Abstract
In neurons, mRNAs can be repressed postinitiation and assembled into granules enabling the transport and later, regulated reactivation of the paused mRNAs. It has been suggested that a large percentage of transcripts in neuronal processes are stored in these stalled polysomes. Given this, it is predicted that nascent peptides should be abundant in these granules. Nascent peptides can be visualized in real time by the SunTag system. Using this system, we observe nascent peptides in neuronal processes that are resistant to runoff with the initiation inhibitor homoharringtonin (HHT) and to release by puromycin, properties expected from RNA granules consisting of stalled polysomes. In contrast, nascent peptides in nonneuronal cells and neuronal cell bodies were not resistant to HHT or puromycin. Stalled polysomes can also be visualized after runoff with ribopuromycylation and the RNA granules imaged with ribopuromycylation were the same as those with SunTag visualized nascent peptides. Accordingly, the ribopuromycylated puncta in neuronal dendrites were also resistant to puromycin. Thus, the SunTag technique corroborates in situ evidence of stalled polysomes and will allow for the live examination of these translational structures as a mechanism for mRNA transport and regulated protein synthesis.
Local translation in neurons plays an important role in determining the proteome of synapses, including during certain forms of synaptic plasticity such as homeostasis, metabotropic glutamate receptor-dependent long-term depression (mGluR-LTD), and long-lasting forms of long-term potentiation (LTP) (Tsokas et al. 2005; Costa-Mattioli et al. 2009; Cajigas et al. 2010; Graber et al. 2013). Local translation requires the transport of mRNAs from the soma in a repressed state, followed by reactivation at the appropriate time and place. Numerous mechanisms have been implicated in this process, separable by the step of translation that is repressed. When translation is blocked at initiation, mRNAs are transported in RNA transport particles, which lack ribosomes. In contrast, ribosomes can be assembled on mRNAs and translation blocked at elongation or termination; these stalled polysomes are stored in large RNA granules (Sossin and DesGroseillers 2006). Stalled polysomes can be identified in situ by their resistance to runoff by inhibitors of new polysome formation, followed by identification of remaining polysomes by labeling nascent peptides with puromycin (ribopuromycylation; RPM) and preventing puromycin-mediated dissociation of nascent peptides with emetine (Graber et al. 2013, 2017; David and Yewdell 2015). Notably, this technique requires fixation and immunocytochemical staining with an antibody against puromycin. Of clear advantage, would be a live imaging technique that allows for the visualization of stalled polysomes in real time.
The SunTag system was developed to allow enhanced live imaging (Tanenbaum et al. 2014). This technique uses a tag consisting of a linear array of a peptide in combination with expression of a green fluorescent protein (GFP)-tagged single chain variable fragment (ScFv) antibody with a high affinity for the peptide. The multiple copies of the peptide lead to many GFPs bound to the same protein and to a large signal relative to the background GFP-tagged antibody. The combination of multiple ribosomes on one mRNA leads to a further amplification of the signal. This technique has been modified to visualize translation by incorporating degradation of the protein after translation has finished, and thus focusing on nascent translation on ribosomes (Pichon et al. 2016; Wang et al. 2016; Wu et al. 2016; Yan et al. 2016).
Here, we use the SunTag technique to corroborate in situ evidence of RNA granules containing polysomes repressed postinitiation. Similar to when using the RPM assay, translating ribosomes can be runoff with initiation inhibitors at which point nascent peptides still attached to ribosomes represent slowly elongating or stalled polysomes. We find that, in contrast to HEK cell lines or neuronal somata, most SunTag puncta observed in distal dendrites are resistant to runoff by either initiation inhibitors or puromycin. In concordance with the resistance of these puncta to puromycin, we find that puromycin also does not cause the disruption of stalled polysomes visualized by RPM, as elongation inhibitors are not required for visualization of ribopuromycylated stalled polysomes in hippocampal or cortical neurites. Thus two independent techniques, RPM and SunTag, both suggest that the majority of polysomes in neuronal processes are stalled.
Results
We used lentivirus generated from plasmids, a kind gift of Dr. Zhuang (Wang et al. 2016), to transduce cultured human embryonic kidney (HEK)293T cells as well as hippocampal and cortical neurons. Specifically, cells were transduced with a lentivirus encoding the ScFv-superfolder GFP (sfGFP) construct which recognizes the GCN4-V4 epitope and a separate lentivirus encoding 24 copies of the GCN4-V4 epitope followed by a degron from the ornithine decarboxylase protein with a 3′untranslated region (UTR) consisting of the woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) and a truncated 5′ long terminal repeat (LTR) or the 3′UTR of Arc followed by the same sequence. The viral titer of the construct containing the V4 epitope could not be measured as this construct had no fluorescent protein, but was considerably less than that encoding the ScFv-GFP construct due to its length (14 vs. 11 kB). Nevertheless, cotransduced cells were easily identified by the puncta not seen when transducing with the ScFv-scGFP construct on its own. Only cells with clear puncta were analyzed.
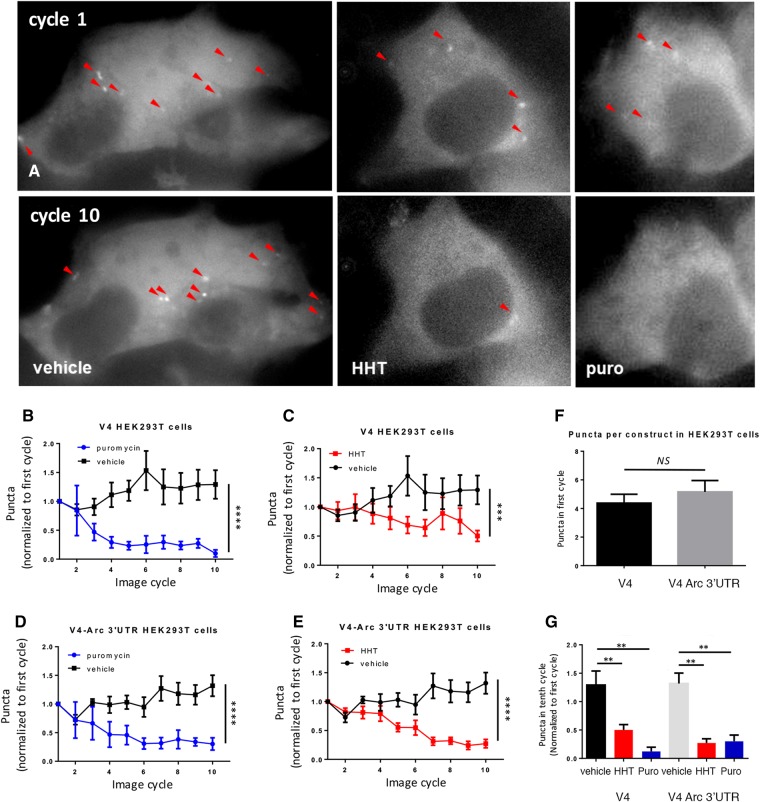
We first examined this system in HEK293T cells, a cell type that has few stalled polysomes (Graber et al. 2013). Similar to a previous report (Wang et al. 2016), addition of puromycin (an aminoacyl tRNA-analog that covalently attaches to, and then allows the dissociation of, the polysome's nascent chain), or homoharringtonin (HHT) (a drug that specifically blocks the first step of elongation, but not subsequent steps and thus prevents the formation of new polysomes while allowing elongating polysomes to finish translation and runoff) led to a significant decrease in the number of puncta over time in HEK293T cells (Fig. 1A–E). There was no difference in puncta with constructs positive (V4-Arc) or negative (V4) for the 3′UTR of Arc (Fig. 1F). We only imaged a single focal plane and thus did not follow individual puncta, but instead simply counted the puncta at each time point (see Materials and Methods). While this does not allow estimation of elongation rates, it serves to demonstrate the loss of puncta by puromycin and HHT. For the time course, we used an automated program for counting puncta, but we also confirmed changes in puncta number with manual puncta counts (see Materials and Methods) from the start and at the end of treatment (Fig. 1G). A similar significant decrease in puncta with puromycin and HHT were seen with automated or manual counting methods.
Figure 1.
Translation visualization in HEK293T cells. (A) HEK293T cells from left to right: vehicle, puromycin, and HHT at first image cycle (top images) and tenth image cycle (bottom images) after treatment (see Materials and Methods for definition of image cycle). Red arrows point to puncta. (B–E) Graphs of number of puncta in V4-expressing cells (B,C) or V4-Arc expressing cells (D,E) normalized to first cycle after treatment (V4; puro n = 5 cells, vehicle n = 8 cells, HHT n = 11 cells. V4-Arc: puro n = 3 cells, vehicle n = 9 cells, HHT n = 10 cells). Experiments are from a minimum of three independent cultures. Puncta were counted with an automated program (Materials and Methods). Vehicle groups from HHT (water) and puromycin (0.0675% Ethanol) were not significantly different from each other and were pooled. The same pooled vehicle groups are used in B,C and D,E. An interaction between time and group was observed for V4 (two-way ANOVA F(18,189) = 2.4, P = 0.002) and V4-Arc (two-way ANOVA F(18,162) = 4.9, P = <0.0001). For V4, control vs. HHT and control vs. Puro, Tukeys post-hoc tests P = 0.0002 and <0.0001, respectively, and for V4-Arc control vs. HHT and control vs. Puro, Tukey's post-hoc tests P < 0.0001 and <0.0001, respectively. (F) Total number of puncta in V4 and V4-Arc cells at first image cycle, P > 0.05 n = 24 (V4 cells), 22 (V4-Arc cells). (G) Number of puncta at tenth image cycle (∼12 min after first image cycle) normalized to first image cycle immediately after treatment (n is same as B–E) with puncta counted manually. Number of puncta was significantly different between controls and cells treated with HHT or puromycin for both V4 (one-way ANOVA F(23,2) P < 0.01), Tukey's post-hoc test P < 0.01 and V4-Arc (one-way ANOVA F(21,2) P < 0.01), Tukey's post-hoc test P < 0.01. All errors are shown as standard errors of the mean. Asterisks are applied as follows: (**) P ≤ 0.01, (****) P ≤ 0.0001.
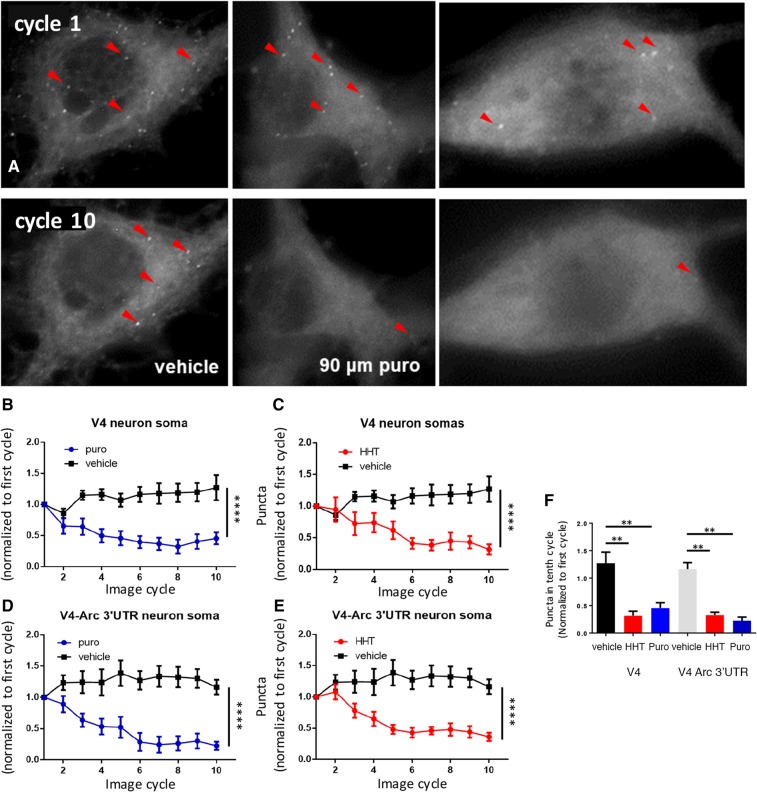
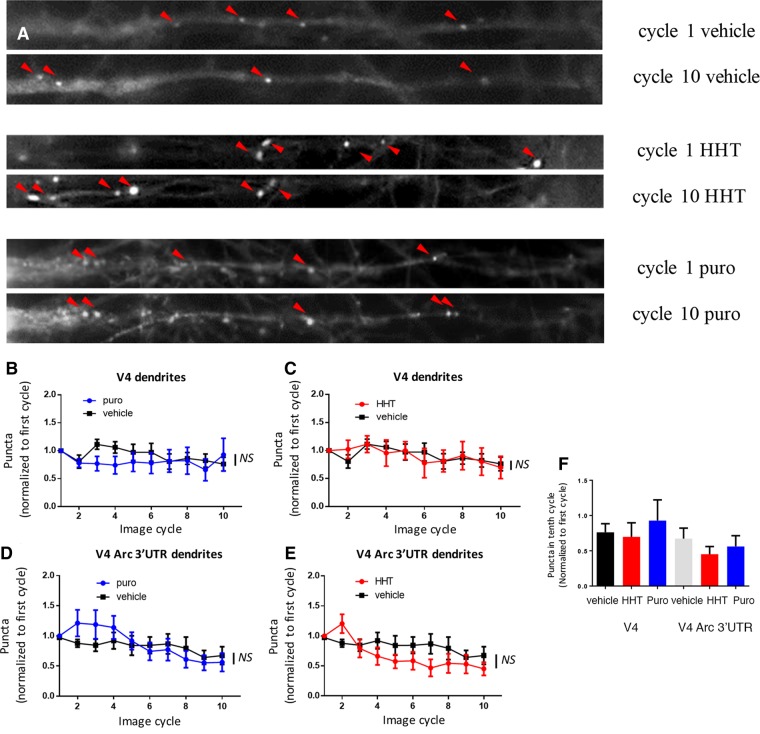
Similar to HEK293T cell lines, a significant loss of puncta in the cell bodies of neurons transduced with either lentivirus was observed after 10 min of treatment with either puromycin or HHT (Fig. 2A) using either automated counting (Fig. 2B–E) or manual counting of puncta (Fig. 2F). Again, as in HEK293T cells there were no differences in the constructs with or without the 3′UTR of Arc. However, in the neurites of hippocampal neurons most puncta were resistant to both puromycin and HHT (Fig. 3A) and there was no significant loss of puncta over time compared to neurons treated with vehicle using either automated counting (Fig. 2B–E) or manual counting of puncta (Fig. 2F). Somewhat unexpectedly, the SunTag puncta with the construct containing a basal 3′UTR were resistant to the same extent as those treated with the construct containing the 3′UTR of Arc.
Figure 2.
Translation visualization in neuronal somas. (A) Neuronal somas: from left to right: vehicle, puromycin, and HHT at first image cycle after treatment (top images) and tenth cycle (bottom images) after treatment was added. Red arrows point to puncta. (B–E) Graphs of number of puncta in V4-expressing cells (B,C) or V4-Arc expressing cells (D,E) normalized to first cycle after treatment (V4; vehicle n = 13 cells, puro n = 7 cells, HHT n = 12 cells; V4-Arc vehicle n = 14 cells, puro n = 8 cells, HHT n = 9 cells. Puncta were counted with an automated program (Materials and Methods). Experiments came from a minimum of three independent cultures. Vehicle groups from HHT (water) and puromycin (0.0675% Ethanol) were not significantly different from each other and were pooled. The same pooled vehicle groups are used in B–C and D–E. An interaction of time and group was observed for V4 (two-way ANOVA F(18,243) = 4.8, P < 0.0001) and V4-Arc (two-way ANOVA F(18,234) = 5.6, P < 0.0001). For V4, control vs. HHT and control vs. Puro, Tukey's post-hoc tests P < 0.0001 and <0.0001, respectively, and for V4-Arc control vs. HHT and control vs. Puro, Tukey's post-hoc tests P < 0.0001 and <0.0001, respectively. (F) Number of puncta at tenth image cycle (∼12 min after first image cycle) normalized to first image cycle immediately after treatment (n is same as B–E) measured using manual counting of puncta. Number of puncta was significantly different between controls and cells treated with HHT or puromycin for both V4 (one-way ANOVA F(31,2) P < 0.001), Tukey's post-hoc test P < 0.01 and V4-Arc (one-way ANOVA F(30,2) P < 0.001), Tukey's post-hoc test P < 0.01. All errors are shown as standard error of the mean. Asterisks are applied as follows: (**) P ≤ 0.01, (****) P ≤ 0.0001.
Figure 3.
Translation visualization in neuronal dendrites. (A) Neuronal somas: from top to bottom: vehicle, HHT and puromycin at first image cycle after treatment (top) and tenth cycle (bottom) after treatment was added. Red arrows point to puncta. (B–E) Graphs of number of puncta in V4-expressing cells (B,C) or V4-Arc expressing cells (D,E) normalized to first cycle after treatment (V4; vehicle n = 13 dendrites, puro n = 8 dendrites, HHT n = 8 dendrites; V4-Arc vehicle n = 14 dendrites puro n = 10 dendrites, HHT n = 12 dendrites. Puncta were counted with an automated program (Materials and Methods). Experiments came from a minimum of three independent cultures. Vehicle groups from HHT (water) and puromycin (0.0675% Ethanol) were not significantly different from each other and were pooled. The same pooled vehicle groups are used in B,C and D,E. No interaction of time and group was observed for V4 (two-way ANOVA F(18,225) = 1.3, P = 0.20) or V4-Arc (two-way ANOVA F(18,279) = 1.5, P = 0.08). For V4, control vs. HHT and control vs. Puro, Tukey's post-hoc tests P = 0.09 and 0.24, respectively, and for V4-Arc control vs. HHT and control vs. Puro, Tukey's post-hoc tests P = 0.24 and 0.82, respectively. (F) Number of puncta at tenth image cycle (∼12 min after first image cycle) normalized to first image cycle immediately after treatment (n is same as B–E) measured using manual counting of puncta. (F) No groups were significantly different V4 (one-way ANOVA F(28,2) P > 0.5), V4-Arc (one-way ANOVA F(35,2) P > 0.5). (NS) not significant. All errors are shown as standard error of the mean.
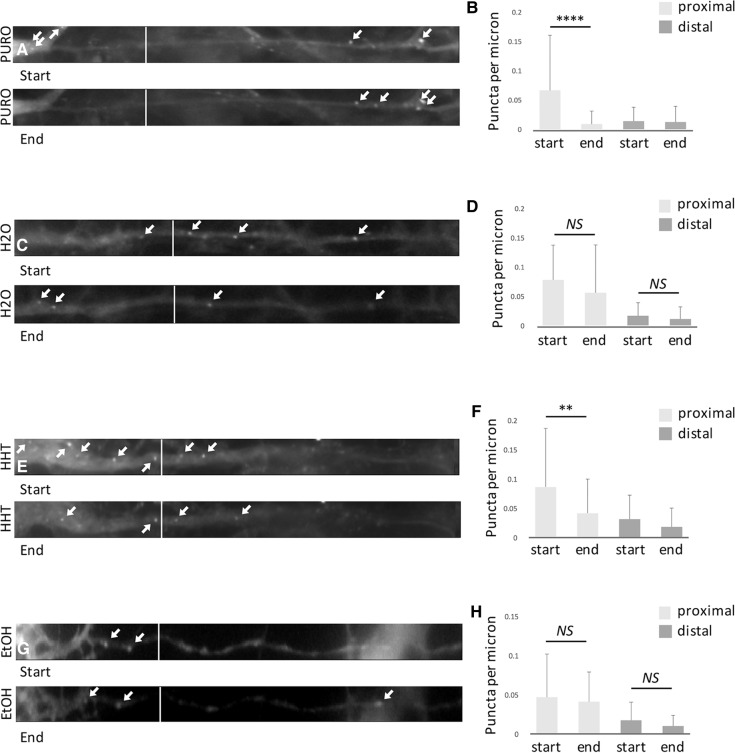
The loss of SunTag puncta with puromycin in dendrites was noted in two previous examinations of SunTag puncta in neurons (Wang et al. 2016; Wu et al. 2016). While one of these (Wu et al. 2016) only reported the loss of puncta in neuronal somas, agreeing with our results (Fig. 2), the other (Wang et al. 2016) claimed that neuronal puncta disappear in dendrites, although this is not quantified and the picture shown (Fig. 7B in Wang et al. 2016) appears to display a proximal dendrite. We thus compared the decrease in the number of puncta with puromycin and HHT between the first and last cycle for proximal and distal dendrites. Indeed, we find that there is a significant loss of puncta with both puromycin and HHT in proximal, but not in distal, dendrites (Fig. 4).
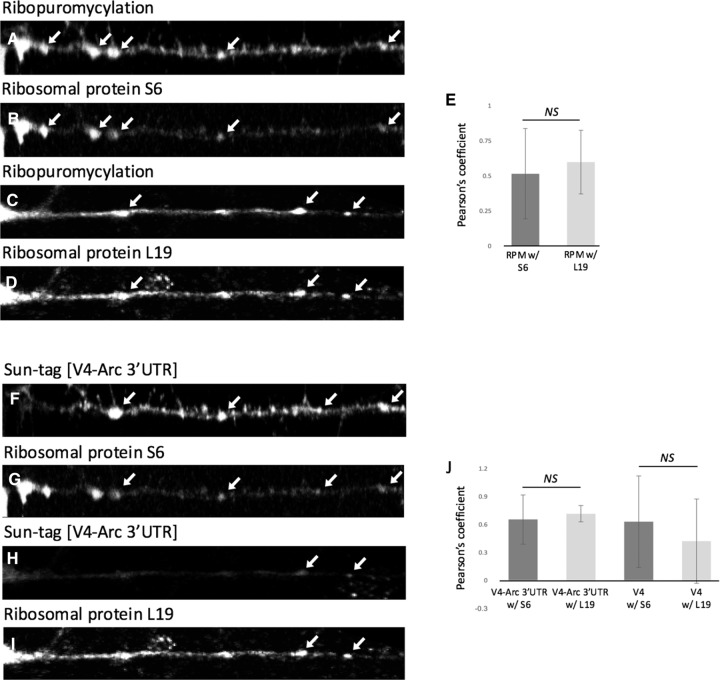
Figure 7.
RPM and SunTag puncta colocalize with small and large ribosomal subunit markers. Shown is the colocalization of RPM (A,C) and V4-Arc 3′UTR (F,H) SunTag puncta with ribosomal protein S6 (B,G) and L19 (D,I) signal; puncta are indicated by white arrows. V4 (not shown) also colocalized with S6 and L19 as indicated by the quantifications in E and J; colocalization is represented as average Pearson's coefficients. A paired, two tailed t-test found no significant difference in colocalization of RPM signal with small and large ribosomal subunits; P = 0.3. Similarly, V4-Arc 3′UTR and V4 SunTag puncta both showed colocalization with the ribosomal subunit proteins; one-way ANOVA P = >0.5. N = 62 RPM and 48 SunTag neurite puncta from a combination of hippocampal and cortical neuron cultures. All errors are shown as standard deviation of the mean.
Figure 4.
Comparison of distal and proximal dendrites. Border between proximal and distal parts of the neurite indicated by white line. Start (first cycle) and End (tenth cycle) throughout. SunTag puncta are indicated by white arrows. (A) A decrease in the density of proximal, but not distal, SunTag puncta is observed between the start and end of imaging in neuronal cultures treated with puromycin. (B) One-way ANOVA with Tukey's post-hoc multiple comparisons tests: F(3,156) = 11.23, (****) P < 0.0001; NS, P > 0.2, n = 40. (C) No decrease in the density of proximal or distal SunTag puncta is observed between the start and end of imaging in neuronal cultures treated with H20. (D) One-way ANOVA with Tukey's post-hoc multiple comparisons tests, NS, P > 0.2, n = 41. (E) A decrease in the density of proximal, but not distal, SunTag puncta is observed between the start and end of imaging in neuronal cultures treated with HHT. (F) One-way ANOVA for HHT treatment with Tukey's post-hoc multiple comparisons tests: F(3,216) = 11.83, (****) P < 0.0001; (**) P < 0.01; NS, P > 0.2, n = 55. (G) No decrease in the density of proximal or distal SunTag puncta is observed between the start and end of imaging in neuronal cultures treated with ethanol. (G) One-way ANOVA for ethanol treatment with Tukey's post-hoc multiple comparisons tests: NS, P > 0.2, n = 34.
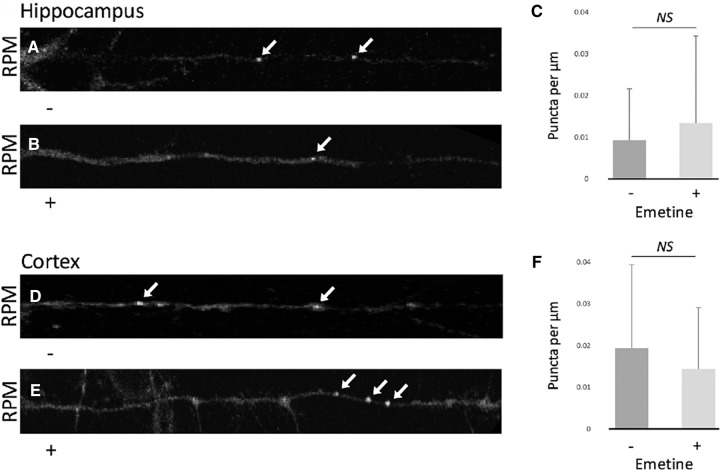
While we had previously described polysomes in neurites that were resistant to runoff by HHT, their resistance to puromycin had not been investigated previously. Although stalled polysomes in hippocampal neurites are ribopuromycylated, these experiments were only performed in the presence of the elongation inhibitor, emetine (Graber et al. 2013, 2017). The resistance of the dendritic SunTag puncta to puromycin suggests that after covalently binding puromycin the nascent chain does not escape the stalled polysome, consistent with a blockade of elongation in stalled polysomes similar to the block normally imposed by emetine. This suggests that addition of the elongation inhibitor emetine should not be necessary for RPM of stalled polysomes. Indeed, we found that there was no difference in the density of stalled polysomes (identified by RPM after running off translating polysomes with HHT) in neuronal processes when puromycin was added in the presence or absence of the elongation inhibitor emetine (Fig. 5A–C). Due to the more robust survival of cortical cultures relative to hippocampal cultures, we also confirmed this result in neurites from cortical neurons (Fig. 5D–F). Cortical neurons, similar to hippocampal neurons, have RPM puncta that are virtually all resistant to runoff by initiation inhibitors (Supplemental Fig. 1).
Figure 5.
Emetine is not necessary for the ribopuromycylation assay. Emetine does not significantly change the number of RPM puncta (indicated by white arrows) in hippocampal (A,B) or cortical (D,E) neurons; quantified in C and F, respectively. A paired, two tailed t-test found P values of 0.3 and 0.2 for emetine vs. no emetine comparisons in hippocampal and cortical neurites, respectively. N = 66 emetine positive and 50 emetine negative hippocampal as well as 70 emetine positive and 52 emetine negative cortical neurites from multiple cultures. All errors are shown as standard deviation of the mean.
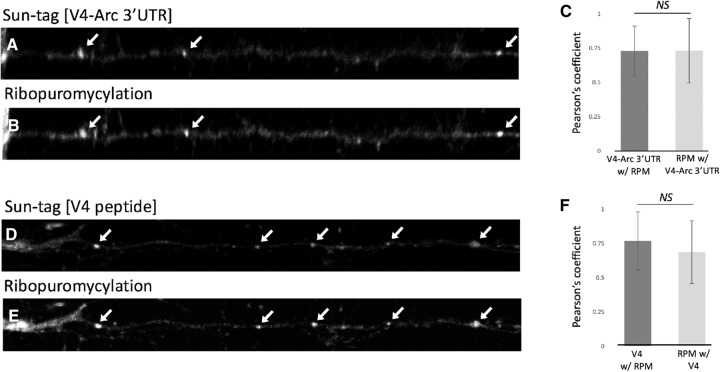
Normally, sensitivity to puromycin is used as the criteria that SunTag fluorescence is due to nascent chains and not to translated, perhaps aggregated, protein. Since the puncta in dendrites were resistant to puromycin, we sought independent evidence that they represented ScFv-scGFP binding to nascent chains. Ribopuromycylation is specific to nascent chains on ribosomes. Thus, if the puncta we observe in dendrites are due to ScFv-scGFP bound to nascent chains, they should colocalize with puncta observed with RPM. We transduced hippocampal and cortical neurons with the two viruses, ran off translating polysomes with HHT, and then performed RPM on these neurons. In these experiments, both V4-Arc 3′UTR and V4 GFP puncta showed very strong colocalization with the puncta observed by RPM (Fig. 6). These findings confirm that SunTag puncta in neurites represent nascent chains on polysomes, as puromycin can only covalently bind to peptides on the ribosome and would not associate with aggregated proteins. Surprisingly, almost all RPM puncta observed were also positive for the ScFv-scGFP (Fig. 6). The implication of this for the complexity of RNA granules is considered in the discussion.
Figure 6.
SunTag and ribopuromycylation label the same initiation inhibitor resistant polysomes. Shown is the colocalization of V4-Arc 3′UTR (A) and V4 (D) SunTag puncta with RPM signal (B,E) (puncta are indicated by white arrows); quantification of V4-Arc 3′UTR-RPM and RPM-V4-Arc 3′UTR as well as V4-RPM and RPM-V4 colocalization, shown as average Pearson's coefficients, is included as C and F, respectively. Paired, two tailed t-tests found P values of 1 for V4-Arc 3′UTR-RPM and RPM-V4-Arc 3′UTR and 0.1 for V4-RPM and RPM-V4 comparisons. N = 72 RPM and 62 SunTag neurite puncta, taken from a combination of hippocampal and cortical neuron cultures. All errors are shown as standard deviation of the mean.
As additional proof that SunTag puncta represent nascent chains of polysomes, we performed colocalization analysis of SunTag and RPM puncta with markers for small and large ribosomal subunits, namely, the ribosomal proteins S6 and L19, respectively. Both markers were observed to colocalize with RPM and SunTag puncta (Fig. 7), confirming these puncta as containing ribosomes.
Discussion
Evidence for stalled polysomes
Our results, consistent with earlier findings using RPM (Graber et al. 2013, 2017) suggest that most polysomes in dendrites are stalled. While there were issues with cell health because of the combination of lentivirus infection, phototoxicity and the cell manipulations required for live imaging (see Materials and Methods; Supplemental Methods), the observed loss of puncta in the neuronal soma combined with the resistance of those in dendrites suggests that polysome stalling was not due to poor health of the cells. The SunTag puncta in dendrites also colocalized with RPM puncta and with small and large ribosomal proteins, and thus could not be simply aggregates of fully translated proteins.
The puncta we observe could represent a single stalled polysome on one mRNA, or a large collection of stalled polysomes in a protected RNA granule. While several studies examining colocalization of mRNAs suggest that transcripts are transported in individual particles (Tubing et al. 2010; Mikl et al. 2011; Amrute-Nayak and Bullock 2012; Batish et al. 2012) these studies may have been viewing mainly mRNA transport particles blocked at initiation, and not mRNAs blocked at elongation. Many of these studies use in situ hybridization and it has also been reported that numerous “masked” granules in RNA dendrites are resistant to this technique (Buxbaum et al. 2014). Other studies have suggested multiple mRNAs in one granule (Carson et al. 2008). A recent study using live imaging with a GFP-tagged RNA granule protein (Fragile X mental retardation protein (FMRP), a protein enriched in RNA granules) showed coalescence and separation of puncta, supporting the notion of multiple polysomes in one granule (El Fatimy et al. 2016). Further, live imaging of RNA granules containing colocalization of two RNA helicases identified in the proteomics of RNA granules (dead box 3 and dead box 1) (Elvira et al. 2006) identified puncta that were visible in differential interference contrast (DIC) microscopy and appeared larger than would be expected for a single polysome (Miller et al. 2009). If each granule was a separate polysome on an individual mRNA, one might expect only a small fraction of RPM puncta would emanate from the mRNA expressing the SunTag nascent peptides. In contrast, we observed that the majority of RPM puncta contained SunTag nascent chains. One explanation for this result would be the presence of many stalled polysomes in each granule. Alternatively, the mRNA encoding the SunTag mRNA may be expressed at such high levels that it completely takes over the stalled polysome formation process and becomes the only mRNA found in stalled polysomes. However, given that only a small percentage of neurons expressed the mRNA (i.e., showed puncta), it seems unlikely that the viral load in any neuron was more than one, and thus expression levels would not be expected to be this high.
Stalled polysomes with a minimal 3′UTR
There was no difference in the resistance of the puncta to puromycin or HHT in the presence or absence of the Arc 3′UTR. While it is clear that the Arc 3′UTR directs its mRNA into transport to dendrites (Kobayashi et al. 2005; Dynes and Steward 2012), whether it is sufficient to direct mRNAs to a stalled polysome is unclear. On one hand, Arc translation has been reported to be initiation-independent (Na et al. 2016). However, this ability was linked to the coding region of Arc, not its 3′UTR. Moreover, Arc mRNA was not in puromycin-resistant ribosomes compared to other FMRP targets in a study using extracts from Neuro2A cells (Darnell et al. 2011). Nevertheless, it is surprising that even the basic 3′UTR was sufficient for localization to a stalled polysome in dendrites. One possibility is that double-stranded sequences in the WPRE or the LTR sequences in the vector bind to double-stranded RNA binding proteins thought to be important for incorporation into stalled polysomes, such as Staufens (Lebeau et al. 2011; Graber et al. 2017). It is also possible that simply overexpressing any mRNA will allow some fraction of the mRNA to travel into dendrites in stalled polysomes.
Stalled polysomes can be labeled by puromycin, but nascent chains in stalled polysomes are not dissociated by puromycin
Puromycin is commonly used as a protein synthesis inhibitor and dissociates ribosomes from translating peptides (Blobel and Sabatini 1971; Rivera et al. 2015). However, we did not see a change in the number of RPM puncta in the absence of emetine, a drug traditionally used to repress translation and prevent puromycin dissociation of nascent peptides (David and Yewdell 2015). The structure of the stalled ribosome in the RNA granule is not clear. The formation of what appears to be organized, tightly packed ribosomes in purified RNA granules suggests some modifications occur during the generation and packing of the RNA granule (Elvira et al. 2006; Darnell et al. 2011; El Fatimy et al. 2016). Our results suggest that this modified structure still allows for RPM, but not for dissociation by puromycin. Emetine binds to the E site of the ribosome and blocks the translocation step of elongation (Wong et al. 2014). Stalled polysomes found during No go decay are stalled at the translocation step (Ikeuchi et al. 2019). The lack of a requirement for emetine, which normally blocks the translocation step, to prevent puromycin dissociation of the nascent chain suggests that neuronal stalled polysomes are naturally blocked at this step, obviating the need for emetine. Interestingly, polysomes in dendrites of dentate gyrus neurons have also been reported to be resistant to puromycin (Dynes and Steward 2012), suggesting this is not just an artifact in cultured neurons.
SunTag puncta in dendrites are stalled polysomes
Our results suggest that SunTag puncta in distal dendrites of neurons are stalled polysomes resistant to runoff and to dissociation by puromycin. This result differs from the initial study using this construct where dendritic puncta were reported to be dissociated by puromycin (Wang et al. 2016). While the reason for this discrepancy is not clear, we suggest it is due to observation of dissociation in proximal dendrites, where we also observe some dissociation by puromycin. The actual percentage of stalled polysomes observed may be affected by our inability to follow granules over time, instead just measuring the number of SunTag puncta at different time points. However, the presence of a large number of SunTag puncta colocalized with ribosomal markers after puromycin or HHT treatment demonstrates a large presence of stalled polysomes, even if the actual percentage of stalled polysomes may not be precise. An additional paper using a distinct construct also states that dendritic SunTag puncta are dissociated by puromycin (Wu et al. 2016). Again, it is not clear that the majority of puncta examined in this study are not somatic or proximal dendritic puncta or nonstalled polysomes that may be a larger fraction of the SunTag puncta with this construct. We believe that this is probably due to the presence of somatic, translating ribosomes in the proximal dendrites. Unlike axons, dendrites do not have a barrier, such as is present at the start of the axon, to prevent diffusion into the dendrite (Britt et al. 2016). Another study examining both the presence of mRNA and SunTag showed an enrichment of mRNAs encoding β-actin that were repressed at initiation in distal dendrites based on the presence of many mRNAs lacking SunTag colocalization (lacking nascent peptides) (Wu et al. 2016). While our studies suggest most polysomes in dendrites are stalled, they do not measure mRNAs stalled at initiation that lack ribosome association and these mRNA transport particles (Sossin and Lacaille 2010) also are likely to play important roles in local translation. In Wu et al. (2016), they also measure dynamics of translation of SunTag puncta in distal dendrites and measure bursting translation. The bursts of translation are inconsistent with stalled polysomes and are more consistent with bursts of translational initiation. We would suggest that under the different conditions used in these experiments (3′UTR of β-actin vs. 3′UTR of Arc, P1 hippocampal cultures from mouse vs. E18 cultures from rats, 14–21 d in vitro vs. 6–11 d in vitro), stalled polysomes make up a smaller fraction of the SunTag puncta. Nevertheless, in Wu et al. (2016) they report that 20% of the puncta have “constitutive translation” or constant intensity of SunTag labeling and this behavior is consistent with what would be expected from a stalled polysome.
We think it is unlikely that our puromycin-resistant puncta were not nascent peptides due to their colocalization with RPM and small and large ribosomal subunits. The equal colocalization of both SunTag and RPM with small and large ribosomal subunits also eliminates the possibility that we were detecting stress granules as these structures lack large ribosomal subunits (Anderson and Kedersha 2006). Moreover, mRNA in stress granules would not be ribopuromycylated as no nascent chains are present under these conditions. Taken together, our research suggests that these structures are polysome complexes stalled postinitiation, which likely operate to transport mRNAs and regulate protein synthesis.
Conclusions
The results presented in this paper provide another independent piece of evidence that many polysomes in neuronal dendrites are stalled. Together with results from RPM and initiation-independent measures of protein synthesis (Graber et al. 2013, 2017), as well as the resistance to runoff of polysomes from neuronal extracts (Darnell et al. 2011), the evidence for stalled polysomes in neuronal granules is accumulating. The confirmation that stalled polysomes can be seen in live imaging paves the way for real-time visualization of stalled polysomes during plasticity to determine if sites of reactivation are linked to sites of translation-dependent plasticity. The present results also raise questions about the specificity of the requirement of particular 3′UTR sequences for inclusion into the stalled polysome. Finally, the strong colocalization between SunTag from a specific message and RPM labeling provides evidence that many stalled polysomes are probably present in an RNA granule.
Materials and Methods
Lentivirus production and application to HEK293T cell cultures
Bacterial cultures
Glycerol stocks of bacterial cultures containing the following target plasmids were grown overnight in autoclaved LB (Lysogeny broth) with 1% ampicillin: ScFv-sfGFP-GB1, pFUGW-24xGCN4_ V4-ODC, and pFUGW-24xV4-GCN_V4-ODC with Arc 3′UTR. For simplicity, we often refer to the transcripts encoded by these plasmids mRNA, as well as protein products of these constructs, as ScFv-GFP, V4, and V4-Arc 3′UTR. Plasmids containing viral packaging components were also grown from glycerol stocks in the same manner, these contained pRSV-Rev, pMD2rg, and pMDLg/pRRE.
HEK293T cell transfection
HEK293T cells were grown to 80%–90% confluency in DMEM (Gibco, Life Technologies) supplemented with 10% fetal bovine serum (Sigma-Aldrich) (DMEM/10% FBS) 15 cm tissue culture plates and then each plate was transfected with 27 µg of plasmid of interest (described above in bacterial cultures), 13 µg pMDLg/pRRE, 8 µg pMD2rg, and 6.5 µg pRSV-Rev using OptiMEM and Lipofectamine 3000 (Invitrogen). Media was collected and replaced with warm DMEM/10% FBS at 24, 36, and 48 h after transfection. Collected media was kept at 4°C until the end of the collection period and was then combined, filtered through 0.45 µm-pore size low protein binding filters, and centrifuged at 7500g for 16 h. Supernatant was removed and disposed of according to lentivirus Standard Operating Procedure (SOP), and pellet was resuspended in 100 µL Neurobasal (Gibco, Thermo Fisher) and frozen at −80°C.
Live imaging experiments in HEK293T cells
Thirty-five millimeter live imaging plates with a #1.5 16 mm glass coverslip (MatTek) were coated overnight in PLL and then rinsed with double distilled autoclaved water and left to dry. HEK293T cells were plated in warm 200 µL DMEM/10% FBS only on the central glass coverslip at a density of 40–50,000 cells per coverslip and incubated at 37°C for an hour or more. After this preliminary incubation 2 mL of DMEM/10% FBS was added to each plate. HEK293T cells were infected 1 d after plating, infection followed protocol described below in the Lentiviral Infection section. These cells were imaged 1 d later (2 d after plating and 1 d after infection), as described below in Live imaging.
Neuronal cell culturing and lentiviral infection
Sprague–Dawley rats were obtained from Charles River Canada. All animal experiments were approved by the Animal Ethics Committees of the Montreal Neurological Institute and Université de Montréal, and abided by the guidelines of the Canadian Council on Animal Care.
Cultures for live imaging experiments
Rat primary hippocampal neurons were cultured from Sprague–Dawley embryos dissected at embryonic day 18 in Hank's Balanced Salt Solution (HBSS) supplemented with 1% Pen-Strep, 1% Glutamax, 1% Sodium Pyruvate, and HEPES (10 mM, pH 7.4) or Leibovitz's L15 Medium (Thermo Fisher). Neurons were then incubated in trypsin-EDTA (0.25%) for 20–30 min at 37°C, and then triturated minimally in DMEM/10%FBS. If there were spots of neural tissue remaining undissociated, then the solution was passed through a cell strainer (mesh size 70 µM). Neuron density was then counted using Trypan blue (Sigma-Aldrich) and was diluted appropriately and plated in DMEM on 35 mm MatTek dishes, with a #1.5 16 mm glass coverslip. Dishes were coated in 100 µg/mL PLL overnight and rinsed in double distilled sterile water and left to dry. Neurons were plated at a density of 50–65,000 cells, depending on the condition of neurons counted at plating, in 250 µL of DMEM/10% FBS only on the central glass coverslip of the MatTek plate. DMEM/10%FBS was then removed at 2 h or more after plating and replaced with 2 mL per plate of Neurobasal supplemented with N2, B27, Pen Strep, and Glutamax (Gibco, Life Technologies). Neurons were grown in vitro at 37°C with CO2 at 5% for 8–11 d.
For lentiviral infection, media was removed from live imaging plates and saved in 15 mL tubes incubated at 37°C. Lentiviruses frozen in Neurobasal medium were thawed and diluted in 250 µL supplemented Neurobasal (neurons) or DMEM with 10% FBS (HEK293T cells) and applied to the central well of live imaging plates. Cells were then incubated for 3–4 h at 37°C. Viral multiplicity of infection for ScFv-GFP was calculated from titres and applied to cells at approximately 0.6 to ensure the infection of a large portion of the culture but to decrease the likelihood of doubly infecting cells. Viral multiplicity for the V4 and V4-Arc could not be calculated and approximately 5× to 10× the volume of purified viral particles were added compared to the ScFv-GFP virus. Following Lentiviral incubation, media which had been previously removed from the cells was reintroduced to the live imaging plates, and the plates were then returned to the incubator at 37°C.
In fixed culture experiments, primary hippocampal and cortical neurons were plated at a density of 80–100,000 per each well of a 12 well plate in DMEM/FBS followed by a media switch to supplemented Neurobasal, as described above. Transduction with ScFv-GFP, V4 and V4-Arc 3′UTR plasmids occurred on day I as follows: plasmids were applied to wells with half of the original media for 2 h before being switched to a 50% old, 50% new, supplemented Neurobasal media and cultured to 6–8 d before undergoing RPM and fixation.
Ribopuromycylation and immunocytochemistry
Ribopuromycylation assay
Cells were assigned to HHT or no HHT conditions and exposed to supplemented Neurobasal solutions with 4 mM HHT or 0.05% ethanol (the vehicle in which HHT is carried) for 10 min at 37°C. Preincubation in HHT allows sufficient time for actively translating ribosomes to runoff (average speed of 4 aa/sec; 240 aa/min, or 3600 amino acids in 15 min) and impedes the creation of new translational sites. Thus, remaining RPM staining represents stalled or slowly elongating ribosomes. After 15 min, cells in each well of a 12 well plate, with 1 mL of total solution per well, were exposed to 2 mg/mL HHT, 25 mg/mL emetine (in a subset of experiments emetine was replaced by 50% ethanol) and 10 mg/mL puromycin. Cells remained in this RPM solution for 5 min, which has proven to be sufficient for the incorporation of puromycin with nascent chains, at 37°C before undergoing a 2 min wash in 0.0003% digitonin on ice (to extract any unbound puromycin) and washed 3 × 5 min in 1× HBS before being fixed for 30 min in 4% paraformaldehyde with 20% sucrose at room temperature. Paraformaldehyde was removed at 30 min and replaced with 1× PBS and cultures were stored at 4°C until undergoing immunohistochemical treatment.
Immunocytochemistry
Cultures fixed and stored in 1× PBS were treated with Triton-X for 10 min to allow for permeabilization of cell membranes followed by 15 min of quench solution (55 mM ammonium chloride in 1× PBS). Permeabilizing and quench solutions are washed out by 3 × 5 min 1× PBS washes before application of a blocking agent, 5% BSA in PBS, for 30 min to reduce nonspecific IgG binding. Cells were then treated for 60 min with a primary antibody solution (1:1000 mouse anti-puromycin (deposited to the Developmental Studies Hybridoma Bank by Yewdell, Jonathan (DSHB Hybridoma Product PMY-2A4) and either 1:1000 rabbit anti-ribosomal protein S6 (Cell Signaling TECHNOLOGY, catalog number: 2217) or anti-L19 (LifeSpan BioSciences, catalog number: LS-C499680) in 5% BSA and 1× PBS). Following 3 × 10 min 1× PBS washes to remove residual primary antibody cells were kept in the secondary antibody solution (1:1000 anti-mouse Alexa Fluor 647 and 1:1000 anti-rabbit 568) for 60 min. Following 3 × 10 min 1× PBS washes to remove residual secondary antibody coverslips were mounted using 10 µL of Dako mounting medium/coverslip and kept at room temperature in the dark overnight before being placed at 4° for long-term storage. All steps in the immunohistochemical treatment occurred at room temperature (∼21°C).
Microscopy
Live imaging
Cells were imaged in HEPES-buffered saline containing 25 mM HEPES (pH 7.4), 119 mM NaCl, 5 mM KCl, 2 mM CaCl2 2 mM MgCl2, 30 mM glucose, 400 nM ascorbic acid, and 0.2 mM Trolox (Sigma-Aldrich), pH adjusted to 7.4 with NaOH and osmolarity adjusted to 300 osm/L using double distilled H2O. Plates were imaged at 37°C on a Zeiss Axio Observer fluorescent microscope using the 63× objective, LED illumination, and the Zeiss Axiocam 506 camera and Zeiss's ZEN imaging software. A number of modifications were necessary for successful live imaging and these are described in the Supplemental Methods. LED intensity and duration was held constant for each experiment. There was an issue with focus drift and blurring during imaging; automated and manual focusing was used to mitigate these issues. In order to facilitate cell survival, long exposure times were selected with lower laser intensity to reduce bleaching, but in HEK293T cells puncta were highly mobile and this long exposure time resulted occasionally in some blurring of the SunTag signal, and may have led to the loss of some puncta which moved too rapidly in and out of frame. Videos of cells were taken in the following manner: Five images were taken at a frequency of one image every 2 sec (which will be referred to as an “imaging cycle”), followed by a 1-min delay before the next imaging cycle. Five image cycles were completed before the addition of treatment or vehicle, after which 10 more image cycles were completed. Once a cell was selected for imaging, only cells that showed persistent signal after five cycles of imaging were selected for the experiment. Since some cells was discarded due to the rapid disappearance of puncta upon live imaging; this may confound our data if those cells which displayed imaging-resistant puncta were in some way different from the average cell. However, since this decision was made before treatments, it should not affect the effect of the treatments. Treatment or vehicle was added directly before an imaging cycle commenced. In the figures, this is the first imaging cycle. Treatments were the following: HHT (Tocris Bioscience) in 50% ethanol, at a concentration of 4 and 25 µM to final ethanol concentration of 0.0675% in solution; 90 µM puromycin (Thermo Fisher) in H2O. There was no difference with the two concentrations of HHT and results were pooled. “Vehicle” refers to either addition of a 50% ethanol concentration to a final concentration of 0.0675% ethanol or water corresponding to puromycin. There was no difference between the two vehicles and they were pooled as vehicle.
Quantification of live imaging
The number of puncta was averaged at each image cycle of five images to give number of puncta used in future calculations. For quantification, the numbers of puncta at tenth image cycle posttreatment were normalized to number of puncta at first image cycle posttreatment. The normalization to the first image cycle posttreatment was due to the change in focus which occurred after the addition of treatment or vehicle; as the microscope's heating chamber had to be opened and the condenser lifted in order to add the substance to the live imaging dish, it was difficult to reattain the exact same plane of focus in the consequent image cycle and it was often shifted, which could reduce the accuracy of the normalization. However, as the first image cycle began directly after the addition of treatment or vehicle, the effects of the treatment would be small and thus the effects of treatment could still be captured. For automated counting of puncta, the number of puncta per frame in live imaging experiments was calculated using TrackMate (Tinevez et al. 2017) plugin for ImageJ in conjunction with a macro written in Visual Basic for Excel. Videos were cropped to isolate the selected cell or neuron cell body; dendrites were straightened using ImageJ's straighten function. A TrackMate “threshold” (a parameter of the program) was manually set for each cell to best captured number of puncta, along with a size limit of 2.5 μm in diameter. The threshold was constant over the course of each video. Bleaching was adjusted using ImageJ's bleaching adjustment function Brightness and contrast was adjusted in videos to facilitate the manual spotting of puncta necessary for selecting an appropriate threshold. In cases where there was blurring or any other issue with a particular imaging cycle, the averaged values of the five images of imaging cycle before and after were averaged and used instead. For manual counting of puncta video brightness, contrast, and bleaching were adjusted as necessary for visualization. Puncta were identified as circular or elliptical shapes that were differentiable from background signal by brightness and regularity, these structures were typically >0.3 and <2.5 μm in diameter. Cell counts were captured by Cell Counter, which counted mouse clicks per frame.
Confocal imaging
In fixation experiments, photomicrographs were obtained at 63× magnification using a LSM 710 or 880 confocal microscope equipped with ZEN imaging software.
Quantification of confocal imaging
For fixation experiments puncta were manually counted (see above) and the total count was divided by neurite length, resulting in a puncta per micron value.
Colocalization analysis
Colocalization of SunTag GFP, RPM, ribosomal protein S6 and L19 puncta was measured using a homemade plugin designed to quantify colocalization with Pearson's coefficients and Costes’ automatic thresholding. The plugin opted for here was modified from Graber et al. (2013) to use an ∼3 µm by 3 µm detection box.
Statistical analysis
T-tests and one-way ANOVAs with post-hoc Tukey's honest significant difference tests were used for two and three or more group comparisons, respectively. Time series data was analyzed using two-way ANOVAs to determine if there was an interaction between group and time followed by post-hoc Tukey's honest significant difference tests between control and treated groups.
Supplementary Material
Acknowledgments
This work was supported by CIHR project grant 374967 to W.S.S. W.S.S. is a James McGill Professor. J.J.L. is supported by a Jeanne Timmins Costello award and the Joan and Warren Chippindale outstanding Student award. We thank Maran Ma for MACRO help and Xiaotang Fan for technical support.
Footnotes
[Supplemental material is available for this article.]
Article is online at http://www.learnmem.org/cgi/doi/10.1101/lm.049965.119.
References
- Amrute-Nayak M, Bullock SL. 2012. Single-molecule assays reveal that RNA localization signals regulate dynein-dynactin copy number on individual transcript cargoes. Nat Cell Biol 14: 416–423. 10.1038/ncb2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. 2006. RNA granules. J Cell Biol 172: 803–808. 10.1083/jcb.200512082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batish M, van den Bogaard P, Kramer FR, Tyagi S. 2012. Neuronal mRNAs travel singly into dendrites. Proc Natl Acad Sci 109: 4645–4650. 10.1073/pnas.1111226109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G, Sabatini D. 1971. Dissociation of mammalian polyribosomes into subunits by puromycin. Proc Natl Acad Sci 68: 390–394. 10.1073/pnas.68.2.390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt DJ, Farías GG, Guardia CM, Bonifacino JS. 2016. Mechanisms of polarized organelle distribution in neurons. Front Cell Neurosci 10: 88 10.3389/fncel.2016.00088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum AR, Wu B, Singer RH. 2014. Single β-actin mRNA detection in neurons reveals a mechanism for regulating its translatability. Science 343: 419–422. 10.1126/science.1242939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajigas IJ, Will T, Schuman EM. 2010. Protein homeostasis and synaptic plasticity. EMBO J 29: 2746–2752. 10.1038/emboj.2010.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson JH, Gao Y, Tatavarty V, Levin MK, Korza G, Francone VP, Kosturko LD, Maggipinto MJ, Barbarese E. 2008. Multiplexed RNA trafficking in oligodendrocytes and neurons. Biochim Biophys Acta 1779: 453–458. 10.1016/j.bbagrm.2008.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. 2009. Translational control of long-lasting synaptic plasticity and memory. Neuron 61: 10–26. 10.1016/j.neuron.2008.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. 2011. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146: 247–261. 10.1016/j.cell.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David A, Yewdell JW. 2015. Applying the ribopuromycylation method to detect nuclear translation. Methods Mol Biol 1228: 133–142. 10.1007/978-1-4939-1680-1_11 [DOI] [PubMed] [Google Scholar]
- Dynes JL, Steward O. 2012. Arc mRNA docks precisely at the base of individual dendritic spines indicating the existence of a specialized microdomain for synapse-specific mRNA translation. J Comp Neurol 520: 3105–3119. 10.1002/cne.23073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Fatimy R, Davidovic L, Tremblay S, Jaglin X, Dury A, Robert C, De Koninck P, Khandjian EW. 2016. Tracking the fragile X mental retardation protein in a highly ordered neuronal ribonucleoparticles population: a link between stalled polyribosomes and RNA granules. PLoS Genet 12: e1006192 10.1371/journal.pgen.1006192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvira G, Wasiak S, Blandford V, Tong XK, Serrano A, Fan X, del Rayo Sánchez-Carbente M, Servant F, Bell AW, Boismenu D, et al. 2006. Characterization of an RNA granule from developing brain. Mol Cell Proteomics 5: 635–651. 10.1074/mcp.M500255-MCP200 [DOI] [PubMed] [Google Scholar]
- Graber TE, Hebert-Seropian S, Khoutorsky A, David A, Yewdell JW, Lacaille JC, Sossin WS. 2013. Reactivation of stalled polyribosomes in synaptic plasticity. Proc Natl Acad Sci 110: 16205–16210. 10.1073/pnas.1307747110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber TE, Freemantle E, Anadolu MN, Hébert-Seropian S, MacAdam RL, Shin U, Hoang HD, Alain T, Lacaille JC, Sossin WS. 2017. UPF1 governs synaptic plasticity through association with a STAU2 RNA granule. J Neurosci 37: 9116–9131. 10.1523/JNEUROSCI.0088-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi K, Tesina P, Matsuo Y, Sugiyama T, Cheng J, Saeki Y, Tanaka K, Becker T, Beckmann R, Inada T. 2019. Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways. EMBO J 38: e100276 10.15252/embj.2018100276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Yamamoto S, Maruo T, Murakami F. 2005. Identification of a cis-acting element required for dendritic targeting of activity-regulated cytoskeleton-associated protein mRNA. Eur J Neurosci 22: 2977–2984. 10.1111/j.1460-9568.2005.04508.x [DOI] [PubMed] [Google Scholar]
- Lebeau G, Miller LC, Tartas M, McAdam R, Laplante I, Badeaux F, DesGroseillers L, Sossin WS, Lacaille JC. 2011. Staufen 2 regulates mGluR long-term depression and Map1b mRNA distribution in hippocampal neurons. Learn Mem 18: 314–326. 10.1101/lm.2100611 [DOI] [PubMed] [Google Scholar]
- Mikl M, Vendra G, Kiebler MA. 2011. Independent localization of MAP2, CaMKIIalpha and β-actin RNAs in low copy numbers. EMBO Rep 12: 1077–1084. 10.1038/embor.2011.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LC, Blandford V, McAdam R, Sanchez-Carbente MR, Badeaux F, DesGroseillers L, Sossin WS. 2009. Combinations of DEAD box proteins distinguish distinct types of RNA: protein complexes in neurons. Mol Cell Neurosci 40: 485–495. 10.1016/j.mcn.2009.01.007 [DOI] [PubMed] [Google Scholar]
- Na Y, Park S, Lee C, Kim DK, Park JM, Sockanathan S, Huganir RL, Worley PF. 2016. Real-time imaging reveals properties of glutamate-induced Arc/Arg 3.1 translation in neuronal dendrites. Neuron 91: 561–573. 10.1016/j.neuron.2016.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon X, Bastide A, Safieddine A, Chouaib R, Samacoits A, Basyuk E, Peter M, Mueller F, Bertrand E. 2016. Visualization of single endogenous polysomes reveals the dynamics of translation in live human cells. J Cell Biol 214: 769–781. 10.1083/jcb.201605024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera MC, Maguire B, Lake JA. 2015. Dissociation of ribosomes into large and small subunits. Cold Spring Harb Protoc 2015: 363–367. 10.1101/pdb.prot081372 [DOI] [PubMed] [Google Scholar]
- Sossin WS, DesGroseillers L. 2006. Intracellular trafficking of RNA in neurons. Traffic 7: 1581–1589. 10.1111/j.1600-0854.2006.00500.x [DOI] [PubMed] [Google Scholar]
- Sossin WS, Lacaille JC. 2010. Mechanisms of translational regulation in synaptic plasticity. Curr Opin Neurobiol 20: 450–456. 10.1016/j.conb.2010.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. 2014. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159: 635–646. 10.1016/j.cell.2014.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez JY, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, Eliceiri KW. 2017. TrackMate: an open and extensible platform for single-particle tracking. Methods 115: 80–90. 10.1016/j.ymeth.2016.09.016 [DOI] [PubMed] [Google Scholar]
- Tsokas P, Grace EA, Chan P, Ma T, Sealfon SC, Iyengar R, Landau EM, Blitzer RD. 2005. Local protein synthesis mediates a rapid increase in dendritic elongation factor 1A after induction of late long-term potentiation. J Neurosci 25: 5833–5843. 10.1523/JNEUROSCI.0599-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubing F, Vendra G, Mikl M, Macchi P, Thomas S, Kiebler MA. 2010. Dendritically localized transcripts are sorted into distinct ribonucleoprotein particles that display fast directional motility along dendrites of hippocampal neurons. J Neurosci 30: 4160–4170. 10.1523/JNEUROSCI.3537-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Han B, Zhou R, Zhuang X. 2016. Real-time imaging of translation on single mRNA transcripts in live cells. Cell 165: 990–1001. 10.1016/j.cell.2016.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong W, Bai XC, Brown A, Fernandez IS, Hanssen E, Condron M, Tan YH, Baum J, Scheres SH. 2014. Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine. Elife 3: e03080 10.7554/eLife.03080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Eliscovich C, Yoon YJ, Singer RH. 2016. Translation dynamics of single mRNAs in live cells and neurons. Science 352: 1430–1435. 10.1126/science.aaf1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Hoek TA, Vale RD, Tanenbaum ME. 2016. Dynamics of translation of single mRNA molecules in vivo. Cell 165: 976–989. 10.1016/j.cell.2016.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.