Abstract
Background/Aims
We have previously identified a tight junction protein claudin-9 (CLDN9) as an upregulated gene in hepatocellular carcinoma (HCC) through an immunohistochemistry analysis. Here, we explore its function and clinical relevance in human HCC.
Materials and Methods
Stable transfection of the hepatocyte line HL7702 with CLDN9 was confirmed by the real-time polymerase chain reaction (PCR), western blotting, and immunofluorescence. The impact of CLDN9 on the cell invasion and migration was assessed in vitro by a transwell assay and wound-healing experiment. Western blotting was used to determine the activation state of the Tyk2 (tyrosine kinase 2)/Stat3 (signal transducer and activator of transcription 3) pathway. Moreover, we used a Tyk2-RNAi assay to silence the expression of Tyk2 in CLDN9 expressing hepatocytes; subsequently, the impact of the Tyk2/Stat3 signaling pathway on the cell invasion and migration in vitro was assessed by a transwell assay and a wound-healing experiment. Furthermore, an immunohistochemistry method was utilized to explore the expression levels of CLDN9 and p-Stat3 in the HCC tissues and histologically non-neoplastic hepatic tissues.
Results
We confirmed that the expression of CLDN9 significantly enhanced the metastatic ability of hepatocytes in vitro, and the activation of the Stat3 pathway by Tyk2 was an important mechanism by which CLDN9 promoted hepatocyte aggressiveness in HCC.
Conclusion
As an HCC proto-oncogene, CLDN9 affected the Stat3 signaling pathway via Tyk2 and ultimately enhanced the metastatic ability of hepatocytes.
Keywords: hepatocellular carcinoma, tight junction, metastasis, claudin-9, tyrosine kinase 2, signal transducer and activator of transcription 3
INTRODUCTION
Recent studies have revealed that the metastasis of epithelium-derived tumors is accompanied by abnormalities in the tight junction structure and function (1,2). Claudin (CLDN) is a structural protein that forms tight junctions (3). The common viewpoint is that the expression of CLDNs is downregulated in tumors (4,5). Abnormalities in the cell tight junction structure and function caused by the lack of the CLDN expression have been suggested to be essential for the invasiveness and metastasis of epithelium-derived tumors (6,7). For instance, CLDN1 is reported to be a metastasis suppressor in lung adenocarcinoma, and its lack of expression is associated with a poor patient prognosis (8). However, recently, there has been growing evidence showing that the CLDNs expression in human tumor tissues is notably upregulated compared to the adjacent normal tissues (9–11). A study of gene sequence analysis expression showed that CLDN3 and CLDN4 were upregulated in ovarian, breast, prostate, and pancreatic tumors (12,13). Specific expression patterns of CLDNs in various tumor tissues indicate that CLDNs may be used as molecular tumors markers (14,15).
Moreover, CLDNs have been revealed to be involved in the regulation of cell growth, differentiation, invasion, and migration through participating in transduction of cellular signals (16,17). For exanple, CLDN3 increases the malignant potential of colorectal cancer cells as modulators of the epidermal growth factor receptor signaling (18). These pieces of evidence suggest that the expression and function of CLDNs in the occurrence and progression of a tumor may be tissue specific. In a preliminary work, we found that the CLDN9 expression was upregulated in the HCC cell lines, but downregulated in hepatocyte lines, revealing that upregulated CLDN9 expression may have an effect on the tumorigenesis of HCC. A gene chip screening revealed that the CLDN9 overexpression activated the Tyk2/Stat3 signaling pathway. At present, there are no reports on the impact and mechanism of CLDN9 on the metastatic phenotype of hepatocytes. In this study, we used molecular biology and other techniques to study the impact and mechanisms of CLDN9 in hepatocyte metastasis and to identify novel targets for the HCC treatment and control of early metastasis.
MATERIALS AND METHODS
Cell culture
The human embryonic kidney cells 293 (HEK293T), the HCC cell lines (MHCC97H, HepG2, Hep3B, and Huh1), and human hepatocyte line (HL7702) used in this study were procured from Shanghai Cell Bank of the Chinese Academy of Sciences. The cells were cultured in Dulbecco’s modified Eagle’s medium with added 10% fetal bovine serum (FBS) in a humidified incubator containing 5% CO2 at 37°C.
Plasmid construction and transfection
A full-length CLDN9 cDNA fragment (GeneBank accession no. NM 9080) was amplified by Nanjing KeyGen Biotech Co. Ltd. The fragments, digested by XhoI/EcoRI, were ligated into the response plasmid p-EGFP-C1 (+) to create the expression vector p-EGFP-C1/CLDN9. The final construct was confirmed by a direct sequence analysis. The cloned CLDN9 expression vector was amplified and purified according to the current protocol in molecular biology. A total of 5 μg of plasmid were transfected into cells using a SuperFect Transfection Reagent (TaKaRa, Japan). Clones resistant to G418 (Sigma, St. Louis, Missouri, USA) were expanded in culture as a monoclonal population. Cells transfected with the empty vector p-EGFP1-C1 (+) were used as a vector control.
Reverse transcription-quantitative polymerase chain reaction
Total RNA was extracted using a Perfect Pure RNA Cultured Cell kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer’s protocol. Typical reverse transcription-quantitative polymerase chain reaction (RT-qPCR) reactions were conducted as previously described (19). The primer pairs used for CLDN9 and glyceraldehyde phosphate dehydrogenase (GAPDH) were as follows: CLDN9 forward, 5′-CCTTCATCGGCAACAGCATC-3′, and reverse, 5′-CGTACACCTTGCACTGCATC-3′; GAPDH forward, 5′-AACGTGTCAGTCGTGGACCTG-3′; and reverse, 5′-AGTGGGTGTCGCTGTFGAAGT-3′. The relative expression was based on the expression ratio of a certain gene versus that of GAPDH.
Western blotting materials
Rabbit polyclonal antibodies against CLDN9 (ab192398) and mouse anti-human β-actin (ab8226) were purchased from Abcam (Massachusetts, US). The rabbit anti-human phospho-Stat1 antibody (#7649), rabbit anti-human phospho-Stat3 antibody (#9145), rabbit anti-human Stat1 antibody (#14994), rabbit anti-human Stat3 antibody (#9139), rabbit anti-human phospho-Tyk2 antibody (#68790), and rabbit anti-human Tyk2 antibody (#13531) were purchased from Cell Signaling Technology (Boston, USA).
Western blotting
The protein concentration of cell lysates was determined using a bicinchoninic acid Protein Assay Kit (Pierce Chemical Co., Rockford, Illinois, USA). The total protein (30 μg) was detached via 10% sodium dodecyl sulfate-polyacrylamide electrophoresis gel. Then, the total protein was transferred onto a nitrocellulose membrane (Millipore, Temecula, California, USA), and the membrane was blocked and investigated with rabbit anti-human phospho-Stat1 antibody, rabbit anti-human Stat1 antibody, rabbit anti-human phospho-Stat3 antibody, rabbit anti-human Stat3, rabbit anti-human phospho-Tyk2 antibody, rabbit anti-human Tyk2 antibody, rabbit anti-human CLDN9 antibody, and mouse anti-human β-actin antibody at a dilution rate of 1:1000 overnight at 4°C. After three washes with phosphate-buffered saline (PBS), the membrane was incubated with horseradish-peroxidase-conjugated secondary antibody (Santa Cruz Biotechnologies, California, USA) at a dilution rate of 1:1000 overnight at 4°C. Immunoreactive bands were detected using the ECL western blotting reagents (GE, Fairfield, Connecticut, USA) and analyzed with the Image Lab 6.0.1 software (Bio-Rad Laboratories).
Immunofluorescence method
The cells were incubated with 4% paraformaldehyde at 20°C for 10 min and permeabilized with 0.1% Triton X-100 (Sigma-Aldrich, cat. no. 9002-93-1) at room temperature for 10 min. Next, after blocking with 2% bovine serum albumin (Bote Biotechnological Corporation, Jilin, China) diluted in PBS at 20°C for 15 min, the cells were probed with a primary rabbit anti-human CLDN9 antibody, diluted in the blocking solution (1:1000 dilution) for 30 min at 20°C. Then, the cells were reacted with Alexa Fluor 647-conjugated anti-rabbit IgG antibody (ab150093, Santa Cruz Biotechnologies, California, USA) at a dilution rate of 1:1000 overnight at 4°C.
Cell counting kit-8 assay
The cells were seeded into 96-well plates in triplicate, and a cell proliferation curve was determined by the colorimetric water-soluble tetrazolium salt assay (Cell Counting Kit-8; Dojindo, Kumamoto, Japan) according to the manufacturer’s protocol.
Wound-healing assay
The cells were maintained in a monolayer at 70% confluence on 24-well plastic dishes, and the monolayer was scratched with a 100 μL pipette tip. The wounds were photographed by a light microscope (E100, Nikon Instruments Inc, Japan) (magnification ×200) at the same location at 0 h, 12 h, and 24 h.
Transwell chamber method
The cells were grown on a monolayer at 90% convergence and were maintained in an FBS-free medium for 12 h. Matrigel (BD Biosciences, cat. no. 356234) was added to the upper Boyden chamber (Millipore, Bedford, MA) in 24-well plates, and they were maintained in a cell incubator at 37°C for 15 min. Then, a medium containing chemotactic factors, which had been collected from the cell culture, was added to the 24-well plate, and a cell suspension was added to the Matrigel and incubated at 37°C in a humidified 5% CO2 atmosphere for 6 h.
RNA interference (RNAi) method
The pGCSIL-scramble, pGCSIL-green fluorescent protein, pGCSIL-Tyk2-RNAi, vesicular stomatitis virus glycoprotein (VSVG) expression plasmid, and virion-packaging elements (pHelper 1.0) frozen glycerol bacterial stocks were purchased from Nanjing KeyGen Biotech Co, Ltd. (Nanjing, China). The target was Tyk2-RNAi (29473), and the control insert sequence was pGCSIL-scramble. HEK293T cells (0.2×107) were seeded and maintained for 24 hours to achieve a 70%–80% confluence in 6-well dishes (Costar, Cambridge, MA). Four plasmids, including pGCSIL-Tyk2-RNAi or pGCSIL-scramble, 5 μg of pGCSIL- GFP, 5 μg of the pHelper 1.0 (packaging vector), and 5 μg of a VSVG expression plasmid vector were added to the Opti-MEM, with a final volume of 1.0 mL. Then, 50 μL of lipofectamine were supplemented to 950 μL of FBS-free medium. These two solutions were mixed and added to the cells. Lentiviral particles were harvested 48 h after transfection, and the viral titer was determined with a fluorescence microscopy (Nikon Diaphot 300) with filters 96 h after transfection.
Patients and tissue samples
Biopsies were gathered from 50 patients diagnosed with HCC who received treatment at First Hospital of Jilin University between May 2007 and June 2012. The cases were carefully chosen on the basis of the following principles: no history of radiotherapy or chemotherapy and no prior malignant disease. The HCC patients were classified and graded on the basis of the American Joint Committee on Cancer tumor node metastasis staging system. Histologically, non-neoplastic hepatic tissues were also obtained from patients infected with hepatitis B virus who received treatment at First Hospital of Jilin University between September 2006 and October 2011.
Immunohistochemistry
Immunohistochemistry was used to explore the expression levels of CLDN9 and p-Stat3 in tissues from 50 HCC tissue samples and 50 histologically non-neoplastic hepatic tissues. The HCC cases included 18 cases of highly differentiated cancer tissues and 32 cases of low-to-moderately differentiated tissues. Of the 50 cases, 26 also exhibited distant metastasis. The experimental method was the same as previously described (20), and the antibodies used were the rabbit anti-human CLDN9 antibody and p-Stat3 antibody. The evaluation of protein expression levels depended on the ratio of stained tumor cells and the staining intensity as previously described (21).
Statistical analysis
All of the experiments were repeated three times, and all of the data are presented as the mean±standard deviation of at least three experimental results. The experimental results were analyzed using Student’s t-test or analysis of variance with Dunnett’s multiple comparisons test, and p<0.05 was considered to indicate statistical significance. The chi-squared test/chi-squared goodness of fit test was utilized to analyze the correlation between the p-Stat3 and CLDN9 expression levels and clinicopathological indicators.
RESULTS
The expression of CLDN9 was upregulated in human HCC
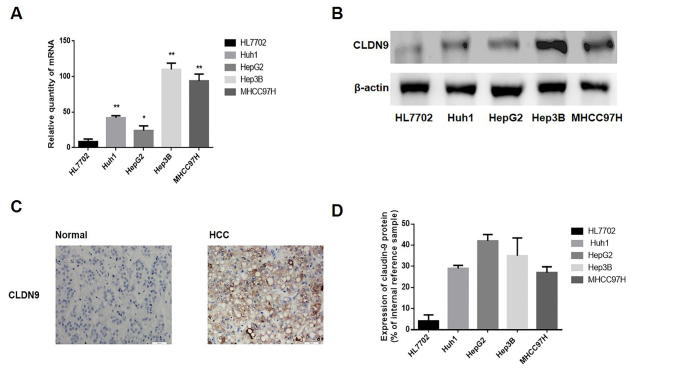
A RT-qPCR and a western blotting analysis were utilized to detect the expression of CLDN9 in human hepatocyte (HL7702) and HCC cell lines (HepG2, Hep3B, Huh1, and MHCC97H). As it is revealed in Figures 1a, b, and d, the mRNA and protein expression of CLDN9 was low or absent in the human hepatocyte line HL7702 but highly expressed in the HCC cell lines Huh1, HepG2, Hep3B, and MHCC97H.
Figure 1. a–d.
The expression levels of CLDN9 in the HCC cell lines and tissues (a). The relative mRNA level of CLDN9 in hepatocyte lines and HCC cell lines (b). The relative protein expression of CLDN9 in hepatocyte lines and HCC cell lines (c). The protein expression of CLDN9 in hepatocyte tissues and HCC cell tissues (d). The corresponding statistical analysis of protein expression in hepatocyte lines and HCC cell lines.
*represents p<0.05; **represents p<0.01; compared with the empty vector groups
Moreover, the CLDN9 expression was explored in the 50 HCC tissues and 50 non-neoplastic hepatic tissues. As shown in Figure 1c, the expression of CLDN9 in hepatic tissues was mainly located in the membrane. A high expression of CLDN9 was seen in 68.0% (34/50 of HCC tissues) and in 24.0% (12/50) of hepatic tissues (p=0.0001) (Table 1). The relationship between CLDN9 and clinical pathological indicators was also analyzed, and the CLDN9 expression was found to be unassociated with patient age (p=1.000), degree of differentiation (p=0.678), clinical staging (p=1.000), or Ki67 expression (p=1.000) of HCC patients, but it was associated with distant metastasis (p=0.001) (Table 1).
Stable transfection of hepatocyte lines with CLDN9
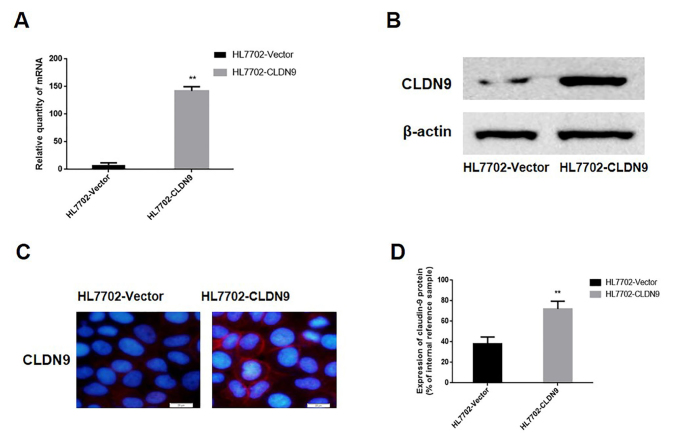
A p-EGFP-C1/CLDN9 plasmid was used to transfect HL7702 cells. After the G418 screening, a monoclonal strain of HL7702 cells was obtained and termed HL7702-CLDN9. The expression of CLDN9 was explored by real-time PCR and western blotting in cultured cells. The data showed that the mRNA and protein expression of CLDN9 in the HL7702-CLDN9 group were notably higher than those in the empty vector groups (p=0.0001; p=0.0001, respectively) (Figure 2a, b, d). Immunofluorescence was used to detect the localization of CLDN9 in HL7702-CLDN9 cells. The results showed that the expression of CLDN9 was primarily localized on cell membranes (Figure 2c). These results demonstrated that the clonal HL7702 cell line that expressed CLDN9 steadily was successfully established.
Figure 2. a–d.
Characterization of the stable expression levels of CLDN9 (a). Detection of CLDN9 in the HL7702 cell line by real-time PCR (b). Detection of CLDN9 expression in the HL7702 cell line by western blotting (c). Detection of CLDN9 expression in the HL7702 cell line by immunofluorescence (d). The corresponding statistical analysis of protein expression in the HL7702 cell line.
**represents p<0.01; compared with the empty vector groups
The impact of CLDN9 on the proliferation and metastasis of hepatocyte lines
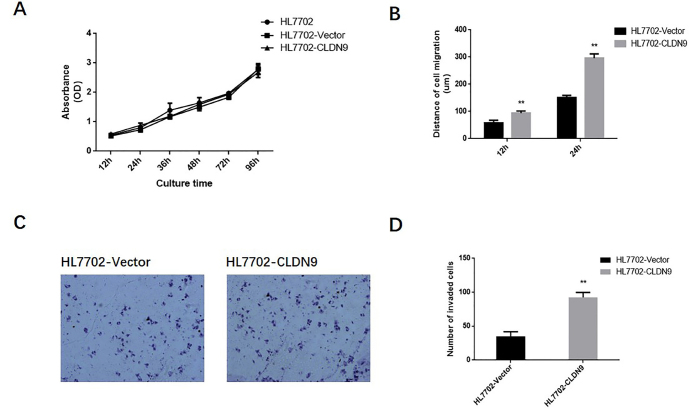
To determine the impact of CLDN9 on the malignant phenotype of hepatocyte lines, the growth curve of HL7702 cell line was drawn by the CCK-8 method. Data revealed that there was no significant difference in the proliferation rate between the HL7702-CLDN9 cells and empty vector groups (Figure 3a). A wound-healing experiment was used to detect the impact of CLDN9 on the migration ability of hepatocytes (Figure 3b). The results showed that the migration distances of HL7702-CLDN9 cells were significantly longer than those of the empty vector groups at 12 h and 24 h (p=0.0022). The transwell chamber method was also used to detect the metastasis ability of hepatocytes. Six hours after the cells were seeded, we quantified the cells that invaded the chamber membrane. These observations displayed that there were notably more invasive cells in the HL7702-CLDN9 groups than in the empty vector groups, suggesting that CLDN9 significantly promotes invasive behavior in hepatocytes in vitro (Figure 3c, d).
Figure 3. a–d.
The impact of CLDN9 on the proliferation and metastasis ability of cells in vitro (a). The growth curve of the HL7702 cell line was drawn by the CCK-8 method (b). The wound-healing assay was used to detect the migration ability of the HL7702 cell line in vitro (c). The transwell chamber method was used to detect the invasive ability of the HL7702 cell line in vitro (d). The corresponding statistical analysis of invasive cells.
**represents p<0.01 compared with the empty vector groups
The impact of CLDN9 on the Tyk2/Stat3 signaling pathway of hepatocytes
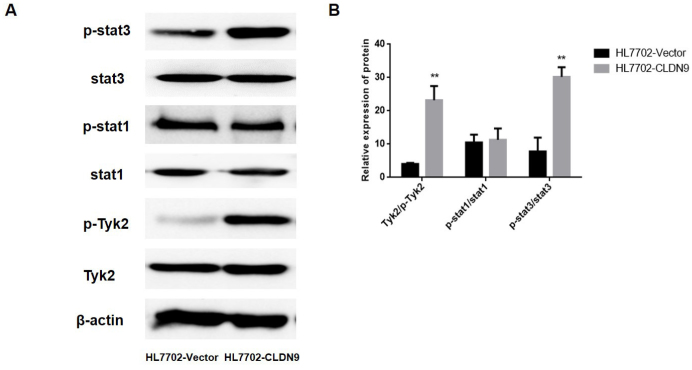
The western blotting was utilized to explore the activation state of the Tyk2/Stat3 pathway. The results showed that after the overexpression of CLDN9, the phosphorylation level of Stat3 and Tyk2, an upstream regulator of Stat3, were significantly increased (Figure 4a, b). These data suggest that CLDN9 significantly enhances the activation of the Tyk2/Stat3 signaling pathway.
Figure 4. a, b.
The impact of CLDN9 on the Tyk2/stat3 signaling pathway (a). Western blotting was used to detect the activation of the Stat3 signaling pathway in the HL7702 cell line (b). The corresponding statistical analysis of the activation status of various Stat3 pathway components.
**represents p<0.01 compared with the empty vector groups
The impact of Tyk2/Stat3 signaling pathway activation on the invasion and migration ability of hepatocytes
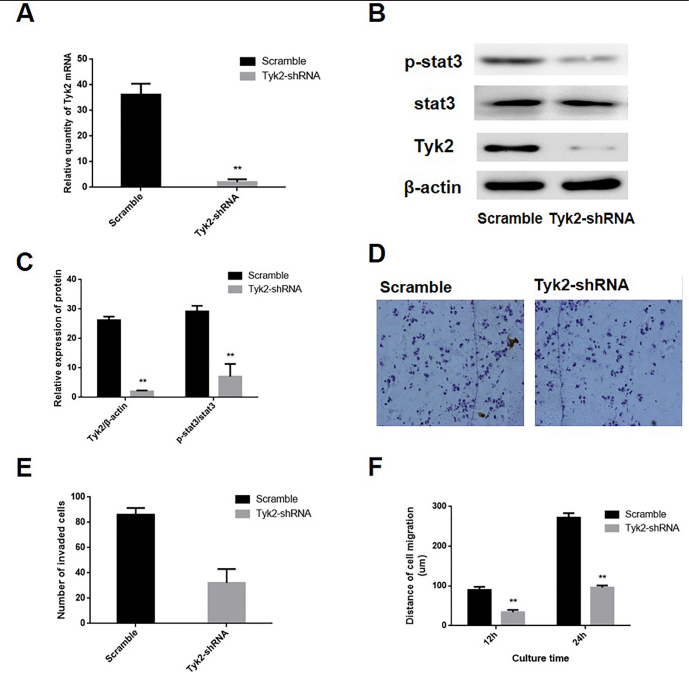
To determine the impact of Tyk2/Stat3 signaling pathway on hepatocyte invasion and migration ability, we transfected HL7702-CLDN9 cells with a pGCSIL-scramble plasmid and a pGCSIL-Tyk2-RNAi plasmid. The real-time PCR and western blotting were utilized to determine the expression level of Tyk2 and changes in the activation state of Stat3 in these cells, and the results showed that compared with the expression in the scramble group, the expression of Tyk2 was significantly reduced in Tyk2-RNAi cells (Figure 5a–c). The phosphorylation level of Stat3 were also explored, and the results displayed that the phosphorylation level of Stat3 was notably downregulated following the knockdown of Tyk2 in the HL7702 cell line that overexpressed CLDN9 (Figure 5b, c).
Figure 5. a–f.
RNAi was used to silence the Tyk2 expression in CLDN9-expressing cells (a). Real-time PCR was used to examine the effects of silencing Tyk2 in the HL7702 cell line (b). Western blotting was used to examine the effects of silencing Tyk2 and the activation of the Stat3 signaling pathway in the HL7702 cell line (c). The corresponding statistical analysis of Stat3 signaling pathway activation (d). The transwell chamber method was used to detect the impact of Tyk2 silencing on the invasive ability of cells in vitro (e). The corresponding statistical analysis of invasive cells (f). The wound-healing assay was used to detect the migration ability of the HL7702 cell line in vitro.
*represents p<0.05; **represents p<0.01 compared with the scramble group
The transwell chamber assay and wound-healing assay were utilized to determine the impact of Tyk2 on the invasive and migration ability of the cells. The results showed that the numbers of invasive HL7702 cell lines in the CLDN9-expressing cells were significantly reduced following Tyk2 silencing (Figure 5d, e). The migration distances of Tyk2-RNAi cells were significantly shorter than those of the scramble group at 12 h and 24 h (Figure 5f).
Expressions of p-Stat3 in human HCC
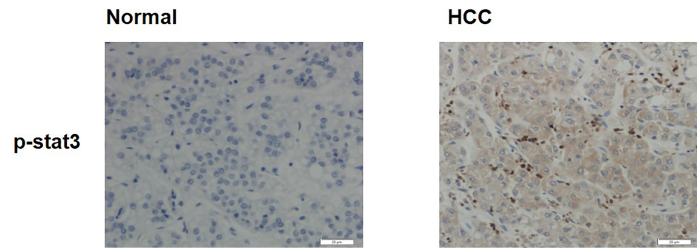
The expression of p-Stat3 was explored in the 50 HCC sections and 50 non-neoplastic hepatic sections. As revealed in Figure 6, the expression of p-Stat3 was highly expressed in 62.0% (31/56) of HCC tissues and in 32.0% (16/50) of non-neoplastic hepatic tissues (p=0.0001) (Table 1). The results suggested that the p-Stat3 expression was increased in human HCC tissues.
Figure 6.
Expressions of p-Stat3 in human HCC and hepatic tissue
The relationship between p-Stat3 and clinical pathological indicators was analyzed, and the expression of p-Stat3 was found to be unassociated with the age (p=0.451), tumor Ki67 expression (p=1.000), or clinical staging (p=0.296) of HCC patients, but it was associated with the degree of differentiation and distant metastasis (p=0.002 and p=0.0258, respectively) (Table 1).
CLDN9 and p-stat3 were concurrently expressed in HCC tissues
In addition, the relevance of CLDN9 and p-stat3 expression in HCC tissues was also explored. As shown in Table 2, the CLDN9 and p-stat3 were concurrently expressed in 44.0% (22/50 of HCC tissues). The expression of CLDN9 was found to be positively correlated with p-stat3 in HCC tissues by the chi-squared test/chi-squared goodness of fit test (ϕ=0.852, p<0.01).
DISCUSSION
To date, certain studies have characterized the CLDN expression in HCC tumorigenesis and progression (22,23). For instance, the CLDN1 and CLDN7 protein expression was suggested to be notably upregulated in cirrhotic tissues versus non-cirrhotic liver tissues (24). Moreover, CLDN1 has been defined as a crucial feature in the entrance of hepatitis C virus into hepatocytes, and upregulated expression of CLDN1 was revealed to serve as a trigger of the epithelial-to-mesenchymal transition through the c-Abl/Raf/Ras/ERK signaling pathway (25). Given these correlations of the claudin protein expression and HCC, these proteins may be established as novel targets in the diagnosis and treatment of HCC (26). However, few studies have explored the etiologic association between the CLDN9 and hepatic carcinogenesis.
The current understanding of the biological functions of CLDN9 is primarily limited to epithelial and epidermal permeability, barrier protection, and cell connections, with reports on the relationship between CLDN9 and HCC being rare. Our research group found that the CLDN9 expression was upregulated in HCC tissues, and we speculated that the expression of this gene may play a part in the HCC tumorigenesis. At present, our data indicated that the upregulation of CLDN9 significantly promoted the invasion and migration abilities of hepatocytes. To determine the character of the Tyk2/Stat3 signaling pathway in this effect, we used Tyk2-RNAi assays to silence the expression of Tyk2 in the hepatocytes that overexpressed CLDN9 and assessed the impact of the Tyk2/Stat3 signaling pathway on cell metastasis ability in vitro. Our data showed that the Tyk2 silencing resulted in the inhibition of the Stat3 signaling pathway and metastasis abilities in hepatocytes. To date, few studies have shown that the Tyk2/Stat3 signaling impacts the role of various CLDNs in human tumorigenesis. One of the most interesting findings in our research is the evidence that upregulated CLDN9 contributed to an enhancement of cell invasion and migration through the Tyk2/Stat3 signaling pathway in HCC. To further confirm our findings, we explored the association between the CLDN9 and p-Stat3 expression in HCC tissues. Our data displayed that the CLDN9 expression was positively correlated with p-Stat3, suggesting that the expression of CLDN9 may be negatively correlated with the activation of Stat3 during the HCC progression. Hence, the CLDN9 and p-Stat3 have the potential to be established as molecular markers for HCC diagnosis.
At present, we confirmed that, as an HCC proto-oncogene, CLDN9 notably promotes the metastatic ability of the HL7702 hepatocyte line. We also performed an initial investigation into the molecular mechanism of this effect and found that CLDN9 affected the Stat3 signaling pathway via Tyk2, ultimately enhancing the metastatic ability of hepatocytes. In view of the insufficient therapeutic selections in HCC, the CLDN9 role as a therapeutic target needs to be further explored. However, the precise mechanisms of the signal transduction from the membrane-anchored CLDN9 to Tyk2 protein located in the cell interior remains to be elucidated.
In conclusion, we confirm that the expression of CLDN9 significantly enhances the metastatic ability of hepatocytes in vitro, and the activation of the Stat3 pathway by Tyk2 may be one of the important mechanisms by which CLDN9 promotes hepatocyte aggressiveness in HCC.
Acknowledgements
The authors would like to thank American Journal Experts (AJE) for English language editing.
Footnotes
Ethics Committee Approval: The ethics committee approval was received for this study from the Ethics Committee of Jilin University hospital (No. JLCH01224).
Informed Consent: Written informed consent was obtained from all patients and their parents who participated in this study.
Author Contributions: Concept - H.L.; Design - M.W.; Supervision - L.G.; Data Collection and/or Processing - H.L.; Analysis and/or Interpretation - L.G.; Writing Manuscript - L.G.; Critical Reviews - L.G., H.L., M.W., N.L.
Conflict of Interest: The authors declare that there was no conflict of interest.
Financial Disclosure: The authors declared that this study has received no financial support.
References
- 1.Escudero-Esparza A, Jiang WG, Martin TA. The Claudin family and its role in cancer and metastasis. Front Biosci (Landmark Ed) 2011;16:1069–83. doi: 10.2741/3736. [DOI] [PubMed] [Google Scholar]
- 2.Cunniffe C, Brankin B, Lambkin H, Ryan F. The Role of Claudin-1 and Claudin-7 in Cervical Tumorigenesis. Anticancer Res. 2014;34:2851–7. [PubMed] [Google Scholar]
- 3.Turksen K, Troy TC. Junctions gone bad: claudins and loss of the barrier in cancer. Biochim Biophys Acta. 2011;1816:73–9. doi: 10.1016/j.bbcan.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Osanai M, Takasawa A, Murata M, Sawada N. Claudins in cancer: bench to bedside. Pflugers Archiv Eur J Physiol. 2017;469:55–67. doi: 10.1007/s00424-016-1877-7. [DOI] [PubMed] [Google Scholar]
- 5.Tabaries S, Siegel PM. The role of claudins in cancer metastasis. Oncogene. 2017;36:1176–90. doi: 10.1038/onc.2016.289. [DOI] [PubMed] [Google Scholar]
- 6.Oliveira S, Morgado-Diaz J. Claudins: multifunctional players in epithelial tight junctions and their role in cancer. Cell Mol Life Sci. 2007;64:17–28. doi: 10.1007/s00018-006-6314-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ouban A, Ahmed A. Claudins in human cancer: a review. Histol Histopathol. 2010;25:83–90. doi: 10.14670/HH-25.83. [DOI] [PubMed] [Google Scholar]
- 8.Chao Y-C, Pan S-H, Yang S-C, et al. Claudin-1 is a metastasis suppressor and correlates with clinical outcome in lung adenocarcinoma. Am J Respir Crit Care Med. 2009;179:123–33. doi: 10.1164/rccm.200803-456OC. [DOI] [PubMed] [Google Scholar]
- 9.Kwon MJ. Emerging roles of claudins in human cancer. Int J Mol Sci. 2013;14:18148–80. doi: 10.3390/ijms140918148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dahiya N, Becker KG, Wood WH, Zhang Y, Morin PJ. Claudin-7 is frequently overexpressed in ovarian cancer and promotes invasion. PloS One. 2011;6:e22119. doi: 10.1371/journal.pone.0022119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heinzelmann-Schwarz VA, Gardiner-Garden M, Henshall SM, et al. Overexpression of the cell adhesion molecules DDR1, Claudin 3, and Ep-CAM in metaplastic ovarian epithelium and ovarian cancer. Clin Cancer Res. 2004;10:4427–36. doi: 10.1158/1078-0432.CCR-04-0073. [DOI] [PubMed] [Google Scholar]
- 12.Worst TS, von Hardenberg J, Gross JC, et al. Database-augmented Mass Spectrometry Analysis of Exosomes Identifies Claudin 3 as a Putative Prostate Cancer Biomarker. Mol Cell Proteomics. 2017;16:998–1008. doi: 10.1074/mcp.M117.068577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maeda T, Murata M, Chiba H, et al. Claudin-4-targeted therapy using Clostridium perfringens enterotoxin for prostate cancer. Prostate. 2012;72:351–60. doi: 10.1002/pros.21436. [DOI] [PubMed] [Google Scholar]
- 14.Ouban A, Ahmed AA. Claudins in human cancer: a review. Histol Histopathol. 2010;25:83–90. doi: 10.14670/HH-25.83. [DOI] [PubMed] [Google Scholar]
- 15.Singh AB, Sharma A, Dhawan P. Claudin family of proteins and cancer: an overview. J Oncol. 2010;1957;54 doi: 10.1155/2010/541957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.González-Mariscal L, Tapia R, Chamorro D. Crosstalk of tight junction components with signaling pathways. Biochim Biophys Acta. 2008;1778:729–56. doi: 10.1016/j.bbamem.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 17.Tsukita S, Furuse M, Itoh M. Structural and signalling molecules come together at tight junctions. Current opinion in cell biology. 1999;11:628–33. doi: 10.1016/S0955-0674(99)00016-2. [DOI] [PubMed] [Google Scholar]
- 18.de Souza WF, Fortunato-Miranda N, Robbs BK, et al. Claudin-3 overexpression increases the malignant potential of colorectal cancer cells: roles of ERK1/2 and PI3K-Akt as modulators of EGFR signaling. PloS One. 2013;8:e74994. doi: 10.1371/journal.pone.0074994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Ruan Y, Li Y, Lin D, Liu Z, Quan C. Expression of apoptosis signal-regulating kinase 1 is associated with tight junction protein claudin-6 in cervical carcinoma. International journal of clinical and experimental pathology. 2015;8:5535–41. [PMC free article] [PubMed] [Google Scholar]
- 21.Gao M, Li W, Wang H, Wang G. The distinct expression patterns of claudin-10, -14, -17 and E-cadherin between adjacent non-neoplastic tissues and gastric cancer tissues. Diagn Pathol. 2013;8:205. doi: 10.1186/1746-1596-8-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang L, Yang YD, Fu L, et al. CLDN3 inhibits cancer aggressiveness via Wnt-EMT signaling and is a potential prognostic biomarker for hepatocellular carcinoma. Oncotarget. 2014;5:7663–76. doi: 10.18632/oncotarget.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tabaries S, Dong Z, Annis MG, et al. Claudin-2 is selectively enriched in and promotes the formation of breast cancer liver metastases through engagement of integrin complexes. Oncogene. 2011;30:1318–28. doi: 10.1038/onc.2010.518. [DOI] [PubMed] [Google Scholar]
- 24.Holczbauer A, Gyongyosi B, Lotz G, et al. Increased expression of claudin-1 and claudin-7 in liver cirrhosis and hepatocellular carcinoma. Pathology oncology research: Pathol Oncol Res. 2014;20:493–502. doi: 10.1007/s12253-013-9683-4. [DOI] [PubMed] [Google Scholar]
- 25.Suh Y, Yoon CH, Kim RK, et al. Claudin-1 induces epithelial-mesenchymal transition through activation of the c-Abl-ERK signaling pathway in human liver cells. Oncogene. 2017;36:1167–8. doi: 10.1038/onc.2016.294. [DOI] [PubMed] [Google Scholar]
- 26.Hashimoto Y, Fukasawa M, Kuniyasu H, Yagi K, Kondoh M. Claudin-targeted drug development using anti-claudin monoclonal antibodies to treat hepatitis and cancer. Ann N Y Acad Sci. 2017;1397:5–16. doi: 10.1111/nyas.13337. [DOI] [PubMed] [Google Scholar]