SUMMARY
To ensure normal development and maintenance of homeostasis, the extensive developmental potential of stem cells must be functionally distinguished from the limited developmental potential of transit amplifying cells. Yet the mechanisms that restrict the developmental potential of transit amplifying cells are poorly understood. Here we show that the evolutionarily conserved transcription factor dFezf/Earmuff (Erm) functions cell-autonomously to maintain the restricted developmental potential of the intermediate neural progenitors generated by type II neuroblasts in Drosophila larval brains. Although erm mutant intermediate neural progenitors are correctly specified and show normal apical-basal cortical polarity, they can dedifferentiate back into a neuroblast state, functionally indistinguishable from normal type II neuroblasts. Erm restricts the potential of intermediate neural progenitors by acti-vating Prospero to limit proliferation and by antagonizing Notch signaling to prevent dedifferentiation. We conclude that Erm dependence functionally distinguishes intermediate neural progenitors from neuroblasts in the Drosophila larval brain, balancing neurogenesis with stem cell maintenance.
INTRODUCTION
Tissue development and homeostasis often require stem cells to transiently expand the progenitor pool by producing transit amplifying cells. Yet the developmental potential of transit amplifying cells must be tightly restricted to ensure generation of differentiated progeny and to prevent unrestrained proliferation that might lead to tumorigenesis (Morrison and Kimble, 2006; Pontious et al., 2008; Vescovi et al., 2006). Transit amplifying cells are defined by their limited developmental capacity, a feature specified during fate determination (Farkas et al., 2008; Hodge et al., 2008; Sessa et al., 2008). It is unknown whether an active mechanism is required to maintain restricted developmental potential in transit amplifying cells after specification. Here we use intermediate neural progenitors (INPs) in developing Drosophila larval brains as a genetic model to investigate how restricted developmental potential is regulated in transit amplifying cells.
A fly larval brain hemisphere contains eight type II neuroblasts that undergo repeated asymmetric divisions to self-renew and to generate immature INPs (Figure 1A) (Bello et al., 2008; Boone and Doe, 2008; Bowman et al., 2008). Immature INPs are unstable in nature and are mitotically inactive, and they lack the expression of Deadpan (Dpn) and Asense (Ase) (Figure S1A). Immature INPs commit to the INP fate through maturation, a differentiation process necessary for specification of the INP identity (Figure 1A). INPs express Dpn and Ase, and undergo 8-10 rounds of asymmetric divisions to self-renew and to produce ganglion mother cells (GMCs) that typically generate two neurons (Figure S1A) (Bello et al., 2008; Boone and Doe, 2008; Bowman et al., 2008). While 5-6 immature INPs and 1-2 young INPs are always in direct contact with their parental neuroblasts, the older INPs become progressively displaced from their parental neuroblasts over time (Bowman et al., 2008).
Figure 1. erm Mutant Brains Show Ectopic Type II Neuroblasts.
(A) A summary of the type II neuroblast lineage.
(B-H) While wild-type (+/+) and erm mutant brains contained a similar number of type I neuroblasts (Dpn+CycE+Ase+EdU+; white arrows), erm mutant brains contained ectopic type II neuroblasts (Dpn+CycE+Ase-EdU+; white arrowheads). In (H), wild-type brains contained 85 ± 5.2 type I neuroblasts and 8.0 ± 0 type II neuroblasts, whereas erm mutant brains contained 83.7 ± 6.4 type I neruoblasts and 159 ± 19.7 type II neuroblasts. Scale bar, 20 μm.
(I and J) In erm mutant brains expressing GFP driven by Ase-Gal4, Prospero (Pros) always colocalized with Numb (Nb) in metaphase type I neuroblasts (GFP+; white circle), but never in type II neuroblasts (GFP-; white circle). Scale bar, 2 μm.
(K and L) erm mutant type I neuroblast clones (white circle) always contained a single neuroblast (white arrow), but erm mutant type II neuroblast clones (white circle) always contained multiple neuroblasts (white arrowheads).
During asymmetric divisions of type II neuroblasts, the basal proteins Brain tumor and Numb are exclusively segregated into immature INPs, and function cooperatively, but nonredundantly, to ensure that immature INPs undergo maturation and commit to the INP fate (Boone and Doe, 2008; Bowman et al., 2008). brain tumor or numb mutant type II neuroblasts generate immature INPs that fail to mature and do not commit to the INP fate. Instead, brain tumor or numb mutant immature INPs adopt their parental neuroblast fate, leading to supernumerary type II neuroblasts. Thus, brain tumor and numb specify the INP fate, and the ectopic expansion of type II neuroblasts in these mutant genetic backgrounds occurs due to failure to properly specify the INP fate. Although Brain tumor is also asymmetrically segregated into GMCs during asymmetric divisions of INPs, the mosaic clones in brain tumor mutant INPs contain only differentiated neurons (Bowman et al., 2008). This result indicates that Brain tumor is dispensable for maintaining the restricted developmental potential of INPs. How restricted developmental potential is maintained in INPs is currently unknown.
To identify genes that regulate self-renewal of neuroblasts, we conducted a genetic screen for mutants exhibiting ectopic larval brain neuroblasts (C.-Y.L. and C.Q. Doe, unpublished data). One mutation, l(2)5138, specifically resulted in massive expansion of neuroblasts in the brain but did not affect neuroblasts on the ventral nerve cord (Figures S1B-S1D). We mapped the l(2)5138 mutation to the 22B4-7 chromosomal interval that contains the earmuff (erm) gene (Pfeiffer et al., 2008). The erm transcripts are first detected at embryonic stage 4-6 in the specific domain preceding formation of the embryonic brain and remain highly expressed in the brain throughout development (Chintapalli et al., 2007; Pfeiffer et al., 2008). Here, we report that Erm functions to restrict the developmental potential of INPs by promoting Prospero-dependent termination of proliferation and suppressing Notch-mediated dedifferentiation. By restricting their developmental potential, Erm ensures that INPs generate only differentiated neurons during Drosophila neurogenesis.
RESULTS
Earmuff Prevents Abnormal Expansion of Neural Progenitors in Type II Neuroblast Lineages
All neuroblasts in l(2)5138 homozygous mutant brains were proliferative, expressed all known neuroblast markers, and lacked neuronal and glial markers (Figures 1B-1G; Figures S1B-S1D; data not shown). We mapped the l(2)5138 mutation to the erm gene, which encodes a homolog of the vertebrate Forebrain embryonic zinc-finger family (Fezf) transcription factors (Hashimoto et al., 2000; Matsuo-Takasaki et al., 2000). The l(2)5138 mutants contained a single A→T nucleotide change in the erm coding region, leading to the substitution of a leucine for a conserved histidine in the third C2H2 zinc-finger domain (data not shown). Consistent with its predicted molecular function, ectopic expression of Erm transgenic proteins tagged with a HA epitope at the amino- or carboxyl-terminus driven by neuroblast-specific Wor-Gal4 was detected in the nuclei of neuroblasts (data not shown). However, the expression of the HA-tagged Erm transgenic protein bearing the identical leucine-tohistidine substitution as in the l(2)5138 mutant was undetectable, suggesting that the mutant Erm protein is unstable (data not presented). We conclude that l(2)5138 is a mutant allele of erm.
To determine whether erm mutant brains have ectopic type I and/or type II neuroblasts, we analyzed the expression pattern of Ase and Prospero (Pros), which are only expressed in type I neuroblasts (Figure S1A) (Bello et al., 2008; Boone and Doe, 2008; Bowman et al., 2008). We found that erm mutant brains contained over 20-fold more type II neuroblasts (Dpn+Ase-) than wild-type brains, with no significant change in the number of type I neuroblasts (Dpn+Ase+) (Figures 1F-1H). Next, we analyzed the localization of Prospero in mitotic neuroblasts in larval brains expressing GFP induced by Ase-Gal4 (Ase > GFP), which mimicked the expression pattern of the endogenous Ase protein (Bowman et al., 2008). In erm mutant larval brains, all mitotic type I neuroblasts (GFP+) showed formation of basal Prospero crescents, but none of the mitotic type II neuroblasts (GFP-) showed the expression of Prospero (Figures 1I and 1J; n = 20). Furthermore, GFP-marked erm mutant type II neuroblast clones consistently contained multiple type II neuroblasts, whereas erm mutant type I neuroblast clones always contained single type I neuroblasts and neurons (Figures 1K and 1L). We conclude that erm mutant brains exhibit an abnormal expansion of type II neuroblasts.
erm Regulates the Developmental Potential of INPs
To determine the cellular origin of ectopic type II neuroblasts in erm mutant brains, we analyzed the identity of cells in the GFP-marked clones derived from wild-type or erm mutant type II neuroblasts using specific cell fate markers. At 30 hr after clone induction, wild-type and erm mutant neuroblast clones appeared indistinguishable, containing single parental neuroblasts (Dpn+Ase-; ≥10 μm) in direct contact with 5-6 immature INPs (Dpn-Ase-), while most of the INPs (Dpn+Ase+; >6 μm) were 1 cell or more away from the parental neuroblasts (Figures 2A and 2B). At 48 hr after clone induction, the overall size of both wild-type and erm mutant neuroblast clones increased significantly due to an increase in cell number, reflecting continuous asymmetric divisions of the parental neuroblasts. In both wild-type and erm mutant clones, the parental neuroblasts remained surrounded by 5-6 immature INPs, while INPs and differentiated neurons (Dpn-Ase-Pros+) were found several cells away from the parental neuroblasts (Figures 2C-2F; Figures S2A-S2F). However, erm mutant clones contained fewer INPs (16 ± 4; n = 10 brains) than the wild-type clones (21 ± 4; n = 10 brains). Importantly, erm mutant clones consistently contained 4-6 smaller ectopic type II neuroblasts (Dpn+Ase- 6-8 αm in diameter) (Figure 2F; Figure S2F). Thus, Erm is dispensable for both the generation and maturation of immature INPs.
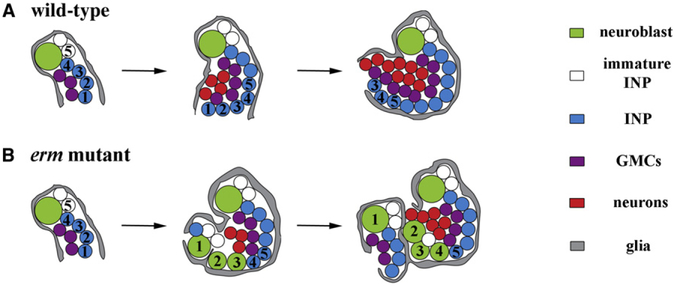
Figure 2. Erm Maintains the Limited Developmental Potential of INPs.
(A and B) At 30 hr after clone induction, both wild-type (+/+) and erm mutant neuroblast clones (yellow circles) contained a single parental neuroblast (white arrows) directly surrounded by immature INPs (white arrowheads) and 1–2 young INPs (Dpn+Ase+).
(C-F) At 48 hr after clone induction, wild-type (+/+) neuroblast clones (yellow circles) contained a single parental neuroblast (white arrows) in direct contact with immature INPs (white arrowheads) and young INPs (Dpn+Ase+). Older INPs were away from their parental neuroblasts and were surrounded by GMCs (white asterisks) and neurons(Dpn_Ase_). In contrast, the erm mutant clones contained ectopic type II neuroblast-like cells ([F], yellow arrows) further from the parental neuroblasts than most INPs and neurons. A summary diagram is shown below.
(G) R9D11-Gal4 (Erm-Ga14) was undetectable in type II neuroblasts (white arrow) and immature INPs (white arrowheads), but was clearly detected in INPs. All scale bars, 10 μm.
Ectopic type II neuroblasts in 48 hr erm mutant clones were always several cells away from the parental neuroblasts (Figure 2F; Figure S2F). This result strongly suggests that ectopic type II neuroblasts in erm mutant clones likely originate from INPs and Erm likely functions in INPs. However, we could not assess the spatial expression pattern of the endogenous Erm protein in larval brains due to lack of a specific antibody and low signals by fluorescent RNA in situ (data not shown). Alternatively, we analyzed the expression of the R9D series of Gal4 transgenes in which Gal4 is expressed under the control of overlapping erm promoter fragments (Pfeiffer et al., 2008). The expression of R9D11-Gal4 was clearly detected in INPs, but was undetectable in type II neuroblasts and immature INPs even when two copies of the UAS-mCD8-GFP transgenes were driven by two copies of R9D11-Gal4 at 32°C for 72 hr after larval hatching (Figure 2G; Figure S2G). Consistently, the expression of Erm-Gal4 was virtually undetectable in brain tumor mutant brains that contain thousands of type II neuroblasts and immature INPs (Figure S2H). While the expression of UAS-erm induced by the neuroblast-specific Wor-Gal4 driver led to premature loss of type II neuroblasts, expression of UAS-erm driven by Erm-Gal4 failed to exert any effect on type II neuroblasts (data not shown). Importantly, targeted expression of the fly Erm or mouse Fezf1 or Fezf2 transgenic protein driven by R9D11-Gal4 restored the function of Erm and efficiently rescued the ectopic neuroblast phenotype in erm mutant brains (Figures S2I-S2L). Therefore, R9D11-Gal4 (Erm-Gal4) contains the enhancer element sufficient to restore the Erm function in INPs leading to suppression of ectopic type II neuroblasts in erm mutant brains.
erm Mutant INPs Dedifferentiate Back into Type II Neuroblasts
Mutant clonal analyses and overexpression studies strongly suggest that Erm functions to suppress reversion of INPs back into a neuroblast state. Here, we directly tested whether INPs in erm mutant brains can dedifferentiate back into type II neuroblasts. We induced βgal-marked lineage clones originating exclusively from INPs via FRT-mediated recombination. We targeted a short pulse of flipase (FLP) expression in INPs by heat-shocking larvae carrying a UAS-flp transgene under the control of Erm-Gal4 and tub-Gal80ts at 30°C for 1 hr (see Experimental Procedures for details). At 72 hr after heat shock, INP clones in wild-type brains contained only differentiated neurons (Dpn-Ase-) (Figure 3A). In contrast, INP clones in erm mutant brains contained one or more type II neuroblasts as well as immature INPs, INPs, GMCs, and neurons (Figures 3B-3C). This result indicates that while INPs in wild-type larval brains can only give rise to neurons, INPs in erm mutant brains can dedifferentiate into type II neuroblasts that can give rise to all cell types found in a normal type II neuroblast lineage. We conclude that Erm functions to maintain the restricted developmental potential of INPs and prevents them from dedifferentiating back into a neuroblast state.
Figure 3. erm Suppresses the Dedifferentiation of INPs.
(A-C) A wild-type (+/+) INP only generated neurons (Dpn_Ase_), but an erm mutant INP generated dedifferentiated neuroblasts (white arrows), immature INPs (white arrowheads) and INPs (Dpn+Ase+), GMCs ([B], white asterisks), and neurons ([C], white asterisks). A lineage clone is circled in yellow, and a summary diagram is shown on the right.
(D-I) Similar to wild-type type II neuroblasts, ectopic type II neuroblasts in erm mutant brains lost incorporated EdU (neuroblasts, white arrows; INPs, whitearrow-heads) (D and E), did not express Pros-Gal4 and Erm-Gal4 (type I neuroblast, white arrowheads; type II neuroblasts, white arrows) (F and G), and established ectopic neuroblast lineages (white asterisks) surrounded by glial membrane (H and I). All scale bars, 10 μm.
We further assessed whether the dedifferentiated type II neuroblasts in erm mutant brains displayed multiple functional characteristics of normal type II neuroblasts.
Apical-Basal Cell Polarity
All mitotic type II neuroblasts in wild-type and erm mutant brains showed normal establishment and maintenance of cortical polarity by asymmetrically localizing and segregating atypical Protein Kinase C (aPKC), Pins, Miranda, and Numb (data not shown).
Proliferation Profile
All wild-type and erm mutant type II neuroblasts could be labeled with a 3 hr pulse of the thymidine analog EdU (Figures 1F’ and 1FG’), and incorporated EdU can be chased into INPs following a 12 hr EdU-free chase (Figures 3D and 3E).
prospero and earmuff Promoter Activity
While all type I neuroblasts in wild-type and erm mutant brains expressed Pros-Gal4 but lacked Erm-Gal4 expression, none of the type II neuroblasts in wild-type and erm mutant brains showed detectable expression of Pros-Gal4 or Erm-Gal4 (Figures 3F and 3G; data not shown).
Formation of Glial Chambers
Individual neuroblast lineages are surrounded by the cortex glial membrane forming distinct chambers (Pereanu et al., 2005). A wild-type brain hemisphere contained eight glial chambers encapsulating eight individual type II neuroblast lineages (Figure 3H). In contrast, an erm mutant brain hemisphere contained more than 50 glial chambers, each containing one or more type II neuroblasts and their presumptive progeny (Figure 3I).
Taken together, INPs in erm mutant brains dedifferentiate back into apparently normal neuroblasts that can establish ectopic type II neuroblast lineages.
erm Mutant INPs Exhibit Normal Apical-Basal Cortical Polarity
Dysregulation of apical-basal polarity can lead to failure in differentiation and result in ectopic neuroblasts at the expense of GMC formation (Betschinger et al., 2006; Lee et al., 2006a, 2006b, 2006c; Wang et al., 2006). To determine whether the dedifferentiation of INPs in erm mutant brains might be due to defects in cortical polarity, we assayed apical-basal polarity by examining the localization of aPKC, Miranda, Prospero, and Numb in larval brains expressing GFP driven by Ase-GAL4 (Ase > GFP). Mitotic INPs (GFP+) in erm mutant brains showed the same asymmetric localization of aPKC, Miranda, Prospero, and Numb as in wild-type brains (Figures 4A and 4B; data not shown). Thus, we conclude that INPs in erm mutant brains dedifferentiate while displaying normal cortical polarity.
Figure 4. erm Mutant INPs Show Normal Apical-Basal Polarity.
(A and B) Metaphase INPs in erm mutant brains expressing GFP induced by Ase-Gal4 showed asymmetric localization of aPKC, Miranda (Mira), Pros, and Numb (Nb). The scale bar, 5 μm.
Erm Restricts Proliferation by Activating Prospero-Dependent Cell Cycle Exit
To determine how Erm maintains the restricted developmental potential of INPs, we performed microarray analyses and found that prospero mRNA was drastically reduced in erm mutant brains compared to the control brains (M.W. and C.-Y.L., unpublished data). We confirmed that the relative level of prospero mRNA was indeed reduced by 60%-70% in erm mutant brain extracts by using real-time PCR (data not shown). These data supported that Erm is necessary for proper transcription of prospero, and prompted us to test if overexpression of Erm might be sufficient to induce ectopic Prospero expression. We induced a short pulse of Erm expression in brain neuroblasts by shifting larvae carrying a UAS-erm transgene under the control of Wor-Gal4 and tub-Gal80ts to from 25°C to 30°C. A 3.5 hr pulse of Erm expression was sufficient to induce nuclear localization of Prospero in larval brain neuroblasts (Figure 5A). Consistent with nuclear Prospero promoting termination of neuroblast proliferation, ectopic expression of Erm induced by Wor-Gal4 resulted in decreased neuroblasts compared to wild-type brains (Figure 5B). Thus, we conclude that overexpression of Erm can restrict neuroblast proliferation by triggering nuclear localization of Pros.
Figure 5. Erm Restricts the Proliferation of INPs by Promoting Nuclear Prospero.
(A) A 3.5 hr pulse of Erm expression induced by Wor-Gal4 was sufficient to trigger Pros localization in neuroblast nuclei (white arrows).
(B) Ectopic expression of Erm (57.9 ± 8.6) or Pros (17.4 ± 4.4 neuroblasts) driven by Wor-Gal4 was sufficient to terminate neuroblast proliferation prematurely (98.0 ± 8.4 neuroblasts in wild-type brains).
(C and D) Ectopic expression of Erm induced by Erm-Gal4 triggered a significant increased in INPs that exhibited nuclear Pros (white arrows), likely leading them to exit cell cycle prematurely and resulting in some type II neuroblasts (white circle) surrounded only by neurons. Scale bar, 10 μm.
(E and F) Overexpression of Pros induced by Erm-Gal4 suppressed ectopic neuroblasts and restored neuronal differentiation in erm mutant brains. Scale bar, 20 μm.
(G and H) pros mutant type I neuroblast clones contained ectopic neuroblasts (white arrows). pros mutant type II neuroblast clones contained a single type II neuroblast (white arrow) but showed dramatic overproliferation of INPs (white arrowheads).
(I) Overexpression of Erm failed to suppress overproliferation of INPs in pros mutant type II neuroblast clones. Scale bar, 10 μm.
Our data suggest that Erm might restrict the developmental potential of INPs in part by limiting their proliferation by activating Prospero-dependent cell cycle exit. If so, we predict that overexpression of Erm should induce ectopic nuclear Prospero in INPs and overexpression of Prospero should suppress ectopic neuroblasts in erm mutant brains. In wild-type brains, 9.6% of INPs (32/325) showed nuclear localization of Prospero. However, overexpression of Erm driven by Erm-Gal4 led to nuclear localization of Prospero in 41.5% of INPs (105/253), likely restricting their proliferation potential and resulting in some parental type II neuroblasts surrounded only by differentiated neurons (Figures 5C and 5D). Importantly, ectopic expression of Prospero induced by Erm-Gal4 efficiently suppressed ectopic neuroblasts and restored neuronal differentiation in erm mutant brains (Figures 5E and 5F). Thus, Erm likely restricts the proliferation of INPs by promoting nuclear localization of Prospero. To confirm that Prospero indeed functions downstream of Erm to restrict the proliferation of INPs, we performed genetic epistatic analyses. Consistent with previously published results, prospero mutant type I neuroblast clones contained ectopic type I neuroblasts (Figure 5G) (Bowman et al., 2008). In contrast, prospero mutant type II neuroblast clones exhibited accumulation of ectopic INPs while maintaining single parental neuroblasts (Figure 5H). Furthermore, overexpression of Erm failed to suppress ectopic INPs in prospero mutant type II neuroblast clones, consistent with Prospero functioning downstream of Erm (Figure 5I). These results indicate that blocking differentiation is not sufficient to trigger the dedifferentiation of INPs back into type II neuroblasts. Thus, Erm’s restriction on the proliferation of INPs is dependent on Prospero function, but its suppression of the dedifferentiation of INPs is independent of Prospero.
Erm Suppresses Dedifferentiation by Antagonizing Notch Signaling
Previous studies showed that overexpression of constitutively active Notch (Notchintra) in both type I and II neuroblasts is sufficient to trigger ectopic neuroblasts (Bowman et al., 2008; Wang et al., 2006). Here, we tested whether Erm suppresses the dedifferentiation of INPs by inhibiting Notch signaling. Indeed, knockdown of Notch function by RNAi in erm mutant brains led to a dramatic reduction in ectopic type II neuroblasts compared to erm mutant brains alone (Figures 6A and 6B). Complementary, ectopic expression of constitutively active Notch (Notchintra) induced by Erm-Gal4 transforms INPs into ectopic type II neuroblasts (Figure 6C). Thus, reduced Notch function suppresses the dedifferentiation of INPs in erm mutant brains whereas ectopic activation of Notch induces the dedifferentiation of INPs. We next tested if Erm suppresses the dedifferentiation of INPs by antagonizing a Notch-activated mechanism. Coexpression of Erm under the control of Erm-Gal4 is sufficient to suppress ectopic neuroblasts induced by the expression of Notchintra (Figure 6D). Thus, we conclude that Erm can suppress the dedifferentiation of INPs by negatively regulating a Notch-activated signaling mechanism.
Figure 6. Erm Suppresses the Dedifferentiation of INPs by Negatively Regulating Notch Signaling.
(A and B) Knocking down Notch function by RNAi suppressed ectopic neuroblasts (white arrows) in erm mutant brains.
(C and D) Ectopic expression of Erm underthe control of Erm-Gal4 suppressed ectopic neuroblasts induced by constitutive activation of Notch signaling. Scale bar, 20 μm.
DISCUSSION
The limited developmental potential of transit amplifying cells is generally thought to be specified during fate determination (Farkas et al., 2008; Hodge et al., 2008; Sessa et al., 2008). In this study, we report a mechanism that actively maintains the restricted developmental potential of transit amplifying cells after specification of their identity. We show that the evolutionarily conserved transcription factor Erm/Fezf functions to maintain the restricted developmental potential of INPs by limiting their proliferation potential and suppressing their dedifferentiation capacity (Figure 7). Combining proper specification of the transit amplifying cell identity and active maintenance of their restricted developmental potential ensures the generation of differentiated progeny and prevents aberrant expansion of stem cells.
Figure 7. erm Maintains the Restricted Developmental Potential of INPs.
(A) Wild-type INPs undergo limited rounds of asymmetric divisions to generate neurons prior to exiting from the cell cycle, and they remain in the same glial chamber as their parental type II neuroblasts.
(B) Some erm mutant INPs fail to terminate proliferation and dedifferentiate back into their parental type II neuroblast fate. These dedifferentiated neuroblasts can establish ectopic type II neuroblast lineages and form ectopic glial chambers.
The lineage clones derived from single INPs in erm1/erm2 mutant brains contain dedifferentiated neuroblasts, immature INPs, INPs, GMCs, and neurons (Figures 3B and 3C). Several mechanisms could lead to the diversity of cells within the clones. First, INPs in erm mutant brains might generate GMCs and neurons initially due to the presence of maternally deposited Erm. However, erm transcripts are undetectable in both adult male and female germlines by microarray analyses and in stage 1-3 embryos by RNA in situ (Chintapalli et al., 2007; http://flybase.org/reports/FBgn0031375.html; data not shown). Furthermore, the erm1/erm2 allelic combination resulted in little to no zygotic Erm in the brain because the erm1 mutation likely leads to the production of an unstable Erm protein, whereas the erm2 mutation deletes the entire erm open reading frame (data not shown). Additionally, the ectopic neuroblast phenotype in erm1/erm2 mutant brains can be observed as early as 36-48 hr after larval hatching (data not presented). Thus, generation of GMCs and differentiated neurons by INPs in erm1/erm2 mutant brains is unlikely due to the maternal effect. Alternatively, erm may promote GMC differentiation in the type II neuroblast lineage, and in erm mutant brains, GMCs might dedifferentiate back into neuroblasts. If so, we would predict an ectopic accumulation of INPs in similarly staged mosaic clones derived from erm mutant type II neuroblasts as compared to wild-type clones. However, 48 hr erm mutant single neuroblast clones consistently contained fewer INPs when compared to the wild-type clones (Figures 2C-2F). In addition, blocking GMC differentiation by removing Prospero function resulted in ectopic accumulation of INPs but did not lead to ectopic neuroblast formation (Figure 5H). Therefore, the diversity of cells within erm mutant clones is also unlikely due to blocking GMC differentiation. We favor the interpretation that erm mutant INPs dedifferentiate into apparently normal neuroblasts that can give rise to all cell types found in a type II neuroblast lineage. Consistently, the dedifferentiated neuroblasts in erm mutant brains exhibited normal cortical polarity and proliferation potential (Figures 3 and 4). Furthermore, the dedifferentiated neuroblasts in erm mutant brains also lost the expression of Pros-Gal4 and Erm-Gal4 and established ectopic type II neuroblast lineages encapsulated by the cortex glial membrane (Figures 3 and 4). Thus, we conclude that Erm likely restricts the developmental potential of INPs by limiting proliferation and suppressing dedifferentiation.
Although mutations in erm, brain tumor, and numb genes all lead to ectopic type II neuroblasts, the proteins appear to regulate INPs at distinct steps in the type II neuroblast lineage (Figure S3). Numb and Brain tumor function cooperatively, but nonredundantly, to ensure that immature INPs undergo maturtion and commit to the INP fate (Boone and Doe, 2008; Bowman et al., 2008). While ectopic expression of Numb induces premature differentiation of type II neuroblasts and immature INPs (J. Haenfler, K.L.G., and C.-Y.L., unpublished data), overexpression of Numb is not sufficient to suppress ectopic neuroblasts in brain tumor mutant brains (H. Komori and C.-Y.L., unpublished data). Thus, Numb likely promotes differentiation of immature INPs whereas Brain tumor likely prevents immature INPs, which are unstable in nature, from adopting their parental neuroblast fate. More studies will be necessary to discern whether ectopic neuroblasts in brain tumor mutant brains arise from dedifferentiation of partially differentiated immature INPs or failure of immature INPs to initiate differentiation. In contrast, immature INPs in erm mutant brains mature into functional INPs that exhibit normal cortical polarity and proliferation potential and can generate GMCs and neurons (Figures 2A-2F, 3D, 3E, and 4; Figure S3). Additionally, overexpression of Brain tumor or Numb in INPs was not sufficient to suppress ectopic neuroblasts in erm mutant brains (data not shown). Finally, lineage clones derived from single INPs in erm mutant brains always contain ectopic type II neuroblasts, multiple immature INPs, INPs, GMCs, and neurons (Figures 3B and 3C). These results indicate that Erm is dispensable for maturation of immature INPs and is not within the genetic hierarchy specifying the INP identity. Instead, Erm maintains the restricted developmental potential of INPs after specification of their identity.
Prospero encodes a homeodomain transcription factor, and nuclear Prospero has been shown to trigger cell cycle exit and GMC differentiation (Choksi et al., 2006; Doe et al., 1991; Maurange et al., 2008). In the wild-type brain, 9.6% of INPs showed nuclear Prospero and were likely undergoing differentiation (data not shown). prospero mutant type II neuroblast clones showed ectopic accumulation of INPs but contained single neuroblasts, indicating that blocking differentiation is not sufficient to trigger the dedifferentiation of INPs (Figure 5H). Thus, Prospero restricts the proliferation potential of INPs but does not suppress dedifferentiation of INPs.
While ectopic expression of Prospero in INPs can restore neuronal differentiation in erm mutant brains, targeted expression of Erm in neuroblasts or INPs was sufficient to induce rapid nuclear localization of Prospero in these cells and terminate their proliferation (Figure 5). In wild-type brains, Prospero is sequestered in a basal crescent by the adaptor protein Miranda in mitotic neural progenitors (Ikeshima-Kataoka et al., 1997; Shen et al., 1997). Interestingly, mitotic neural progenitors including neuroblasts and INPs transiently overexpressing Erm also showed basal localization and segregation of Miranda and Prospero (data not shown). As such, Erm likely restricts the proliferation potential of INPs by indirectly promoting nuclear localization of Prospero. Therefore, Prospero does not localize in the nuclei of mitotically active INPs, which express Miranda, but does localize in the nuclei of GMCs that do not express Miranda.
How does Erm suppress the dedifferentiation of INPs? Our results show that reduced Notch function can efficiently suppress ectopic neuroblasts in erm mutant brains while constitutive activation of Notch signaling induced the dedifferentiation of INPs (Figures 6A–6C). Importantly, coexpression of Erm is sufficient to suppress the dedifferentiation of INPs triggered by expression of constitutively active Notchintra (Figure 6D). Together, these results strongly suggest that Erm prevents the dedifferentiation of INPs by antagonizing a Notch-activated mechanism through interfering with the assembly of the Notch transcriptional activator complex or inhibiting the expression of Notch targets. Intriguingly, the amino terminus of all Fezf proteins contains an engrailed homology 1 domain. This domain can mediate direct interaction with the conserved transcriptional corepressor Groucho that can function as a corepressor of Notch signaling (Cinnamon and Paroush, 2008; Copley, 2005; Jeong et al., 2006; Levkowitz et al., 2003; Shimizu and Hibi, 2009). Additional experiments will be needed to discern how Erm antagonizes Notch-activated dedifferentiation of INPs.
EXPERIMENTAL PROCEDURES
Fly Genetics and Transgenes
A total of six erm alleles were recovered from EMS mutagenesis following a standard protocol. erm2 was generated by a FRT-based high-resolution deletion method and verified by PCR (Parks et al., 2004). The cDNA for CG31670 was obtained from the Drosophila Genome Resource Center, sequenced, and cloned into the pUAST-HA vector for germline transformation. Mouse fezf1 and fezf2 cDNAs were sequenced (M. Hibi) and cloned into the pUAST-HA vector for germline transformation. Drosophila cultures were kept at 25°C on standard cornmeal food. Other mutant alleles and transgenes used in this study include brat11 (Lee et al., 2006c), pros17, FRT82B (Lee et al., 2006c), aPKCk06403 (Lee et al., 2006b), pins62 (Lee et al., 2006b), UAS-pros (Hirata et al., 1995), Wor-gal4 (Lee et al., 2006b), Ase-gal4 (Zhu et al., 2006), and R9D-Gal4 lines (Pfeiffer et al., 2008). The UAS-NotchRNAi lines were obtained from the Vienna Drosophila Resource Center. Oregon R, elav-gal4 (C155), hs-flp, UAS-mCD8-GFP, FRT40A, tub-gal80, FRT82B, hs-flp(F38), act-FRT-Stop-FRT-lacZ, UAS-flp, tub-GAL80ts, UAS-dcr-2, UAS-Notchintra, Repo-Gal4 flies were obtained from Bloomington Drosophila Stock Center.
Immunofluorescent Staining and Antibodies
Antibody staining was performed as previously described (Lee et al., 2006b). The rabbit Ase antibody was raised against a previously described synthetic peptide (Brand and Perrimon, 1993). Other antibodies used in this study include guinea pig Ase (1:100; J. Knoblich), rat Wor (1:1), rat Dpn (1:1), guinea pig Dpn (1:2500, J. Skeath), mouse Pros (1:100), rat Mira (1:100); guinea pig Mira (1:400), guinea pig Numb (1:3000, J. Skeath); rat Pins (1:500), rabbit Scrib (1:2500), mouse Elav(1:50, DSHB), mouse Dlg (1:100, DSHB), mouse Repo (1:50, DSHB), mouse BrdU (1:50, Roche), rabbit β-gal (1:1000, ICN/Cappel), rat μ-Tub (1:100, Sigma), rat mCD8 (1:100, Caltag), rabbit GFP (1:1000, Torrey-pine), mouse HA (1:1000, Covance), rat HA (1:2000, Roche). Secondary antibodies were from Molecular Probes (details are available upon request). The confocal images were acquired on a Leica SP5 scanning confocal microscope with AOBS.
Edu Pulse-Chase
Larvae were aged for 72 hr after hatching, and were pulse labeled for 3 hr by feeding on the Kankel-White media containing 50 μg/ml EdU (5-ethynyl-2’deoxyuridine) (Lee et al., 2006c). Half of the larvaewere processed for staining immediately following the pulse; remaining larvae were transferred to standard media for a 12 hr EdU-free chase. Larvae were dissected and processed for antibody staining as previously described (Lee et al., 2006b). Incorporated EdU was detected by Click-iT fluorescent dye azide reaction as described in the Click-iT product literature (Invitrogen).
Lineage Clonal Analysis
We initially performed genetic clonal analyses of INPs using Ase-Gal4 by crossing erm1, Actin-FRT-Stop-FRT-lacZ/CyO, Actin-GFP flies to erm2, Ase-Gal4/CyO, Actin-GFP; UAS-flp, tub-Gal80ts flies. At 24 hr after hatching, erm1/erm2 larvae were shifted to 31ºC for 48 hr to inactivate Gal80ts, allowing FRT-mediated recombination to induce permanently marked lineage clones. The expression level of Ase-Gal4 is very low (Bowman et al., 2008), allowing us to induce genetic clones at a very low frequency. However, due to the prolonged incubation time atthe nonpermissive temperature, clonesderived from two neighboring INPs sometimes became overlapped, resulting in appearance of a “large” clone. We repeated this experiment by using Erm-Gal4, whose expression level was significantly higher compared to Ase-Gal4 (M.W. and C.-Y.L., data not shown). We crossed erm1, Actin-FRT-Stop-FRT-lacZ/CyO, Actin-GFP; Erm-Gal4 flies to erm2/CyO, Actin-GFP; UAS-flp, tub-Gal80ts flies. At 24 hr after hatching, erm1/erm2 larvae were shifted to 31ºC for 1 hr to induce positively marked genetic clones derived from single INP. Larvae were returned back to 25ºC for 72 hr prior to processing larval brains for antibody staining.
Mutant Clonal Analyses
We induced mosaic clones derived from erm1 and pros17 mutant neuroblasts by following a previously established protocol (Lee et al., 2006c; Lee and Luo, 2001).
Overexpression of Notchintra
Overexpression of Notchintra in INPs in larval brains was accomplished by crossing UAS-Notchintra/CyO, Actin-GFP; tub-Gal80ts flies to Erm-Gal4 flies. GFP- larvae were allowed to hatch at 25ºC, and were then shifted to 31ºC for 72 hr. Larval brains were dissected and processed for antibody staining. Co-overexpression of Erm and Notchintra was carried out following an identical protocol.
Real-Time PCR
Late third instar larval brains were dissected free of surrounding tissues. Total RNA was extracted following the standard Trizol RNA isolation protocol and cleaned by the QIAGEN RNeasy kit. cDNA was transcribed using First Strand cDNA Synthesis Kit for RT-PCR (AMV) (Roche). Quantitative PCR was performed by using SYBR-green. Resulting data were analyzed by the comparative CT method, and the relative mRNA expression is presented.
Supplementary Material
ACKNOWLEDGMENTS
We thank Chris Q. Doe in whose lab the l(2)51381 was isolated when C.-Y.L was a postdoctoral fellow. We thank Kristin Wildermuth and Caitlin Gamble for the technical assistance in the isolation and the initial phenotypic characterization of the l(2)51381 mutant allele. We thank C.Q. Doe, J.A. Knoblich, J.B. Skeath, M. Hibi, and J. Lin for fly stocks, antibody reagents, cDNAs, and advice with real-time PCR. We thank the Bloomington Drosophila Stock Center and Vienna Drosophila RNAi Center for fly stocks. We thank C.Q. Doe and Sean J. Morrison and the members of the Lee lab for reading the manuscript and providing critical comments. C.-Y.L was initially supported by a Damon Runyon postdoctoral fellowship and is currently supported by Burroughs Wellcome Fund Career Award in the Biomedical Sciences (1006160.01), a Sontag Foundation Distinguished Scientist Award, and the University of Michigan start-up fund.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes three figures and can be found with this article online at doi:10.1016/j.devcel.2009.12.007.
REFERENCES
- Bello BC, Izergina N, Caussinus E, and Reichert H (2008). Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 3, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J, Mechtler K, and Knoblich JA (2006). Asymmetric segregation of the tumor suppressor Brat regulates self-renewal in Drosophila neural stem cells. Cell 124, 1241–1253. [DOI] [PubMed] [Google Scholar]
- Boone JQ, and Doe CQ (2008). Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev. Neuro-biol 68, 1185–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman SK, Rolland V, Betschinger J, Kinsey KA, Emery G, and Kno-blich JA (2008). The tumor suppressors Brat and Numb regulate transiamplifying neuroblast lineages in Drosophila. Dev. Cell 14, 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, and Perrimon N (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, and Dow JAT (2007). Using FlyAtlas to identify better Drosophila models of human disease. Nat. Genet. 39, 715–720. [DOI] [PubMed] [Google Scholar]
- Choksi SP, Southall TD, Bossing T, Edoff K, deWit E, Fischer BE, van Steensel B, Micklem G, and Brand AH (2006). Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev. Cell 11, 775–789. [DOI] [PubMed] [Google Scholar]
- Cinnamon E, and Paroush Z (2008). Context-dependent regulation of Groucho/TLE-mediated repression. Curr. Opin. Genet. Dev. 18, 435–440. [DOI] [PubMed] [Google Scholar]
- Copley RR (2005). The EH1 motif in metazoan transcription factors. BMC Genomics 6, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CQ, Chu-LaGraff Q, Wright DM, and Scott MP (1991). The prospero gene specifies cell fates in the Drosophila central nervous system. Cell 65, 451–464. [DOI] [PubMed] [Google Scholar]
- Farkas LM, Haffner C, Giger T, Khaitovich P, Nowick K, Birchmeier C, Pääbo S, and Huttner WB (2008). Insulinoma-associated 1 has a panneurogenic role and promotes the generation and expansion of basal progenitors in the developing mouse neocortex. Neuron 60, 40–55. [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Yabe T, Hirata T, Shimizu T, Bae Y, Yamanaka Y, Hirano T, and Hibi M (2000). Expression of the zinc finger gene fez-like in zebrafish forebrain. Mech. Dev. 97, 191–195. [DOI] [PubMed] [Google Scholar]
- Hirata J, Nakagoshi H, Nabeshima Y, and Matsuzaki F (1995). Asymmetric segregation of the homeodomain protein Prospero during Drosophila development. Nature 377, 627–630. [DOI] [PubMed] [Google Scholar]
- Hodge RD, Kowalczyk TD, Wolf SA, Encinas JM, Rippey C, Enikolopov G, Kempermann G, and Hevner RF (2008). Intermediate progenitors in adult hippocampal neurogenesis: Tbr2 expression and coordinate regulation of neuronal output. J. Neurosci. 28, 3707–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeshima-Kataoka H, Skeath JB, Nabeshima Y, Doe CQ, and Matsuzaki F (1997). Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature 390, 625–629. [DOI] [PubMed] [Google Scholar]
- Jeong JY, Einhorn Z, Mercurio S, Lee S, Lau B, Mione M, Wilson SW, and Guo S (2006). Neurogenin1 is a determinant of zebrafish basal forebrain dopaminergic neurons and is regulated by the conserved zinc finger protein Tof/Fezl. Proc. Natl. Acad. Sci. USA 103, 5143–5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, and Luo L (2001). Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 24, 251–254. [DOI] [PubMed] [Google Scholar]
- Lee CY, Andersen RO, Cabernard C, Manning L, Tran KD, Lanskey MJ, Bashirullah A, and Doe CQ (2006a). Drosophila Aurora-A kinase inhibits neuroblast self-renewal by regulating aPKC/Numb cortical polarityand spindle orientation. Genes Dev. 20, 3464–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Robinson KJ, and Doe CQ (2006b). Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature 439, 594–598. [DOI] [PubMed] [Google Scholar]
- Lee CY, Wilkinson BD, Siegrist SE, Wharton RP, and Doe CQ (2006c). Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev. Cell 10, 441–449. [DOI] [PubMed] [Google Scholar]
- Levkowitz G, Zeller J, Sirotkin HI, French D, Schilbach S, Hashimoto H, Hibi M, Talbot WS, and Rosenthal A (2003). Zinc finger protein too few controls the development of monoaminergic neurons. Nat. Neurosci. 6, 28–33. [DOI] [PubMed] [Google Scholar]
- Matsuo-Takasaki M, Lim JH, Beanan MJ, Sato SM, and Sargent TD (2000). Cloning and expression of a novel zinc finger gene, Fez, transcribed in the forebrain of Xenopus and mouse embryos. Mech. Dev. 93, 201–204. [DOI] [PubMed] [Google Scholar]
- Maurange C, Cheng L, and Gould AP (2008). Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cell 133, 891–902. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, and Kimble J (2006). Asymmetric and symmetric stem-cell divisions in development and cancer. Nature 441, 1068–1074. [DOI] [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, et al. (2004). Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36, 288–292. [DOI] [PubMed] [Google Scholar]
- Pereanu W, Shy D, and Hartenstein V (2005). Morphogenesis and proliferation of the larval brain glia in Drosophila. Dev. Biol. 283, 191–203. [DOI] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, et al. (2008). Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. USA 105, 9715–9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontious A, Kowalczyk T, Englund C, and Hevner RF (2008). Role of intermediate progenitor cells in cerebral cortex development. Dev. Neurosci. 30, 24–32. [DOI] [PubMed] [Google Scholar]
- Sessa A, Mao CA, Hadjantonakis AK, Klein WH, and Broccoli V (2008). Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron 60, 56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen CP, Jan LY, and Jan YN (1997). Miranda is required for the asymmetric localization of Prospero during mitosis in Drosophila. Cell 90, 449–458. [DOI] [PubMed] [Google Scholar]
- Shimizu T, and Hibi M (2009). Formation and patterning of the forebrain and olfactory system by zinc-finger genes Fezf1 and Fezf2. Dev. Growth Differ. 51, 221–231. [DOI] [PubMed] [Google Scholar]
- Vescovi AL, Galli R, and Reynolds BA (2006). Brain tumour stem cells. Nat. Rev. Cancer 6, 425–436. [DOI] [PubMed] [Google Scholar]
- Wang H, Somers GW, Bashirullah A, Heberlein U, Yu F, and Chia W (2006). Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts. Genes Dev. 20, 3453–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Lin S, Kao CF, Awasaki T, Chiang AS, and Lee T (2006). Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell 127, 409–422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.