Abstract
In Brassica oleracea, heterosis is the most efficient tool providing impetus to hybrid vegetable industry. In this context, we presented the first report on identifying superior heterotic crosses for yield and commercial traits in cauliflower involving cytoplasmic male sterile (CMS) and doubled haploid (DH) lines as parents. We studied the suitability of genomic-SSRs and EST-SSRs based genetic distance (GD) and agronomic trait based phenotypic distance (PD) for predicting heterosis in F1 hybrids using CMS and DH based parents. 120 F1 hybrids derived from 20Ogura based CMS lines and 6 DH based testers were evaluated for 16 agronomic traits along with the 26 parental lines and 4 commercial standard checks. The genomic-SSRs and EST-SSRs based genetic structure analysis grouped the 26 parental lines into 4 distinct clusters. The CMS lines Ogu118-6A, Ogu33A, Ogu34-1A were good general combiner for developing early maturity hybrids. The SCA effects were significantly associated with heterosis suggesting non-additive gene effects for the heterotic response of hybrids. Less than unity value of σ2A/D coupled with σ2gca/σ2sca indicated the predominance of non-additive gene action in the expression of studied traits. The correlation analysis of genetic distance with heterosis for commercial traits suggested that microsatellites based genetic distance estimates can be helpful in heterosis prediction to some extent.
Introduction
In the plant kingdom, the family Brassicaceae holds a great agronomic, scientific and economic significance and comprises of more than 372 genera and 4060 species [1]. Brassica oleracea (CC, 2n = 18) constitutes a diverse group of economically and nutritionally important morphotypes known as cole vegetables (kale, kohlrabi, cabbage, cauliflower, broccoli, brussels sprout) [2]. The Brassica vegetables are also termed as ‘super-food’ as they are vital source of secondary metabolites, antioxidants, vitamins and minerals [3, 4, 5, 6]. Among the cultivated B. oleracea morphotypes, cauliflower (B. oleracea var. botrytis L.) is an important vegetable crop grown worldwide. Great efforts have been made to improve the productivity and quality of this crop because of its huge economic value and quality attributes [7]. The replacement of open-pollinated varieties with F1 hybrids become more pronounced in cole vegetables because of their high uniformity, better quality, tolerance to various biotic and abiotic stresses [5, 8]. The genetic mechanisms namely, sporophytic self-incompatibility (SI) and cytoplasmic male-sterility (CMS) have been used widely in hybrid breeding programme of B. oleracea [5, 8, 9, 10, 11, 12]. However, frequent breakdown of self-incompatibility has been reported in Brassica vegetables due to the high temperature sensitivity of S-alleles. Thus, SI lines are not always stable and result in ‘sibbed’ seed in hybrid population [11]. Moreover, maintenance of S-allele lines is time-consuming and expensive. In snowball cauliflower, SI system is very poor or not present at all [11, 13]. Under these circumstances, the CMS provides a better alternative for the heterosis breeding in cole crops [5, 8, 14].
Heterosis or hybrid vigor, is manifested as superior performance of F1 hybrids as compared to the parents [15, 16, 17]. Heterosis is highly complex phenomenon and different hypothesis and genetic basis have been suggested to explain the basis of heterosis. [15, 16, 18, 19, 20]. Further, the application of genomics tools has suggested the role of epigenetic regulations in explaining the heterosis phenomenon across the crops [16, 17, 19, 20]. Proper selection of inbreds and identification of superior heterotic combinations is crucial for exploiting heterosis in crop improvement. The traditional approaches of quantitative genetics like diallel, generation mean, line × tester analysis and estimating genetic components revealing various gene effects are effective in unraveling genetic basis of heterosis [5, 15, 21, 22]. The measures of both general combining ability (GCA) and specific combining ability (SCA) are necessary for selection of parental lines to develop heterotic combinations [23]. Estimation of GCA provides information on breeding value and additive genetic variance while, SCA is associated with non-additive effects (dominance effects, additive×dominant, and dominant×dominant interactions). Among different biometrical approaches, line × tester analysis is very efficient for estimating GCA effects of lines and testers, SCA effects of cross combinations and revealing information about the nature of gene actions [8, 21, 24]. The extent of heterosis has been reported to vary with the mode of reproduction, genetic distance of parents, traits under investigation, developmental stage of plant and prevailing environment [23, 25, 26, 27, 28, 29]. The pair-wise parental GD has been suggested as a good indicator of per se hybrid performance and recognition of heterotic groups [23, 25, 26, 27, 28, 29].
Different approaches are available to determine genetic distance depending upon morphological traits, horticultural data, biochemical characteristics and DNA markers based genotypic data [30, 31]. The SSR (simple sequence repeat) markers have been regarded as the markers of choice owing to their co-dominant inheritance, whole-genome coverage, abundance and high reproducibility [27, 32]. However, contradictory results have been reported with respect to the relationship between genetic distance and heterosis in different crops [15, 24, 25, 26, 27, 28, 29, 30, 33, 34]. According to Cress [35], the extent of parental genetic distance is essential but is not enough to assure the significant heterosis. It was also suggested that the better forecasting of heterosis is possible only when genetic distance is less than a definite threshold level [36]. The association of genetic distance and heterosis also depends upon the germplasm, population structure under investigation and methods of calculation.[29, 37]. The parents with lesser genetic distance can also display a high level of heterosis when closely related ecotypes of Arabidopsis are used to develop hybrids [16, 38]. Contrasting results are reported about relationship between phenotypic distance (PD), genetic distance (GD), heterosis and specific combining ability (SCA) in different crops [30, 34, 39, 40]. Teklewold and Becker [39] have reported significant positive association of PD with mid-parent heterosis (MPH), general combining ability (GCA) and hybrid performance for seed yield in Ethiopian mustard (Brassica carinata), while Hale et al. [26] found no correlation between PD and heterosis in broccoli.
The development of homozygous inbred lines is tedious and time consuming process in B. oleracea as these crops are highly heterozygous in nature [41]. Availability of completely homozygous inbred lines is pre-requisite in hybrid development. Development of inbreds through conventional approaches requires several generation selfing. Presence of high inbreeding depression makes the process of inbred development more complicated. On the other hand, development of doubled haploids (DHs) enables to produce large number of completely homozygous lines within a very short period of time. DH technology has been used widely in B. oleracea in different genetic and genomic research, such as QTL mapping, construction of high density genetic linkage maps and genome sequencing [41–51]. Our group has developed large number of DH lines on Brassica vegetables through isolated microspore culture [41, 52, 53]. The improved CMS lines of cauliflower have also been developed indigenously through protoplast fusion followed by recurrent backcrossing [9, 54]. Till date, only limited information is available regarding combining ability, gene action and heterosis using CMS systems and DH lines to improve yield and quality traits in B. oleracea. Moreover, no report is available in cauliflower regarding the association of GD with heterosis and combining ability for important traits. Several contrasting results have been reported in different crops in this context as heterosis is a complex biological phenomenon [16, 17, 19, 20]. Hence, the present investigation was conducted with the objectives (i) to identify heterotic combinations using CMS and DH lines and to analyze combining ability, nature of gene action and heritability for important traits. (ii) to find out correlation between microsatellites based genetic distance and morphological traits based phenotypic distance with heterosis and combining ability. The present investigation is the first report of heterosis and combining ability study based on CMS and DH lines in cauliflower.
Materials and methods
Plant materials
The field experiment was carried out at Baragram Experimental Farm of ICAR-Indian Agricultural Research Institute (IARI), Regional Station, Katrain, Kullu Valley, Himachal Pradesh, India. The experimental farm is located at 32.12N latitude and 77.13E longitudes with an altitude of 1,560 m above mean sea level. The basic plant material comprised of 20 genetically diverse Ogura CMS lines developed after more than nine generations of backcrossing (Table 1). These CMS lines were used as female parent in the breeding programme. Six DH based inbred lines of snowball cauliflower with abundant pollen production developed through isolated microspore culture (IMC) were used as testers (Table 1).
Table 1. Characteristics of parental lines (cytoplasmic male-sterile lines) and testers (doubled haploid lines) used in the study.
| Code | Line | Curd Color | Curd compactness | Curd covering by inner leaves | Riceyness | Anthocyanin pigmentation | Origin of germplasm |
|---|---|---|---|---|---|---|---|
| L1 | Ogu122-5A | White | Compact | PC | Absent | Absent | IARI, Katrain, India |
| L2 | Ogu115-33A | White | Compact | PC | Absent | Absent | IARI, Katrain, India |
| L3 | Ogu118-6A | White | Compact | PC | Absent | Absent | IARI, Katrain, India |
| L4 | Ogu307-33A | Creamy White | Compact | NC | Absent | Absent | IARI, Katrain, India |
| L5 | Ogu309-2A | Creamy White | Compact | PC | Absent | Absent | IARI, Katrain, India |
| L6 | Ogu33A | White | Compact | FC | Absent | Absent | IARI, Katrain, India |
| L7 | OguKt-2-6A | White | Compact | PC | Absent | Absent | IARI, Katrain, India |
| L8 | Ogu1A | White | Compact | PC | Absent | Absent | IARI, Katrain, India |
| L9 | Ogu13-85-6A | White | Compact | NC | Absent | Absent | IARI, Katrain, India |
| L10 | Ogu1-6A | White | Compact | PC | Absent | Absent | IARI, Katrain, India |
| L11 | Ogu2A | White | Compact | PC | Absent | Absent | IARI, Katrain, India |
| L12 | OguKt-9-2A | White | Compact | PC | Absent | Absent | IARI, Katrain, India |
| L13 | Ogu22-1A | Creamy White | Compact | PC | Absent | Absent | IARI, Katrain, India |
| L14 | Ogu122-1A | White | Compact | PC | Absent | Absent | IARI, Katrain, India |
| L15 | Ogu126-1A | White | Compact | PC | Absent | Absent | IARI, Katrain, India |
| L16 | Ogu12A | White | Compact | PC | Absent | Absent | IARI, Katrain, India |
| L17 | Ogu119-1A | Creamy White | Compact | PC | Absent | Absent | IARI, Katrain, India |
| L18 | Ogu34-1A | White | Compact | PC | Absent | Absent | IARI, Katrain, India |
| L19 | Ogu125-8A | White | Compact | FC | Absent | Absent | IARI, Katrain, India |
| L20 | Ogu33-1A | Creamy White | Compact | PC | Absent | Absent | IARI, Katrain, India |
| T1 | DH-18-8-1 | White | Compact | PC | Absent | Absent | IARI, Katrain, India |
| T2 | DH-18-8-3 | White | Compact | PC | Absent | Absent | IARI, Katrain, India |
| T3 | DH-53-1 | White | Compact | FC | Absent | Absent | IARI, Katrain, India |
| T4 | DH-53-6 | White | Compact | FC | Absent | Absent | IARI, Katrain, India |
| T5 | DH-53-9 | White | Compact | PC | Absent | Absent | IARI, Katrain, India |
| T6 | DH-53-10 | White | Compact | PC | Absent | Absent | IARI, Katrain, India |
L: lines, T: testers, PC: partly covered, FC: fully covered, NC: not covered
Experimental design
All the recommended package of practices suggested for raising cauliflower crop at IARI- Regional station, Katrain, were followed to grow a healthy crop [55]. The size of the plot was kept 3.0 x 3.0 m2 with an inter and intra-row spacing of 45 cm. The 20 CMS lines of cauliflower were crossed with 6 DH male fertile testers to generate 120 testcross progenies by following the line × tester mating design [21]. To avoid any natural pollination, CMS lines were grown under net house. The fully opened flowers of CMS lines were pollinated with the freshly dehisced pollen from DH testers. The 120 testcross progenies along with their 26 parental lines were evaluated for various morphological, horticultural and yield -related traits along with 4 CMS based commercial hybrids (HVCF-18, HVCF-29, HVCF-16 from Acsen HyVeg and Pahuja from Pahuja Seeds) as standard checks. The 10×15 alpha lattice experimental design with three replications was used for conducting this study.
For recording the data on various agronomic traits, five well-established plants were randomly selected in each plot/block/replication.
Phenotypic measurements
The 20 Ogura based CMS lines, 6 DH testers along with their 120 F1 hybrids were evaluated for sixteen phenotypic traits viz. (i) days to 50% curd initiation (CI); (ii) days to 50% curd maturity (CM); (iii) plant height(PH) (cm) (iv) gross plant weight(GPW) (g) (v) marketable curd weight (MCW); (g) (vi) net curd weight (NCW); (g) (vii) leaf length (LL) (cm); (viii) leaf width(LW) (cm); (ix) number of leaves (NoL); (x) curd length(CL) (cm); (xi) curd diameter(CD) (cm); (xii) core length (CoL) (cm); (xiii) curd size index (CSI) (cm2); (xiv) leaf size index (LSI) (cm2); (xv) harvest index (HI) (%); (xvi) Total marketable yield (TMY) (t/ha) [8–10, 56].
DNA extraction and PCR amplification
The parental CMS lines and DH testers were grown in pro-trays under glasshouse conditions in a soilless mixture of cocopeat, perlite and vermiculite in the ratio of 3:1:1. The fully expanding leaves (100mg) from 25–30 days old seedlings were used for genomic DNA isolation using cetyltrimethyl ammonium bromide (CTAB) method with slight modifications [57]. The Genomic DNA samples were adjusted to 25–50 ng DNA/μl and the DNA samples were stored at -80°C till use. For the genetic diversity analysis in parental CMS and DH lines, 350 microsatellite primers (comprising of genomic-SSRs and EST-SSRs), distributed throughout C genome the B.oleracea [58, 59] were used. Out of 350 microsatellite primers, 145 primers were found to be polymorphic. However, only 87 SSRs and EST-SSRs which showed clear amplification and polymorphism were used for final molecular analysis. The PCR reaction mixture comprised of 1 μl of each forward and reverse primers, 2 μl of genomic DNA template (50 ng), 12.50 μl of 2× PCR Green master mix (GoTaq DNA polymerase; Promega, USA) and 8.50 μl nuclease free water. The Eppendorf Mastercycler Nexus GSX1 was used for PCR amplification. The PCR cycling programme was set up as follow: an initial denaturation of 94°C for 4 min followed by 35 cycles of denaturation at 94°C for 30s, annealing of primers at 50 to 60°C for 30s depending upon appropriate primer annealing temperatures and extension at 72°C for 1min, then final extension of 72°C for 7min. Amplified PCR products were separated by 3.0% agarose gel electrophoresis in 1X TBE buffer (pH 8.0) and the gel was run at 100 mA voltage for 120 min. Ethidium bromide (EtBr) of 0.5 mg/ml was used for gel staining and gel pictures were captured using digital gel documentation unit (BioSpectrum Imaging System, UK). The determination of fragment sizes was done using Promega 50 bp DNA step ladder.
Genetic analysis
Among the 350 microsatellite markers, 87 polymorphic genomic-SSR and EST-SSRs loci depicting genetic diversity (S1 Table) were used for cluster analysis, Principal component analysis (PCA) and clustering through neighbor-joining (NJ) UPGMA method using DARwin software version 6.0.017 [60]. For testing the reliability of NJ dendrogram, a bootstrap value of 1000 replicates was used. For the allelic diversity analysis, estimation of observed number of alleles (N) per loci, observed heterozygosity (Ho), expected heterozygosity (He) and polymorphism information content (PIC) was computed through software CERVUS version 3.0 [61]. The estimation of PIC for each locus using CERVUS 3.0 was calculated according to the formula; PIC = 1-Ʃ Pi2, where Pi represents the ith allele frequency in a locus for the genotypes P under study [62].
The genetic structure analysis of the parental population of the testcross progenies was studied with Bayesian model-based clustering approach implemented in STRUCTURE version 2.3.4 software [63] to assign individuals to k clusters and sub-clusters. For the estimation of the proportion of ancestral contribution in each parental line, all simulations were performed by parameter setting as: “admixture model” with “correlated allele frequencies”. The algorithm was implemented with 10,000 lengths of the burn-in period followed by 100000 Markov Chain Monte Carlo (MCMC) repetitions and a plausible range of putative k values was kept from k = 1 to k = 10 run independently with 15 iterations for each k. The optimum value of k for determining most likely number of sub-populations was predicted according to the simulation method of DeltaK (ΔK) [64] with the help of web-based STRUCTURE HARVESTER version v0.6.94 [65].
Statistical analysis
The agronomic data recorded for each parent, 120 F1 hybrids and 4 commercial checks in alpha lattice design were subjected to analysis of variance (ANOVA) using GLM procedure of SAS (statistical analysis system) software version 9.4 [66]. The line × tester statistical analysis of GCA, SCA, heterosis, heritability, variance and mean performance for was performed as per Kempthorne [21] through SAS version 9.4. The testing of significance of GCA and SCA effects was done at 5%, 1%, and 0.1% probability through F test. Heterosis estimates for different traits were computed as per Xie et al. [67] based on formulae: MPH% (Mid- parent heterosis) = [(F1-MP)/MP] x 100, BPH% (Better parent heterosis) = [(F1-BP)/BP] x 100, where MP is mid-parent and BP is better-parent performance and testing of significance was done at probability of p < 0.05, p < 0.01 and p < 0.01 through F test. The narrow-sense heritability (h2ns = VA/VP; VP = VG + VE) estimates were categorized into three classes viz., high (> 30%), medium (10–30%) and low (< 10%) [68]. The GA was calculated as = H2b× phenotypic standard deviation × K, where K value is 2.06, which is a standardized selection differential constant at 5% selection intensity [69]. The parental lines and testers were clustered into different groups based on sixteen agronomic traits using R software [70]. They were grouped through principal component analysis (PCA) to estimate the explained variance in the first two axes. Pooled data from five randomly selected plants of each genotype per plot per block per replication for all the sixteen morphological and commercial traits were taken for statistical analysis.
The Euclidean distance (ED), hereafter referred as phenotypic distance (PD) was calculated based on sixteen phenotypic traits using R software [70]. The simple matching dissimilarity coefficient (hereafter referred as genetic distance: GD) was computed based on g-SSR and EST-SSRs data analysis using DARwin software version 6.0.017. The association among genetic distances, heterosis and combining ability was computed by Pearson’s correlation coefficients (r) (pearson product moment correlation coefficient: PPMCC) by using R software packages version 3.5.1 in Rstudio 1.1.456 [70] and testing of significance at p < 0.05 and p < 0.01. The corrplot displaying correlation among distances, heterosis and combining ability was demonstrated via Rcorrplot package in Rstudio [71].
Results
Analysis of variance
The mean square estimates for different vegetative and commercial traits revealed significant differences among treatments for all the characters except curd diameter, core length and curd size index and curd diameter at 0.01% probability (S2 Table). Similarly, block effects were significant for all the traits except days to 50% curd initiation, curd diameter, core length and curd size index at the probability of 0.01% (S2 Table). The coefficient of determination (R2) indicated high variability percentage (>70%) for all the traits except curd length, curd diameter, core length and curd size index. The higher R2 value also suggests a higher significance of the model. The line × tester analysis of variance (ANOVA) for combining ability revealed highly significant differences among the treatments and parents for all the studied traits (Table 2). The mean squares of the lines were found significant for all the traits, while the mean squares for testers were for all except curd length (Table 2). The significant differences were also observed with respect to lines versus testers for all the traits except leaf width, curd length, curd diameter, core length, curd size index and harvest index, while the mean squares of parents versus crosses were significant for all the traits except for number of leaves (Table 2). The variance analysis for combining ability also revealed highly significant differences among the120 testcross progenies for all the 16 traits at 0.1% probability, while no significant differences were found among three replications for all the traits except leaf width, suggesting the presence of inherent variability among all the crosses (Table 2). The line × tester interaction effects were also significant for all 16 phenotypic traits.
Table 2. Line × Tester Analysis of variance (ANOVA) for combining ability for yield and horticultural traits in cauliflower.
| Source of variation | df | Days to 50% CI | Days to 50% CM | PH (cm) | GPW (g) | MCW (g) | NCW (g) | LL (cm) | LW (cm) |
| Replicates | 2 | 0.25 | 0.83 | 8.75 | 23153.58 | 11384.77 | 2565.82 | 1.65 | 6.04* |
| Treatments | 145 | 6.08*** | 299.66*** | 176.18*** | 1095365.25*** | 266871.75*** | 106378.93*** | 175.89*** | 43.16*** |
| Parents | 25 | 13.28*** | 563.97*** | 113.34*** | 403231.69*** | 157665.16*** | 56107.28*** | 106.85*** | 29.26*** |
| Parents (Line) | 19 | 15.26*** | 598.47*** | 116.59*** | 324516.00*** | 129477.23*** | 47982.00*** | 104.48*** | 34.83*** |
| Parents (Testers) | 5 | 7.26*** | 35.52*** | 32.52* | 93011.02*** | 92945.55*** | 59482.50*** | 72.38*** | 13.26*** |
| Parents (L vs T) | 1 | 5.75* | 2550.53*** | 455.84*** | 3449932.75*** | 1016833.69*** | 193611.64*** | 324.21*** | 3.48 |
| Parents vs Crosses | 1 | 29.58*** | 1020.72*** | 5878.71*** | 31342492.00*** | 4855010.50*** | 1962991.75*** | 2546.75*** | 330.86*** |
| Crosses | 119 | 4.37*** | 238.08*** | 141.46*** | 986594.00*** | 251258.53*** | 101338.41*** | 170.48*** | 43.66*** |
| Line Effect | 19 | 5.53 | 1054.16*** | 222.35* | 1737689.88* | 335689.69 | 114123.88 | 270.94* | 86.83** |
| Tester Effect | 5 | 5.06 | 33.96 | 65.61 | 694204.25 | 153128.39 | 47422.77 | 180.44 | 49.83 |
| Line * Tester Eff. | 95 | 4.10*** | 85.61*** | 129.27*** | 851763.75*** | 239537.05*** | 101618.98*** | 149.86*** | 34.71*** |
| Error | 290 | 1.20 | 4.94 | 11.04 | 20306.63 | 10387.84 | 3296.66 | 4.67 | 1.96 |
| Total | 437 | 2.82 | 102.71 | 65.82 | 377032.50 | 95495.77 | 37496.81 | 61.47 | 15.65 |
| Source of variation | df | NoL | CL (cm) | CD (cm) | CoL (cm) | CSI (cm2) | LSI (cm2) | HI % | TMY (t/ha) |
| Replicates | 2 | 2.92 | 1.28 | 1.07 | 0.01 | 181.72 | 3407.57 | 6.81 | 18.22 |
| Treatments | 145 | 29.54*** | 2.35*** | 5.11*** | 1.23*** | 1325.33*** | 348863.53*** | 238.79*** | 426.99*** |
| Parents | 25 | 38.75*** | 1.07* | 3.70*** | 0.95*** | 736.22*** | 169525.92*** | 305.09*** | 252.26*** |
| Parents (Line) | 19 | 40.96*** | 1.21* | 4.14*** | 1.08*** | 820.98*** | 190813.19*** | 323.77*** | 207.16*** |
| Parents (Testers) | 5 | 15.33*** | 0.78 | 2.77*** | 0.61*** | 561.34** | 88306.48*** | 292.54*** | 148.71*** |
| Parents (L vs T) | 1 | 113.78*** | 0.00 | 0.00 | 0.01 | 0.17 | 171165.05*** | 12.92 | 1626.93*** |
| Parents vs Crosses | 1 | 0.79 | 61.33*** | 168.99*** | 16.64*** | 43832.41*** | 3937259.75*** | 1887.12*** | 7768.02*** |
| Crosses | 119 | 27.84*** | 2.12*** | 4.03*** | 1.16*** | 1091.89*** | 356384.91*** | 211.01*** | 402.01*** |
| Line Effect | 19 | 33.27 | 2.20 | 3.84 | 1.39 | 1097.96 | 670563.13** | 226.60 | 537.10 |
| Tester Effect | 5 | 23.30 | 0.10 | 3.10 | 0.58 | 264.71 | 413817.59 | 132.45 | 245.01 |
| Line * Tester Eff. | 95 | 27.00*** | 2.21*** | 4.11*** | 1.15*** | 1134.21*** | 290526.47*** | 212.03*** | 383.26*** |
| Error | 290 | 4.07 | 0.64 | 0.61 | 0.11 | 157.99 | 8677.53 | 27.14 | 16.62 |
| Total | 437 | 12.51 | 1.21 | 2.11 | 0.48 | 545.43 | 121529.77 | 97.27 | 152.79 |
*, **, ***, ****significant at 5%, 1%, 0.1%, 0.01% probability respectively through F test
Genetic components of variance
The estimation of genetic components of variance revealed that nature of gene action, heritability, genetic advance and degree of dominance is presented in Table 3. The GCA variance (σ2gca) for both lines and testers was found lower as compared to the SCA variance (σ2sca) for all the studied traits except for Days to 50% curd maturity. The value of dominance variance (σ2D) was greater as compared to the additive component of variance (σ2A) for all the traits except for days to 50% curd maturity. The greater than unity value of degree of dominance for all the traits indicated dominant nature of these traits except for days to 50% curd maturity. The ratio of additive to dominance variance (σ2A/D) coupled with predictability ratio (σ2gca/σ2sca) was less than unity for all the traits suggesting preponderance of non-additive gene action. The estimation of heritability is associated with selection efficiency. We observed the lowest estimate of narrow-sense heritability (h2ns) for curd length (3.44%) and the highest h2ns value was recorded for days to 50% curd maturity (49.21%). Generally, moderate level of h2ns estimates was found for majority of the traits except for CL, CD, CSI, and HI. The higher estimates of genetic advance (GA) at 5% selection intensity were observed for GPW, MCW, NCW and LSI, while lower estimates of GA were recorded for all the remaining traits.
Table 3. Estimates of genetic components of variance, heritability, genetic advance and predictability ratio for sixteen vegetative and commercial traits.
| Variance components | Days to 50% CI | Days to 50% CM | PH (cm) | GPW (g) | MCW (g) | NCW (g) | LL (cm) | LW (cm) | NoL | CL (cm) | CD (cm) | CoL (cm) | CSI (cm2) | LSI (cm2) | HI % | TMY (t/ha) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| σ2gcaline | 0.24 | 58.29 | 11.74 | 95410.18 | 18072.32 | 6157.07 | 14.79 | 4.71 | 1.62 | 0.09 | 0.18 | 0.07 | 52.22 | 36771.42 | 11.08 | 28.92 |
| σ2gca tester | 0.06 | 0.48 | 0.91 | 11231.63 | 2379.01 | 735.44 | 2.93 | 0.80 | 0.32 | -0.01 | 0.04 | 0.01 | 1.78 | 6752.33 | 1.76 | 3.81 |
| sl2 GCA (Average) HS | 0.10 | 13.82 | 3.41 | 30657.45 | 6000.54 | 1986.58 | 5.67 | 1.70 | 0.62 | 0.01 | 0.07 | 0.02 | 13.42 | 13679.82 | 3.91 | 9.60 |
| σ2sca | 0.97 | 26.89 | 39.41 | 277152.38 | 76383.07 | 32774.11 | 48.40 | 10.92 | 7.64 | 0.52 | 1.17 | 0.34 | 325.41 | 93949.65 | 61.63 | 122.21 |
| σ2A | 0.21 | 27.65 | 6.82 | 61314.89 | 12001.09 | 3973.16 | 11.33 | 3.40 | 1.24 | 0.03 | 0.15 | 0.04 | 26.84 | 27359.63 | 7.81 | 19.20 |
| σ2D | 0.97 | 26.89 | 39.41 | 277152.38 | 76383.07 | 32774.11 | 48.40 | 10.92 | 7.64 | 0.52 | 1.17 | 0.34 | 325.41 | 93949.65 | 61.63 | 122.21 |
| σ2A/D | 0.22 | 1.03 | 0.17 | 0.22 | 0.16 | 0.12 | 0.23 | 0.31 | 0.16 | 0.05 | 0.13 | 0.13 | 0.08 | 0.29 | 0.13 | 0.16 |
| Degree of Dominance | 2.15 | 0.99 | 2.40 | 2.13 | 2.52 | 2.87 | 2.07 | 1.79 | 2.48 | 4.46 | 2.82 | 2.78 | 3.48 | 1.85 | 2.81 | 2.52 |
| Heritability (Narrow Sense) % | 13.31 | 49.21 | 13.66 | 17.76 | 13.07 | 10.50 | 18.49 | 22.73 | 12.13 | 3.44 | 9.67 | 10.48 | 6.63 | 22.03 | 9.96 | 13.07 |
| Genetic Advance 5% | 0.34 | 7.60 | 1.99 | 214.97 | 81.57 | 42.07 | 2.98 | 1.81 | 0.80 | 0.06 | 0.25 | 0.14 | 2.75 | 159.92 | 1.82 | 3.26 |
| Predictability Ratio | 0.18 | 0.51 | 0.15 | 0.18 | 0.14 | 0.11 | 0.19 | 0.24 | 0.14 | 0.05 | 0.11 | 0.11 | 0.08 | 0.23 | 0.11 | 0.14 |
σ2A = additive genetic variance, σ2D = dominance genetic variance, σ2gca = estimate of GCA variance, σ2sca = estimate of SCA variance
Combining ability effects
The estimates of combining ability are effective for early generation selection of inbred lines and identifying heterotic crosses. The GCA estimates of parental lines and testers are summarized in Table 4 and Table 5. The GCA estimates revealed that the CMS lines Ogu118-6A, Ogu33A, Ogu34-1A and Ogu33-1A had significantly high GCA in desirable direction for traits related to earliness. The CMS lines Ogu307-33A, Ogu119-1A, Ogu125-8A and tester DH-53-10 also showed significantly high GCA for days to 50% curd maturity in desirable direction. For core length, the significantly high GCA in the desirable negative direction was observed in CMS lines Ogu122-5A, Ogu118-6A, Ogu1A, Ogu13-85-6A, Ogu1-6A, Ogu122-1A and tester DH-53-10. Among the 20 female parents, 6 and 9 numbers of CMS lines exhibited high GCA for plant height and gross plant weight, respectively. Similarly, 6 and 9 numbers of CMS lines were found to good combiner for the marketable curd weight and net curd weight\ respectively. Among the 20 female parents, 8 numbers of CMS line for plant height, 9 numbers of CMS lines for gross plant weight, 6 numbers of CMS lines for marketable curd weight and 8 numbers of CMS lines for net curd weight exhibited significantly high GCA in desirable direction. The results pertaining to SCA effects of 120 cross combinations are presented in S3 Table. Among the 120 hybrids, 9 and 28 crosses showed significantly negative SCA effects for earliness traits CI and CM, respectively (S3 Table).
Table 4. Estimates of general combining ability (GCA) effects of lines and testers.
| Lines/testers | Days to 50% CI | Days to 50% CM | PH (cm) | GPW (g) | MCW (g) | NCW (g) | LL (cm) | LW (cm) |
|---|---|---|---|---|---|---|---|---|
| Ogu122-5A | 0.73** | 6.78*** | -0.10 | 278.71*** | 213.54*** | 31.59* | 0.14 | 1.06** |
| Ogu115-33A | 0.62* | 5.56*** | -2.29** | 330.54*** | 26.21 | 24.64 | 5.09*** | 1.59*** |
| Ogu118-6A | -0.60* | -7.99*** | 0.68 | 274.38*** | 23.93 | 148.70*** | 1.19* | 2.26*** |
| Ogu307-33A | -0.49 | -13.16*** | -0.03 | 209.32*** | 11.21 | -14.02 | 5.17*** | 2.61*** |
| Ogu309-2A | 0.17 | 6.39*** | -5.51*** | -267.13*** | -5.07 | 77.64*** | -4.70*** | -1.80*** |
| Ogu33A | -0.71** | -13.27*** | 0.74 | 402.88*** | 179.27*** | 50.42*** | -1.23* | -0.24 |
| OguKt-2-6A | -0.16 | 4.84*** | 7.55*** | 609.65*** | 223.04*** | 77.20*** | 6.84*** | 5.56*** |
| Ogu1A | -0.27 | 4.73*** | -2.50** | 19.88 | 18.27 | -27.74* | -0.79 | 0.39 |
| Ogu13-85-6A | 0.06 | -0.77 | -3.32*** | 64.04 | -41.46 | 40.31** | -6.09*** | -3.54*** |
| Ogu1-6A | -0.44 | 5.95*** | -0.32 | -40.40 | 62.66** | 66.81*** | -5.78*** | -2.02*** |
| Ogu2A | 0.56* | 4.51*** | -0.84 | -266.40*** | -159.57*** | -68.63*** | -2.00*** | -1.74*** |
| OguKt-9-2A | 0.68** | 8.34*** | 2.87*** | -282.07*** | -135.73*** | -56.63*** | -0.18 | 0.84* |
| Ogu22-1A | 0.12 | 2.95*** | 3.36*** | -109.96** | -39.34 | -53.13*** | -2.38*** | -2.14*** |
| Ogu122-1A | 0.56* | 6.34*** | 3.31*** | -148.29*** | -105.84*** | -84.13*** | 4.09*** | -0.22 |
| Ogu126-1A | 0.56* | 4.01*** | 1.83* | 76.99* | 115.16*** | 67.81*** | 4.55*** | 0.65* |
| Ogu12A | 0.56* | 6.89*** | -0.89 | -459.68*** | -131.51*** | -60.47*** | -3.57*** | -1.59*** |
| Ogu119-1A | -0.05 | -8.05*** | 1.36 | -339.13*** | -206.07*** | -115.13*** | 0.27 | -0.81* |
| Ogu34-1A | -1.10*** | -11.61*** | 1.70* | 78.49* | 25.99 | -0.97 | 1.78*** | 1.05** |
| Ogu125-8A | -0.10 | -2.99*** | 1.42 | 171.71*** | 170.43*** | 71.14*** | 2.30*** | 1.30*** |
| Ogu33-1A | -0.71** | -9.44*** | -9.03*** | -603.51*** | -245.12*** | -175.41*** | -4.70*** | -3.22*** |
| Testers | ||||||||
| DH-18-8-1 | 0.25 | 1.29*** | -0.70 | -145.43*** | -67.08*** | -39.01*** | -2.78*** | -1.39*** |
| DH-18-8-3 | 0.16 | 0.28 | -0.59 | -27.38 | -2.74 | -25.86*** | 0.85*** | 0.25 |
| DH-53-1 | -0.40** | -0.36 | 1.38** | 183.02*** | 78.32*** | 31.56*** | 2.12*** | 1.29*** |
| DH-53-6 | -0.34* | -0.72* | 0.86* | -3.38 | -35.79** | -3.36 | -1.09*** | -0.62*** |
| DH-53-9 | 0.11 | 0.14 | -1.35** | -38.20* | -1.61 | 11.89 | 0.95*** | 0.25 |
| DH-53-10 | 0.21 | -0.64* | 0.40 | 31.37 | 28.91* | 24.76*** | -0.06 | 0.22 |
| CD 95% GCA(Line) | 0.51 | 1.03 | 1.54 | 66.17 | 47.32 | 26.66 | 1.00 | 0.65 |
| CD 95% GCA(Tester) | 0.28 | 0.57 | 0.84 | 36.24 | 25.92 | 14.60 | 0.55 | 0.36 |
*, **, ***, ****Significance at P ≤ 0.05, P ≤ 0.01, P ≤ 0.001, P ≤ 0.0001, respectively, CD: critical difference
Table 5. Estimates of general combining ability effects of lines and testers.
| Lines/testers | NoL | CL (cm) | CD (cm) | CoL (cm) | CSI (cm2) | LSI (cm2) | HI % | TMY (t/ha) |
|---|---|---|---|---|---|---|---|---|
| Ogu122-5A | -0.48 | -0.06 | -0.17 | -0.30*** | -2.18 | 42.94 | 3.07* | 8.54*** |
| Ogu115-33A | 0.79 | 0.15 | 0.18 | 0.26** | 2.84 | 198.54*** | -5.79*** | 1.05 |
| Ogu118-6A | 1.63*** | 0.05 | -0.07 | -0.23** | -1.02 | 132.96*** | -5.21*** | 0.96 |
| Ogu307-33A | 0.29 | -0.34 | 0.17 | -0.07 | -3.82 | 261.97*** | -3.71** | 0.45 |
| Ogu309-2A | -1.54** | -0.41* | -0.72*** | -0.05 | -12.58*** | -200.01*** | 6.05*** | -0.20 |
| Ogu33A | 1.13* | 0.33 | 0.01 | 0.00 | 3.38 | -40.80 | -1.78 | 7.17*** |
| OguKt-2-6A | -0.15 | 0.21 | 0.38* | -0.15 | 5.62 | 445.65*** | -4.05** | 8.92*** |
| Ogu1A | 0.96* | 0.31 | 0.48** | -0.48*** | 8.43** | -4.68 | -0.23 | 0.73 |
| Ogu13-85-6A | 1.41** | -0.47* | -0.70*** | -0.22** | -12.56*** | -281.45*** | -1.56 | -1.66 |
| Ogu1-6A | -2.43*** | 0.25 | 0.77*** | -0.26** | 9.57** | -231.80*** | 2.47* | 2.51** |
| Ogu2A | 1.91*** | -0.12 | 0.20 | -0.11 | 0.13 | -143.18*** | -2.02 | -6.38*** |
| OguKt-9-2A | -3.15*** | 0.35 | -0.24 | 0.22** | 2.37 | 28.93 | -0.53 | -5.43*** |
| Ogu22-1A | -1.48** | -0.49* | -0.46* | 0.70*** | -3.55 | -142.26*** | 1.93 | -1.57 |
| Ogu122-1A | -0.59 | -0.09 | -0.16 | -0.22** | -2.54 | 79.39*** | -0.97 | -4.23*** |
| Ogu126-1A | 0.29 | 0.48* | 0.25 | 0.15 | 7.58* | 139.82*** | 3.39** | 4.61*** |
| Ogu12A | -0.65 | -0.06 | -0.22 | 0.23** | -3.34 | -163.41*** | 5.69*** | -5.26*** |
| Ogu119-1A | 0.96* | 0.12 | 0.64*** | 0.26*** | 8.00** | -48.09* | -2.46* | -8.24*** |
| Ogu34-1A | 0.91 | -0.34 | -0.18 | -0.06 | -5.98* | 103.02*** | -1.81 | 1.04 |
| Ogu125-8A | 0.68 | 0.66*** | 0.66*** | 0.36*** | 14.24*** | 116.42*** | 3.52** | 6.82*** |
| Ogu33-1A | -0.48 | -0.54** | -0.81*** | -0.04 | -14.60*** | -293.94*** | 4.00** | -9.80*** |
| Testers | ||||||||
| DH-18-8-1 | -0.01 | 0.01 | 0.36*** | 0.17*** | 3.35* | -135.48*** | 1.04 | -2.68*** |
| DH-18-8-3 | -0.98*** | -0.07 | -0.19 | -0.07 | -0.99 | 36.61** | 0.58 | -0.11 |
| DH-53-1 | 0.14 | 0.03 | 0.16 | -0.02 | 1.53 | 100.79*** | -1.71* | 3.13*** |
| DH-53-6 | 0.02 | 0.03 | -0.25* | 0.00 | -2.57 | -54.16*** | -1.87** | -1.43** |
| DH-53-9 | 0.97*** | -0.02 | -0.04 | 0.04 | -0.77 | 39.02** | 1.78** | -0.06 |
| DH-53-10 | -0.14 | 0.03 | -0.05 | -0.12** | -0.56 | 13.21 | 0.19 | 1.16* |
| CD 95% GCA(Line) | 0.94 | 0.37 | 0.36 | 0.16 | 5.84 | 43.25 | 2.42 | 1.89 |
| CD 95% GCA(Tester) | 0.51 | 0.20 | 0.20 | 0.09 | 3.20 | 23.69 | 1.32 | 1.04 |
*, **, ***, ****Significance at P ≤ 0.05, P ≤ 0.01, P ≤ 0.001, P ≤ 0.0001, respectively, CD: critical difference
The highest SCA effect in the desirable negative direction for curd initiation was recorded in the hybrid Ogu307-33A × DH-18-8-3 followed by Ogu34-1A × DH-53-1 and Ogu309-2A × DH-53-9. For the days to 50% CM, the highest SCA effect in desirable negative direction was observed in the cross Ogu22-1A × DH-53-10 followed by Ogu125-8A × DH-53-6 and Ogu1A × DH-18-8-3. Among the 120 testcross progenies, 31 exhibited high significant positive SCA effects for PH. The highest positive significant SCA effect for PH was observed in the cross Ogu122-1A × DH-18-8-3 followed by Ogu13-85-6A × DH-53-1, Ogu13-85-6A × DH-18-8-3 and Ogu118-6A × DH-53-9. For the commercial traits viz. GPW, MCW, NCW, CL, CD, CoL and CSI, out of 120 crosses, 39, 32, 38, 10, 26, 33 and 24 crosses exhibited significantly high SCA effects in the desirable direction. For the vegetative traits, among 120 crosses, 44, 37, 33 and 48 crosses showed significantly high positive SCA effects for LL, LW, NoL and LSI, respectively. For the HI and TMY, among 120 hybrids, 18 and 32 hybrids displayed significantly high positive SCA effects, respectively. The cross combination Ogu22-1A × DH-53-6 exhibited highest positive significant SCA effect for GPW followed by Ogu307-33A × DH-18-8-3 and Ogu122-5A × DH-53-10. For the MCW, hybrid Ogu33A × DH53-1 showed the highest positive significant SCA effects followed by Ogu122-5A × DH-53-10 and Ogu1-6A × DH-53-1. The highly significant positive SCA estimate for NCW was observed in the cross Ogu33A × DH-53-1 followed by Ogu1A × DH-53-9 and Ogu22-1A × DH-53-6. For the total marketable yield, the highest significant SCA estimate in desirable positive direction was observed in the cross, Ogu33A × DH-53-1 (good combiner × good combiner) followed by Ogu122-5A × DH-53-10 (good combiner × good combiner) and Ogu1-6A × DH-53-1 (good combiner × good combiner) (S3 Table).
Phenotypic characterization
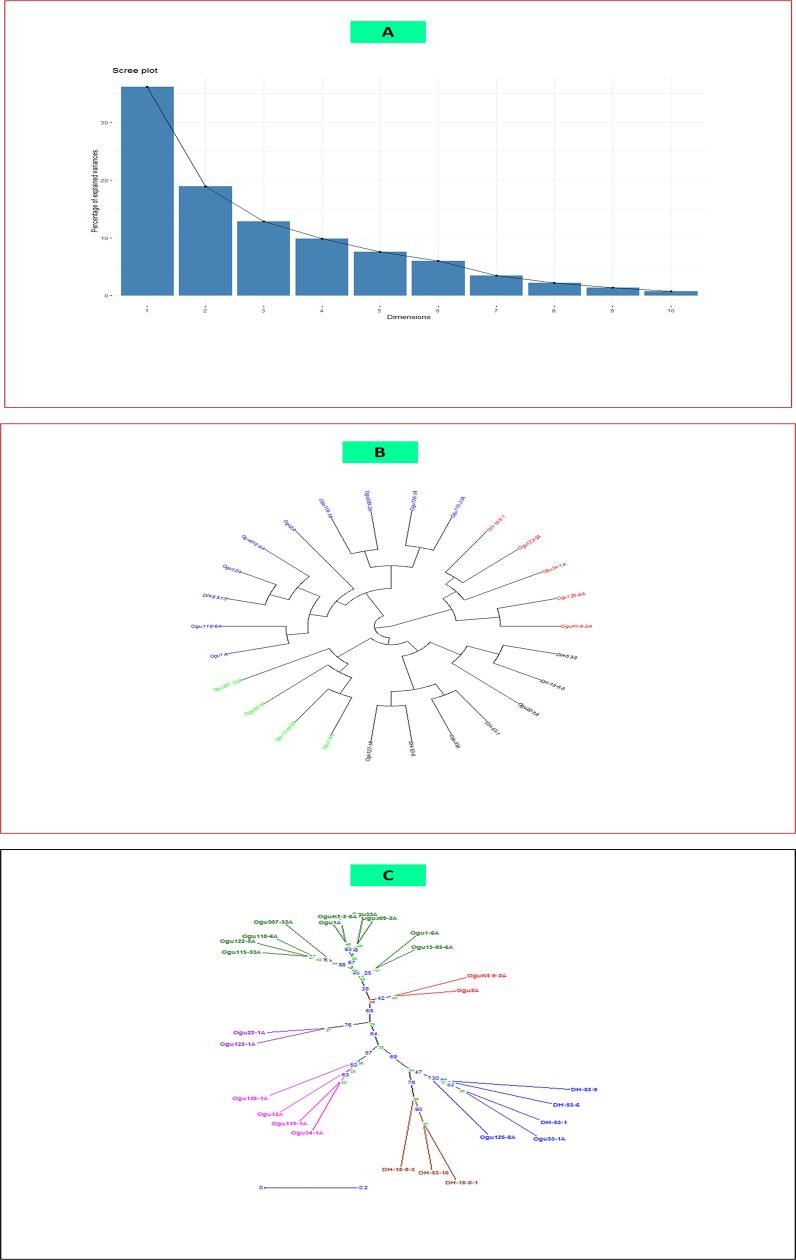
The mean performance of the parental CMS and DH lines along with standard checks is presented in S4 Table and S1A–S1H Fig. On the basis of curd initiation, the CMS lines Ogu307-33A and Ogu13-85-6A were earliest to initiate curd. Similarly, CMS lines Ogu307-33A and Ogu33-1A were earliest for curd maturity. The CMS line Ogu22-1A, OguKt-9-2A, Ogu12A and DH lines DH-53-1, DH-53-10 had the highest plant height. While CMS lines Ogu115-33A and Ogu309-2A were dwarf as compared to other parental lines. The highest number of leaves was observed in CMS lines Ogu34-1A and Ogu309-2A. The shortest core length was recorded in genotype Ogu13-85-6A and Ogu309-2A. The tester DH-53-1 was having highest MCW. The highest total marketable yield was recorded in CMS line Ogu33A and Ogu125-8A, whereas the tester DH-18-8-1 was having the highest TMY among all the parental genotypes and standard checks. The PCA revealed that the first two dimensions (PC1 and PC2) captured 36.2% and 18.9% of the total existing variation among the parental lines. The HCA based dendrogram depicting inter-relationships displayed high genetic divergence among 26 CMS and DH lines based upon Euclidean distance matrix (Fig 1B). On the basis of HCA, the 26 parental lines were classified into 3 major clusters with varying extent of divergence within internal sub-clusters. The DH lines DH-53-10 and DH-18-8-1 were distantly placed from the rest of DH testers in two different major clusters.
Fig 1. Morphological and molecular diversity in parental CMS and DH lines.
(A) Percentage of explained variance among parental genotypes in different principal components (B) Dendrogram illustrating the genetic relationships among parental genotypes based on morphological traits (C) UPGMA cluster analysis illustrating the genetic relationships among parental genotypes based on g-SSR and EST-SSR analysis.
Microsatellites based polymorphism, allelic diversity and genetic distances
Overall 511 alleles were amplified through 87 SSRs (Table 6) with the mean number of alleles per locus was 5.87. The allele numbers per locus ranged from 2 (1 primer BoSF1640) to 10 (1 primer: BRAS011). The observed heterozygosity (Ho) ranged from 0.03 (for the loci BoSF2232, BoSF062, BRAS011, BoESSR080, BoSF2406 and BoSF2421) to 0.19 (for the loci BoSF2294a). The mean expected heterozygosity (He) was 0.68, with a range of 0.27 (primer cnu107) to 0.83 (for primer Na12F03a and BoESSR041) and had a higher mean value than Ho. The mean polymorphic information content (PIC) for 87 loci was 0.63. The PIC content ranged from 0.24 for the primer cnu107 to 0.80 for the primer Na12F03a (Table 6). Further, the PCA and Neighbour joining (NJ) cluster analysis based on molecular data for 87 loci, revealed distinct clusters and sub-clusters of parental CMS and DH lines based on their phylogeny (Fig 1C). The PCA revealed that the first two major coordinate axes 1 and 2 (PC1 and PC2) explained 61.41% of the total existing variation among CMS and DH lines. The dendrogram constructed revealed 3 main clusters of parental lines with internal sub-clusters showing varying degree of diversity. The DH testers remained in 2 different sub-clusters of the single main cluster. The CMS lines Ogu2A and OguKt-9-2A placed distantly from the rest of CMS lines. The CMS lines Ogu33-1A and Ogu125-8A were in close affinity with DH lines.
Table 6. Characteristics of 87 polymorphic SSR and EST-SSRs loci depicting diversity.
| Locus | LG | Ho | He | N | PIC | Locus | LG | Ho | He | N | PIC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BoSF2304b | C09 | 0.00 | 0.48 | 5 | 0.43 | BoESSR303 | C04 | 0.00 | 0.33 | 4 | 0.31 |
| BoSF1740 | C08 | 0.00 | 0.63 | 5 | 0.57 | BoESSR333 | C04 | 0.00 | 0.81 | 9 | 0.77 |
| BoSF378 | C08 | 0.00 | 0.72 | 4 | 0.65 | BoESSR338 | C08 | 0.00 | 0.77 | 8 | 0.72 |
| BoSF2680 | C08 | 0.00 | 0.78 | 5 | 0.73 | BoESSR403 | C08 | 0.00 | 0.72 | 7 | 0.66 |
| BoSF2054 | C06 | 0.00 | 0.52 | 3 | 0.41 | BoESSR409 | C04 | 0.00 | 0.78 | 6 | 0.73 |
| BoSF1215 | C06 | 0.00 | 0.59 | 6 | 0.54 | BoESSR576 | C06 | 0.00 | 0.71 | 6 | 0.65 |
| BoSF250 | C06 | 0.00 | 0.67 | 3 | 0.58 | BoESSR581 | C06 | 0.00 | 0.67 | 7 | 0.62 |
| BoSF2505 | C06 | 0.00 | 0.60 | 4 | 0.54 | BoESSR632 | C01 | 0.00 | 0.71 | 6 | 0.67 |
| BoSF2374 | C05 | 0.00 | 0.62 | 4 | 0.54 | BoESSR901 | C09 | 0.00 | 0.77 | 7 | 0.73 |
| BoSF1846 | C05 | 0.00 | 0.69 | 4 | 0.62 | BoESSR766 | C03 | 0.00 | 0.69 | 7 | 0.63 |
| BoSF2878 | C05 | 0.00 | 0.57 | 3 | 0.48 | BoESSR825 | 0.00 | 0.69 | 7 | 0.63 | |
| BoSF912 | C01 | 0.00 | 0.72 | 6 | 0.67 | BoESSR673 | C03 | 0.00 | 0.66 | 5 | 0.58 |
| BoSF063 | C01 | 0.00 | 0.74 | 8 | 0.69 | BoESSR758 | C07 | 0.00 | 0.68 | 7 | 0.64 |
| BoSF2294a | C02 | 0.19 | 0.73 | 6 | 0.67 | BoESSR763 | C04 | 0.00 | 0.80 | 8 | 0.76 |
| BoSF2615 | C02 | 0.00 | 0.61 | 4 | 0.55 | BoESSR863 | C06 | 0.00 | 0.69 | 4 | 0.62 |
| BoSF1167 | C02 | 0.00 | 0.79 | 6 | 0.75 | BoESSR903 | C06 | 0.00 | 0.50 | 5 | 0.46 |
| BoSF2248 | C03 | 0.00 | 0.60 | 5 | 0.55 | Na12F03a | C07 | 0.07 | 0.83 | 8 | 0.80 |
| BoSF2232 | C03 | 0.03 | 0.49 | 3 | 0.38 | O110B11 | C05 | 0.00 | 0.65 | 4 | 0.57 |
| BoSF184 | C04 | 0.00 | 0.73 | 7 | 0.68 | BoSF2406 | C07 | 0.03 | 0.67 | 7 | 0.62 |
| BoSF1640 | C09 | 0.00 | 0.43 | 2 | 0.33 | BoSF2313 | C07 | 0.07 | 0.75 | 8 | 0.70 |
| BoSF2612 | C08 | 0.00 | 0.67 | 4 | 0.59 | BoSF2033 | C07 | 0.00 | 0.81 | 9 | 0.77 |
| BoSF2860 | C07 | 0.04 | 0.76 | 8 | 0.71 | BoSF317 | C05 | 0.00 | 0.70 | 6 | 0.66 |
| BoSF2345 | C01 | 0.00 | 0.76 | 5 | 0.71 | BoSF2421 | C09 | 0.03 | 0.61 | 6 | 0.57 |
| BoSF1207 | C01 | 0.07 | 0.77 | 8 | 0.73 | BoSF1957 | C04 | 0.00 | 0.82 | 8 | 0.77 |
| BoSF042 | C03 | 0.00 | 0.73 | 6 | 0.68 | Na12B09 | C03 | 0.00 | 0.70 | 6 | 0.66 |
| BoSF062 | C03 | 0.03 | 0.51 | 5 | 0.46 | cnu107 | C02 | 0.00 | 0.27 | 3 | 0.24 |
| BoSF2985 | C03 | 0.00 | 0.68 | 4 | 0.60 | BoSF1131 | C03 | 0.00 | 0.78 | 7 | 0.73 |
| BoE862 | C04 | 0.00 | 0.75 | 6 | 0.70 | BoSF966 | C03 | 0.00 | 0.80 | 7 | 0.75 |
| BRAS011 | C02 | 0.03 | 0.77 | 10 | 0.73 | CB10258 | C01 | 0.00 | 0.78 | 8 | 0.73 |
| BrBAC214 | C03 | 0.00 | 0.48 | 5 | 0.43 | BoESSR920 | C09 | 0.00 | 0.79 | 6 | 0.73 |
| BoESSR080 | C07 | 0.03 | 0.78 | 9 | 0.73 | BoESSR041 | C06 | 0.00 | 0.83 | 8 | 0.79 |
| BoESSR086 | C03 | 0.00 | 0.63 | 6 | 0.58 | BoESSR934 | C08 | 0.00 | 0.65 | 4 | 0.57 |
| BoESSR087 | C04 | 0.00 | 0.70 | 5 | 0.65 | Ni4D12 | C02 | 0.00 | 0.57 | 3 | 0.48 |
| BoESSR089 | C01 | 0.00 | 0.80 | 7 | 0.76 | cnu149 | C05 | 0.00 | 0.81 | 8 | 0.76 |
| BoESSR105 | C04 | 0.00 | 0.76 | 6 | 0.71 | BoESSR482 | C02 | 0.00 | 0.79 | 7 | 0.75 |
| BoESSR108 | C04 | 0.00 | 0.67 | 5 | 0.60 | O112G04a | C08 | 0.00 | 0.44 | 4 | 0.40 |
| BoESSR122 | C02 | 0.00 | 0.82 | 8 | 0.78 | BoESSR492 | C03 | 0.00 | 0.76 | 8 | 0.72 |
| BoESSR151 | C02 | 0.00 | 0.70 | 6 | 0.65 | BoESSR510 | C03 | 0.00 | 0.59 | 4 | 0.51 |
| BoESSR206 | C05 | 0.00 | 0.53 | 4 | 0.47 | BoESSR523 | C07 | 0.00 | 0.76 | 7 | 0.71 |
| BoESSR207 | C05 | 0.00 | 0.78 | 5 | 0.72 | BoESSR560 | C03 | 0.00 | 0.73 | 6 | 0.67 |
| BoESSR208 | C04 | 0.00 | 0.71 | 8 | 0.66 | BoESSR736 | C05 | 0.00 | 0.66 | 5 | 0.61 |
| BoESSR212 | C07 | 0.00 | 0.56 | 4 | 0.51 | BoESSR030 | C03 | 0.00 | 0.76 | 8 | 0.71 |
| BoESSR216 | C01 | 0.00 | 0.76 | 5 | 0.70 | BoESSR073 | C03 | 0.00 | 0.50 | 6 | 0.46 |
| BoESSR248 | C04 | 0.00 | 0.62 | 5 | 0.57 |
LG: linkage group, Ho: observed heterozygosity, He: expected heterozygosity, PIC: polymorphic information content
The Euclidean distance (PD) between lines and testers were computed from 16 phenotypic traits (S5 Table) and GD was calculated from molecular data based on 87 microsatellite markers (genomic-SSR and EST-SSRs) used for assessment of genetic diversity between parents (S5 Table). The PD was ranged from 2.07 for the cross L16 × T6 (Ogu12A × DH-53-10) to 8.27 for the cross combination L5 × T1 (Ogu309-2A × DH-18-8-1) with a mean of 5.52. The GD was ranged from 0.44 for the cross L20 × T1 (Ogu33-1A × DH-18-8-1) to 0.98 for the cross combinations L4 × T5 (Ogu307-33A × DH-53-9) with the average GD of 0.83.
Genetic structure analysis
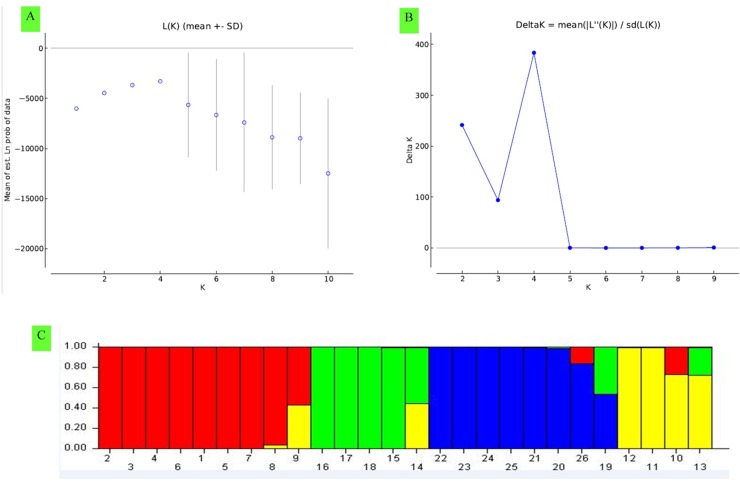
The result of analysis by ‘Structure Harvester version v0.6.94’ revealed that second-order likelihood, ΔK reached to peak at k = 4 (Fig 2A to 2C), hence, optimal k value should be 4. This indicated that the 26 parental CMS and DH inbred lines could be grouped into 4 genetic sub-clusters (Fig 2C). All the DH testers placed in cluster III, along with 2 CMS lines Ogu125-8A and Ogu33-1A. Although there is minor admixture in DH-53-10 and Ogu125-8A from the genotypes of cluster I and cluster II, respectively. The other CMS lines placed themselves in separate clusters. The 20 parental CMS lines were grouped into 4 sub-clusters. The majority of the CMS lines were placed in cluster I. There is admixture from cluster IV to the cluster I (Ogu1A, Ogu13-85-6A) and cluster II (Ogu122-1A) genotypes. Similarly, there was minor admixture from cluster I and cluster II to cluster IV genotypes (Ogu1-6A and Ogu22-1A). Thus, there were four distinct four sub-clusters including minor gene flow within some genotypes of respective clusters from each other.
Fig 2. Genetic structure analysis of parental CMS and DH lines by STRUCTURE v2.3.4 and STRUCTURE HARVESTER based upon 87 g-SSR, EST-SSR loci.
(a) Mean L(k) ± SD over 15 runs for each k value from 1 to 10. (b) ΔK calculated as ΔK = m|L′′ (K)|/s [L(K)], reached peak at k = 4. (c) Q-plot clustering. Inferred ancestries of CMS and DH lines based on 4 genetic groups. Each cluster is represented by different color and each column represent respective genotype allotted to respective cluster. Different color of each column depicts the percent of membership (vertical values on the left of cluster) of each genotype for four clusters.
Analysis of heterosis
The heterotic response of all the 120 testcross progenies varied in magnitude and highly significant heterosis was observed for all the 16 traits in both directions. The top ten cross combinations based on significant mid-parent heterosis in desirable direction along with their better-parent heterosis and SCA effects are presented in S6 and S7 Tables. The cross combinations, Ogu34-1A×DH-53-1 and Ogu33A×DH-53-6 showed significantly high MPH in the desirable direction for days to 50% curd initiation and days to 50% curd maturity (S6 Table). For CI, the testers DH-53-1 and DH-53-9 were involved in 4 crosses individually out of top 10 crosses. For CM the CMS line Ogu33A as a female parent was involved in 6 hybrids for earliness among top 10 hybrids. Ogu33A was also involved as female parent in one of the top 10 cross combinations related to CI. This line had significantly highest GCA for earliness among all the CMS lines used in the study. For the plant height, among the top 10 heterotic crosses, the cross combination Ogu118-6A×DH-53-9 exhibited highest significant positive heterosis over mid-parent followed by OguKt-2-6A×DH-53-9 and Ogu34-1A×DH-53-9. The highest significant positive heterosis for GPW was observed in the cross Ogu118-6A×DH-53-10 over mid-parent followed by Ogu126-1A×DH-53-1 and Ogu307-33A×DH-18-8-3. The top ten crosses having significant positive MPH for 8 commercial traits are presented in S7 Table.
The hybrid Ogu122-1A×DH-53-6 exhibited significantly highest MPH for CoL in desirable negative direction followed by Ogu1A×DH-53-6 and Ogu1A×DH-53-10. The CMS line Ogu1A was involved in 3 crosses as female parent among top 4 crosses with respect to core length, and it had significantly highest GCA in the desirable negative direction for core length. Thus, Ogu1A could be used as a parent for developing hybrids with short core.
For the commercial traits curd length and curd diameter, the cross combinations Ogu119-1A×DH-18-8-1 followed by Ogu126-1A×DH-53-1 and Ogu122-1A×DH-53-10 for CL, and Ogu1-6A×DH-53-1 followed by Ogu122-1A×DH-53-10 and Ogu22-1A×DH-53-6 for CD had highest MPH in the desirable positive direction. The cross Ogu122-1A×DH-53-10 showed the highest significant positive heterosis over mid-parent for CSI. For economic trait MCW, the hybrid Ogu126-1A×DH-18-8-3 followed by Ogu122-5A×DH-53-10 and Ogu307-33A×DH-18-8-3 exhibited significantly high positive MPH at P ≤ 0.001. Likewise, for the NCW, the significant highest heterosis over mid parent in the desirable positive direction was observed in the hybrid Ogu118-6A×DH-53-10 followed by Ogu1A×DH-53-9 and Ogu119-1A×DH-53-10. The cross combination Ogu122-5A×DH-53-9 (56.64%) showed highest significant MPH for percent HI followed by Ogu126-1A×DH-18-8-3 (49.16%) and Ogu12A×DH-18-8-3 (41.99%). For the total marketable yield, the highest significant heterosis over mid-parent in desirable positive direction was observed in the testcross Ogu126-1A×DH-18-8-3 followed by Ogu122-5A×DH-53-10 and Ogu307-33A×DH-18-8-3. Among the top ten crosses also wide range of MPH was recorded for TMY from 92.76% (Ogu33A×DH-53-1) to 172.31% (Ogu126-1A×DH-18-8-3).
Association of genetic distances, heterosis and combining ability
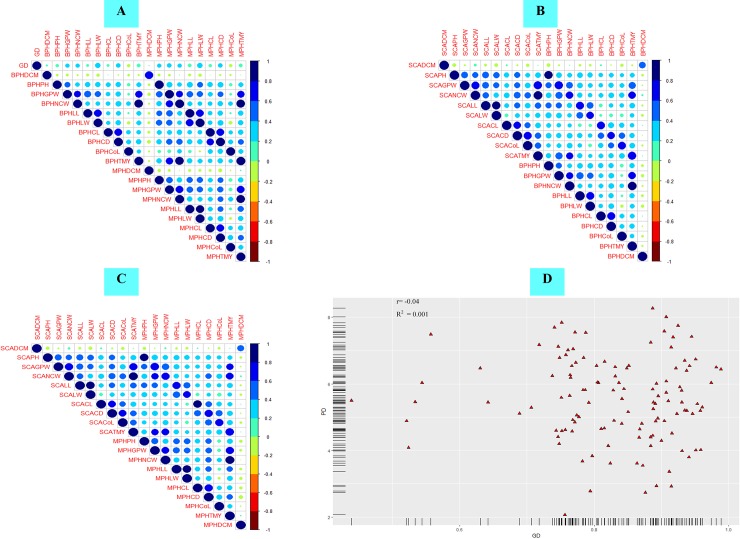
The Pearson correlation coefficient of genetic distances with heterosis, combining ability, and combining ability with heterosis for ten commercial traits is presented in Table 7. The GD and PD exhibited no significant correlation coefficient with SCA for any of the traits (Table 7 and S2B Fig, S2C Fig). SCA showed significantly positive correlation with MPH and BPH for all the traits at P ≤ 0.01 (Table 6, Fig 3B and 3C). No significant association of GD with MPH and BPH was observed with respect to days to 50% CM, leaf length, curd length and core length (Table 7, Fig 3A). For the commercial traits viz. plant height, gross plant weight, net curd weight, leaf width, curd diameter and total marketable yield, significant correlation was observed between GD and MPH in desirable direction for the respective traits (Table 7, Fig 3A). The highest magnitude of correlation coefficient of GD with MPH and BPH in desirable direction was observed for LW. However, PD exhibited no significant correlation with heterosis for majority of traits (Table 7, S2A Fig). The significant correlation of phenotypic distance with mid-parent heterosis was observed only for leaf length. PD exhibited a significant correlation in undesirable direction with core length. However, no significant association was observed between parental genetic distances based on phenotypic traits (PD) and molecular data (GD) (r = -0.04) (Fig 3D).
Table 7. Pearson correlation coefficients among parental genetic distance (GD, PD), combining ability and heterosis in cauliflower for ten morphological and commercial traits.
| Traits | Days to 50% CM | PH | GPW | NCW | LL | LW | CL | CD | CoL | TMY |
|---|---|---|---|---|---|---|---|---|---|---|
| GD | ||||||||||
| MPH | 0.02 | 0.23* | 0.27** | 0.21* | 0.17 | 0.34** | 0.13 | 0.25** | -0.15 | 0.18* |
| BPH | 0.07 | 0.15 | 0.21* | 0.22* | 0.12 | 0.36** | 0.12 | 0.26** | -0.15 | 0.18* |
| SCA | -0.02 | -0.01 | 0.02 | -0.01 | 0.01 | 0.03 | -0.01 | 0.01 | 0.03 | 0.01 |
| PD | ||||||||||
| MPH | 0.12 | 0.14 | 0.07 | -0.07 | 0.22* | 0.14 | 0.01 | -0.04 | 0.26** | -0.04 |
| BPH | 0.11 | 0.01 | -0.07 | -0.20* | 0.16 | 0.07 | -0.07 | -0.14 | 0.24** | -0.25** |
| SCA | 0.06 | 0.11 | 0.02 | -0.05 | 0.09 | 0.12 | 0.07 | 0.02 | 0.03 | -0.06 |
| SCA | ||||||||||
| MPH | 0.52** | 0.83** | 0.79** | 0.74** | 0.73** | 0.68** | 0.81** | 0.75** | 0.74** | 0.77** |
| BPH | 0.53** | 0.82** | 0.78** | 0.69** | 0.69** | 0.63** | 0.74** | 0.65** | 0.69** | 0.76** |
*, **significance at P < 0.05 and P < 0.01, respectively, GD: genetic distance, PD: phenotypic distance, MPH: mid parent heterosis, BPH: better parent heterosis, SCA: specific combining ability
Fig 3. Pearson’s correlation matrix depicting association among genetic distances and heterosis.
(A) Corrplot depicting pearson’s correlation coefficient of genetic distance based on molecular data with better-parent heterosis (BPH) and mid-parent heterosis (MPH) for 10 commercial traits, (B) Corrplot depicting pearson’s correlation coefficient of specific combining ability with BPH (C) Corrplot depicting pearson’s correlation coefficient of specific combining ability with MPH (D) Scatter plot depicting no correlation between genetic distance based on molecular data and phenotypic distance. GD is on X-axis and PD on Y-axis.
Discussion
Genetic components of variance and combining ability
The analysis of variance depicted highly significant differences among all the treatments for all the 16 agronomic traits, indicating considerable genetic differences among parents and their testcross progenies. The success of any crop breeding programme relies on genetic variation present in the germplasm. High genetic divergence among genotypes was reported by Garg and Lal [72], Verma and Kalia [73] for yield and related traits in early maturity Indian cauliflower, and for antioxidant traits in snowball cauliflower [5]. All the studied traits were found to be under the genetic control of both additive and non-additive gene effects, as revealed by the significant mean squares of lines, testers and line × tester interactions. The results are in agreement with Singh et al. [5] for antioxidant traits in cauliflower and Verma and Kalia [73] for days to 50% CM, leaf area, PH, MCW, NCW, curd compactness, GPW and HI in cauliflower using SI inbred lines. The analysis of genetic components of variance indicated the importance of SCA in developing heterotic crosses as revealed by the higher value of σ2sca than σ2gca of lines and testers for the majority of traits. As the response to natural and artificial selection relies on additive genetic variance, the narrow sense heritability (h2ns) holds a great promise in plant breeding as it provides the basis to precise selection of genotypes based on phenotypic variance because of additive genetic components [74]. In this study, the low to intermediate level of h2ns was observed for majority of vegetative and commercial traits suggesting non-additive genetic control of these traits, which might be due to large epistatic effects. We also observed moderate estimates of h2ns for antioxidant traits in cauliflower in our previous study [5], the results are also in agreement with Xie et al. [67] for mineral content in Chinese cabbage. Thus, the early generation selection for these vegetative and commercial traits would be difficult due to dominance effects in the expression of phenotypic variance, and hence selection must be practiced in later generations. The combining ability analysis has been successfully utilized in crop breeding for evaluating parental performance and understanding the dynamics of genes involved in trait expression. The parental GCA estimates in desirable direction also indicate potentiality of parents in generating promising breeding populations. In the present investigation, the significantly high GCA effects of parental lines in desirable direction for the respective vegetative and commercial traits are due to the predominance of additive genetic effects of genes and additive × additive interactions [5, 10]. It depicts a desirable gene flow from parents to progeny at high frequency and these parental lines exhibiting significantly high GCA for the respective traits in desirable direction can be utilized to accumulate favorable alleles via recombination and selection [5, 9, 10, 75, 76]. Further, our results revealed that none of the parents was good general combiner for all the studied vegetative and commercial traits. Similar findings were also reported inself-incompatible (SI) and CMS lines in cauliflower for yield and quality traits [5, 9, 73]. The findings indicate requirement of multiple breeding programmes in suitable mating designs for the development of productive cultivars with the accumulation of positive alleles of genes. On the other hand, the parental lines depicting GCA in opposite direction for the respective traits can be utilized to generate desirable mapping population to study the genetics of respective traits [76]. The SCA, which reflects the loci having non-additive and epistatic gene effects, can be utilized to determine specific heterotic crosses for the respective trait of interest. The inconsistent association of GCA and SCA of respective crosses for respective traits is the indication of complex interaction of genes for quantitative traits [29]. The majority of testcross progenies manifesting significantly high SCA in desirable direction had at least one of the parents reflecting poor GCA effects (poor × good general combiner or good × poor general combiner). It may be attributed to good combiner parent depicting favorable additive effects and poor combiner parent displaying epistatic effects [5, 77]. Few crosses with significantly high SCA in desirable direction for various traits had both parents with good GCA. (Such results suggested the role of cumulative effects of additive × additive interaction of positive alleles [5, 77]. Concurrently, some of the crosses had poor SCA effects for the respective traits, despite involving parents with significant GCA. It might be ascribed to the absence of any interaction among the positive alleles of genes. Thus, our results have clearly indicated that breeders must pay attention to both GCA and SCA in the selection of elite parents for the development of heterotic hybrids. Further, the recombination breeding and random mating in conjunction with selection among segregates (recurrent selection), synthetics and composites, may be exploited to harness utility of both additive and non-additive gene effects in cauliflower [73]. The high SCA effects are not always correlated with significantly high heterosis and concurrently, the heterotic crosses exhibiting high mid parent and better parent heterosis.
Cluster analysis, allelic diversity, and genetic structure
The morphological and molecular diversity is vital in selecting desirable parents for hybrid breeding. The identification of heterotic groups and analyzing existing genetic variation in CMS lines is the preliminary requisite for efficient use of elite CMS lines in heterosis breeding. Study of genetic diversity at morphological and molecular level has been regarded as potential tool in identification of promising parental lines for developing heterotic hybrids in B.oleracea [8, 78–80]. Based upon PCA and HCA of 26 parental lines for 16 phenotypic traits, it was evident that all the parental lines had sufficient genetic variation. The PCA and NJ clustering based on molecular data represent better informative results for correct analysis and to be useful in crop improvement programme [8]. The female parents with significantly high GCA in the desirable direction for the traits related to earliness and high could be useful in developing early maturity and high yielding hybrids. Thus, the information pertaining to morphological and genetic diversity along with GCA could be useful in selecting desirable CMS lines as female parent for the development of cultivars with desirable traits.
We also observed high allele frequency of overall 511 alleles through 87 genomic-SSR and EST-SSRs loci in 26 parental CMS and DH lines with an average allelic frequency of 5.87 alleles per locus. Allelic diversity could be due to variation in germplasm used in the study, difference in methods of detection of markers and number of markers from transcribed regions of genome etc. [78, 79, 81, 82]. Varied allele frequency has been reported by various workers in Brassica crops with different set of germplasm and molecular markers [78, 79]. The PIC in genetic studies is utilized as a measure of informativeness of a marker locus for linkage analysis [78, 83] and it categorizes informative markers as highly informative (PIC ≥ 0.5), reasonably informative (0.5 < PIC >0.25) and slightly informative (PIC < 0.25) [78, 83]. In the present study the PIC content of 87 polymorphic loci ranged from 0.24–0.80, which classified all the 87 loci (g-SSR and EST-SSRs) as slightly informative (1 primer cnu107), reasonably informative (12 primers) and highly informative markers (74 primers). The mean PIC content of 0.63 in present investigation based on 87 genomic-SSR and EST-SSRs was higher than the mean PIC of 0.316 observed for 165 cauliflower inbred lines by Zhu et al. [84] and 0.60 as recorded for 57 genotypes of Brassica oleracea comprising 51 cultivars of cauliflower by Zhao et al. [85]. The DH lines developed through microspore culture are very diverse from most of the CMS lines. Therefore, these DH lines will be instrumental in developing heterotic combinations in snowball cauliflowers where genetic base is very narrow. Moreover, the diverse DH lines indicated the role of DH technology in creation of more diversity in different plant species. In all the clusters minor admixture was observed from each of clusters among themselves, which indicated the gene flow among the parental lines of different groups.
Association of genetic distances and combining ability with heterosis
Numerous studies in different crops have revealed the utility of the genetic distances in prediction of heterotic crosses [23, 25–29], assuming positive correlation of genetic distances with heterosis [86]. But the correlation between GD and heterosis is not absolute and significantly high level of heterosis may result when parents with low, intermediate or high genetic distance are used. Genetic distances based on both phenotypic and genotypic data are utilized to study the genetic variation among different genotypes or parental inbred lines. The contrasting reports are available regarding correlations of genetic distances, heterosis and combining ability. In the present study, no correlation was observed between two distance measurements, based on morphological data (PD) and molecular data (GD). This is in contrary to the findings of Gupta et al. [30] who reported a significantly positive correlation of GD and PD (r = 0.2) at P < 0.001 in pearl millets. Absence of correlation between two distance measures might be due to the fact that morphological traits showing continuous variation are largely influenced by environment and polygenic inheritance besides linkage disequilibrium [30, 87–89]. Both the distance measures displayed no significant correlation with SCA of all the traits, suggesting genetic distances might not be effective in predicting SCA effects. Su et al. [29] also reported no significant association between genetic distances and SCA in chrysanthemum. However, Tian et al. [34], Lariepe et al. [28] reported a significant correlation between total GD and SCA for the length of terminal raceme in rapeseed and for grain yield and plant height in maize, respectively. Thus, the association of GD with SCA is not absolute. Further, our results suggested that SCA effects had a stronger significant positive correlation with MPH and BPH for all the studied traits. Similar findings were reported by Zhang et al. [90], Su et al. [29], Tian et al. [34] in barley, chrysanthemum and rapeseed, respectively they indicated non-additive gene effects for heterosis. The GD and PD differed in their ability to predict MPH and BPH for different traits. Neither GD nor PD displayed any significant correlation with MPH and BPH for days to 50% CM, and CL. GD also exhibited no significant correlation with heterosis for LL and CoL. Similarly, PD showed no significant association with heterosis for majority of traits except LL. However, GD was significantly correlated with MPH and BPH for important commercial traits viz. PH, GPW, NCW, LW, CD, and TMY in the desirable direction. These results are in line with the theory proposed by Falconer and Mackay [86]. In general, GD had a greater magnitude of correlation than PD with heterosis for all the traits under study. The variability in correlation coefficients between heterosis for respective traits and genetic distances may reflect allele numbers controlling the trait expression [26]. Similar findings were reported in Maize by Wegary et al. [89] where they have highlighted the importance of GD in contrast to PD for predicting hybrid performance. They have also reported a significant correlation of GD with heterosis for grain yield, plant height and ear height and morphological distance with heterosis for certain traits in quality protein maize. Jarosz [33] has reported a significant association of GD (based on RAP and AFLP markers) with heterosis for total and marketable yield in carrot. On the other hand, Tian et al. [34] and Su et al. [29] reported no significant correlation of PD and GD with MPH and BPH for any traits in rapeseed and chrysanthemum, respectively. Our results are also in contrary to the findings of Geleta et al. [91] and Kawamura et al. [27] in pepper and Chinese cabbage, respectively, suggesting no utility of GD in prediction of heterosis. While Krishnamurthy et al. [92] suggested selection of parents with intermediate divergence based on AFLP markers for getting a greater number of heterotic hybrids using CMS lines for yield in the hot pepper. Incole group of vegetables, we found only a single report describing interrelationships between genetic distances and heterosis in broccoli (B. oleracea var. italic L.) by Hale et al. [26] using DH based population. They observed a significantly negative correlation between total GD (based on SRAP, AFLP, SSR markers) and heterosis for all the traits, suggesting the reduction in heterosis with the increase in genetic distances. Thus, our study is the first comprehensive report regarding interrelationships between GD (based on SSR, EST-SSRs) and heterosis for commercial traits in snowball cauliflower, suggesting significant correlation in desirable direction for important yield related traits. Hence, based on this study, we recommend the application of genomic-SSR and EST-SSRs based genetic distances in prediction of heterosis for yield and commercial traits. The non-significant or poor correlation between GD and heterosis for certain traits might be due to the lack of linkage between different alleles responsible for expression of particular trait and molecular marker used for estimating GD, inadequate coverage of entire genome and epistasis. Besides, lack of correlation may also be due to use of DNA markers from unexpressed region of genome having no interaction with commercial traits and heterosis [26, 40, 62, 93]. The molecular marker-based GD would be more predictive of heterosis, when there are strong dominance effects among hybrids, high heritability, linkage of molecular markers and QTLs of traits of interest [26, 40, 62, 93]. Hence, from our findings, it is quite evident that significance of genetic distances in prediction of heterosis inevitably depends upon the methods used to calculate genetic distances, type of molecular markers, genome coverage, region of genome, crop, breeding system, traits under consideration, type of germplasm and environmental conditions. High correlation among the GD and heterosis among important traits may also because of completely homozygous DH lines as male parent. Presence of minor heterozygosity among the conventionally developed inbreds also results in poor correlation among GD and heterosis in most of the earlier study.
Conclusions
The current study is first of its kind in determining the heterotic groups based on combining ability for morphological, yield and commercial traits using Ogura cytoplasm-based CMS lines and DH based testers. We also presented the first comprehensive report on predicting the association of genomic SSRs and EST-SSRs based GD and morphological traits-based PD with heterosis involving CMS and DH parental lines in snowball cauliflower. Analysis of variance of parents and their testcross progenies revealed the presence of significant genetic variability. Present investigation also emphasizes the relevance of both GCA and SCA in the selection of elite parents for the improvement of yield and commercial traits and predicting appropriate breeding strategies for the crop genetic improvement, developing high yielding hybrids, synthetics and composites in cauliflower. High and significant correlation among SCA with heterosis suggested the role of non-additive gene effects in heterosis. It was also evident that development of DH lines could broaden the genetic base of any crop through creating more diversity in the existing population. The findings of our study further suggested that genetic distances based on genomic and EST-SSRs can be used as a predictor of heterosis for commercial traits in CMS and DH based F1cauliflower. Contrasting results obtained in different earlier studies regarding the efficacy of genetic distances in the prediction of heterosis, invites further investigation with different sets of large number of molecular markers covering entire genome, and a different set of parental germplasm, in multiple standard environments.
Supporting information
DCI: days to 50% curd initiation, DCM: days to 50% curd maturity, PH: plant height, LL: leaf length, LW: leaf width, GPW: gross plant weight, MCW: marketable curd weight, NCW: net curd weight, CSI: curd size index, NoL: number of leaves, HI: harvest index, TMY: total marketable yield, CL: curd length, CD: curd diameter, CoL: core length, LSI: leaf size index. The upper and lower lines outside the box stand for maximum and minimum adjacent value, respectively. The median value is represented by line inside the box. The lower and upper hinge of the box stands for 25% and 75% percentile, respectively.
(TIF)
(A) Corrplot depicting correlation of phenotypic distance with heterosis, (B) Corrplot of association of genetic distance with combining ability, (C) Corrplot depicting association of phenotypic distance with combining ability.
(TIF)
Among the 350 microsatellites, 87 primers showed clear cut polymorphism and used for allelic diversity analysis.
(DOCX)
* = significant at 5% probability, ** = significant at 1% probability, *** = significant at 0.1%, **** = significant at 0.01% probability through F test, Rep = Replication, Blk = Block, Trt = Treatment, Rep (Blk)Adj = Rep (Blk) Adjustable Days to 50% CI = Days to 50% curd initiation, Days to 50%CM = Days to 50% curd maturity, PH = Plant height, GPW = Gross plant weight, MCW = marketable curd weight, NCW = net curd weight, LL = leaf length, LW = leaf width, NoL = No of leaves, CL = curd length, CD = curd diameter, CoL = core length, CSI = curd size index, LSI = leaf size index, HI = harvest index, TMY = total marketable yield, R2: coefficient of determination.
(DOCX)
* = significant at 5% probability, ** = significant at 1% probability, *** = significant at 0.1%, **** = significant at 0.01% probability through F test, CD = critical difference, Days to 50% CI = Days to 50% curd initiation, Days to 50%CM = Days to 50% curd maturity, PH = Plant height, GPW = Gross plant weight, MCW = marketable curd weight, NCW = net curd weight, LL = leaf length, LW = leaf width, NoL = No of leaves, CL = curd length, CD = curd diameter, CoL = core length, CSI = curd size index, LSI = leaf size index, HI = harvest index, TMY = total marketable yield.
(DOCX)
*commercial standard checks.
(DOCX)
L: CMS lines; T: DH testers.
(DOCX)
* = significant at 5% probability, ** = significant at 1% probability, *** = significant at 0.1%, **** = significant at 0.01% probability through F test, MPH: Mid parent heterosis, BPH: better parent heterosis, SCA: specific combining ability (value in parenthesis).
(DOCX)
* = significant at 5% probability, ** = significant at 1% probability, *** = significant at 0.1%, **** = significant at 0.01% probability through F test, MPH: Mid parent heterosis, BPH: better parent heterosis, SCA: specific combining ability (value in parenthesis).
(DOCX)
Acknowledgments
First author is thankful to Head ICAR-IARI, Regional Station, Katrain (H.P.) for providing necessary facilities during research period and ICAR-IARI, New Delhi, during Ph.D. research programme. We also acknowledge the help of Dr. S. Dash, IASRI in statistical analysis using SAS software.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Financial support was provided by the NAHEP/CAAST research grant by Indian Council of Agricultural Research.
References
- 1.Bayer PE, Golicz AA, Tirnaz S, Chan CK, Edwards D, Batley J. Variation in abundance of predicted resistance genes in the Brassica oleracea pangenome. Plant Biotechnol J. 2018; 10.1111/pbi.13015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maggioni L, von Bothmer R, Poulsen G, Lipman E. Domestication, diversity and use of Brassica oleracea L., based on ancient Greek and Latin texts. Genet Resour Crop Evol. 2018; 65: 137–159. [Google Scholar]
- 3.Dey SS, Bhatia R, Parkash C, Sharma S, Dabral M, Mishra V et al. Alteration in important quality traits and antioxidant activities in Brassica oleracea with Ogura cybrid cytoplasm. Plant Breed. 2017a; 136: 400–409. [Google Scholar]
- 4.Samec D, Urlic B, Salopek-Sondi B. Kale (Brassica oleracea var. acephala) as a superfood: Review of the scientific evidence behind the statement. Crit Rev Food Sci Nutr. 2018; 20: 1–12. https://doi.org/0.1080/10408398.2018. [DOI] [PubMed] [Google Scholar]
- 5.Singh S, Bhatia R, Kumar R, Sharma K, Dash S, Dey SS. Cytoplasmic male sterile and doubled haploid lines with desirable combining ability enhances the concentration of important antioxidant attributes in Brassica oleracea. Euphytica. 2018a; 214: 207 10.1007/s10681-018-2291-3. [DOI] [Google Scholar]
- 6.Sotelo T, Cartea ME, Velasco P, Soengas P. Identification of antioxidant-capacity related QTLs in Brassica oleracea. PLOS ONE. 2014; 9: e107290 10.1371/journal.pone.0107290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh S, Singh R, Thakur P, Kumar R. Phytochemicals, functionality and breeding for enrichment of cole vegetables (Brassica oleracea L.) In: Petropoulos SA, Ferreira ICFR, Barros L, editors. Phytochemicals in vegetables: a valuable source of bioactive compounds. Bentham Science Publishers, UAE; 2018. Pp. 256–295. [Google Scholar]
- 8.Dey SS, Bhatia R, Bhardwaj I, Mishra V, Sharma K, Parkash C et al. Molecular-agronomic characterization and genetic study reveals usefulness of refined Ogura cytoplasm based CMS lines in hybrid breeding of cauliflower (Brassica oleracea var. botrytis L.). Sci Hortic. 2017b; 224: 27–36. [Google Scholar]
- 9.Dey SS, Sharma SR, Bhatia R, Parkash C, Barwal RN. Superior CMS (Ogura) lines with better combining ability improved yield and maturity in cauliflower (Brassica oleracea var. botrytis). Euphytica. 2011; 182: 187–197. [Google Scholar]
- 10.Dey SS, Singh N, Bhatia R, Parkash C, Chandel C. Genetic combining ability and heterosis for important vitamins and antioxidant pigments in cauliflower (Brassica oleracea var. botrytis L.). Euphytica. 2014; 195: 169–181. [Google Scholar]
- 11.Sehgal N, Singh S. Progress on deciphering the molecular aspects of cell-to-cell communication in Brassica self-incompatibility response. 3Biotech. 2018; 8: 347 10.1007/s13205-018-1372-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thakur P, Vidyasagar Singh S. Evaluation of cytoplasmic male sterile (CMS) progenies and maintainer lines for yield and horticultural traits in cabbage (Brassica oleracea var. capitataL.). SABRAO J Breed Genet. 2015; 47: 29–39. [Google Scholar]
- 13.Singh S Vidyasagar. Effect of common salt (NaCl) sprays to overcome the self-incompatibility in the S-allele lines of Brassica oleracea var. capitataL. SABRAO J Breed Genet. 2012; 44: 339–348. [Google Scholar]
- 14.Singh S, Dey SS, Bhatia R, Kumar R, Behera TK. Current understanding of male sterility systems in vegetable Brassicas and their exploitation in hybrid breeding. Plant Reprod. 2019; 32: 231–256. 10.1007/s00497-019-00371-y [DOI] [PubMed] [Google Scholar]
- 15.Bansal P, Banga S, Banga SS. Heterosis as investigated in terms of polyploidy and genetic diversity using designed Brassica junceaamphidiploids and its progenitor diploid species. PLOS ONE. 2012; 7: e29607 10.1371/journal.pone.0029607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujimoto R, Uezono K, Ishikura S, Osabe K, Peacock WJ, Dennis ES. Recent research on the mechanism of heterosis is important for crop and vegetable breeding systems. Breed Sci. 2018; 68: 145–158. 10.1270/jsbbs.17155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lauss K, Wardenaar R, Oka R, van Hulten MHA, Guryev E, Keurentjes JJB, et al. Parental DNA methylation states are associated with heterosis in epigenetic hybrids. Plant Physiol. 2018; 176: 1627–1645. 10.1104/pp.17.01054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lippman ZB, Zamir D. Heterosis: revisiting the magic. Trends Genet. 2007; 23: 60–66. 10.1016/j.tig.2006.12.006 [DOI] [PubMed] [Google Scholar]
- 19.Groszmann M, Greaves IK, Fujimoto R, Peacock WJ, Dennis ES. The role of epigenetics in hybrid vigour. Trends Genet. 2013; 29: 684–690. 10.1016/j.tig.2013.07.004 [DOI] [PubMed] [Google Scholar]
- 20.Li H, Yuan J, Wu M, Han Z, Li L, Jiang H, et al. Transcriptome and DNA methylome reveal insights into yield heterosis in the curds of broccoli (Brassica oleracea L. var. italica). BMC Plant Biol. 2018; 18: 168 10.1186/s12870-018-1384-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kempthorne O. An introduction to genetic statistics. John Wiley & Sons, Inc, New York; 1957. [Google Scholar]
- 22.Melchinger AE, Piepho HP, Utz HF, Muminovic J, Wegenast T, Torjek O, et al. Genetic basis of heterosis for growth-related traits in Arabidopsis investigated by testcross progenies of near-isogenic lines reveals a significant role of epistasis. Genetics. 2007; 177: 1827–1837. 10.1534/genetics.107.080564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaushik P, Plazas M, Prohens J, Vilanova S, Gramazio P. Diallel genetic analysis for multiple traits in eggplant and assessment of genetic distances for predicting hybrids performance. PLOS ONE. 2018; 13: e0199943 10.1371/journal.pone.0199943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esposito MA, Bermejo C, Gatti I, Guindon MF, Cravero V, Cointry EL. Prediction of heterotic crosses for yield in Pisum sativum L. Sci Hortic. 2014; 177: 53–62. [Google Scholar]
- 25.Fujimoto R, Uezono K, Ishikura S, Osabe K, Peacock WJ, Dennis ES. Recent research on the mechanism of heterosis is important for crop and vegetable breeding systems. Breed Sci. 2018; 68: 145–158. 10.1270/jsbbs.17155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hale AL, Farnham MW, Nzaramba MN, Kimbeng CA. Heterosis for horticultural traits in broccoli. TheorAppl Genet. 2007; 115: 351–360. [DOI] [PubMed] [Google Scholar]
- 27.Kawamura K, Kawanabe T, Shimizu M, Nagano AJ, Saeki N, Okazaki K, et al. Genetic distance of inbred lines of Chinese cabbage and its relationship to heterosis. Plant Gene. 2016; 5: 1–7. [Google Scholar]
- 28.Lariepe A, Moreau L, Laborde J, Bauland C, Mezmouk S, Decousset L,et al. General and specific combining abilities in a maize (Zea mays L.) test-cross hybrid panel: relative importance of population structure and genetic divergence between parents. TheorAppl Genet. 2017; 130: 403–417. [DOI] [PubMed] [Google Scholar]
- 29.Su J, Zhang F, Yang X, Feng Y, Yang X, Wu Y,et al. Combining ability, heterosis, genetic distance and their intercorrelations for waterlogging tolerance traits in chrysanthemum. Euphytica. 2017; 213: 42. [Google Scholar]
- 30.Gupta SK, Nepolean T, Shaikh CG, Rai K, Hash CT, Das RR, et al. Phenotypic and molecular diversity-based prediction of heterosis in pearl millet (PennisetumglaucumL. (R.) Br.). Crop J. 2018; 6: 271–281. [Google Scholar]
- 31.Meyer RC, Torjek O, Becher M, Altmann T. Heterosis of biomass production in Arabidopsis. Establishment during early development. Plant Physiol. 2004; 134: 1813–1823. 10.1104/pp.103.033001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thakur AJ, Singh KH, Singh L, Nanjundan J, Khan YJ, Singh D. SSR marker variations in Brassica species provide insight into the origin and evolution of Brassica amphidiploids. Hereditas. 2018; 155: 6 10.1186/s41065-017-0041-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jagosz B. The relationship between heterosis and genetic distances based on RAPD and AFLP markers in carrot. Plant Breed. 2011; 130: 574–579. [Google Scholar]
- 34.Tian HY, Channa SA, Hu SW. Relationships between genetic distance, combining ability and heterosis in rapeseed (Brassica napusL.). Euphytica. 2017; 213: 1. [Google Scholar]
- 35.Cress CE. Heterosis of the hybrid related to gene frequency differences between two populations. Genetics. 1966; 53: 269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Betran FJ, Ribaut JM, Beck D, de Leon DG. Genetic diversity, specific combining ability, and heterosis in tropical maize under stress and nonstress environments. Crop Sci. 2003; 43: 797–806. [Google Scholar]
- 37.Tams SH, Bauer E, Oettler G, Melchinger AE, Schon CC. Prospects for hybrid breeding in winter triticale: II. Relationship between parental genetic distance and specific combining ability. Plant Breed. 2006; 125: 331–336. [Google Scholar]
- 38.Groszmann M, Gonzalez-Bayon R, Greaves IK, Wang L, Huen AK, Peacock WJ, et al. Intraspecific Arabidopsis hybrids show different patterns of heterosis despite the close relatedness of the parental genomes. Plant Physiol. 2014; 166(1): 265–80. 10.1104/pp.114.243998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teklewold A, Becker HC. Comparison of phenotypic and molecular distances to predict heterosis and F1 performance in Ethiopian mustard (Brassica carinataA. Braun.). 2006; 112 (4): 752–9. 10.1007/s00122-005-0180-3. [DOI] [PubMed] [Google Scholar]
- 40.Yu CY, Hu SW, Zhao HX, Guo AG, Sun GL. Genetic distances revealed by morphological characters, isozymes, proteins and RAPD markers and their relationships with hybrid performance in oilseed rape (Brassica napus L.). TheorAppl Genet. 2005; 110: 511–518. [DOI] [PubMed] [Google Scholar]
- 41.Bhatia R, Dey SS, Parkash C, Sharma K, Sood S, Kumar R. Modification of important factors for efficient microspore embryogenesis and doubled haploid production in field grown white cabbage (Brassica oleraceavar. capitataL.) genotypes in India. SciHortic. 2018; 233: 178–187. [Google Scholar]
- 42.Ferrie AMR, Mollers C. Haploids and doubled haploids in Brassica spp. for genetic and genomic research. Plant Cell Tiss Organ Cult. 2011; 104: 375–386. [Google Scholar]
- 43.Liu X, Han F, Kong C, Fang Z, Yang L, Zhang Y,et al. Rapid introgression of fusarium wilt resistance gene into the elite cabbage line through the combined application of a microspore culture, genome background analysis, and disease resistance-specific marker assisted foreground selection. Front Plant Sci. 2017; 8: 354 10.3389/fpls.2017.00354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh S, Dey SS, Bhatia R, Batley J, Kumar R. Molecular breeding for resistance to black rot [Xanthomonas campestris pv. campestris (Pammel) Dowson] in Brassicas: recent advances. Euphytica. 2018b; 214: 196 10.1007/s10681-018-2275-3. [DOI] [Google Scholar]
- 45.Hasan Y, Briggs W, Matschegewski C, Ordon F, Stützel H, Zetzsche H, et al. Quantitative trait loci controlling leaf appearance and curd initiation of cauliflower in relation to temperature. TheorAppl Genet. 2016; 129: 1273–1288. [DOI] [PubMed] [Google Scholar]
- 46.Lin YR, Lee JY, Tseng MC, Lee CY, Shen CH, Wang CS, et al. Subtropical adaptation of a temperate plant (Brassica oleraceavar. italica) utilizes non-vernalization-responsive QTLs. Sci Rep. 2018; 8: 13609 10.1038/s41598-018-31987-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lv H, Wang Q, Liu X, Han F, Fang Z, Yang L,et al. Whole-genome mapping reveals novel QTL cluster associated with main agronomic traits of cabbage (Brassica oleraceavar. capitataL.). Front Plant Sci. 2016; 7: 989 10.3389/fpls.2016.00989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosen A, Hasan Y, Briggs W, Uptmoor R. Genome-based prediction of time to curd induction in cauliflower. Front Plant Sci. 2018; 9: 78 10.3389/fpls.2018.00078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Su Y, Liu Y, Li Z, Fang Z, Yang L, Zhuang M,et al. QTL analysis of head splitting resistance in cabbage (Brassica oleraceaL. var. capitata) using SSR and InDel markers based on whole genome re-sequencing. PLOS ONE. 2015; 10: e0138073 10.1371/journal.pone.0138073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X, Su Y, Liu Y, Fang Z, Yang L, Zhuang M,et al. Genetic analysis and QTL mapping of traits related to head shape in cabbage (Brassica oleraceavar. capitataL.). SciHortic. 2016; 207: 82–88. [Google Scholar]
- 51.Zhao Z, Gu H, Sheng X, Yu H, Wang J, Huang L,et al. Genome-wide single-nucleotide polymorphisms discovery and high-density genetic map construction in cauliflower using specific-locus amplified fragment sequencing. Front Plant Sci. 2016; 7: 334 10.3389/fpls.2016.00334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhatia R, Dey SS, Sood S, Sharma K, Sharma VK, Parkash C, et al. Optimizing protocol for efficient microspore embryogenesis and doubled haploid development in different maturity groups of cauliflower (Brassica oleraceavar. botrytis L.) in India. Euphytica. 2016; 212: 439–454. [Google Scholar]
- 53.Bhatia R, Dey SS, Sood S, Sharma K, Parkash C, Kumar R. Efficient microspore embryogenesis in cauliflower (Brassica oleraceavar. botrytis L.) for development of plants with different ploidy level and their use in breeding programme. SciHortic. 2017; 216: 83–92. [Google Scholar]
- 54.Bhatia R, Dey SS, Sharma PC, Kumar R. In vitro maintenance of CMS lines of Indian cauliflower: an alternative for conventional CMS-based hybrid seed production. J HorticSciBiotechnol. 2015; 90: 171–179. 10.1080/14620316.2015.11513169. [DOI] [Google Scholar]
- 55.Sharma SR. Cabbage In: Sirohi PS (ed) Vegetable crop production. Division of Vegetable Science, IARI, New Delhi, 2003. [Google Scholar]
- 56.Dey SS, Bhatia R, Sharma SR, Parkash C, Sureja AK. Effects of chloroplast substituted Ogura male sterile cytoplasm on the performance of cauliflower (Brassica oleraceavar. botrytis L.) F1 hybrids. SciHortic. 2013; 157: 45–51. [Google Scholar]
- 57.Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acid Res. 1980; 8: 4321 10.1093/nar/8.19.4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Chen X, Yang Y, Xu J, Gu J, Fu J, et al. Development and genetic mapping of microsatellite markers from whole genome shortgun sequences in Brassica oleracea. Mol Breed. 2011; 28: 585–596. [Google Scholar]
- 59.Wang W, Huang S, Liu Y, Fang Z, Yang L, Hua W, et al. Construction and analysis of a high-density genetic linkage map in cabbage (Brassica oleraceaL. var. capitata). BMC Genomics. 2012; 13: 523 10.1186/1471-2164-13-523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perrier X, Jacquemoud-Collet JP. DARwin software. 2006; http://darwin.cirad.fr/ [Google Scholar]
- 61.Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol. 2007; 16: 1099–1106. 10.1111/j.1365-294X.2007.03089.x [DOI] [PubMed] [Google Scholar]
- 62.Makumbi D, Betran JF, Banziger M, Ribaut JM. Combining ability, heterosis and genetic diversity in tropical maize (Zea mays L.) under stress and non-stress conditions. Euphytica. 2011; 180: 143–162. [Google Scholar]
- 63.Pritchard J, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Evanno G, Regnaut S, Goude J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14: 2611–2620. 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- 65.Earl DA, vonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genet Resour. 2012; 4: 359–361. 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- 66.SAS Institute Inc. SAS Online Doc, Version 9.4. Cary, NC; 2013.
- 67.Xie F, Zha J, Tang H, Xu Y, Liu X, Wan Z. Combining ability and heterosis analysis for mineral elements by using cytoplasmic male-sterile systems in non-heading Chinese cabbage (Brassica rapa). Crop Pasture Sci. 2018; 69: 296–302. [Google Scholar]
- 68.Robinson HS. Quantitative genetics in relation to breeding on the central of mendalism. Indian J Genet. 1966; 26: 171–187. [Google Scholar]
- 69.Johnson HW, Robinson HF, Comstock RE. Estimation of genetic and environmental variability in soybeans. Agron J. 1955; 47: 314–318. [Google Scholar]
- 70.RStudio Team. RStudio: Integrated Development for R. RStudio, Inc., Boston, MA, 2015; URL: http://www.rstudio.com/. [Google Scholar]
- 71.Wei T, Simko VR. Package “corrplot”: Visualization of a Correlation Matrix (Version 0.84). 2017; https://github.com/taiyun/corrplot. [Google Scholar]
- 72.Garg N, Lal T. Heterosis for growth and curd characters in Indian cauliflower (Brassica oleraceavar. botrytis L.). Crop Improv. 2005; 32: 193–199. [Google Scholar]
- 73.Verma VK, Kalia P. Combining ability analysis and its relationship with gene action and heterosis in early maturity cauliflower. Proc Natl AcadSci, India, Sect B Biol Sci. 2017; 87: 877–884. [Google Scholar]
- 74.Evans LM, Tahmasbi R, Jones M, Vrieze SI, Abecasis GR, Das S, et al. Narrow-sense heritability estimation of complex traits using identy-by-descent information. Heredity. 2018; 10.1038/s41437-018-0067-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Arashida FM, Maluf WR, Carvalho RC. General and specific combining ability in tropical winter cauliflower. Hortic Bras. 2017; 35: 167–173. 10.1590/s0102-053620170203. [DOI] [Google Scholar]
- 76.Gao W, Baars JJP, Dolstra O, Visser RGF, Sonnenberg ASM. Genetic variation and combining ability analysis of bruising sensitivity of Agaricusbisporus. PLOS ONE. 2013; 8: e76826 10.1371/journal.pone.0076826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fasahat P, Rajabi A, Rad MJ, Derera J. Principles and utilization of combining ability in plant breeding. BiomBiostatInt J. 2016; 4: 1–24. 10.15406/bbij.2016.04.00085. [DOI] [Google Scholar]
- 78.El-Esawi MA, Germaine K, Bourke P, Malone R. Genetic diversity and population structure of Brassica oleraceagermplasm in Ireland using SSR markers. C R Biol. 2016; 339: 133–140. 10.1016/j.crvi.2016.02.002 [DOI] [PubMed] [Google Scholar]
- 79.Parkash C, Kumar S, Singh R, Kumar A, Kumar S, Dey SS,et al. ‘Ogura’-based ‘CMS’ lines with different nuclear backgrounds of cabbage revealed substantial diversity at morphological and molecular levels. 3Biotech. 2018; 8: 27 10.1007/s13205-017-1047-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yousef EAA, Muller T, Borner A, Schmid KJ. Comparative analysis of genetic diversity and differentiation of cauliflower (Brassica oleraceavar. botrytis) accessions from two ex situ gene banks. PLOS ONE. 2018; 13: e0192062 10.1371/journal.pone.0192062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Taheri S, Abdullah TL, Yusop MR, Hanafi MM, Sahebi M, Azizi P,et al. Mining and development of novel SSR markers using next generation sequencing (NGS) data in plants. Molecules. 2018; 23: 399 10.3390/molecules23020399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Varshney RK, Graner A, Sorrells MW. Genic microsatellite markers in plants: features and applications. Trends Biotechnol. 2005; 23: 48–55. 10.1016/j.tibtech.2004.11.005 [DOI] [PubMed] [Google Scholar]
- 83.Moges AD, Admassu B, Belew D, Yesuf M, Nijuguna J, Kyalo M, et al. Development of microsatellite markers and analysis of genetic diversity and population structure of Colletotrichumgloeosporioidesfrom Ethiopia. PLOS ONE. 2016; 11: e0151257 10.1371/journal.pone.0151257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhu S, Zhang X, Liu Q, Luo T, Tang Z, Zhou Y. The genetic diversity and relationships of cauliflower (Brassica oleraceavar. botrytis) inbred lines assessed by using SSR markers. PLOS ONE. 2018; 13: e0208551 10.1371/journal.pone.0208551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao Z, Gu H, Sheng X, Yu H, Wang J, Zhao J, et al. Genetic diversity and relationships among loose-curd cauliflower and related varieties as revealed by microsatellite markers. SciHortic. 2014; 166: 105–110. [Google Scholar]
- 86.Falconer DS, Mackay TFC. Introduction to quantitative genetics, 4th edn Longman, London; 1996. [Google Scholar]
- 87.Burstin J, Charcosset A. Relationship between phenotypic and marker distances: theoretical and experimental investigations. J Hered. 1997; 79: 477–483. [Google Scholar]
- 88.Camussi A, Ottaviano E, Calinski T, Kaczmarek Z. Genetic distances based on quantitative traits. Genetics. 1985; 111: 945–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wegary D, Vivek B, Labuschagne M. Association of parental genetic distance with heterosis and specific combining ability in quality protein maize. Euphytica. 2013; 191: 205–216. [Google Scholar]
- 90.Zhang X, Lv L, Lv C, Guo B, Xu R. Combining ability of different agronomic traits and yield components in hybrid barley. PLOS ONE. 2015; 10: e0126828 10.1371/journal.pone.0126828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Geleta LF, Labuschagne MT, Viljoen CD. Relationship between heterosis and genetic distance based on morphological traits and AFLP markers in pepper. Plant Breed. 2004; 123: 467–473. [Google Scholar]
- 92.Krishnamurthy SL, Rao AM, Reddy KM, Ramesh S, Hittalmani S, Rao MG. Limits of parental divergence for the occurrence of heterosis through morphological and AFLP marker in chilli (Capsicum annuumL.).Curr Sci. 2013; 104: 738–746. [Google Scholar]
- 93.Bernardo R. Relationship between single-cross performance and molecular marker heterozygosity. TheorAppl Genet. 1992; 83: 628–634. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DCI: days to 50% curd initiation, DCM: days to 50% curd maturity, PH: plant height, LL: leaf length, LW: leaf width, GPW: gross plant weight, MCW: marketable curd weight, NCW: net curd weight, CSI: curd size index, NoL: number of leaves, HI: harvest index, TMY: total marketable yield, CL: curd length, CD: curd diameter, CoL: core length, LSI: leaf size index. The upper and lower lines outside the box stand for maximum and minimum adjacent value, respectively. The median value is represented by line inside the box. The lower and upper hinge of the box stands for 25% and 75% percentile, respectively.
(TIF)
(A) Corrplot depicting correlation of phenotypic distance with heterosis, (B) Corrplot of association of genetic distance with combining ability, (C) Corrplot depicting association of phenotypic distance with combining ability.
(TIF)
Among the 350 microsatellites, 87 primers showed clear cut polymorphism and used for allelic diversity analysis.
(DOCX)
* = significant at 5% probability, ** = significant at 1% probability, *** = significant at 0.1%, **** = significant at 0.01% probability through F test, Rep = Replication, Blk = Block, Trt = Treatment, Rep (Blk)Adj = Rep (Blk) Adjustable Days to 50% CI = Days to 50% curd initiation, Days to 50%CM = Days to 50% curd maturity, PH = Plant height, GPW = Gross plant weight, MCW = marketable curd weight, NCW = net curd weight, LL = leaf length, LW = leaf width, NoL = No of leaves, CL = curd length, CD = curd diameter, CoL = core length, CSI = curd size index, LSI = leaf size index, HI = harvest index, TMY = total marketable yield, R2: coefficient of determination.
(DOCX)
* = significant at 5% probability, ** = significant at 1% probability, *** = significant at 0.1%, **** = significant at 0.01% probability through F test, CD = critical difference, Days to 50% CI = Days to 50% curd initiation, Days to 50%CM = Days to 50% curd maturity, PH = Plant height, GPW = Gross plant weight, MCW = marketable curd weight, NCW = net curd weight, LL = leaf length, LW = leaf width, NoL = No of leaves, CL = curd length, CD = curd diameter, CoL = core length, CSI = curd size index, LSI = leaf size index, HI = harvest index, TMY = total marketable yield.
(DOCX)
*commercial standard checks.
(DOCX)
L: CMS lines; T: DH testers.
(DOCX)
* = significant at 5% probability, ** = significant at 1% probability, *** = significant at 0.1%, **** = significant at 0.01% probability through F test, MPH: Mid parent heterosis, BPH: better parent heterosis, SCA: specific combining ability (value in parenthesis).
(DOCX)
* = significant at 5% probability, ** = significant at 1% probability, *** = significant at 0.1%, **** = significant at 0.01% probability through F test, MPH: Mid parent heterosis, BPH: better parent heterosis, SCA: specific combining ability (value in parenthesis).
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.