Abstract
Alkaline proteases have several industrial applications. In the present study, newly isolated Neocosmospora sp. N1 was screened as hyper producer of serine protease. A multimeric protease of the fungus was purified to homogeneity till 96.78 fold purification with 22.51% recovery. The homogeneity of purified enzyme was checked by native PAGE and its molecular weight was found to be 198.03 kDa by MALDI-TOF. On SDS-PAGE analysis, enzyme was found to be a hetero oligomer of 17.66 kDa and 20.89 kDa subunits. The purified enzyme showed maximum activity with casein as substrate at 60 °C and pH 8.5. The Km and Vmax values were found to be 0.015 mg/ml and 454.45 U/ml, respectively. The enzyme was completely inhibited by PMSF, while the activity was 40% enhanced using β-mercaptoethanol, suggesting that it is a thiol-dependent serine protease. The purified protease was active over an alkaline pH range from 7 to 12 and temperatures from 20 °C to 60 °C. The enzyme exhibited excellent stability, almost 100% towards organic solvents such as toluene, benzene and hexane, surfactants such as Triton X-100, Tween-20, Tween-80 and SDS, as well as commercial detergents. The significant properties of purified enzyme assure that it could be a potential candidate for commercial purposes.
Keywords: Microbiology, Biotechnology, Enzyme kinetics, Enzymology, Microbial biotechnology, Serine protease, MALDI-TOF, Superdex 200
1. Introduction
Proteases (peptidases or proteinases) are found in all living organisms playing a vital role in metabolic and physiological processes [1]. They catalyze the hydrolysis of peptide bonds in protein molecules [2]. Proteases not always fit clearly into the international system for the classification and nomenclature of enzymes (EC number) because of their diversified and complex nature [3]. However, they can be classified according to their source (animal, plant or microbial), catalytic action (endo or exopeptidases), pH, molecular size, charge or substrate specificity and nature of active site [4]. Ever since the advent of protein structure and homology modelling, other forms of classification such as MEROPS databases have been proposed which is based upon the chemical structure and information about evolutionary relationship of proteins [5].
The worldwide sale of industrial enzymes accounts to US $ 300–600 million per annum, 75% share is held by hydrolytic enzymes, of which two thirds are proteolytic enzymes [6]. Microorganisms largely contribute to the production of intracellular and extracellular enzymes utilized in biotechnological and industrial applications [7]. Proteases exploited commercially are mostly derived from microorganisms like bacteria, yeast and fungi [8]. Although, there are many microbial sources available for producing proteases, only few are recognized as commercial producers. In recent years, proteases from other sources like insects, plants, mushrooms etc. have also been reported [9, 10, 11]. Extracellular alkaline proteases contribute to 25% of total microbial enzyme sales [12] encompassing widespread applications in industrial sectors such as laundry [13], leather [14], silk [15], pharmaceutical [16], nutrition [17], silver recovery [18] etc.
The escalating demand of alkaline proteases in various industries entails the need of definite properties of enzyme like thermal stability profile, catalytic efficiency at higher pH, substrate specificity, kinetic studies, activity and stability in presence of organic solvents, surfactants etc. [19, 20, 21]. Bacillus derived alkaline proteases are stable at elevated temperatures and pH, but a majority are incompatible with detergent matrices [22, 23]. Therefore, high performance alkaline proteases are being sought for commercial exploitations, especially for detergents. Proteases from microbial origin have long been used in industry. Nowadays, filamentous fungi are preferred over other microbial sources because of their biochemical diversity, growth on cost effective substrates such as those used in SSF, bulk production of extracellular enzymes, ease in recovery of product from fungal biomass and suitability to genetic manipulation [24, 25]. Many fungal species secrete extracellular proteases to confer pathogenicity in plants. Proteases from plant pathogenic fungi have been reported earlier [26, 27, 28].
The present study aimed to screen a hyper producing fungus from nature for production of alkaline protease, possessing novel characteristics. Therefore, we isolated a plant pathogenic fungus Neocosmospora sp. N1 secreting a novel high molecular weight thiol-dependent serine protease from soil. The biochemical characterization of an enzyme is necessary to evaluate their biotechnological potential, hence attempts were made to purify and characterize the protease produced by the microrganism. To the best of authors’ knowledge, there is no such work reported on Neocosmospora genus yet.
2. Materials and methods
2.1. Chemicals
Alkali soluble casein (Hi-Media) was used for performing alkaline protease assay. All other chemicals used were from Hi-Media and Sigma-Aldrich. Agro-industrial waste materials were procured from local market of Indore, India.
2.2. Isolation and identification of microorganism
Samples were collected in sterile polythene bags from various regions including soil, rotten fruits, meat waste, effluent of slaughter house, dairy etc. from Indore, India. These samples were serially diluted and inoculated on skim milk agar medium, pH 9 at 30 °C for 7 days. The fungal colonies giving a zone of hydrolysis on skim milk agar plate were isolated on PDA and purified. On the basis of the magnitude of the zone of hydrolysis, the isolate N1 was selected for further studies. Czapek Dox medium consisting of (g/l) Sucrose, 10.0; NaNO3, 1.0; K2HPO4, 1.0; MgSO4·7H2O, 0.5; KCl, 0.5; FeSO4·7H2O, 0.01; casein, 1.0; pH 9 [29] was used for growth and production of protease enzyme from the selected isolate N1. Flask was incubated for 4 days at 30 °C and supernatant was collected by centrifugation at 100xg and was assayed for its proteolytic potential by well diffusion assay. 1% agarose was copolymerised with 0.03% substrate in a sterile petri dish and pH was adjusted to 9 with 1N NaOH. Wells were made with a sterile cup borer, and 40 μl of cell-free culture was dispensed aseptically into each well and incubated at 37 °C. After 18 h, plates were flooded with Coomassie Brilliant Blue dye and clear zones of protein hydrolysis were observed around the wells. Proteolytic zone diameter was measured.
The fungal strain N1 was identified on the basis of morphological and molecular characteristics. Preliminary identification of the isolate was done by determining the growth characteristics on PDA plate and microscopic observations. The structure of fungal isolate N1 was also studied using JEOL–JSM 5600 Scanning Electron Microscope (SEM).
Molecular identification of the selected fungal isolate N1 was done at National Fungal Culture Collection of India (NFCCI), Pune, India. The ITS region of rDNA was amplified using fungal universal primers ITS4 & ITS5 and PCR was set up with ABI-BigDye® Terminatorv3.1 Cycle Sequencing Kit. The sequence obtained was edited manually to avoid inconsistency and was compared with 16S rDNA sequences using NCBI-BLAST program. The sequence showed maximum similarity with Neocosmospora ramosa SF56 (MG682504) and was submitted to GenBank under accession number MK417797. Clustal W software was used to align closely related sequences and phylogenetic tree was constructed based on neighbour joining (NJ) method using MEGA X program. Branch support of the trees was assessed by bootstrap analysis with 1000 replications using the heuristic search option.
2.3. Inoculum preparation
Spores were harvested from 5 days old fungal culture raised on PDA slant using sterile distilled water; pH 8 adjusted with 1N NaOH solution. The inoculum was used after adjusting the desired spore count (1 × 107 spores/ml) using Neubauer's chamber.
2.4. Solid state fermentation
Wheat bran and Custard apple seed powder in the ratio of 4:1 (15 g) was taken in a 250 ml Erlenmeyer flask and moisture content was adjusted to 50% (v/w) by distilled water. This media was autoclaved at 121 °C, 15 lbs for 20 min. The sterile medium was inoculated by 1 ml of spore suspension and the flask was incubated at 30 °C for 96 h.
2.5. Enzyme extraction
The fermented medium was thoroughly mixed with 50 mM Tris-HCl buffer, pH 8.5 (1:20 dilution, w/v) and incubated at 30 °C on a shaker at 120 rpm for 1 h. The slurry was centrifuged at 5000xg for 15 min at 4 °C. The supernatant obtained was used as crude enzyme extract.
2.6. Enzyme assay and protein estimation
Alkaline protease activity was determined according to the modified method of Charles et al. [30] using alkali soluble casein as substrate. Reaction mixture contained 1 ml of 2% (w/v) casein prepared in Tris-HCl buffer, pH 8.5 and 0.1 of the enzyme and incubated for 30 min at 60 °C. The reaction was stopped by adding 2 ml of 0.4 M TCA to precipitate undigested protein. The tubes were centrifuged at 100xg for 5 min to remove precipitate. A 0.5 ml aliquot of supernatant was neutralized with 5.0 ml of 0.4 M Na2CO3 to which 1 ml of 0.5 N Folin Ciocalteau reagent was added. Absorbance was read at 660nm using UV–visible spectrophotometer. One unit of enzyme activity (U) was defined as the amount of enzyme required to liberate 1 μmol of tyrosine per minute under standard assay conditions. All experiments were performed in triplicates and standard error was calculated.
Protein concentration was determined using Bovine Serum Albumin as standard by the method of Lowry et al. [31]. Protein concentration in various steps of purification was estimated by absorbance at 280 nm.
2.7. Purification of enzyme
2.7.1. Ammonium sulphate precipitation
Ammonium sulfate till 90% saturation was added to supernatant and placed for overnight incubation at 4 °C. The precipitate was collected by centrifugation at 7000xg at 4 °C, for 15 min. Enzyme was recovered by re-suspending protein pellet in 25 mM Tris–HCl buffer, pH 8.5. The suspension was desalted by passing it through Sephadex G-25 column pre-equilibrated with the same buffer.
2.7.2. Anion exchange chromatography
Desalted sample was applied to DEAE-cellulose column (bed volume, 150 ml) previously equilibrated with 25 mM Tris-HCl buffer, pH 8.5. Unbound fractions were collected at a flow-rate of 1.0 ml/min by eluting with the same buffer. Bound proteins were eluted with a linear 0–1 M NaCl gradient, prepared in equilibration buffer. The eluted fractions were monitored at 280 nm for estimation of protein and assayed for enzyme activity. The fractions containing maximum protease activity were pooled and concentrated by reverse dialysis against solid sucrose. Dialysis also removed NaCl from the protein by selective diffusion.
2.7.3. Gel filtration chromatography
The concentrated fraction was further purified by Sephadex G-200 gel filtration chromatography. The column (bed volume, 100 ml) was previously equilibrated with 25 mM Tris–HCl buffer, pH 8.5. Elution was performed with the same buffer at a flow rate of 8 ml/h. Active fractions showing proteolytic activity were concentrated by reverse dialysis and then injected on Superdex 200 increase 10/300 GL high performance column (AKTA purification system). Elution was performed using 20 mM Tris-HCl buffer, pH 8.5 containing 1M NaCl. The protein content was determined by measuring absorbance at 280nm. The enzyme solution was concentrated against solid sucrose for further analysis.
2.8. Molecular weight determination & casein zymography
The purity, homogeneity and molecular mass of enzyme preparation were confirmed by native PAGE according to the method of Laemmli [32] using 5.0% stacking and12.0% resolving gel. Relative molecular mass of the protease was calculated using standard protein markers (GENEi). Protein bands were visualized on the gel by Coomassie Blue staining.
Zymographic analysis was performed using modified method of Heussen and Dowdle [33]. 0.5% casein was copolymerized with 12% polyacrylamide gel. Sample was mixed with Laemmli buffer and electrophoresis was carried out at 4 °C, 100V. After electrophoresis, gel was rinsed with distilled water, immersed in 100 mM Tris–HCl buffer, pH 8.5 and incubated at 50 °C for 1 h. The gel was stained with 0.25% CBB R-250 followed by destaining with methanol: acetic acid: water (10:10:80) in order to remove excess stain. Proteolytic activity was observed as a clear lysis band of degraded protein on a uniform blue background.
2.9. MALDI-TOF
The purified protein band was excised from gel and protein was eluted in Tris-HCl buffer. Molecular mass of the purified protease was analyzed in the linear mode by Matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF/MS) using a Voyager DE-RP instrument (Applied Biosystems/PerSeptive Biosystems, Inc., Framingham, MA, USA). Data was collected with a Tektronix TDS 520 numeric oscillograph and analyzed using the GRAMS/386 software (Galactic Industries Corporation, Salem, NH, USA).
2.10. SDS-PAGE
SDS-PAGE was carried out to determine subunit composition of purified enzyme according to the method of Laemmli [32] using 4% stacking gel and 10% resolving polyacrylamide gel. Purified protease sample was mixed with Laemmli sample buffer, heated at 100 °C for 3 min and immediately cooled in ice-bath. The molecular weight of the subunits was estimated using a low molecular weight calibration kit (Bio-Rad) containing prepared SDS-PAGE standards.
2.11. Protein identification by tandem mass spectrometry
The two protein bands obtained on SDS-PAGE gel corresponding to the two subunits of protease enzyme were excised separately, destained and subjected to in-gel trypsin digestion according to the method described by Riffel et al. [34]. The generated peptides were subjected to mass fingerprinting using matrix-assisted laser desorption ionization-time of flight–MS (MALDI-TOF/MS), sequences were obtained using quadrupole/time of flight (Q-TOF) tandem mass spectrometry (ESI-MS/MS) and analyzed using the Mascot Search Software. Multiple sequence alignment of the peptide sequences was carried out by Clustal Omega using EMBL databases.
2.12. Biochemical characterization of enzyme
2.12.1. Effect of temperature and thermal stability
The effect of temperature on enzyme activity was examined from 10–90 °C with interval of 5 °C at pH 8.5 using casein as a substrate for 30 min. Thermal stability of protease was determined after incubation of enzyme alone at 40 °C, 50 °C, 60 °C and 70 °C for 4 h. Aliquots were withdrawn at 15 min interval to test relative activity under standard conditions.
2.12.2. Effect of pH and pH stability
The enzyme activity was measured at different pH using three buffer systems between pH 6 and 12 with the interval of 0.5. The buffer system was prepared as follows: 0.1 M of sodium phosphate buffer (pH range, 6–7.5), 0.1 M of Tris-HCl buffer (pH range, 8–9.5) and 0.1 M Glycine-NaOH buffer (pH range, 10–12). The pH stability of the alkaline protease was determined by incubating enzyme preparation in buffers of different pH in the range of 5.0–12.0 (with interval of 1.0 pH unit) for 24 h, at 37 °C. Aliquots were withdrawn and proteolytic activity was determined at 60 °C. Enzyme activity at the beginning of experiment was taken as control (100%).
2.12.3. Determination of Km and Vmax
Km and Vmax values of the purified enzyme were determined by measuring the activity with casein concentrations ranging from 1 to 20 mg/ml. Kinetic constants were calculated from Lineweaver-Burk plot.
2.12.4. Effect of inhibitors
The effect of various inhibitors on protease activity was studied by pre-incubating enzyme with different inhibitors like EDTA, β-mercapto ethanol, Phenyl methyl sulphonyl flouride (PMSF), Urea, para chloro mercuribenzoic acid (pCMB), HgCl2 at 2 and 5 mM effective concentration for 1 h at 37 °C. The residual activity was measured at 60 °C, pH 8.5. Enzyme activity without any inhibitor was considered as control (100%).
2.12.5. Effect of metal ions
The effect of metal ions on protease activity was examined using different ions such as Cu2+, Fe2+, Mn2+, Ca2+, Na+, Mg2+, Zn2+, K+, Hg2+ and Ni2+ at 2 and 5 mM concentration. Protease activity was evaluated after pre-incubating the enzyme with respective ions at room temperature for 1 h under standard assay conditions. Activity of enzyme in the absence of any metal ion was considered as 100%.
2.12.6. Substrate specificity
Substrate specificity of enzyme was studied using different protein substrates, such as casein, gelatin, soybean, BSA, haemoglobin, egg albumin and skimmed milk. Each substrate (2%) was dissolved in 50 mM Tris-HCl buffer, pH 8.5 and enzyme assay was performed under standard conditions.
2.12.7. Effect of organic solvents
Organic solvents including chloroform, acetone, hexane, isoamyl alcohol, toluene, benzene, isopropanol and DMSO at 30% concentration were used to evaluate the effect on protease activity. In practice, these solvents were incorporated into the reaction mixture at above mentioned concentration in screw capped tubes. Subsequently, enzyme activity was measured as described before. Activity of enzyme in the absence of any solvent was considered as control (100%).
2.12.8. Effect of detergents and surfactants
The effect of some surfactants (Triton X-100, Tween-20, Tween-80 and SDS) and commercial detergents on enzyme stability was studied by pre-incubating enzyme for 1 h at 40 °C in presence of 1% and 5% final concentration of each agent. The relative activity was measured at pH 8.5 and 60 °C. Activity of the enzyme without any agent was considered as 100%.
3. Results and discussion
3.1. Isolation and identification of the microorganism
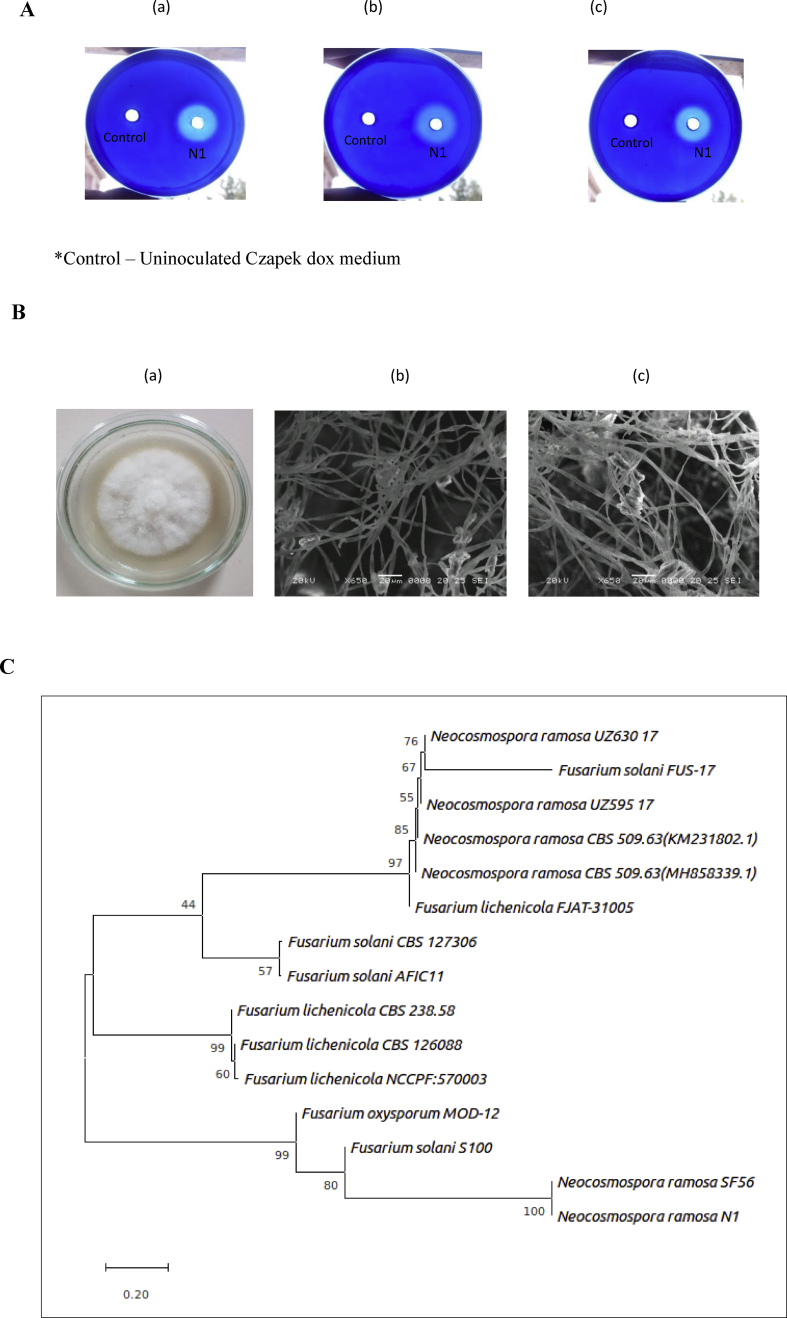
A hyper producer of alkaline protease was identified on skim milk agar plate giving a zone of protein hydrolysis (6 mm). The isolate was subsequently grown on Czapek Dox medium for production of protease for four days. The culture was centrifuged and the supernatant was used for well diffusion assay on casein, gelatin and soybean agarose plates exhibiting a maximum zone diameter of 26, 27 and 24 mm, respectively (Fig. 1A).
Fig. 1.
(A) Isolate N1 grown in liquid medium and supernatant obtained was loaded on agarose plates containing casein (a), gelatin (b) and soybean (c), respectively. Plates were stained with 0.3% CBB R-250 to observe clear zone of protein hydrolysis and zone diameter was measured subsequently. (B) Morphological characteristics of isolate N1. (a) Growth of isolate N1 on Potato Dextrose Agar. (b) Scanning electron microscopic image of isolate showing fungal hyphae and (c) fascicle arrangement of macroconidia. (C) Phylogenetic tree constructed by the neighbor-joining method (MEGA X software) showing the position of Neocosmospora sp. N1 with the sum of branch length = 3.72569419. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches.
The morphological characteristics of the isolate N1 were studied by growing the fungus on PDA (Fig. 1B). The colony appeared pale white to buff in colour; expanding flat, thin and punctate by the production of ascomata. Scanning electron microscopy (SEM) revealed further details about the structure of isolate (Fig. 1B). Ascomata is perithecial and smooth-walled; globose to pyriform, with hyaline inner layers and have periphysoids. The macroconidial cells seem to be thick walled, hyaline, elongate to cylindrical often aggregating in fascicles, typically divided into 2-4 septa. Chlamydospores were present. The 18S rDNA sequence of the fungal strain was aligned with similar sequences using Clustal W. Phylogenetic tree was constructed using 14 similar sequences. The branch lengths were in the same units as those of the evolutionary distances computed using the Maximum Composite Likelihood method and were present in the units of the number of base substitutions per site (Fig. 1C). The phylogenetic analysis confirmed a close resemblence of the strain with Neocosmospora ramosa SF56 (bootstrap- 100%) and also indicated ancestral relation with the Fusarium solani S100 (bootstrap- 80%).
On the basis of these results, the isolate was named as Neocosmospora sp. N1 and alkaline protease production was optimized (data not shown) subsequently. To the best of authors’ knowledge, the organism has not yet been exploited for this kind of study.
3.2. Purification of alkaline protease
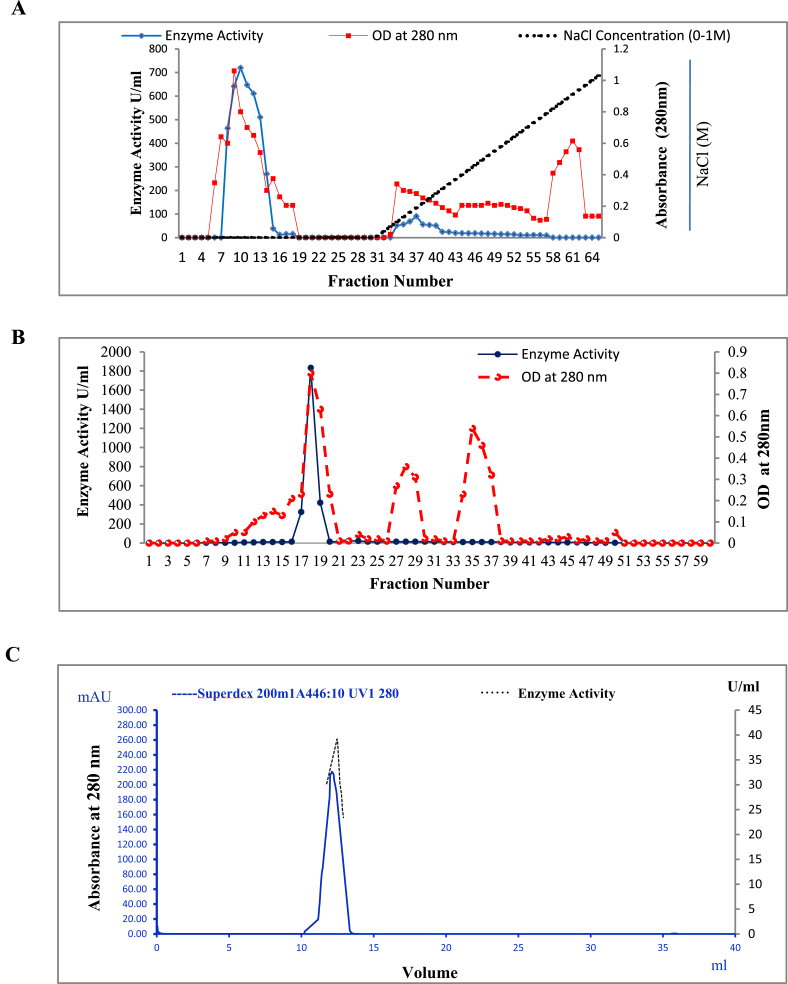
The purification steps are summarized in Table 1. A 2.94 fold purification of enzyme was achieved with a recovery of 88.56% after ammonium sulphate precipitation. The enzyme was further purified by DEAE cellulose anion exchange chromatography. Two peaks of proteolytic activity were obtained, as shown in Fig. 2A, wherein maximum enzyme was eluted in the unbound fractions in absence of NaCl indicating that the enzyme did not bind to DEAE cellulose. As accounted by its higher molecular weight, there could be some amount of glycosylation or may be a higher pI of the enzyme, which could result in impaired binding to the column [35]. Sephadex G-200 gel filtration chromatography of the unbound fraction exhibited one major peak of protease enzyme as seen in the elution profile (Fig. 2B). The purification fold increased to 96.78 with 22.51% recovery of the enzyme. The purity of enzyme was further analysed by AKTA purifier (Fig. 2C). The purified enzyme preparation exhibited a unique peak with an elution volume 12.8 ml, corresponding to a protein of nearly 205 kDa on Superdex 200 increase 30/100 GL AKTA purification system.
Table 1.
Purification steps of protease from Neocosmospora sp. N1.
| Purification steps | Total activity (U) | Total protein (mg) | Specific activity (U/mg) | Fold purification | Yield (%) |
|---|---|---|---|---|---|
| Crude | 34391.97 | 1712.04 | 20.08 | – | 100 |
| Ammonium Sulphate (0–90%) | 30459.17 | 515.45 | 59.09 | 2.94 | 88.56 |
| DEAE Cellulose | 19302 | 18.6 | 1037.74 | 51.68 | 56.12 |
| Sephadex G-200 | 7743 | 3.984 | 1943.52 | 96.78 | 22.51 |
Fig. 2.
Purification of alkaline protease from Neocosmospora sp. N1 (A) Elution profile of enzyme on DEAE cellulose anion exchange column, equilibrated with 25 mM Tris-HCl buffer, pH 8.5. Bound fractions were eluted with the same buffer in a linear salt gradient of NaCl (0–1M) at a flow-rate of 1.0 ml/min. All fractions were assayed for protease activity (⋄-blue fill) and monitored for protein content (□-red fill). (B) Chromatographic profile of purified protease on Sephadex G-200 gel filtration column. Fractions showing protease activity were pooled, concentrated and applied to Sephadex G-200 column, equilibrated with 25 mM Tris-HCl buffer. Elution was performed with the same buffer at a flow rate of 8 ml/h and protease activity (○) and protein content (○) was determined subsequently. (C) Purification profile of enzyme on Superdex 200 increase 30/100 GL column. Active enzyme was injected in AKTA purifier (10 × 300mm) equilibrated with 20 mM Tris-HCl buffer containing 1M NaCl. Protein was eluted with the same buffer at a flow rate of 0.75 ml/min and detected using a UV spectrometric detector at 280 nm. The peak obtained at elution volume of 12.8 ml contains protease activity.
The enzyme purified in this study was recovered up to 22.5% with a specific activity of 1943.52 U/mg. In an earlier study, Salihi et al. [25] obtained 1.32 fold purification with 18.13 % recovery and 15.86 U/mg specific activity of an alkaline protease produced from Aspergillus oryzae CH93 while Ueda et al. [28] reported 47.9 fold purification of a protease purified by Fusarium with 7% recovery and specific activity of 665 U/mg. Riffel et al. [34] reported 14.26 fold purification with only 1.18% recovery of a protease produced by Chryseobacterium having specific activity of 2406.68 U/mg.
3.3. Molecular weight determination and zymogram
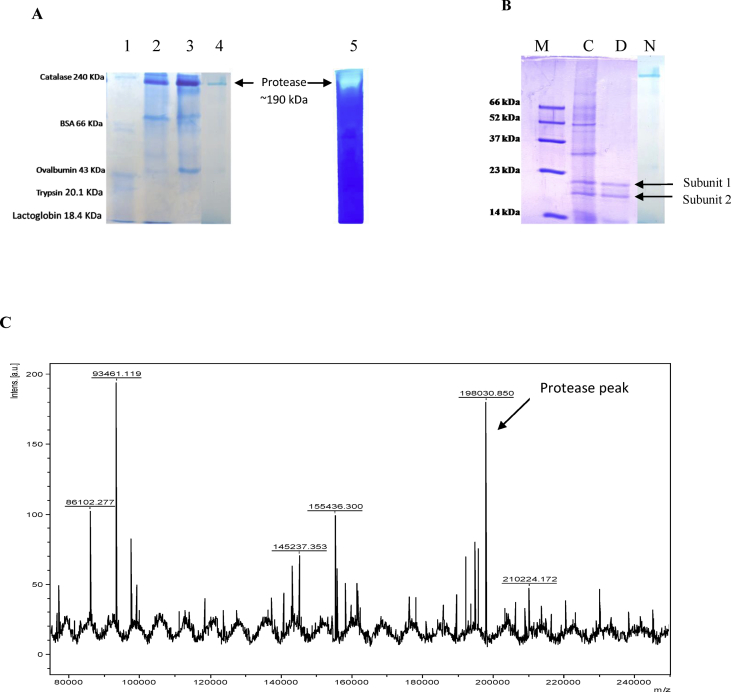
Molecular weight and homogeneity of purified protease was checked on native PAGE. The enzyme migrated as a single band of approximate molecular weight 190 kDa, suggesting that the enzyme was homogeneously pure (Fig. 3A). Zymogram analysis revealed a clear hydrolytic zone at the same level (Fig. 3A). The purified enzyme also migrated as a single band on SDS-PAGE under non-denaturing conditions (Fig. 3B). The exact molecular mass of the purified protein, as confirmed by MALDI-TOF mass spectrometry was found to be 198030.85Da (Fig. 3C). To determine subunit composition, purified enzyme was denatured and subjected to SDS-PAGE which revealed that the protease is composed of two major subunits of molecular weight 20.89 kDa (subunit 1) and 17.66 kDa (subunit 2), respectively (Fig. 3B). This suggested that the enzyme is a hetero-oligomer having multiple subunits.
Fig. 3.
Electrophoretic profile of purified protease and molecular weight determination. (A) Native-PAGE analysis. Lane 1: standard protein marker; lane 2: enzyme after ammonium sulphate fractionation (0–90%); lane 3: protease fraction after ion exchange chromatography; lane 4: purified enzyme obtained after Sephadex G-200 chromatography; lane 5: zymogram of purified enzyme showing caseinolytic activity. (B) 12% SDS PAGE. M: standard marker; C: crude enzyme; D: purified enzyme run under denaturing conditions; N: purified enzyme run under non-denaturing conditions. (C) MALDI-TOF spectrum of purified enzyme. The mass spectrum shows a series of protonated molecular ions. The molecular mass of the enzyme was found to be 198030.850 Da.
The molecular mass of microbial alkaline proteases usually ranges from 15 to 40 kDa [4]. Thiol-dependent serine proteases have been isolated previously from Paecilomyces lilacinus, 33 kDa [36], Bacillus mojavensis, 30 kDa [37],Streptomyces thermovulgaris, 32 kDa [38]. To the best of our knowledge, no previous work is reported on a high molecular weight thiol-dependent protease from Neocosmospora species. However, few higher molecular weight proteases were reported from Bacillus halotolerans, 250 kDa serine protease [39], Aspergillus fumigatus, 124 kDa serine protease [40] and Bacillus laterosporus, 86.29 kDa metal dependent protease [41], which are not thiol-dependent.
3.4. Characterization of purified protease by mass spectrometry
Eight peptide sequences of subunit 1 and six peptide sequences of subunit 2 obtained by mass spectrometry matched with subtilisin-like serine protease pepD from Aspergillus niger CBS 513.88 with overall sequence coverage of 49.5% (Table 2). These peptide sequences were aligned in UNIPROT with protein sequences obtained from BLAST. Proteases from Aspergillus niger strain CBS 513.88 (A2QTZ2), Aspergillus niger ATCC 13496 (A0A370BNF1), Aspergillus neoniger CBS 115656 (A0A318ZA13), Aspergillus vadensis CBS 113365 (A0A319BAX9) and Aspergillus eucalypticola CBS 122712 belong to subtilisin like serine peptidase family having peptidase S8 domain in their protein sequence. Most of the peptide sequences from Neocosmospora sp. were aligned with Peptidase S8 domain in the given sequences (Fig. 4). This confirms that the protease under study can also be considered as a member of subtilisin family.
Table 2.
Peptide sequences of both subunits obtained by ESI/MS and analyzed by Mascot Software. The peptide sequences matched with subtilisin-like serine protease pepD from Aspergillus niger strain CBS 513.88 (A2QTZ2) with 100% similarity.
| Purified protease | Peptide sequence | Sequence coverage with A2QTZ2 (%) |
|---|---|---|
| Subunit 1 | TIGGKTYGVSKNAHLLSVK | 30.04 |
| HDHVAYVEQDQVWYLDTLVTERR | ||
| ASLAYNAAGGEHVDDVGHGTHVAGTIGGK | ||
| MAFLKR | ||
| VENAFDEGVLSCVAAGNENR | ||
| TERRAPWGLGSISHR | ||
| ATEQVPHPTIQTIPGKYIVTFK | ||
| ENAFDEGVLSCVAAGNENRDAAR | ||
| Subunit 2 | VFVGESSSTSVILDGFNWAANDIVSK | 30.7 |
| HDHVAYVEQDQVWYLDTLVTERR | ||
| ASLAYNAAGGEHVDDVGHGTHVAGTIGGK | ||
| TYRIANFAGYAGSFDEK | ||
| MAFLKR | ||
| LATRNAVTNVAGSPNLLAYNGNSGVSK |
Fig. 4.
Multiple sequence alignment of the peptide sequences obtained from protease derived from Neocosmospora sp. N1 with those of subtilisin-like serine protease pepD from Aspergillus niger strain CBS 513.88 (A2QTZ2), subtilisin-like serine protease from Aspergillus niger ATCC 13496 (A0A370BNF1), protease from Aspergillus neoniger CBS 115656 (A0A318ZA13), protease from Aspergillus vadensis CBS 113365 (A0A319BAX9), protease from Aspergillus eucalypticola CBS 122712 (A0A317VKS4). The asterisk (*) sign indicates fully conserved amino acid residue; colon (:) indicates conservation between groups of strongly similar properties and period (.) represents conservation between groups of weakly similar properties. Peptidase S8 domain in the protein sequence is marked by grey color.
3.5. Biochemical characterization of enzyme
3.5.1. Effect of temperature and thermal stability
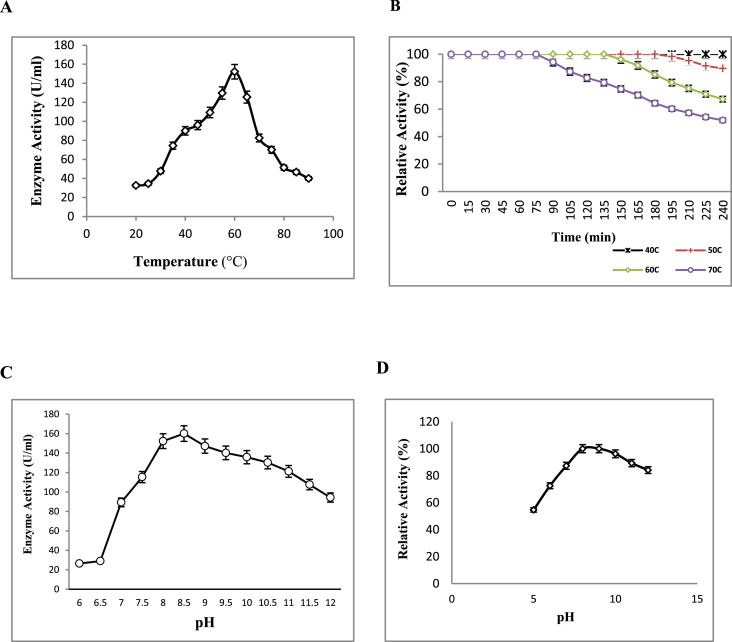
Optimum temperature of protease enzyme was investigated and enzyme was found to be active over a wide temperature range from 35 °C to 75 °C. A linear increase in protease activity was observed with increase in temperature, optimum being at 60 °C (Fig. 5A). About 70% enzyme activity was retained at temperature 50 °C and 65 °C. There was a sharp decrease in activity at temperature above 70 °C while the enzyme still retained 30% activity at 85 °C. Commercially exploited alkaline proteases like Alcalase, Savinase and Maxatase have temperature optima between 50–60 °C [37] which is comparable with the protease in study. Looking to the wide thermal stability, this enzyme preparation can find utility in detergent industries, where normal to mild washing conditions are required. Alkaline protease from Fusarium sp. displayed maximum activity at 50 °C [28, 42]. Deng et al. [43] reported 60 °C optimum temperature of a protease derived from Bacillus. Similar results were reported by Mechri et al. [44] on a protease from Aeribacillus pallidus having optimum temperature 60 °C. On the contrary, a protease from Aspergillus flavus was completely inactivated at 65 °C [45].
Fig. 5.
(A) Effect of temperature on protease activity. The reaction mixture was incubated at respective temperatures and assayed by standard assay method. (B) Effect of temperature on protease stability. Enzyme was pre-incubated at respective temperatures viz.40 °C, 50 °C, 60 °C and 70 °C for 4 h and samples were assayed after 15 min interval. Relative activity was calculated against non-heated enzyme assayed at 60 °C (100%). (C) Effect of pH on protease activity. The enzyme activity was measured at a pH range of 6–12 with the interval of 0.5 using three buffer systems. (D) Effect of pH on enzyme stability. The enzyme was pre-incubated with buffers of different pH (5–12) for 24 h at 37 °C and relative activity was determined under standard assay conditions.
Thermal stability profile revealed that the enzyme retained total activity at 40 °C while there was 11% reduction in activity at 50 °C after 4 h of incubation (Fig. 5B). Half of the initial activity was lost when the enzyme was incubated at 70 °C for 210 min. Protease from Beauveria sp. was stable at 30 °C when incubated for 60 min with optimum temperature for enzyme activity at 50 °C [46]. Ueda et al. [28] reported a protease from Fusarium sp. which was stable below 50 °C, its optimum temperature for enzyme activity being 50 °C. Thermo stable enzymes derived from mesophilic microorganisms find utility in various commercial applications because of their low cost preparations. The appreciable thermo stability of protease can be ascribed to the presence of the carbohydrate moiety [12, 47].
3.5.2. Effect of pH on activity and stability
The effect of pH on protease activity was studied at a pH range of 6.0–12.0 using casein as substrate at 60 °C (Fig. 5C). The enzyme was active over a wide range of pH from pH 7.0–12.0, exhibiting maximum activity at pH 8.5. Relative activity at pH 10 and 11 were 84 and 75%, respectively. There was around 80% fall in activity at pH below 7 suggesting its alkaline nature. These results were in agreement with earlier reports of alkaline protease from fungi Botrytis cinerea [48], Fusarium culmorum [42] and Aspergillus oryzae [25]. pH optima of purified protease was comparable to commercial detergent enzymes, AlcalaseTM and SavinaseTM having pH optima in the range of 7–10 and 8–11, respectively [37].
The pH stability of the enzyme was assayed by measuring residual activity after overnight incubation at various pH (Fig. 5D). Enzyme retained more than 80% activity when incubated at pH 7 to 12. Others have reported optimum pH of 11, 10 and 10.6 and stability in the range of 8–12 for bacterial proteases [49, 50, 51]. The pH stability displayed by the purified protease exceeded the ones reported for commercialized detergent proteases, subtilisin BPN’ and SavinaseTM, which generally have a pH optimum of 10.5 and pH stability at 8–10 [52].
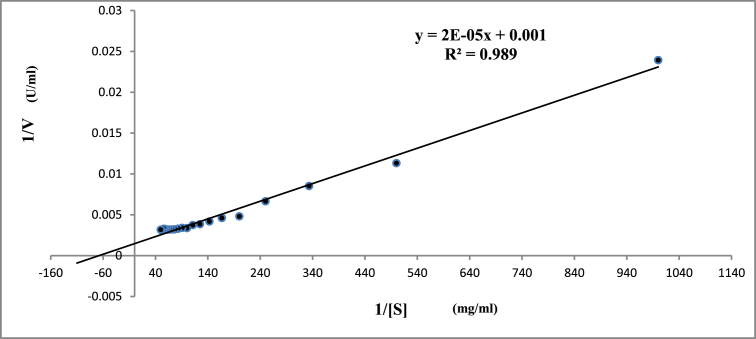
3.5.3. Kinetic studies
An increasing concentration of casein was used to determine Km and Vmax values for the purified protease. The values of Km and Vmax were found to be 0.015 mg/ml and 454.45 U/ml, respectively (Fig. 6). Relatively low value of Km indicates a higher affinity of the enzyme for the substrate while a higher Vmax/Km ratio demonstrates a powerful catalytic activity [53]. Km value for the purified protease was lower than that reported by Yadav et al. [45] who showed Km 1.95 mg/ml for a protease from Aspergillus flavus and Tremacoldi et al. [54] who reported Km 2.9 mg/ml for a protease of Aspergillus clavatus. High Km values of 0.96 mg/ml and 2 mg/ml of protease from Bacillus species were also reported by Sari et al. [55] and Gupta et al. [56], respectively.
Fig. 6.
Lineweaver-Burk plot for alkaline protease under varying substrate (casein) concentration (1–20 mg/ml).
3.5.4. Effect of protease inhibitors on protease activity
Proteases can be classified on the basis of their sensitivity towards inhibitors [57]. The enzyme activity was checked in presence of various inhibitors at 2 mM and 5 mM concentration and relative activity was calculated subsequently (Table 3). Phenyl methyl sulfonyl fluoride (PMSF) completely inhibited enzyme activity perhaps by sulfonating serine residues present at the active site suggesting that the enzyme belongs to serine family [2]. Other inhibitors like EDTA, pCMB, urea and HgCl2 used in the study had little or no effect on enzyme activity while 2- mercaptoethanol increased the enzyme activity by 40% suggesting the presence of SH groups on active site. Thus, the enzyme was classified as thiol-dependent serine protease. Beg and Gupta [37] reported a thiol-dependent serine protease from Bacillus mojavensis while Moradian et al. [58] also reported similar enzyme from Bacillus sp. However, no such report exists on thiol-dependent serine protease from Neocosmospora species.
Table 3.
Effect of various inhibitors and metal ions on protease activity. The non-treated enzyme was considered as control (100%).
| Inhibitor/Metal ion | Relative activity (%) |
|
|---|---|---|
| 2mM | 5mM | |
| Control | 100 ± 2.16 | 100 ± 2.16 |
| PMSF | 5 ± 0.11 | 0 ± 0.0 |
| EDTA | 111 ± 2.16 | 102 ± 1.90 |
| HgCl2 | 70 ± 1.23 | 64 ± 1.24 |
| Urea | 125 ± 1.27 | 120 ± 2.43 |
| pCMB | 100 ± 1.98 | 95 ± 1.86 |
| DTNB | 80 ± 1.42 | 74 ± 1.72 |
| 2-ME | 126 ± 2.54 | 143 ± 2.18 |
| FeSO4 | 84 ± 1.86 | 98 ± 2.67 |
| CaCl2 | 102 ± 2.43 | 105 ± 2.55 |
| KCl | 99 ± 2.54 | 101 ± 2.34 |
| NaCl | 68 ± 1.64 | 70 ± 1.62 |
| MgSO4 | 92 ± 2.0 | 94 ± 1.78 |
| MnCl2 | 91 ± 2.03 | 86 ± 1.71 |
| CuSO4 | 110 ± 2.64 | 96 ± 2.08 |
| HgCl2 | 73 ± 1.54 | 68 ± 1.45 |
| ZnCl2 | 94 ± 1.78 | 83 ± 1.88 |
| NiCl2 | 93 ± 2.04 | 84 ± 1.78 |
3.5.5. Effect of metal ions
The effect of some metal ions at concentration of 2 and 5 mM on protease activity was investigated at pH 8.5 and 60 °C by the addition of the respective cations to the reaction mixture. Table 3 indicates that the enzyme did not require any specific ion for catalytic activity. The result further correlates with the fact that the enzyme is a metal independent protease belonging to serine enzyme family. No significant effect on enzyme activity was found in the presence of Ca2+, Mn2+ and K+ at both the concentrations. Cu2+ at 2 mM concentration slightly enhanced enzyme activity while other metal ions like Hg2+, Ni2+, Mg2+ reduced enzyme activity at 5 mM concentration. Beg and Gupta [37] also reported a marginal increase in activity of a thiol-dependent serine protease in presence of Cu2+.
Inhibition of proteases by heavy metals is well established in literature, for instance, Hg2+ ions interact with protein thiol groups (converting them to mercaptides) as well as histidine and tryptophan residues present at active site, thus, impairing enzyme activity [59]. A serine protease from Bacillus safensis was activated by Ca2+, Co2+, and Mg2+ and inhibited by Ni2+ and Hg2+ [60]. Alkaline protease from Aeribacillus pallidus was activated by Ca2+, Cu2+ and Fe2+ [44] while a trypsin-like protease from Fusarium oxysporum was slightly activated by Ca2+ and Mg2+ [61]. Similar results were reported by Hadjidj et al. [50], Jaouadi et al. [51]. However, an alkaline protease from Botrytis cinerea did not require any metal ion for its activity [48]. The requirement of metal ions for activation and inhibition of enzyme activity varies widely with the type of enzyme. Metals ions play an important role in enhancing thermal stability and maintaining active conformation of the enzyme apart from influencing enzyme activity by binding at catalytic site [44].
3.5.6. Substrate specificity
Most of the alkaline proteases have broad substrate specificity and are active against a number of synthetic substrates and natural proteins [7]. The purified protease was assayed using different substrates as reported in Table 4. Casein (100% relative activity) was the most preferred substrate followed by soybean (90.03%), gelatin (86.95%), BSA (73.9%) and haemoglobin (23.4%). This data supports the fact that the enzyme can be considered as a promising additive in detergent industry. Similarly, Yadav et al. [45] also reported maximum activity of a seine protease from Aspergillus flavus using casein as substrate. The present results are also in accordance with Mechri et al. [44], Touioui et al. [62] and Anandan et al. [63].
Table 4.
Substrate specificity of protease against different substrates. Enzyme activity in presence of casein was taken as control (100%).
| Substrate | Relative activity (%) |
|---|---|
| Casein | 100 ± 2.87 |
| Gelatin | 86.95 ± 2.32 |
| Soybean | 90.03 ± 2.15 |
| BSA | 73.9 ± 1.65 |
| Haemoglobin | 23.4 ± 0.32 |
| Albumin | 20.54 ± 0.31 |
| Skimmed milk | 17.7 ± 0.27 |
3.5.7. Effect of organic solvents
The organic solvent stability of the enzyme depends on its nature. This property of enzyme is well utilized in pharmaceutical industry for the biosynthesis of small peptides. The effect of various organic solvents on protease activity was investigated at 30% concentration (Table 5). Enzyme retained 100% activity in toluene, benzene and hexane while there was a slight decrease in activity in presence of chloroform and DMSO. However, there was 50% decrease in enzyme activity when checked with acetone and isoamylalcohol. The possible reason could be hydrophobic interference of the organic solvent on the enzyme's catalytic site. A protease from Aspergillus flavus also retained its activity in benzene and chloroform [43] while Mothe and Sultanpuram [64] reported reduced activity of a protease from Bacillus caseinilyticus in presence of chloroform, benzene and toluene. Reddy et al. [65] reported enhanced enzyme activity in presence of hexane, toluene and benzene at 25% effective concentration.
Table 5.
Effect of organic solvents on the activity of purified protease. The non-treated enzyme was considered as 100%.
| Organic solvent (30%, v/v) | Relative activity (%) |
|---|---|
| Control | 100 ± 2.32 |
| Chloroform | 93 ± 1.94 |
| Acetone | 50 ± 1.42 |
| Isoamylalcohol | 57 ± 0.91 |
| Toluene | 100 ± 2.15 |
| Hexane | 107 ± 2.0 |
| Benzene | 109 ± 2.47 |
| DMSO | 90 ± 2.13 |
3.5.8. Effect of surfactants and detergents
The effect of various laboratory and commercial detergents on protease activity was examined and reported in Table 6. Enzyme presented a small loss in activity in presence of Triton X-100 and Tween-80, while Tween-20 triggered activity upto 20%. A similar finding was reported by Zanphorlin et al. [35] on a serine protease obtained from Myceliophthora species. Hadder et al. [18] and Tremacoldi et al. [54] reported SDS to be a strong inhibitor of protease enzyme. However, there was only 30% loss of purified protease enzyme activity in presence of SDS. Also, the remarkable stability exhibited by the enzyme towards various commercial detergents demonstrates its compatibility as a potential bioadditive in detergent industries.
Table 6.
Effect of detergents and surfactants on protease activity at 1 and 5% concentration. The enzyme activity was taken as 100% in absence of any of these agents.
| Surfactant/detergent | Relative activity (%) |
|
|---|---|---|
| 1% | 5% | |
| Control | 100 ± 1.91 | 100 ± 2.21 |
| Triton X-100 | 94 ± 2.12 | 82 ± 1.91 |
| Tween-20 | 120 ± 2.60 | 103 ± 2.47 |
| Tween-80 | 80 ± 1.84 | 86 ± 1.79 |
| SDS | 73 ± 1.77 | 51 ± 1.42 |
| Surf Excel | 84 ± 1.68 | 56 ± 1.64 |
| Tide | 79 ± 1.93 | 63 ± 2.91 |
| Aerial | 91 ± 1.99 | 76 ± 2.36 |
| Wheel | 85 ± 2.1 | 65 ± 1.78 |
| Chameli | 80 ± 1.87 | 66 ± 2.10 |
| Ghadi | 78 ± 1.56 | 107 ± 2.89 |
4. Conclusion
In the present study, a thiol-dependent serine protease was purified and characterised from Neocosmospora sp. N1. The enzyme has a high molecular weight of 198 kDa possessing multiple subunits. The novel characteristics of the enzyme like stability at high temperatures and wide pH range, lower Km value, superior activity in presence of various organic solvents, detergents and surfactants pave a way for its utility in numerous commercial applications. The relative high molecular weight protease from unexplored fungal species Neocosmospora sp. N1 enlightens the path of finding novel proteases which can be exploited in industries.
Declarations
Author contribution statement
Fatema Matkawala, Sadhana Nighojkar, Anil Kumar, Anand Nighojkar: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
Data associated with this study has been deposited at GenBank under the accession number MK417797.
Acknowledgements
The authors acknowledge the facilities provided by the Department of Biotechnology, Ministry of Science and Technology, Government of India, New Delhi (DBT) in School of Biotechnology under M.Sc. Biotechnology Program and Bioinformatics sub-center. The authors thank Dr. D.M. Phase, UGC-DAE Consortium for Scientific Research, Indore for helping in Scanning Electron Microscopic analysis. The authors acknowledge facilities of Indian Institute of Science Education and Research, Bhopal for MALDI-TOF analysis and R.R. Centre for Advanced Technology, Indore for AKTA Purification System. The authors also acknowledge the facilities of Department of Biosciences, Maharaja Ranjit Singh College of Professional Sciences, Indore used in the present study.
References
- 1.Vojcic L., Pitzler C., Korfer G., Jakob F., Martinez R., Maurer K.H., Schwaneberg U. Advances in protease engineering for laundry detergents. N. Biotech. 2015;32(6):629–634. doi: 10.1016/j.nbt.2014.12.010. [DOI] [PubMed] [Google Scholar]
- 2.Elliah P., Srinivasalu B., Adinarayana K. A review on microbial alkaline proteases. J. Sci. Ind. Res. 2002;61:690–704. [Google Scholar]
- 3.Jisha V.N., Smitha R.B., Pradeep S., Sreedevi S., Unni K.N., Sajith S., Priji P., Josh M.S., Benjamin S. Versatility of microbial proteases. Adv. Enzym. Res. 2013;1(3):39–51. [Google Scholar]
- 4.Sharma K.M., Kumar R., Panwar S., Kumar A. Microbial alkaline proteases: optimization of production parameters and their properties. J. Genet. Eng. Biotechnol. 2017;15:115–126. doi: 10.1016/j.jgeb.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tavano O.L. Protein hydrolysis using proteases: an important tool for food biotechnology. J. Mol. Catal. B Enzym. 2013;90:1–11. [Google Scholar]
- 6.Anonyme World enzymes to 2015 (Ref: 2824) Focus Catal. 2012;2012:2. [Google Scholar]
- 7.Gupta R., Beg Q.K., Khan S., Chauhan B. An overview on fermentation, downstream processing and properties of microbial alkaline proteases. Appl. Microbiol. Biotechnol. 2002;60:381–395. doi: 10.1007/s00253-002-1142-1. [DOI] [PubMed] [Google Scholar]
- 8.Anwar A., Saleemuddin M. Alkaline protease from Spilosoma oblique: potential applications in bio-formulations. Biotechnol. Appl. Biochem. 2000;31:85–89. doi: 10.1042/ba19990078. [DOI] [PubMed] [Google Scholar]
- 9.Stofko-Hahn R.E., Carr D.W., Scott J.D. A single step purification for recombinant proteins: characterization of a microtubule associated protein (MAP2) fragment which associates with the type II cAMP-dependent protein kinase. FEBS Lett. 1992;302:274–278. doi: 10.1016/0014-5793(92)80458-s. [DOI] [PubMed] [Google Scholar]
- 10.Alici E.H., Arabaci G. A novel serine protease from strawberry (Fragaria ananassa): purification and biochemical characterization. Int. J. Biol. Macromol. 2018;114:1295–1304. doi: 10.1016/j.ijbiomac.2018.03.165. [DOI] [PubMed] [Google Scholar]
- 11.Cui L., Liu Q.H., Wang H.X., Ng T.B. An alkaline protease from fresh fruiting bodies of the edible mushroom Pleurotus citrinopileatus. Appl. Microbiol. Biotechnol. 2007;75:81–85. doi: 10.1007/s00253-006-0801-z. [DOI] [PubMed] [Google Scholar]
- 12.Niyonzima F.N., More S. Detergent-compatible proteases: microbial production, properties, and stain removal analysis, Prep. Biochem. Biotechnol. 2015;45:233–258. doi: 10.1080/10826068.2014.907183. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J., Zhang J. Study on the interaction of alkaline protease with main surfactants in detergent. Colloid Polym. Sci. 2016;294:247–255. [Google Scholar]
- 14.Franco D.G., Spalanzani R.N., Lima E.E., Marchetti C.R., Silva P.O., Masui D.C., Giannesi G.C., Zanoelo F.F. Biochemical properties of a serine protease from Aspergillus flavus and application in dehairing. Biocatal. Biotransform. 2017;35:249–260. [Google Scholar]
- 15.Kanehisa K. Woven or knit fabrics manufactured using yarn dyed raw silk. US Patent. 2000;6(080):689. [Google Scholar]
- 16.Jellouli K., Ghorbel-Bellaaj O., Ayed H.B., Manni L., Agrebi R., Nasri M. Alkaline protease from Bacillus licheniformis MP1: purification, characterization and potential application as a detergent additive and for shrimp waste deproteinization. Process Biochem. 2011;46(6):1248–1256. [Google Scholar]
- 17.Bah C.S.F., Carne A., McConnell M.A., Mros S., Bekhit A.E.D.A. Production of bioactive peptide hydrolysates from deer, sheep, pig and cattle red blood cell fractions using plant and fungal protease preparations. Food Chem. 2016;202:458–466. doi: 10.1016/j.foodchem.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 18.Haddar A., Agrebi R., Bougatef A., Hmidet N., Sellamikamoun A., Nasri M. Two detergent stable alkaline serine proteases from Bacillus mojavensis A21: purification, characterization and potential application as a laundry detergent additive. Bioresour. Technol. 2009;100:3366–3373. doi: 10.1016/j.biortech.2009.01.061. [DOI] [PubMed] [Google Scholar]
- 19.Hadj-Ali N.E., Agrebi R., Ghorbel-Frikha B., Sellami-Kamoun A., Kanoun S., Nasri M. Biochemical and molecular characterization of a detergent stable alkaline serine-protease from a newly isolated Bacillus licheniformis NH1. Enzym. Microb. Technol. 2007;40:515–523. [Google Scholar]
- 20.Kamran A., Rehman H.U., Qader S.A.U., Baloch A.H., Kamal M. Purification and characterization of thiol-dependent, oxidation-stable serine alkaline protease from thermophilic Bacillus sp. J. Genet. Eng. Biotechnol. 2015;13(1):59–64. doi: 10.1016/j.jgeb.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benmrad M.O., Moujehed E., Elhoul M.B., Mechri S., Bejar S., Zouari R., Baffoun A., Jaouadi B. Production, purification, and biochemical characterization of serine alkaline protease from Penicillium chrysogenium strain X5 used as excellent bio-additive for textile processing. Int. J. Biol. Macromol. 2018;119:1002–1016. doi: 10.1016/j.ijbiomac.2018.07.194. [DOI] [PubMed] [Google Scholar]
- 22.Gupta R., Beg Q., Lorenz P. Bacterial alkaline proteases: molecular approaches and industrial applications. Appl. Microbiol. Biotechnol. 2002;59:15–32. doi: 10.1007/s00253-002-0975-y. [DOI] [PubMed] [Google Scholar]
- 23.Saeki K., Ozaki K., Kobayashi T., Ito S. Detergent alkaline proteases: enzymatic properties, genes, and crystal structures. J. Biosci. Bioeng. 2007;103:501–508. doi: 10.1263/jbb.103.501. [DOI] [PubMed] [Google Scholar]
- 24.Sumantha A., Larroche C., Pandey A. Microbiology and industrial biotechnology of food-grade proteases: a perspective. Food Technol. Biotechnol. 2006;44(2):211–220. [Google Scholar]
- 25.Salihi A., Asoodeh A., Aliabadian M. Production and biochemical characterization of an alkaline protease from Aspergillus oryzae CH93. Int. J. Biol. Macromol. 2016;94:827–835. doi: 10.1016/j.ijbiomac.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 26.Dobinson K.F., Lecomte N., Lazarovits G. Production of extracellular trypsin-like protease by the fungal plant pathogen Verticillium dahlia. Can. J. Microbiol. 1997;43:227–233. doi: 10.1139/m97-031. [DOI] [PubMed] [Google Scholar]
- 27.Abidi F., Limam F., Neji M.M. Production of alkaline proteases by Botrytis cinerea using economic raw materials: assay as biodetergent. Process Biochem. 2008;43:1202–1208. [Google Scholar]
- 28.Ueda M., Kubo T., Miyatake K., Nakamura T. Purification and characterization of fibrinolytic alkaline protease from Fusarium sp. BLB. Appl. Microbiol. Biotechnol. 2007;74:331–338. doi: 10.1007/s00253-006-0621-1. [DOI] [PubMed] [Google Scholar]
- 29.Oyeleke S.B., Egwim E.C., Auta S.H. Screening of Aspergillus flavus and Aspergillus fumigatus strains for extracellular protease enzyme production. J. Microbiol. Antimicrob. 2010;2:83–87. [Google Scholar]
- 30.Charles P., Devanathan V., Anbu P., Ponnuswamy M.N., Kalaichelvan P.T., Hur B.K. Purification, characterization and crystallization of an extracellular alkaline protease from Aspergillus nidulans HA-10. J. Basic Microbiol. 2008;48:347–352. doi: 10.1002/jobm.200800043. [DOI] [PubMed] [Google Scholar]
- 31.Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.L. Protein measurement with Folin phenol reagent. J. Biol. Chem. 1951;193:265–273. [PubMed] [Google Scholar]
- 32.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 33.Heussen C., Dowdle E.B. Electrophoretic analysis of plasminogen activators in polyacrylamide gels containing sodium dodecyl sulfate and copolymerized substrates. Anal. Biochem. 1980;102:196–202. doi: 10.1016/0003-2697(80)90338-3. [DOI] [PubMed] [Google Scholar]
- 34.Riffel A., Brandelli A., Bellato C.M., Souza G.H.M.F., Eberlin M.N., Tavar F.C.A. Purification and characterization of a keratinolytic metalloprotease from Chryseobacterium sp. kr6. J. Biotechnol. 2007;128:693–703. doi: 10.1016/j.jbiotec.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 35.Zanphorlin L.M., Cabral H., Arantes E., Assis D., Juliano M.A., Da-Silva R., Bonilla-Rodriguez G.O. Purification and characterization of a new alkaline serine protease from the thermophilic fungus Myceliophthora sp. Process Biochem. 2011;46:2137–2143. [Google Scholar]
- 36.Kotlova E.K., Ivanova N.M., Yusupova M.P., Voyushina T.L., Ivanushkina N.E., Chestukhina G.G. Thiol-dependent serine proteinase from Paecilomyces lilacinus: purification and catalytic properties. Biochem. 2007;72(1):117–123. doi: 10.1134/s0006297907010142. [DOI] [PubMed] [Google Scholar]
- 37.Beg Q.K., Gupta R. Purification and characterization of an oxidation-stable, thiol-dependent serine alkaline protease from Bacillus mojavensis. Enzym. Microb. Technol. 2003;32:294–304. [Google Scholar]
- 38.Khaĭdarova N.V., Rudenskaia G.N., Revina L.P., Stepanov V.M., Egorov N.S. Thiol- dependent serine proteinase from Streptomyces thermovulgaris. Biokhimiia. 1990;55(6):1110–1119. [PubMed] [Google Scholar]
- 39.Dorra G., Ines K., Imen B.S., Laurent C., Sana A., Tabbene O., Pascal C., Thierry J., Ferid L. Purification and characterization of a novel high molecular weight alkaline protease produced by an endophytic Bacillus halotolerans strain CT2. Int. J. Biol. Macromol. 2018;111:342–351. doi: 10.1016/j.ijbiomac.2018.01.024. [DOI] [PubMed] [Google Scholar]
- 40.Wang S.L., Chen Y.H., Wang C.L., Yen Y.H., Chern M.K. Purification and characterization of a serine protease extracellularly produced by Aspergillus fumigatus in a shrimp and crab shell powder medium. Enzym. Microb. Technol. 2005;36:660–665. [Google Scholar]
- 41.Arulmani M., Aparanjini K., Vasanthi K., Arumugam P., Arivuchelvi M., Kalaichelvan P.T. Purification and partial characterization of serine protease from thermostable alkalophilic Bacillus laterosporus-AK1. World J. Microbiol. Biotechnol. 2006;23:475–481. [Google Scholar]
- 42.Pekkarinen A.I., Jones B.L., Niku P., Marja L. Purification and properties of an alkaline proteinase of Fusarium culmorum. Eur. J. Biochem. 2002;269:798–807. doi: 10.1046/j.0014-2956.2001.02697.x. [DOI] [PubMed] [Google Scholar]
- 43.Deng A., Wua J., Zhang Y., Zhang G., Wen T. Purification and characterization of a surfactant-stable high-alkaline protease from Bacillus sp. B001. Bioresour. Technol. 2010;101:7100–7106. doi: 10.1016/j.biortech.2010.03.130. [DOI] [PubMed] [Google Scholar]
- 44.Mechri S., Ben Elhoul Berrouina M., Omrane Benmrad M., Zaraî Jaouadi N., Rekik H., Moujehed E., Chebbi A., Sayadi S., Chamkha M., Bejar S., Jaouadi B. Characterization of a novel protease from Aeribacillus pallidus strainVP3 with potential biotechnological interest. Int. J. Biol. Macromol. 2017;94:221–232. doi: 10.1016/j.ijbiomac.2016.09.112. [DOI] [PubMed] [Google Scholar]
- 45.Yadav S.K., Bisht D., Tiwari S., Darmwal N.S. Purification, biochemical characterization and performance evaluation of analkaline serine protease from Aspergillus flavus MTCC 9952 mutant. Biocatal. Agric. Biotechnol. 2015;4:667–677. [Google Scholar]
- 46.Shankar S., Rao M., Laxman R.S. Purification and characterization of an alkaline protease by a new strain of Beauveria sp. Process Biochem. 2011;46:579–585. [Google Scholar]
- 47.Niyonzima F.N., More S.S. Purification and properties of detergent compatible extracellular alkaline protease from Scopulariopsis spp., Prep. Biochem. Biotechnol. 2014;44:738–759. doi: 10.1080/10826068.2013.854254. [DOI] [PubMed] [Google Scholar]
- 48.Abidi F., Chobert J.M., Haertlé T., Marzouki M.N. Purification and biochemical characterization of stable alkaline protease Prot-2 from Botrytis cinerea. Process Biochem. 2011;46:2301–2310. [Google Scholar]
- 49.Li F., Yang L., Lv X., Liu D., Xia H., Chen S. Purification and characterization of a novel extracellular alkaline protease from Cellulomonas bogoriensis. Protein Expr. Purif. 2016;121:125–132. doi: 10.1016/j.pep.2016.01.019. [DOI] [PubMed] [Google Scholar]
- 50.Hadjidj R., Badis A., Mechri S., Eddouaouda K., Khelouia L., Annane R., El Hattab M., Jaouadi B. Purification, biochemical, and molecular characterization of novel protease from Bacillus licheniformis strain K7A. Int. J. Biol. Macromol. 2018;114:1033–1048. doi: 10.1016/j.ijbiomac.2018.03.167. [DOI] [PubMed] [Google Scholar]
- 51.Jaouadi B., Ellouz-Chaabouni S., Rhimi M., Bejar S. Biochemical and molecular characterization of a detergent-stable serine alkaline protease from Bacillus pumilus CBS with high catalytic efficiency. Biochimie. 2008;90:1291–1305. doi: 10.1016/j.biochi.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 52.Maurer K.H. Detergent proteases. Curr. Opin. Biotechnol. 2004;15:330–334. doi: 10.1016/j.copbio.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 53.Yildirim V., Baltaci M.O., Ozgencli I., Sisecioglu M. Purification and biochemical characterization of a novel thermostable serine alkaline protease from Aeribacillus pallidus C10: a potential additive for detergents. J. Enzym. Inhib. Med. Chem. 2017;32:468–477. doi: 10.1080/14756366.2016.1261131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tremacoldi C.R., Monti R., Selistre-de-Araujo H.S., Carmona E.C. Purification and properties of an alkaline protease of Aspergillus clavatus. World J. Microbiol. Biotechnol. 2007;23:295–299. [Google Scholar]
- 55.Sari E., Logoglu E., Oktemer A. Purification and characterization of organic solvent stable serine alkaline protease from newly isolated Bacillus circulans M34. Biomed. Chromatogr. 2015;29:1356–1363. doi: 10.1002/bmc.3431. [DOI] [PubMed] [Google Scholar]
- 56.Gupta A., Roy I., Patel R.K., Singh S.P., Khare S.K., Gupta M.N. One step purification and characterization of an alkaline protease from haloalkaliphilic Bacillus sp. J. Chromatogr. A. 2005;1075:103–108. doi: 10.1016/j.chroma.2005.03.127. [DOI] [PubMed] [Google Scholar]
- 57.Rao M.B., Tankasale A.M., Ghatge M.S., Desphande V.V. Molecular and biotechnological aspects of microbial proteases. Microbiol. Mol. Biol. Rev. 1998;62:597–634. doi: 10.1128/mmbr.62.3.597-635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moradian F., Khajeh K., Naderi-Manesh H., Ahmadvand R., Sajedi R.H., Sadeghizadeh M. Thiol-dependent serine alkaline proteases from Bacillus sp. HR-08 and KR-8102. Appl. Biochem. Biotechnol. 2006;134:77–87. doi: 10.1385/abab:134:1:77. [DOI] [PubMed] [Google Scholar]
- 59.Jaouadi B., Abdelmalek B., Fodil D., Ferradji F.Z., Rekik H., Zaraî N., Bejar S. Purification and characterization of a thermostable keratinolytic serine alkaline proteinase from Streptomyces sp. strain AB1 with high stability in organic solvents. Bioresour. Technol. 2010;101:8361–8369. doi: 10.1016/j.biortech.2010.05.066. [DOI] [PubMed] [Google Scholar]
- 60.Rekik H., Jaouadi N.Z., Gargouri F., Bejar W., Frikha F., Jmal N., Bejar S., Jaouadi B. Production, purification and biochemical characterization of a novel detergent-stable serine alkaline protease from Bacillus safensis strain RH12. Int. J. Biol. Macromol. 2019;121:1227–1239. doi: 10.1016/j.ijbiomac.2018.10.139. [DOI] [PubMed] [Google Scholar]
- 61.Barata R.A., Andrade M.H.G., Rodrigues R.D., Castro I.M. Purification and characterization of an extracellular trypsin-like protease of Fusarium oxysporum var. lini. J. Biosci. Bioeng. 2002;94(4):304–308. doi: 10.1263/jbb.94.304. [DOI] [PubMed] [Google Scholar]
- 62.Touioui S.B., Jaouadi N.Z., Bouacem K., Ayed R.B., Rekik H., Zenati B., Kourdali S., Boudjella H., Sabaou N., Bejar S., El Hattab M., Badis A., Annane R., Jaouadi B. Biochemical and molecular characterization of a novel metalloprotease from Pseudomonas fluorescens strain TBS09. Int. J. Biol. Macromol. 2018;107:2351–2363. doi: 10.1016/j.ijbiomac.2017.10.116. [DOI] [PubMed] [Google Scholar]
- 63.Anandan D., Marmer W.N., Dudley R.L. Isolation, characterization and optimization of culture parameters for production of an alkaline protease isolated from Aspergillus tamarii. J. Ind. Microbiol. Biotechnol. 2007;34:339–347. doi: 10.1007/s10295-006-0201-5. [DOI] [PubMed] [Google Scholar]
- 64.Mothe T., Sultanpuram V.R. Production, purification and characterization of a thermo tolerant alkaline serine protease from a novel species Bacillus caseinilyticus. 3 Biotech. 2016;6(53):2–10. doi: 10.1007/s13205-016-0377-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reddy L., Wee Y.J., Ryu H.W. Purification and characterization of an organic solvent and detergent-tolerant novel protease produced by Bacillus sp. RKY3. J. Chem. Technol. Biotechnol. 2008;83:1526–1533. [Google Scholar]