Abstract
Storage of long-term memory requires not only protein synthesis but also protein degradation. In this article, we overview recent publications related to this issue, stressing that the balanced actions of protein synthesis and degradation are critical for long-term memory formation. We particularly focused on the brain-derived neurotrophic factor signaling that leads to protein synthesis; proteasome- and autophagy-dependent protein degradation that removes molecular constraints; the role of Fragile X mental retardation protein in translational suppression; and epigenetic modifications that control gene expression at the genomic level. Numerous studies suggest that an imbalance between protein synthesis and degradation leads to intellectual impairment and cognitive disorders.
The brain stores and recalls information throughout life in the form of memories. For long-term storage, learned information should be “consolidated” into neural circuits through dynamic gene regulatory events in the brain (Kandel 2001). Through de novo protein synthesis during certain time windows (∼1–3 h after learning) (Bourtchouladze et al. 1998), learned information is believed to be solidified into the brain and permanently stored as a long-term memory that is relatively insensitive to disruption (Kandel 2001).
Long-term potentiation (LTP), initially identified as a sustained increase in synaptic efficacy at the hippocampal synapses (Bliss and Gardner-Medwin 1973; Schwartzkroin and Wester 1975; Bliss and Collingridge 1993; Huang et al. 1996), has been suggested as the cellular substrate of hippocampal memory formation. LTP is triggered by activation of the N-methyl-D-aspartate receptors (NMDAR). NMDAR-dependent LTP can be classified as early-phase LTP (E-LTP) and late-phase LTP (L-LTP), each including mechanistically distinct features. High-frequency stimulation to the hippocampal Schaffer collateral pathway induces short-term potentiation that is rapidly decayed in several minutes, followed by a sustained phase of elevated synaptic transmission lasting for hours in vitro and days to months in vivo (Abraham et al. 2002; Abraham 2003). This sustained phase of synaptic potentiation can be subdivided based on dependency on protein kinase A (PKA) and protein synthesis (Park et al. 2014b). E-LTP, which is insensitive to inhibitors of PKA and protein synthesis, involves modifications of preexisting synapses through NMDAR-dependent Ca2+ signaling. L-LTP requires PKA-dependent signaling, subsequent activation of gene transcription, and new protein synthesis (Huang et al. 1996; Matsushita et al. 2001; Abraham 2003). Due to its sensitivity to protein synthesis blockers, the molecular mechanisms underlying L-LTP are similar to those of long-term memory formation (Bourtchouladze et al. 1998).
Despite these notions, another line of studies has provided evidence that protein degradation is also involved in LTP. In 1993, Bliss laboratory demonstrated that in vivo LTP induction in the rat hippocampus results in either an increase or decrease in protein expression levels by utilizing two-dimensional gel electrophoresis technique (Fazeli et al. 1993). Consistent with findings of proteolysis-dependent Aplysia long-term facilitation (LTF) (Hegde et al. 1993, 1997; Chain et al. 1999), treatments with the proteasome inhibitor such as lactacystin could produce impaired long-term memory (Lopez-Salon et al. 2001) and disrupted L-LTP (Fonseca et al. 2006) in the rodent brain. Since then, the importance of the suppression of protein synthesis or protein degradation for synaptic plasticity and memory formation has been supported by accumulating evidence (Lee et al. 2008a, 2012).
Long-term synaptic plasticity and increased protein level through BDNF signaling
Similar to that in other nonneuronal cells, protein synthesis in neuronal cells is regulated by general translation processes. During new protein synthesis in response to learning or L-LTP triggering stimuli, the role of the brain-derived neurotrophic factor (BDNF) is prominent. BDNF is a member of the neurotrophin family of secretory proteins that not only acts as a survival factor for peripheral neurons but also mediates diverse activity-dependent processes, such as neuronal differentiation and growth, synapse formation and plasticity, and learning and memory, through signaling pathways dependent on a high-affinity receptor, tyrosine receptor kinase B (TrkB) (Chao 2003; Reichardt 2006). Since neural activity directly controls either the expression or secretion of BDNF through Ca2+-dependent signaling (Poo 2001; Matsuda et al. 2009; Park and Poo 2013; Park et al. 2014a), L-LTP-inducing stimuli can elicit activity-dependent BDNF secretion at the synapses, leading to structural and functional enhancements of the synapses, such as enlargement of the synaptic structures correlated with L-LTP maintenance and sustained enhancement of the presynaptic release, through local TrkB activation (Aicardi et al. 2004; Tanaka et al. 2008; Harward et al. 2016). Conversely, retrograde transport of the neurotrophin-Trk complexes to the soma could exert their transcriptional regulation of activity-dependent genes, such as Arc (Cohen et al. 2011; Yamashita and Kuruvilla 2016), indicating the capability of local BDNF signaling to induce a cell-wide enhancement of neural functions.
After BDNF secretion, the subsequent activation of TrkB stimulates the phosphoinositide 3-kinase (PI3K) and extracellular-signal-regulated kinase (ERK) signaling pathways (Fig. 1; Costa-Mattioli et al. 2009). The PI3K-Akt pathway can induce either the eukaryotic translation initiation factor 4F (eIF4F)- or the mechanistic target of rapamycin-ribosomal protein S6 (mTOR-S6) kinase-dependent initiation of translation processes (Roux and Topisirovic 2012). ERK up-regulates translation processes by phosphorylation of the eIF4B or eIF4E, leading to an increased translation rate of 5′CAP-mRNAs (Roux and Topisirovic 2012). In addition, phosphorylation of the eIF4E-binding protein 1 (4EBP1) through the Akt- and ERK-signaling pathways leads to its decreased binding with eIF4E and a consequent decrease in the 4EBP1-mediated inhibition of translation, resulting in increased protein synthesis (Ruvinsky and Meyuhas 2006; Roux and Topisirovic 2012). Translation elongation steps could also be influenced by BDNF signaling. The eukaryotic elongation factor 2 (eEF2) kinase, whose activity is controlled by the S6 kinase or mitogen-activated protein kinase (MAPK), down-regulates the actions of eEF2, a member of the guanosine triphosphate (GTP)-binding translation elongation factor family, by phosphorylating eEF2 (Heise et al. 2014). In vivo BDNF infusion into the hippocampus could promote a rapid and transient increase in eEF2 phosphorylation (Kanhema et al. 2006), which can decrease overall protein synthesis. In contrast, activation of eEF2 in the cortex was induced by BDNF treatment (Takei et al. 2009), suggesting that the effect of BDNF signaling on translation elongation is brain area-specific.
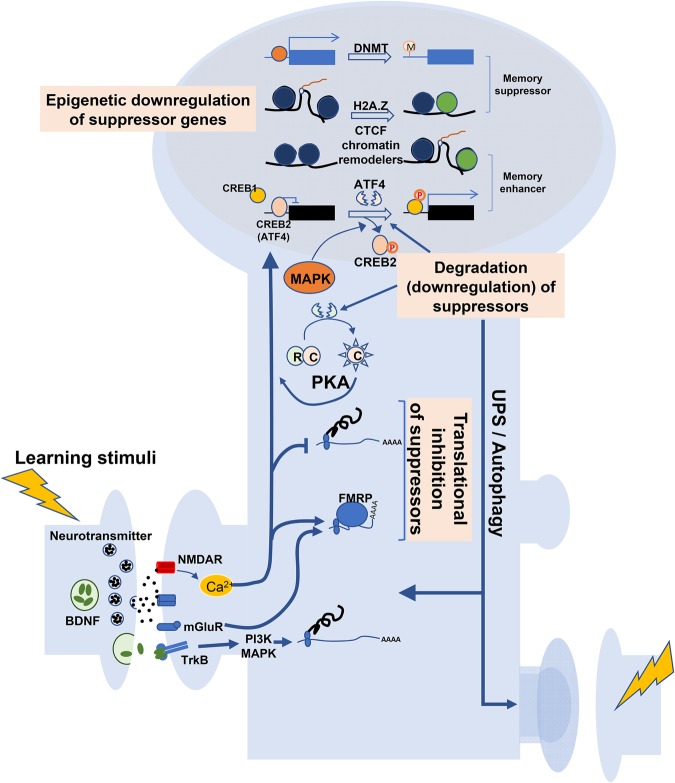
Figure 1.
Schematic diagram showing protein synthesis and degradation involved in memory consolidation. Learning stimuli trigger the presynaptic release of neurotransmitters and BDNF, leading to activation of NMDARs and TrkB receptors, respectively. Secreted BDNF activates TrkB signaling linked with PI3K and MAPK, resulting in increased de novo protein synthesis. Presynaptic neurotransmitters activate NMDARs and subsequent NMDAR-dependent Ca2+ elevation, which not only activates signaling pathways increasing gene transcription and translation but also induces activity-dependent translational suppression of mRNAs that encode memory suppressor proteins, by reducing translation rates or FMRP-dependent inhibition of mRNA translation (Translational inhibition of suppressors). On the other hand, learning stimuli enhance expression and activities of proteolysis mechanisms mediated by UPS or autophagy, leading to degradation of the PKA regulatory subunit (R), ATF4, or other memory suppressor proteins. Degradation of PKA R subunit produces the constitutively active form of PKA catalytic (C) subunit, which triggers diverse molecular mechanisms required for long-term synaptic plasticity. Down-regulation of CREB2 activity is known to induce CREB1-dependent transcription of memory enhancer genes (Degradation or down-regulation of suppressors). Finally, learning stimuli lead to epigenetic down-regulation of memory suppressor genes by suppressing transcription via DNMT-dependent DNA methylation, H2A.Z incorporation to histone complexes, or CTCF-mediated alteration of gene expression. In some cases, such epigenetic modulation was reported to produce an increase in expression of memory enhancer genes, suggesting that epigenetic modulation of transcription is gene-specific (Epigenetic down-regulation of suppressor genes).
Rather than affecting global protein synthesis, BDNF signaling seems to play a selective role in regulating protein expression. When rapid changes in the synaptic proteome were analyzed using a multidimensional protein identification technology, BDNF-induced proteins were classified as translation factors, proteins involved in synaptic structure maintenance and remodeling, and vesicle formation and movement (Liao et al. 2007). Confirming the alteration of the expression level of the translation machinery, another study additionally demonstrated BDNF-induced up-regulation of the ubiquitin-proteasome system (UPS)-related proteins such as 26S proteasome subunits (Psmd7, Psma2) or ubiquitin carboxy-terminal hydrolase 14 (USP14) (Manadas et al. 2009). Since ubiquitination of synaptic proteins could be tightly regulated by neural activity or BDNF signaling (Jia et al. 2008), these results suggest that BDNF-dependent L-LTP involves not only new protein synthesis but also protein degradation.
Memory formation by disinhibiting molecular constraints
Memory formation and disruption have been correlated with protein synthesis and degradation, respectively. Indeed, many studies have demonstrated that the decrease in memory is dependent on protein degradation. Memory destabilization during memory retrieval required proteasome-dependent protein degradation (Lee et al. 2008a, 2012), and synaptic destabilization by autophagy induction was sufficient for disrupting already consolidated memories (Shehata et al. 2018).
However, to store learned information in the neural circuits, the inhibitory mechanisms hindering activity-dependent facilitation of structural and functional enhancements of synapses need to be removed (Abel et al. 1998). These inhibitory molecular constraints appear to be conserved mechanisms. One possible way of removing such “memory suppressor proteins” is to relieve their inhibitory functions without affecting protein expression levels (Fig. 1). In invertebrates, such as Aplysia and Drosophila, inhibitory forms of cAMP response element binding protein (CREB2) suppress the CREB1-dependent transcription during basal states; however, repetitive activation of specific Aplysia 5-HT receptors (Lee et al. 2009) triggers either PKA-dependent CREB1 phosphorylation (Bartsch et al. 1998) or mitogen-activated protein kinase (MAPK)-dependent phosphorylation of CREB2 (Bartsch et al. 1995; Michael et al. 1998), leading to up-regulation of CREB1-dependent transcription that is necessary for LTF (Liu et al. 2008, 2011). Similarly, a mammalian homolog of inhibitory CREB2, activating transcription factor 4 (ATF4), was also reported to repress the transcription of memory genes, and this repression is relieved by LTP- or learning-inducing stimuli (Chen et al. 2003; Pasini et al. 2015).
Degradation of memory suppressor proteins has also been shown to be effective for long-term memory formation. During serotonin-induced LTF of the sensory-motor neuron synapses in Aplysia, facilitation of PKA-dependent synaptic release, activation of CREB1, and induction of gene transcription (Kaang et al. 1993; Kandel 2001; Lee et al. 2008b), as well as an UPS-dependent degradation of the regulatory subunits of cAMP-dependent PKA were demonstrated (Hegde et al. 1993). Since the rapid induction of ubiquitin carboxy-terminal hydrolase, which interacts with the proteasome, was elicited by an LTF-triggering neurotransmitter, serotonin (Hegde et al. 1993, 1997), learning stimuli could evoke such protein degradation mechanism during memory formation. Such protein degradation is able to induce constitutive activation of the catalytic subunits of PKA without an increase in cAMP concentration (Chain et al. 1999), leading to facilitation of PKA-dependent presynaptic release and activation of CREB1 (Greenberg et al. 1987; Hegde et al. 1993; Hegde 2017).
UPS-dependent memory formation has also been observed in the mammalian brain. Infusion of lactacystin, a UPS inhibitor, into the CA1 area of the rat hippocampus impaired long-term memory formation (Lopez-Salon et al. 2001). Accordingly, treatment with lactacystin enhanced the induction but inhibited the maintenance of L-LTP (Dong et al. 2008). Collectively, these results clearly demonstrate that the UPS-dependent protein degradation of memory suppressor proteins is essential for maintaining of synaptic plasticity that is crucial for long-term memory. The detailed mechanisms of memory consolidation targeted by the protein degradation pathways remain to be elusive; however, there is evidence that synaptic plasticity is dependent on the UPS proteolytic activity. Proteasome-mediated augmentation of the early phase of L-LTP required activation of NMDAR and PKA as well as protein synthesis, whereas CREB-dependent transcription of BDNF was blocked by the same treatment due to inhibition of the degradation of ATF4, a CREB repressor (Dong et al. 2008). These results indicate that the LTP maintenance in the hippocampal slices was dependent on proteasome activation (Fonseca et al. 2006; Dong et al. 2008). In addition, an NMDAR- and Ca2+/calmodulin-dependent protein kinase II (CaMKII)-dependent increase in local protein degradation was required for activity-induced new spine growth (Hamilton et al. 2012). Since the activity-dependent remodeling of the molecular composition of the postsynaptic density was UPS-dependent (Ehlers 2003), it is possible that the molecular constraints existing at the synaptic regions are the main target of the UPS-dependent down-regulation during memory formation.
In addition to UPS, other protein degradation pathways are also involved in memory consolidation. Another line of evidence suggests a calpain-mediated removal of inhibitory constraints for long-term memory formation. At the hippocampal CA3-CA1 synapses, it was reported that activity-induced accumulation of proteolytic fragments in the stimulated dendrites is induced by calpain, a protein that belongs to the family of Ca2+-dependent nonlysosomal systemic proteinases (Vanderklish et al. 1995). Theta burst stimulation (TBS)-induced LTP in the cultured or acute hippocampal slices was dependent on calpain expression (Vanderklish et al. 1995; Zhu et al. 2015), raising the possibility that long-term memory formation requires the actions of calpain at the synaptic level. The molecular targets of calpains are largely unknown, but µ-calpain-mediated degradation of suprachiasmatic nucleus circadian oscillatory protein (SCOP), a negative ERK regulator, was demonstrated to be induced during the early phase of TBS-induced LTP (Wang et al. 2014).
Additionally, an autophagy-dependent regulation of memory formation was recently demonstrated. The autophagy flux was increased by learning stimuli, and inhibition of this activity-induced autophagy in the hippocampal neuron was sufficient for producing deficits in TBS-induced LTP and long-term memory formation, with no alteration of the memory stability (Glatigny et al. 2019). These studies collectively indicate that memory formation is mediated by well-balanced actions of activity-dependent protein synthesis and degradation, although the list of molecular targets remains incomplete (Ehlers 2003; Fonseca et al. 2006; Bingol and Sheng 2011; Jarome and Helmstetter 2014; Hegde 2017).
Translational suppression for memory formation
The reduction in protein expression could be also mediated by translational suppression, and there is evidence that such translational suppression is implicated in memory consolidation. A fragile X metal retardation protein (FMRP) protein, encoded by the FMR1 gene, is known to be a key negative regulator of the mRNA translation (Laggerbauer et al. 2001; Li et al. 2001). As an RNA binding protein, FMRP represses either the initiation or the elongation of translation, and its activity is modulated by bidirectional regulation of phosphorylation through the activity-dependent actions of S6 kinase and protein phosphatase 2A (PP2A) (Bagni and Greenough 2005; Bhakar et al. 2012). Moreover, exposure to enriched environments as well as learning stimuli could transiently increase the expression and dendritic localization of FMRP via metabotropic glutamate receptor (mGluR) signaling (Weiler et al. 1997; Irwin et al. 2000; Gabel et al. 2004), suggesting that the FMRP protein plays a role in long-term synaptic plasticity and memory formation (Fig. 1).
To screen mRNAs that are translationally suppressed during memory consolidation, a recent study utilized the technique that quantitatively measures translating mRNA levels by ribosome profiling (RPF)-mediated RNA deep sequencing (Cho et al. 2015). Contextual fear conditioning elicits translational suppression of a subset of genes in the hippocampus, including the ribosomal protein-coding genes, as well as Nrsn1 and Mapk6. Overexpression of the Nrsn1 gene resulted in deficits in hippocampal long-term memory tasks (Cho et al. 2015), supporting the idea that long-term memory consolidation requires reduced translation of a subset of mRNAs (Fig. 1). This translational suppression was mediated by NMDAR-dependent, but mTOR-independent signaling (Cho et al. 2015). The exact mechanisms underlying NMDAR-dependent translational repression during memory consolidation remain to be elucidated.
Epigenetic inhibition for memory consolidation
Although changes in the rates of protein synthesis/degradation directly contribute to the expression level of proteins during memory formation, the relatively high protein turnover rates (hours) induced by these mechanisms do not fully explain the lifelong endurance of memory traces. Hence, epigenetic regulation of gene expression is believed to be as a part of the mechanism achieving sustained up- or down-regulation of the gene products affecting memory storage. Two major epigenetic modifications are known to be involved in memory formation and maintenance: DNA methylation and posttranslational modification of histone proteins. There are numerous reports showing a sustained increase in the memory-promoting genes via epigenetic mechanisms (Levenson and Sweatt 2005; Day and Sweatt 2011; Yu et al. 2011; Peixoto and Abel 2013; Zovkic et al. 2013; Kim and Kaang 2017). In this review, we will focus on epigenetic inhibition of the memory suppressor proteins during memory formation (Fig. 1).
One of the epigenetic modifications of DNA, methylation, refers to the covalent addition of a methyl group to the cytosine base of DNA and is mediated by actions of DNA methyltransferases (DNMTs). Hippocampal Dnmt gene expression was shown to be up-regulated following contextual fear conditioning, and the inhibition of DNMT activation was sufficient for blocking memory formation; however, it did not affect memory retrieval (Miller and Sweatt 2007). Among the target genes of DNMT is a protein phosphatase 1 (PP1), which plays an inhibitory role in learning and memory (Genoux et al. 2002; Miller and Sweatt 2007). Learning-induced methylation of another suppressor, calcineurin (CaN), was also reported (Malleret et al. 2001), and persistent methylation of CaN in the cortex was necessary for consolidation of remote memory in the prefrontal cortex (Miller et al. 2010). These findings are consistent with the notion that the removal of molecular constraints is essential for memory consolidation and suggest the importance of epigenetic down-regulation of memory suppressor proteins for inducing and maintaining long-term memory.
In addition to DNA methylation, the exchange of histone proteins in the nucleosome plays a role in memory consolidation by regulating the gene expression. A replacement of canonical histones with other variants regulates the DNA accessibility of transcription factors, through alterations of the chromatin structures around the transcription initiation sites, caused by the incorporation of histone variants. Among histone proteins, the histone 2A (H2A) family consists of the largest number of variants with the highest sequence divergence at their carboxy-terminal domain (Bönisch and Hake 2012). One of the H2A variants, H2A.Z, is included in the nucleosomes near the transcription start site (TSS) and plays either a positive or a negative role in transcription by influencing the accessibility of genomic DNA to the transcription regulator proteins (Marques et al. 2010; Bargaje et al. 2012). Whether the H2A.Z. is associated with transcriptional activation or repression during memory consolidation appears to be gene-specific. In the mouse brain, chromatin immunoprecipitation (ChIP) analysis demonstrated that the learning stimulus by contextual fear conditioning induces dissociation of H2A.Z from the first nucleosome (+1 nucleosome), upstream of the TSS of memory-promoting genes, such as Npas4, Arc, and Egr1. In contrast, the same fear conditioning stimulus elicited incorporation of H2A.Z into +1 nucleosome of the memory suppressor genes, such as Ppp3ca (the α isoform of a subunit of calcineurin), resulting in reduced gene expression (Zovkic et al. 2014). Since the binding of H2A.Z to the +1 nucleosome was transient and returned to the baseline level within ∼2 h after learning, the repressive role of H2A.Z in memory suppressor gene expression seems to be selective to the memory consolidation process (Zovkic et al. 2014).
Alteration of the 3D chromatin architecture is also reported to be required for remote memory formation. The binding of CCCTC-binding factor (CTCF) to DNA is involved in transcriptional regulation, insulator activity, and regulation of the chromatin architecture (Phillips and Corces 2009). Conditional knockout of CTCF in the forebrain excitatory neurons of adult male mice caused selective deficits in the L-LTP and spatial memory (Sams et al. 2016; Kim et al. 2018), and the effect of CTCF in remote memory was more profound than that in recent memory (Kim et al. 2018). Because CTCF conditional knockout mice exhibited disrupted cortical, but normal hippocampal LTP (Kim et al. 2018), a major function of CTCF in the cortex appears to be to preserve remote memory. RNA sequencing of neurons with deficient CTCF expression revealed that target genes that are implicated in synaptogenesis were down-regulated, whereas the expression of several genes, such as RIN1 or HDAC3, both of which were reported as negative regulators for LTP and memory formation, was up-regulated (Dhaka et al. 2003; McQuown et al. 2011). These findings suggest that the CTCF-dependent chromatin remodeling and regulation of gene expression are critical for regulating remote memory formation. Other chromatic remodeling factor such as neuronal Brm/Brg-associated factor (nBAF) complexes, a mammalian homolog of the yeast SWI/SNF complex, and its regulatory functions in memory formation or drug addiction behavior have been also demonstrated (Wu et al. 2007; Vogel-Ciernia et al. 2013; Alaghband et al. 2018). Neurobiological mechanisms underlying nBAF complex-dependent gene expression and memory regulation were discussed recently (Choi et al. 2015).
Abnormal learning and memory by imbalanced protein synthesis and degradation
We have discussed the importance of balanced synthesis and degradation of proteins for LTP and memory consolidation (Fig. 1). A representative example for an imbalanced protein expression-mediated memory disruption could be found in studies about FMRP-mediated regulation of the translational control. Since FMR1 gene is silenced in Fragile X syndrome, the symptoms of which are intellectual disability and autism, the Fmr1 KO mouse model has been utilized for studying how learning and memory are altered by the FMRP-mediated translational control (The Dutch-Belgian Fragile X Consortium et al. 1994). Fmr1 KO results in increased levels of protein synthesis, due to the inhibitory actions of FMRP on the translation process. Indeed, protein synthesis- and mGluR-dependent long-term depression (LTD) was selectively enhanced in Fmr1 KO mice, with no alteration of NMDAR-dependent LTD (Huber et al. 2002; Hou et al. 2006). Furthermore, a recent study demonstrated that a rat with Fmr1 KO generated by CRISPR/Cas9 displayed impaired synaptic plasticity and spatial memory, as well as a deficit in social interaction (Tian et al. 2017).
Abnormal protein degradation is also linked to neurodegenerative diseases and related cognitive defects. It appears that postmitotic neurons are more vulnerable to misaccumulated proteins than the proliferating cells, due to the lack of dilution of misaccumulated proteins by cell division. Thus, proper protein turnover is not only critical for cellular health but also essential for remodeling of synapses during long-term synaptic plasticity. Regulated proteolysis in the neuron is mediated by various proteolytic pathways, including UPS, lysosome-, and autophagy-dependent protein degradation, and defects in any of these proteolytic pathways can lead to generating abnormally large protein complexes composed of aggregate-prone proteins, such as β-amyloid, tau, α-synuclein, and polyglutamine-repeat proteins. For example, malfunctioning of the UPS is known to be linked with an accumulation of intracellular inclusions enriched with aggregations of ubiquitinated proteins and associated with the development of Alzheimer's disease (AD), Parkinson's disease (PD), and Huntington's disease (HD) (Ciechanover and Kwon 2015; Gentier and van Leeuwen 2015; Zheng et al. 2016; Hegde 2017).
Molecular evidence for neurodegeneration due to abnormal proteolysis has been well established. For instance, mice with ablation of Atg7 or Atg5, essential autophagy genes, showed behavioral defects and reduced lifespan (Komatsu et al. 2006). Deletion of such autophagy genes not only caused impaired axonal membrane trafficking and turnover (Hara et al. 2006; Komatsu et al. 2007) but also resulted in accumulation of polyubiquitinated proteins in the inclusion bodies, without loss of proteasome function. Similarly, blocking the lysosomal proteolysis by inhibiting the lysosomal acidification of cathepsin triggered a slowed cargo-specific axonal transport of endosomes and lysosomes, accompanied by dystrophic swelling of the axons (Lee et al. 2011a,b). This lysosome-dependent axonal dystrophy was found to be associated with accumulation of ubiquitin, APP, and hyperphosphorylated neurofilament proteins, all of which are also found in mouse models of AD (Lee et al. 2011a). In accordance with these findings, it was recently shown that neurons derived from human patients with AD displayed accumulated autophagosomes in the axons, due to the amyloid β oligomer-mediated disruption of the interaction between dynein motor proteins and autophagosomes, leading to a delayed axonal retrograde transport of the autophagosome for fusion with the mature lysosomes (Tammineni et al. 2017).
A defective turnover of certain synaptic proteins is also known to be implicated in the development of pathological sensitization of nociception. Abnormal chronic pain, induced by a peripheral injury and subsequent cortical synaptic plasticity, shares similar features with those of protein synthesis-dependent L-LTP maintenance (Bliss et al. 2016). Similar to learning stimuli, a peripheral injury could induce alterations of the protein turnover rates in the anterior cingulate cortex (ACC), resulting in protein synthesis-dependent spine structural changes and synaptic potentiation (Ko et al. 2018). Among the candidate proteins in the ACC showing rapid synthesis and degradation induced by the peripheral injury, the neuronal cell adhesion molecule 1 (NCAM1) played an essential role in regulating the structural plasticity involved in behavioral sensitization in response to the peripheral nerve injury (Ko et al. 2018).
Age-related memory loss is also suggested to be controlled by autophagy. An age-related memory decline is found during normal aging, and this is believed to be mediated by the reduced hippocampal plasticity (Burke and Barnes 2010; Grady 2012). As discussed earlier, autophagy is induced by learning stimuli and is required for memory formation (Glatigny et al. 2019); however, reduced autophagy biogenesis and reduced expression of autophagy essential genes were found to be correlated with age-related memory decline (Rubinsztein et al. 2011; Stavoe and Holzbaur 2018). Consistently, selective impairment of autophagy in the Drosophila mushroom body produced similar alterations of associative olfactory memory, neuronal structures, and neuropeptide signaling (Bhukel et al. 2019). Boosting autophagy levels in the hippocampus of the aged mice was sufficient for reversing memory deficits (Glatigny et al. 2019), suggesting that memory impairment can be restored by manipulating the autophagy levels.
Summary and future perspectives
Accumulating evidence suggests that protein degradation as well as protein synthesis are not only critical for memory formation but also underlie cognitive impairments and numerous psychiatric disorders. Any intervention to specifically control the protein synthesis/degradation could be a good therapeutic target to restore cognitive disorders. Future studies should provide greater insight for the understanding of the following: (1) how protein synthesis/degradation are regulated in molecular terms, both in the local synaptic structures and in the cell nucleus; and (2) how these two regions, that is, nucleus and synapse, communicate to produce activity-dependent Hebbian plasticity (Choi et al. 2018).
Acknowledgments
H.P. received support from the KBRI basic research program through Korea Brain Research Institute, and this study is funded by the Ministry of Science and ICT (19-BR-01-03). B.-K.K. received support from the National Honor Scientist Program (NRF-2012R1A3A1050385).
Footnotes
Article is online at http://www.learnmem.org/cgi/doi/10.1101/lm.048785.118.
References
- Abel T, Martin KC, Bartsch D, Kandel ER. 1998. Memory suppressor genes: inhibitory constraints on the storage of long-term memory. Science 279: 338–341. 10.1126/science.279.5349.338 [DOI] [PubMed] [Google Scholar]
- Abraham WC. 2003. How long will long-term potentiation last? Philos Trans R Soc Lond B Biol Sci 358: 735–744. 10.1098/rstb.2002.1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham WC, Logan B, Greenwood JM, Dragunow M. 2002. Induction and experience-dependent consolidation of stable long-term potentiation lasting months in the hippocampus. J Neurosci 22: 9626–9634. 10.1523/JNEUROSCI.22-21-09626.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aicardi G, Argilli E, Cappello S, Santi S, Riccio M, Thoenen H, Canossa M. 2004. Induction of long-term potentiation and depression is reflected by corresponding changes in secretion of endogenous brain-derived neurotrophic factor. Proc Natl Acad Sci 101: 15788–15792. 10.1073/pnas.0406960101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaghband Y, Kramár E, Kwapis JL, Kim ES, Hemstedt TJ, López AJ, White AO, Al-Kachak A, Aimiuwu OV, Bodinayake KK, et al. 2018. CREST in the nucleus accumbens core regulates cocaine conditioned place preference, cocaine-seeking behavior, and synaptic plasticity. J Neurosci 38: 9514–9526. 10.1523/JNEUROSCI.2911-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagni C, Greenough WT. 2005. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat Rev Neurosci 6: 376–387. 10.1038/nrn1667 [DOI] [PubMed] [Google Scholar]
- Bargaje R, Alam MP, Patowary A, Sarkar M, Ali T, Gupta S, Garg M, Singh M, Purkanti R, Scaria V, et al. 2012. Proximity of H2A.Z containing nucleosome to the transcription start site influences gene expression levels in the mammalian liver and brain. Nucleic Acids Res 40: 8965–8978. 10.1093/nar/gks665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch D, Ghirardi M, Skehel PA, Karl KA, Herder SP, Chen M, Bailey CH, Kandel ER. 1995. Aplysia CREB2 represses long-term facilitation: relief of repression converts transient facilitation into long-term functional and structural change. Cell 83: 979–992. 10.1016/0092-8674(95)90213-9 [DOI] [PubMed] [Google Scholar]
- Bartsch D, Casadio A, Karl KA, Serodio P, Kandel ER. 1998. CREB1 encodes a nuclear activator, a repressor, and a cytoplasmic modulator that form a regulatory unit critical for long-term facilitation. Cell 95: 211–223. 10.1016/S0092-8674(00)81752-3 [DOI] [PubMed] [Google Scholar]
- Bhakar AL, Dölen G, Bear MF. 2012. The pathophysiology of fragile X (and what it teaches us about synapses). Annu Rev Neurosci 35: 417–443. 10.1146/annurev-neuro-060909-153138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhukel A, Beuschel CB, Maglione M, Lehmann M, Juhász G, Madeo F, Sigrist SJ. 2019. Autophagy within the mushroom body protects from synapse aging in a non-cell autonomous manner. Nat Commun 10: 1318 10.1038/s41467-019-09262-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingol B, Sheng M. 2011. Deconstruction for reconstruction: the role of proteolysis in neural plasticity and disease. Neuron 69: 22–32. 10.1016/j.neuron.2010.11.006 [DOI] [PubMed] [Google Scholar]
- Bliss TV, Collingridge GL. 1993. A synaptic model of memory: long-term potentiation in the hippocampus. Nature 361: 31–39. 10.1038/361031a0 [DOI] [PubMed] [Google Scholar]
- Bliss TV, Gardner-Medwin AR. 1973. Long-lasting potentiation of synaptic transmission in the dentate area of the unanaestetized rabbit following stimulation of the perforant path. J Physiol 232: 357–374. 10.1113/jphysiol.1973.sp010274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss TVP, Collingridge GL, Kaang B-K, Zhuo M. 2016. Synaptic plasticity in the anterior cingulate cortex in acute and chronic pain. Nat Rev Neurosci 17: 485–496. 10.1038/nrn.2016.68 [DOI] [PubMed] [Google Scholar]
- Bönisch C, Hake SB. 2012. Histone H2A variants in nucleosomes and chromatin: more or less stable? Nucleic Acids Res 40: 10719–10741. 10.1093/nar/gks865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtchouladze R, Abel T, Berman N, Gordon R, Lapidus K, Kandel ER. 1998. Different training procedures recruit either one or two critical periods for contextual memory consolidation, each of which requires protein synthesis and PKA. Learn Mem 5: 365–374. [PMC free article] [PubMed] [Google Scholar]
- Burke SN, Barnes CA. 2010. Senescent synapses and hippocampal circuit dynamics. Trends Neurosci 33: 153–161. 10.1016/j.tins.2009.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chain DG, Casadio A, Schacher S, Hegde AN, Valbrun M, Yamamoto N, Goldberg AL, Bartsch D, Kandel ER, Schwartz JH. 1999. Mechanisms for generating the autonomous cAMP-dependent protein kinase required for long-term facilitation in Aplysia. Neuron 22: 147–156. 10.1016/S0896-6273(00)80686-8 [DOI] [PubMed] [Google Scholar]
- Chao MV. 2003. Neurotrophins and their receptors: a convergence point for many signalling pathways. Nat Rev Neurosci 4: 299–309. 10.1038/nrn1078 [DOI] [PubMed] [Google Scholar]
- Chen A, Muzzio IA, Malleret G, Bartsch D, Verbitsky M, Pavlidis P, Yonan AL, Vronskaya S, Grody MB, Cepeda I, et al. 2003. Inducible enhancement of memory storage and synaptic plasticity in transgenic mice expressing an inhibitor of ATF4 (CREB-2) and C/EBP proteins. Neuron 39: 655–669. 10.1016/S0896-6273(03)00501-4 [DOI] [PubMed] [Google Scholar]
- Cho J, Yu NK, Choi JH, Sim SE, Kang SJ, Kwak C, Lee SW, Kim JI, Choi DI, Kim VN, et al. 2015. Multiple repressive mechanisms in the hippocampus during memory formation. Science 350: 82–87. 10.1126/science.aac7368 [DOI] [PubMed] [Google Scholar]
- Choi K-Y, Yoo M, Han J-H. 2015. Toward understanding the role of the neuron-specific BAF chromatin remodeling complex in memory formation. Exp Mol Med 47: e155 10.1038/emm.2014.129 [DOI] [PubMed] [Google Scholar]
- Choi J-H, Sim S-E, Kim J-I, Choi DI, Oh J, Ye S, Lee J, Kim T, Ko H-G, Lim C-S, et al. 2018. Interregional synaptic maps among engram cells underlie memory formation. Science 360: 430–435. 10.1126/science.aas9204 [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Kwon YT. 2015. Degradation of misfolded proteins in neurodegenerative diseases: therapeutic targets and strategies. Exp Mol Med 47: e147 10.1038/emm.2014.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MS, Orth CB, Kim HJ, Jeon NL, Jaffrey SR. 2011. Neurotrophin-mediated dendrite-to-nucleus signaling revealed by microfluidic compartmentalization of dendrites. Proc Natl Acad Sci 108: 11246–11251. 10.1073/pnas.1012401108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. 2009. Translational control of long-lasting synaptic plasticity and memory. Neuron 61: 10–26. 10.1016/j.neuron.2008.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. 2011. Epigenetic mechanisms in cognition. Neuron 70: 813–829. 10.1016/j.neuron.2011.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaka A, Costa RM, Hu H, Irvin DK, Patel A, Kornblum HI, Silva AJ, O'Dell TJ, Colicelli J. 2003. The RAS effector RIN1 modulates the formation of aversive memories. J Neurosci 23: 748–757. 10.1523/JNEUROSCI.23-03-00748.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Upadhya SC, Ding L, Smith TK, Hegde AN. 2008. Proteasome inhibition enhances the induction and impairs the maintenance of late-phase long-term potentiation. Learn Mem 15: 335–347. 10.1101/lm.984508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD. 2003. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci 6: 231–242. 10.1038/nn1013 [DOI] [PubMed] [Google Scholar]
- Fazeli MS, Corbet J, Dunn MJ, Dolphin AC, Bliss TV. 1993. Changes in protein synthesis accompanying long-term potentiation in the dentate gyrus in vivo. J Neurosci 13: 1346–1353. 10.1523/JNEUROSCI.13-04-01346.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca R, Vabulas RM, Hartl FU, Bonhoeffer T, Nägerl UV. 2006. A balance of protein synthesis and proteasome-dependent degradation determines the maintenance of LTP. Neuron 52: 239–245. 10.1016/j.neuron.2006.08.015 [DOI] [PubMed] [Google Scholar]
- Gabel LA, Won S, Kawai H, McKinney M, Tartakoff AM, Fallon JR. 2004. Visual experience regulates transient expression and dendritic localization of fragile X mental retardation protein. J Neurosci 24: 10579–10583. 10.1523/JNEUROSCI.2185-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genoux D, Haditsch U, Knobloch M, Michalon A, Storm D, Mansuy IM. 2002. Protein phosphatase 1 is a molecular constraint on learning and memory. Nature 418: 970–975. 10.1038/nature00928 [DOI] [PubMed] [Google Scholar]
- Gentier RJ, van Leeuwen FW. 2015. Misframed ubiquitin and impaired protein quality control: an early event in Alzheimer's disease. Front Mol Neurosci 8: 47 10.3389/fnmol.2015.00047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatigny M, Moriceau S, Rivagorda M, Ramos-Brossier M, Nascimbeni AC, Lante F, Shanley MR, Boudarene N, Rousseaud A, Friedman AK, et al. 2019. Autophagy is required for memory formation and reverses age-related memory decline. Curr Biol 29: 435–448.e8. 10.1016/j.cub.2018.12.021 [DOI] [PubMed] [Google Scholar]
- Grady C. 2012. The cognitive neuroscience of ageing. Nat Rev Neurosci 13: 491–505. 10.1038/nrn3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg SM, Castellucci VF, Bayley H, Schwartz JH. 1987. A molecular mechanism for long-term sensitization in Aplysia. Nature 329: 62–65. 10.1038/329062a0 [DOI] [PubMed] [Google Scholar]
- Hamilton AM, Oh WC, Vega-Ramirez H, Stein IS, Hell JW, Patrick GN, Zito K. 2012. Activity-dependent growth of new dendritic spines is regulated by the proteasome. Neuron 74: 1023–1030. 10.1016/j.neuron.2012.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441: 885–889. 10.1038/nature04724 [DOI] [PubMed] [Google Scholar]
- Harward SC, Hedrick NG, Hall CE, Parra-Bueno P, Milner TA, Pan E, Laviv T, Hempstead BL, Yasuda R, McNamara JO. 2016. Autocrine BDNF-TrkB signalling within a single dendritic spine. Nature 538: 99–103. 10.1038/nature19766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde AN. 2017. Proteolysis, synaptic plasticity and memory. Neurobiol Learn Mem 138: 98–110. 10.1016/j.nlm.2016.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde AN, Goldberg AL, Schwartz JH. 1993. Regulatory subunits of cAMP-dependent protein kinases are degraded after conjugation to ubiquitin: a molecular mechanism underlying long-term synaptic plasticity. Proc Natl Acad Sci 90: 7436–7440. 10.1073/pnas.90.16.7436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde AN, Inokuchi K, Pei W, Casadio A, Ghirardi M, Chain DG, Martin KC, Kandel ER, Schwartz JH. 1997. Ubiquitin C-terminal hydrolase is an immediate-early gene essential for long-term facilitation in Aplysia. Cell 89: 115–126. 10.1016/S0092-8674(00)80188-9 [DOI] [PubMed] [Google Scholar]
- Heise C, Gardoni F, Culotta L, di Luca M, Verpelli C, Sala C. 2014. Elongation factor-2 phosphorylation in dendrites and the regulation of dendritic mRNA translation in neurons. Front Cell Neurosci 8: 35 10.3389/fncel.2014.00035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Antion MD, Hu D, Spencer CM, Paylor R, Klann E. 2006. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron 51: 441–454. 10.1016/j.neuron.2006.07.005 [DOI] [PubMed] [Google Scholar]
- Huang YY, Nguyen P V, Abel T, Kandel ER. 1996. Long-lasting forms of synaptic potentiation in the mammalian hippocampus. Learn Mem 3: 74–85. 10.1101/lm.3.2-3.74 [DOI] [PubMed] [Google Scholar]
- Huber KM, Gallagher SM, Warren ST, Bear MF. 2002. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc Natl Acad Sci 99: 7746–7750. 10.1073/pnas.122205699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin SA, Swain RA, Christmon CA, Chakravarti A, Weiler IJ, Greenough WT. 2000. Evidence for altered Fragile-X mental retardation protein expression in response to behavioral stimulation. Neurobiol Learn Mem 74: 87–93. 10.1006/nlme.1999.3914 [DOI] [PubMed] [Google Scholar]
- Jarome TJ, Helmstetter FJ. 2014. Protein degradation and protein synthesis in long-term memory formation. Front Mol Neurosci 7: 61 10.3389/fnmol.2014.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J-M, Chen Q, Zhou Y, Miao S, Zheng J, Zhang C, Xiong Z-Q. 2008. Brain-derived neurotrophic factor-tropomyosin-related kinase B signaling contributes to activity-dependent changes in synaptic proteins. J Biol Chem 283: 21242–21250. 10.1074/jbc.M800282200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaang BK, Kandel ER, Grant SG. 1993. Activation of cAMP-responsive genes by stimuli that produce long-term facilitation in Aplysia sensory neurons. Neuron 10: 427–435. 10.1016/0896-6273(93)90331-K [DOI] [PubMed] [Google Scholar]
- Kandel ER. 2001. The molecular biology of memory storage: a dialogue between genes and synapses. Science 294: 1030–1038. 10.1126/science.1067020 [DOI] [PubMed] [Google Scholar]
- Kanhema T, Dagestad G, Panja D, Tiron A, Messaoudi E, Håvik B, Ying SW, Nairn AC, Sonenberg N, Bramham CR. 2006. Dual regulation of translation initiation and peptide chain elongation during BDNF-induced LTP in vivo: evidence for compartment-specific translation control. J Neurochem 99: 1328–1337. 10.1111/j.1471-4159.2006.04158.x [DOI] [PubMed] [Google Scholar]
- Kim S, Kaang B-K. 2017. Epigenetic regulation and chromatin remodeling in learning and memory. Exp Mol Med 49: e281 10.1038/emm.2016.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Yu N-K, Shim K-W, Kim J, Kim H, Han DH, Choi JE, Lee S-W, Choi DI, Kim MW, et al. 2018. Remote memory and cortical synaptic plasticity require neuronal CCCTC-binding factor (CTCF). J Neurosci 38: 5042–5052. 10.1523/JNEUROSCI.2738-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko H-G, Choi J-H, Park DI, Kang SJ, Lim C-S, Sim S-E, Shim J, Kim J-I, Kim S, Choi T-H, et al. 2018. Rapid turnover of cortical NCAM1 regulates synaptic reorganization after peripheral nerve injury. Cell Rep 22: 748–759. 10.1016/j.celrep.2017.12.059 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441: 880–884. 10.1038/nature04723 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Wang QJ, Holstein GR, Friedrich VL, Iwata J, Kominami E, Chait BT, Tanaka K, Yue Z. 2007. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci 104: 14489–14494. 10.1073/pnas.0701311104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B, Ostareck D, Keidel E-M, Ostareck-Lederer A, Fischer U. 2001. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum Mol Genet 10: 329–338. 10.1093/hmg/10.4.329 [DOI] [PubMed] [Google Scholar]
- Lee SH, Choi JH, Lee N, Lee HR, Kim JI, Yu NK, Choi SL, Lee SH, Kim H, Kaang BK. 2008a. Synaptic protein degradation underlies destabilization of retrieved fear memory. Science 319: 1253–1256. 10.1126/science.1150541 [DOI] [PubMed] [Google Scholar]
- Lee Y-S, Bailey CH, Kandel ER, Kaang B-K. 2008b. Transcriptional regulation of long-term memory in the marine snail Aplysia. Mol Brain 1: 3 10.1186/1756-6606-1-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-S, Choi S-L, Lee S-H, Kim H, Park H, Lee N, Lee S-H, Chae Y-S, Jang D-J, Kandel ER, et al. 2009. Identification of a serotonin receptor coupled to adenylyl cyclase involved in learning-related heterosynaptic facilitation in Aplysia. Proc Natl Acad Sci 106: 14634–14639. 10.1073/pnas.0907502106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Sato Y, Nixon RA. 2011a. Lysosomal proteolysis inhibition selectively disrupts axonal transport of degradative organelles and causes an Alzheimer's-like axonal dystrophy. J Neurosci 31: 7817–7830. 10.1523/JNEUROSCI.6412-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Sato Y, Nixon RA. 2011b. Primary lysosomal dysfunction causes cargo-specific deficits of axonal transport leading to Alzheimer-like neuritic dystrophy. Autophagy 7: 1562–1563. 10.4161/auto.7.12.17956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-H, Kwak C, Shim J, Kim J-E, Choi S-L, Kim HF, Jang D-J, Lee J-A, Lee K, Lee C-H, et al. 2012. A cellular model of memory reconsolidation involves reactivation-induced destabilization and restabilization at the sensorimotor synapse in Aplysia. Proc Natl Acad Sci 109: 14200–14205. 10.1073/pnas.1211997109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson JM, Sweatt JD. 2005. Epigenetic mechanisms in memory formation. Nat Rev Neurosci 6: 108–118. 10.1038/nrn1604 [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang Y, Ku L, Wilkinson KD, Warren ST, Feng Y. 2001. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res 29: 2276–2283. 10.1093/nar/29.11.2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao L, Pilotte J, Xu T, Wong CC, Edelman GM, Vanderklish P, Yates JR. 2007. BDNF induces widespread changes in synaptic protein content and up-regulates components of the translation machinery: an analysis using high-throughput proteomics. J Proteome Res 6: 1059–1071. 10.1021/pr060358f [DOI] [PubMed] [Google Scholar]
- Liu R-Y, Fioravante D, Shah S, Byrne JH. 2008. cAMP response element-binding protein 1 feedback loop is necessary for consolidation of long-term synaptic facilitation in Aplysia. J Neurosci 28: 1970–1976. 10.1523/JNEUROSCI.3848-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R-Y, Cleary LJ, Byrne JH. 2011. The requirement for enhanced CREB1 expression in consolidation of long-term synaptic facilitation and long-term excitability in sensory neurons of Aplysia. J Neurosci 31: 6871–6879. 10.1523/JNEUROSCI.5071-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Salon M, Alonso M, Vianna MR, Viola H, Mello e Souza T, Izquierdo I, Pasquini JM, Medina JH. 2001. The ubiquitin-proteasome cascade is required for mammalian long-term memory formation. Eur J Neurosci 14: 1820–1826. 10.1046/j.0953-816x.2001.01806.x [DOI] [PubMed] [Google Scholar]
- Malleret G, Haditsch U, Genoux D, Jones MW, Bliss TVP, Vanhoose AM, Weitlauf C, Kandel ER, Winder DG, Mansuy IM. 2001. Inducible and reversible enhancement of learning, memory, and long-term potentiation by genetic inhibition of calcineurin. Cell 104: 675–686. 10.1016/S0092-8674(01)00264-1 [DOI] [PubMed] [Google Scholar]
- Manadas B, Santos AR, Szabadfi K, Gomes JR, Garbis SD, Fountoulakis M, Duarte CB. 2009. BDNF-induced changes in the expression of the translation machinery in hippocampal neurons: protein levels and dendritic mRNA. J Proteome Res 8: 4536–4552. 10.1021/pr900366x [DOI] [PubMed] [Google Scholar]
- Marques M, Laflamme L, Gervais AL, Gaudreau L. 2010. Reconciling the positive and negative roles of histone H2A.Z in gene transcription. Epigenetics 5: 267–272. 10.4161/epi.5.4.11520 [DOI] [PubMed] [Google Scholar]
- Matsuda N, Lu H, Fukata Y, Noritake J, Gao H, Mukherjee S, Nemoto T, Fukata M, Poo MM. 2009. Differential activity-dependent secretion of brain-derived neurotrophic factor from axon and dendrite. J Neurosci 29: 14185–14198. 10.1523/JNEUROSCI.1863-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita M, Tomizawa K, Moriwaki A, Li ST, Terada H, Matsui H. 2001. A high-efficiency protein transduction system demonstrating the role of PKA in long-lasting long-term potentiation. J Neurosci 21: 6000–6007. 10.1523/JNEUROSCI.21-16-06000.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuown SC, Barrett RM, Matheos DP, Post RJ, Rogge GA, Alenghat T, Mullican SE, Jones S, Rusche JR, Lazar MA, et al. 2011. HDAC3 is a critical negative regulator of long-term memory formation. J Neurosci 31: 764–774. 10.1523/JNEUROSCI.5052-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael D, Martin KC, Seger R, Ning MM, Baston R, Kandel ER. 1998. Repeated pulses of serotonin required for long-term facilitation activate mitogen-activated protein kinase in sensory neurons of Aplysia. Proc Natl Acad Sci 95: 1864–1869. 10.1073/pnas.95.4.1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. 2007. Covalent modification of DNA regulates memory formation. Neuron 53: 857–869. 10.1016/j.neuron.2007.02.022 [DOI] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, Rivera IM, Rubio MD, Rumbaugh G, Sweatt JD. 2010. Cortical DNA methylation maintains remote memory. Nat Neurosci 13: 664–666. 10.1038/nn.2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Poo M. 2013. Neurotrophin regulation of neural circuit development and function. Nat Rev Neurosci 14: 7–23. 10.1038/nrn3379 [DOI] [PubMed] [Google Scholar]
- Park H, Popescu A, Poo MM. 2014a. Essential role of presynaptic NMDA receptors in activity-dependent BDNF secretion and corticostriatal LTP. Neuron 84: 1009–1022. 10.1016/j.neuron.2014.10.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park P, Volianskis A, Sanderson TM, Bortolotto ZA, Jane DE, Zhuo M, Kaang B-K, Collingridge GL. 2014b. NMDA receptor-dependent long-term potentiation comprises a family of temporally overlapping forms of synaptic plasticity that are induced by different patterns of stimulation. Philos Trans R Soc Lond B Biol Sci 369: 20130131 10.1098/rstb.2013.0131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini S, Corona C, Liu J, Greene LA, Shelanski ML. 2015. Specific downregulation of hippocampal ATF4 reveals a necessary role in synaptic plasticity and memory. Cell Rep 11: 183–191. 10.1016/j.celrep.2015.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixoto L, Abel T. 2013. The role of histone acetylation in memory formation and cognitive impairments. Neuropsychopharmacology 38: 62–76. 10.1038/npp.2012.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. 2009. CTCF: master weaver of the genome. Cell 137: 1194–1211. 10.1016/j.cell.2009.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poo M. 2001. Neurotrophins as synaptic modulators. Nat Rev Neurosci 2: 24–32. 10.1038/35049004 [DOI] [PubMed] [Google Scholar]
- Reichardt LF. 2006. Neurotrophin-regulated signalling pathways. Philos Trans R Soc B Biol Sci 361: 1545–1564. 10.1098/rstb.2006.1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux PP, Topisirovic I. 2012. Regulation of mRNA translation by signaling pathways. Cold Spring Harb Perspect Biol 4: a012252 10.1101/cshperspect.a012252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Mariño G, Kroemer G. 2011. Autophagy and aging. Cell 146: 682–695. 10.1016/j.cell.2011.07.030 [DOI] [PubMed] [Google Scholar]
- Ruvinsky I, Meyuhas O. 2006. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem Sci 31: 342–348. 10.1016/j.tibs.2006.04.003 [DOI] [PubMed] [Google Scholar]
- Sams DS, Nardone S, Getselter D, Raz D, Tal M, Rayi PR, Kaphzan H, Hakim O, Elliott E. 2016. Neuronal CTCF is necessary for basal and experience-dependent gene regulation, memory formation, and genomic structure of BDNF and arc. Cell Rep 17: 2418–2430. 10.1016/j.celrep.2016.11.004 [DOI] [PubMed] [Google Scholar]
- Schwartzkroin PA, Wester K. 1975. Long-lasting facilitation of a synaptic potential following tetanization in the in vitro hippocampal slice. Brain Res 89: 107–119. 10.1016/0006-8993(75)90138-9 [DOI] [PubMed] [Google Scholar]
- Shehata M, Abdou K, Choko K, Matsuo M, Nishizono H, Inokuchi K. 2018. Autophagy enhances memory erasure through synaptic destabilization. J Neurosci 38: 3809–3822. 10.1523/JNEUROSCI.3505-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavoe AK, Holzbaur EL. 2018. Expression of WIPI2B counteracts age-related decline in autophagosome biogenesis in neurons. bioRxiv. 10.1101/325449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei N, Kawamura M, Ishizuka Y, Kakiya N, Inamura N, Namba H, Nawa H. 2009. Brain-derived neurotrophic factor enhances the basal rate of protein synthesis by increasing active eukaryotic elongation factor 2 levels and promoting translation elongation in cortical neurons. J Biol Chem 284: 26340–26348. 10.1074/jbc.M109.023010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammineni P, Ye X, Feng T, Aikal D, Cai Q. 2017. Impaired retrograde transport of axonal autophagosomes contributes to autophagic stress in Alzheimer's disease neurons. Elife 6: e21776 10.7554/eLife.21776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka J-I, Horiike Y, Matsuzaki M, Miyazaki T, Ellis-Davies GCR, Kasai H. 2008. Protein synthesis and neurotrophin-dependent structural plasticity of single dendritic spines. Science 319: 1683–1687. 10.1126/science.1152864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Dutch-Belgian Fragile X Consorthium, et al. 1994. Fmr1 knockout mice: a model to study fragile X mental retardation. The Dutch-Belgian Fragile X Consortium. Cell 78: 23–33. [PubMed] [Google Scholar]
- Tian Y, Yang C, Shang S, Cai Y, Deng X, Zhang J, Shao F, Zhu D, Liu Y, Chen G, et al. 2017. Loss of FMRP impaired hippocampal long-term plasticity and spatial learning in rats. Front Mol Neurosci 10: 269 10.3389/fnmol.2017.00269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderklish P, Saido TC, Gall C, Arai A, Lynch G. 1995. Proteolysis of spectrin by calpain accompanies θ-burst stimulation in cultured hippocampal slices. Brain Res Mol Brain Res 32: 25–35. 10.1016/0169-328X(95)00057-Y [DOI] [PubMed] [Google Scholar]
- Vogel-Ciernia A, Matheos DP, Barrett RM, Kramár EA, Azzawi S, Chen Y, Magnan CN, Zeller M, Sylvain A, Haettig J, et al. 2013. The neuron-specific chromatin regulatory subunit BAF53b is necessary for synaptic plasticity and memory. Nat Neurosci 16: 552–561. 10.1038/nn.3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhu G, Briz V, Hsu Y-T, Bi X, Baudry M. 2014. A molecular brake controls the magnitude of long-term potentiation. Nat Commun 5: 3051 10.1038/ncomms4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler IJ, Irwin SA, Klintsova AY, Spencer CM, Brazelton AD, Miyashiro K, Comery TA, Patel B, Eberwine J, Greenough WT. 1997. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc Natl Acad Sci 94: 5395–5400. 10.1073/pnas.94.10.5395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, Graef IA, Crabtree GR. 2007. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron 56: 94–108. 10.1016/j.neuron.2007.08.021 [DOI] [PubMed] [Google Scholar]
- Yamashita N, Kuruvilla R. 2016. Neurotrophin signaling endosomes: biogenesis, regulation, and functions. Curr Opin Neurobiol 39: 139–145. 10.1016/j.conb.2016.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N-K, Baek SH, Kaang B-K. 2011. DNA methylation-mediated control of learning and memory. Mol Brain 4: 5 10.1186/1756-6606-4-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Huang T, Zhang L, Zhou Y, Luo H, Xu H, Wang X. 2016. Dysregulation of ubiquitin-proteasome system in neurodegenerative diseases. Front Aging Neurosci 8: 303 10.3389/fnagi.2016.00303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Liu Y, Wang Y, Bi X, Baudry M. 2015. Different patterns of electrical activity lead to long-term potentiation by activating different intracellular pathways. J Neurosci 35: 621–633. 10.1523/JNEUROSCI.2193-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovkic IB, Guzman-Karlsson MC, Sweatt JD. 2013. Epigenetic regulation of memory formation and maintenance. Learn Mem 20: 61–74. 10.1101/lm.026575.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovkic IB, Paulukaitis BS, Day JJ, Etikala DM, Sweatt JD. 2014. Histone H2A.Z subunit exchange controls consolidation of recent and remote memory. Nature 515: 582–586. 10.1038/nature13707 [DOI] [PMC free article] [PubMed] [Google Scholar]