Abstract
The epigenome serves as a signal integration platform that encodes information from experience and environment that adds tremendous complexity to the regulation of transcription required for memory, beyond the directions encoded in the genome. To date, our understanding of how epigenetic mechanisms integrate information to regulate gene expression required for memory is primarily obtained from male derived data despite sex-specific life experiences and sex differences in consolidation and retrieval of memory, and in the molecular mechanisms that mediate these processes. In this review, we examine the contribution of chromatin modification to learning and memory in both sexes. We provide examples of how exposure to a number of internal and external factors influence the epigenome in sex-similar and sex-specific ways that may ultimately impact transcription required for memory processes. We also pose a number of key open questions and identify areas requiring further investigation as we seek to understand how histone modifying mechanisms shape memory in females.
The prevalence of memory-related disorders such as posttraumatic stress disorder (PTSD) and Alzheimer's disease (AD) in women is at a rate twice that of men (Kessler et al. 1995, 2012; Kim et al. 2015; Neu et al. 2017), whereas in neurodevelopmental disorders the opposite is observed (Fombonne 2009; Zablotsky and Black 2015). In PTSD, women are diagnosed at higher rates even in studies that reveal greater exposure to traumatic events in men (Tolin and Foa 2006). Associated with the increased prevalence of AD in women, there is also an acceleration of cognitive decline and increased severity of cognitive impairment (Lin and Doraiswamy 2015). However, it is important to note that clinical care for AD symptomology varies greatly by sex with women being more likely to be prescribed certain classes of psychotropic medications associated with increased risk of cognitive impairment (Moga et al. 2017). As these brief examples illustrate, there is significant research needed to understand differences in vulnerability, susceptibility, and resilience to various disorders between sexes, and develop appropriate sex-specific treatment and care. Fundamental to disorders associated with cognitive function is understanding how mechanisms of memory formation are different and similar between females and males.
The ability to consolidate information and form long-term memories is largely reliant on changes in gene expression which are coordinated, in part, through epigenetic mechanisms that function to regulate transcriptional processes (Burgess-beusse et al. 2002; Horn and Petersen 2002; Mozzetta et al. 2014; Sartor et al. 2015; Korb et al. 2016). A large number of studies report sex differences in expression patterns of memory-relevant genes. However, how epigenetic mechanisms regulate gene expression required for memory formation in females and males is less understood. With the recent National Institutes of Health (NIH) mandate to include sex as a biological variable and incorporate both sexes in biomedical research (Clayton and Collins 2014), the number of studies examining the epigenetic mechanisms of memory in females and males has been increasing. As might be currently predicted though, our understanding of how epigenetic mechanisms regulate memory function is largely shaped from data collected from male animals, emphasizing that we are at the very beginning of understanding how these mechanisms impact memory processing in the female.
In this review, we discuss the critical role that histone modifying enzymes play in the formation of memory in both sexes. We primarily focus on histone modifications in the hippocampus, as this region is critically involved in spatial memory formation and retrieval in both sexes. We focus on histone modifying enzymes as there is more literature on these enzymes, more studies involving females, and as one of the main epigenetic mechanisms that regulate gene expression in coordination with other mechanisms (e.g., nucleosome remodeling, DNA modification, histone variants), information from histone modifying mechanisms may inform and indicate where future studies on other epigenetic mechanisms may be most fruitful. We also discuss examples of internal and external factors capable of influencing the epigenome in sex-similar and sex-specific ways that regulate gene expression required for long-term memory. Last, we pose key open questions in understanding histone modifying mechanisms in females and males, and how knowledge of these epigenetic mechanisms may impact our understanding of sex differences in learning and memory.
A brief primer on chromatin modification and memory
The term “epigenetics” refers to the ability of both external and internal factors to influence gene expression without modifying or altering the DNA sequence. DNA is tightly wrapped around an octamer of two pairs of four proteins called histones (H2A, H2B, H3, and H4) which make up a nucleosome, the repeating unit of chromatin. Modification and remodeling of chromatin structure can dynamically regulate gene expression in numerous ways including transcriptional initiation, elongation, repression, chromosomal looping (to bring enhancers and promoters together for example), etc. The ability to form and consolidate new memories is dependent upon new transcription and activity-dependent protein synthesis (Kang and Schuman 1996; Alberini 2009). Studies of learning and memory continue to determine how chromatin modifying enzymes function to coordinately regulate gene expression required for the consolidation of memory. Yet, this is only the tip of the iceberg when considering the epigenome serves not only to regulate gene expression, but also as an incomprehensibly complex signal integration platform capable of encoding information from experience including exposure to the environment, stress, exercise, diet, etc. One key place to begin to understand the epigenome is the histone tail.
The amino-terminal end of a histone protein is referred to as the histone tail, which is the most common site of posttranslational modification (e.g., acetylation, methylation, phosphorylation). Histone modifications alter interactions within and between nucleosomes, and also serve as sites of interaction for proteins with specialized motifs (e.g., bromodomain recognizes acetylated residues). Although there are a multitude of histone modifications, the best studied ones we will discuss here include acetylation, methylation and phosphorylation, all of which dynamically regulate chromatin, and thus gene expression, and all have been implicated in memory processes. These modifications are regulated by histone modifying enzymes including acetyltransferases, deacetylases, methyltransferases, demethylases, kinases, and phosphatases (for reviews, see Kouzarides 2007; Barrett and Wood 2008). Importantly, although these enzymes are frequently called “histone acetyltransferases” (HATs), they also modify nonhistone protein residues.
Despite many similarities between females and males, sex differences are present in learning and memory (e.g., for reviews, see Cahill 2006; Andreano and Cahill 2009). Sex differences have been observed in the consolidation of memory and in the molecular mechanisms that mediate consolidation (for reviews, see Shors et al. 2000; Mizuno and Giese 2010; Keiser and Tronson 2016). The process of transcription in itself is a dynamic and complex orchestration of dozens and dozens of transcription factors and coregulators induced by various converging signaling pathways that ultimately coordinate the expression of a specific profile of genes required for lasting memory formation. While research on the molecular mechanisms of memory in females is sparse, studies including female animals report sex differences in activation of kinases (Mizuno et al. 2006, 2007; Gresack et al. 2009; Sase et al. 2019), transcription factors (Kudo et al. 2004), and more recently, histone modifying enzymes (Tsai et al. 2009; Benoit et al. 2015; Tyler et al. 2015; Sase et al. 2019), which all function to regulate gene expression. Furthermore, there are a number of internal (e.g., hormones) and external (e.g., stress) stimuli that partially modulate molecular mechanisms of memory in both sexes. Therefore, sex differences in the effects of internal and external stimuli likely have a significant impact on the function of histone posttranslational modifications in females and males (Fig. 1). We next review the role of histone modifications in the regulation of memory-related gene expression in females and males.
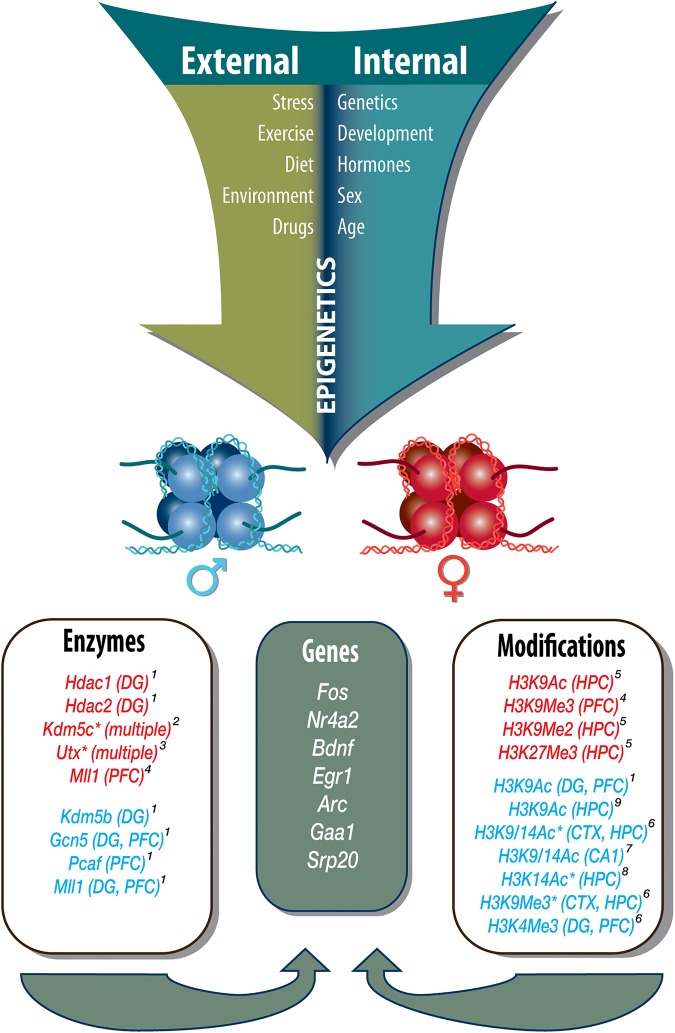
Figure 1.
External and internal factors influence modification of the epigenome, potentially leading to sex differences in gene expression required for memory consolidation and retrieval. A number of external and internal factors are important modulators of histone modification in the female and male brain. A subset of histone modifications and enzymes listed reflect either a sex difference, with higher levels of a histone modification or a histone modifying enzyme, or reflect changes in a histone modification or histone modifying enzyme by one sex but not the other. Histone modifications or enzymes that are only observed in males, or reflect higher levels compared with females, are denoted in blue. Histone modifications or enzymes that are only affected in females are denoted in red. Differences between females and males are labeled with an asterisk. Histone modifications or enzymes without an asterisk reflect changes in one sex, but not the other, when compared to same-sex controls. Superscript numbers above brain regions denote references. Together, these modifications affect expression of memory-relevant genes; examples of genes associated with memory consolidation and retrieval are listed in the middle, many of which are found to be differentially expressed between females and males. (CTX) cortex, (HPC) hippocampus, (DG) dentate gyrus, (PFC) prefrontal cortex. References: (1Tyler et al. 2015; 2Xu et al. 2008a; 3Xu et al. 2008b; 4Huang et al. 2007; 5Sobolewski et al. 2018; 6Tsai et al. 2009; 7Sase et al. 2019; 8Benoit et al. 2015; 9Glendining and Jasoni 2019).
Examples of sex similarities and differences in histone modifications involved in regulating gene expression required for memory processes
Histone acetylation
Histone acetylation involves the addition of an acetyl group to lysine tails, which generally facilitates gene expression. More specifically, acetylation occurs through enzymes called histone acetyltransferases (HATs) which utilize acetyl CoA as a cofactor and transfer an acetyl group to a lysine residue. Because lysine is a positively charged amino acid, it naturally interacts with the negative DNA phosphate backbone. Thus, lysine acetylation neutralizes that charge and is thought to relax chromatin structure and facilitate transcription by also providing docking sites for bromodomain containing proteins involved in transcriptional regulation. There are two major types of HATs (type A and type B) based in general on their presumed localization within the cell. Type B HATs are primarily found in the cytoplasm and acetylate numerous proteins as well as free histones that are newly synthesized. Interestingly, type B HATs have not been well investigated in the learning and memory field. Type A HATs on the other hand, are located in the nucleus and directly modify multiple sites on histone tails. Type A HATs can be categorized into three families: CBP/p300, MYST, and GNAT (Hodawadekar and Marmorstein 2007). Histone Deacetylases (HDACs) remove acetyl groups and return chromatin structure back to its repressed state, therefore restricting gene expression. HDACs are broken down into four classes: Class I HDACs are comprised of HDACs 1, 2, 3, and 8; class II are separated into IIa (HDACs 4, 5, 7, and 9) and IIb (HDACs 6 and 10); and class IV is comprised of HDAC 11. In general, an experience that is capable of being encoded into long-term memory will prompt histone acetylation by increasing activity of HATs and decreasing activity of HDACs, resulting in gene expression required for memory consolidation (for reviews, see Marmorstein and Roth 2001; Barrett and Wood 2008; Gräff and Tsai 2013).
Histone acetylation is the most prominently studied histone modification involved in hippocampus-dependent learning and memory (see brief history described in Campbell and Wood 2019). In males, numerous reports indicate that hippocampus-dependent memory and synaptic plasticity depend on HAT and HDAC activity. For example, our laboratory and others have demonstrated that the HAT CREB-Binding Protein (CBP) is required for memory formation in male mice and that HDAC3 is a negative regulator of memory formation in males (McQuown et al. 2011; for review, see Kwapis and Wood 2015). Similarly, histone acetylation at several sites (e.g., H3K14Ac, H4K8Ac, H4K12Ac) is increased during memory consolidation, correlating with increased gene expression (e.g., Fos, Nr4a2, Egr1, Bdnf, Arc). Mutations in HATs, like CBP, result in developmental abnormalities and intellectual disability disorders in humans as well as mice. Conversely, manipulations that inhibit HDACs dramatically enhance memory processes (for reviews, see Korzus 2017; Campbell and Wood 2019). Together, these studies highlight the powerful role of these enzymes in regulating gene expression and memory processes.
While the majority of studies implicating HATs and HDACs as critical regulators of long-term memory formation have primarily used male animals, histone acetylation is necessary for hippocampus-dependent spatial memory in females as well. For example, a study using cbp+/− female mice demonstrated that reducing CBP protein levels results in impairments in spatial memory and hippocampal neurogenesis compared with same sex-controls (Lopez-Atalaya et al. 2011). In both sexes, intrahippocampal HDAC inhibition results in a memory that is longer lasting in mice (Frick 2013). Additionally, global knock out of a transcriptional coactivator that also acts as a HAT, the p300/CBP-associated factor (PCAF), leads to impaired memory in object recognition and water maze tasks in both female and male mice (Maurice et al. 2008). Despite these and many other similarities between males and females, HATs and HDACs also play distinct roles in the male and female brain. For example, hormones are important regulators of histone acetylation and other modifying enzymes (Zhao et al. 2012; Fortress et al. 2014; Frick et al. 2015) and histone acetylation is a fundamental process for brain sexual differentiation (Murray et al. 2009; Matsuda et al. 2011). How histone acetylation impacts development and memory processing in females and males will be discussed in more detail in upcoming sections.
Histone methylation
Histone methylation involves the addition of methyl groups to lysine or arginine residues of histone tails that depending on the position of the modification and number of methyl groups added to a specific site (referred to as mono, di, or tri-methylation), can either activate or repress transcription. For example, in males, tri-methylation of lysines such as H3K4, H3K36, and H3K79 is associated with gene expression, whereas di- or tri-methylation at H3K9 and at H3K27 is associated with repression of transcription (for review, see Dong and Weng 2014). Histone methylation occurs through enzymes called methyltransferases, which transfer methyl groups to lysine or arginine residues of histone tails. There are two different types of histone methyltransferases: those that are arginine-specific and those that are lysine specific. Lysine-specific histone methyltransferases consist of two main classes: the SET domain containing family and the DOT1/KMT4 family (which only methylate nucleosomal substrates, not free histones). Demethylases, on the other hand, function to remove methyl groups from lysine or arginine residues of histone tails.
Histone methylation serves an important function for consolidation of memory and several methyltransferases have been recognized as being activated following a learning event. A study by Gupta et al. (2010) found that in male rats, 1 h following contextual fear conditioning tri-methylation of H3K4 (a mark associated with active transcription) was found to be up-regulated in the hippocampus, at specific promoters of memory-related genes such as Egr1 and Bdnf. Up-regulation of tri methyl-H3K4 at these promoters was not sustained 24 h later, suggesting that a demethylase may have facilitated removal of the methyl group. Additional studies using male rats have also demonstrated changes in H3K4 methylation in retrieval of cued fear-related memory. H3K4Me3 was increased in the CA1 region of the hippocampus at the promoter of the gene Npas4 (implicated in memory formation) compared with controls (Webb et al. 2017). Further, direct manipulation of histone methylation via knockdown of the methyltransferase Kmt2a in the CA1 region of the hippocampus prevented the retrieval-induced increases in CA1 Npas4 mRNA levels and resulted in impaired fear memory (Webb et al. 2017). These brief examples demonstrate that similar to histone acetylation mechanisms, histone methylation mechanisms likely play an important role in transcriptional regulation required for memory.
Also similar to histone acetylation, the majority of studies on histone methylation have exclusively used males. Thus, it is not well understood whether similar histone methylation mechanisms in females similarly impact memory processes. There is intriguing evidence to suggest that histone methyltransferases and demethylases may function differently in the female brain to affect memory. For example, the histone demethylase KDM5C and UTX are coded by genes that are X-linked and escape X-inactivation in females (Fig. 1; Agulnik et al. 1994; Wu et al. 1994a,b; Greenfield et al. 1998), which result in higher expression of these histone demethylases (Xu et al. 2002, 2008a,b), and may mediate sex differences in brain development, memory and behavior. Although a paralogue of KDM5C, called KDM5D is expressed from the Y chromosome, it was found to be expressed only at nearly 1/3 the level of KDM5C (Xu et al. 2008a), therefore not compensating for the higher levels of KDM5C observed in females. We discuss this in more detail and discuss how sex differences in histone methylation during development and sexual differentiation may impact memory processing (see upcoming X-chromosome inactivation section).
Histone phosphorylation
Histone phosphorylation, similar to the other modifications above, is critical for transcription required for memory consolidation. Generally, histone phosphorylation involves the addition of a phosphate group on serines, threonines and tyrosines, mainly occurring on the N-terminal histone tails. Histone phosphorylation occurs via the action of kinases and phosphatases that function to add or remove phosphate groups, respectively (Oki et al. 2007). The act of transferring a phosphate group to the target amino acid provides a negative charge, as well as a docking site for proteins with phospho-binding domains.
In males, a number of kinases have been shown to facilitate histone phosphorylation and be critical for memory formation. For example, following context fear conditioning, extracellular signal-regulated kinase/mitogen-activated protein kinase (ERK/MAPK) signaling has been shown to positively correlate with histone H3 phosphorylation in the hippocampus of male rats (Chwang et al. 2006). Specifically, the authors of this study found that activation of ERK/MAPK led to an increase in histone H3 phosphorylated at serine 10 (H3S10p), H3K14 acetylation (H3K14Ac) and phospho-acetyl histone H3 at these same sites in the CA1 region of the hippocampus. Further, blocking ERK/MAPK activity, via inhibition of the kinase upstream of ERK, MAP kinase/ERK kinase (MEK), prevented the increase in histone H3 phosphorylation normally observed in the hippocampus following context fear conditioning. Therefore, following acquisition of context fear conditioning, histone phosphorylation in males is regulated by ERK/MAPK, presumably in the service of regulating gene expression required for memory.
Few studies of learning and memory have examined histone phosphorylation in females and to our knowledge, no studies have determined upstream kinases critical for regulating histone phosphorylation in the female brain. However, a number of studies that have included both sexes find that during consolidation expression of some kinases, necessary for memory in males, are expressed at different levels in females and may not be required for consolidation (for reviews, see Shors et al. 2000; Mizuno and Giese 2010; Keiser and Tronson 2016). For example, following acquisition of context fear conditioning male rats display higher levels of hippocampal phosphorylated ERK (pERK) compared with females (Gresack et al. 2009). Given that in males, ERK regulates H3S10p (Chwang et al. 2006), higher levels of hippocampal ERK in males may lead to greater phosphorylation at H3S10 in males compared with females. Additionally, following acquisition of context fear, female rats show lower hippocampal levels of cAMP response element-binding protein (CREB), a known target of ERK/MAPK signaling in addition to lower levels of freezing compared with male rats (Kudo et al. 2004). CREB is a well-studied transcription factor necessary for memory processes (Kogan et al. 1996; Pittenger et al. 2002; Josselyn et al. 2004; Alberini 2009). Therefore, sex differences in the expression and requirement of upstream kinases during memory consolidation also likely lead to female- and male-specific differences in histone modification, potentially resulting in sex differences in expression of memory-relevant genes. Before we can begin to understand how histone phosphorylation is regulated to impact memory in the female brain, foundational work must first begin to unravel the critical brain regions, enzyme expression, and patterns of histone modifications in consolidation of memory.
Other histone modifications
In addition to histone acetylation, methylation, and phosphorylation there are many additional histone modifications critical for transcription. These include, but are not limited to, ubiquitination (Goldknopf et al. 1975), sumoylation (Nathan et al. 2003; Shiio and Eisenman 2003), ribosylation (Fontan-Lozano et al. 2010), citrullination (Christophorou et al. 2016), and serotonylation (Farrelly et al. 2019). To date, the number of studies examining the impact of these other histone modifications in memory formation in the female, in addition to the male brain are largely nonexistent. Thus, we cannot discuss these modifications here in detail but to say that more research is needed in this area. For the remainder of this review we discuss how both internal and external factors (Fig. 1) may differentially impact histone modification associated with memory processing in the female and male brain.
The interplay between internal factors (X-chromosome inactivation, brain sexual differentiation, hormonal regulation), histone modifications, and memory processes
The majority of studies examining the role of histone modifying enzymes in the female in addition to the male brain have focused on development and brain sexual differentiation. Histone acetylation and methylation and a number of other modifications are implicated not just in brain development but a number of brain disorders, most of which have an effect on learning and memory function. Therefore, engagement of histone modification mechanisms early in life in females and males has lasting consequences for learning and memory function into adulthood. Here, we highlight how epigenetic modifications that influence development and brain sexual differentiation strongly impacts memory processing in females and males. We also discuss how memory processing is impacted by epigenetic mechanisms as a consequence of activational effects of hormones beyond development.
X-chromosome inactivation
Recent evidence indicates that X chromosome-dependent mechanisms play a role in observed sex differences in memory. The process of X-inactivation refers to transcriptional silencing of one X chromosome in females to provide dosage equivalence or compensation of the X chromosome between the sexes (Nguyen and Disteche 2006). Which X chromosome undergoes inactivation is unpredictable, making the female brain (and body) a mosaic of cells, some expressing the maternally inherited X chromosome and the others expressing the paternally inherited X chromosome (Tan et al. 1995). While the majority of genes on the inactive X-chromosome in females are silenced, around 3% of X-linked genes have been shown to escape X inactivation (Yang et al. 2010). As mentioned prior, one such gene that escapes X-inactivation is a gene that codes for a chromatin modifying enzyme called Kdm5c (also known as Jarid1c), which is a histone 3, lysine 4 (H3K4Me2/3) demethylase (Wu et al. 1994a; Agulnik et al. 1994). Zamurrad et al. (2018) demonstrated in a recent study that Drosophila harboring a missense allele of Kdm5, which is analogous to a mutation in Kdm5c in patients with intellectual disability are impaired in learning and/or memory. Additionally, the authors demonstrate that this mutation disrupts the transcription of a number of genes important for cognitive function (Zamurrad et al. 2018). However, to our knowledge, the sex of flies was not reported in this study. In male and female mice, KDM5C also plays a critical role in learning and memory as KDM5C knockout mice exhibit impairments in learning and memory in a variety of different tasks (Iwase et al. 2016; Scandaglia et al. 2017).
KDM5C also plays a critical role in brain development (Lingenfelter et al. 1998) and mouse studies have demonstrated greater mRNA expression of Kdm5c in a number of tissues in the female compared with the male, including the hippocampus (Fig. 1; Xu et al. 2002, 2008a). Further, it was observed that the greater levels of Kdm5c mRNA was due to the presence of two X chromosomes, independent of gonadal sex (testes versus ovaries) (Xu et al. 2008a). Studies using mouse models with either one or two X chromosomes but the same gonads, allow for sex chromosome complement to be unrelated to biological sex (for review, see Arnold and Chen 2009). Therefore, higher levels of Kdm5c mRNA expression in the XX female hippocampus may result in lower levels of H3K4Me3 at specific promoters in females compared with males. As H3K4Me3 is associated with active transcription (Klose and Zhang 2007), the possibility of greater demethylase activity in females compared with males may result in decreased transcription, or perhaps a higher threshold for transcriptional activation, of target genes in females, which may ultimately impact learning and memory differentially in females versus males. Conversely, the single copy of Kdm5c may leave males particularly vulnerable to mutations in Kdm5c, resulting in developmental and intellectual disability.
In humans, males are diagnosed with neurodevelopmental and intellectual disability disorders at a rate more than 2–3 times higher than females (Fombonne 2009; Zablotsky and Black 2015, Zablotsky et al. 2017), and one potential mechanism involves X-linked genes, as these genes are more vulnerable to mutation in males than females. Kdm5c is thought to be one of the more frequently mutated genes in X-linked Intellectual Disability disorders (Jensen et al. 2005; Vallianatos et al. 2015). Mutations that affect the start or stop codon or frameshift mutations cause severe developmental and cognitive impairments in men (Gonçalves et al. 2014). However, women carriers usually have mild learning and memory impairments (e.g., Simensen et al. 2012). Overall, these results are intriguing and give rise to many open questions including: do expression level differences hold at the protein level and are protein levels reflective of enzymatic activity; are the same genes regulated by KDM5C and KDM5D in the adult female and male brain during memory consolidation; and how do developmental changes in expression of KDM5C/KDM5D ultimately affect cognition in intellectual disability disorders?
Brain sexual differentiation
During sensitive periods of development, the brain undergoes sexual differentiation (Phoenix et al. 1959). In the perinatal period testosterone is secreted in male rodents and enters the brain where it is then converted to estradiol via the enzyme aromatase, resulting in masculinization of the brain (Naftolin et al. 1975; Maclusky and Naftolin 1981; McEwen 1981). Brain masculinization has been demonstrated to be important for sex differences observed in studies of learning and memory. For example, nearly three decades ago, a study by Williams et al. (1990) demonstrated that male rats performed better than females when tested for spatial memory recall in a radial arm maze task. However, females that were masculinized in early development performed better in the task than their same-sex counterparts, suggesting that brain sexual differentiation contributes to sex differences observed in spatial memory performance. A number of other important studies demonstrate the critical role of brain masculinization and spatial memory performance (Roof and Havens 1992; Galea et al. 1996). More recent research points to the fundamental role that histone modifying enzymes play in brain sexual differentiation.
The principal nucleus of the bed nucleus of the stria terminalis (BNSTp) is a sexually dimorphic brain region that is larger in volume and cell number in males compared with females (Hines et al. 1985, 1992; Guillamon et al. 1988; Forger et al. 2004). In rodents, the observed sex difference in cell number in the BNSTp is due to sexually dimorphic cell death occurring during the first week after birth (Chung et al. 2000; Forger et al. 2004; Gotsiridze et al. 2007) and testosterone treatment in females at birth, or castration in males, prevents this sex difference (Guillamon et al. 1988). Epigenetic mechanisms have also been shown to play a critical role in masculinization of the BNSTp. Inhibition of HDAC activity on the day of birth has been shown to increase acetylation of histone H3 in the brain the following day and results in reduced volume and cell number of the BNSTp in male mice (as well as testosterone-treated females), therefore preventing masculinization of the sexually dimorphic BNSTp (Murray et al. 2009). These findings suggest that histone acetylation plays a key role in masculinization of the BNSTp. Epigenetic modulation of BNST masculinization likely affects memory processing in males and females due to memory circuits that interact with this region (e.g., Gungor et al. 2015; Kruger et al. 2015; Asok et al. 2018; Gorka et al. 2018) and the role of BNSTp in social (Dumais et al. 2016) and fear memory (Verma et al. 2018; for review, see Goode and Maren 2017). However, it is not clear how BNST masculinization in early development impacts social and fear memory in females and males into adulthood. Further, it is difficult to prescribe histone modifying enzyme activity in the developing brain, and in the BNSTp in particular, with effects on adult learning and memory as even the hippocampus exhibits differences in histone modifying enzymatic activity during development. There are few, if any, studies directly linking sexual differentiation of specific brain regions during development with sex differences in brain regions and circuitry necessary for learning and memory. Thus, an exciting area for future investigation is to understand how histone modifying enzymes may be involved in establishing sexual differentiation and subsequently adult brain cognitive function.
In the mouse hippocampus, sex differences in histone H3 modifications have been observed early in development on embryonic day 18, the day of birth and 6 d later (Fig. 1; Tsai et al. 2009). After observing greater levels of acetylation at H3K9/14 and tri-methylation of H3K9 in the hippocampus of males compared with females, the authors sought out to determine whether masculinization was critical in mediating sex differences in histone modifications. For the last 4 d prior to gestation, pregnant dams received either testosterone or vehicle and pup brains were collected at birth. Despite hormone treatment, H3K9Me3 was still greater in the male hippocampus as compared with the female, but H3K9/14Ac increased to male levels (Tsai et al. 2009). These findings suggest that histone H3 modifications may be sexually dimorphic in the hippocampus and suggest that the H3K9Me3 modification is masculinized in females by testosterone in utero. Much further work needs to be done to confirm and extend these results, but overall they indicate that epigenetic mechanisms engaged during development in a sexually dimorphic manner in a region like the hippocampus may have important implications for how we understand learning and memory processes in females and males.
Hormonal regulation of memory and epigenetic mechanisms beyond development
Beyond development, memory is also modulated in females and males through activational effects of hormones throughout the lifespan. Epigenetic mechanisms play a fundamental role in hormonal modulation of learning and memory and cognitive function from adulthood to aging. For example, the sex-steroid hormone and primary estrogen produced by the ovaries, 17β-oestradiol, infused into the dorsal hippocampus has been shown to enhance object recognition memory in young (Fortress et al. 2014) and middle aged (Fan et al. 2010) female mice. Administration of 17β-oestradiol was also observed to correlate with increased pan-acetyl histone H3 at brain derived neurotrophic factor (BDNF) promoters pII and pIV, a critical neurotrophic factor necessary for hippocampus-dependent memory (Fortress et al. 2014; Frick et al. 2015). Additionally, the memory-enhancing effects of 17β-oestradiol on object recognition memory correlated with acetylation of histone H3 in dorsal hippocampus as bilateral dorsal hippocampal infusion of the histone acetyltransferase inhibitor, garcinol, prevented the ability of 17β-oestradiol to enhance object recognition memory in female mice (Zhao et al. 2012). These findings suggest a key role for the involvement of histone modifying mechanisms in regulating gene expression required for memory by 17β-oestradiol within the dorsal hippocampus.
The above examples clearly point to an important role for epigenetic mechanisms in hormonal modulation of learning and memory, but many open questions remain. For example, which additional sex steroid hormones in adulthood (other than estradiol) regulate histone modifications in brain regions important for memory processing, and if this is the case, which histone modifying enzymes are involved and which residues are being modified? Additionally, what is the consequence of histone modifications that are regulated by sex-steroid hormones, and do epigenetic mechanisms such as histone acetylation always become engaged when hormonal regulation leads to enhanced memory or is it selective for specific types of memory? Regardless, the current evidence strongly indicates that hormones serve an internal regulation of learning and memory by impacting how epigenetic modifications exert their effects in females and males. In the next section, we provide examples of a number of external factors that impact memory through histone modifications in the female and male brain.
The interplay between external factors (stress, environmental exposure, exercise), histone modifications, and memory processes
A variety of external factors are responsible for influencing when and which histone modifying enzymes are engaged. Below we will discuss external factors (i.e., stimuli from our environment) including stress, environmental exposure, and exercise, and how they engage epigenetic machinery and ultimately affect memory. While there is a wealth of literature on how environmental stimuli are encoded at the level of the epigenome, it is less clear how such external stimuli impact memory processing via epigenetic mechanisms (the lens with which we focus this review). With the majority of literature predominantly using males to address these questions, even less is understood about how external stimuli impact the female epigenome. Here, we briefly highlight emerging literature on how exposure to external factors such as stress, environmental toxins and exercise influence learning and memory through epigenetic modifications in manners both similar and distinct between the sexes.
Stress
Stress as an external stimulus has been shown to modulate memory in both sexes via epigenetic mechanisms. Before we discuss alterations in histone modifications as a result of stress, we must first briefly lay out how stress can differentially impact memory in females and males behaviorally and mechanistically (for a more in-depth reviews, see Luine 2002; Shors 2004; Bowman 2005; Shansky et al. 2013; Maeng and Milad 2015; Bangasser and Wicks 2017; Luine et al. 2017; Merz and Wolf 2017). In males, acute stress is capable of enhancing memory consolidation (Shors et al. 1992; Radulovic et al. 1999; Roozendaal et al. 2002). In studies using both sexes chronic stress has been observed to impair spatial memory in males and either lead to small enhancements of memory or to cause no effect in females (for review, see Luine et al. 2017). In addition, exposure to the same stressor in male and female rats leads to an NMDA-dependent decrease in dendritic spines in males, whereas in females an NMDA-dependent increase in dendritic spines is observed (Shors et al. 2004). These findings suggest that exposure to the exact same type, intensity and duration of stress in males and females can lead to sex differences in dendritic morphology and behavioral measures of memory. Furthermore, stress has been shown to differentially affect central transmitter levels of female and male rats in regions important for memory function such as the hippocampus, frontal cortex, and amygdala, which may also impact memory function (Bowman et al. 2003). Exposure to a number of external stimuli, including stress, also impacts intracellular signaling events that influence the activity of multiple histone acetyltransferases, deacetylases, methyltransferases, and demethylases (Graff et al. 2011). Therefore, sex differences in the effects of external stimuli, likely have a significant impact on the function of posttranslational modifications of histones in females and males.
In males, stress has been shown to engage histone modifying enzymes, and alter histone modifications, but the effects are largely dependent on the type and intensity of stress, age when the stressor was experienced, and brain region examined (for review, see Bagot et al. 2014). For example, regulation of corticotropin-releasing hormone (CRH) is critically important for central and peripheral responses to stress. CRH is regulated in part by cyclic-AMP response element (CRE) via interaction with the transcription factor CREB. CREB in turn is known to recruit the HAT CBP. Using a genetically modified knock-in mouse that expresses a mutant form of CBP that can no longer interact with CREB, a study found that in males, the recruitment of CBP was not necessary under basal CRH expression (Cope et al. 2014). However, the interaction and recruitment of CBP contributed to stress-induced CRH expression in the adult mouse, suggesting that CBP and its HAT activity may be important for the orchestration of gene regulation under specific stress-induced conditions.
Stress has been shown to alter histone modifications where exposure to chronic social defeat stress in male mice induced hippocampal histone H3 dimethylation of H3K27 (thought to be associated with transcriptional repression) at BDNF promoter regions, which correlated with a reduction in BDNF transcripts in the hippocampus of male mice (Tsankova et al. 2006). Additionally, the authors of this study found that hyperacetylation at BDNF promoter regions as a result of chronic administration of the tricyclic antidepressant imipramine reduced susceptibility to chronic social defeat stress (Tsankova et al. 2006). Therefore, in males, histone modifications are altered as a result of stress exposure and may play an important role in mediating response to stress, which would likely impact memory function. In mice, examining whether chronic social defeat induces the same epigenetic modifications in females becomes challenging with lack of aggression in female mice when using this model (Jacobson-Pick et al. 2013). However, female voles are as highly social and aggressive as males, and thus voles may be a more effective model to study female and male social defeat (Smith et al. 2013; Wang et al. 2018).
Additional studies, using males, have demonstrated that the severity of stress differentially impacts histone H3 methylation marks in the dentate gyrus and CA1 region of the hippocampus. Acute stress was associated with increased the levels of H3K9Me3 (associated with repression of transcription) in the dentate gyrus (DG) and CA1, while it reduced levels of H3K9Me (associated with active transcription) and H3K27Me3 (associated with repression of transcription) in the same regions. Seven days of restraint stress in male rats reduced levels of H3K4Me3 (associated with active transcription) in the CA1 and H3K27Me3 in the DG and CA1, while increasing basal levels of H3K9Me3; and chronic restraint stress for 21 d mildly increased levels of H3K4mMe3 and reduced H3K9Me3 levels in the DG (Hunter et al. 2009). Therefore, stress severity differentially affects histone methylation modifications in male hippocampus, likely affecting transcription required for long-term memory processes. Future studies are required to link stress severity with epigenetic modifications and transcription in females.
Although the majority of studies examining the impact of stress on histone modifications and memory use male subjects, there is some evidence that stress impacts histone modifications in females in ways that are distinct from males. An example where stress can lead to sex-specific consequences for histone modifications and memory can be observed in a model of prenatal stress (PNS). PNS such as sparse maternal care can lead to cognitive impairments and exaggerated stress responses and epigenetic mechanisms may underlie changes in the brain as a result of PNS (for review, see Maccari et al. 2014). A 2015 study by Benoit et al. (2015) examined how PNS impacted hippocampal-dependent spatial memory performance and histone modifications in the hippocampus of adult female and male mouse offspring. The researchers found that while both sexes that underwent PNS were impaired in the spatial water maze task, females exhibited greater changes in histone modifications in dorsal hippocampus as a result of PNS. More specifically, male mice that were exposed to PNS exhibited decreased acetylation of histone H3, lysine 14 (H3K14Ac; associated with transcriptional activation) in the hippocampus as compared with male mice that were not exposed to PNS. Females that underwent PNS showed an even further decrease in H3K14Ac when compared with female mice that did not undergo PNS (Fig. 1; Benoit et al. 2015). These studies suggest that PNS can result in similar effects on memory performance between females and males, yet histone modifications may be more impacted by PNS in one sex than the other. Due to sex differences in response to stress, it is likely that experiencing the same form of stress will differentially influence which enzymes are engaged and where those histone modifying enzymes exert their effects in females. However, much is left to be determined regarding how different intensities and types of stressors differentially influence epigenetic mechanisms that impact learning and memory.
Environmental exposure
Just as stress impacts the epigenome, so can direct exposure to compounds found in the environment from early in development and throughout the lifespan. While few studies have examined how changes in the epigenome as a result of environmental influence directly impact learning and memory in either sex, we will discuss studies suggesting that similar environmental exposure in females and males can lead to sex-specific changes in histone modifications to impact learning and memory.
Exposure to the same compounds during development in females and males can result in differences in the function of epigenetic modifying enzymes in brain regions that play a role in memory. For example, developmental exposure to arsenic during all three trimesters of fetal/neonatal development leads to opposing histone modifications in the dentate gyrus (DG) region of the hippocampus of female and male mice at postnatal day 70 (Fig. 1; Tyler et al. 2015). The authors find that developmental exposure was associated with an increase in H3K4Me3 and H3K9Ac (both associated with active transcription) in the DG of males compared with same-sex controls. In contrast, in females this same developmental exposure to arsenic led to a decrease in H3K4Me3 and H3K9Ac compared to same-sex controls. Further, the histone methyltransferase responsible for trimethylation of H3K4, MLL was also found to be increased in the DG of male mice, but decreased in females, exposed to arsenic during development. Females exposed to arsenic during development exhibited greater levels of HDAC1 and 2 in the DG, whereas in males the levels were unchanged as a result of arsenic exposure (Tyler et al. 2015). A more recent study demonstrates sex-dependent effects of the class I-IIa HDAC inhibitor sodium valproate on reversal learning following exposure to arsenic during development. Here, the authors find that arsenic exposure during development leads to male-specific impairments in memory into adulthood that were reversed with systemic HDAC inhibition over a period of 2 wk (Tyler et al. 2018). In contrast, the same HDAC inhibition treatment had no effect in females. Together, these findings suggest that identical exposure to the same compound in males and females during development can lead to distinct histone acetylation patterns in the brain likely enhancing transcription in males compared with females. Therefore, it is possible that sex differences in expression of genes important for memory function early in life are partially responsible for sex differences in memory performance into adulthood. These results also have important sex-specific implications for the use of HDAC inhibitors as a therapeutic intervention.
The above example demonstrates how exposure to a toxin or toxic environment can result in sex-specific epigenetic modifications in the brain to potentially impact memory. Another study suggests that in females, exposure to an enriched environment may also involve or require epigenetic mechanisms. Environmental enrichment (EE) has clear benefits for memory function and neurogenesis in both females and males (e.g., Kempermann et al. 1997; Frick and Fernandez 2003; Bruel-Jungerman et al. 2005). A study performed in females only by Lopez-Atalaya et al. (2011) found that the beneficial effects of EE on memory and hippocampal neurogenesis are occluded in female CBP mutant mice, suggesting that the HAT CBP is likely an important mediator of the cognitive benefits associated with EE. Future research is needed to precisely determine epigenetic mechanisms involved and required for mediating EE in females and whether epigenetic mechanisms regulate pro-cognitive effects in males.
Exercise
Several studies have demonstrated that in males exercise facilitates induction of hippocampal brain-derived neurotrophic factor (BDNF), which plays a critical role in synaptic plasticity and memory (Neeper et al. 1995, 1996; Adlard et al. 2004; Berchtold et al. 2005; Intlekofer et al. 2013). In males, expression of BDNF is regulated by epigenetic mechanisms, including histone modification. In a collaboration led by the Cotman lab, we previously demonstrated that in males, exercise enables hippocampus-dependent learning in conditions that are normally subthreshold for long-term memory formation. Similar effects are observed with males receiving an HDAC inhibitor, sodium butyrate (NaB)—the transformation of a subthreshold learning event into long-term memory (Stefanko et al. 2009; Roozendaal et al. 2010; Haettig et al. 2011; Intlekofer et al. 2013). In male mice, exercise and NaB increased BDNF transcripts I and IV in the hippocampus, and the increases were associated with BDNF promoter acetylation on H4K8 (associated with transcriptional activation), but not H4K12 (also associated with active promoters; Intlekofer et al. 2013). These results support the idea that in males, exercise leads to enhanced memory formation via opening chromatin structure and facilitating gene expression required for long-term memory.
Although studies on the effects of exercise and epigenetics in females are sparse, there is evidence to suggest that exercise works in both similar and distinct ways to enhance memory in females and males. One study by Marlatt et al. (2012) demonstrated that like males, physical activity enhances learning in female mice, as observed with improved retention in the Morris water maze task in females undergoing voluntary wheel running compared with same-sex sedentary controls. However, in studies assessing the impact of exercise on cognitive function in women and men, exercise was shown to improve object location memory in men, but not women (Coleman et al. 2018), suggesting that a similar amount of exercise may have a differential impact for learning and memory in human females and males. Sex-similar and sex-specific patterns of hippocampal Bdnf mRNA as a result of exercise have also been observed. Twenty weeks of voluntary wheel running resulted in higher Bdnf IV mRNA expression in the hippocampus of both sexes compared with same-sex sedentary mice. However, total Bdnf mRNA expression was only significantly greater in male mice that underwent exercise, but not observed to be significantly elevated in females when compared with sedentary controls (Venezia et al. 2016). This study may also be consistent with reports demonstrating that expression of BDNF in dorsal hippocampus is observed in male, but not female mice during consolidation (Mizuno et al. 2012). Estrogen regulates Bdnf and induces Bdnf expression following activation of estrogen receptors. Bdnf contains a canonical estrogen response element that allows for ER complexes to bind and modulate transcriptional activation (Sohrabji et al. 1995). Additionally, ovary removal reduces Bdnf mRNA levels in both the hippocampus and cortex, which is partially restored with estrogen supplementation (Singh et al. 1995; Sohrabji et al. 1995; Berchtold et al. 2001). Therefore, fluctuations in hormones throughout the lifespan, among a number of other factors, may modulate the effects of exercise in the female and male brain. While these findings clearly speak to the ability of exercise to facilitate gene expression in both sexes, potentially through epigenetic modification, a number of questions remain such as whether the amount of exercise required to enhance cognitive function is similar between the sexes, the mechanism by which exercise enhances memory in females and how BDNF is regulated by epigenetic mechanisms in females.
Considerations for the interpretation of sex differences in epigenetic mechanisms involved in memory
Thus far we have touched on a number of important factors which similarly and distinctly influence the epigenome in females and males. However, it is important to note that sex-similar changes in the epigenome do not necessitate similarities in learning and memory between females and males and the reverse is true (for example, see Sase et al. 2019, discussed below). With this in mind, we briefly highlight additional factors to consider when interpreting sex differences in epigenetic regulation of memory (see Fig. 2).
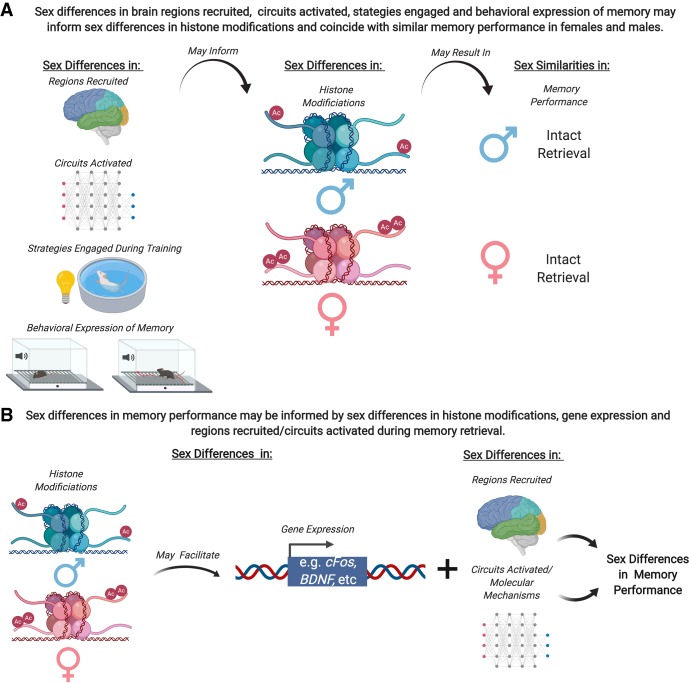
Figure 2.
Conceptual framework for understanding how sex differences in histone modifications can coincide with similarities or differences in memory performance. (A) Sex differences in epigenetic modifications may be due in part, to sex differences in brain regions recruited (e.g., Gasbarri et al. 2007; Cahill 2011; Bellace et al. 2013; Keiser et al. 2017) and circuits activated during consolidation or retrieval of memory or in behavioral strategies engaged during learning and/or expression of memory (Rodríguez et al. 2011; Bettis and Jacobs 2013; Keeley et al. 2013; Shah et al. 2013; Gruene et al. 2015). Intact memory retrieval in both females and males may exist despite sex differences in histone modifications (e.g., Sase et al. 2019). (B) Sex differences in epigenetic modifications may facilitate sex differences in expression of genes involved in memory (see Fig. 1) which may impact sex differences in brain regions/circuits activated during retrieval, potentially influencing sex differences in memory performance. Not presented here, but important to note is also the possibility of similar histone modifications between females and males coinciding with differences in memory performance.
Memory circuitry: similar epigenetic modification ≠ similar behavior
In studies of learning and memory similar behavioral outcomes between the sexes may still involve different neural mechanisms (for review, see Becker and Koob 2016) and the reverse. For this reason, it will be important to step away from comparing data obtained from females to previously established male findings when examining how epigenetic modifications shape learning and memory.
A recent study by Sase et al. (2019) underscores this concept, demonstrating that the same hippocampal histone modifications in females and males may be causally associated with opposing effects on memory and biological function. In this study, the authors examined the impact of histone H3 acetylation at the Cdk5 promoter in dorsal hippocampus of female and male mice after context fear memory retrieval. First, the authors find that retrieval of context fear results in markedly different patterns of histone H3 acetylation in the CA1 region of the dorsal hippocampus of male and female mice where histone H3 lysine 9/14 acetylation (H3K9/14Ac) is enriched at the Cdk5 promoter in males compared with same-sex context only controls, correlating with higher freezing observed in males compared with females. Next, the authors assess the causal role of histone H3 lysine 9/14 acetylation (H3K9/14Ac) at the Cdk5 promoter in males and females by viral expression of zinc finger proteins (ZFPs) targeting histone acetylation to the Cdk5 promoter. Forcing histone H3 acetylation at the Cdk5 promoter in both sexes resulted in impaired memory performance (as assessed by freezing behavior) in females compared to same-sex control, but freezing in males did not differ from control (Fig. 1; Sase et al. 2019). The demonstration that recruiting histone modifying enzymes at the same promoter region in both sexes oppositely affects memory in females and males (and also led to female-specific phosphorylation of tau protein) opens up a number of critical questions regarding the mechanism by which histone modifying enzymes regulate chromatin to affect memory in females. Next, we briefly discuss how sex differences in strategies used to learn or retrieve information can be viewed in light of understanding how histone modifications influence memory acquisition, consolidation and retrieval.
Sex differences in learning strategy and brain region recruitment
As we begin to interpret sex differences in epigenetic mechanisms of learning and memory, it is important that we avoid assumptions that a similar behavior or epigenetic signature in the female is equivalent to that of the male and vice versa. To avoid this pitfall, we must first understand how behavioral strategies can be sex-specific and involve sex differences in regional recruitment for a given memory task (Fig. 2). In spatial memory tasks for an example, different navigational strategies are adapted by males and females to reach the same goal (Andreano and Cahill 2009; Rodríguez et al. 2011; Bettis and Jacobs 2013; Keeley et al. 2013; Shah et al. 2013).
In spatial tasks such as the Morris water maze, males have been reported to outperform females. However, when strategy is taken into account and a landmark such as a wall cue is included as a navigation tool, these sex differences become no longer apparent (Saucier et al. 2002; Chai and Jacobs 2010). This is because males are shown to rely predominantly on distal cues on tasks of spatial navigation, whereas females rely on landmarks or proximal cues (Rodríguez et al. 2011; Bettis and Jacobs 2013; Keeley et al. 2013; Shah et al. 2013). Similarly, in tasks examining memory for a tone that was previously paired with footshock, a “typical” response observed in males upon reintroduction to the tone is lack of movement other than respiration or freezing (Blanchard and Blanchard 1969; Fanselow 1980) however, more recent studies highlight a female-prone response to the tone associated with footshock where female rats engage in an active “darting” behavior (Gruene et al. 2015), demonstrating that a strong memory by both sexes may be expressed in sex-specific ways. Together, these examples highlight the need for considering sex differences in strategies used to learn that may lead to a similar behavioral outcome when assessing sex differences in memory performance, while at the same time understanding the possibility that similarly strong memory may be expressed in sex-biased ways.
As we interpret sex differences in epigenetic mechanisms of learning and memory, it is also important that we consider the possibility of sex differences in regional recruitment for a given learning and memory task. For example, in instances where females and males demonstrate strong memory for a fear-conditioned context, male mice show activation of hippocampus, whereas females show activation of amygdala following retrieval (Keiser et al. 2017). In memory tests with an emotional component, men and women show differences in lateralization of a region important for fear-related and emotionally charged memories, the amygdala (Gasbarri et al. 2007; Cahill 2011), and women have been shown to have increased recruitment of hippocampal circuitry for cues with an emotional component (Bellace et al. 2013). Therefore, efforts to broadly examine interactions between brain regions and circuits engaged in a given learning and memory task, rather than restricting analysis to one region of interest will allow for a more complete picture when interpreting how epigenetic mechanisms in the female, in addition to the male brain influence memory.
Technical innovations in epigenetics: approaches to examine sex differences in the epigenome
A number of technical innovations over recent years are now providing researchers the tools to better examine and understand causal molecular mechanisms involved in learning and memory. The advent of epigenetic editing tools such as clustered regularly interspaced short palindromic repeats (CRISPR–Cas9 and CRISPR–dCas9) enable researchers to directly target and edit specific genomic regions and genes of interest with a desired chromatin modification, methods, which begin to elucidate causality between epigenetic modification (by a specific enzyme) and memory (for reviews, see Kwapis et al. 2018; Xie et al. 2018; Campbell and Wood 2019; Savell et al. 2019; Xu and Heller 2019). Zinc finger protein based systems also serve as a great tool, which enables targeting of transcriptional activators to specific genomic regions (for reviews, see Hamilton et al. 2018a; see also Heller et al. 2016; Hamilton et al. 2018b).
One example of this can be found in the aforementioned study by Sase et al. (2019). The authors observed that Cdk5 expression was increased in the hippocampus of males, but not females, following retrieval of context fear conditioning, which correlated with higher levels of freezing in males compared with females. This suggested that Cdk5 expression/activity is necessary for retrieval of memory in males, but potentially not females. To further examine sex differences in this mechanism, the authors used a ZFP based system packaged into HSV and delivered to the CA1 region of the hippocampus. ZFPs implemented in this study were designed to bind an 18-base pair motif in the Cdk5 promoter and fused to the p65 transcriptional activation domain. Cdk5-ZFP-p65 was used to drive Cdk5 expression in the hippocampus of males and females to determine whether increasing Cdk5 expression, and activity, could result in similar retrieval effects in both sexes. Perhaps by increasing Cdk5 in females, one would now observe memory retrieval similar to males. However, the results demonstrated the opposite. Increased Cdk5 in females decreased retrieval of long-term memory for fear, demonstrating that the role of Cdk5 in males and females with regard to memory retrieval is different between sexes. Without taking this promoter-specific approach to regulate Cdk5, the data could not have been interpreted past a simple conclusion that level of freezing (interpreted as memory strength) is dependent on level of Cdk5 expression, which would lead to a misunderstanding of what Cdk5 does in the female hippocampus. One caveat with any of these site-directed approaches is that the endogenous transcriptional machinery may be disrupted by the recruitment of ZFP constructs or CRISPR–dCas9, leading to novel molecular events.
Additional methods enable researchers to take nonbiased approaches to uncovering novel epigenetic mechanisms in females. Single-cell RNA sequencing for example, will enable researchers to examine how memory formation and retrieval impact specific cell types in the brains of females and males (Tang et al. 2010; Jiang et al. 2018). Tools such as INTACT (isolation of nuclei tagged in specific cell types) and TRAP (translating ribosome affinity purification) add to this by enabling specific cell types to be isolated and manipulated (Palovaara and Weijers 2019). Adapting this approach could shine light on female mosaicism as transcriptional silencing of one X chromosome in females (X chromosome-inactivation) leaves the female brain (and body) a mosaic of cells, some expressing the maternally inherited X chromosome and the others expressing the paternally inherited X chromosome (Tan et al. 1995). Therefore, implementing this approach may inform whether a specific cell type expresses the maternally or paternally inherited X chromosome and whether this impacts learning and memory function.
Gene regulation is also influenced by the spatial organization of chromatin and chromosomal interactions. Chromosomal conformation capture serves as an excellent tool to examine higher order chromatin structure and chromosomal interactions in females and males. This method works by using specific restriction enzymes to cross-link, isolate and digest chromatin and the remaining ligated fragments are assessed with real-time polymerase chain reaction (PCR) or sequencing. Correlation of the abundance of the fragments with frequency of interactions between two regions can define the three dimensional (3D) structure, organization and boundaries of the genome inside a nucleus (e.g., Quinodoz et al. 2018). Since these factors directly impact gene expression, chromosomal conformation capture serves as an important tool to specifically identify how higher order chromatin structure and chromosomal interactions affect gene expression and impact learning and memory in females and males.
Conclusions and a few open questions
Throughout this review, we have highlighted a number of factors that influence epigenetic modifications critical for expression of genes implicated in memory consolidation and retrieval in females and males (Fig. 1). While the literature on sex-similarities and differences in a number of learning and memory tasks continues to grow, and the inclusion of females in studies of epigenetics and memory begins to emerge, the connection between these areas and the factors that influence them are largely absent. Internal and external factors such as the ones discussed here lead to epigenetic changes in key brain regions important for learning and memory and can also impact females and males in sex-specific ways. Sex differences in the influence of these factors may be attributed to sex differences in susceptibility to a number of external factors such as stress or differences in exposure to begin with; while other factors simply impact the female and male epigenome differently, despite similar exposure. As we move forward, it is important that we avoid assumptions that sex-specific epigenetic modifications must always lead to sex-specific changes in learning and memory and vice versa (Fig. 2).
As the field begins to elucidate how learning and memory is impacted in females in addition to males by epigenetic modifications that are largely influenced by a number of factors, we are left with more questions than answers. For example, which male or female specific factors that affect memory processing contribute to sex-specific patterns of histone modifications and how do those patterns affect gene expression required for memory and retrieval? How does hormonal milieu during both development and throughout the lifespan impact how histone modifying enzymes act to regulate memory function? How might sex differences in strategies used to learn or retrieve memories impact the function of histone modifying enzymes? While much research is still needed to answer these questions, multiple studies incorporating both sexes into research examining histone modifying enzymes reveal similar and distinct roles for memory function. Understanding how histone modifications function to impact memory in females as well as males is critically important in regards to improving our understanding of increased prevalence of disorders that directly affect memory such as PTSD and AD which afflict women at a rate more than twice that of men (Kessler et al. 1995, 2012).
Acknowledgments
This work was supported by NIH grants (to M.A.W.: NIA AG051807, AG050787, AG054349; NIMH MH101491; NIDA DA025922; to A.A.K.: NIA T32 AG00096). Illustrations designed by Paul Schiffmacher of Schiffmacher Illustration & Design. We apologize to our colleagues whose work we could not include due to space restrictions.
Footnotes
Article is online at http://www.learnmem.org/cgi/doi/10.1101/lm.048850.118.
References
- Adlard PA, Perreau VM, Engesser-cesar C, Cotman CW. 2004. The timecourse of induction of brain-derived neurotrophic factor mRNA and protein in the rat hippocampus following voluntary exercise. Neurosci Lett 363: 43–48. 10.1016/j.neulet.2004.03.058 [DOI] [PubMed] [Google Scholar]
- Agulnik AI, Mitchell MJ, Mattei MG, Borsani G, Avner PA, Lerner JL, Bishop CE. 1994. A novel X gene with a widely transcribed Y-linked homologue escapes X-inactivation in mouse and human. Hum Mol Genet 3: 879–884. 10.1093/hmg/3.6.879 [DOI] [PubMed] [Google Scholar]
- Alberini CM. 2009. Transcription factors in long-term memory and synaptic plasticity. Physiol Rev 89: 1–46. 10.1152/physrev.00017.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano JM, Cahill L. 2009. Sex influences on the neurobiology of learning and memory. Learn Mem 16: 248–266. 10.1101/lm.918309 [DOI] [PubMed] [Google Scholar]
- Arnold AP, Chen X. 2009. What does the “four core genotypes” mouse model tell us about sex differences in the brain and other tissues? Front Neuroendocr 30: 1–9. 10.1016/j.yfrne.2008.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asok A, Draper A, Hoffman AF, Schulkin J, Lupica CR, Rosen JB. 2018. Optogenetic silencing of a corticotropin-releasing factor pathway from the central amygdala to the bed nucleus of the stria terminalis disrupts sustained fear. Mol Psychiatry 23: 914–922. 10.1038/mp.2017.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagot RC, Labonté B, Peña C, Nestler EJ. 2014. Epigenetic signaling in psychiatric disorders: stress and depression. Dialogues Clin Neurosci 16: 281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangasser DA, Wicks B. 2017. Sex-specific mechanisms for responding to stress. J Neurosci Res 95: 75–82. 10.1002/jnr.23812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RM, Wood MA. 2008. Beyond transcription factors: the role of chromatin modifying enzymes in regulating transcription required for memory. Learn Mem 15: 460–467. 10.1101/lm.917508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JB, Koob GF. 2016. Sex differences in animal models: focus on addiction. Pharmacol Rev 68: 242–263. 10.1124/pr.115.011163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellace M, Williams JM, Mohamed FB, Faro SH. 2013. An fMRI study of the activation of the hippocampus by emotional memory. Int J Neurosci 123: 121–127. 10.3109/00207454.2012.742894 [DOI] [PubMed] [Google Scholar]
- Benoit J, Rakic P, Frick KM. 2015. Prenatal stress induces spatial memory deficits and epigenetic changes in the hippocampus indicative of heterochromatin formation and reduced gene expression. Behav Brain Res 71: 3831–3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold NC, Kesslak JP, Pike CJ, Adlard PA, Cotman CW. 2001. Estrogen and exercise interact to regulate brain-derived neurotrophic factor mRNA and protein expression in the hippocampus. Eur J Neurosci 14: 1992–2002. 10.1046/j.0953-816x.2001.01825.x [DOI] [PubMed] [Google Scholar]
- Berchtold NC, Chinn G, Chou M, Kesslak JP, Cotman CW. 2005. Exercise primes a molecular memory for brain-derived neurotrophic factor protein induction in the rat. Neuroscience 133: 853–861. 10.1016/j.neuroscience.2005.03.026 [DOI] [PubMed] [Google Scholar]
- Bettis TJ, Jacobs LF. 2013. Sex differences in memory for landmark arrays in C57BL/J6 mice. Anim Cogn 16: 873–882. 10.1007/s10071-013-0619-x [DOI] [PubMed] [Google Scholar]
- Blanchard RJ, Blanchard DC. 1969. Crouching as an index of fear. J Comp Physiol Psychol 67: 370–375. 10.1037/h0026779 [DOI] [PubMed] [Google Scholar]
- Bowman RE. 2005. Stress-induced changes in spatial memory are sexually differentiated and vary across the lifespan. J Neuroendocrinol 17: 526–535. 10.1111/j.1365-2826.2005.01335.x [DOI] [PubMed] [Google Scholar]
- Bowman RE, Beck KD, Luine VN. 2003. Chronic stress effects on memory: sex differences in performance and monoaminergic activity. Horm Behav 43: 48–59. [DOI] [PubMed] [Google Scholar]
- Bruel-Jungerman E, Laroche S, Rampon C. 2005. New neurons in the dentate gyrus are involved in the expression of enhanced long-term memory following environmental. Eur J Neurosci 21: 513–521. 10.1111/j.1460-9568.2005.03875.x [DOI] [PubMed] [Google Scholar]
- Burgess-beusse B, Farrell C, Gaszner M, Litt M, Mutskov V, Recillas-targa F, Simpson M, West A, Felsenfeld G. 2002. The insulation of genes from external enhancers and silencing chromatin. Proc Natl Acad Sci 99: 16433–16437. 10.1073/pnas.162342499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill L. 2006. Why sex matters for neuroscience. Nat Rev Neurosci 6: 477–484. 10.1038/nrn1909 [DOI] [PubMed] [Google Scholar]
- Cahill L. 2011. Oversimplifying sex differences in the brain review: man and woman: an inside story. Cerebrum 2011: 7. [PMC free article] [PubMed] [Google Scholar]
- Campbell RR, Wood MA. 2019. How the epigenome integrates information and reshapes the synapse. Nat Rev Neurosci 20: 133–147. 10.1038/s41583-019-0121-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai XJ, Jacobs LF. 2010. Effects of cue types on sex differences in human spatial memory. Behav Brain Res 208: 336–342. 10.1016/j.bbr.2009.11.039 [DOI] [PubMed] [Google Scholar]
- Christophorou MA, Castelo-branco G, Halley-stott RP, Oliveira S, Loos R, Radzisheuskaya A, Mowen K, Bertone P, Silva J, Zernicka-Goetz M, et al. 2016. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature 507: 104–108. 10.1038/nature12942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WCJ, Swaab DF, De Vries GJ. 2000. Apoptosis during sexual differentiation of the Bed Nucleus of the Stria Terminalis in the rat brain. J Neurobiol 43: 234–243. [DOI] [PubMed] [Google Scholar]
- Chwang WB, Riordan KJO, Levenson JM, Sweatt JD. 2006. ERK/MAPK regulates hippocampal histone phosphorylation following contextual fear conditioning. Learn Mem 13: 322–328. 10.1101/lm.152906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton JA, Collins FS. 2014. Policy: NIH to balance sex in cell and animal studies. Nature 509: 282–283. 10.1038/509282a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman M, Offen K, Markant J. 2018. Exercise similarly facilitates men and women's selective attention task response times but differentially affects memory task performance. Front Psychol 9: 1–19. 10.3389/fpsyg.2018.01405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope JL, Regev L, Chen Y, Korosi A, Rice CJ. 2014. Differential contribution of CBP:cREB binding to corticotropin- releasing hormone expression in the infant and adult hypothalamus. Stress 17: 1–23. 10.3109/10253890.2013.794450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Weng Z. 2014. The correlation between histone modifications and gene expression. Epigenomics 5: 113–116. 10.2217/epi.13.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumais KM, Alonso AG, Immormino MA, Bredewold R, Veenema AH. 2016. Involvement of the oxytocin system in the bed nucleus of the stria terminalis in the sex-specific regulation of social recognition. Psychoneuroendocrinology 64: 79–88. 10.1016/j.psyneuen.2015.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Zhao Z, Orr PT, Chambers CH, Lewis MC, Frick KM. 2010. Estradiol-induced object memory consolidation in middle-aged female mice requires dorsal hippocampal extracellular signal-regulated kinase and phosphatidylinositol 3-kinase activation. J Neurosci 30: 4390–4400. 10.1523/JNEUROSCI.4333-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanselow MS. 1980. Conditional and unconditional components of post-shock freezing. Pavlov J Biol Sci 15: 177–182. [DOI] [PubMed] [Google Scholar]
- Farrelly LA, Thompson RE, Zhao S, Lepack AE, Lyu Y, Bhanu NV, Zhang B, Loh YE, Ramakrishnan A, Vadodaria KC, et al. 2019. Histone serotonylation is a permissive modification that enhances TFIID binding to H3K4me3. Nature 567: 535–539. 10.1038/s41586-019-1024-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fombonne E. 2009. Epidemiology of pervasive developmental disorders. Pediatr Res 65: 591–598. 10.1203/PDR.0b013e31819e7203 [DOI] [PubMed] [Google Scholar]
- Fontan-Lozano A, Suarez-Pereira I, Horrillo A, Del-Pozo-Martin Y, Hmadcha A, Carrio AM. 2010. Histone H1 Poly [ADP]-ribosylation regulates the chromatin alterations required for learning consolidation. J Neurosci 30: 13305–13313. 10.1523/JNEUROSCI.3010-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forger NG, Rosen GJ, Waters EM, Jacob D, Simerly RB, De Vries GJ. 2004. Deletion of Bax eliminates sex differences in the mouse forebrain. Proc Natl Acad Sci 101: 13666–13671. 10.1073/pnas.0404644101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortress AM, Kim J, Poole RL, Gould TJ, Frick KM. 2014. 17β-estradiol regulates histone alterations associated with memory consolidation and increases Bdnf promoter acetylation in middle-aged female mice. Learn Mem 21: 457–467. 10.1101/lm.034033.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick KM. 2013. Epigenetics, oestradiol and hippocampal memory consolidation. J Neuroendocrinol 25: 1151–1162. 10.1111/jne.12106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick KM, Fernandez SM. 2003. Enrichment enhances spatial memory and increases synaptophysin levels in aged female mice. Neurobiol Aging 24: 615–626. 10.1016/S0197-4580(02)00138-0 [DOI] [PubMed] [Google Scholar]
- Frick KM, Kim J, Tuscher JJ, Fortress AM. 2015. Sex steroid hormones matter for learning and memory: estrogenic regulation of hippocampal function in male and female rodents. Learn Mem 22: 472–493. 10.1101/lm.037267.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galea LAM, Kavaliers M, Ossenkopp K. 1996. Sexually dimorphic spatial learning in meadow voles Microtus pennsylvanicus and deer mice Peromyscus maniculatus. J Exp Biol 199: 195–200. [DOI] [PubMed] [Google Scholar]
- Gasbarri A, Arnone B, Pompili A, Pacitti F, Pacitti C, Cahill L. 2007. Sex-related hemispheric lateralization of electrical potentials evoked by arousing negative stimuli. Brain Res 1138: 178–186. 10.1016/j.brainres.2006.12.073 [DOI] [PubMed] [Google Scholar]
- Glendining KA, Jasoni CL. 2019. Maternal high fat diet-induced obesity modifies histone binding and expression of Oxtr in offspring hippocampus in a sex-specific manner. Int J Mol Sci 20: 1–11. 10.3390/ijms20020329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldknopf I, Taylor CW, Baum RM, Yeoman LC, Olson J, Prestayko AW, Busch H. 1975. Isolation and characterization of protein A24, a “histone-like” non-histone chromosomal protein. J Biol Chem 250: 7182–7187. [PubMed] [Google Scholar]
- Gonçalves TF, Gonçalves AP, Rodrigues NF, Mendonça J, Mattos M, Pimentel G, Santos-rebouças CB. 2014. KDM5C mutational screening among males with intellectual disability suggestive of X-Linked inheritance and review of the literature. Eur J Med Genet 57: 138–144. 10.1016/j.ejmg.2014.02.011 [DOI] [PubMed] [Google Scholar]
- Goode TD, Maren S. 2017. Role of the bed nucleus of the stria terminalis in aversive learning and memory. Learn Mem 24: 480–491. 10.1101/lm.044206.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorka AX, Torrisi S, Shackman AJ, Grillon C, Ernst M. 2018. Intrinsic functional connectivity of the central nucleus of the amygdala and bed nucleus of the stria terminalis. Neuroimage 168: 392–402. 10.1016/j.neuroimage.2017.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotsiridze T, Kang N, Jacob D, Forger NG. 2007. Development of sex differences in the principal nucleus of the bed nucleus of the stria terminalis of mice: role of Bax-dependent cell death. Dev Neurobiol 67: 355–362. 10.1002/dneu.20353 [DOI] [PubMed] [Google Scholar]
- Gräff J, Tsai LH. 2013. Histone acetylation: molecular mnemonics on the chromatin. Nat Rev Neurosci 14: 97–111. 10.1038/nrn3427 [DOI] [PubMed] [Google Scholar]
- Graff J, Kim D, Dobbin M, Tsai L. 2011. Epigenetic regulation of gene expression in physiological and pathological brain processes. Physiol Rev 91: 603–649. 10.1152/physrev.00012.2010 [DOI] [PubMed] [Google Scholar]
- Greenfield A, Carrel L, Pennisi D, Philippe C, Quaderi N, Siggers P, Steiner K, Tam PPL, Willard HF, Koopman P. 1998. The UTX gene escapes X inactivation in mice and humans. Hum Mol Genet 7: 1–6. 10.1093/hmg/7.4.737 [DOI] [PubMed] [Google Scholar]
- Gresack JE, Schafe G, Orr P, Frick K. 2009. Sex differences in contextual fear conditioning are associated with differential ventral hippocampal extracellular signal-regulated kinase activation. Neuroscience 159: 451–67. 10.1016/j.neuroscience.2009.01.009 [DOI] [PubMed] [Google Scholar]
- Gruene TM, Flick K, Stefano A, Shea SD, Shansky RM. 2015. Sexually divergent expression of active and passive conditioned fear responses in rats. Elife 4: 1–9. 10.7554/eLife.11352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillamon A, Segovia S, Abril A. 1988. Early effects of gonadal steroids on the neuron number in the medial posterior region and the lateral division of the bed nucleus of the stria terminalis in the rat. Dev Brain Res 44: 281–290. 10.1016/0165-3806(88)90226-X [DOI] [PubMed] [Google Scholar]
- Gungor NZ, Yamamoto R, Paré D. 2015. Optogenetic study of the projections from the bed nucleus of the stria terminalis to the central amygdala. J Neurophysiol 114: 2903–2911. 10.1152/jn.00677.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, David J, Paylor RE, Lubin FD. 2010. Histone methylation regulates memory formation. J Neurosci 30: 3589–3599. 10.1523/JNEUROSCI.3732-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haettig J, Stefanko DP, Multani M, Figueroa DX, McQuown S, Wood MA. 2011. Object location in a CBP-dependent manner HDAC inhibition modulates hippocampus-dependent long-term memory for object location in a CBP-dependent manner. Learn Mem 18: 71–79. 10.1101/lm.1986911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton PJ, Lim CJ, Nestler EJ, Heller EA. 2018a. Neuroepigenetic editing. Methods Mol Biol 1767: 113–136. 10.1007/978-1-4939-7774-1_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton PJ, Burek DJ, Lombroso SI, Neve RL, Robison AJ, Nestler EJ, Heller EA. 2018b. Cell-type specific epigenetic editing at the fosb gene controls susceptibility to social defeat stress. Neuropsychopharmacology 2: 272–284. 10.1038/npp.2017.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller EA, Hamilton PJ, Burek DD, Lombroso SI, Pena CJ, Neve RL, Nestler EJ. 2016. Targeted epigeentic remodeling of the Cdk5 gene in nucleus accumbens regulates cocaine- and stress-evoked behavior. J Neurosci 17: 4690–4697. 10.1523/JNEUROSCI.0013-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines M, Davis F, Coquelin A, Goy R, Gorski R. 1985. Sexually dimorphic regions in the medial preoptic area and the bed nucleus of the tria terminalis of the guinea pig brain: a description and investigation of their relationship to gonadal steroids in adulthood. J Neurosci 1: 40–47. 10.1523/JNEUROSCI.05-01-00040.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines M, Allen LS, Gorski RA. 1992. Sex differences in subregions of the medial nucleus of the amygdala and the bed nucleus of the stria terminalis of the rat. Brain Res 579: 321–326. 10.1016/0006-8993(92)90068-K [DOI] [PubMed] [Google Scholar]
- Hodawadekar S, Marmorstein R. 2007. Chemistry of acetyl transfer by histone modifying enzymes: structure, mechanism and implications for effector design. Oncogene 26: 5528–5540. 10.1038/sj.onc.1210619 [DOI] [PubMed] [Google Scholar]
- Horn P, Petersen C. 2002. Chromatin higher order folding: wrapping up transcription. Science 297: 1824–1827. 10.1126/science.1074200 [DOI] [PubMed] [Google Scholar]
- Huang H, Matevossian A, Whittle C, Kim SY, Schumacher A, Baker SP, Akbarian S. 2007. Prefrontal dysfunction in schizophrenia involves mixed-lineage leukemia 1-regulated histone methylation at GABAergic gene promoters. Neurobiol Dis 27: 11254–11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter RG, Mccarthy KJ, Milne TA, Pfaff DW, McEwen BS. 2009. Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proc Natl Acad Sci 106: 3–8. 10.1073/pnas.0911143106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer KA, Berchtold NC, Malvaez M, Carlos AJ, McQuown SC, Cunningham MJ, Wood MA, Cotman CW. 2013. Exercise and sodium butyrate transform a subthreshold learning event into long-term memory via a brain-derived neurotrophic factor-dependent mechanism. Neuropsychopharmacology 38: 2027–2034. 10.1038/npp.2013.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S, Brookes E, Agarwal S, Badeaux AI, Ito H, Vallianatos CN, Tomassy GS, Kasza T, Lin G, Thompson A, et al. 2016. A mouse model of X-linked intellectual disability associated with impaired removal of histone methylation. Cell Rep 5: 1000–1009. 10.1016/j.celrep.2015.12.091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson-Pick S, Marie-Claude A, McQuaid RJ, Kalvapalle R, Anisman H. 2013. Social agonistic distress in male and female mice: changes of behavior and brain monoamine functioning in relation to acute and chronic challenges. PLoS One 8: e60133 10.1371/journal.pone.0060133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LR, Amende M, Gurok U, Moser B, Gimmel V, Tzschach A, Janecke AR, Tariverdian G, Chelly J, Fryns J, et al. 2005. Mutations in the JARID1C gene, which Is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am J Hum Genet 76: 227–236. 10.1086/427563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Williams K, Kong X, Zeng W, Ma X, Tawil R, Yokomori K, Mortazavi A. 2018. Single-nucleus RNA-seq identifies divergent populations of FSHD2 myotube nuclei. bioRxiv 10.1101/478636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josselyn SA, Kida S, Silva AJ. 2004. Inducible repression of CREB function disrupts amygdala-dependent memory. Neurobiol Learn Mem 82: 159–163. 10.1016/j.nlm.2004.05.008 [DOI] [PubMed] [Google Scholar]
- Kang H, Schuman EM. 1996. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science 273: 1402–1406. 10.1126/science.273.5280.1402 [DOI] [PubMed] [Google Scholar]
- Keeley RJ, Tyndall AV, Scott GA, Saucier DM. 2013. Sex difference in cue strategy in a modified version of the morris water task: correlations between brain and behaviour. PLoS One 8: e69727 10.1371/journal.pone.0069727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiser AA, Tronson NC. 2016. Molecular mechanisms of memory in males and females. In Sex differences in the central nervous system (ed. Shansky RM), pp. 27–51. Elsevier Academic Press, Boston. [Google Scholar]
- Keiser AA, Turnbull LM, Darian MA, Feldman DE, Song I, Tronson NC. 2017. Sex differences in context fear generalization and recruitment of hippocampus and amygdala during retrieval. Neuropsychopharmacology 42: 397–407. 10.1038/npp.2016.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempermann G, Kuhn HG, Gage FH. 1997. More hippocampal neurons in adult mice living in an enriched environment. Nature 386: 493–495. 10.1038/386493a0 [DOI] [PubMed] [Google Scholar]
- Kessler RC, Sonnega A, Bromet E, Hughes M, Nelson CB. 1995. Posttraumatic stress disorder in the national comorbidity survey. Arch Gen Psychiatry 52: 1048–1060. 10.1001/archpsyc.1995.03950240066012 [DOI] [PubMed] [Google Scholar]
- Kessler RC, Petukhova M, Sampson NA, Zaslavsky AM, Wittchen HU. 2012. Twelve-month and lifetime prevalence and lifetime morbid risk of anxiety and mood disorders in the United States. Int J Methods Psychiatr Res 21: 169–184. 10.1002/mpr.1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim MJ, Kim S, Kang HS, Lim SW, Myung W, Lee Y, Hong CH, Choi SH, Na DL, et al. 2015. Gender differences in risk factors for transition from mild cognitive impairment to Alzheimer's disease: a CREDOS study. Compr Psychiatry 62: 114–122. 10.1016/j.comppsych.2015.07.002 [DOI] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y. 2007. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol cell Biol 8: 307–318. 10.1038/nrm2143 [DOI] [PubMed] [Google Scholar]
- Kogan JH, Frankland PW, Blendy JA, Coblentz J, Marowitz Z, Schütz G, Silva AJ. 1996. Spaced training induces normal long-term memory in CREB mutant mice. Curr Biol 7: 1–11. 10.1016/S0960-9822(06)00022-4 [DOI] [PubMed] [Google Scholar]
- Korb E, Herre M, Zucker-scharff I, Darnell RB, Allis CD. 2016. BET protein Brd4 activates transcription in neurons and BET inhibitor Jq1 blocks memory in mice. Nat Neurosci 18: 1464–1473. 10.1038/nn.4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzus E. 2017. Rubinstein-Taybi Syndrome and epigenetic alterations. In Neuroepigenomics in aging and disease (ed. Delgado-Morales R), pp. 39–62. Springer International Publishing, Cham. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell 128: 693–705. 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Kruger O, Shiozawa T, Kreifelts B, Scheffler K, Ethofer T. 2015. Three distinct fiber pathways of the bed nucleus of the stria terminalis to the amygdala and prefrontal cortex. Cortex 66: 60–68. 10.1016/j.cortex.2015.02.007 [DOI] [PubMed] [Google Scholar]
- Kudo K, Qiao C-X, Kanba S, Arita J. 2004. A selective increase in phosphorylation of cyclic AMP response element-binding protein in hippocampal CA1 region of male, but not female, rats following contextual fear and passive avoidance conditioning. Brain Res 1024: 233–243. 10.1016/j.brainres.2004.08.007 [DOI] [PubMed] [Google Scholar]
- Kwapis JL, Wood MA. 2015. Epigenetic mechanisms in fear conditioning: implications for treating post-traumatic stress disorder. Trends Neurosci 37: 706–720. 10.1016/j.tins.2014.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwapis JL, Alaghband Y, Kramár EA, López AJ, Vogel Ciernia A, White AO, Shu G, Rhee D, Michael CM, Montellier E, et al. 2018. Epigenetic regulation of the circadian gene Per1 contributes to age-related changes in hippocampal memory. Nat Commun 9: 3323 10.1038/s41467-018-05868-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin KA, Doraiswamy PM. 2015. When mars versus venus is not a cliché: gender differences in the neurobiology of alzheimer's disease. Front Neurol 5: 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingenfelter P, Adler D, Poslinski D, Thomas S, Elliot R, Chapman V, Disteche CM. 1998. Escape from X inactivation of Smcx is preceded by silencing during mouse development. Nat Genet 3: 212–213. 10.1038/ng0398-212 [DOI] [PubMed] [Google Scholar]
- Lopez-Atalaya JP, Ciccarelli A, Viosca J, Valor LM, Jimenez-Minchan M, Canals S, Giustetto M, Barco A. 2011. CBP is required for environmental enrichment-induced neurogenesis and cognitive enhancement. EMBO J 20: 4287–4298. 10.1038/emboj.2011.299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luine V. 2002. Sex differences in chronic stress effects on memory in rats. Stress 5: 205–216. 10.1080/1025389021000010549 [DOI] [PubMed] [Google Scholar]
- Luine V, Gomez J, Beck K, Bowman R. 2017. Sex differences in chronic stress effects on cognition in rodents. Pharmacol Biochem Behav 152: 13–19. 10.1016/j.pbb.2016.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccari S, Krugers HJ, Morley-Fletcher S, Szyf M, Brunton PJ. 2014. The consequences of early-life adversity: neurobiological, behavioural and epigenetic adaptations. J Neuroendocrinol 10: 707–723. 10.1111/jne.12175 [DOI] [PubMed] [Google Scholar]
- Maclusky NJ, Naftolin F. 1981. Sexual differentiation of the central nervous system. Science 211: 1294–1303. 10.1126/science.6163211 [DOI] [PubMed] [Google Scholar]
- Maeng LY, Milad MR. 2015. Sex differences in anxiety disorders: interactions between fear, stress, and gonadal hormones. Horm Behav 76: 106–117. 10.1016/j.yhbeh.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlatt M, Potter M, Lucassen P, van Praag H. 2012. Running throughout middle-age improves memory function, hippocampal neurogenesis and BDNF levels in female C57Bl/6J mice. Dev Neurobiol 72: 943–952. 10.1002/dneu.22009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R, Roth SY. 2001. Histone acetyltransferases: function, structure, and catalysis. Curr Opin Genet Dev 11: 155–161. 10.1016/S0959-437X(00)00173-8 [DOI] [PubMed] [Google Scholar]
- Matsuda KI, Mori H, Nugent BM, Pfaff DW, McCarthy MM, Kawata M. 2011. Histone deacetylation during brain development is essential for permanent masculinization of sexual behavior. Endocrinology 152: 2760–2767. 10.1210/en.2011-0193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice T, Duclot F, Meunier J, Naert G, Givalois L, Meffre J, Celerier A, Jacquet C, Copois V, Mechti N, et al. 2008. Altered memory capacities and response to stress in p300/CBP-associated dactor (PCAF) histone acetylase knockout mice. Neuropsychopharmacology 33: 1584–1602. 10.1038/sj.npp.1301551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BS. 1981. Sexual differentiation of the brain. Nature 291: 610 10.1038/291610a0 [DOI] [PubMed] [Google Scholar]
- McQuown SC, Barrett RM, Matheos DP, Post RJ, Rogge GA, Alenghat T, Mullican SE, Jones S, Rusche JR, Lazar MA, et al. 2011. HDAC3 is a critical negative regulator of long-term memory formation. J Neurosci 31: 764–774. 10.1523/JNEUROSCI.5052-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz CJ, Wolf OT. 2017. Sex differences in stress effects on emotional learning. J Neurosci Res 105: 93–105. 10.1002/jnr.23811 [DOI] [PubMed] [Google Scholar]
- Mizuno K, Giese KP. 2010. Towards a molecular understanding of sex differences in memory formation. Trends Neurosci 33: 285–291. 10.1016/j.tins.2010.03.001 [DOI] [PubMed] [Google Scholar]
- Mizuno K, Ris L, Sánchez-Capelo A, Godaux E, Giese KP. 2006. Ca2+/ calmodulin kinase kinase α is dispensable for brain development but is required for distinct memories in male, though not in female, mice. Mol Cell Biol 26: 9094–9104. 10.1128/MCB.01221-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K, Antunes-Martins A, Ris L, Peters M, Godaux E, Giese KP. 2007. Calcium/calmodulin kinase kinase β has a male-specific role in memory formation. Neuroscience 145: 393–402. 10.1016/j.neuroscience.2006.11.056 [DOI] [PubMed] [Google Scholar]
- Mizuno K, Dempster E, Mill J, Giese KP. 2012. Long-lasting regulation of hippocampal Bdnf gene transcription after contextual fear conditioning. Genes Brain Behav 11: 651–659. 10.1111/j.1601-183X.2012.00805.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moga DC, Taipale H, Tolppanen A, Tanskanen A, Hartikainen S, Wu Q, Jicha GA, Gnjidic D, Hospital N, Centre CP. 2017. A comparison of sex differences in psychotropic medication use in older people with Alzheimer's Disease in the US and Finland. Drugs Aging 34: 55–65. 10.1007/s40266-016-0419-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozzetta C, Pontis J, Fritsch L, Robin P, Portoso M, Proux C, Margueron R, Ait-Si-Ali S. 2014. The histone H3 lysine 9 methyltransferases G9a and GLP regulate polycomb repressive complex 2-mediated gene silencing. Mol Cell 53: 277–289. 10.1016/j.molcel.2013.12.005 [DOI] [PubMed] [Google Scholar]
- Murray EK, Hien A, De Vries GJ, Forger NG. 2009. Epigenetic control of sexual differentiation of the bed nucleus of the stria terminalis. Endocrinology 150: 4241–4247. 10.1210/en.2009-0458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naftolin F, Ryan KJ, Davies IJ, Reddy VV, Flores F, Petro Z, Kuhn M, White RJ, Takaoka Y, Wolin L. 1975. The formation of estrogens by central neuroendocrine tissues. Recent Prog Horm Res 31: 295–319. [DOI] [PubMed] [Google Scholar]
- Nathan D, Sterner DE, Berger SL. 2003. Histone modifications: now summoning sumoylation. Proc Natl Acad Sci 100: 13118–13120. 10.1073/pnas.2436173100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neeper SA, Gomez-Pinilla F, Choi J, Cotman C. 1995. Exercise and brain neurotrophins. Nature 373: 109 10.1038/373109a0 [DOI] [PubMed] [Google Scholar]
- Neeper SA, Gdmez-pinilla F, Choi J, Cotman CW. 1996. Physical activity increases mRNA for brain-derived neurotrophic factor and nerve growth factor in rat brain. Brain Res 726: 49–56. 10.1016/0006-8993(96)00273-9 [DOI] [PubMed] [Google Scholar]
- Neu SC, Pa J, Kukull W, Beekly D, Kuzma A, Gangadharan P, Wang LS, Romero K, Arneric SP, Redolfi A, et al. 2017. Apolipoprotein E genotype and sex risk factors for Alzheimer Disease: a meta-analysis. JAMA Neurol 74: 1178–1189. 10.1001/jamaneurol.2017.2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DK, Disteche CM. 2006. Dosage compensation of the active X chromosome in mammals. Nat Genet 38: 47–53. 10.1038/ng1705 [DOI] [PubMed] [Google Scholar]
- Oki M, Aihara H, Ito T. 2007. Role of histone phosphorylation in chromatin dynamics and its implications in diseases. Subcell Biochem 41: 319–336. [PubMed] [Google Scholar]
- Palovaara J, Weijers D. 2019. Adapting INTACT to analyse cell-type specific transcriptomes and nucleoplasmic mRNA dynamics in the Arabidopsis embryo. Plant Reprod 32: 113–121. 10.1007/s00497-018-0347-0 [DOI] [PubMed] [Google Scholar]
- Phoenix CH, Goy RW, Gerall AA, Young WC. 1959. Organizing action of prenatally administered testosterone propionate on the tissues mediating mating behavior in the female guinea pig. Endocrinology 65: 369–382. [DOI] [PubMed] [Google Scholar]
- Pittenger C, Huang YY, Paletzki RF, Bourtchouladze R, Scanlin H, Vronskaya S, Kandel ER. 2002. Reversible inhibition of CREB/ATF transcription factors in region CA1 of the dorsal hippocampus disrupts hippocampus-dependent spatial memory. Neuron 34: 447–462. 10.1016/S0896-6273(02)00684-0 [DOI] [PubMed] [Google Scholar]
- Quinodoz SA, Ollikainen N, Tabak B, Palla A, Schmidt JM, Detmar E, Lai MM, Shishkin AA, Bhat P, Takei Y, et al. 2018. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell 174: 744–757. 10.1016/j.cell.2018.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radulovic J, Ru A, Liepold T, Spiess J. 1999. Modulation of learning and anxiety by corticotropin-releasing factor (CRF) and stress: differential roles of CRF receptors 1 and 2. J Neurosci 19: 5016–5025. 10.1523/JNEUROSCI.19-12-05016.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez CA, Chamizo VD, Mackintosh NJ. 2011. Overshadowing and blocking between landmark learning and shape learning: the importance of sex differences. Learn Behav 39: 324–335. 10.3758/s13420-011-0027-5 [DOI] [PubMed] [Google Scholar]
- Roof RL, Havens MD. 1992. Testosterone improves maze performance and indues development of a male hippocampus in females. Brain Res 572: 310–313. 10.1016/0006-8993(92)90491-Q [DOI] [PubMed] [Google Scholar]
- Roozendaal B, Brunson KL, Holloway BL, Mcgaugh JL, Baram TZ. 2002. Involvement of stress-released corticotropin-releasing hormone in the basolateral amygdala in regulating memory consolidation. Proc Natl Acad Sci 99: 13908–13913. 10.1073/pnas.212504599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roozendaal B, Hernandez A, Cabrera SM, Hagewoud R, Stefanko DP, Haettig J, Wood MA. 2010. Membrane-associated glucocorticoid activity is necessary for modulation of long-term memory via chromatin modification. J Neurosci 30: 5037–5046. 10.1523/JNEUROSCI.5717-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor GC, Powell XSK, Brothers SP, Wahlestedt C. 2015. Epigenetic readers of lysine acetylation regulate cocaine-induced plasticity. J Neurosci 35: 15062–15072. 10.1523/JNEUROSCI.0826-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sase AS, Lombroso SI, Santhumayor BA, Wood RR, Lim CJ, Neve RL, Heller EA. 2019. Sex-specific regulation of fear memory by targeted epigenetic editing of Cdk5. Biol Psychiatry 85: 623–634. 10.1016/j.biopsych.2018.11.022 [DOI] [PubMed] [Google Scholar]
- Saucier DM, Green SM, Leason J, MacFadden A, Bell S, Elias LJ. 2002. Are sex differences in navigation caused by sexually dimorphic strategies or by differences in the ability to use the strategies? Behav Neurosci 116: 403–410. 10.1037/0735-7044.116.3.403 [DOI] [PubMed] [Google Scholar]
- Savell KE, Bach SV, Zipperly ME, Revanna JS, Goska NA, Tuscher JJ, Duke CG, Sultan FA, Burke JN, Williams D, et al. 2019. A neuron- optimized CRISPR/dCas9 activation system for robust and specific gene regulation. eNeuro 1: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scandaglia M, Lopez-Atalaya JP, Medrano-Fernandez A, Lopez-Cascales MT, del Blanco B, Lipinski M, Benito E, Olivares R, Iwase S, Shi Y, et al. 2017. Loss of Kdm5c causes spurious transcription and prevents the fine-tuning of activity-regulated enhancers in neurons. Cell Rep 21: 47–59. 10.1016/j.celrep.2017.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah DS, Prados J, Gamble J, De Lillo C, Gibson CL. 2013. Sex differences in spatial memory using serial and search tasks. Behav Brain Res 257: 90–99. 10.1016/j.bbr.2013.09.027 [DOI] [PubMed] [Google Scholar]
- Shansky RM, Lipps J, Shackman AJ, Emrich S. 2013. Stress-induced cognitive dysfunction: hormone-neurotransmitter interactions in the prefrontal cortex. Front Hum Neurosci 7: 123 10.3389/fnhum.2013.00123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiio Y, Eisenman RN. 2003. Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci 100: 13225–13230. 10.1073/pnas.1735528100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shors TJ. 2004. Learning during stressful times. Learn Mem 11: 137–144. 10.1101/lm.66604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shors T, Weiss C, Thompson R. 1992. Stress-induced facilitation of classical conditioning. Science 257: 537–539. 10.1126/science.1636089 [DOI] [PubMed] [Google Scholar]
- Shors TJ, Beylin AV, Wood GE, Gould E. 2000. The modulation of Pavlovian memory. Behav Brain Res 110: 39–52. 10.1016/S0166-4328(99)00183-7 [DOI] [PubMed] [Google Scholar]
- Shors TJ, Falduto J, Leuner B. 2004. The opposite effects of stress on dendritic spines in male vs. female rats are NMDA receptor-dependent. Eur J Neurosci 19: 145–150. 10.1046/j.1460-9568.2003.03065.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simensen R, Rogers R, Collins J, Abidi F, Schwartz C, Stevenson R. 2012. Short-term memory defecits in carrier femaleswith KDM5C mutations. Genet Couns 1: 31–40. [PubMed] [Google Scholar]
- Singh M, Meyer EM, Simpkins JW. 1995. The effect of ovariectomy and estradiol replacement on brain-derived neurotrophic factor messenger ribonucleic acid expression in cortical and hippocampal brain regions of female Sprague-Dawley rats. Endocrinology 136: 2320–2324. 10.1210/endo.136.5.7720680 [DOI] [PubMed] [Google Scholar]
- Smith AS, Lieberwirth C, Wang Z. 2013. Behavioral and physiological responses of female prarie voles (Microtus ochrogaster) to various stressful conditions. Stress 5: 531–539. 10.3109/10253890.2013.794449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolewski M, Singh G, Schneider JS, Cory-slechta DA, Stanwood G. 2018. Different behavioral experiences produce distinctive parallel changes in, and correlate with, frontal cortex and hippocampal global post-translational histone levels. Front Integr Neurosci 12: 1–12. 10.3389/fnint.2018.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrabji F, Miranda R, Toran-Alleand C. 1995. Identification of a putative estrogen response element in the gene encoding brain-derived neurotrophic factor. Proc Natl Acad Sci 92: 11110–11114. 10.1073/pnas.92.24.11110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanko DP, Barrett RM, Ly AR, Reolon GK, Wood MA. 2009. Modulation of long-term memory for object recognition via HDAC inhibition. PNAS 106: 5–10. 10.1073/pnas.0903964106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S, Breen SJ, Walsh M, Bertram JF, Reese BE. 1995. Cell dispersion patterns in different cortical regions studied with an X-inactivated transgenic marker. Development 121: 1029–1039. [DOI] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Nordman E, Li B, Xu N, Bashkirov VI, Lao K, Surani MA. 2010. RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat Protoc 5: 516–535. 10.1038/nprot.2009.236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolin DF, Foa EB. 2006. Sex differences in trauma and posttraumatic stress disorder: a quantitative review of 25 years of research. Psychol Bull 131: 959–992. 10.1037/0033-2909.132.6.959 [DOI] [PubMed] [Google Scholar]
- Tsai H, Grant PA, Rissman EF. 2009. Sex differences in histone modifications in the neonatal mouse brain. Epigenetics 4: 47–53. 10.4161/epi.4.1.7288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. 2006. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci 9: 519–525. 10.1038/nn1659 [DOI] [PubMed] [Google Scholar]
- Tyler CR, Hafez AK, Solomon ER, Allan AM. 2015. Developmental exposure to 50 parts-per-billion arsenic influences histone modifications and associated epigenetic machinery in a region- and sex-specific manner in the adult mouse brain. Toxicol Appl Pharmacol 288: 40–51. 10.1016/j.taap.2015.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler CR, Smoake JJW, Solomon ER, Villicana E, Caldwell KK, Allan AM. 2018. Sex-dependent effects of the histone deacetylase inhibitor sodium valproate, on reversal learning after developmental arsenic exposure. Front Genet 9: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallianatos CN, Iwase S, Arbor A. 2015. Disrupted intricacy of histone H3K4 methylation in neurodevelopmental disorders. Epigenomics 7: 503–519. 10.2217/epi.15.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venezia A, Guth L, Sapp R, Spangenburg E, Roth S. 2016. Sex-dependent and independent effects of long-term voluntary wheel running on Bdnf mRNA and protein expression. Physiol Behav 156: 8–15. 10.1016/j.physbeh.2015.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma D, Tasan R, Sperk G, Pape H. 2018. Neurobiology of Learning and Memory Neuropeptide Y2 receptors in anteroventral BNST control remote fear memory depending on extinction training. Neurobiol Learn Mem 149: 144–153. 10.1016/j.nlm.2018.01.011 [DOI] [PubMed] [Google Scholar]
- Wang L, Hou W, He Z, Yuan W, Yang J, Jia R, Zhu Z, Zhou Y, Tai F. 2018. Effects of chronic social defeat on social behaviors in adult female mandrin voles (Microtus mandarinus): involvement of the oxytocin system in the nucleus accumbens. Prog Neuropsychopharmacol Biol Psychiatry 82: 278–288. 10.1016/j.pnpbp.2017.11.002 [DOI] [PubMed] [Google Scholar]
- Webb WM, Sanchez RG, Perez G, Butler AA, Hauser RM, Rich MC, O'Bierne AL, Jarome TJ, Lubin FD. 2017. Dynamic association of epigenetic H3K4me3 and DNA 5hmC marks in the dorsal hippocampus and anterior cingulate cortex following reactivation of a fear memory. Neurobiol Learn Mem 142: 66–78. 10.1016/j.nlm.2017.02.010 [DOI] [PubMed] [Google Scholar]
- Williams CL, Barnett AM, Meck WH. 1990. Organizational effects of early gonadal secretions on sexual differentiation in spatial memory. Behav Neurosci 104: 84–97. 10.1037/0735-7044.104.1.84 [DOI] [PubMed] [Google Scholar]
- Wu J, Ellison J, Salido E, Yen P, Mohandas T, Shapiro LJ. 1994a. Isolation and characterization of XE169, a novel human gene that escapes x-inactivation. Hum Mol Genet 3: 153–160. 10.1093/hmg/3.1.153 [DOI] [PubMed] [Google Scholar]
- Wu J, Salido E, Yen P, Mohandas T, Heng H, Tsui L, Park J, Chapman V, Sharpiro L. 1994b. The murine Xe169 gene escapes X-inactivation like its human homologue. Nat Genet 7: 136–140. 10.1038/ng0694-136 [DOI] [PubMed] [Google Scholar]
- Xie N, Zhou Y, Sun Q, Tang B. 2018. Novel epigenetic techniques provided by the CRISPR/ Cas9 system. Stem Cells Int 2018: 7834175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Heller EA. 2019. Recent advances in neuroepigenetic editing. Curr Opin Neurobiol 59: 26–33. 10.1016/j.conb.2019.03.010 [DOI] [PubMed] [Google Scholar]
- Xu J, Burgoyne PS, Arnold AP. 2002. Sex differences in sex chromosome gene expression in mouse brain. Hum Mol Genet 11: 1409–1419. 10.1093/hmg/11.12.1409 [DOI] [PubMed] [Google Scholar]
- Xu J, Deng X, Disteche CM. 2008a. Sex-specific expression of the X-linked histone demethylase gene Jarid1c in brain. PLoS One 3: e2553 10.1371/journal.pone.0002553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Deng X, Watkins R, Disteche CM. 2008b. Sex-specific differences in expression of histone demethylases Utx and Uty in mouse brain and neurons. J Neurosci 28: 4521–4527. 10.1523/JNEUROSCI.5382-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Babak T, Shendure J, Disteche CM. 2010. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res 20: 614–622. 10.1101/gr.103200.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zablotsky B, Black LI, Maenner MJ, Schieve LA, Blumberg SJ. 2015. Estimated prevalence of autism and other developmental disabilities following questionnaire changes in the 2014 National Health Interview Survey. Natl Health Stat Report, Number 87: 1–21. [PubMed] [Google Scholar]
- Zablotsky B, Black LI, Blumberg SJ. 2017. Estimated prevalence of children with diagnosed developmental disabilities in the United States, 2014–2016. NCHS Data Brief 291: 1–8. [PubMed] [Google Scholar]
- Zamurrad S, Hatch HAM, Drelon C, Belalcazar HM, Zamurrad S, Hatch HAM, Drelon C, Belalcazar HM, Secombe J. 2018. A Drosophila model of intellectual disability caused by mutations in the histone demethylase KDM5. Cell Rep 22: 2359–2369. 10.1016/j.celrep.2018.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Fan L, Fortress AM, Boulware MI, Frick KM. 2012. Recognition and the estradiol-induced enhancement of object recognition. J Neurosci 32: 2344–2351. 10.1523/JNEUROSCI.5819-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]