Abstract
Genome-scale single-cell biology has recently emerged as a powerful technology with important implications for both basic and medical research. There are urgent needs for the development of computational methods or analytic pipelines to facilitate large amounts of single-cell RNA-Seq data analysis. Here, we present a detailed protocol for SINCERA (SINgle CEll RNA-Seq profiling Analysis), a generally applicable analytic pipeline for processing single-cell data from a whole organ or sorted cells. The pipeline supports the analysis for the identification of major cell types, cell type-specific gene signatures, and driving forces of given cell types. In this chapter, we provide step-by-step instructions for the functions and features of SINCERA together with application examples to provide a practical guide for the research community. SINCERA is implemented in R, licensed under the GNU General Public License v3, and freely available from CCHMC PBGE website, https://research.cchmc.org/pbge/sincera.html.
Keywords: Single-cell, RNA-Seq, Pipeline, Cell type, Signature gene, Driving force
1. Introduction
Single cells are the fundamental units of life. Recent advances in high-throughput cell isolation and sequencing at the single-cell level enable studying individual transcriptomes of large numbers of cells in parallel, providing new insights into the diversity of cell types, rare cells and cell lineage relationships that has been difficult to resolve in genomic data from bulk tissue samples [1, 2, 3, 4, 5, 6, 7, 8]. While the single cell research field is still in its early stages, it has already made a strong impact on many fields in biology and led to great improvements in our fundamental understanding of human diseases [9, 10, 11, 12, 13, 14, 15, 16, 17]. We believe that the demand of single cell analytic tools will continue to grow in the future as broad applications of single cell transcriptomics in biological and medical researches.
While the future of single-cell next-generation sequencing based genomic/transcriptomic studies is promising, it comes with new and specific analytical challenges including the identification and characterization of unknown cell types, handling the confounding factors such as batch and cell cycle effects, and addressing the cellular heterogeneity in complex biological systems, just to name a few [18, 19, 20, 21, 22]. Recently, a number of methods specifically designed for single-cell RNA-Seq (scRNA-Seq) analysis have been introduced including BackSPIN [15], SNN-Cliq [23], and RaceID [24] for cell cluster identification; scLVM [22] for confounding factor handling; Seurat [25] for spatial reconstruction of scRNA-Seq data, cell cluster identification, and expression pattern visualization; SAMstrt [26] and SCDE [20] for single-cell differential expression analysis; and Monocle [21], Wanderlust [27], SCUBA [28], Waterfall [29], StemID [16], and SLICE [30] for extracting lineage relationships from scRNA-Seq and modeling the dynamic changes associated with cellular biological processes. Here, we present SINCERA [31], a top-to-bottom single cell analytic tool set designed for the practical usages of the research community. Specifically, the pipeline enables investigators to analyze scRNASeq data using standard desktop/laptop computers to conduct data filtering, normalization, clustering, cell type identification, gene signature prediction, transcriptional regulatory network construction, and identification of driving forces (key nodes) for each cell type. We have successfully applied SINCERA to multiple scRNA-Seq datasets from normal developmental lung and various pathological states from both mouse and human, demonstrating SINCERA’s general utility and accuracy [31, 32, 33].
2. Materials
The entire SINCERA pipeline was implemented in R. The execution requires the following hardware and software.
A standard desktop or laptop computer with Windows, Mac OS X, or Linux operating system.
R statistical computing environment (version 3.2.0 or later) from The Comprehensive R Archive Network (https://cran.r-project.org/).
Install R and Bioconductor packages into the R environment, including Biobase [34], ROCR [35], RobustRankAggreg [36], G1DBN [37], igraph [38], ggplot2 [39], ggdendro (https://cran.r-project.org/web/packages/ggdendro), plyr [40], and zoo [41].
Download SINCERA scripts from https://research.cchmc.org/pbge/sincera.html.
3. Methods
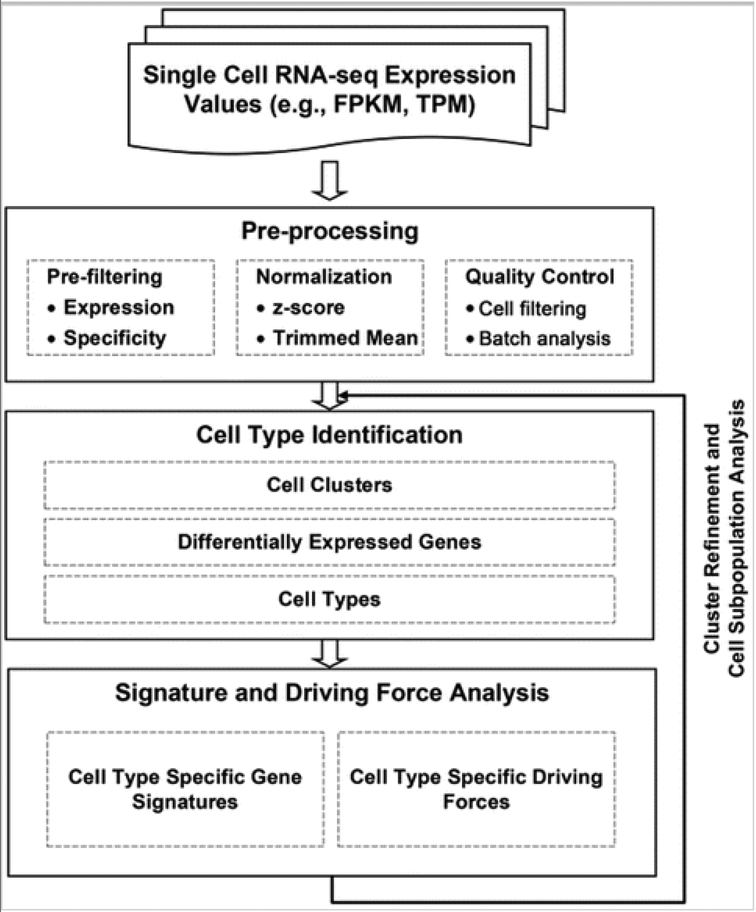
SINCERA consists of four major analytic components: preprocessing, cell type identification, gene signature prediction, and driving force analysis (Fig 1). The pipeline takes RNA-Seq expression values (e.g., FPKM [42] or TPM [43]) from heterogeneous single cell populations as inputs, and it outputs a clustering scheme of cells, differentially expressed genes for each cell cluster, enriched cell type annotations for each cluster, refined cell type-specific gene signature, and cell type-specific rankings of transcription factors. SINCERA is a comprehensive toolset with a variety of options for key analytic steps, many of which can be run independently of one another. To facilitate ease of reference for beginner users, we have marked essential steps with *. In the rest of this chapter, we dissect the functional features of SINCERA into the four components and describe the usages of each component step by step. R functions in SINCERA are depicted in italic font.
FIG 1.
Schematic flow of the SINCERA protocol (Adapted from Fig. 1 in Guo et al. [31])
3.1. Preprocessing
The preprocessing steps include data transformation and normalization, prefiltering cells with low quality, and prefiltering genes with low expression abundancy and selectivity as described below.
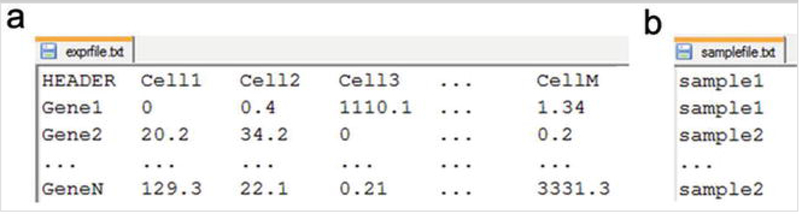
*The analysis starts with running the construct function to create an R S4 object, which will hold all the data and analysis results. The function takes two parameters as inputs: “exprfile” and “samplefile”. The “exprfile” specifies the full path to a gene expression profile matrix where rows are genes and columns are individual cells (see Note 1). The “samplefile” parameter specifies the full path to a table that contains a single column describing the sample information (e.g., biological replicates or batch difference) of individual cells. Figure 2 shows the required formats of the two input files.
The CCHMC single cell core inspects each individual cell under microscope after capture and prior to lysis. This quality control (QC) step is important in filtering out libraries made from empty wells or wells with excess debris. In addition, we run the filterLowQualityCells function of SINCERA to further identify and remove low quality cells. The key parameters of running this function include: “min.expression”, which specifies the minimum expression value for a gene to be considered an expressed gene, and “min.genes”, which specifies the lower bound of the number of expressed genes in a cell. This function identifies and removes cells with few expressed genes. The default value for the “min.expression” parameter is 1 FPKM/TPM and for the “min.genes” parameter is 500.
Use filterContaminatedCells function to remove potential contaminated cells based on the coexpression of known marker genes of two distinct cell types, such as the coexpression of mouse lung epithelial marker Epcam and mouse lung endothelial cell marker Pecam1. Users can specify the marker genes of the first cell type and of the second cell type in the “markers.1” and “markers.2” parameter, respectively. This step can repeat multiple times. For each cell type, we suggest using only highly specific markers for contamination detection.
*Use prefilterGenes function to filter out non- or low-expressed genes, as well as genes that are expressed in less than a certain number of cells per sample preparation. By default, genes expressed (>5 FPKM/TPM) in less than two cells will be filtered out by this function.
*Use expr.minimum function to set a minimum expression value. As part of the preprocessing step, we transformed FPKM/TPM values less than or equal to 0.01–0.01 in order to eliminate “zero”s from the follow up data transformation and analysis. The default minimum value is 0.01 FPKM/TPM.
Run batch.analysis function to identify batch differences. This function plots the quantiles of gene expression in individual cells from different batches, and compares the distribution of gene expression among batches using MA plot, Q–Q plot, and cell correlation and distance measure [31].
*Normalization methods are applied to reduce batch effect and enable expression level comparisons within or across sample preparations. SINCERA provides both gene level and cell level normalizations. For gene level normalization, normalization.zscore function is applied to each gene expression profile for per-sample z-score transformation (see Note 2). For cell level normalizations, we use the trimmed mean. If starting with normalized expression data (e.g., FPKM or TPM), cell level normalization is not always necessary.
*Run cluster.geneSelection function to select genes with a certain level of expression specificity for cell type identification. This specificity filter [31] removes genes unselectively expressed across all cell types (e.g., housekeeping genes) and keeps genes with a certain degree of cell type selective expression. The default specificity threshold is set as 0.7. The main purpose of this step is to select expression profiles that are potentially informative about cell types/states and remove genes that may increase noise in the cell type identification step (see Note 3).
FIG 2.
Formats of the input files to the SINCERA pipeline. (a) Format of expression profile table. (b) Format of sample description table. The number of rows in the sample description table is the same as the number of cells in the expression profile table. Both files are tab delimited text files
3.2. Cell Type Identification
Cell clustering and cell type identification is a key step in the pipeline and directly influences all downstream analysis. SINCERA starts with an unsupervised hierarchical clustering of the cells using the selected expression profiles. Use of an unsupervised hierarchical clustering approach does not impose prerequisite external biological knowledge, nor does it require preset knowledge of the number of clusters; therefore, it is capable of discovering novel cell types. Multiple iterations using more than one clustering methods are usually required for cell cluster refinement (see Note 4).
*Run cluster.assignment function to assign cells to initial clusters. The default algorithm uses hierarchical clustering with average linkage, Pearson’s correlation based distance measurement, and z-score transformed expression values of the selected genes.
*Run plotMarkers function to check the quality of the obtained clustering scheme and inspect the expression patterns of a number of known markers across cell clusters. A scattered and/or overlapping expression pattern of cell type marker genes across different cell clusters may suggest a low quality clustering scheme. In this case, we recommend using cluster.assignment function with a different parameter setting to redefine cell clusters. This process may need to be iterated several times to achieve better separation.
Run the cluster.permutation.analysis function to perform a cluster membership permutation analysis [31] to determine cluster significance. SINCERA implements several quality control or internal validation steps; this is one of them, used to check quality of clustering schemes.
*Once cell clusters have been defined, use cluster.diffgenes function to identify differentially expressed genes in each cluster. For each cell cluster, this function uses one-tailed Welch’s t test or Wilcoxon test to compare the gene expression in a given cell cluster to the corresponding gene expression in all other cells, and genes with p-value less than a threshold are identified as differentially expressed genes for the cluster. One can also choose binomial or negative-binomial probability test in this step.The default threshold is 0.05.
Next, run celltype.enrichment function to predict cell type for each cluster (see Note 5). SINCERA has built a precompiled cell type and gene association table using experimental expression data obtained from EBI expression atlas (https://www.ebi.ac.uk/gxa). Cell type prediction is based on the enrichment of cell type annotations significantly associated with differentially expressed genes of the given cluster using a one-tailed Fisher’s exact test.
Once cell clusters have been defined, use plotMarkers function to visualize the expression patterns of known cell type markers in order to cross validate the predicted cell type, i.e., to check whether they are selectively expressed in their defined cell clusters.
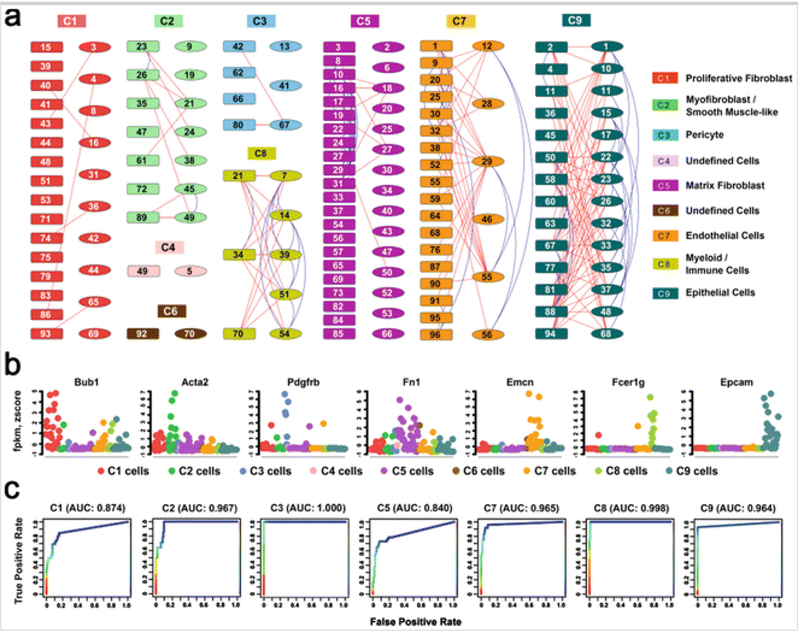
Run celltype.validation function to perform a rank-aggregation-based quantitative assessment of the consistency between mapped cell type and the expression pattern of known cell type marker genes. Figure 3 demonstrates the application of SINCERA to identify major cell types at E16.5 mouse lung and to validate the cell type assignment using known markers.
FIG 3.
Identification and validation of major lung cell types at E16.5 mouse lung (Adapted from Figs. 2 and 3 in Guo et al. [31]). (a) Cells (n = 148) from two sample preparations from fetal mouse lung at E16.5 [31] were assigned into nine clusters via hierarchical clustering using average linkage and centered Pearson’s correlation. Each color represents a distinct cell cluster, labeled as C1–C9. The rectangles represent single lung cells from the first preparation and the ellipses consist of single cells from a second independent preparation. Connection lines indicate the z-score correlation between the two cells ≥0.05. The blue lines connect cells within the same preparation, while the red lines connect cells across preparations. (b) Expression patterns of representative known cell type markers were used to validate the correct assignment of major lung cell types at E16.5. Expression levels were normalized by per-sample z-score transformation. (c) Receiver Operating Characteristic curves of the rank-aggregation-based validation showed a high consistency between the cell type assignments and the expression patterns of known cell type-specific markers
3.3. Cell Type-Specific Signature Gene Analysis
We define cell type-specific gene signature as a group of genes uniquely or selectively expressed in a given cell type. Once cell types have been defined, the analysis proceeds with the identification of cell type-specific gene signatures using the following functions.
Collect positive and negative marker genes for each mapped cell type. Use setCellTypeMarkers function to add the collected markers into SINCERA.
*Run the signature.prediction function to predict cell type signature genes. The basic level of prediction defines differentially expressed genes of the given cell type as the signature genes. For more advanced prediction, the signature.prediction function uses four features [31] to define cell type-specific signature genes, including common gene metric (genes shared by the cluster cells), unique gene metric (genes selectively expressed in the cluster cells), test statistic metric (group mean comparison between cluster cells and all the other cells), and synthetic profile similarity (genes correlating with the model profile of the given cluster). When the marker genes of a cell type are available, the signature.prediction function uses a logistic regression model to integrate the four metrics for ranking prediction of cell-specific signatures [31]. Nevertheless, marker genes may not be always available, especially for novel cell types. In such cases, the signature.prediction function predicts signature by using additional filters to refine differentially expressed genes, including a frequency filter and a fold change filter. The frequency filter selects genes expressed in at least a certain percentage of the cells within the defined cluster. The fold change filter selects genes with a certain degree of average expression enrichment in the given cluster compared to the cluster with its second highest average expression. The default frequency and fold change threshold is 30% and 1.5, respectively.
Use plotHeatmap function to visualize the expression of the predicted signature genes across cell types (clusters). This allows a visual inspection of the selective expression of the predicted signature genes in the defined cell types.
Run signature.validation function to validate the signature prediction using a repeated random subsampling approach [31]. Essentially, this approach validates the predicted signature by assessing its classification accuracy in distinguishing the cells of the given cell type from cells of other types.
3.4. Cell Type-Specific Key Regulator Prediction
Identification of the key regulators controlling cell fate is essential for understanding complex biological systems. SINCERA utilizes a transcriptional regulatory network (TRN) approach to establish the relationships between transcription factors (TFs) and target genes (TGs) based on their expression-based regulatory potential and identify the key TFs for a given cell type by measuring the importance of each node in the constructed TRN.
Run drivingfoce.selectTFs function to select candidate transcription factors for the prediction. The function selects the union of cell type-specific differentially expressed TFs (e.g., p-value of one-tailed Welch’s t test <0.05) and commonly expressed TFs (e.g., expressed in at least 80% of the cell type) as candidates. Note that here we do not require a key regulator for a given cell type to be differentially expressed in the cell type.
Use drivingforce.selectTGs function to select cell type-specific differentially expressed genes or signature genes as candidate target genes (TGs).
Use drivingforce.inferTRN function to infer a TRN using the cell type-specific expression patterns of the selected candidate TFs and TGs. The “edge.threshold” parameter is used to select significant TF-TF or TF-TG interactions (see Note 6) for building the network. The default threshold is set to 0.05 (see Note 7).
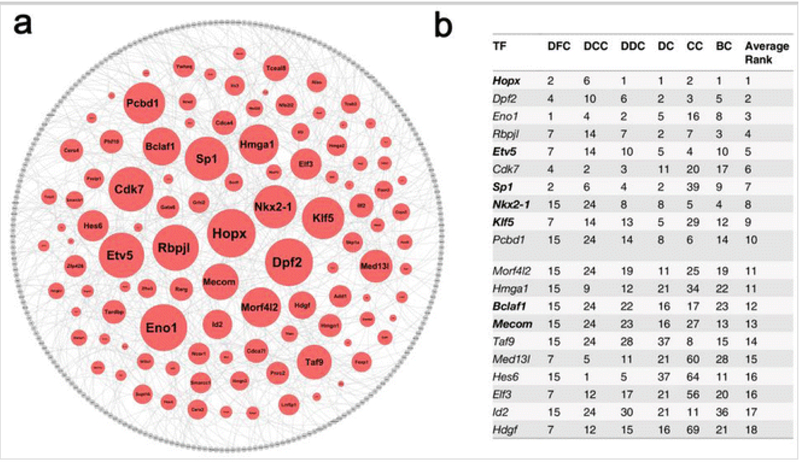
Use drivingforce.rankTFs function to rank TFs based on their importance to the inferred TRN (see Note 8). Top ranked TFs are predicted as key regulators (driving force) for the given cell type. Figure 4 demonstrates of usingSINCERA to predict key TFs in E16.5 mouse lung epithelial cells.
Fig. 4.
Prediction of E16.5 mouse lung epithelial specific driving force (Adapted from Fig. 6 and Table 1 in Guo et al. [31]). (a) Rank importance of transcription factors (TFs) in the largest connected component (LCC) of epithelial specific transcriptional regulatory network (TRN). The sizes of the TF nodes are proportional to their average-ranked node importance. The LCC of epithelial TRN is comprised of 348 nodes and 432 edges. The nodes in red are TFs and the nodes in grey are differentially expressed genes in epithelial cells and are not TFs. The edges were established using the first-order conditional dependence approach described in the Guo et al. [31] with a cutoff at 0.05. (b) Top 20 predicted key TFs for lung epithelial cells at E16.5 based on the integration of six TF importance metrics. DC ranking based on degree centrality, CC ranking based on closeness centrality, BC ranking based on betweenness centrality, DFC ranking based on disruptive fragmentation centrality, DCC ranking based on disruptive connection centrality, DDC ranking based on disruptive distance centrality. All ranks are in decreasing order of the TF importance values. TFs in bold font are associated with lung-related mouse phenotypes. TRN is plotted using cytoscape 2.8 (http://www.cytoscape.org/)
4. Notes
The pipeline takes aligned and quantified RNA-Seq expression values (e.g., FPKM or TPM) as inputs. Functions related to sequencing data mapping, alignment, quantification, and annotation are not part of the pipeline, and they can be processed using widely available software such as Tophat [44, 45], BWA [46], Cufflinks [42], and RSEM [43].
We noticed that, typically, in a scRNA-Seq dataset, individual genes can have different levels of baseline expression, which means that a cell type selective marker may have nonzero expression in cells other than its defined cell type, but its expression amplitude is usually much higher in the selective cell type than in other cell types. The normalization.zscore function scales the expression of individual genes using a z-score transformation in order to better reveal their major expression patterns and suppress the unnecessary variations associated with the scRNA-Seq data. Performing within-sample z-score transformation is based on the assumption that cell type distribution is roughly the same among replicates. If this assumption cannot be guaranteed (e.g., there is a large batch difference among different replicates), a global z-score transformation should be used. Of note, the z-score transformed expression values are mainly used in the cell type identification step and the visualization of gene expression patterns, but not in differential expression analysis.
The cluster.geneSelection function also provides other criteria for informative gene selection, including coefficient of variance and average expression across all cells, which have been utilized in existing scRNA-Seq analyses [12, 22]. The specificity.thresholdSelection function in SINCERA can be used to determine the specificity threshold. This function measures the per-sample specificity of a set of ribosomal genes based on Ribosome pathway annotation (KEGG PATHWAY: hsa03010), and then chooses a criterion that can filter out at least 95% of the ribosomal genes.
We compared multiple clustering algorithms using a variety of independent scRNA-Seq datasets [31] and showed that hierarchical clustering, while may not always be the best way, is generally applicable and easy to use. Therefore, hierarchical clustering is suitable for biologists to use as one of the tools for initial cell clustering identification [31]. In addition to the default clustering method, we also include hierarchical clustering with ward linkage [47], consensus clustering [48, 49], and tight clustering [50] as optional cluster determination methods in the pipeline. Users can choose different clustering methods for cell cluster identification by setting the “clustering.method” parameter in the cluster.assignment function. For advanced users, comparing different methods and adjusting parameters to achieve optimized results are encouraged.
The cell cluster identification and cell type assignment are the bottlenecks in current scRNA-Seq analysis. It requires us to extract cell type relevant information from multiple sources, including the expression patterns of known marker genes and functional annotations enriched by the cluster specific differentially expressed genes. Knowledge integration by an expert is usually required to determine the cell type of a given cell cluster at the end. To our knowledge, there are multiple tools for gene sets enrichment analysis, e.g., DAVID [51] and ToppGene [52], but lack of tools for cell type enrichment analysis. To facilitate the general usage of the pipeline, we implemented celltype.enrichment function in SINCERA as an attempt to automate the cell type prediction. The current version of cell type annotations is based on the open source gene expression data from EBI Expression Atlas (https://www.ebi.ac.uk/gxa); bias and incompleteness from the collection of individual experimental sources are inevitable. We recommend the use of it for initial cell type screening, together with functional enrichment analysis using cluster specific differentially expressed genes, and curation and knowledge integration by experts to refine the cell type mapping. We foresee that single cell transcriptome analyses will largely improve cell type prediction by providing a high resolution and unbiased cell type separation and associated signature identification for lung and other organs.
For the transcriptional regulatory network (TRN) construction, we focus on identifying the relationships between TF-TF (transcription factor and its partners/cofactors) and TF-TG (transcription factor and its target genes). The possible feedback regulations from target genes to TFs and TF autoregulations are not considered in the present implementation of SINCERA. Regulatory relationships are established based on first-order conditional dependence of gene expression [31], adapted from the inference of first-order conditional dependence Directed Acyclic Graph (DAG) in [37].
The inferred TRN may consist of multiple connected components. The largest connected component (LCC) is the one that has the largest number of nodes among all connected components. If the LCC of the inferred TRN is not large enough, which means that the number of nodes in LCC is less than a certain percentage (e.g., 80%) of the total number of selected TFs and TGs for TRN inference, this indicates that the number of interactions is insufficient to build the TRN. The drivingforce.inferTRN function needs to be reexecuted with a higher threshold to build the TRN using more interactions. The drivingforce.getLCC function can be used to assess whether a large enough LCC exists in the inferred TRN.
To identify cell type-specific driving force, we measure and rank the importance of TFs in the cell type-specific TRN based on the integration of six TF importance metrics, including degree centrality, closeness centrality, betweenness centrality, disruptive fragmentation centrality, disruptive connection centrality, and disruptive distance centrality. Details about the six metrics can be found in Guo et al. [31]. Individual metrics provide local views of the importance of a node to the network, and their integration can provide a better global view of the node importance in the network. In the current setting, only the TFs in the LCC of the inferred TRN are included in the TF ranking, and only the LCC is used to calculate the values of the six metrics for each TF.
Acknowledgment
This work was supported by the National Heart, Lung, and Blood Institute of National Institutes of Health (http://www.nhlbi.nih.gov, grants U01HL122642 (LungMAP), U01 HL110967 (LRRC), and R01 HL105433). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A et al. (2009) mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 6:377–382 [DOI] [PubMed] [Google Scholar]
- 2.Tang F, Barbacioru C, Bao S, Lee C, Nordman E, Wang X, Lao K, Surani MA (2010) Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell 6:468–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qiu S, Luo S, Evgrafov O, Li R, Schroth GP, Levitt P, Knowles JA, Wang K (2012) Single-neuron RNA-Seq: technical feasibility and reproducibility. Front Genet 3:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramskold D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC et al. (2012) Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol 30:777–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Islam S, Kjallquist U, Moliner A, Zajac P, Fan JB, Lonnerberg P, Linnarsson S (2011) Characterization of the single-cell transcriptional landscape by highly multiplex RNA-Seq. Genome Res 21:1160–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Narsinh KH, Sun N, Sanchez-Freire V, Lee AS, Almeida P, Hu S, Jan T, Wilson KD, Leong D, Rosenberg J et al. (2011) Single cell transcriptional profiling reveals heterogeneity of human induced pluripotent stem cells. J Clin Invest 121:1217–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, Raychowdhury R, Schwartz S, Yosef N, Malboeuf C, Lu D et al. (2013) Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 498:236–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wills QF, Livak KJ, Tipping AJ, Enver T, Goldson AJ, Sexton DW, Holmes C (2013) Single-cell gene expression analysis reveals genetic associations masked in whole-tissue experiments. Nat Biotechnol 31:748–752 [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Waters J, Leung ML, Unruh A, Roh W, Shi X, Chen K, Scheet P, Vattathil S, Liang H et al. (2014) Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature 512:155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanchate NK, Kondoh K, Lu Z, Kuang D, Ye X, Qiu X, Pachter L, Trapnell C, Buck LB (2015) Single-cell transcriptomics reveals receptor transformations during olfactory neurogenesis. Science 350:1251–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Ferrante TC, Terry R, Turczyk BM, Yang JL, Lee HS, Aach J et al. (2015) Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat Protoc 10:442–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM et al. (2015) Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161:1202–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saadatpour A, Lai S, Guo G, Yuan GC (2015) Single-cell analysis in cancer genomics. Trends Genet 31:576–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaughan AE, Brumwell AN, Xi Y, Gotts JE, Brownfield DG, Treutlein B, Tan K, Tan V, Liu FC, Looney MR et al. (2015) Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature 517:621–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, Marques S, Munguba H, He L, Betsholtz C et al. (2015) Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-Seq. Science 347:1138–1142 [DOI] [PubMed] [Google Scholar]
- 16.Grün D, Muraro MJ, Boisset JC, Wiebrands K, Lyubimova A, Dharmadhikari G, van den Born M, van Es J, Jansen E, Clevers H et al. (2016) De novo prediction of stem cell identity using single-cell transcriptome data. Cell Stem Cell 19:266–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M et al. (2016) Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell 166(1308–1323):e1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim JK, Marioni JC (2013) Inferring the kinetics of stochastic gene expression from single-cell RNA-sequencing data. Genome Biol 14:R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brennecke P, Anders S, Kim JK, Kolodziejczyk AA, Zhang X, Proserpio V, Baying B, Benes V, Teichmann SA, Marioni JC et al. (2013) Accounting for technical noise in single-cell RNA-Seq experiments. Nat Methods 10:1093–1095 [DOI] [PubMed] [Google Scholar]
- 20.Kharchenko PV, Silberstein L, Scadden DT (2014) Bayesian approach to single-cell differential expression analysis. Nat Methods 11:740–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL (2014) The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32:381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buettner F, Natarajan KN, Casale FP, Proserpio V, Scialdone A, Theis FJ, Teichmann SA, Marioni JC, Stegle O (2015) Computational analysis of cell-to-cell heterogeneity in single-cell RNA-Sequencing data reveals hidden subpopulations of cells. Nat Biotechnol 33:155–160 [DOI] [PubMed] [Google Scholar]
- 23.Xu C, Su Z (2015) Identification of cell types from single-cell transcriptomes using a novel clustering method. Bioinformatics 31:1974–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grun D, Lyubimova A, Kester L, Wiebrands K, Basak O, Sasaki N, Clevers H, van Oudenaarden A (2015) Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 525:251–255 [DOI] [PubMed] [Google Scholar]
- 25.Satija R, Farrell JA, Gennert D, Schier AF, Regev A (2015) Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33:495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katayama S, Tohonen V, Linnarsson S, Kere J (2013) SAMstrt: statistical test for differential expression in single-cell transcriptome with spike-in normalization. Bioinformatics 29:2943–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bendall SC, Davis KL, Amirel AD, Tadmor MD, Simonds EF, Chen TJ, Shenfeld DK, Nolan GP, Pe’er D (2014) Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell 157:714–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marco E, Karp RL, Guo G, Robson P, Hart AH, Trippa L, Yuan GC (2014) Bifurcation analysis of single-cell gene expression data reveals epigenetic landscape. Proc Natl Acad Sci U S A 111:E5643–E5650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shin J, Berg DA, Zhu YH, Shin JY, Song J, Bonaguidi MA, Enikolopov G, Nauen DW, Christian KM, Ming GL et al. (2015) Single-cell RNA-seq with waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell 17:360–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo M, Bao EL, Wagner M, Whitsett JA, Xu Y (2017) SLICE: determining cell differentiation and lineage based on single cell entropy. Nucleic Acids Res 45(7):e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo M, Wang H, Potter SS, Whitsett JA, Xu Y (2015) SINCERA: a pipeline for single-cell RNA-seq profiling analysis. PLoS Comput Biol 11:e1004575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du Y, Guo M, Whitsett JA, Xu Y (2015) ‘LungGENS’: a web-based tool for mapping single-cell gene expression in the developing lung. Thorax 70:1092–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu Y, Mizuno T, Sridharan A, Du Y, Guo M, Tang J, Wikenheiser-Brokamp KA, Perl A-KT, Funari VA, Gokey JJ et al. (2016) Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight 1:e90558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T et al. (2015) Orchestrating high-throughput genomic analysis with bioconductor. Nat Methods 12:115–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sing T, Sander O, Beerenwinkel N, Lengauer T (2005) ROCR: visualizing classifier performance in R. Bioinformatics 21:3940–3941 [DOI] [PubMed] [Google Scholar]
- 36.Kolde R, Laur S, Adler P, Vilo J (2012) Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics 28:573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lebre S (2009) Inferring dynamic genetic networks with low order independencies. Stat Appl Genet Mol Biol 8:Article 9 [DOI] [PubMed] [Google Scholar]
- 38.Csardi G, Nepusz T (2006) The igraph software package for complex network research. InterJ Comp Syst 1695:1–9 [Google Scholar]
- 39.Wickham H (2009) ggplot2: elegant graphics for data analysis. Springer, New York, NY [Google Scholar]
- 40.Wickham H (2011) The split-apply-combine strategy for data analysis. J Stat Softw 40:1–29 [Google Scholar]
- 41.Zeileis A, Grothendieck G (2005) zoo: S3 infrastructure for regular and irregular time series. J Stat Softw 14 10.18637/jss.v014.i06 [DOI] [Google Scholar]
- 42.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ward JH Jr (1963) Hierarchical grouping to optimize an objective function. J Am Stat Assoc 58:236–244 [Google Scholar]
- 48.Monti S, Tamayo P, Mesirov JP, Golub TR (2003) Consensus clustering: a resampling-based method for class discovery and visualization of gene expression microarray data. Mach Learn 52:91–118 [Google Scholar]
- 49.Wilkerson MD, Hayes DN (2010) ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 26:1572–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tseng GC, Wong WH (2005) Tight clustering: a resampling-based approach for identifying stable and tight patterns in data. Biometrics 61:10–16 [DOI] [PubMed] [Google Scholar]
- 51.Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57 [DOI] [PubMed] [Google Scholar]
- 52.Chen J, Bardes EE, Aronow BJ, Jegga AG (2009) ToppGene suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37:W305–W311 [DOI] [PMC free article] [PubMed] [Google Scholar]