325 | 188 | 125 | 231 | 157.2 | 0.0001 | N |
| Hussein et al. | 2011 | Caucasian | PCR-RFLP | P-B | Breast Cancer | 19 | 21 | 60 | 35 | 23 | 6 | 0.58 | 0.44 | Y |
| Naidu et al. | 2010 | Asian | PCR-RFLP | P-B | Breast Cancer | 159 | 178 | 50 | 126 | 109 | 17 | 1.04 | 0.308 | Y |
| Tang et al. | 2017 | Asian | TaqMan | P-B | Esophagogastric Cancer | 971 | 69 | 1 | 1573 | 99 | 2 | 0.12 | 0.73 | Y |
| Uluocak et al. | 2017 | Caucasian | PCR-RFLP | H-B | Prostate Cancer | 19 | 24 | 6 | 43 | 45 | 10 | 0.13 | 0.72 | Y |
| Wu et al. | 2017 | Asian | TaqMan | H-B | Breast Cancer | 284 | 72 | 9 | 346 | 30 | 2 | 3.24 | 0.064 | Y |
| Akkız et al. | 2013 | Caucasian | PCR-RFLP | P-B | Hepatocellular Carcinoma | 105 | 81 | 31 | 101 | 89 | 27 | 1.12 | 0.29 | Y |
| Geng R et al. | 2014 | Asian | TaqMan | H-B | Metastatic Gastric Cancer | 11 | 7 | 0 | 82 | 7 | 0 | 0.15 | 0.7 | Y |
| Vasconcelos et al. | 2014 | Mixed | TaqMan | H-B | Embryonal Tumors | 85 | 56 | 15 | 177 | 134 | 25 | 0.023 | 0.95 | Y |
| Metin et al. | 2013 | Caucasian | PCR-RFLP | H-B | Ovarian Cancer | 33 | 22 | 0 | 33 | 19 | 2 | 0.13 | 0.72 | Y |
| Vecka et al. | 2012 | Caucasian | PCR-RFLP | H-B | Pancreatic Cancer | 24 | 39 | 10 | 28 | 37 | 8 | 0.67 | 0.41 | Y |
| De Aguiar Goncalves et al. | 2012 | Mixed | TaqMan | H-B | Acute Leukemia | 104 | 99 | 34 | 131 | 75 | 19 | 2.91 | 0.09 | Y |
| Kokouva et al. | 2012 | Caucasian | PCR-RFLP | H-B | Hematologic Cancer | 117 | 139 | 60 | 142 | 159 | 50 | 0.26 | 0.61 | Y |
| Aksoy-Sagirli et al. | 2011 | Caucasian | PCR-RFLP | H-B | Lung Cancer | 119 | 94 | 10 | 118 | 102 | 14 | 1.75 | 0.19 | Y |
| Uyar et al. | 2011 | Caucasian | PCR-RFLP | P-B | Renal Cell Cancer | 29 | 25 | 6 | 21 | 29 | 10 | 4.96 | 0.998 | Y |
| Lurie et al. | 2008 | Mixed | TaqMan | P-B | Ovarian Cancer | 14 | 65 | 192 | 24 | 145 | 276 | 0.74 | 0.39 | Y |
| Ergen et al. | 2010 | Caucasian | PCR-RFLP | H-B | Osteosarcoma | 24 | 23 | 3 | 21 | 20 | 9 | 1.14 | 0.29 | Y |
| Martínez et al. | 2010 | Caucasian | TaqMan | H-B | Brain Tumor | 11 | 32 | 30 | 38 | 94 | 88 | 2.15 | 0.14 | Y |
| Arpaci et al. | 2009 | Caucasian | PCR-RFLP | H-B | Ovarian Cancer | 27 | 19 | 5 | 25 | 27 | 2 | 2.65 | 0.103 | Y |
| Van der Logt et al. | 2005 | Caucasian | PCR-RFLP | P-B | Colorectal Cancer | 139 | 166 | 59 | 140 | 162 | 50 | 0.08 | 0.78 | Y |
| Antognelli et al. | 2005 | Caucasian | PCR-RFLP | H-B | Prostate Cancer | 120 | 197 | 67 | 148 | 169 | 43 | 0.65 | 0.35 | Y |
| Herrera et al. | 2015 | Mixed | TaqMan | H-B | Brain Tumor | 46 | 17 | 4 | 42 | 14 | 2 | 0.37 | 0.56 | Y |
| J. De Roos et al. | 2006 | Mixed | TaqMan | P-B | Hematologic Cancer | 299 | 307 | 100 | 282 | 260 | 69 | 0.59 | 0.44 | Y |
| Stevens et al. | 2008 | Mixed | TaqMan | P-B | Prostate Cancer | 481 | 609 | 165 | 498 | 575 | 189 | 1.18 | 0.28 | Y |
| Wang et al. | 2012 | Asian | PCR-RFLP | P-B | Lung Cancer | 307 | 47 | 2 | 166 | 18 | 0 | 0.49 | 0.49 | Y |
| Antognelli et al. | 2013 | Caucasian | PCR-RFLP | H-B | Prostate Cancer | 180 | 291 | 100 | 497 | 540 | 131 | 0.75 | 0.39 | Y |
| Hemati et al. | 2019 | Asian | PCR-RFLP | H-B | Gastric Cancer | 41 | 40 | 9 | 34 | 49 | 7 | 0.027 | 0.87 | Y |
Abbreviations: PCR-RFlP, polymerase chain reaction-restriction fragment length polymorphism; HWE, Hardy–Weinberg equilibrium; Y, polymorphisms conforming to HWE in the control group; N, polymorphisms not conforming to HWE in the control group; H-B, hospital based; P-B, population based.
3.2. Meta-Analysis
3.2.1. Association between PON1-Q192R and Cancer Susceptibility
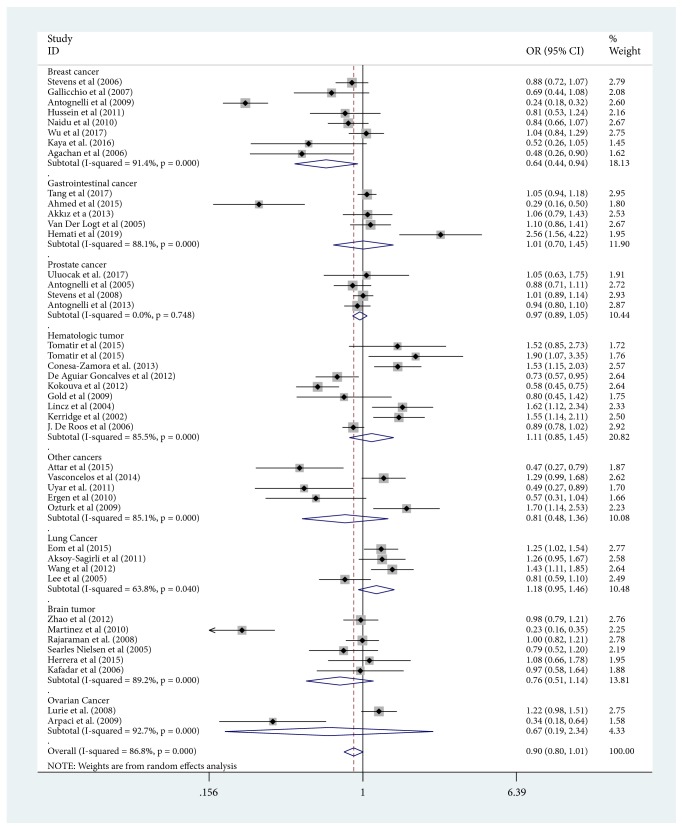
In summary, in allele contrast model, we have found that there were not association between PON1-Q192R allele and reduced overall cancer risk (Table 3). In the subgroup analysis of cancer type, we identified a decreased risk in breast cancer (R versus Q: OR=0.643, 95%CI=0.440-0.942; RR versus QQ: OR=0.542, 95%CI=0.331-0.886; RQ versus QQ: OR=0.529, 95%CI=0.325-0.861; and RR+RQ versus QQ: OR=0.534, 95%CI=0.330-0.865). Nevertheless, an increased risk was confirmed in prostate cancer in the dominant model (RR+RQ versus QQ: OR=1.249, 95%CI=1.030-1.514). Furthermore, by racial stratification, there was a reduced risk of cancer in recession model (RR+RQ versus QQ: OR=0.744, 95%CI=0.557-0.993) among Caucasian population. Similarly, in the stratification analysis of control source, the overall risk of cancer is reduced in the heterozygote comparison and dominant model (RQ versus QQ: OR= 0.793, 95%CI=0.638-0.984; RR+RQ versus QQ: OR=0.789, 95%CI=0.630-0.988) in the population-based group. In addition, we did not observe any risk factor by stratified analysis using genotyping method. Figure 2 showed the meta-analysis of the association between PON1-Q192R polymorphism and cancer risk (R versus Q)
Table 3.
Results of meta-analysis for PON1-Q192R polymorphism and cancer risk.
| Variables | Case/control | R vs. Q | RR vs. QQ | RQ vs. QQ | RR+RQ vs. QQ | RR vs. RQ+QQ | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR(95% CI) | pa | I2(%) | OR(95% CI) | pa | I2(%) | OR(95% CI) | pa | I2(%) | OR(95% CI) | pa | I2(%) | OR(95% CI) | pa | I2(%) | ||
| Total | 11412/13936 | 0.897(0.798-1.008) | 0 | 86.8 | 0.855(0.683-1.073) | 0 | 81.1 | 0.861(0.724-1.023) | 0 | 86.7 | 0.857(0.730-1.008) | 0 | 86.7 | 0.914(0.758-1.102) | 0 | 78.1 |
| Breast cancer | 2005/2748 | 0.643(0.440-0.942) ∗ | 0 | 91.4 | 0.542(0.331-0.886)∗ | 0 | 74 | 0.529(0.325-0.861)∗ | 0 | 89.1 | 0.534(0.330-0.865)∗ | 0 | 90.6 | 0.720(0.492-1.054) | 0.010 | 62.3 |
| Gastrointestinal cancer | 1752/2356 | 1.008(0.700-1.450) | 0 | 88.1 | 0.969(0.463-2.025) | 0.000 | 80.2 | 1.079(0.761-1.529) | 0.002 | 75.7 | 1.038(0.682-1.580) | 0.000 | 85.0 | 0.968(0.547-1.711) | 0.013 | 68.5 |
| Prostate cancer | 2261/2891 | 0.967(0.886-1.055) | 0.748 | 0 | 0.563(0.313-1.015) | 0.001 | 83 | 1.544(0.969-2.458) | 0 | 90.9 | 1.249(1.030-1.514)∗ | 0.083 | 55 | 0.498(0.235-1.053) | 0 | 90.3 |
| Hematologic tumor | 2303/2355 | 1.113(0.852-1.453) | 0 | 85.5 | 1.358(0.787-2.341) | 0 | 82 | 0.943(0.740-1.202) | 0.007 | 62.3 | 1.040(0.774-1.397) | 0 | 78.2 | 1.364(0.863-2.157) | 0 | 77.8 |
| Lung cancer | 1172/1011 | 1.179(0.949-1.464) | 0.04 | 63.8 | 1.244(0.665-2.326) | 0.005 | 76.6 | 1.214(0.704-2.094) | 0.004 | 77.5 | 1.262(0.741-2.149) | 0.003 | 78.6 | 1.201(0.997-1.449) | 0.442 | 0 |
| Brain tumor | 1173/1395 | 0.759(0.505-1.140) | 0 | 89.2 | 0.576(0.266-1.248) | 0 | 85.9 | 0.778(0.539-1.124) | 0.006 | 69 | 0.696(0.431-1.123) | 0 | 84.4 | 0.698(0.388-1.256) | 0 | 79.4 |
| Ovarian cancer | 322/496 | 0.665(0.189-2.337) | 0 | 92.7 | 0.940(0.314-2.813) | 0.087 | 65.8 | 0.328(0.030-3.572) | 0 | 94.6 | 0.443(0.060-3.283) | 0 | 94.5 | 1.359(0.985-1.875) | 0.658 | 0 |
| Other cancers | 424/684 | 0.809(0.481-1.362) | 0 | 85.1 | 1.264(0.515-3.101) | 0.022 | 65 | 0.670(0.250-1.798) | 0 | 89.4 | 0.741(0.299-1.838) | 0 | 89.1 | 1.352(0.797-2.294) | 0.209 | 31.8 |
| Ethnicities | ||||||||||||||||
| Caucasian | 4424/6292 | 0.815(0.658-1.011) | 0 | 90.1 | 0.784(0.516-1.193) | 0 | 84.5 | 0.731(0.528-1.010) | 0 | 91.1 | 0.744(0.557-0.993)∗ | 0 | 90.3 | 0.893(0.608-1.311) | 0 | 83.5 |
| Asian | 3253/3629 | 0.840(0.840-1.244) | 0 | 89.9 | 1.019(0.689-1.506) | 0 | 78.5 | 1.020(0.779-1.337) | 0 | 76.3 | 1.022(0.758-1.377) | 0 | 83.0 | 1.052(0.850-1.303) | 0.025 | 54.4 |
| Mixed | 3735/4015 | 0.981(0.877-1.098) | 0.035 | 51.6 | 0.913(0.727-1.145) | 0.062 | 46.2 | 1.012(0.873-1.174) | 0.109 | 38.8 | 0.990(0.851-1.153) | 0.057 | 47 | 0.929(0.770-1.119) | 0.115 | 38.1 |
| Control source | ||||||||||||||||
| Population based | 6871/8354 | 0.849(0.717-1.004) | 0 | 88.8 | 0.802(0.604-1.065) | 0 | 79.1 | 0.793(0.638-0.984)∗ | 0 | 85 | 0.789(0.630-0.988)∗ | 0 | 87.9 | 0.920(0.749-1.130) | 0 | 70.3 |
| Hospital based | 4309/5667 | 0.897(0.798-1.008) | 0 | 85.1 | 0.948(0.655-1.374) | 0 | 83.2 | 0.927(0.704-1.221) | 0 | 87.3 | 0.923(0.726-1.175) | 0 | 85.5 | 0.967(0.696-1.342) | 0 | 83.3 |
| Genotype method | ||||||||||||||||
| PCR-RFLP | 5445/6900 | 0.884(0.735-1.064) | 0 | 88.4 | 0.888(0.735-1.064) | 0 | 88.4 | 0.815(0.607-1.094) | 0 | 90.4 | 0.839(0.646-1.091) | 0 | 89.4 | 0.938(0.692-1.271) | 0 | 80.5 |
| TaqMan | 5967/7036 | 0.922(0.801-1.060) | 0 | 83 | 0.834(0.622-1.117) | 0 | 81.3 | 0.952(0.824-1.099) | 0.001 | 61.1 | 0.906(0.762-1.078) | 0 | 76.7 | 0.915(0.736-1.139) | 0 | 73.5 |
Notes: ∗ statistically significant (P<0.05); P valuea: P value of Q test for heterogeneity test; I2: 0%–25% means no heterogeneity, 25%–50% means modest heterogeneity, and 50% means high heterogeneity. Abbreviations: CI, confidence interval; OR, odds ratio; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism.
Figure 2.
Meta-analysis of the association between PON1-Q192R polymorphism and cancer risk (R versus Q). Abbreviations: ID, identification; CI, confidence interval; NA, not available; OR, odds ratio; weights come from random effects analysis.
3.2.2. Association between PON1-L55M and Cancer Susceptibility
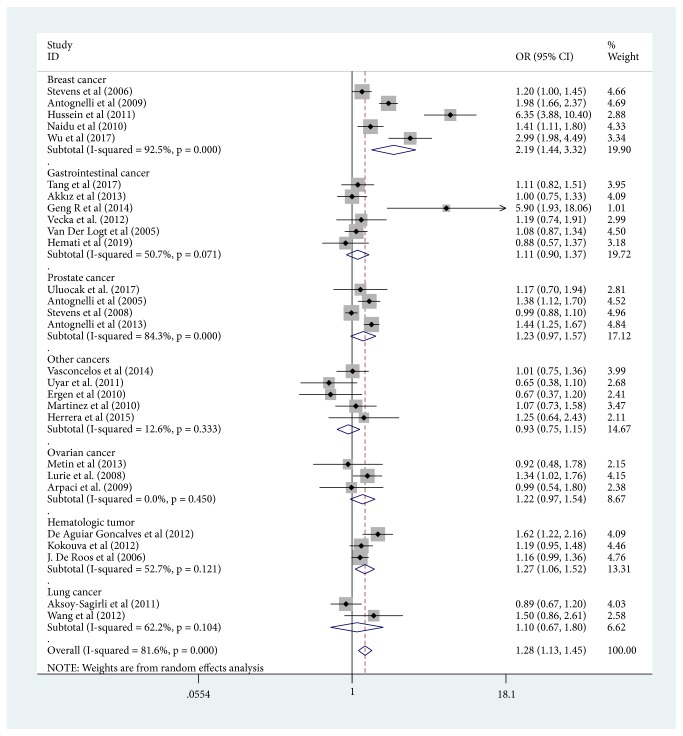
Our study had uncovered that the PON1-L55M polymorphism was significantly associated with an increased risk of the overall cancers under all the genetic models (Table 4) (M versus L: OR =1.277, 95% CI =1.127-1.448; MM versus LL: OR =1.507, 95% CI =1.205-1.885; ML versus LL: OR =1.192, 95%CI =1.064-1.337; MM versus ML+LL: OR =1.288, 95%CI =1.120-1.408; ML+MM versus LL: OR =1.417, 95%CI =1.176-1.708). Furthermore, we found an increased risk of breast cancer under all the five models when conducting the cancer type subgroup analysis (M versus L: OR =2.186, 95%CI =1.438-3.323; MM versus LL: OR =3.215, 95%CI=1.756-5.886; ML versus LL: OR =1.579, 95%CI=1.145-2.177; MM versus ML+LL: OR =2.727, 95%CI=1.563-4.756; ML+MM versus LL: OR =2.110, 95%CI =1.397-3.188), prostate cancer in the dominant and heterozygote comparison model (ML versus LL: OR =1.291, 95% CI =1.071-1.557; ML+MM versus LL: OR =1.341, 95%CI=1.024-1.756), and hematologic tumor in the allele contrast model (M versus L: OR =1.271, 95% CI =1.059-1.525), homozygote model (MM versus LL: OR =1.514, 95%CI =1.178-1.946), recessive model (MM versus ML+LL: OR =1.405, 95%CI =1.111-1.778), and dominant model (ML+MM versus LL: OR =1.299, 95%CI =1.017-1.661). Figure 3 showed the meta-analysis of the association between PON1-L55M polymorphism and cancer risk (M versus L).
Table 4.
Results of meta-analysis for PON1-L55M polymorphism and cancer risk.
| Variables | Case/control | M vs. L | MM vs. LL | ML vs. LL | ML+MM vs. LL | MM vs. ML+LL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR(95% CI) | pa | I2(%) | OR(95% CI) | pa | I2(%) | OR(95% CI) | pa | I2(%) | OR(95% CI) | pa | I2(%) | OR(95% CI) | pa | I2(%) | ||
| Total | 8565/9996 | 1.277(1.127-1.448)∗ | 0 | 81.6 | 1.507(1.205-1.885)∗ | 0 | 68.5 | 1.192(1.064-1.337)∗ | 0.001 | 50.6 | 1.288(1.120-1.480)∗ | 0 | 70.9 | 1.417(1.176-1.708)∗ | 0 | 64.2 |
| Breast cancer | 1882/1731 | 2.186(1.438-3.323) ∗ | 0 | 92.5 | 3.215(1.756-5.886)∗ | 0 | 81.8 | 1.579(1.145-2.177) ∗ | 0.01 | 69.9 | 2.110(1.397-3.188)∗ | 0 | 85.3 | 2.727(1.563-4.756)∗ | 0 | 81.9 |
| Gastrointestinal cancer | 1803/2495 | 1.111(0.898-1.375) | 0.071 | 50.7 | 1.165(0.848-1.601) | 0.988 | 0 | 1.097(0.794-1.515) | 0.023 | 61.5 | 1.120(0.829-1.512) | 0.032 | 59.0 | 1.185(0.881-1.594) | 0.996 | 0 |
| Prostate cancer | 2259/2888 | 1.233(0.971-1.566) | 0 | 84.3 | 1.496(0.876-2.556) | 0 | 85.5 | 1.291(1.071-1.557)∗ | 0.144 | 44.6 | 1.341(1.024-1.756)∗ | 0.008 | 74.6 | 1.284(0.838-1.966) | 0.002 | 80.5 |
| Hematologic tumor | 1259/1187 | 1.271(1.059-1.525)∗ | 0.121 | 52.7 | 1.514(1.178-1.946)∗ | 0.376 | 0 | 1.212(0.954-1.540) | 0.172 | 43.1 | 1.299(1.017-1.661)∗ | 0.124 | 52 | 1.405(1.111-1.778)∗ | 0.622 | 0 |
| Ovarian cancer | 377/553 | 1.219(0.965-1.539) | 0.450 | 0 | 1.208(0.648-2.253) | 0.393 | 0 | 0.833(0.536-1.296) | 0.579 | 0 | 0.952(0.623-1.454) | 0.802 | 0 | 1.482(0.800-2.743) | 0.316 | 13.2 |
| Lung cancer | 579/418 | 1.095(0.666-1.801) | 0.104 | 62.2 | 0.781(0.344-1.771) | 0.404 | 0 | 1.074(0.711-1.622) | 0.215 | 34.8 | 1.089(0.670-1.767) | 0.146 | 52.7 | 0.806(0.361-1.799) | 0.432 | 0 |
| Other cancers | 406/724 | 0.932(0.753-1.155) | 0.333 | 12.6 | 0.884(0.505-1.548) | 0.215 | 31 | 0.907(0.682-1.206) | 0.801 | 0 | 0.910(0.696-1.190) | 0.625 | 0 | 0.930(0.584-1.481) | 0.252 | 25.4 |
| Ethnicities | ||||||||||||||||
| Caucasian | 3616/4392 | 1.231(1.028-1.474)∗ | 0 | 83.8 | 1.737(1.519-1.986)∗ | 0 | 72 | 1.170(1.034-1.324)∗ | 0.199 | 22.4 | 1.334(1.215-1.465)∗ | 0 | 70.7 | 1.407(1.092-1.813)∗ | 0 | 67.8 |
| Asian | 2257/2667 | 1.604(1.089-2.363)∗ | 0 | 80.7 | 2.093(1.295-3.381)∗ | 0.441 | 0 | 1.550(0.995-2.417) | 0.000 | 79.7 | 1.624(1.041-2.535)∗ | 0 | 80.8 | 1.967(1.238-3.125)∗ | 0.656 | 0 |
| Mixed | 2692/2937 | 1.177(1.004-1.379)∗ | 0.019 | 63.1 | 1.137(0.955-1.354) | 0.088 | 47.8 | 1.112(0.953-1.297) | 0.268 | 22.1 | 1.126(1.006-1.261)∗ | 0.165 | 36.3 | 1.262(0.957-1.665) | 0.034 | 58.4 |
| Control source | ||||||||||||||||
| Population based | 5787/6158 | 1.325(1.085-1.618)∗ | 0 | 88.7 | 1.568(1.091-2.253)∗ | 0 | 81.9 | 1.275(1.051-1.548)∗ | 0.401 | 4.5 | 1.275(1.051-1.548)∗ | 0 | 75.7 | 1.503(1.110-2.034)∗ | 0 | 80.5 |
| Hospital based | 2778/3838 | 1.240(1.056-1.456)∗ | 0 | 68.9 | 1.531(1.199-1.955)∗ | 0.132 | 29.8 | 1.255(1.020-1.543)∗ | 0.000 | 62.3 | 1.288(1.120-1.480)∗ | 0 | 66.7 | 1.411(1.173-1.698)∗ | 0.324 | 11.6 |
| Genotype method | ||||||||||||||||
| PCR-RFLP | 4376/4698 | 1.243(1.053-1.466)∗ | 0 | 82.2 | 1.571(1.183-2.087)∗ | 0 | 70.1 | 1.164(1.033-1.311)∗ | 0.145 | 26.5 | 1.246(1.045-1.487)∗ | 0 | 69.3 | 1.483(1.167-1.884)∗ | 0 | 62.7 |
| TaqMan | 4189/5298 | 1.330(1.091-1.622)∗ | 0 | 79.5 | 1.309(0.988-1.735) | 0.091 | 41.4 | 1.307(1.026-1.665)∗ | 0.001 | 71.5 | 1.370(1.073-1.748)∗ | 0 | 74.7 | 1.264(0.986-1.620)∗ | 0.05 | 48.5 |
Notes: ∗ statistically significant (P<0.05); P valuea: P value of Q test for heterogeneity test; I2: 0%–25% means no heterogeneity, 25%–50% means modest heterogeneity, and 50% means high heterogeneity.
Abbreviations: CI, confidence interval; OR, odds ratio; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism.
Figure 3.
Meta-analysis of the association between PON1-L55M polymorphism and cancer risk (M versus L). Abbreviations: ID, identification; CI, confidence interval; NA, not available; OR, odds ratio; weights come from random effects analysis.
Similarly, an increased risk was observed in the Caucasian population under the five genetic models: M versus L: OR =1.231, 95% CI = 1.028-1.474; MM versus LL: OR =1.737, 95% CI =1.519-1.986; ML versus LL: OR =1.170, 95% CI =1.034-1.324; MM versus ML+LL: OR =1.407, 95%CI =1.092-1.813; ML+MM versus LL: OR =1.334, 95%CI =1.215-1.465, the Asian population (M versus L: OR =1.604, 95% CI =1.089-2.363; MM versus LL: OR =2.093, 95% CI =1.295-3.381; ML versus LL: OR =1.550, 95% CI =0.995-2.417; MM versus ML+LL: OR =1.624, 95%CI =1.041-2.535; ML+MM versus LL: OR =1.967, 95%CI =1.238-3.125), the mixed population (M versus L: OR =1.177, 95%CI =1.004-1.379; ML+MM versus LL: OR =1.126, 95%CI =1.006-1.261) (Table 4), hospital-based group (M versus L: OR =11.240, 95%CI=1.056-1.456; MM versus LL: OR =1.531, 95%CI =1.199-1.955; ML versus LL: OR =1.255, 95%CI =1.020-1.543; MM versus ML+LL: OR =1.288, 95%CI =1.120-1.480; ML+MM versus LL: OR =1.411, 95%CI=1.173-1.698), and population-based group (M versus L: OR =1.325, 95%CI=1.085-1.618; MM versus LL: OR =1.568, 95%CI =1.091-2.253; ML versus LL: OR =1.275, 95%CI =1.051-1.548; MM versus ML+LL: OR =1.503, 95%CI =1.110-2.034; ML+MM versus LL: OR =1.222, 95%CI=1.122-1.331). In addition, we identified an increased risk by stratified analysis using genotyping method.
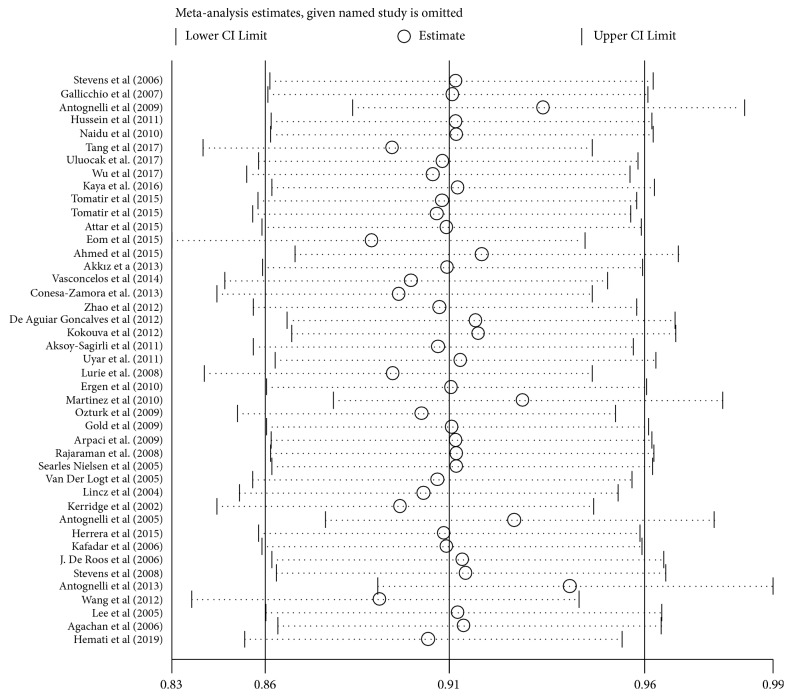
3.2.3. Publication Bias and Sensitivity Analysis
A sensitivity analysis was carried out to detect the impact of individual papers on whole data by getting rid of one report at a time from the pooled analysis. And no individual report has been significantly affected by the pooled OR. Figure 4 showed the plot of the sensitivity analysis for evaluating the association between PON1-Q192R and cancer risk (RR versus QQ). Besides, we perform Egger's test and Begg's funnel plot to evaluate publication bias (Figure 5). And the results of Egger's test and Begg's funnel plot did not uncover publication bias in PON1 (Q192R and L55M) gene polymorphisms (PON1 Q192R: R versus Q: Begg's test: z=1.74 P=0.082; Egger's test: t= -1.26 P=0.216; PON1-L55M: M versus L: Begg's test: z=0.06 p=0.953; Egger's test: t= 0.66; P=0.516). Thus, our results are believable due to the absence of significant publication bias in our meta-analysis.
Figure 4.
Sensitivity analysis of PON1-Q192R in overall OR coefficients (RR versus QQ). Abbreviations: OR, odds ratio CI, confidence interval. Sequentially calculated results of each study are omitted. Both ends of the broken line represent 95% of the CI.
Figure 5.
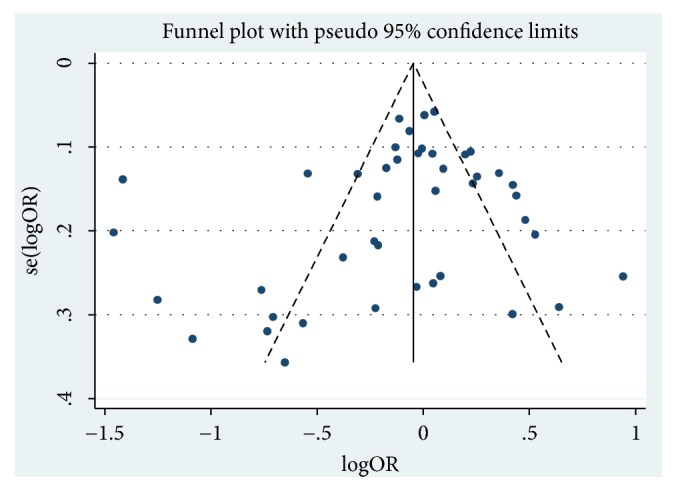
Funnel figure of PON1-Q192R in overall OR coefficients (RR versus QQ). Abbreviations: OR, odds ratio.
4. Discussion
Several studies have indicated that PON1, which is one of xenobiotic metabolising enzymes, plays a crucial role in the detoxification of carcinogenic compounds and decreases oxidative stress. Genetic polymorphisms can influence the enzyme and modify its activity, resulting in an impact on individual sensitivity to certain pathologies [61]. Indeed, a great deal of researches have showed that polymorphisms encoding the gene of these enzymes have been linked to the progression of cancer [49, 62]. Furthermore, several variants of PON1, including Q192R and L55M, have been found to be a biologically reasonable candidate which has an obvious influence on cancer. PON1 (Q192R and L55M) gene polymorphisms were related to many types of cancer, such as breast, prostate, and hepatocellular carcinoma [20, 50, 63]. For instance, PON1-L55M polymorphism may increase the risk in multiple cancer types, such as prostate and breast cancers but decrease renal cell carcinoma and ovarian cancer risk. As for PON1-Q192R, it has been revealed to suppress expression in lung [64] and pancreatic cancer [65] and reduce the risk of breast and prostate cancers. And the results of these researches were inconclusive and controversial.
In our work, in all genetic models we have identified the significant association between PON1-L55M polymorphism and overall cancer risk, while PON1-Q192R allele was not associated with reduced overall cancer risks. In the stratified analysis, we observed an increased risk in the Caucasian population and the Asian population, as well as the hospital-based group and population-based group under all the five genetic models in the PON1-L55M polymorphism. Similarly, a significantly increased risk of the overall cancers under the homozygote, allele contrast, recessive, and dominant models was uncovered in hematological tumor in the PON1-L55M polymorphism. Nevertheless, in the PON1-Q192R polymorphism, we also observe a reduced risk of the overall cancers in the allele contrast and dominant models. Meanwhile, we could obtain an interesting phenomenon that PON1-L55M polymorphism acts as a risk factor in all the five genetic models and there was an association between Q192R polymorphism and a reduced risk for cancer progression (except recessive model) after stratified analyses by cancer type, especially breast cancer. Thus, we can obtain that PON1 (Q192R and L55M) gene polymorphisms play a vital role in the development of breast cancer, whose mechanism maybe as follows: there was a critical association between L allele and higher PON1 serum concentrations while M variant decreased the stability of this enzyme. Therefore, the blood concentration of PON1 was reduced in this way; then, the activity of the enzyme was influenced, which may increase the vulnerability to genomic damage by reducing the inflammatory oxidant and the detoxifying ability of dietary carcinogens, thereby increasing the risk of breast cancer [5]. Furthermore, breast cancer becomes more susceptible to genomic damage as a result of lower levels of PON1 which could decrease the ability to detoxify inflammatory oxidants and dietary carcinogens [5]. Similarly, the exchange of Q and R could produce an enzyme which has a higher detoxification activity when there were potential carcinogenic products of oxidative stress and lipid peroxidation [66, 67]. In addition, not only genetic factors but also other contributors including nutrition and lifestyle can significantly affect PON1 enzyme activity, thereby reducing the risk of breast cancer [68]. To sum up, PON1, as a member of lipid peroxidation scavenging systems, may have an impact on malignant transformation and cell proliferation in the progression of breast cancer [69]. In the ethnographic analysis, we found ethnic groups having different results, which may be due to ethnic living habits, living environment, and genetic factors.
Previous meta-analysis also reported the association of PON1 polymorphism with cancer risk [10, 70]. As far as we know, we are the first of the typical functional polymorphism of the PON1 gene including all the published and defined case-control studies that have been conducted in a comprehensive meta-analysis. Compared with previous researches, our report was more persuasive and we have carried out a more detailed analysis to demonstrate our results. First and most obviously, the data we collected in our study was up-to-date, and we could keep up with the research front. Secondly, we included more qualified studies and larger sample size, which indicates that we are relatively more accurate in assessing that association between the PON1 gene SNPs and the risk of cancer.
Despite the association between PON1 (Q192R and L55M) gene polymorphism and cancer risk which has been studied in detail, we should note some limitations at the same time. First of all, the quantity of publications collected in our study was limited and there was a relatively small sample size of the report. What is more, Caucasian accounted for the most of the registered publications and there were no Africans. Furthermore, some of publications would only publish positive results, which could make the meta-analysis less credible. Lastly, our results were based on the estimates of single-factor, which could lead to serious confusion and bias due to the lack of raw data, and there is a need to adjust the effect size with possible confounders related to lifestyle risk factors, such as age, obesity, alcohol consumption, and smoking.
In conclusion, our study has demonstrated that PON1-Q192R can significantly reduce the risk of cancer and the polymorphism of PON1-L55M is a risk factor leading to cancer, especially breast cancer. Next, we need a larger sample size at protein levels to confirm whether PON1 polymorphisms may be potential genetic markers of tumor prognosis and identify its role in the risk of women developing breast cancer.
Acknowledgments
This work was supported by grants from National Science Foundation of China (81760530) and National Science Foundation of Guangxi (2017GXNSFBA198047).
Contributor Information
Lingsha Huang, Email: huanglinshagx@126.com.
Jinfeng Liu, Email: rainbowgxnn@yeah.net.
Bo Zhu, Email: zhubogxnn@126.com.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Authors' Contributions
Bo Zhu, Lingsha Huang, and Jinfeng Liu conceived and designed the experiments; Xiaolan Pan, Lei Huang, and Meiqin Li conduced literature review and data abstraction; Xiaolan Pan and Bo Zhu analyzed data; and Dan Mo, Yihua Liang, and Zhaodong Huang conducted a hand search for extra studies. Xiaolan Pan, Bo Zhu, Lingsha Huang, and Meiqin Li wrote the manuscript. All authors reviewed and approved the manuscript. Xiaolan Pan, Lei Huang, and Meiqin Li have contributed equally to this work.
References
- 1.Bredberg A. Cancer: more of polygenic disease and less of multiple mutations? A quantitative viewpoint. Cancer. 2011;117(3):440–445. doi: 10.1002/cncr.25440. [DOI] [PubMed] [Google Scholar]
- 2.Ivanišević J., Kotur-Stevuljević J., Stefanović A., et al. Association of paraoxonase 1 and oxidative stress with acute kidney injury in premature asphyxiated neonates. Chemico-Biological Interactions. 2017;272:47–52. doi: 10.1016/j.cbi.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 3.Assis R., Arcaro C., Gutierres V., et al. Combined effects of curcumin and lycopene or bixin in yoghurt on inhibition of LDL oxidation and increases in HDL and paraoxonase levels in streptozotocin-diabetic rats. International Journal of Molecular Sciences. 2017;18(4) doi: 10.3390/ijms18040332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farinati F., Piciocchi M., Lavezzo E., Bortolami M., Cardin R. Oxidative stress and inducible nitric oxide synthase induction in carcinogenesis. Digestive Diseases. 2010;28(4-5):579–584. doi: 10.1159/000320052. [DOI] [PubMed] [Google Scholar]
- 5.Hussein Y. M., Gharib A. F., Etewa R. L., ElSawy W. H. Association of L55M and Q192R polymorphisms in paraoxonase 1 (PON1) gene with breast cancer risk and their clinical significance. Molecular and Cellular Biochemistry. 2011;351(1-2):117–123. doi: 10.1007/s11010-011-0718-4. [DOI] [PubMed] [Google Scholar]
- 6.Eroglu M., Yilmaz N., Yalcinkaya S., Ay N., Aydin O., Sezer C. Enhanced HDL-cholesterol-associated anti-oxidant PON-1 activity in prostate cancer patients. Kaohsiung Journal of Medical Sciences. 2013;29(7):368–373. doi: 10.1016/j.kjms.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Brophy V. H., Jampsa R. L., Clendenning J. B., McKinstry L. A., Jarvik G. P., Furlong C. E. Effects of 5′ regulatory-region polymorphisms on paraoxonase-gene (PON1) expression. American Journal of Human Genetics. 2001;68(6):1428–1436. doi: 10.1086/320600.61053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aviram M., Hardak E., Vaya J., et al. Human serum paraoxonases (PON1) Q and R selectively decrease lipid peroxides in human coronary and carotid atherosclerotic lesions: PON1 esterase and peroxidase-like activities. Circulation. 2000;101(21):2510–2517. doi: 10.1161/01.CIR.101.21.2510. [DOI] [PubMed] [Google Scholar]
- 9.Chen L., Lu W., Fang L., et al. Association between L55M polymorphism in Paraoxonase 1 and cancer risk: a meta-analysis based on 21 studies. OncoTargets and Therapy. 2016;9:1151–1158. doi: 10.2147/OTT.S96990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang M., Xiong H., Fang L., et al. Paraoxonase 1 (PON1) Q192R gene polymorphism and cancer risk: a meta-analysis based on 30 publications. Asian Pacific Journal of Cancer Prevention. 2015;16(10):4457–4463. doi: 10.7314/APJCP.2015.16.10.4457. [DOI] [PubMed] [Google Scholar]
- 11.Eom S., Yim D., Lee C., et al. Interactions between paraoxonase 1 genetic polymorphisms and smoking and their effects on oxidative stress and lung cancer risk in a korean population. Plos One. 2015;10(3) doi: 10.1371/journal.pone.0119100.0119100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H., Li L., Ding L., Zhang Z., Pu C. Association of genetic polymorphisms in the paraoxonase 1 gene with the risk and prognosis of non-small cell lung cancer in Chinese Han population. Journal of Investigative Medicine. 2012;60(3):592–597. doi: 10.2310/JIM.0b013e318245d557. [DOI] [PubMed] [Google Scholar]
- 13.Vasconcelos G. M., Aguiar Alves Gonçalves B., Montalvão-de-Azevedo R., et al. PON1 Q192R polymorphism (rs662) is associated with childhood embryonal tumors. Molecular Biology Reports. 2014;41(9):6111–6115. doi: 10.1007/s11033-014-3489-7. [DOI] [PubMed] [Google Scholar]
- 14.Lau J., Ioannidis J. P. A., Schmid C. H. Quantitative synthesis in systematic reviews. Annals of Internal Medicine. 1997;127(9):820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 15.Higgins J. P. T., Thompson S. G. Quantifying heterogeneity in a meta-analysis. Statistics in Medicine. 2002;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 16.DerSimonian R., Laird N. Meta-analysis in clinical trials. Controlled Clinical Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 17.Tobias A., Campbell M. J. Modelling influenza epidemics in the relation between black smoke and total mortality. A sensitivity analysis. Journal of Epidemiology & Community Health. 1999;53(9):583–584. doi: 10.1136/jech.53.9.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Begg C. B., Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. doi: 10.2307/2533446. [DOI] [PubMed] [Google Scholar]
- 19.Ahmed N. S., Shafik N. M., Elraheem O. A., Abou-Elnoeman S. A. Association of paraoxonase-1(Q192R and L55M) gene polymorphisms and activity with colorectal cancer and effect of surgical intervention. Asian Pacific Journal of Cancer Prevention. 2015;16(2):803–809. doi: 10.7314/APJCP.2015.16.2.803. [DOI] [PubMed] [Google Scholar]
- 20.Akkız H., Kuran S., Akgöllü E., et al. Effect of PON1 gene polymorphisms in Turkish patients with hepatocellular carcinoma. Meta Gene. 2013;1:93–101. doi: 10.1016/j.mgene.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antognelli C., Del Buono C., Ludovini V., et al. CYP17, GSTP1, PON1 and GLO1 gene polymorphisms as risk factors for breast cancer: an Italian case-control study. BMC Cancer. 2009;9, article no. 115 doi: 10.1186/1471-2407-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antognelli C., Mearini L., Talesa V. N., Giannantoni A., Mearini E. Association of CYP17, GSTP1, and PON1 polymorphisms with the risk of prostate cancer. The Prostate. 2005;63(3):240–251. doi: 10.1002/pros.20184. [DOI] [PubMed] [Google Scholar]
- 23.Antognelli C., Mezzasoma L., Mearini E., Talesa V. N. Glyoxalase 1-419C>a variant is associated with oxidative stress: implications in prostate cancer progression. Plos One. 2013;8(9) doi: 10.1371/journal.pone.0074014.e74014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng T. D., Makar K. W., Neuhouser M. L., et al. Folate-mediated one-carbon metabolism genes and interactions with nutritional factors on colorectal cancer risk: Women's Health Initiative Observational Study. Cancer. 2015;121(20):3684–3691. doi: 10.1002/cncr.29465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Roos A. J. Metabolic gene variants and risk of non-hodgkin's lymphoma. Cancer Epidemiology Biomarkers & Prevention. 2006;15(9):1647–1653. doi: 10.1158/1055-9965.EPI-06-0193. [DOI] [PubMed] [Google Scholar]
- 26.Ergen A., Kılıcoglu O., Ozger H., Agachan B., Isbir T. Paraoxonase 1 192 and 55 polymorphisms in osteosarcoma. Molecular Biology Reports. 2011;38(6):4181–4184. doi: 10.1007/s11033-010-0538-8. [DOI] [PubMed] [Google Scholar]
- 27.Gallicchio L., McSorley M. A., Newschaffer C. J., et al. Body mass, polymorphisms in obesity-related genes, and the risk of developing breast cancer among women with benign breast disease. Cancer Detection and Prevention. 2007;31(2):95–101. doi: 10.1016/j.cdp.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 28.Geng R., Chen Z., Zhao X., et al. Oxidative stress-related genetic polymorphisms are associated with the prognosis of metastatic gastric cancer patients treated with epirubicin, oxaliplatin and 5-fluorouracil combination chemotherapy. Plos One. 2014;9(12) doi: 10.1371/journal.pone.0116027.e116027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gold L. S., De Roos A. J., Brown E. E., et al. Associations of common variants in genes involved in metabolism and response to exogenous chemicals with risk of multiple myeloma. Cancer Epidemiology. 2009;33(3-4):276–280. doi: 10.1016/j.canep.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gonzalez-Herrera L., Gamas-Trujillo P. A., Medina-Escobedo G., et al. The paraoxonase 1 Gene c.-108C>T SNP in the promoter is associated with risk for glioma in mexican patients, but not the p.L55M or p.Q192R polymorphisms in the coding region. Genetic Testing and Molecular Biomarkers. 2015;19(9):494–499. doi: 10.1089/gtmb.2014.0322. [DOI] [PubMed] [Google Scholar]
- 31.Hussein Y. M., Gharib A. F., Etewa R. L., ElSawy W. H. Association of L55M and Q192R polymorphisms in paraoxonase 1 (PON1) gene with breast cancer risk and their clinical significance. Molecular and Cellular Biochemistry. 2011;351(1-2):117–123. doi: 10.1007/s11010-011-0718-4. [DOI] [PubMed] [Google Scholar]
- 32.Kaya M. O., Sinan S., Güler Ö. Ö., Arslan O. Is there a relation between genetic susceptibility with cancer? A study about paraoxanase (PON1) enzyme activity in breast cancer cases. Journal of Enzyme Inhibition and Medicinal Chemistry. 2015;31(6):1349–1355. doi: 10.3109/14756366.2015.1134523. [DOI] [PubMed] [Google Scholar]
- 33.Kokouva M., Koureas M., Dardiotis E., et al. Relationship between the paraoxonase 1 (PON1) M55L and Q192R polymorphisms and lymphohaematopoietic cancers in a Greek agricultural population. Toxicology. 2013;307:12–16. doi: 10.1016/j.tox.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 34.Lurie G., Wilkens L. R., Thompson P. J., et al. Genetic polymorphisms in the paraoxonase 1 gene and risk of ovarian epithelial carcinoma. Cancer Epidemiology Biomarkers & Prevention. 2008;17(8):2070–2077. doi: 10.1158/1055-9965.EPI-08-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martínez C., Molina J. A., Alonso-Navarro H., Jiménez-Jiménez F. J., Agúndez J. A., García-Martín E. Two common nonsynonymous paraoxonase 1 (PON1) gene polymorphisms and brain astrocytoma and meningioma. BMC Neurology. 2010;10, article no. 71 doi: 10.1186/1471-2377-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Metin Z. B., Aydin S., Unur M., et al. Oral squamous cell carcinoma and serum paraoxonase 1. The Journal of Laryngology & Otology. 2013;127(12):1208–1213. doi: 10.1017/S0022215113002533. [DOI] [PubMed] [Google Scholar]
- 37.Naidu R., Har Y. C., Taib N. A. M. Genetic polymorphisms of paraoxonase 1 (PON1) gene: Association between L55M or Q192R with breast cancer risk and clinico-pathological parameters. Pathology & Oncology Research. 2010;16(4):533–540. doi: 10.1007/s12253-010-9267-5. [DOI] [Google Scholar]
- 38.Nielsen S. S., Mueller B. A., De Roos A. J., A. Viernes H., Farin F. M., Checkoway H. Risk of brain tumors in children and susceptibility to organophosphorus insecticides: the potential role of paraoxonase (PON1) Environmental Health Perspectives. 2005;113(7):909–913. doi: 10.1289/ehp.7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stevens V. L., Rodriguez C., Pavluck A. L., Thun M. J., Calle E. E. Association of polymorphisms in the paraoxonase 1 gene with breast cancer incidence in the CPS-II nutrition cohort. Cancer Epidemiology Biomarkers & Prevention. 2006;15(6):1226–1228. doi: 10.1158/1055-9965.EPI-05-0930. [DOI] [PubMed] [Google Scholar]
- 40.Stevens V. L., Rodriguez C., Talbot J. T., Pavluck A. L., Thun M. J., Calle E. E. Paraoxonase 1 (PON1) polymorphisms and prostate cancer in the CPS-II nutrition cohort. The Prostate. 2008;68(12):1336–1340. doi: 10.1002/pros.20796. [DOI] [PubMed] [Google Scholar]
- 41.Uluocak N., Atilgan D., Parlaktas B. S., Erdemir F., Ates O. A pilot study assessing the association between paraoxonase 1 gene polymorphism and prostate cancer. Turkish Journal of Urology. 2017;43(3):279–283. doi: 10.5152/tud.2017.74151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uyar O., Kara M., Erol D., Ardicoglu A., Yuce H. Investigating paraoxonase-1 gene Q192R and L55M polymorphism in patients with renal cell cancer. Genetics and Molecular Research. 2011;10(1):133–139. doi: 10.4238/vol10-1gmr927. [DOI] [PubMed] [Google Scholar]
- 43.Arpaci A., Görmüş U., Dalan B., Berkman S., Isbir T. Investigation of PON1192 and PON1 55 polymorphisms in ovarian cancer patients in Turkish population. In Vivo. 2009;23(3):421–424. [PubMed] [Google Scholar]
- 44.Tomatir A. G., Pehlivan S., Sahin H. H., Balci S. O., Budeyri S., Pehlivan M. Q192R and L55M polymorphisms of paraoxonase 1 gene in chronic myelogenous leukemia and chronic lymphocytic leukemia. Anticancer Reseach. 2015;35(9):4807–4812. [PubMed] [Google Scholar]
- 45.Kafadar A. M., Ergen A., Zeybek U., Agachan B., Kuday C., Isbir T. Paraoxonase 192 gene polymorphism and serum paraoxonase activity in high grade gliomas and meningiomas. Cell Biochemistry & Function. 2006;24(5):455–460. doi: 10.1002/cbf.1284. [DOI] [PubMed] [Google Scholar]
- 46.Agachan B., Yilmaz H., Ergen H. A., Karaali Z. E., Isbir T. Paraoxonase (PON1) 55 and 192 polymorphism and its effects to oxidant-antioxidant system in Turkish patients with type 2 diabetes mellitus. Physiological Research. 2005;54(3):287–293. [PubMed] [Google Scholar]
- 47.De Aguiar Gonçalves B. A., Vasconcelos G. M., Thuler L. C. S., Andrade C., Faro A., Pombo-De-Oliveira M. S. NQO1 rs1800566 (C609T), PON1 rs662 (Q192R), and PON1 rs854560 (L55M) polymorphisms segregate the risk of childhood acute leukemias according to age range distribution. Cancer Causes & Control. 2012;23(11):1811–1819. doi: 10.1007/s10552-012-0060-5. [DOI] [PubMed] [Google Scholar]
- 48.Van Der Logt E. M. J., Janssen C. H. J. M., Van Hooijdonk Z., et al. No association between genetic polymorphisms in NAD(P)H oxidase p22 phox and paraoxonase 1 and colorectal cancer risk. Anticancer Reseach. 2005;25(2 B):1465–1470. [PubMed] [Google Scholar]
- 49.Kerridge I., Lincz L., Scorgie F., Hickey D., Granter N., Spencer A. Association between xenobiotic gene polymorphisms and non-Hodgkin's lymphoma risk. British Journal of Haematology. 2002;118(2):477–481. doi: 10.1046/j.1365-2141.2002.03606.x. [DOI] [PubMed] [Google Scholar]
- 50.Wu J., Fang M., Zhou X., Zhu B., Yang Z. Paraoxonase 1 gene polymorphisms are associated with an increased risk of breast cancer in a population of Chinese women. Oncotarget . 2017;8(15):25362–25371. doi: 10.18632/oncotarget.15911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lincz L. F., Kerridge I., Scorgie F. E., Bailey M., Enno A., Spencer A. Xenobiotic gene polymorphisms and susceptibility to multiple myeloma. Haematologica. 2004;89(5):628–629. [PubMed] [Google Scholar]
- 52.Vecka M., Jachymova M., Vavrova L., Kodydkova J., Macasek J., Urbanek M. Paraoxonase-1 (PON1) status in pancreatic cancer: relation to clinical parameters. Folia Biologica. 2012;58(6):231–237. [PubMed] [Google Scholar]
- 53.Öztürk O., Kağnici Ö. F., Öztürk T., et al. 192R allele of paraoxanase 1 (PON1) gene as a new marker for susceptibility to bladder cancer. Anticancer Reseach. 2009;29(10):4041–4046. [PubMed] [Google Scholar]
- 54.Aksoy-Sagirli P., Cakmakoglu B., Isbir T., et al. Paraoxonase-1 192/55 polymorphisms and the risk of lung cancer in a Turkish population. Anticancer Reseach. 2011;31(6):2225–2229. [PubMed] [Google Scholar]
- 55.Conesa-Zamora P., Ruiz-Cosano J., Torres-Moreno D., et al. Polymorphisms in xenobiotic metabolizing genes (EPHX1, NQO1 and PON1) in lymphoma susceptibility: a case control study. BMC Cancer. 2013;13(228):1471–2407. doi: 10.1186/1471-2407-13-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rajaraman P., Hutchinson A., Rothman N., et al. Oxidative response gene polymorphisms and risk of adult brain tumors. Neuro-Oncology. 2008;10(5):709–715. doi: 10.1215/15228517-2008-037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao P., Zhao L., Zou P., et al. Genetic oxidative stress variants and glioma risk in a Chinese population: a hospital-based case–control study. BMC Cancer. 2012;12(617):1471–2407. doi: 10.1186/1471-2407-12-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Attar R., Atasoy H., Inal-Gültekin G., et al. The effects of PON1 gene Q192R variant on the development of uterine leiomyoma in Turkish patients. In Vivo. 2015;29(2):243–246. [PubMed] [Google Scholar]
- 59.Tang W., Liu J., Wang Y., et al. Association between Paraoxonase 1 polymorphisms and risk of esophagogastric junction adenocarcinoma: A case-control study involving 2,740 subjects. Oncotarget . 2017;8(60):101095–101102. doi: 10.18632/oncotarget.20104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hemati M., Mansourabadi A. H., Bafghi M. K., Moradi A. Association between paraoxonase-1 gene Q192R and L55M polymorphisms and risk of gastric cancer: a case-control study from Iran. Nucleosides, Nucleotides and Nucleic Acids. 2019;38(7):521–532. doi: 10.1080/15257770.2019.1573371. [DOI] [PubMed] [Google Scholar]
- 61.Ouerhani S., Ben Bahria I., Rouissi K., Cherni L. Distribution of xenobiotic metabolising enzyme genotypes in different Tunisian populations. Annals of Human Biology. 2017;44(4):366–372. doi: 10.1080/03014460.2016.1272714. [DOI] [PubMed] [Google Scholar]
- 62.De Roos A. J., Gold L. S., Wang S., et al. Metabolic gene variants and risk of non-hodgkin's lymphoma. Cancer Epidemiology Biomarkers & Prevention. 2006;15(9):1647–1653. doi: 10.1158/1055-9965.EPI-06-0193. [DOI] [PubMed] [Google Scholar]
- 63.Uluocak N., Atilgan D., Parlaktas B. S., Erdemir F., Ates O. A pilot study assessing the association between paraoxonase 1 gene polymorphism and prostate cancer. Türk Üroloji Dergisi/Turkish Journal of Urology. 2017;43(3):279–283. doi: 10.5152/tud.2017.74151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elkiran E. T., Mar N., Aygen B., Gursu F., Karaoglu A., Koca S. Serum paraoxonase and arylesterase activities in patients with lung cancer in a Turkish population. BMC Cancer. 2007;7, article 48 doi: 10.1186/1471-2407-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Akçay M. N., Polat M. F., Yilmaz I., Akçay G. Serum paraoxonase levels in pancreatic cancer. Hepato-Gastroenterology. 2003;50(2):ccxxv–ccxxvii. [PubMed] [Google Scholar]
- 66.Antognelli C., Del Buono C., Ludovini V., et al. CYP17, GSTP1, PON1 and GLO1 gene polymorphisms as risk factors for breast cancer: an Italian case-control study. BMC Cancer. 2009;9(115):1471–2407. doi: 10.1186/1471-2407-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gallicchio L., McSorley M. A., Newschaffer C. J., et al. Body mass, polymorphisms in obesity-related genes, and the risk of developing breast cancer among women with benign breast disease. Cancer Epidemiology. 2007;31(2):95–101. doi: 10.1016/j.cdp.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 68.Ferrè N., Camps J., Fernández-Ballart J., et al. Regulation of serum paraoxonase activity by genetic, nutritional, and lifestyle factors in the general population. Clinical Chemistry. 2003;49(9):1491–1497. doi: 10.1373/49.9.1491. [DOI] [PubMed] [Google Scholar]
- 69.Delimaris I., Faviou E., Antonakos G., Stathopoulou E., Zachari A., Dionyssiou-Asteriou A. Oxidized LDL, serum oxidizability and serum lipid levels in patients with breast or ovarian cancer. Clinical Biochemistry. 2007;40(15):1129–1134. doi: 10.1016/j.clinbiochem.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 70.Lu W., Fang L., Xiong H., et al. Association between L55M polymorphism in Paraoxonase 1 and cancer risk: a meta-analysis based on 21 studies. OncoTargets and Therapy. 2016:p. 1151. doi: 10.2147/OTT.S96990. [DOI] [PMC free article] [PubMed] [Google Scholar]