Abstract
Immunologic responses to anti-PD-1 therapy in melanoma patients occur rapidly with pharmacodynamic T cell responses detectable in blood by 3 weeks. It is unclear, however, whether these early blood-based observations translate to the tumor microenvironment. We conducted a study of neoadjuvant/adjuvant anti-PD-1 therapy in stage III/IV melanoma. We hypothesized that immune reinvigoration in the tumor would be detectable at 3 weeks and this response would correlate with disease-free survival. We identified a rapid and potent anti-tumor response, with 8/27 patients experiencing a complete or major pathological response after a single dose of anti-PD-1, all of whom remain disease-free. These rapid pathologic and clinical responses were associated with accumulation of exhausted CD8 T cells in the tumor at 3 weeks with reinvigoration in the blood observed as early as 1 week. Transcriptional analysis demonstrated a pre-treatment immune signature (Neoadjuvant Response Signature) that was associated with clinical benefit. In contrast, patients with disease recurrence displayed mechanisms of resistance including immune suppression, mutational escape, and/or tumor evolution. Neoadjuvant anti-PD-1 treatment is effective in high-risk resectable stage III/IV melanoma. Pathological response and immunological analyses after a single neoadjuvant dose can be used to predict clinical outcome and to dissect underlying mechanisms in checkpoint blockade.
Introduction
Clinical responses to anti-PD-1 therapies can occur rapidly1,2. A pharmacodynamic response including reinvigoration of exhausted-phenotype CD8 T cells (TEX) can be detected in blood of cancer patients after a single dose3,4. However, the precise type(s) of T cells in the tumor that respond to anti-PD-1 remains poorly understood. Moreover, whereas early immunological responses to checkpoint blockade are observed at 3 weeks in blood, the kinetics of immune reinvigoration in the tumor and the relationship to pathological response and clinical outcomes are unclear.
We conducted a neoadjuvant/adjuvant anti-PD-1 clinical trial in stage III/IV resectable melanoma. This approach provided early on-treatment tumor tissue at resection and insights into the mechanisms of PD-1 blockade. Our study demonstrated the clinical feasibility of neoadjuvant/adjuvant anti-PD-1 therapy in melanoma, and identified a rapid pathological and immunologic response in tumors. Complete pathological responses could be identified by 3 weeks and correlated with disease-free survival. Data from early on-treatment resected tumor indicate that TEX, but not bystander cells, are a major responding cell type. Studies in an additional cohort identified reinvigoration of TEX as early as day 7 after the first dose of anti-PD-1. Finally, in patients who developed disease recurrence, potential mechanisms of resistance were identified.
Results
A pharmacodynamic immune response can be detected in blood 3 weeks after initiation of PD-1 blockade3,4. To understand the early effects of anti-PD-1 in tumors, we conducted an investigator initiated clinical trial of neoadjuvant anti-PD-1 (pembrolizumab) in stage IIIB/C or IV melanoma. All patients underwent baseline pre-treatment biopsy and received a single dose of pembrolizumab (200 milligrams), followed by complete resection three weeks later and adjuvant therapy (Figure 1A). Twenty-nine patients were enrolled and treated; (Supplemental Table 1). Patients proceeded to surgical resection at three weeks (median 21 days, range 17-42). Median interval between surgery and initiation of adjuvant pembrolizumab was 23 days (range 13-39). There were no unexpected adverse events (Supplemental Table 2); the rate of grade 3 or higher adverse events not attributed to pembrolizumab or to surgery alone was not higher than 30%, the prespecified safety endpoint (observed rate was 0%, p = 0.0002, z test). There were no unexpected delays in surgery, adjuvant pembrolizumab, or unexpected surgical complications.
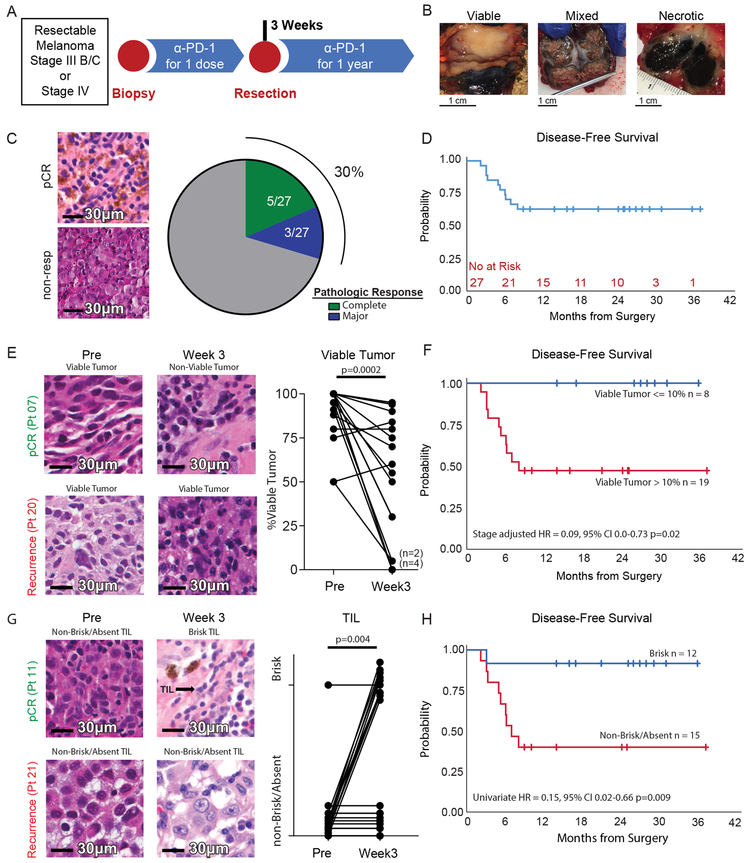
Figure 1: Pathologic response and tumor infiltrating lymphocytes are predictive of clinical outcome after a single dose of anti-PD-1.
A, Schema of the neoadjuvant and then adjuvant pembrolizumab clinical trial. B, Representative images of viable, mixed, and necrotic tumors resected at the three-week post-treatment time point. C, Representative H&E images of pathologic complete response (pCR) and non-response (non-resp) (left) and fraction of patients with complete pathologic response and major pathologic response (right). D, Kaplan Meier estimate of disease-free survival. E, Representative H&E images (left) and changes in the percent of viable tumor in pre-treatment and post-treatment tumors (right, n = 20); p value calculated using two-sided Wilcoxon matched-pairs test. F, Kaplan Maier estimate of disease-free survival stratified according to pathologic response. Cox proportional hazards regression modeling was used to calculate hazard ratio. G, Changes in TIL infiltration in pre-treatment and post-treatment tumors (n = 20); p value calculated using McNemar’s Test. H, shows Kaplan Maier estimate of disease-free survival stratified according to TIL score. Cox proportional hazards regression modeling was used to calculate hazard ratio.
We assessed the pathologic response after one dose of pembrolizumab in the resection specimens of 27 patients available at time of data cut-off. Resected tumor had features of completely viable, mixed viable and necrotic, or completely necrotic tumor upon gross examination (Figure 1B). On histologic assessment, 8 of 27 patients (29.6%, 95% CI 13.8–50.2%) had a complete (no residual tumor identified; n=5) or major (10% or less viable tumor cells; n=3) pathologic response7,8 (Figure 1C). The median duration of follow-up for disease-free survival (DFS) was 25 months; median DFS has not been reached. The 1-year DFS rate ± SE was 63% ± 9% (Figure 1D).
Approximately 30% of patients had a complete or major pathologic response after one dose of pembrolizumab, consistent with a recent neoadjuvant anti-PD-1 clinical trial in melanoma that observed a pCR of 25% after 4 doses of nivolumab9. In 20 patients with paired pre- and post-treatment tumor samples, the percentage of viable tumor significantly decreased after treatment (Figure 1E). All patients with complete or major pathologic responses remain disease-free (Figure 1F); viable tumor ≤10% at resection was associated with a low risk of recurrence independent of stage (Supplemental Table 3). LDH, BRAF status, and tumor burden (unidimensional tumor diameter) were not associated with recurrence (Supplemental Table 3). In 20 patients with paired samples, a single dose of pembrolizumab significantly increased the fraction of patients with brisk TIL (Supplemental Table 4; Figure 1G) which was also associated with complete or major pathologic response (Supplemental Table 5; Extended Data Figure 1) and significantly improved disease-free survival (Supplemental Table 3; Figure 1H).
We examined a subset of 6 patients that had early imaging at 3 weeks (trial UPCC11615). 18F-fluorodeoxyglucose positron emission tomography - computed tomography (FDG PET-CT) scans were performed at baseline and prior to surgical resection; images were analyzed per RECIST 1.1. Consistent with the pathological responses, radiographic responses were observed after one dose of anti-PD-1. In 3 patients, we observed a ≥20% decrease in tumor diameter and these patients remained recurrence free (Figure 2A). In contrast, the 3 patients without evidence of tumor shrinkage at 3 weeks had recurrences. Changes in tumor size at 3 weeks correlated with the percentage of viable tumor observed histologically (Figures 2B-C). However, changes in FDG avidity were not associated with response (Data not shown). The percentage of viable tumor at 3 weeks was correlated with a change in tumor dimensions by imaging and recurrence status which were both inversely correlated with TIL infiltration (Figures 2D-E, Supplemental Table 5).
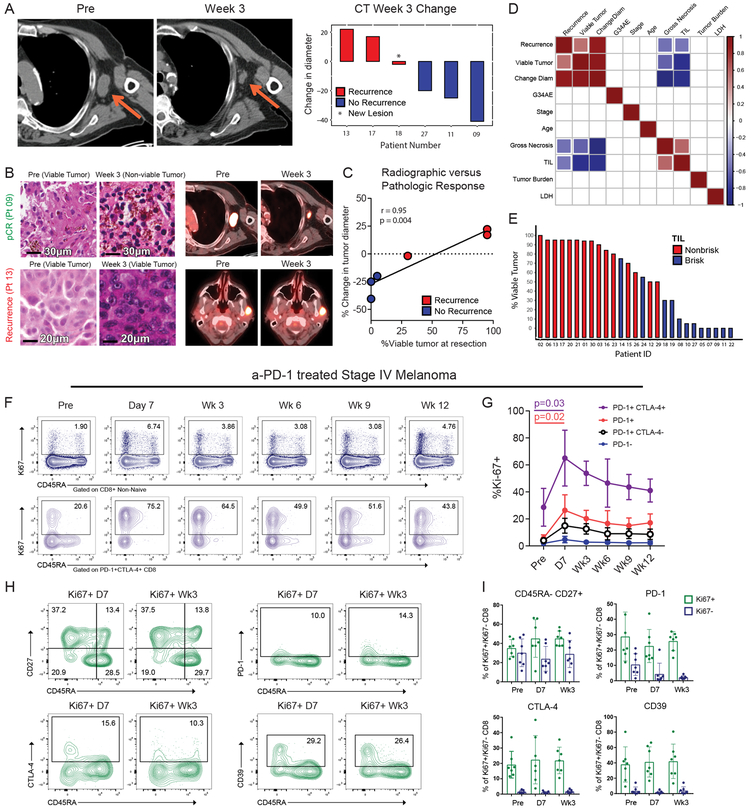
Figure 2: Early radiographic, pathologic, and immune response to anti-PD-1.
A, Changes in tumor diameter based on the CT portion of FDG PET-CT imaging at three weeks compared to pre-treatment colored by recurrence status. B, Paired histology and radiographic images from a patient with pCR (top) and a patient with recurrence (bottom). C, Correlation of the change in tumor diameter on CT imaging after 1 dose of pembrolizumab versus percent viable tumor at resection (n = 6). R-score and p-value generated using Pearson’s correlation. D, Correlation matrix of selected variables for 27 trial patients with data available at data cut-off. Colored boxes represent Pearson’s correlations with a significance of p<0.05. Red to blue represents correlation coefficients ranging from 1 to −1 respectively. E, Waterfall plot of percent viable tumor colored by TIL infiltration. F, Representative flow plots of 7 independent patients. G, Ki67 expression in CD8 T cell subsets at indicated times. (n = 7); p value calculated using two-sided Wilcoxon matched-pairs test. Error bar denotes mean +/− sd. H, Representative flow plots of 7 independent patients; gated on Ki67+ CD8. I, Percent expression of markers in Ki67+ (green), or Ki67- (blue) CD8 T cells (n = 7 independent patient samples). Error bar denotes mean +/− sd.
Given the rapid pathological effects of anti-PD-1, we hypothesized that the reinvigoration of anti-tumor immune responses might occur before 3 weeks. Indeed, there is anecdotal evidence of expansion of CD39+ exhausted phenotype CD8 T cells 9 days after anti-PD-15. We conducted an additional trial of anti-PD-1 (UPCC02616) with blood collected at day 7 after initiation of therapy. We observed robust increases in Ki67+ CD8 T cells at day 7; the responding cells were PD1+ and enriched for cells that co-expressed PD-1 and CTLA-4 (Figures 2F-G). This response peaked at day 7 and then declined. Analysis of the Ki67+ cells at day 7 and week 3 showed similarity in expression of markers of differentiation state (CD45RA, CD27), inhibitory receptors (PD-1, CTLA-4), and other markers of T cell exhaustion (CD39) (Figures 2H-I), indicating similar qualitative responses at day 7 and 3 weeks. Ki67+ cells pre-treatment also had similar phenotype, suggesting that that PD-1 blockade reinvigorates a pre-existing pool of TEX cells (Figure 2I). The robust response of TEX-like CD8 T cells to anti-PD-1 as early 7 days post treatment is consistent with the robust tumor eradiation in many patients by 3 weeks.
To further investigate the nature of the rapid intratumoral response to anti-PD-1, we compared PBMC to TIL at week 3. Increased proliferation of CD8, CD4, and FoxP3+ regulatory T cells (Tregs) was detected in the blood after anti-PD-1 (Extended Data Figure 2A) as described3. In the tumor, CD8 T cell proliferation increased (Extended Data Figure 2B) and as expected10,11 pembrolizumab treatment increased CD8 T cell infiltration (Figure 3A). The majority of CD8 T cells in the tumor were CD45RAlo CD27hi. PD-1 was co-expressed with Tim-3, CTLA-4, LAG3, TIGIT and CD39, and many of these TIL CD8 T cells expressed high Eomes, but low Tbet (Figures 3B-C), consistent with the phenotype of TEX12,13. In addition, the majority of CD8 T cells in the tumor were bound by pembrolizumab (Figure 3D).
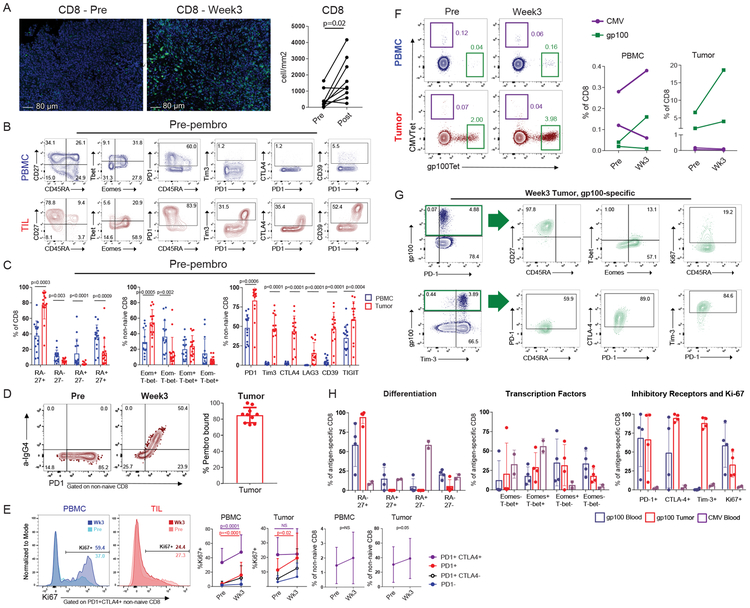
Figure 3: Pembrolizumab targets TEX in tumor.
A, Changes in tumor CD8 T cells pre- versus post-treatment using immunofluorescence (IF) staining (n = 9 independent paired patient samples; one outlier removed due to pre-treatment value >2 sd above the mean). P value calculated using two-sided Wilcoxon matched-pairs test. B, Representative flow plots of pre-treatment PBMC and TIL for 15 independent patient samples. Gated on CD8 (CD45RA versus CD27), and non-naïve CD8 (Eomes versus Tbet, CD45RA versus PD-1, PD-1 versus Tim-3, PD-1 versus CTLA4, PD-1 versus CD39). C, Percent of memory markers, transcription factors, and inhibitory receptors in non-naïve CD8 T cells (n = 15 independent patient samples). Comparisons for all CD45RA versus CD27 subsets performed using two-sided Wilcoxon matched-pairs test. Eomes+Tbet- comparison performed using two-sided paired t-test. Eomes-Tbet- comparison performed using two-sided Wilcoxon matched-pairs test. Comparisons of PD-1 and Lag-3 performed using Wilcoxon matched-pairs test. Comparisons of Tim-3, CTLA-4, CD39, and TIGIT performed using two-sided t test. D, Percent of pembrolizumab bound, calculated by % of IgG4+/%PD-1(EH12)+ (n = 10 independent patient samples). Error bar denotes mean +/− sd. E, Percent Ki67 and frequency of CD8 subsets, pre and at week 3. (n = 28 independent paired patient samples). P values calculated using two-sided Wilcoxon matched-pairs test. Error bars denote mean +/− sd. F, Representative flow plots of two paired CMV and gp100-specific responses pre and post-treatment, in blood and tumor (left). Frequencies of CMV and gp100 tetramer+ CD8 T cells pre and at week 3 in the blood and tumor (right). G, Representative flow plots of memory markers, transcription factors, and IRs for gp100-specific CD8 T cells. H, Expression of selected markers in gp100-specific and CMV-specific CD8 T cells at week 3 post-treatment timepoint. (n = 4 for gp100-specfic responses in blood and tumor; n = 2 for CMV specific responses). Error bar denotes mean +/− sd.
At 3 weeks there was a robust increase in Ki67 in PD-1+ and PD-1+CTLA4+ CD8 T cells in the blood. Although there was an increase in Ki67 in PD-1+ CD8 T cells in the tumor, the PD-1+CTLA4+ subset lacked a consistent increase in Ki67 whereas the proportion of PD1+CTLA4+ CD8 T cells in the tumor increased following treatment (Figure 3E). This finding suggested that: 1) Immune reinvigoration in the tumor may be early and transient, while systemic reinvigoration may be sustained, and/or 2) T cells may be reinvigorated in the periphery before trafficking to the tumor.
We observed a high frequency of PD-1+CD39+ CD8 T cells in the TIL pre-treatment (Figure 3B), consistent with tumor-reactive T cells5. To examine tumor specificity in more detail, we examined gp100 and CMV-specific CD8 T cells using HLA/peptide tetramers pre and post-treatment in two patients. There was no consistent change for these populations in the blood following treatment. However, in both patients, gp100-specific CD8 T cells were present at higher frequencies in the tumor than blood pre-treatment and these cells increased in frequency after anti-PD-1 (Figure 3F). Conversely, CMV-specific CD8 T cells in the tumor did not expand after treatment. At week 3, gp100-specific CD8 T cells in the tumor were CD45RAloCD27hi, expressed high PD-1, Tim-3, and CTLA-4, and most were EomeshiT-betlo (Figures 3G-H). Compared to blood, the gp100-specific CD8 T cells in the tumor had similar PD-1, more Tim-3 and CTLA-4, but less Ki67. In contrast, CMV specific CD8 T cells in the blood were CD45RA+CD27-, coexpressed Eomes and T-bet and had lower inhibitor receptors (Figure 3H). Thus, melanoma-specific CD8 T cells in the blood and tumor of a subset of patients had a phenotype consistent with TEX in the tumor and increased numerically after anti-PD-1.
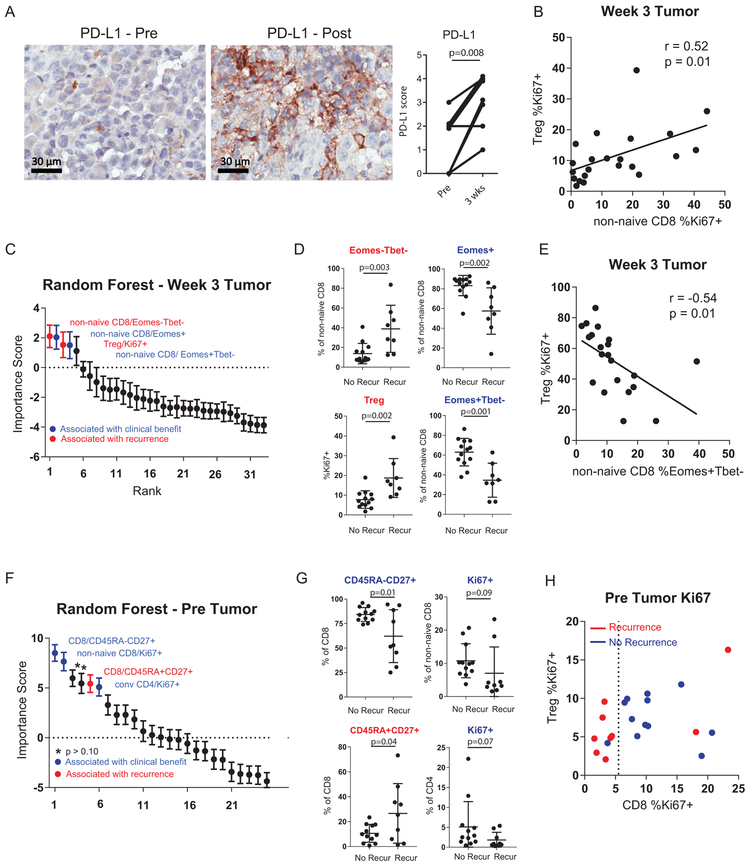
We next studied the cellular determinants of anti-PD-1 response versus resistance in this cohort. Anti-PD-1 increased CD8 T cells in the tumor (Figure 3A), but also increased PD-L1 and Tregs (Extended Data Figure 3A; Figure 4A). Proliferation of CD8 T cells in the tumor also correlated with Treg proliferation (Extended Data Figure 3B), suggesting rapid upregulation of immunoregulatory feedback mechanisms. Random forest analysis in the tumor post-treatment revealed Eomes expression and Treg proliferation as strong correlates of recurrence-free survival (Extended Data Figure 3C). A high percentage of EomeshiTbetlow TEX was associated with clinical benefit and Treg proliferation was associated with recurrence and poor disease-free survival (Extended Data Figure 3D; Figure 4B). Treg proliferation was inversely correlated with the frequency of EomeshiTbetlow TEX (Extended Data Figure 3E). Random forest analysis of pre-treatment immune parameters identified baseline Ki67 expression by non-naïve CD8 T cells as associated with clinical benefit (Figure 4C; Extended Data Figures 3F-H), suggesting pre-existing CD8 T cell responses drive clinical responses to anti-PD-111,14.
Figure 4: Mechanisms of response and resistance to anti-PD-1 therapy.
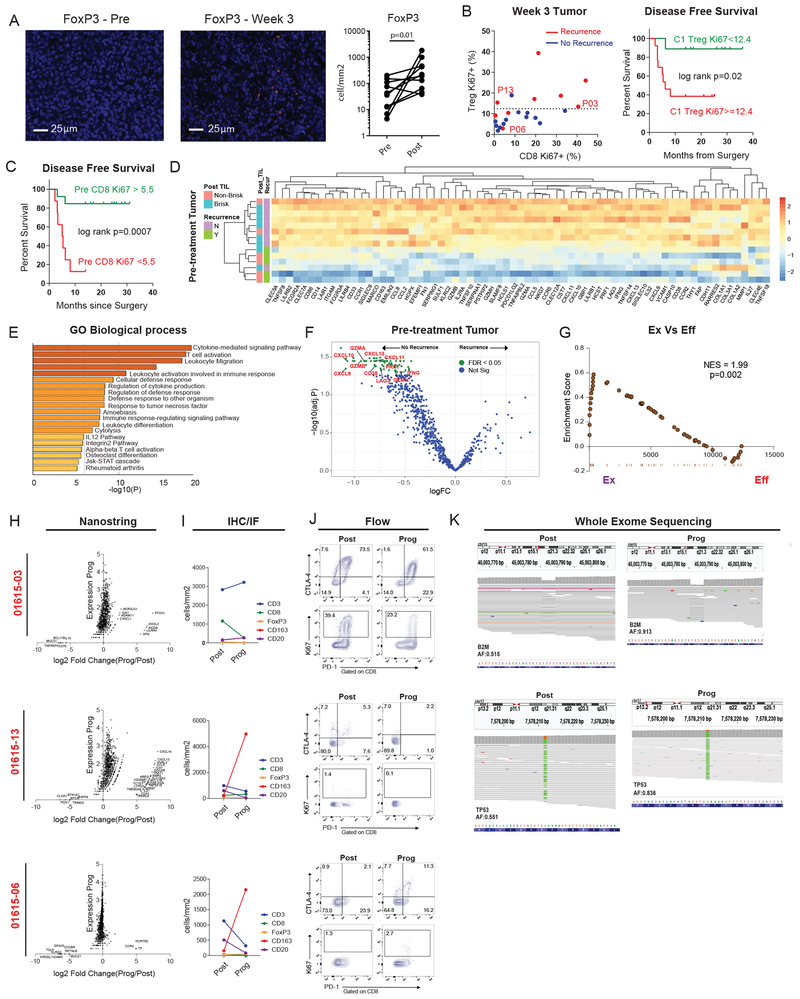
A. Changes in tumor FoxP3+ cells pre- versus post-treatment using immunofluorescence (IF) staining (n = 10). P value calculated using two-sided Wilcoxon matched-pairs test. B. Scatter plot of %Ki67+ in non-naïve CD8 versus %Ki67+ in FoxP3+ CD4 (Tregs) at week 3 post treatment stratified by recurrence status. P13, P06, and P03 represent patients with recurrence tumors samples analyzed below. Dotted line denotes Treg Ki67+ of 12.4 calculated by CART analysis as the optimal cutpoint separating recurrence versus no recurrence. (Left, n = 22). Kaplan Maier estimate of disease-free survival stratified according to CART defined cutoff for Treg Ki67 (Right, Ki67<12.4, n = 9, Ki67>=12.4, n = 13). P value calculated using log-rank test. C. Kaplan Maier estimate of disease-free survival stratified according to CART defined cutoff for non-naïve CD8 Ki67 at baseline (Ki67>5.5, n = 13; Ki67>=12.4, n = 8). P value calculated using log-rank test. D. Heatmap of 69 differentially expressed genes (Neoadjuvant Response Signature) at the pre-treatment time point between patients with no recurrence (n = 9 patients) and recurrence (n = 5 patients). Differentially expressed genes identified using FDR cutoff of p = 0.05. E, Pathways identified using gene ontology analysis. F, Volcano plot of differentially expressed genes. G, Gene set enrichment analysis of the Neoadjuvant Response Signature (NRS); genes that were enriched in TEX versus TEFF -cell signatures from Doering et al 19. H, NanoString gene expression data showing log2 fold change between progression versus post samples (x axis), and expression at progression (y axis). I, Lymphocyte subsets at post and progression time points by immunohistochemistry (IHC), and immunofluorescence (IF). J, Flow plots of selected markers at post and progression time points for three individual patients. K, Integrative Genomics Viewer (IGV) images corresponding to B2M and TP53 mutations at post and progression time points.
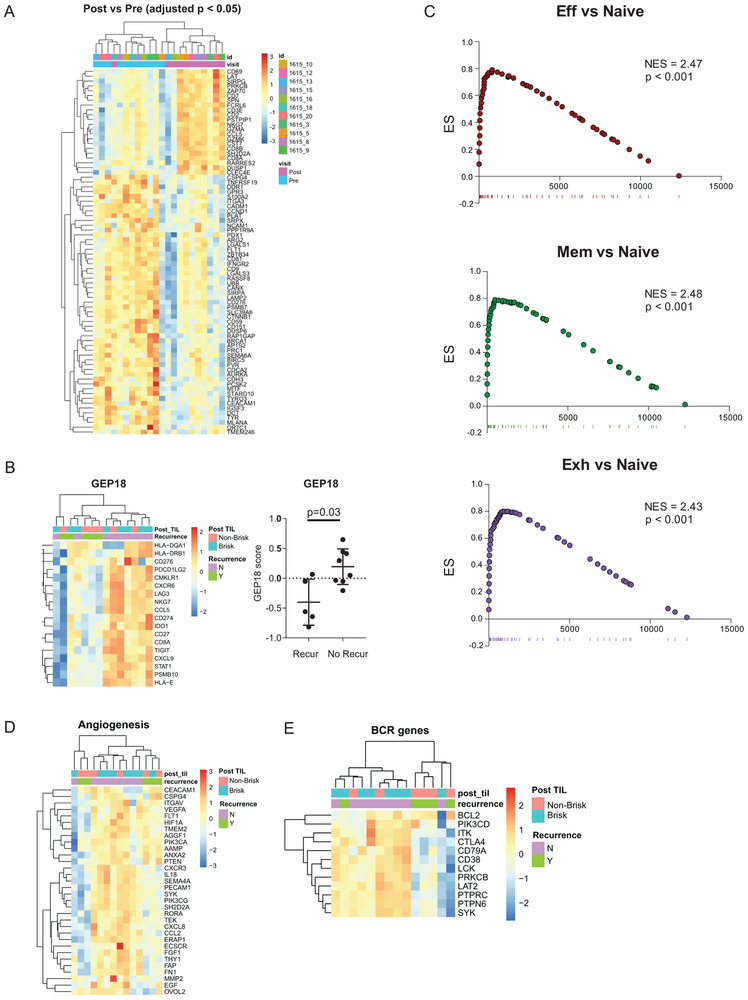
NanoString analysis of post-treatment tumor revealed a distinct signature of T cell activation compared to pre-treatment including CD8A, CD8B, GZMA, GZMK, ZAP70, LAT, and CD69 (Extended Data Figure 4A). Pre-treatment, we also identified a strong “Neoadjuvant Response Signature” (NRS) including genes involved in T cell activation, adaptive immune response, and T cell migration (Figures 4D-F) that correlated with post-treatment TIL responses and recurrence-free survival. This finding is consistent with T cell inflamed tumors described in Stage IV disease11,15-17. Indeed, an 18-gene IFNγ T cell-inflamed signature, GEP18, predictive of clinical response to pembrolizumab in unresectable locally advanced and metastatic cancers15,18, was also associated with clinical response in our largely stage III setting (Extended Data Figure 4B). The NRS strongly enriched for the transcriptional signatures19 from either effector (TEFF) or memory (TMEM) CD8 T cells compared to naïve CD8 T cells (TN) and also enriched strongly for the signature of TEX from mice19 (Extended Data Figure 4C). Moreover, there was a stronger enrichment of the TEX signature when compared directly to TEFF (Figure 4G), suggesting the importance of pre-existing TEX. In addition, high expression of genes involved in angiogenesis and B cell receptor pathways were associated with clinical benefit (Extended Data Figures 4D-E) implicating possible T cell independent mechanisms of anti-PD-1 response and changes in the tumor microenvironment.
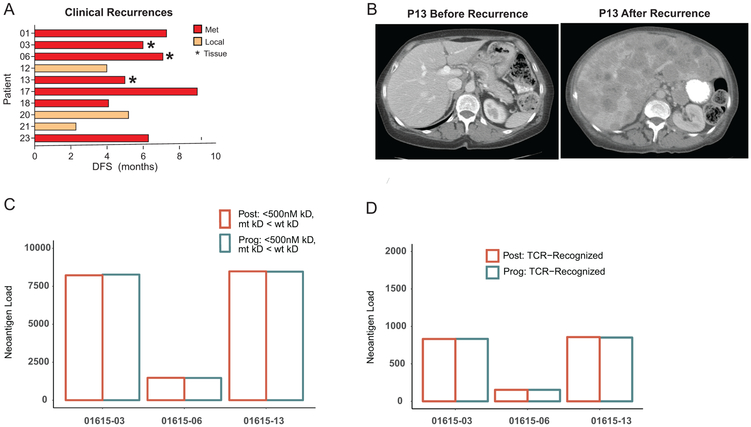
Ten patients have recurred thus far, seven of whom developed distant metastatic disease, two of whom have died (Extended Data Figures 5A-B). To study mechanisms of resistance, we collected recurrence tumor samples from three patients and compared them to the resection samples (Figures 4H-K). There was no significant difference in the number of predicted total, or high-quality, neoantigens likely to be recognized by TCR (Extended Data Figures 5C-D). Patients 01615-06 and 01615-13, however, displayed few CD8 T cells by immunofluorescence (IF), low Ki67 and a low percentage of PD-1+ or PD-1+CTLA4+ CD8 T cells in the resection tumor and at progression (Figures 4I-J). Furthermore, there was a prominent increase in CD163+ myeloid cells with a concomitant decrease in CD3+ cells in both patients (Figure 4I). Although patient 01615-13 had evidence of T cell activation upon progression based on NanoString, there was a concomitant signature of myeloid chemotaxis and activation, including CD14, CCL8, CXCL14, CLEC5A and CSF1R (Figure 4H). Moreover, analysis of whole exome sequencing data revealed a deleterious single-nucleotide variant (SNV) at the TP53 locus (encoding p53) that was heterozygous at resection (allele frequency of 0.55), but homozygous at recurrence (allele frequency of 0.84; Figure 4K). P53, a tumor suppressor, may also play a role in immunogenic cell death, CD8 T cell responses, and suppressive myeloid subsets such at MDSCs20. Thus, p53 loss may lead to a more aggressive tumor at recurrence and decreased anti-tumor immunity, including increased suppressive myeloid cells. In patient 01615-06, there was little transcriptomic change between resection and recurrence (Figure 4H); the underlying drivers of progression are unknown. Finally, patient 01615-03 had prominent T cell infiltration post treatment including many PD-1+CTLA4+ cells with a high percentage of Ki67+ cells (Figures 4I-J). This population of cells phenotypically resembled TEX cells that are associated with positive clinical responses in other settings3,4,21, suggesting a disconnect between this reinvigorated TEX-like population and the lack of tumor control. Whole exome sequencing, however, revealed loss of heterozygosity in B2M upon recurrence, with an increase in mutant allele frequency from 0.52 to 0.91 (Figure 4K), providing a possible explanation for tumor progression despite a vigorous CD8 T cell response to anti-PD-1. Thus, although mechanisms of adaptive resistance have been described that involve mutation, including loss of B2M22-24, suppressive myeloid cells25, and “cold tumor” microenvironments17,26,27; these data highlight the importance of a neoadjuvant approach to potentially identify these mechanisms early on during treatment.
Discussion
Despite the clinical success of checkpoint blockade, we still understand relatively little about the precise mechanism(s) of response or resistance to these treatments. A neoadjuvant approach with anti-PD-1 therapy allowed us to address this question and was feasible and effective in stage IIIB/C and IV melanoma with 63% disease-free survival and 93% overall survival at 2 years. There are several major findings. First, we observed a rapid immune response after PD-1 blockade with T cell reinvigoration peaking 7 days post treatment initiation and complete or major pathologic response in 30% of patients within 3 weeks. Notably, these patients with early complete or major pathological tumor response have 100% disease-free survival at 24 months. In contrast, patients without robust pathological responses at surgery had a poor prognosis with greater than 50% risk of recurrence despite adjuvant therapy. A neoadjuvant approach might allow for early identification of high-risk patients and a change to more effective adjuvant therapy. Second, the data from TIL pre and post-treatment support a role for CD8 T cells with characteristics of TEX. Finally, these studies provide evidence for mechanisms of response and adaptive resistance including immune failure, immune regulation and immune escape.
Many patients displayed rapid (i.e. 3 weeks) complete or nearly complete pathologic responses. These data support the notion that anti-PD-1 revitalizes an already-existing T cell response. In support of this hypothesis are the observations that baseline proliferation of CD8 T cells was associated with clinical benefit and that the immunologic pharmacodynamic response to PD-1 blockade peaked at day 7 after treatment initiation. Although new T cell priming cannot be excluded, this rapidity and robustness favors a model where previously primed CD8 T cells become exhausted and then, upon PD-1 blockade, get rapidly reinvigorated. These immune response data are consistent with the rapid anti-tumor responses observed, but also suggest that preventing or avoiding early concomitant immunoregulatory (e.g. Treg) and/or later acquired resistance may require combination immunotherapies delivered at times that synergize with this early response to anti-PD-1 therapy.
An obvious question relates to the specificity of the CD8 T cells responding to PD-1 blockade. Previous work has shown that the TEX phenotype cells in the blood responding to PD-1 blockade were enriched for TCRs also found in TILs3. Although it is possible that some of these cells are bystander cells that can be found in blood and tumor, our current studies suggest that CMV-specific CD8 T cells are not likely to respond to anti-PD-1 whereas tumor-specific CD8 T cells targeting gp100 do respond. Moreover, recent work indicates that the PD-1+CD39+ subset of TEX are enriched for tumor-specific T cells compared to the CD39- subset5 and we find strong enrichment for the CD39+ subset among the cells responding to PD-1 blockade in this cohort. It will be important to extend these types of studies to larger cohorts with detectable shared or neo-antigen specific CD8 T cell populations. However, based on these previous studies, the cells responding to PD-1 blockade in the cohorts analyzed here are likely to be enriched for tumor-specific CD8 T cell populations.
We identified an association between accumulation of TEX phenotype (EomeshiTbetlow) CD8 T cells and clinical benefit. Minimal changes in Ki67 at 3 weeks contrasted with an increased frequency of TEX-like cells in the tumor after anti-PD-1. These observations suggest that either reinvigoration in the tumor occurs before week 3, and/or that TEX are reinvigorated peripherally and migrate to the tumor. Indeed, our data demonstrate a peak of TEX reinvigoration in the blood by day 7 and data in mice suggest that immune activity in the tumor occurs early and transiently, whereas a systemic immune response may persist longer28. Ki67+ Tregs were also observed in the tumor and correlated with Ki67+ CD8 T cells at week 3. Moreover, proliferation of Tregs was associated with poor disease-free survival. PD-1 blockade may therefore reinvigorate TEX in the tumor but also activate Tregs; the relationship between these changes may be an important feature influencing clinical outcome and an opportunity for Treg-modulating drugs. Analysis of pre-treatment tumor also revealed Ki67 expression by non-naïve CD8 T cells as a potential pre-treatment biomarker of response. This observation is consistent with published data in Stage IV melanoma where a pre-existing CD8 T cells in the tumor are associated with clinical benefit following PD-1 blockade11. Our data demonstrate that this pre-existing CD8 T cell immune response is initiated earlier in cancer development (i.e. Stage III disease). Consistent with this notion, NanoString data demonstrated a distinct inflamed signature prior to therapy. These observations support the idea that even in localized disease, the tumor microenvironment is already primed for response or non-response to checkpoint blockade. In patients who recurred, distinct genomic and cellular resistance factors were identified, including B2M and P53 loss of heterozygosity, as well as increased myeloid cell accumulation. Future neoadjuvant studies will allow further study of mechanisms of resistance that may help tailor therapies and/or clinical trials in patients.
METHODS
Study Design and Patient Population
This phase 1b clinical trial (), was a single institution investigator-initiated study sponsored by the University of Pennsylvania. The protocol and its amendments were approved by the Institutional Review Board at the University of Pennsylvania, and all patients provided written informed consent. Patients were eligible for enrolment if they were 18 years of age or older, had an Eastern Cooperative Group performance status of 0 or 1, adequate organ function according to protocol criteria, and had measurable resectable clinical stage III or resectable stage IV melanoma. Patients had to have an adequate tumor size, in the opinion of the surgical oncologist and study team, to allow for collection of biopsy tissue equivalent to at least two to four core biopsies for the pre-treatment biopsy, with an expectation that an equal or greater amount of tissue would remain after biopsy. Patients could not have received prior ipilimumab or other immune therapies. Prior BRAF directed therapies were permitted. Patients with uveal or mucosal melanoma were not eligible, nor were patients on systemic steroids or immunosuppression, patients who had received radiation to the resectable tumor, or patients with active brain metastasis.
Treatment and Assessment
After obtaining informed consent, all patients underwent a baseline pre-treatment biopsy, which consisted of the equivalent amount of tissue of at least two to four core biopsies. Patients then received a single flat dose of pembrolizumab 200 mg intravenously, followed by complete resection three weeks later. Patients also provided paired blood samples at the pre-treatment and post-treatment time points. After resection and upon surgical recovery, patients continued to receive adjuvant pembrolizumab every 3 weeks for up to one year, or until the time of recurrence or any unacceptable treatment related toxicity (Figure 1A). Both the biopsies and the resection specimens were processed in the Department of Pathology and Laboratory Medicine, Hospital of University of Pennsylvania and immediately snap or formalin fixed and paraffin embedded, with a portion of unfixed tissue allocated for extraction of tumor infiltrating lymphocytes (TIL) for translational studies. Gross photos were taken of resection specimens. Patients were assessed for pathologic response at the 3-week resection time point, with the resection tumor assessed for percentage of viable tumor by a melanoma pathologist. Complete pathologic response was defined as the absence of viable tumor based on hematoxylin and eosin staining. A major pathologic response was defined as less than 10% of viable tumor. Pre- and post-treatment tumors were also assessed for TIL infiltration and scored as either brisk (diffuse lymphocytes throughout the tumor), non-brisk (only foci of lymphocytes), or absent.29 Imaging (PET CT or CT of the chest, abdomen, and pelvis) was performed at baseline and on study. Patients were assessed for recurrence and followed for disease-free survival and overall survival. Grading of adverse events was performed using the Common Terminology Criteria– of Adverse Events version 4. The objective of this report is to examine the pathologic complete response rate at the time of resection after 1 dose of pembrolizumab, and to evaluate pathologic response as a predictor of disease-free survival. To date, 29 patients have received treatment on the trial and are included in this analysis.
Statistical Analysis
The primary objectives of the clinical trial are to establish safety and obtain paired tissue samples for analysis of immunologic effects. The target sample size was 30 patients. If 5 or fewer of 30 patients experienced a severe (grade 3 or higher) adverse event that was not attributable to pembrolizumab or to surgery alone, then safety was established since the true toxicity rate is likely no higher than 30% based on the upper bound of the 2-sided 95% confidence interval.
A secondary objective of the trial was to study the biological changes in paired tumor tissue specimens prior to and in the presence of anti-PD-1. For this report, the primary endpoint was disease-free survival (DFS) from the date of definitive surgery to first documented disease recurrence, death due to any cause, or last patient contact documenting disease-free status.The rate of pathologic response (defined as complete or major pathologic response7,8) and exact 95% confidence interval were estimated for 29 patients who have enrolled and undergone surgery on the trial. Median potential follow-up was estimated by the reverse Kaplan-Meier method. Disease-free survival (DFS) from landmark date of definitive surgery to first documented disease recurrence, death due to any cause, or last patient contact documenting disease-free status was estimated by the Kaplan-Meier method. Cox proportional hazards regression modeling was employed to estimate the magnitude of effect of pathologic response on DFS. Multivariable models were employed to adjust for established prognostic factors. Unadjusted and adjusted hazard ratios and their 95% confidence intervals were estimated. To address the low event rate in the group with pathologic CR and the group with brisk TIL post-treatment, we applied Firth’s penalized regression method for the Cox proportional hazard model, with the penalized likelihood ratio test.30 For normal data, parametric Student’s t-test and paired t-test were used for unpaired and paired analyses, respectively. For non-normal data, nonparametric Mann-Whitney or Wilcoxon matched-pairs signed rank tests were used for unpaired and paired analyses, respectively. Correlations between continuous variables were determined by Pearson’s r coefficient. Fisher’s exact test was employed to test for association between two categorical variables. McNemar’s test was used to test concordance between paired categorical variables to determine treatment effect (i.e., pre-treatment and post-treatment TIL score). Statistical analyses were performed using either IBM SPSS v23, R, or Graphpad Prism.
Sample Processing: PBMC and Tumor
Patient blood was drawn in sodium heparin tubes, kept at room temperature and processed within 18 hours of blood draw. Whole blood was centrifuged and plasma samples were collected, aliquoted in the volume of 1 ml per cryotube and stored at −80°C. Blood samples were reconstituted by adding an equal volume of Hank’s balanced salt solution (HBSS, Corning MT21021CM) to the volume of collected plasma. Reconstituted blood samples were diluted 2-fold in HBSS and Ficoll (Ficoll-Paque PLUS density gradient medium, GE Healthcare Life Sciences, 17-1440-03) was layered underneath. The volume of Ficoll-Paque PLUS used was equal to that of the undiluted blood sample. The buffy coat was collected and washed twice with HBSS. ACK lysis buffer (Lonza 10-548E) was used to lyse the red blood cells, if necessary. Melanoma tumor samples were processed within 4 hours after excision from the patient. Samples were cut into 2-3 mm pieces with a scalpel in a RPMI 1640 (Corning 10-040-CM) containing petri dish. A 70 μm cell strainer (Falcon 352350) was placed on top of a 50 ml conical tube and the tissue fragments were transferred to the cell strainer with a pipette. The cells were released by gently grinding the tissue fragments using the thumb depressor of a sterile syringe plunger placed against the cell strainer. Cells were washed twice and counted in a hemocytometer. The numbers of lymphocytes and melanoma cells were distinguished by their morphology and recorded separately.
Flow Cytometry
Fresh trial PBMC and tumor suspension with stained with a master mix of antibodies for surface stains including CD4 (Biolegend, OKT4), CD8 (BD, RPA-T8), CD45RA (Biolegend, HI100), Tim-3 (Biolegend, F38-2E2), Lag3 (3DS223H, ebioscience), CD39 (Biolegend, A1), CD27 (BD, L128), and PD-1(BD, EH-12) and intracellular stains for FoxP3 (BD, 259D/C7), CTLA4 (BD, BNI3), Eomes (ebioscience, WD1928), Tbet (Biolegend, 4B10), and Ki67 (BD, B56). HLA-A2 tetramers were generated at the NIH tetramer facility for the melanoma gp100 peptide G280 (YLEPGPVTV) and CMV peptide pp65 (NLVPMVATV). Permeabilization was performed using the Foxp3 Fixation/Permeabilization Concentrate and Diluent kit (eBioscience). PD-1 on post pembrolizumab specimens was detected using anti-human IgG4 PE (Southern Biotec) as previously described3,31. Cells were resuspended in 1% para-formaldehyde until acquisition on a BD Biosciences LSR II cytometer and analyzed using FlowJo (Tree Star).
Immunohistochemical Staining and Scoring of PD-L1 (Single Chromogenic)
Whole tissue sections cut from FFPE tissue blocks were deparaffinized and rehydrated with serial passage through changes of xylene and graded ethanols. All slides were subjected to heat induced epitope retrieval in Envision FLEX Target Retrieval Solution, High pH (Dako, Carpineteria CA). Endogenous peroxidase in tissues was blocked by incubation of slides in 3% hydrogen peroxide solution prior to incubation with primary antibody (anti-PD-L1, clone 22C3, Merck Research Laboratories, Palo Alto CA or anti-PD-1 clone NAT105, Cell Marque, Rocklin CA) for 60 minutes. Antigen-antibody binding was visualized via application of the FLEX+ polymer system (Dako, Carpineteria CA) via application of mouse Envision system, and application of 3, 3’ diaminobenzidine (DAB) chromogen (Dako, Carpineteria CA). Stained slides were counterstained with hematoxylin and cover slipped for review. Scoring of PD-L1 was conducted by a pathologist blinded to patient characteristics and clinical outcomes.32 A semi-quantitative 0–5 scoring system was applied; Negative: 0, Rare: 1 = individuated positive cells or only very small focus within or directly adjacent to tumor tissue, Low: 2 = infrequent small clusters of positive cells within or directly adjacent to tumor tissue, Moderate: 3 = single large cluster, multiple smaller clusters, or moderately dense diffuse infiltration, within or directly adjacent to tumor tissue, High: 4 = single very large dense cluster, multiple large clusters, or dense diffuse infiltration, Very high: 5 Coalescing clusters, dense infiltration throughout the tumor tissue.
Immunofluorescent Staining and Image Acquisition.
Multiplexed immunofluorescent staining was performed on Dako autostainer (Agilent, Carpinteria, CA). The FFPE sections were rehydrated in a series of graded alcohols to distilled water. Antigen retrieval was performed in TRS citrate buffer (pH 6.0) using a pressure cooker. The slides were blocked in 1% casein blocking buffer for 30 minutes, followed by incubation with primary antibody for 1 hour. Slides were incubated with polymer HRP-secondary antibody for 30 minutes followed by application of fluorophore - 4 tyramide signal amplification (TSA)-dye (Opal 7 color kit, Perkin Elmer, Hopkington, MA). After detection of first primary antibody, the slides were stripped off any primary and secondary antibodies by treating the slides in AR6 (Perkin Elmer) antibody stripping buffer in a microwave oven. Sequentially, the slides were stained with second or third primary antibody and the process was repeated. Nuclear staining was carried out with DAPI followed by cover slipping. CD8xFoxP3xCD163 triplex staining was accomplished with PerkinElmer Opal kit (Perkin-Elmer, Waltham, MA). CD8, FoxP3 and CD163, labeled with Opal fluorophores 690, 570 and 520, respectively were sequentially applied to the tissue sections. CD8xCD3 duplex immunofluorescent staining was accomplished with ThermoFisher Scientific Tyramide Signal Amplification Kit (ThermoScientific, Waltham, MA). The sequence for antibodies staining is CD8 and CD3, labeled with Alexa fluorophores 488 and 568, respectively. Epifluorescence multispectral whole slides images of all sections were acquired through the Vectra 3.0 Automated Quantitative Pathology Imaging System (PerkinElmer) at 10x and 20x magnifications. Quantitative image analyses were carried out on10X images using Halo Highplex FL module (Indica labs, Corrales, MN, USA).
Preparation of FFPE RNA Isolation and Gene Expression Analysis using the NanoString nCounter™ System
RNA was isolated from slides of FFPE tissue for analysis on the NanoString nCounter™ gene expression platform (NanoString Technologies, Seattle, WA). Prior to RNA isolation, tissue sections were deparaffinized in xylene for 3 × 5 min, then sequentially rehydrated in 100% ethanol for 2 × 2 min, 95% ethanol for 2 min, 70% ethanol for 2 min and then immersed in dH2O until ready to be processed. Tissue was lysed on the slide by adding 10-50ul of PKD buffer (Qiagen catalog #73504). Tissue was then scraped from the slide and transferred to a 1.5 ml eppendorf tube. Proteinase K (Roche Catalog #03115836001) was added at no more than 10% final volume and the RNA lysate was incubated for 15 min at 55ºC and then 15 min at 80ºC. RNA was isolated using Qiagen RNeasy FFPE kit per manufacturer’s protocol (Qiagen, Valencia, CA). Sample concentration was measured on the NanoDrop Spectrophotometer (Thermo Fisher Scientific, Foster City, CA) per manufacturer protocol. The resulting total RNA was stored at −80ºC until gene expression profiling was performed using the NanoString nCounter™ system. Per sample, 50ng of total RNA isolated from FFPE was mixed with a 3′ biotinylated capture probe and a 5′reporter probe tagged with a fluorescent barcode, from a custom designed gene expression codeset. Probes and lysate were hybridized overnight at 65 ºC for 12-16 hours. Hybridized samples were then run on the NanoString preparation station using their high sensitivity protocol per manufacturer’s instructions (NanoString Technologies, Seattle, WA). The samples were scanned at maximum scan resolution capabilities using the nCounter™ Digital Analyzer (NanoString Technologies, Seattle, WA). All sample and data normalization occurred within the nCounter™ digital analyzer software, nSolver™. Specifically, the raw code count data was normalized using a positive control normalization factor based on the spiked-in positive control raw counts and also a content normalization factor derived from the raw counts of a set of relevant housekeeping genes. Transcripts with counts less than or equal to the highest embedded negative controls (background noise) in that sample are first set to its background. The gene count for each gene is then subtracted from this background so that each sample has the same footing where zero numbers represent undetectable noise.
NanoString Gene Expression Analysis.
R package ‘limma_3.34.9’ was used to do pairwise comparisons of the normalized NanoString gene expression data. R package ‘pheatmap_1.0.10’ was used for creating heatmaps to display NanoString gene expression data. Metascape.org was used to enrich genes for GO biological processes. Gene Set Enrichment Analysis (GSEA) using Broad Institute software (https://software.broadinstitute.org/gsea/) was used to check for enrichment of gene signatures from microarray data in the NanoString data. The 18-gene T-cell inflamed Gene Expression Profile (GEP) was developed as described by Ayers et al. and consists of genes related to antigen presentation, chemokine expression, cytolytic activity and adaptive immune resistance15. These genes are as follows: CCL5, CD27, CD274 (PD-L1), CD276 (B7-H3), CD8A, CMKLR1, CXCL9, CXCR6, HLA-DQA1, HLA-DRB1, HLA-E, IDO1, LAG3, NKG7, PDCD1LG2 (PDL2), PSMB10, STAT1, TIGIT. The GEP score was computed by taking a weighted sum of the housekeeping normalized values of the 18 genes on the GEP18 signature.
DNA Isolation and Exome Sequencing
Whole exome sequencing was performed on formalin-fixed paraffin-embedded (FFPE) tumor tissue with matched germline DNA. Manual macrodissection was performed on FFPE slides, if necessary, using a scalpel and H&E stained slide as a guide. Tissue deparaffinization and DNA extraction were performed using standard methods. DNA was quantified using Qubit dsDNA BR Assay (Invitrogen) and sheared to an average of 250 bp with the Covaris LE220 ultrasonicator (Covaris Inc., Woburn, MA). For genomic library preparation, paired-end libraries were prepared using the NEBNext Ultra Kit (New England Biolabs, Ipswich, MA) and DNA library quality and fragment size were measured with an Agilent 2100 Bioanalyzer. Exome sequences were enriched from the genomic libraries according to the manufacturer’s protocol with the SureSelectXT2 Human All Exon V6+COSMIC (Agilent, Santa Clara, CA). Samples were pooled and sequenced on the Illumina HiSeq4000 with 150 bp paired end reads with an average target depth of 87x.
Bioinformatic Analysis of Whole Exome Sequencing Data
Fastq data for tumor and matched normal DNA were aligned to the Genome Reference Consortium Human Build 37(GRCh37) using the Burrows-Wheeler Aligner34. Resultant bam files were further processed following Genome Analysis Toolkit (GATK) “Best Practices” 35,36. Somatic SNV and indels were detected using Mutect237 included as part of GATK v4.0.7.0. Lower confidence variants, determined from cross-sample contamination estimates and sequence context-dependent artifacts, were filtered out from the somatic VCF files. Copy number alterations and regions displaying allele-specific loss of heterozygosity (LOH) were determined using Sequenza38. Unique somatic SNV and indel mutations in post-therapy versus progression samples were determined by pairwise overlap analysis implemented in data.table in R. Further, allele frequencies for mutations common to both post and progression samples were investigated for changes in zygosity. Unique somatic CNVs across post-therapy versus progression tumors were determined through an analysis of overlapping chromosomal coordinates using the IRanges Bioconductor (v. 3.7) package implemented in R.
Neoantigen Analysis
VCF files containing somatic non-synonymous SNVs and indels passing two-stage filtering from Mutect2 were annotated for wildtype peptide sequence using the Wildtype plugin from ensembl-Variant Effect Predictor (VEP, v. 92.0). Further, the downstream effects of frameshift variants on the protein sequence were determined using the Downstream plugin from ensembl-VEP. Prediction of HLA-binding neoantigens ranging from 9-11 amino acids in length was accomplished using NetMHCcons39 implemented within pVacSeq40. Neoantigens were subsequently filtered by the predicted binding dissociation constant (kD <= 500nM) and by agretopicity, requiring that mutant peptides have greater binding affinities than their corresponding wild type sequences. All 9-mers were then analyzed for their likelihood of T-cell receptor (TCR) recognition using a thermodynamic neoantigen fitness model, as previously described33. Neoantigen load for all 9-11 mers filtered by dissociation constant and agretopicity described above as well as neoantigens likely to be recognized by TCRs were plotted using the ggplot2 package within R (v 3.5.0). Further, loss of TCR-recognized neoantigens from post-therapy to progression was determined by pairwise overlap analysis implemented in the data.table package in R.
Code Availability
Custom code used to analyzed tumor whole exome sequencing data are available at https://zenodo.org/badge/latestdoi/162582612
Data Availability
Nanostring data that support the findings have been deposited in NCBI Gene Expression Omnibus and are accessible through GEO Series accession number GSE123728. DNA whole exome sequencing data have been deposited in SRA and is accessible under SRA accession number PRJNA510621. All other relevant data are available from the corresponding author upon reasonable request.
Extended Data
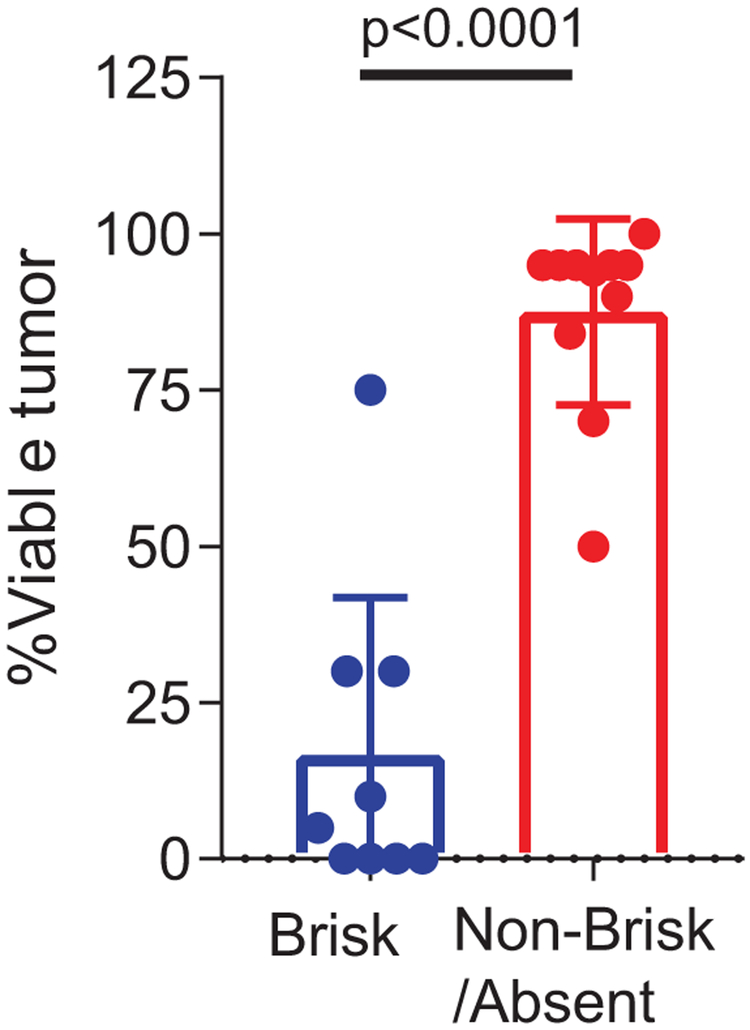
Extended Data Fig. 1. TIL score associated with pathologic response.
Percent viable tumor between brisk (n = 9) versus non-brisk/absent tumors (n = 11). P value calculated using two-sided Mann Whitney test.
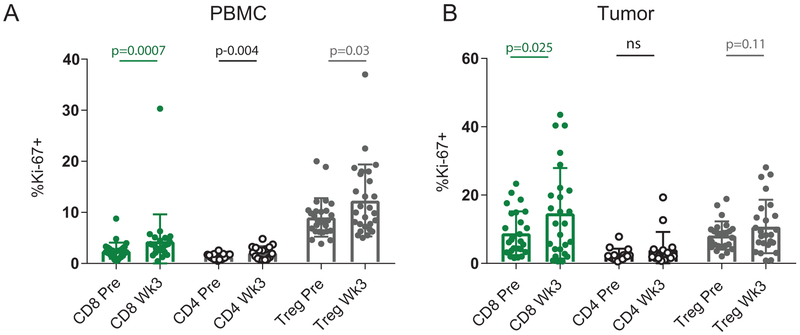
Extended Data Fig. 2. Immune response to anti-PD-1 for T cell subsets in blood and tumor.
A, %Ki67 expression in CD8, conventional CD4, and Treg (FoxP3+ CD4) T cells pre and post in blood (n = 28 independent paired patient samples for CD8 comparisons, n = 17 independent paired patient samples for CD4 comparisons, and n = 27 independent patient samples for Treg comparisons). Two-sided Wilcoxon matched-pairs test was performed for CD8 and Treg comparisons. Two-sided t-test was performed for CD4 comparison. B, %Ki67 expression in CD8, conventional CD4, and Treg (FoxP3+ CD4) T cells pre and post in tumor (n = 26 independent paired patient samples for CD8 comparisons, n = 15 independent paired patient samples for CD4 comparisons, and n = 25 independent paired patient samples for Treg comparisons). Two-sided Wilcoxon matched-pairs test was performed for CD4 and Treg comparisons. Two-sided t-test was performed for CD8 comparison.
Extended Data Fig. 3. Cellular determinants of response and resistance to anti-PD-1.
A, Changes in tumor PD-L1 pre versus post-treatment using immunohistochemistry staining (n = 9 independent paired patient samples). ** p<0.01 using two-sided Wilcoxon matched-pairs test. B, Correlation of % Ki67+ in non-naïve CD8 T cells versus %Ki67+ in Tregs (FoxP3+ CD4) (n = 21 independent patient samples), R-score and p-value generated using Pearson’s correlation. C, 33 post-treatment immune parameters classified by recurrence using random forest analysis and ranked by importance score (n = 21 independent patient samples). Error bar denotes mean +/− sd for 1000 random forest iterations. D, % expression of selected markers in tumor between patients with recurrence (9 independent patient samples) and no recurrence (12 independent patient samples). P value calculated using two-sided Mann-Whitney test. E, Correlation of % Ki67+ in Tregs (FoxP3+ CD4) versus %Eomes+Tbet- in non-naïve CD8 (n = 21 independent patient samples), R-score and p-value generated using Pearson’s correlation. F, 25 pre-treatment immune parameters classified by recurrence using random forest analysis and ranked by importance score (n = 21 independent patient samples). Error bar denotes mean +/− sd for 1000 random forest iterations. G, % expression of selected markers in tumor between patients with recurrence (9 independent patient samples) and no recurrence (12 independent patient samples). Two-sided t test was used for CD45RA-CD27+ and CD45RA+CD27+ comparisons. Two-sided Mann Whitney test was used for CD8 Ki67+ and CD4 Ki67+ comparisons. Error bar denotes mean +/− sd. H, Scatter plot of %Ki67+ in non-naïve CD8 versus %Ki67+ in FoxP3+ CD4 (Tregs) at pre-treatment stratified by recurrence status. Dotted line denotes non-naïve CD8 Ki67+ of 5.5 calculated by CART analysis as the optimal cutpoint separating recurrence vs no recurrence. (n = 21 independent patient samples).
Extended Data Fig. 4. Immune signatures associated with clinical response.
A, Heatmap of differentially expressed genes between pre-treatment and post treatment tumor (n = 11 independent paired patient samples). Differentially expressed genes identified using a FDR cutoff of p=0.05 after adjusting for multiple comparisons. B, Heatmap and GEP score between patients with recurrence (n = 5 independent patient samples) and no recurrence (n = 8 independent patient samples). P value calculated using two-sided Mann-Whitney test. Error bar denotes mean +/− sd. C, Gene set enrichment analysis of NRS genes that were enriched in TEFF, TMEM, and TEX versus TNAIVE cell signatures from19. D, Heatmap of angiogenesis associated genes from Gene Ontology. E, Heatmap of B cell receptor associated genes from Gene Ontology.
Extended Data Fig. 5. Clinical progression and neoantigen quantity and quality.
A, Disease-free survival of patients that recurred. B, CT image before and after of a patient with recurrent metastatic disease. C, Neoantigen load based on predicted binding (predicted kD of <500nM and mutant kD< WT kD). D, Number of high quality neoantigens that are likely to be recognized by TCR based on neoantigen fitness model33 at post treatment versus recurrence timepoints.
Supplementary Material
Acknowledgements:
Clinical and correlative studies were supported in part by the SPORE in Skin Cancer: P50-CA174523 (X.X., K.L.N., L.M.S., R.K.A., R.M., G.K.), P01-CA114046 (X.X.), T32-2T32CA009615 (A.C.H.), The NIH/NCI Cancer Center Support Grant P30-CA016520 (R.K.A, K.L.N, L.M.S, R.M.), and NIH grants to E.J.W. AI105343, AI108545, AI117950, AI082630, CA210944, AI114852 (R.S.H), The Tara Miller Foundation (A.C.H), the Melanoma Research Alliance (E.J.W.), the David and Hallee Adelman Immunotherapy Research Fund (E.J.W.), the Heisenberg program BE5496/2-1 of the DFG (B.B), and The Parker Institute for Cancer Immunotherapy Bridge Scholar Award (A.C.H). Merck, Inc. supplied drug and supported clinical and translational aspects of this study. The Human Immunology Core (HIC) and the Tumor Tissue and Biospecimen Bank (TTAB) of the University of Pennsylvania (supported by P30-CA016520) assisted in tissue collection, processing, and storage.
Footnotes
Competing Interests Statement. Merck provided funding and drug for the clinical trial. Merck performed immunohistochemistry, immunofluorescence, and Nanostring assays, and played a role in analysis of these data. Merck played no role in the design, data collection, decision to publish, or preparation of the manuscript. R.J.O, L.A., Q.Z, R.M., W.B, and J.H.Y is currently or was employed at Merck when engaged in this project. E.J.W. is a member of the Parker Institute for Cancer Immunotherapy which supported the UPenn cancer immunotherapy program. E.J.W. has consulting agreements with and/or is on the scientific advisory board for Merck, Roche, Pieris, Elstar, and Surface Oncology. E.J.W. has a patent licensing agreement on the PD-1 pathway with Roche/Genentech.
References
- 1.Fridman WH, Pages F, Sautes-Fridman C & Galon J The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 12, 298–306 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Vesely MD, Kershaw MH, Schreiber RD & Smyth MJ Natural innate and adaptive immunity to cancer. Annu Rev Immunol 29, 235–271 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Huang AC, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature 545, 60–65 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamphorst AO, et al. Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1-targeted therapy in lung cancer patients. Proc Natl Acad Sci U S A 114, 4993–4998 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simoni Y, et al. Bystander CD8(+) T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 557, 575–579 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Thommen DS, et al. A transcriptionally and functionally distinct PD-1(+) CD8(+) T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat Med (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hellmann MD, et al. Pathological response after neoadjuvant chemotherapy in resectable non-small-cell lung cancers: proposal for the use of major pathological response as a surrogate endpoint. Lancet Oncol 15, e42–50 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forde PM, et al. Neoadjuvant PD-1 Blockade in Resectable Lung Cancer. N Engl J Med 378, 1976–1986 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amaria RN, et al. Neoadjuvant immune checkpoint blockade in high-risk resectable melanoma. Nat Med (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herbst RS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 515, 563–567 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tumeh PC, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blackburn SD, et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol 10, 29–37 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paley MA, et al. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science 338, 1220–1225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zappasodi R, Merghoub T & Wolchok JD Emerging Concepts for Immune Checkpoint Blockade-Based Combination Therapies. Cancer Cell 33, 581–598 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayers M, et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 127, 2930–2940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harlin H, et al. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer Res 69, 3077–3085 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taube JM, et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med 4, 127ra137 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cristescu R, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 362(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doering TA, et al. Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity 37, 1130–1144 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo G, Yu M, Xiao W, Celis E & Cui Y Local Activation of p53 in the Tumor Microenvironment Overcomes Immune Suppression and Enhances Antitumor Immunity. Cancer Res 77, 2292–2305 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daud AI, et al. Tumor immune profiling predicts response to anti-PD-1 therapy in human melanoma. J Clin Invest 126, 3447–3452 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Restifo NP, et al. Loss of functional beta 2-microglobulin in metastatic melanomas from five patients receiving immunotherapy. J Natl Cancer Inst 88, 100–108 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sucker A, et al. Genetic evolution of T-cell resistance in the course of melanoma progression. Clin Cancer Res 20, 6593–6604 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaretsky JM, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med 375, 819–829 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engblom C, Pfirschke C & Pittet MJ The role of myeloid cells in cancer therapies. Nat Rev Cancer 16, 447–462 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Ribas A Adaptive Immune Resistance: How Cancer Protects from Immune Attack. Cancer Discov 5, 915–919 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teng MW, Ngiow SF, Ribas A & Smyth MJ Classifying Cancers Based on T-cell Infiltration and PD-L1. Cancer Res 75, 2139–2145 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spitzer MH, et al. Systemic Immunity Is Required for Effective Cancer Immunotherapy. Cell 168, 487–502 e415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-Only References
- 29.Mihm MC Jr., Clemente CG & Cascinelli N Tumor infiltrating lymphocytes in lymph node melanoma metastases: a histopathologic prognostic indicator and an expression of local immune response. Lab Invest 74, 43–47 (1996). [PubMed] [Google Scholar]
- 30.Heinze G & Schemper M A solution to the problem of monotone likelihood in Cox regression. Biometrics 57, 114–119 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Brahmer JR, et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J Clin Oncol 28, 3167–3175 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen M, et al. Development and validation of a novel clinical fluorescence in situ hybridization assay to detect JAK2 and PD-L1 amplification: a fluorescence in situ hybridization assay for JAK2 and PD-L1 amplification. Mod Pathol 30, 1516–1526 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Luksza M, et al. A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature 551, 517–520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2 [q-bio.GN] [Google Scholar]
- 35.A framework for variation discovery and genotyping using next-generation DNA sequencing data DePristo M, Banks E, Poplin R, Garimella K, Maguire J, Hartl C, Philippakis A, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell T, Kernytsky A, Sivachenko A, Cibulskis K, Gabriel S, Altshuler D, Daly M, 2011. NATURE GENETICS 43:491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline Van der Auwera GA, Carneiro M, Hartl C, Poplin R, del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, Banks E, Garimella K, Altshuler D, Gabriel S, DePristo M, 2013. CURRENT PROTOCOLS IN BIOINFORMATICS 43:11.10.1–11.10.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MuTect2 is a somatic SNP and indel caller that combines the DREAM challenge-winning somatic genotyping engine of the original MuTect (Cibulskis et al., 2013) with the assembly-based machinery of HaplotypeCaller.
- 38.“Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data.” Favero F, Joshi T, Marquard AM, Birkbak NJ, Krzystanek M, Li Q, Szallasi Z and Eklund AC (2015). Annals of Oncology, 26, pp. 64–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karosiene E, Lundegaard C, Lund O & Nielsen M NetMHCcons: a consensus method for the major histocompatibility complex class I predictions. Immunogenetics 64, 177–186 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Hundal J, et al. pVAC-Seq: A genome-guided in silico approach to identifying tumor neoantigens. Genome Med 8, 11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Nanostring data that support the findings have been deposited in NCBI Gene Expression Omnibus and are accessible through GEO Series accession number GSE123728. DNA whole exome sequencing data have been deposited in SRA and is accessible under SRA accession number PRJNA510621. All other relevant data are available from the corresponding author upon reasonable request.