Abstract
As agriculture industrializes, concentrated animal feeding operations (CAFOs) are becoming more common. Feces from CAFOs is often used as fertilizer on fields. However, little is known about the effects manure has on the soil microbiome, which is an important aspect of soil health and fertility. In addition, due to the subtherapeutic levels of antibiotics necessary to keep the animals healthy, CAFO manure has elevated levels of antibiotic resistant bacteria. Using 16s rRNA high-throughput sequencing and qPCR, this study sought to determine the impact of swine CAFO manure application on both the soil microbiome and abundance of select antibiotic resistance genes (ARGs) and mobile element genes (erm(B), erm(C), sul1, str(B), intI1, IncW repA) in agricultural soil over the fall and spring seasons. We found the manure community to be distinct from the soil community, with a majority of bacteria belonging to Bacteroidetes and Firmicutes. The soil samples had more diverse communities dominated by Acidobacteria, Actinobacteria, Proteobacteria, Verrucomicrobia, and unclassified bacteria. We observed significant differences in the soil microbiome between all time points, except between the spring samples. However, by tracking manure associated taxa, we found the addition of the manure microbiome to be a minor driver of the shift. Of the measured genes, manure application only significantly increased the abundance of erm(B) and erm(C) which remained elevated in the spring. These results suggest bacteria in the manure do not survive well in soil and that ARG dynamics in soil following manure application vary by resistance gene.
Introduction
In the past two decades, agriculture has become more industrialized and has shifted toward fewer but larger farm operations. Swine production is no exception to this trend; in the US, nearly three-quarters of swine are grown in concentrated animal feeding operations (CAFOs) containing over 5,000 pigs each [1]. Manure from swine CAFOs is often used as an organic fertilizer on fields to improve soil quality. Organic fertilizer provides nitrogen enrichment, increases soil organic matter, and is often thought to be a better alternative than synthetic fertilizer [2, 3]. The manure microbiome can influence the soil microbiome through direct competition and transfer of antibiotic resistance genes (ARGs) [4]. However, our knowledge of these processes is limited.
The soil microbiome has been linked to overall soil quality because it is involved in nutrient cycling, helps maintain soil water content, and influences soil acidity [5–8]. Manure application can impact the soil microbiome by increasing available nutrients or by introducing the manure microbiome [9, 10]. However, it is unclear how much of a role the introduction of the manure microbiome plays in altering the soil microbiome as members of the manure microbiome may not compete well in the soil environment [11, 12]. While previous studies have found that organic manure application significantly alters the soil microbiome, we still do not fully understand the extent to which manure application shifts the soil microbiome or the duration of the shift [9, 13, 14].
Manure from CAFOs has also been shown to be a reservoir of antibiotic resistant bacteria due to the use of subtherapeutic levels of antibiotics in the feed [15–19]. Because of high levels of antibiotic resistant bacteria in manure, manure application has been shown to significantly increase the abundance of ARGs in soil [20–24]. From the soil, ARGs can be dispersed into surrounding waterways via runoff and drainage [25–27]. As ARGs may be transferred to pathogenic bacteria in the environment through horizontal gene transfer, agricultural dissemination of ARGs into the environment may perpetuate the current increase of antibiotic resistance, which is a public health threat as the number of antibiotics available becomes more limited [15, 28, 29].
In the present study, we investigated the impact of swine CAFO manure fertilization on both the soil microbiome and the abundance of antibiotic resistance and mobile element genes over the fall and spring seasons. The effect of manure fertilization on the soil microbial community was explored by using 16S rRNA gene high-throughput sequencing. qPCR was employed to determine the relative abundances and dynamics of select antibiotic resistance and mobile element target genes (erm(B), erm(C), sul1, str(B), intI1, IncW repA) from soil samples over the fall and spring season. These genes were chosen because they represent resistance to a variety of antibiotics commonly used in the swine industry, including the CAFO in this study, and because they overlap ARGs investigated in several studies, most specifically with those used by Marti et al., allowing for us to begin building comparisons across environments [23]. The present study found that manure application may not be a significant factor in altering the soil community over a five-month period but does affect the gene abundance of select resistance genes.
Materials and methods
Study site
Manure, manure line, and field soil samples were taken from a swine CAFO farm located near Grinnell, IA (41.5896, -92.7569). Permission was granted by the land owner for sample collection. The CAFO houses approximately 6,500 hogs. Manure was collected in a pit below the CAFO for a year before application. The year before sampling the following antibiotics were dispensed in the feed according to a feeding regimen for growing swine, which included: tiamulin, chlortetracycline, sulfamethazine, penicillin, lincomycin, and tylosin phosphate. When infections arose in the barn, swine were additionally treated with penicillin and cycline, separately supplied in water, and 60 swine had Indoflex shots to treat illness.
The agricultural field sampled is a flat section of land at the bottom of the field that slopes up to the farm buildings. The field was split into two sites with each site centered around a tile line that empties into the stream positioned next to the field. The field is primarily silt loam soil (https://websoilsurvey.sc.egov.usda.gov/) [30]. Soil samples were collected before manure application on November 8, 2016. Manure was applied on the corn field for the 2017 growing season on November 12, 2016 at a rate of 4,127 gallons per acre, and the manure was sampled at this time. Manure was injected into knife lines running parallel through the field, and after 2–3 weeks the entire field is tilled. The samples collected just after application, on November 15, 2016, are from the injection lines referred to as “manure lines” and from soil in between the injection lines “soil”. After tilling samples were collected from the mixed soil on February 17, 2017 and March 22, 2017. There were no samples available on this site that had not been impacted by animal manure.
Sample collection
For field soil and manure line samples, soil cores were taken at random locations within each site using a T-sampler sterilized with 70% ethanol between samplings. Samples were contained in a sterile bag and mixed by hand to homogenize the sample. Samples were then transported to the lab and refrigerated within one hour of collection and stored frozen at -20°C. Data from the two sites were combined for analysis for each respective sample type and date.
DNA extraction
For 16S community analysis, DNA was extracted from 250 mg of soil and 250 μl of manure samples using the MagAttract Powersoil DNA EP Kit (384) (Qiagen, Germantown, MD) according to the manufacturer’s instructions. DNA was extracted from ten manure samples, twelve manure line samples from each site, and twelve soil samples from each site for every sampling date.
For qPCR, DNA was extracted from 250 mg of soil and 250 μl of manure samples using the DNeasy PowerSoil Kit (Qiagen, Germantown, MD) using the manufacturer’s instructions with one modification: the samples were placed on a Mini-Beadbeater (Biospec Products, Bartlesville, OK) for 140 seconds to homogenize. In total, DNA was extracted from ten manure samples, ten manure line samples from each site, and 10 soil samples from each site for every sampling date.
16S rRNA community analysis
The V4 region of the 16S rRNA gene was amplified using the Earth Microbiome Project primers (F: 5’GTGYCAGCMGCCGCGGTAA3’, R: 5’GGACTACNVGGGTWTCTAAT3’; fwd-barcoded: 515FB-806RB) and standard protocol [31]. Samples were indexed and sequenced using Illumina MiSeq (Illumina, San Diego, CA) producing 250-bp paired-end reads. Sequences were processed with MOTHUR (version 1.39.5) [32] according to standard operating procedure (http://www.mothur.org/wiki/MiSeq_SOP) [33]. Briefly, paired sequences were joined, primers were trimmed, and sequences were screened. The SILVA 16S rRNA sequence database (Release 132) was used for reference alignment and the RDP 16S rRNA reference (http://rdp.cme.msu.edu) for taxonomic classification [34, 35]. The VSEARCH algorithm in MOTHUR was used to filter chimeras. The OptiClust algorithm in MOTHUR was used to cluster processes sequences into operational taxonomic units (OTUs) using a 97% sequence similarity cutoff [36]. For the purposes of this study, only sequences associated with bacteria were used for analysis.
Statistical analysis and data visualization were carried out in R (version 3.5.0) (https://www.r-project.org/). The package phyloseq (version 1.24.2) was used for rarefying and data cleaning [37]. The packages ape (version 5.1) [38], dplyr (version 0.7.6) [39], and reshape2 (version 0.8.7) [40] functions were used in data transformations. Both alpha diversity indices and relative phyla abundance were produced in phyloseq and analyzed using ANOVA from the package vegan (version 2.5–2) [41]. NMDS ordination was produced and analyzed using the ADONIS function, which is equivalent to a PERMANOVA, using the vegan package. DEseq2 was used to calculate differential abundance of OTUs after singletons were removed (version 1.20.0) [42]. The package ggplot2 (version 3.0.0) [43] was used for visualization along with RColorBrewer (version 1.1–2) [44].
qPCR for gene targets
Quantitative Real-Time PCR amplification was performed using an Applied Biosystems StepOnePlus Real-Time PCR System (ThermoFisher, Waltham, MA). The primers used in this study are described in Table 1, and were obtained from Integrated DNA Technologies in Coralville, IA.
Table 1. qPCR primers and probes used in this study.
| Name | Sequence (5'→3')a | Annealing Temp (C) | Final primer concn (nM) | Target | Reference |
|---|---|---|---|---|---|
| Universal bacteria | 59 | 300 | rrnS gene | [45] | |
| BACT1369F | CGGTGAATACGTTCYCGG | ||||
| PROK1492R | GGWTACCTTGTTACGACTT | ||||
| TM1389F | HEX-CTTGTACACACCGCCCGTC-BHQ1 | ||||
| erm(B) | 65 | 200 | Erythromycin resistance gene locus B | [46] | |
| ermB-F | AAAACTTACCCGCCATACCA | ||||
| ermB-R | TTTGGCGTGTTTCATTGCTT | ||||
| erm(C) | 62 | 200 | Erythromycin resistance gene locus C | [47] | |
| ermC-F | AATCGTGGAATACGGGTTTGC | ||||
| ermC-R | CGTCAATTCCTGCATGTTTTAAGG | ||||
| sul1 | 64 | 200 | Sulfamethazine resistance gene 1 | [23] | |
| sul1-F | GACTGCAGGCTGGTGGTTAT | ||||
| sul1-R | GAAGAACCGCACAATCTCGT | ||||
| str(B) | 61 | 300 | Streptomycin phosphotransferase B | [48] | |
| strB-F | ATCGCTTTGCAGCTTTGTTT | ||||
| strB-R | ATGATGCAGATCGCCATGTA | ||||
| strB-P | HEX-ATGCCTCGGAACTGCGT-BHQ1 | ||||
| intI1 | 62 | 200 | Integrase class 1 | [49] | |
| Int1F2 | TCGTGCGTCGCCATCACA | ||||
| Int1R2 | GCTTGTTCTACGGCACGTTTGA | ||||
| IncW repA | 61 | 300 | repA gene from plasmid incompatibility group W | [23] | |
| IncW-F | GGCCATCGTATCAACGAGAT | ||||
| IncW-R | ATTGGTGCGCTCAAAGTAGC | ||||
| IncW-P | HEX-AGCTGGCTTAGTCGGCTACA-BHQ1 |
a HEX, 2’, 4’, 5’, 7’-tetrachloro-6-carboxy-4,7-dichlorofluorescein succinimidyl ester; BHQ1, black hole quencher 1.
Plasmids containing erm(C) target DNA fragment were provided by the Howe Lab at Iowa State University [24]. Plasmids containing all other target DNA fragments for every primer were provided by Marti et. al [23]. Plasmids were transformed into Escherichia coli One Shot TOP10 Cells (Thermofisher, Waltham, MA) using the manufacturer’s instructions and were extracted using the Wizard Plus SV Minipreps DNA Purification System (Promega, Madison, WI) following the manufacturer’s instructions. Plasmid copy number was calculated using the NanoDrop One Microvolume UV-Vis Spectrophotometer (ThermoFisher, Waltham, MA). The cultivated plasmids were used for standard curves consisting of 10-fold samples.
Each reaction was prepared using 12.5 μl of Takyon ROX SYBR serial dilutions that spanned the range of the target gene amplification from environmental MasterMix blue dTTP (Eurogentec, Fremont, CA) for SYBR green PCR and PrimeTime Gene Expression Master Mix (Integrated DNA Technologies, Coralville, IA) for TaqMan PCR. Two microliters of template DNA and deionized water were added to reach a final volume of 25 μl. Every sample, including a no template DNA control of DNA free water, was run in triplicate. A melting curve afterwards was used to check the purity of the SYBR green assay qPCR product.
Gene abundance analysis
The quantification thresholds and cycle were determined using the StepOne Software (Version 2.0.2) (Applied Biosystems). Standard curves were generated using linear regression analysis of the quantification cycle versus the amount of template DNA. A regression goodness of fit (r2) of above 0.9 was needed for the qPCR run to be used in analysis. Amplification efficiency was calculated from the linear regression as described previously [50]. Amplification efficiency between 90% and 110% was needed in order for the qPCR run to be used in analysis. Limit of detection (LOD) and limit of quantification (LOQ) were determined by serial dilutions of known plasmid amounts. The LOD was the lowest dilution that was distinguishable from the no template control. The LOQ was the lowest dilution that stayed within one standard deviation of the linear regression line and was distinguishable from the no template control. Gene target copy number was calculated using the standard curve and each gene abundance is expressed as a ratio of targeted gene copy per total rrnS gene copy in the reaction.
GraphPad Prism 7 (GraphPad Software, Inc.) was used for statistical analysis and the results were visualized in R. Gene abundances were compared using Kruskal-Wallis test with Dunn’s test for multiple comparisons using Luby et al.’s method to compensate for LOD and LOQ abundances [24]. Briefly, samples with gene abundance below the specified LOQ and above the LOD were assigned the average of the LOQ and LOD for analysis. Sample abundances below the LOD were assigned a value of one for analysis. This does not alter statistical significance because Kruskal-Wallis is a non-parametric rank based test.
Results
Sample collection
Manure, soil, and manure line samples were obtained from a swine CAFO located near Grinnell, IA. Ten manure samples were taken directly from the lagoon beneath the swine CAFO at the same time of its application onto the field, November 12, 2016. Soil samples were obtained from two sites from a commercial agricultural field. Data from the two sites were combined for analysis for each respective sample type and date. Twelve soil samples were taken from each site at four time points: fall pre-manure application (November 8, 2016), fall post-manure application (November 15, 2016), spring time 1 (February 17, 2017) and spring time 2 (March 22, 2017).
General description of DNA sequences
There was an average of 28,129 sequences per an individual soil or manure sample. Sequences were rarefied to 19,239 reads for analysis (the lowest sampling depth of this experiment that provided adequate taxa coverage), and samples below the rarefying depth were discarded. Rarefaction curves suggest that the rarefying depth covered the dominant taxa. After rarefying, 125 out of 130 samples remained, representing a total of 2,404,875 reads, 25,514 Operational Taxonomic Units (OTUs) at 97% similarity, and 33 phyla. The sequences have been deposited in NCBI repository with the accession no. SRP158016.
Characterization of bacterial communities
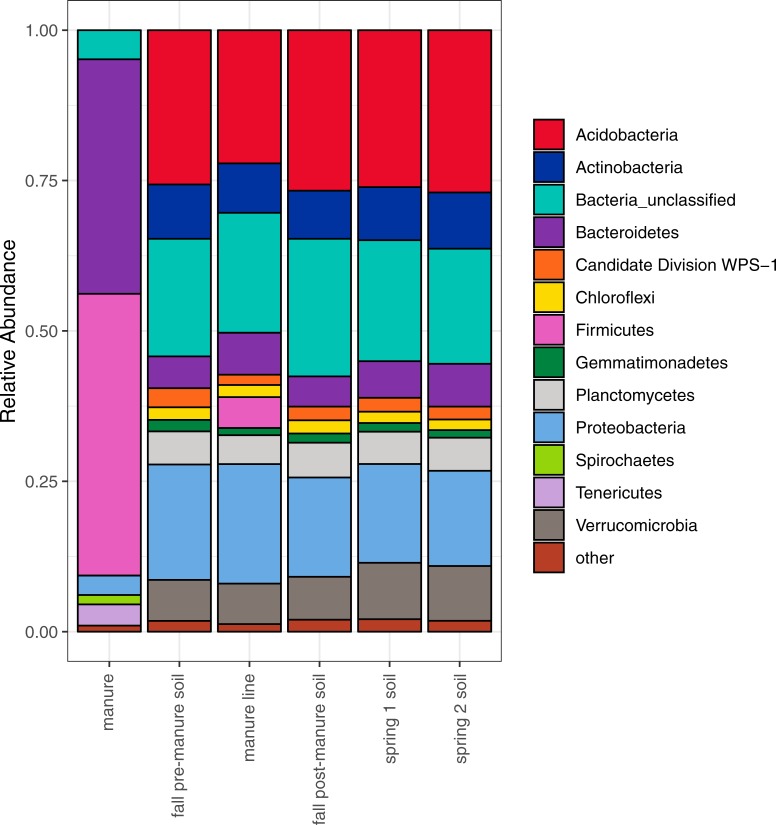
Using 16S rRNA gene high-throughput sequencing, the microbial community structure of the samples was characterized (Fig 1). The manure community was unique compared to the soil communities having significantly more members from the Bacteroidetes and Firmicutes phyla than the soil communities (P<0.05), while also having significantly lower percentages of the remaining phyla compared to soil and manure line samples (P<0.05). Although the most abundant phyla stayed fairly consistent in soil samples across our study (Fig 1), we did observe significant changes with specific phyla. Soil prior to manure application had significantly more candidate division WPS-1 OTUs than all of the other soil and manure line samples (P<0.01). After manure application, manure line soil had significantly more Firmicutes than the rest of the soil sample dates (P<0.01). The manure line also had significantly more Proteobacteria than the soil four months after manure application (P<0.05). Fall post-manure soil had significantly more unclassified bacteria than pre-manure, manure line, and spring time 2 soil (P<0.05). The spring soil samples had significantly more Verrucomicrobia than the rest of the soil samples but were not significantly different between each other (P<0.01).
Fig 1. Average relative abundances of phyla among sample types.
Included in “Other” are phyla that individually make up less than 1% of total abundance: Nitrospirae, Armatimonadetes, Spirochaetes, Synergistetes, Latescibacteria, BRC1, Parcubacteria, Microgenomates, Chlamydiae, Deferribacteres, Fibrobacteres, Candidatus Saccharibacteria, Elusimicrobia, candidate division WPS-2, SR1, Lentisphaerae, Hydrogenedentes, Cloacimonetes, Ignavibacteriae, Deinococcus-Thermus, and Fusobacteria.
Bacterial community α-diversity
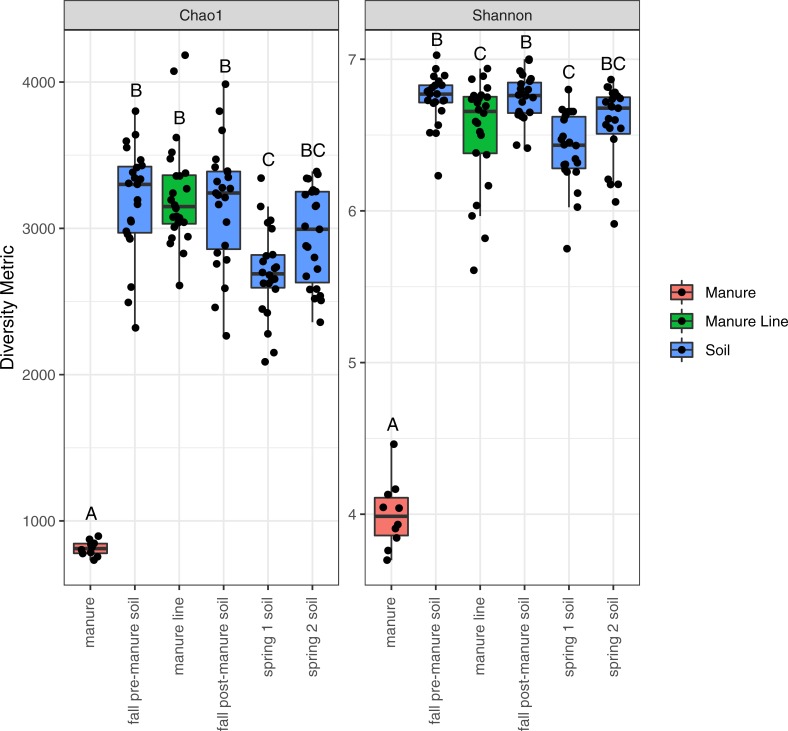
The alpha diversity, the diversity within each sample type, was determined using the Chao1 and Shannon diversity indices (Fig 2). Alpha diversity was significantly lower in the manure compared to the soil and manure line samples. In both the alpha diversity measures, there was an insignificant difference in diversity between fall pre-manure and post-manure soil. There was a significant decrease in diversity between pre-manure and spring time 1 soil, but no significant difference between pre-manure and spring time 2 soil diversity. The manure line alpha diversity was similar to the soil alpha diversity. However, the difference between fall pre-manure soil and manure line diversity was significant using the Shannon index, but not using Chao1.
Fig 2. Alpha diversity indices of microbiome samples.
Boxplots represent 25th to 75th percentiles and whiskers showing a maximum of 1.5x the interquartile range (IQR). Different letters indicate significant differences within the alpha diversity indexes (P<0.05).
Community comparisons (β-diversity)
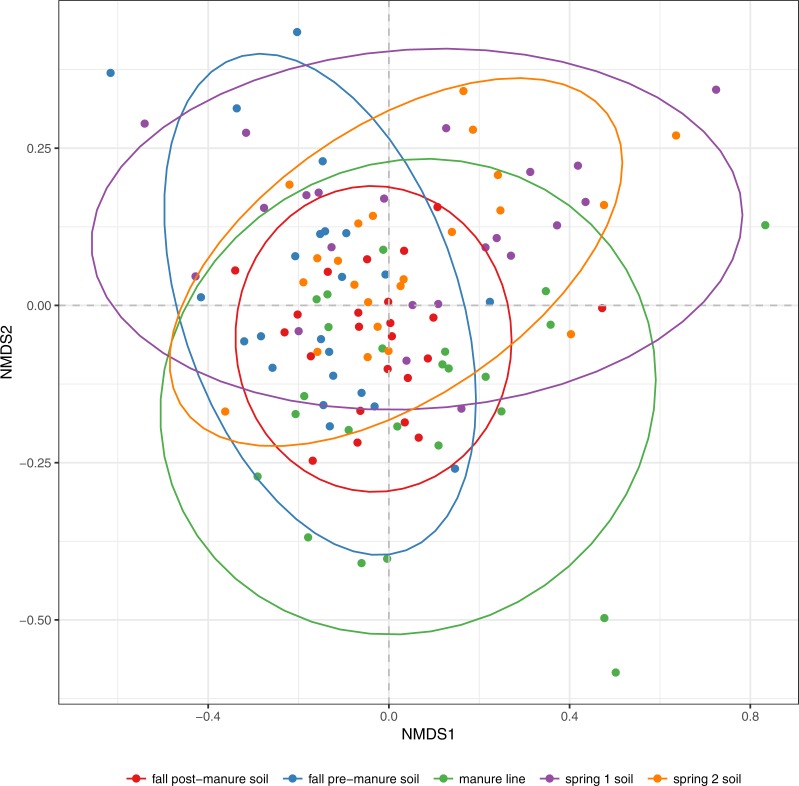
The NMDS ordination and PERMANOVA were utilized to compare the bacterial community between samples. Pairwise PERMANOVA showed that the manure community was significantly different than the soil and manure line samples (P<0.001). While the soil and manure line community compositions shared the same dominant phyla, pairwise PERMANOVA, which considers the whole community composition, showed that the bacterial community differed significantly between all soil and manure line samples, except for the spring samples (P<0.001, Fig 3). Both time and sample type were significant factors in the difference of microbiome composition between soil and manure line samples (P<0.05). Sample date explained 27.6% of the variance while type of sample explained 0.023% of variance.
Fig 3. Comparison of community composition between soil and manure line samples.
Community composition varied significantly between all samples except between the spring samples. Spring time 1 and spring time 2 soil are combined as spring soil. Clustering is based on nonmetric multidimensional scaling analysis of samples according to Bray-Curtis distances. Manure was excluded from the NMDS plot to show variation of soil samples.
Dispersion of manure OTUs
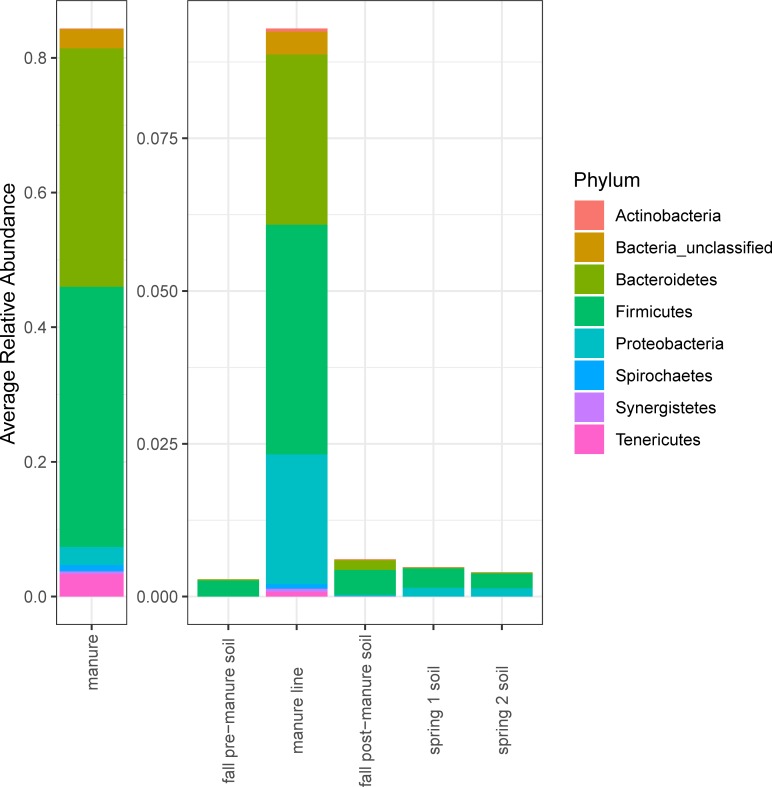
To determine if the changes in the soil community post-manure application were due to the addition of OTUs from manure, 104 OTUs significantly more abundant (P<0.05) in manure than pre-manure soil were identified and tracked (S1 Table). The manure associated OTUs were used as indicators of dispersion from the manure microbiome and their changes in abundance between pre-manure soil and post-manure soil samples were measured. In the manure line samples immediately following manure application, 57 of the 104 identified manure associated OTUs were significantly elevated from pre-manure soil abundance (Fig 4). In the soil samples taken in between injection lines, only seven manure associated OTUs were significantly elevated in the fall post-manure soil from pre-manure soil. In both the spring sampling dates, the same three manure associated OTUs were significantly more abundant than pre-manure soil. Of the three OTUs that remained elevated in the spring, two were of the Proteobacteria phylum and one was of the Firmicutes phylum.
Fig 4. Average relative abundance of manure associated OTUs.
OTUs are classified as manure associated if abundances were significantly greater (P<0.05) in manure compared to fall pre-manure soil.
Of the 104 OTUs identified as being significantly more abundant in manure than pre-manure soil, 69 were only present in manure and not in pre-manure soil. These OTUs were also tracked. Thirty-eight of the manure specific OTUs were found in the manure line. Only one manure specific OTU, a Proteobacteria belonging to the genus Pseudomonas, remained in the fall post-manure soil. This same OTU was the only OTU present in manure and not pre-manure soil that remained significantly elevated in both spring soil samples. Pseudomonas are known to be well adapted to soil and agricultural environments along with many also living as opportunistic pathogens. We propose this OTU could have the ability to grow and survive in both animal and soil environments.
Resistance gene and mobile genetic element abundance
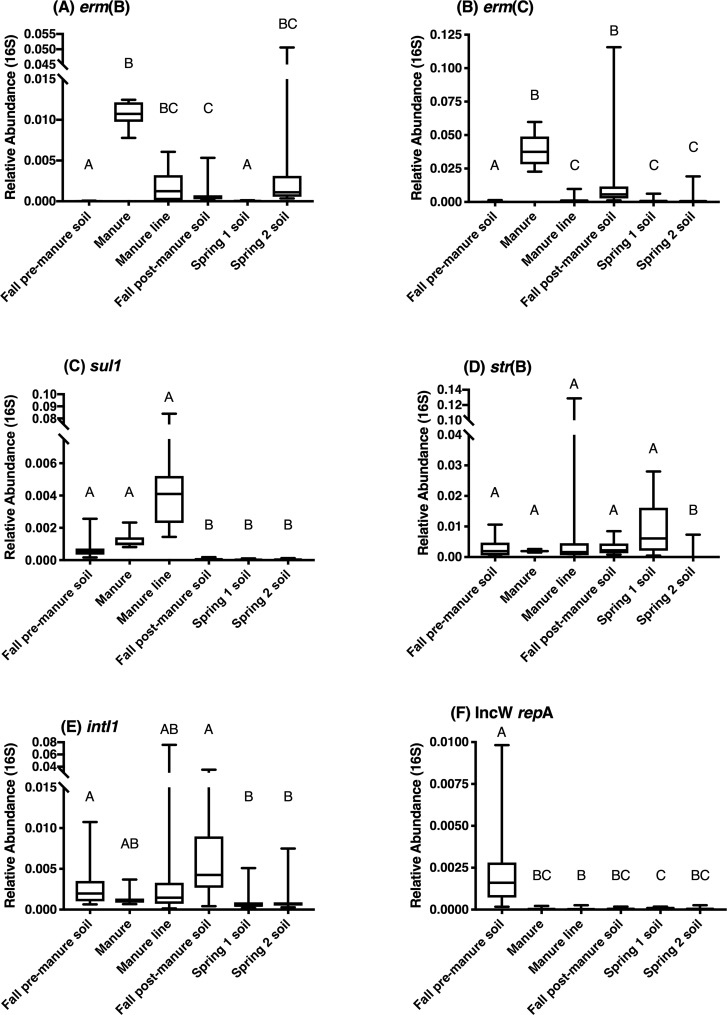
Along with the community characterization, we investigated if the abundance of antibiotic resistance genes and mobile genetic elements in soil were significantly enriched by manure application, as it is possible that even if manure bacteria do not survive in the soil, they may be able to pass along their ARGs to members of the soil microbiome. Abundances of target ARGs and mobile genetic elements were measured in samples relative to 16S rRNA gene. Every gene was detected in at least one sample of every sample type. Manure generally had the highest abundances of the target genes. However, relative abundances of each gene varied considerably in all samples and the abundance pattern of each gene was unique (Fig 5).
Fig 5. Relative abundance of target genes to 16S rRNA copy number.
Different letters indicate significant differences (P<0.05). n = 10 for manure. n = 20 for soil samples.
erm(B) and erm(C) had similar dynamics after manure application, with significantly higher abundance in fall post-manure soil and manure line samples compared to pre-manure soil. erm(C) abundances remained significantly higher in both spring sampling dates compared to pre-manure soil. However, erm(B) abundance lowered to pre-manure levels in the spring time 1 soil, but abundance was significantly elevated again from pre-manure soil in the spring time 2 soil to a level similar to post-manure soil abundance.
The other gene targets were not significantly increased following manure application (Fig 5). sul1 abundance decreased in the fall post-manure soil and remained at a lower level throughout the spring samples. Abundance of str(B) was similar in all the samples, except there was a significant decrease in the soil three months after manure application where str(B) abundance was mostly below the limit of quantification. Like str(B), all the fall soil, manure, and manure line samples had similar abundances of intI1. However, there was a significant decrease in abundance of intI1 in both the spring sample dates. IncW repA abundance significantly dropped in soil and manure line after manure application and stayed significantly lowered throughout the spring.
Discussion
Utilizing high throughput sequencing and quantitative PCR, this study characterized changes in the microbial community and resistance gene abundance following the application of swine manure in the soil of a commercial Iowa farm. The soil microbiome significantly changed throughout the experiment. However, the addition of the manure microbiome likely had limited influence on soil community composition as the microbes in the manure had short persistence in the soil. A similar trend was observed for ARGs as the target antibiotic resistance and mobile genetic element genes did not display uniform dynamics in response to manure application. These results suggest the response of the soil microbiome to manure amendment is complex and dependent on many environmental factors.
Soil microbiome dynamics
High throughput 16S rRNA sequencing was employed to characterize the bacterial microbiome of manure and of the farmland prior to and following manure application. The dominant phyla of both the manure and soil microbiomes in this study were generally characteristic of microbiomes described in prior studies [6, 9, 21, 51–54]. Both the soil alpha diversity and phyla composition were fairly consistent across all sampling dates. Unlike our results, some studies have found that manure amendment increases soil diversity and significantly changes the abundance of the major phyla after manure application [55, 56]. Other studies have found no change in alpha diversity and phyla composition of the bacterial community after manure application [57, 58]. Sample date was a significant factor in microbiome composition suggesting that the microbiome composition changes throughout time.
Our results are comparable to numerous other studies which have found manure application to significantly alter the microbe community in soils [9, 59–61]. Studies that have examined the soil community response after manure application longitudinally have found results similar to the present study in that the soil microbiome shifted away from the community prior to manure application [61, 62]. Two long-term studies found that 40 years of annual manure application significantly altered the soil microbiome compared to control soil [14, 63]. The soil microbiome composition and function have been shown to be very sensitive and, in a majority of studies, never fully recovers from disturbances [64, 65]. These differing results suggest that the response of the soil microbiome to manure amendment may depend on factors like regional soil characteristics and weather [66]. Our study was conducted on farm soil just before and following seasonal frost, ice and snow, which likely impacted dispersion of microbes and microbial survivorship over a three-month winter period.
The significant differences of the bacterial community between soil before and after manure application and between manure line and soil suggest that the introduction of the manure microbes could be a source of the change. To determine the impact of the addition of the manure microbes on the soil microbiome, OTUs significantly more abundant in the manure microbiome compared to the pre-manured soil were measured in post-manure soil samples. About half of the manure associated OTUs, most of which were present in manure and absent in pre-manure soil, were significantly elevated in the manure line samples suggesting that part of the soil microbiome shift is due to the addition of bacteria originating from manure. However, almost all of the manure associated OTUs returned to pre-manure levels of abundance by the spring which suggests that the influence of dispersion originating from the manure microbiome is temporary. The fall post-manure soil samples, which had low levels of manure associated OTUS, were taken between injection lines and thus were not in direct contact with manure. Nutrient addition from manure application may be another major factor in shifting the soil microbiome [67], but the limited duration of the manure associated bacteria in the soil combined with the fact that there was no pattern in the phyla abundance changes suggests that it is unlikely that manure was the sole driving factor in the soil microbiome shifts throughout the time points measured. Analysis at the family level follows what was observed at the phyla level with no clear patterns beyond the significant differences between soil and manure (S1 Fig). Previous studies have found that manure-associated bacteria had only a temporary effect on soil the microbiome and suggested that most manure-associated bacteria are not well adapted to survive in soil [68, 69]. In a microcosm study, Rieke et al. found that most manure associated OTUs which elevated in soil after manure application began to decrease in soil after 24 days [70]. However, it is possible that the manure microbiome is instead dispersing to elsewhere in the environment. Other factors like temperature, soil moisture, and soil pH can also have significant effects on microbiomes [6, 71]. Future research should determine the environmental characteristics that contribute to the composition of the soil microbiome.
Dynamics of ARGs and mobile genetic elements abundance
Abundance of select antibiotic resistance and mobile element target genes were measured to determine their dynamics in soil after manure application. Gene target abundances in almost all the soil samples were low. Within samples ARG abundances varied considerably, which is consistent with the heterogeneous nature of soil and previous studies [6, 72, 73]. Each gene displayed a unique abundance pattern across time points. An increase in gene abundance can be either from the direct addition of bacteria originating from manure, from the proliferation of bacteria already in the soil, or from the spread of genes due to horizontal gene transfer [23, 74]. Likewise, a decrease in gene abundance may be attributed to either the natural decay of the genes or spread of genes to other parts of the environment due to factors such as precipitation and temperature [4, 25, 75–81]. A recent study by Wang and colleagues, determined that after 26 years of manure application, there was not a large accumulation of ARGs in the soils studied and the low fold increases found in ARGs were not consistent across all types of ARGs [66].
The antibiotic resistant gene abundance patterns in the present study may be connected to the previous antibiotic exposure of the hogs. The abundance of erm(B), erm(C), and sul1 all increased after manure application. The manure producing hogs were given sulfamethazine, which sul1 provides resistance, and the macrolide tylosin phosphate, which both erm(B) and erm(C) may provide cross resistance [82, 83]. Abundance of sul1 has also been previously shown to be positively correlated with the amount of sulfonamide in soil [84]. The hogs were not given any streptomycin or other aminoglycosides of which str(B) provides resistance to [85]. It is important to note that str(B) was still detected in all samples. This may mean the str(B) abundances may be a result of naturally occurring antibiotics present in the soil or from a previous course of antibiotics, as resistance is known to persist for years after antibiotics have been administered [86, 87]. However, Udikovic-Kolic et al. found that cow manure amendment increased ARG abundance independent of the previous antibiotic exposure of the manure producing cows suggesting that ARGs may be enriched rather than introduced from manure application [22]. It is unknown whether the changes in gene abundance in this study were directly due to the addition of manure or due to other environmental factors. The extensive use of manure fertilizer along with grazing animals made us unable to obtain control soil that had no animal impact.
ARGs are often found to be linked to mobile element genes in the environment. The genes intI1, an integrase gene which allows exogenous genes to be inserted in the genome [88], and IncW repA, which assists in replication of plasmids carrying resistance genes [89], did not increase in abundance after manure application in our study. However, other studies have found both ARGs and mobile element genes to be increased following manure application [90, 91]. Recent work by Zhao and colleagues has found a link between the presence of metals in soils and ARGs and mobile element gene selection, a new factor to consider for horizontal gene transfer [92].
To our knowledge, the present field study is the first of its kind to examine soil after manure application from a swine CAFO in Iowa, but many studies have shown trends of increase in ARG abundance in soil after manure application [23, 24, 90, 91, 93, 94]. Marti et al. measured the abundance of five of the six genes in the present study in an agricultural field for 304 days after manure application and compared abundance to soil that had no manure exposure [23]. Their measured abundance dynamics of erm(B), str(B), and IncW repA in manure applied soil were similar to the present study and the abundances of the genes were significantly elevated from control soil [23]. Unlike this study, the abundance of sul1 and intI1 in Marti et al. both seemed to be significantly increased by manure application [23]. The differences observed across studies are likely due to many confounding factors including differences in antibiotics given to the animals that produced the manure, water availability, bacterial community composition, and soil pH.
The present study found that the fate of gene abundances depends on the gene. Two genes, erm(B) and erm(C), were significantly elevated in abundance in the spring 2 sampling. erm(B) remained elevated through all time points, while erm(C) was below quantification at spring time 1 and returned to elevated levels at spring time 2. The other four target genes were either at a similar or a lower abundance than pre-manure soil. Marti et al., which followed five of the genes in this study, suggests an offset time of at least one growing period is necessary to allow abundance to safely return to pre-application conditions [23]. In another study, soils treated with swine manure returned to ARG levels in untreated soil within 20 days [95]. However, another study found significantly elevated levels of ARGs in soil after 25 years of swine manure application [58], which can be contrasted with Wang et al. who found relatively low fold changes after 26 years of manure application [66]. Studies have shown that gene abundances are sensitive to factors like soil type [96], application type [97], and the presence of plants [98]. Further studies are needed to better characterize the fate and dynamics of ARGs following manure application, especially because the results of this study suggest that abundance generalizations cannot be made across genes and ARGs have been shown to have different rates of dissipation [73]. Additionally, future research may be able to link certain resistance genes to specific taxa in the environment.
Supporting information
Different letters indicated significant differences within each family by ANOVA with Tukey post-hoc test (P<0.05).
(DOCX)
(DOCX)
Acknowledgments
The authors would like to thank J. Jones, H. Allen, and N. Ricker of the USDA for their assistance with sequencing. We thank the Topp lab for providing control vectors for several resistance genes and a mobile element gene. The authors thank D. Hoksbergen for providing access to the study site. The authors also thank H. O’Neill, K. Vorhies, and S. Young for their assistance with DNA extractions. The authors are very appreciative to the many Grinnell College students who helped with sampling.
Data Availability
The sequences have been deposited in NCBI repository (https://www.ncbi.nlm.nih.gov/sra/) with the accession no. SRP158016.
Funding Statement
This work was funded by the Leopold Center for Sustainable Agriculture (E2016-08) SHL, USDA-NIFA-AFRI (2015-07849) SHL and AH, and Grinnell College SHL, EL, AC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The funders URLs are: https://www.leopold.iastate.edu/leopold-center-sustainable-agriculture; https://nifa.usda.gov/program/agriculture-and-food-research-initiative-afri; https://www.grinnell.edu/.
References
- 1.Agriculture USDA. Census of Agriculture 2017. Available from: https://www.nass.usda.gov/Publications/AgCensus/2017/index.php.
- 2.Das S, Jeong ST, Das S, Kim PJ. Composted cattle manure increases microbial activity and soil fertility more than composted swine manure in a submerged rice paddy. Front Microbiol. 2017;8:1702 10.3389/fmicb.2017.01702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meng L, Ding W, Cai Z. Long-term application of organic manure and nitrogen fertilizer on N2O emissions, soil quality and crop production in a sandy loam soil. Soil Biol Biochem. 2005;37(11):2037–45. [Google Scholar]
- 4.Chee-Sanford JC, Mackie RI, Koike S, Krapac IG, Lin YF, Yannarell AC, et al. Fate and transport of antibiotic residues and antibiotic resistance genes following land application of manure waste. J Environ Qual. 2009;38(3):1086–108. 10.2134/jeq2008.0128 [DOI] [PubMed] [Google Scholar]
- 5.Trivedi P, Delgado-Baquerizo M, Anderson IC, Singh BK. Response of soil properties and microbial communities to agriculture: implications for primary productivity and soil health indicators. Front Plant Sci. 2016;7:990 10.3389/fpls.2016.00990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fierer N. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol. 2017;15(10):579–90. 10.1038/nrmicro.2017.87 [DOI] [PubMed] [Google Scholar]
- 7.Yarwood RR, Rockhold ML, Niemet MR, Selker JS, Bottomley PJ. Impact of microbial growth on water flow and solute transport in unsaturated porous media. Water Resour Res. 2006;42(10). [Google Scholar]
- 8.Morales VL, Parlange JY, Steenhuis TS. Are preferential flow paths perpetuated by microbial activity in the soil matrix? A review. J Hydrol. 2010;393(1):29–36. [Google Scholar]
- 9.Hamm AC, Tenuta M, Krause DO, Ominski KH, Tkachuk VL, Flaten DN. Bacterial communities of an agricultural soil amended with solid pig and dairy manures, and urea fertilizer. Appl Soil Ecol. 2016;103:61–71. [Google Scholar]
- 10.Stocker MD, Pachepsky YA, Hill RL, Shelton DR. Depth-dependent survival of Escherichia coli and enterococci in soil after manure application and simulated rainfall. Appl Environ Microbiol. 2015;81(14):4801–8. 10.1128/AEM.00705-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Unc A, Goss MJ. Transport of bacteria from manure and protection of water resources. Appl Soil Ecol. 2004;25(1):1–18. [Google Scholar]
- 12.Sun R, Zhang X-X, Guo X, Wang D, Chu H. Bacterial diversity in soils subjected to long-term chemical fertilization can be more stably maintained with the addition of livestock manure than wheat straw. Soil Biol and Biochem. 2015;88:9–18. [Google Scholar]
- 13.Hartmann M, Frey B, Mayer J, Mader P, Widmer F. Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 2015;9(5):1177–94. 10.1038/ismej.2014.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soman C, Li D, Wander MM, Kent AD. Long-term fertilizer and crop-rotation treatments differentially affect soil bacterial community structure. Plant Soil. 2017;413(1):145–59. [Google Scholar]
- 15.Gilchrist MJ, Greko C, Wallinga DB, Beran GW, Riley DG, Thorne PS. The potential role of concentrated animal feeding operations in infectious disease epidemics and antibiotic resistance. Environ Health Perspect. 2007;115(2):313–6. 10.1289/ehp.8837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barton MD. Antibiotic use in animal feed and its impact on human healt. Nutr Res Rev. 2000;13(2):279–99. 10.1079/095442200108729106 [DOI] [PubMed] [Google Scholar]
- 17.Zhu YG, Johnson TA, Su JQ, Qiao M, Guo GX, Stedtfeld RD, et al. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc Natl Acad Sci U S A. 2013;110(9):3435–40. 10.1073/pnas.1222743110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chattopadhyay MK. Use of antibiotics as feed additives: a burning question. Front Micro. 2014;5:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Boeckel TP, Brower C, Gilbert M, Grenfell BT, Levin SA, Robinson TP, et al. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci U S A. 2015;112(18):5649 10.1073/pnas.1503141112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou X, Qiao M, Wang F-H, Zhu Y. Use of commercial organic fertilizer increases the abundance of antibiotic resistance genes and antibiotics in soil. Environ Sci Pollut Res Int. 2017;24(1):701–10. 10.1007/s11356-016-7854-z [DOI] [PubMed] [Google Scholar]
- 21.Xiong W, Wang M, Dai J, Sun Y, Zeng Z. Application of manure containing tetracyclines slowed down the dissipation of tet resistance genes and caused changes in the composition of soil bacteria. Ecotoxicol Environ Saf. 2018;147:455–60. 10.1016/j.ecoenv.2017.08.061 [DOI] [PubMed] [Google Scholar]
- 22.Udikovic-Kolic N, Wichmann F, Broderick NA, Handelsman J. Bloom of resident antibiotic-resistant bacteria in soil following manure fertilization. Proc Natl Acad Sci U S A. 2014;111(42):15202–7. 10.1073/pnas.1409836111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marti R, Tien YC, Murray R, Scott A, Sabourin L, Topp E. Safely coupling livestock and crop production systems: how rapidly do antibiotic resistance genes dissipate in soil following a commercial application of swine or dairy manure? Appl Environ Microbiol. 2014;80(10):3258–65. 10.1128/AEM.00231-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luby EM, Moorman TB, Soupir ML. Fate and transport of tylosin-resistant bacteria and macrolide resistance genes in artificially drained agricultural fields receiving swine manure. Sci Total Environ. 2016;550:1126–33. 10.1016/j.scitotenv.2016.01.132 [DOI] [PubMed] [Google Scholar]
- 25.Hruby CE, Soupir ML, Moorman TB, Shelley M, Kanwar RS. Effects of tillage and poultry manure application rates on Salmonella and fecal indicator bacteria concentrations in tiles draining Des Moines Lobe soils. J Environ Manage. 2016;171:60–9. 10.1016/j.jenvman.2016.01.040 [DOI] [PubMed] [Google Scholar]
- 26.Pruden A, Arabi M, Storteboom HN. Correlation between upstream human activities and riverine antibiotic resistance genes. Environ Sci Technol. 2012;46(21):11541–9. 10.1021/es302657r [DOI] [PubMed] [Google Scholar]
- 27.McMurry SW, Coyne MS, Perfect E. Fecal coliform transport through intact soil blocks amended with poultry manure. Plant Soil Sci Faculty Pub. 1998;27(1):86–92. [Google Scholar]
- 28.Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MO, Dantas G. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337(6098):1107–11. 10.1126/science.1220761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Norrby SR, Nord CE, Finch R, European Society of Clinical M, Infectious D. Lack of development of new antimicrobial drugs: a potential serious threat to public health. Lancet Infect Dis. 2005;5(2):115–9. 10.1016/S1473-3099(05)01283-1 [DOI] [PubMed] [Google Scholar]
- 30.Soil Survey Staff NRCS, United States Department of Agriculture. Web Soil Survey 2017. Available from: https://websoilsurvey.sc.egov.usda.gov/.
- 31.Gohl DM, Vangay P, Garbe J, MacLean A, Hauge A, Becker A, et al. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat Biotechnol. 2016;34(9):942–9. 10.1038/nbt.3601 [DOI] [PubMed] [Google Scholar]
- 32.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–41. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79(17):5112–20. 10.1128/AEM.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41(Database issue):D590–6. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–7. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Westcott SL, Schloss PD. OptiClust, an improved method for assigning amplicon-based sequence data to operational taxonomic units. mSphere. 2017;2(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One. 2013;8(4):e61217 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20(2):289–90. 10.1093/bioinformatics/btg412 . [DOI] [PubMed] [Google Scholar]
- 39.Wickham HF R. dplyr: A Grammar of Data Manipulation. R package version 0.7.6. 2015. [Google Scholar]
- 40.Wickham H. Reshaping data with the reshape package. J Stat Softw. 2007;21(12). [Google Scholar]
- 41.Oksanen J, Blanchet G, Friendly, M, Kindt, R, Legendre, P, McGlinn, D, et al. Vegan: community ecology package. R package version 2.5–2. 2018. Available from: https://CRAN.R-project.org/package=vegan.
- 42.Love MH W.; Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wickham H. ggplot2: Elegant graphics for data analysis: Springer-Verlag; New York; 2016. [Google Scholar]
- 44.Neuwirth E. RColorBrewer: ColorBrewer Palettes. 2014. [Google Scholar]
- 45.Suzuki MT, Taylor LT, DeLong EF. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5'-nuclease assays. Appl Environ Microbiol. 2000;66(11):4605–14. 10.1128/aem.66.11.4605-4614.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knapp CW, Dolfing J, Ehlert PA, Graham DW. Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ Sci Technol. 2010;44(2):580–7. 10.1021/es901221x [DOI] [PubMed] [Google Scholar]
- 47.Koike S, Aminov RI, Yannarell AC, Gans HD, Krapac IG, Chee-Sanford JC, et al. Molecular ecology of macrolide-lincosamide-streptogramin B methylases in waste lagoons and subsurface waters associated with swine production. Microb Ecol. 2010;59(3):487–98. 10.1007/s00248-009-9610-0 [DOI] [PubMed] [Google Scholar]
- 48.Walsh F, Ingenfeld A, Zampicolli M, Hilber-Bodmer M, Frey JE, Duffy B. Real-time PCR methods for quantitative monitoring of streptomycin and tetracycline resistance genes in agricultural ecosystems. J Microbiol Methods. 2011;86(2):150–5. 10.1016/j.mimet.2011.04.011 [DOI] [PubMed] [Google Scholar]
- 49.Gaze WH, Zhang L, Abdouslam NA, Hawkey PM, Calvo-Bado L, Royle J, et al. Impacts of anthropogenic activity on the ecology of class 1 integrons and integron-associated genes in the environment. ISME J. 2011;5(8):1253–61. 10.1038/ismej.2011.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–22. 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- 51.Looft T, Johnson TA, Allen HK, Bayles DO, Alt DP, Stedtfeld RD, et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc Natl Acad Sci U S A. 2012;109(5):1691–6. 10.1073/pnas.1120238109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fierer N, Lauber CL, Ramirez KS, Zaneveld J, Bradford MA, Knight R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012;6(5):1007–17. 10.1038/ismej.2011.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li R, Khafipour E, Krause DO, Entz MH, de Kievit TR, Fernando WG. Pyrosequencing reveals the influence of organic and conventional farming systems on bacterial communities. PloS One. 2012;7(12):e51897 10.1371/journal.pone.0051897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chaudhry V, Rehman A, Mishra A, Chauhan PS, Nautiyal CS. Changes in Bacterial Community Structure of Agricultural Land Due to Long-Term Organic and Chemical Amendments. Microb Ecol.2012;64(2):450–60. 10.1007/s00248-012-0025-y [DOI] [PubMed] [Google Scholar]
- 55.Zhen Z, Liu H, Wang N, Guo L, Meng J, Ding N, et al. Effects of manure compost application on soil microbial community diversity and soil microenvironments in a temperate cropland in China. PloS One. 2014;9(10):e108555 10.1371/journal.pone.0108555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen X, Li Z, Liu M, Jiang C, Che Y. Microbial community and functional diversity associated with different aggregate fractions of a paddy soil fertilized with organic manure and/or NPK fertilizer for 20 years. J Soil Sediments. 2015;15(2):292–301. [Google Scholar]
- 57.Riber L, Poulsen PH, Al-Soud WA, Skov Hansen LB, Bergmark L, Brejnrod A, et al. Exploring the immediate and long-term impact on bacterial communities in soil amended with animal and urban organic waste fertilizers using pyrosequencing and screening for horizontal transfer of antibiotic resistance. FEMS Microbiol Ecol. 2014;90(1):206–24. 10.1111/1574-6941.12403 [DOI] [PubMed] [Google Scholar]
- 58.Xie W-Y, Yuan S-T, Xu M-G, Yang X-P, Shen Q-R, Zhang W-W, et al. Long-term effects of manure and chemical fertilizers on soil antibiotic resistome. Soil Biol Biochem. 2018;122:111–9. [Google Scholar]
- 59.Ding J, Jiang X, Ma M, Zhou B, Guan D, Zhao B, et al. Effect of 35 years inorganic fertilizer and manure amendment on structure of bacterial and archaeal communities in black soil of northeast China. Appl Soil Ecol. 2016;105:187–95. [Google Scholar]
- 60.Zhong W, Gu T, Wang W, Zhang B, Lin X, Huang Q, et al. The effects of mineral fertilizer and organic manure on soil microbial community and diversity. Plant Soil. 2010;326(1):511–22. [Google Scholar]
- 61.He J-Z, Zheng Y, Chen C-R, He Y-Q, Zhang L-M. Microbial composition and diversity of an upland red soil under long-term fertilization treatments as revealed by culture-dependent and culture-independent approaches. J Soil Sediments. 2008;8(5):349–58. [Google Scholar]
- 62.Marschner P, Kandeler E, Marschner B. Structure and function of the soil microbial community in a long-term fertilizer experiment. Soil Biol and Biochem. 2003;35(3):453–61. [Google Scholar]
- 63.Zhang YH X.; Alexander T. W.; Thomas B. W.; Shi X.; Lupwayi N. Z. Long-term and legacy effects of manure application on soil microbial community composition. Biol Fertil Soils. 2018;54(2):269–83. [Google Scholar]
- 64.Shade A, Peter H, Allison SD, Baho DL, Berga M, Burgmann H, et al. Fundamentals of microbial community resistance and resilience. Front Microbiol. 2012;3:417 10.3389/fmicb.2012.00417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Allison SD, Martiny JB. Colloquium paper: resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci U S A. 2008;105 Suppl 1: 11512–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang F, Xu M, Stedtfeld RD, Sheng H, Fan J, Liu M, et al. Long-term effect of different fertilization and cropping systems on the soil antibiotic resistome. Environ Sci Technol. 2018;52(22):13037–46. 10.1021/acs.est.8b04330 [DOI] [PubMed] [Google Scholar]
- 67.Cederlund H, Wessén E, Enwall K, Jones CM, Juhanson J, Pell M, et al. Soil carbon quality and nitrogen fertilization structure bacterial communities with predictable responses of major bacterial phyla. Appl Soil Ecol. 2014;84:62–8. [Google Scholar]
- 68.Leclercq SO, Wang C, Sui Z, Wu H, Zhu B, Deng Y, et al. A multiplayer game: species of Clostridium, Acinetobacter, and Pseudomonas are responsible for the persistence of antibiotic resistance genes in manure-treated soils. Environ Microbiol. 2016;18(10):3494–508. 10.1111/1462-2920.13337 [DOI] [PubMed] [Google Scholar]
- 69.Jechalke S, Focks A, Rosendahl I, Groeneweg J, Siemens J, Heuer H, et al. Structural and functional response of the soil bacterial community to application of manure from difloxacin-treated pigs. FEMS Microbiol Ecol. 2014;87(1):78–88. 10.1111/1574-6941.12191 [DOI] [PubMed] [Google Scholar]
- 70.Rieke EL, Soupir ML, Moorman TB, Yang F, Howe AC. Temporal dynamics of bacterial communities in soil and leachate water after swine manure application. Front Microbiol. 2018;9:3197 10.3389/fmicb.2018.03197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Classen AT, Sundqvist MK, Henning JA, Newman GS, Moore JAM, Cregger MA, et al. Direct and indirect effects of climate change on soil microbial and soil microbial-plant interactions: What lies ahead? Ecosphere. 2015;6(8):art130. [Google Scholar]
- 72.Fitzpatrick D, Walsh F. Antibiotic resistance genes across a wide variety of metagenomes. FEMS Microbiol Ecol. 2016;92(2). [DOI] [PubMed] [Google Scholar]
- 73.Wang M, Liu P, Xiong W, Zhou Q, Wangxiao J, Zeng Z, et al. Fate of potential indicator antimicrobial resistance genes (ARGs) and bacterial community diversity in simulated manure-soil microcosms. Ecotoxicol Environ Saf. 2018;147:817–23. 10.1016/j.ecoenv.2017.09.055 [DOI] [PubMed] [Google Scholar]
- 74.Heuer H, Schmitt H, Smalla K. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr Opin Microbiol. 2011;14(3):236–43. 10.1016/j.mib.2011.04.009 [DOI] [PubMed] [Google Scholar]
- 75.Barkovskii AL, Bridges C. Persistence and profiles of tetracycline resistance genes in swine farms and impact of operational practices on their occurrence in farms’ vicinities. Water Air Soil Pollut.2012;223(1):49–62. [Google Scholar]
- 76.Ahmed W, Zhang Q, Lobos A, Senkbeil J, Sadowsky MJ, Harwood VJ, et al. Precipitation influences pathogenic bacteria and antibiotic resistance gene abundance in storm drain outfalls in coastal sub-tropical waters. Environ Int. 2018;116:308–18. 10.1016/j.envint.2018.04.005 [DOI] [PubMed] [Google Scholar]
- 77.Di Cesare A, Eckert EM, Rogora M, Corno G. Rainfall increases the abundance of antibiotic resistance genes within a riverine microbial community. Environ Pollut. 2017;226:473–8. 10.1016/j.envpol.2017.04.036 [DOI] [PubMed] [Google Scholar]
- 78.Tian Z, Zhang Y, Yu B, Yang M. Changes of resistome, mobilome and potential hosts of antibiotic resistance genes during the transformation of anaerobic digestion from mesophilic to thermophilic. Water Res. 2016;98:261–9. 10.1016/j.watres.2016.04.031 [DOI] [PubMed] [Google Scholar]
- 79.Diehl DL, LaPara TM. Effect of temperature on the fate of genes encoding tetracycline resistance and the integrase of class 1 integrons within anaerobic and aerobic digesters treating municipal wastewater solids. Environ Sci Technol. 2010;44(23):9128–33. 10.1021/es102765a [DOI] [PubMed] [Google Scholar]
- 80.Dunivin TK, Shade A. Community structure explains antibiotic resistance gene dynamics over a temperature gradient in soil. FEMS Microbiol Ecol. 2018;94(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sapkota AR, Curriero FC, Gibson KE, Schwab KJ. Antibiotic-resistant enterococci and fecal indicators in surface water and groundwater impacted by a concentrated swine feeding operation. Environ Health Perspect. 2007;115(7):1040–5. 10.1289/ehp.9770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Antunes P, Machado J, Sousa JC, Peixe L. Dissemination of sulfonamide resistance genes (sul1, sul2, and sul3) in Portuguese Salmonella enterica strains and relation with integrons. Antimicrob Agents Chemother. 2005;49(2):836–9. 10.1128/AAC.49.2.836-839.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Leclercq R, Courvalin P. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob Agents Chemother. 1991;35(7):1267–72. 10.1128/aac.35.7.1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao X, Wang J, Zhu L, Ge W, Wang J. Environmental analysis of typical antibiotic-resistant bacteria and ARGs in farmland soil chronically fertilized with chicken manure. Sci Total Enviro. 2017;593–594:10–7. [DOI] [PubMed] [Google Scholar]
- 85.Srinivasan VG B.; Nguyen L.; Headrick S.; Murinda S.; Oliver S. Characterization of antimicrobial resistance patterns and class 1 integrons in Escherichia coli O26 isolated from humans and animals. Int J Antimicrob Agents. 2007;29(3):254–62. [DOI] [PubMed] [Google Scholar]
- 86.Johnsen PJ, Townsend JP, Bøhn T, Simonsen GS, Sundsfjord A, Nielsen KM. Factors affecting the reversal of antimicrobial-drug resistance. Lancet Infect Dis. 2009;9(6):357–64. 10.1016/S1473-3099(09)70105-7 [DOI] [PubMed] [Google Scholar]
- 87.Wellington EMH, Boxall ABA, Cross P, Feil EJ, Gaze WH, Hawkey PM, et al. The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect Dis. 2013;13(2):155–65. 10.1016/S1473-3099(12)70317-1 [DOI] [PubMed] [Google Scholar]
- 88.Gillings MR, Gaze WH, Pruden A, Smalla K, Tiedje JM, Zhu YG. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 2015;9(6):1269–79. 10.1038/ismej.2014.226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fernandez-Lopez R, Garcillan-Barcia MP, Revilla C, Lazaro M, Vielva L, de la Cruz F. Dynamics of the IncW genetic backbone imply general trends in conjugative plasmid evolution. FEMS Microbiol Rev. 2006;30(6):942–66. 10.1111/j.1574-6976.2006.00042.x [DOI] [PubMed] [Google Scholar]
- 90.Wang M, Sun Y, Liu P, Sun J, Zhou Q, Xiong W, et al. Fate of antimicrobial resistance genes in response to application of poultry and swine manure in simulated manure-soil microcosms and manure-pond microcosms. Environ Sci Pollut Res Int. 2017;24(26):20949–58. 10.1007/s11356-017-9623-z [DOI] [PubMed] [Google Scholar]
- 91.Zhao X, Wang J, Zhu L, Wang J. Field-based evidence for enrichment of antibiotic resistance genes and mobile genetic elements in manure-amended vegetable soils. Sci Total Environ. 2019;654:906–13. 10.1016/j.scitotenv.2018.10.446 [DOI] [PubMed] [Google Scholar]
- 92.Zhao Y, Cocerva T, Cox S, Tardif S, Su JQ, Zhu YG, et al. Evidence for co-selection of antibiotic resistance genes and mobile genetic elements in metal polluted urban soils. Sci Total Environ. 2019;656:512–20. 10.1016/j.scitotenv.2018.11.372 [DOI] [PubMed] [Google Scholar]
- 93.Wu N, Qiao M, Zhang B, Cheng W-D, Zhu Y-G. Abundance and diversity of tetracycline resistance genes in soils adjacent to representative swine feedlots in China. Environ Sci Technol. 2010;44(18):6933–9. 10.1021/es1007802 [DOI] [PubMed] [Google Scholar]
- 94.Munir M, Xagoraraki I. Levels of antibiotic resistance genes in manure, biosolids, and fertilized soil. J Environ Qual. 2011;40(1):248–55. [DOI] [PubMed] [Google Scholar]
- 95.Zhang Y-J, Hu H-W, Gou M, Wang J-T, Chen D, He J-Z. Temporal succession of soil antibiotic resistance genes following application of swine, cattle and poultry manures spiked with or without antibiotics. Environ Pollut. 2017;231:1621–32. 10.1016/j.envpol.2017.09.074 [DOI] [PubMed] [Google Scholar]
- 96.Tang X, Lou C, Wang S, Lu Y, Liu M, Hashmi MZ, et al. Effects of long-term manure applications on the occurrence of antibiotics and antibiotic resistance genes (ARGs) in paddy soils: Evidence from four field experiments in south of China. Soil Biol & Biochem. 2015;90:179–87. [Google Scholar]
- 97.Joy SR, Bartelt-Hunt SL, Snow DD, Gilley JE, Woodbury BL, Parker DB, et al. Fate and transport of antimicrobials and antimicrobial resistance genes in soil and runoff following land application of swine manure slurry. Environ Sci Technol. 2013;47(21):12081–8. 10.1021/es4026358 [DOI] [PubMed] [Google Scholar]
- 98.Wang FH, Qiao M, Chen Z, Su JQ, Zhu YG. Antibiotic resistance genes in manure-amended soil and vegetables at harvest. J Hazard Mater. 2015;299:215–21. 10.1016/j.jhazmat.2015.05.028 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Different letters indicated significant differences within each family by ANOVA with Tukey post-hoc test (P<0.05).
(DOCX)
(DOCX)
Data Availability Statement
The sequences have been deposited in NCBI repository (https://www.ncbi.nlm.nih.gov/sra/) with the accession no. SRP158016.