Abstract
Long noncoding RNAs (lncRNAs) are a class of functional non-coding transcripts that are longer than 200 nt and regulate gene expression via diverse mechanisms in eukaryotes. In fact, they have emerged as critical epigenetic and transcriptional regulators of autophagy in mammals in response to various stressors. Autophagy not only plays a crucial role in maintaining cellular homeostasis, but it is also essential to immunity, targets intracellular pathogens for degradation, modulates inflammation, and participates in adaptive immune responses. However, the expression profile of lncRNA and its role in regulating autophagy in macrophages have been poorly defined. Here, we used transcriptomic and bioinformatics to analysis LncRNA expression profile during autophagy and functional studies to evaluate the function of the metastasis-associated lung adenocarcinoma transcript-1 (Malat1) lncRNA in macrophages. A total of 1112 putative lncRNAs (240 novel lncRNAs) were identified, including 831 large intergenic, 129 intronic, and 152 anti-sense lncRNA, of which 59 differentially expressed transcripts exhibited a greater than 1.5-fold change under different conditions. The interaction of Malat1 lncRNA with microRNA (mir)-23-3p and lysosomal-associated membrane protein 1 (Lamp1) was found, Malat1 releases inhibition of Lamp1 expression in macrophages through competitive adsorption of mir-23-3p. The results of this study provide a better understanding of lncRNA function in macrophages and a basis for further investigation into the roles and mechanisms of ncRNA in immunology, particularly the functions of Malat1 and mir-23-3p in the pathogenesis of macrophages.
Introduction
Protein-coding sequences only take up a very small fraction (~1.5%) of the human genome. many Non-coding sequences that are once considered as DNA junk are recently found to be essential for gene regulation by the Encyclopedia of DNA Elements project. Also, non-coding RNAs (ncRNAs) have nearly 58,648 different transcripts in humans [1]. Many ncRNAs in the genome play key roles in regulating gene expression at the transcriptional and post-transcriptional levels [2]. These ncRNAs have housekeeping, regulatory, and unknown functional roles [3]. Regulatory ncRNAs are usually classified into small ncRNAs and long ncRNAs (lncRNAs) according to their transcript lengths [4]. LncRNAs are a large class of transcribed RNA molecules of more than 200 nucleotides (nt) in length that lack of protein encoding potential and function [5]. LncRNA can be classified into different subtypes (antisense, intergenic, overlapping, intronic, bidirectional, and processed) according to the position and direction of gene transcription [6]. LncRNAs regulate biological activities through various mechanisms, including signaling [7], scaffolds [8], decoys [9]and endogenous competitive RNAs (CeRNA) [10,11]. In particular, the lncRNA-microRNA (miRNA)-mRNA CeRNA ternary network may play a key role in gene regulation [12]. Numerous studies have indicated that lncRNAs play critical roles in a wide range of biological processes, including cell differentiation and development [13,14], X-chromosome inactivation [15], the development of neurological diseases [16], cancer progression [17], and the immune response[18,19].
Autophagy is a lysosome-mediated evolutionarily conserved catabolic process in cells that serves to decompose unwanted cytoplasmic content, degenerate organelles and pathogens for high-throughput chemical recycling, and regulate cellular functions under normal and stressed conditions [20]. Similar to macrophages and dendritic cells, autophagy in antigen-presenting cells (APCs) not only acts as a phagocyte, but also plays a crucial role in initial immunity [21]. Increasing evidence on the fundamental role of autophagy in pathogenic microorganisms and cell host interactions has emerged [22]. Macrophages are part of the mononuclear phagocytic system and play crucial roles in the nonspecific immune response through autophagy. Thus, understanding how lncRNA regulates autophagy in macrophages may help to understand pathogen invasion and survival.
Accumulating data have revealed that many aspects of the autophagy process are regulated by lncRNAs [23–25]. Specifically, one of the mechanisms by which lncRNAs regulate cellular functions is by acting as CeRNA [26,27]. For example, the autophagy-promoting factor lncRNA regulates autophagy and myocardial infarction by functioning as a CeRNA sponge for mir-188-5p [28]. In addition, the metastasis-associated lung adenocarcinoma transcript-1 (Malat1) lncRNA also functions as a CeRNA to regulate autophagy. MALAT1 is highly conserved lncRNA, which reported as a nuclear-retained in mammals, also known as NEAT2. It has been reported that MALAT1 promotes gene activation or inhibition in a cell type-specific manner by promoting specific chromatin regulators, and it is likely to be used as a scaffold to recruit proteins in the vicinity of nuclear plaques and exert gene regulation. Recently, some studies also have demonstrated that Malat1 interacts with the RNA-binding protein human antigen R, and silencing of Malat1 greatly enhances the post-transcriptional regulation of TIA-1, further inhibiting autophagy [29]. The Malat1 lncRNA is a potent autophagy inducer that protects brain microvascular endothelial cells against oxygen-glucose deprivation/re-oxygenation-induced injury by sponging miR-26b and upregulating the expression of unk-51-like autophagy activating kinase 2 [30]. Moreover, Malat1 activates autophagy and upregulates stathmin, Ras-related protein Rab-5A, and autophagy related 4D expression in glioma through cavernously adsorption of miR-101, promoting cell proliferation [31]. LncRNA Malat1 expression is upregulated in LPS-activated macrophages, while it is downregulated in IL-4 activated macrophages [32]. In summary, Malat1 is involved in cell autophagy, immunology and may play a sponge in the RNA triplet network consisting of lncRNA-miRNA-mRNA.
Macrophages are important APCs and the main cells of the phagocyte system. However, the expression and function of lncRNAs in macrophages during autophagy are still largely unknown. In this study, we focused on determining the expression profiles of lncRNA and mRNA in murine RAW264.7 macrophages treated under different conditions to induce or inhibit autophagy. Among the differentially expressed lncRNAs (DELs), The possible sponge mechanism of Malat1 lncRNA in regulating autophagy in macrophages was further explored and discussed. The results of this study provide the differential expression profiles of lncRNAs during autophagy, and we confirm that Malat1 lncRNA function as a CeRNA to regulate the expression of lysosomal-associated membrane protein 1 (Lamp1) by sponging mir-23-3p in macrophages.
Materials and methods
Materials
Cell culture
The RAW264.7 cell lines (obtained from ATCC, Beijing, China) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Thermo Fisher Scientific, USA) supplemented with 10% fetal bovine serum (FBS, Gibco) at 37°C with 5% CO2 (vol/vol).
Treatments
The autophagy inducer RAPA and 3-MA were dissolved respectively in DMSO or double-distilled water at stock concentrations of 10 mM. The cells were plated in 6-well plates at the appropriate cell density. When confluence reached about 50–70%, cells were treated with 3-methyladenine (3mM, 12h) or rapamycin (50nm, 2h) and STV in HANKs buffer for 6h before harvesting cells to acquire total RNA.
Methods
High-throughput sequencing
Total RNA quality was checked with electrophoresed formaldehy de-denatured agarose gel and quantified with a spectrophotometer (NanoDrop ND-2000, USA). According to the composite sequencing quality requirements, 10 μg RNA was mixed from each condition with three repeats and then submitted to Biomarker Technologies Co. Ltd. (Beijing, China) for high-throughput transcriptome sequencing. For detecting lncRNAs, the cDNA library was constructed in a ribosome-free manner according to the instructions of the Illumine Kit. All clean reads of this work are available from the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) database (PRJNA544884).
Pipeline for identifying lincRNAs
A computational lncRNA dig pipeline was used to detect putative lncRNAs of each condition from RNA-seq read mapping and transcriptome assembly generated according to published methods [33]. To obtain putative novel non-coding transcripts and filter out known transcripts, the unique assembled transcriptome dataset was compared to the Ensemble Mouse genome annotation by using the Cuffcompare program from the Cufflinks package. Consequently, the assemblies that matched annotations could be identified. Unknown transcripts of more than 200 nt length and one exon were selected as candidate lncRNA transcripts. Moreover, the novel transcripts were clustered into different lncRNAs types, including lincRNA, intronic lncRNA, and anti-sense lncRNA based on their locations relative to the known genes by using Cuffcompare according to the class code. The protein-coding potential of candidate lncRNAs was analyzed by integrating the results of three widespread computational coding tools, namely, CPC, CNCI, and CPAT, to maximize the prediction of true-putative lncRNAs. Finally, putative protein-coding RNAs were filtered out by using minimum length, exon number threshold, and Pfam blast.
Differential expression analysis
For lncRNA sequencing, after library construction, RNA-seq (PE250) and the sequence read went through the data cleaning procedure. The ncRNAs among the different groups were compared as follows: the expression of four samples to obtain the FPKM was normalized, and the FC and FDR were calculated from the normalized expression. The logarithmetics FC formula: FC = Log2 (Treatment/Control).
LncRNA target prediction
Based on the functioning mode between the lncRNA and its target, we adopted two different prediction methods. The cis role of lncRNA is to act on neighboring target genes. We searched for coding genes within 100 kb upstream and downstream of lncRNA to predict putative cis target genes of lncRNA, followed by analysis of the functions of these coding genes to annotate lncRNA. Second, lncRNA and mRNA militate due to the base complementary, so we mainly used lncTar [34] to predict lncRNA targets by calculating the free energy and standardized free energy of mating sites, and below the threshold of the standardized free energy could be deemed as antisense targets of lncRNA. The GO functional and pathway analysis of the putative target genes were performed by using ClueGO. RNA22 and PITA database were used to explore the connection between lncRNA Malat1 and its potential sponge miRNA targets.
Total RNA extraction and qPCR analysis
After stimulating, cells were washed three times with PBS, and 2 ml TriZol reagent (Invitrogen) was added to 6-wells culture plate in each group. RNA extraction followed the manufacturer's instructions (Invitrogen). The RNA quality was checked by NanoDrop ND-2000 (Thermo Fisher Scientific, Waltham, MA, USA), and RNA integrity was assessed by standard electrophoreses in formaldehyde-denatured agarose gel. qPCR was used to confirm the lncRNA, miRNA, and mRNA expression levels. The RAW264.7 cells were treated with RAPA, and the expression levels of eight DELs, Lamp1, and mir-23-3p were measured by qPCR analysis. For lncRNAs, cDNA was synthesized using the Trans Script First-Strand cDNA Synthesis Super Mix Kit (Thermo Fisher Scientific) with random primers at 65°C for 30 min to degenerate the RNA secondary structure. For miRNAs, cDNA was synthesized using the PrimeScript™ RT reagent Kit (Takara Biotech) with specific primer (mir-23-3p stem loop primers). The qPCR reaction was performed using SYBR Green Gene Expression Master Mix (Applied Biosystems, USA) on the StepOne Plus Real-Time PCR system (Applied Biosystems) following the manufacturer's protocols. Small nuclear RNA (U6) was used for normalization. Glyceraldehyde three-phosphate dehydrogenase (GAPDH) and small nuclear RNA (U6) acted as endogenous controls to normalize the relative gene abundance. qPCR experiments and data analysis were determined using the relative quantification 2-ΔΔCT method according to MIQE guidelines [35]. All of the primers for qPCR are shown in S3 Table.
Vector construction and cell transfection
For luciferase reporter assays, mir-23-3p target genes were predicted using in silico software (TargetScan and miRanda) and Malat1 sponge miRNA was obtained by RNA22 and StarBase. The DNA fragment containing the mir-23-3p MRE and MRE-Mut was amplified through PCR in vitro and the sequence was inserted downstream of the pmirGLO vector (Promega) of the renilla luciferase encoding gene between the restriction sites of Nhe I and Sal I. The cDNA generated from Raw264.7 as template to generate luciferase expression vector of pmirGLO-Lamp1-Wt/Mut or PmirGLO-Malat1-Wt/Mut recombinant plasmid. For autophagy flux monitor and Malat1 overexpression assays, the fusion gene of partial CMV-MCS-mRFP-EGFP-LC3 with double restriction sites, Not I and Sal I was designed, optimized in MCS, codon bias and sensitivity to acid, synthesized through PAS (Zoonbio Biology, China), and then inserted into the PCDH-CMV-MCS-EF1a-CopGFP (SBI) vector between the same restriction sites to construct autophagy flux lentivirus expression plasmid, named PCDH-Duo. Malat1 cDNA was amplified through PCR from PmirGLO-Malat1-Wt plasmids with EcoR I and BamH I restriction site, then sub-cloned into PCDH-Duo to generate PCDH-Duo-Malat1 lentivirus over-expression plasmid. All of the sequences, primers and PCDH-Duo plasmid map are shown in S6 Fig. Transfection of the Raw264.7 cells was performed according to instructions of the Lip3000 and P3000 Transfection Agent (Thermo Fisher Scientific). The mix of Malat1-siRNA and ASO was bought from Riboio (Sheng Zheng, China). Mir-23-3p mimics, inhibitor, and relevant NC or NC-FAM were bought from Gene Pharma (Shanghai, China). All of the vector primer and RNA oligo in this work are shown in S3 Table.
Lentivirus production
For Malat1 overexpression, the 600 bp fragment containing mir-23-3p MRE of Malat1 complementary DNA with EcoR I and BamH I restriction sites was sub-cloned into the PCDH-Duo lentivirus vector (Laboratory construction, information of vector was shown in S6 Fig) and co-transfected with plp1, plp2, and vsv.g into 293FT cells to generate PCDH-Duo-Malat1 lentivirus. PCDH-Duo empty lentivirus construct (Laboratory construction) was used as a negative control for Malat1 overexpression. Lentivirus production was conducted according to the manufacturer’s instructions of lip3000.
Dual luciferase report assays
The Raw264.7 cells were cultured in 96-well plates and co-transfected with 50 nM mimics, 100 nM inhibitor or relevant control mir-23-3p (Gene Pharma), and luciferase expression vector carrying wild-type or mutant 3’-UTR of Lamp1, wild-type or mutant Malat1 fragment containing MRE using Lipofectamine 3000 (Invitrogen). At 48 h after transfection, relative renilla luciferase activity was detected using the dual luciferase assay kit (E2910, Promega, Madison, WI) according to the manufacturer’s instructions, and the activity of firefly luciferase was the internal reference. The transfection was repeated in three independent experiments.
Immunofluorescence
After cell transfection for 6 h, the slides of Raw264.7 cells were washed with pre-warmed (37°C) 1× PBS three times, 5 min each. Then, the specimens were fixed in 4% paraformaldehyde (pH 7.5) at room temperature for 30 min. The cells were rinsed three times with 1× PBS for 10 min each. To each group of specimens, 0.2% Triton X-100 was added, followed by incubation at room temperature for 5 min to fully permeate the cell membrane. Then, the cells were rinsed again and blocked at room temperature for 30 min with 5% BSA. After overnight incubation at 4°C with primary LC3 antibody at a dilution of 1:100 (Proteintech), the cells were incubated with FITC-labeled goat anti-mouse secondary antibodies (Beyotime) at 37°C for 80 min after washing with PBS containing 0.05% Tween 20 (PBS-T). The cells were then washed again 5 times for 10 min with PBST, and the nuclei were counterstained with DEPI (Beyotime) for 5 min. Then, cells were visualized under a LSCM (Olympus FV-1000, Tokyo, Japan) using FV10-ASW software. At least triple independent experimental group were performed.
RNA-FISH
The Raw264.7 cells were cultured on slides placing in 24-well plates at appropriate density (~6 × 104/well) until 50% convergence reached. The cells were washed with pre-warmed (37°C) 1× PBS three times for 5 min each, fixed in 4% paraformaldehyde at room temperature for 10 min, rinsed again, and incubated with pre-chilled penetrant at 4°C for 5 min. The permeabilization solution was discarded and the cells were rinsed with PBS three times for 5 min each. Then, 200 μL prehybridization solution was added to each well and incubated at 37°C for 30 min, after which 2.5 μL of 20 μM Malat1 RNA-FISH probe mix (Riboio) or reference FISH probe Mix (18S/U6, Riboio) was added to 100 μL hybridization solution in each well and incubated overnight at 37°C. The cells were washed with Hybrid fluid I/II/III successively three times for 5 min each. The nuclei were counterstained with DAPI (Beyotime) for 5 min. Then cells were visualized under an LSCM (FV-1000; Olympus, Tokyo, Japan) using FV10-ASW software. At least triple independent experimental group were performed.
Western blot analysis and WES
Cells were harvested and lysed in RIPA buffer containing protease inhibitors, and the total cell collected. Proteins were separated by 7.5%, 10%, or 15% SDS-polyacrylamide gel electrophoresis and electro transferred to Hybond membranes (Amersham, Munich, Germany). BSA (5%) was used to block membranes for 2 h at room temperature. After blocking, primary antibodies targeting ATG5, P62 (SQSTM1), LAMP1 (1: 1000, Proteintech Group, USA), LC3 (1: 2000, CST, MA, USA), and β-actin (1:5000; Proteintech Group, Chicago, IL, USA) were incubated with the blot overnight at 4°C. The following day, secondary antibodies were added for 2 h at room temperature after the membrane was washed three times with TBST. Protein was visualized using an enhanced chemiluminescence system according to the manufacturer’s protocol (Santa Cruz Biotechnology, California, CA, USA). Furthermore, WES was used to detect LAMP1 expression in the rescue experiment according to the manufacturer protocols (SM-W004—Protein Simple, San Jose, CA) using a 12–230 kDa WES separation module coupled to a 25-capillary cartridge. The WES system is not a typical immunoblot method where qualitative images are quantified to generate data. Rather, chemiluminescence is used to detect and directly quantify the data.
Statistical analysis
Statistical analyses were performed using SPSS 19.0 (SPSS, Chicago, IL, USA) and the R platform. All of the experiments were repeated at least three times. Data are expressed as the mean ± standard deviation (SD). Two-tailed student’s t test or one-way analysis of variance was used to analyze the statistical difference between different treatments. P < 0.05 was considered statistically significant.
Results
Systematic identification of lncRNA in Raw264.7 cells
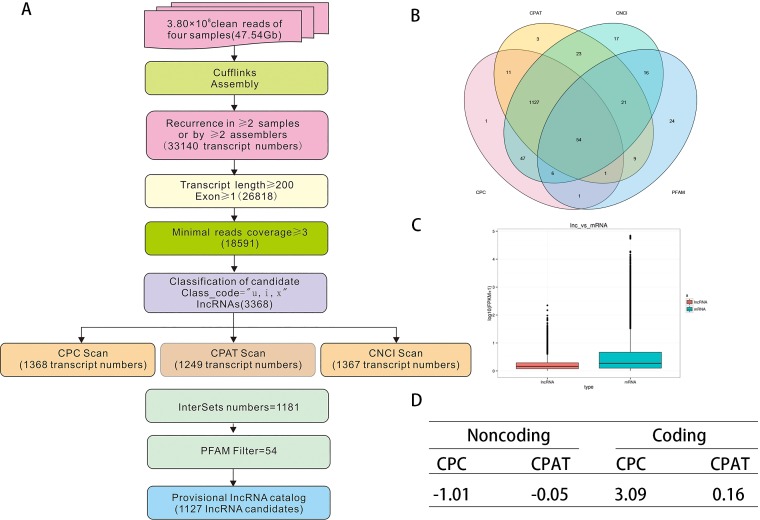
We performed RNA-seq to identify lncRNAs in Raw264.7 cells under different stressors including negative control (NC, dimethyl sulfoxide [DMSO]), 50 nM RAPA 12h, 3 mM 3-MA 6h and amino acid-deprived (starvation [36]) conditions 6h. After quality control (S1 Table), the reliable sequencing data as input for downstream identification. To effectively distinguish between protein-coding and non-coding sequences, coding potential filtering was performed according to an integrative techflow and computational pipeline (Fig 1A). In this way, 1112 lncRNAs were ultimately obtained (Fig 1B). The fragments per kilobase of transcript per million (FPKM) was used to represent the abundance of lncRNAs. In accordance with a previous report [37], we observed that the average lncRNA expression level under different conditions was lower than protein-coding gene expression (Fig 1C). Analysis using computational coding tools showed that noncoding sequences from the unknown transcript had an average CPC score of -1.01 and CPAT score of -0.05 compared with coding sequences, which had scores of 3.09 and 0.06, respectively (Fig 1D). All of the information on the lncRNAs identified by our pipeline is listed in S2 Table.
Fig 1. An integrative techflow and computational pipeline for the systematic identification of LncRNAs in Raw264.7.
(A) Informatics pipeline for the identification of LncRNAs in Raw264.7. (B) Venn chart showing the numbers of candidate LncRNAs filtered by the CNCI, CPC (Coding Potential Calculator), CPAT, and PFAM with the default parameters. (C) Box plots of log10 maximum expression values (FPKM+1) for protein-coding (Blue) and lncRNA (red) genes. Boxes represent first and third quartiles. Whiskers are 1.5-times the interquartile range. (D) Average score of Noncoding and Coding sequences calculated with CPC and CPAT.
Characterization of lncRNAs in Raw264.7 cells
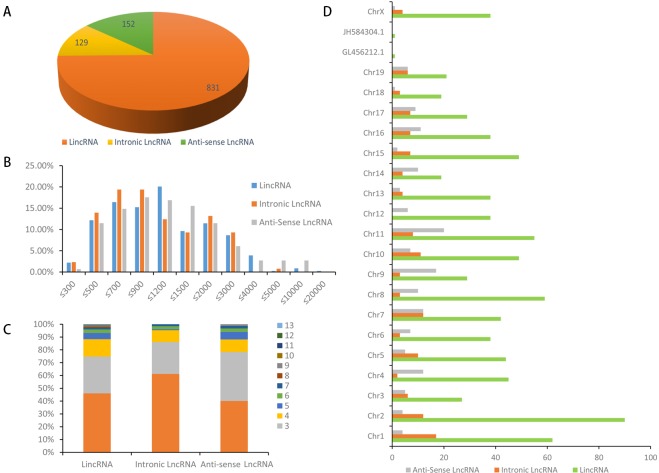
For the first time, the characteristics and transcription patterns of Raw264.7 lncRNAs were investigated in our study. A total of 1112 lncRNAs in Raw264.7 cells were identified, including 831 lincRNAs, 129 intronic lncRNAs, and 152 anti-sense lncRNAs (Fig 2A). Of these lncRNA, 240 were novel, including 158 lincRNAs, 60 intronic lncRNAs, and 22 anti-sense lncRNAs. LincRNAs comprised the major part of total Raw264.7 lncRNAs (75%). Full-length Raw264.7 lncRNA transcripts (median length: 953 nt) were longer than human lncRNA transcripts (median length: 592 nt). Interestingly, almost all of the lincRNA lengths were shorter than 4000 nt, and 90% of intronic RNAs were shorter than 3000 nt in Raw264.7 cells. However, less than 20% of Raw264.7 lincRNAs and anti-sense lncRNAs were shorter than 500 nt. Approximately 75% of the intronic lncRNAs ranged from 500 to 3000 nt, whereas only about 5% of lncRNAs were longer than 3000 nt (Fig 2B). The exon numbers in different types of lncRNAs of Raw264.7 were compared. LincRNA had 13 exons, intronic lncRNA had 7, and anti-sense lncRNA had 9. Among all of the lncRNAs, 45.97% of lincRNAs, 61.24% of intronic lncRNAs, and 40.13% of anti-sense lncRNAs had two exons. Exon numbers of 2–4 accounted for 90% of all of the exon numbers (average: 90.57%; Fig 2C). LncRNAs were evenly distributed in each chromosome, and lincRNAs did not show a significant chromosome location preference (Fig 2D). Properties, such as transcript abundance, lengths, exon number, and open reading frame (ORFs) of lncRNAs and mRNAs in Raw264.7, were also compared under the same conditions (S1 Fig).
Fig 2. Characteristics of Raw264.7 lncRNAs.
(A) Numbers of lincRNAs, intronic lncRNAs and antisense; ncRNAs in Raw264.7. (B) Transcript length distribution of lincRNAs, intronic lncRNAs and antisense lncRNAs. (C) The number of exons per transcript for LncRNAs. (D) Distribution of lincRNAs, intronic lncRNAs and anti-sense along each chromosome.
DEG profiling of lncRNAs in macrophages
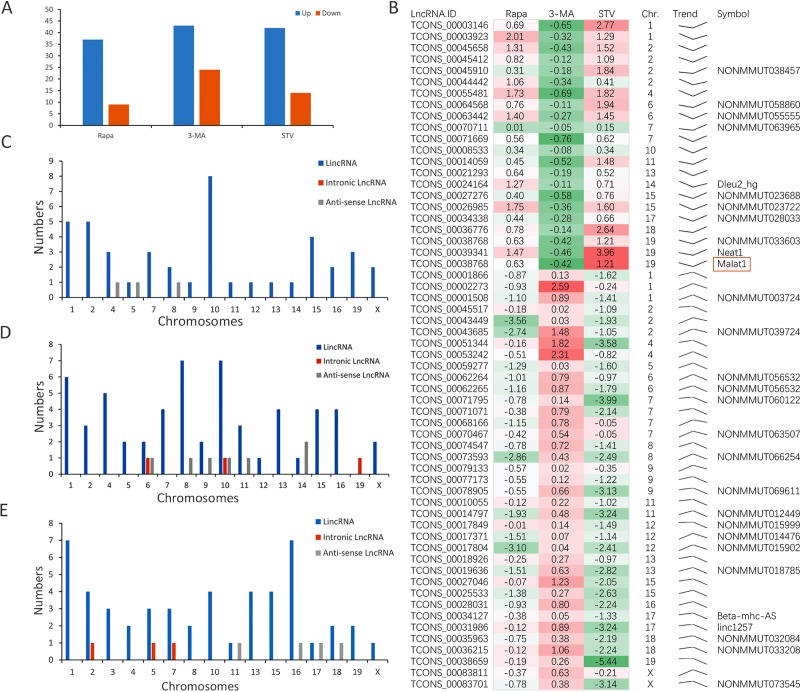
The lncRNA expression profiles under conditions of RAPA, 3-MA, and STV were compared. In order to improve the accuracy of the screening, based on an FPKM ≥ 0.5, DELs were identified by P ≤ 0.05 using Fisher’s exact test, FDR ≤ 0.05, and |log2FC| ≥ 1.5 in either of the two groups. Furthermore, compared with the NC group, 62 lncRNAs were differentially expressed in the RAPA group (32 upregulated and 30 downregulated), 65 lncRNAs were differentially expressed in the 3-MA group (43 upregulated and 24 downregulated), and 55 lncRNAs were differentially expressed in the STV group (43 upregulated and 14 downregulated). The number of lncRNAs that were only expressed in one group was 23 in the RAPA group, 22 in the 3-MA group, and 40 in the STV group. The number of lncRNAs expressed in each groups are shown in Fig 3A. In all 1112 lncRNAs, 240 were novel lncRNAs that were responsive to different conditions (S2 Table), including 158 lncRNAs (65.6%), 60 intronic lncRNAs (24.9%), and 22 anti-sense lncRNAs (9.5%); 12 were consecutively expressed in all three conditions (RAPA, 3-MA, STV). After strict expression filtering, 59 DELs were screened out (Fig 3B), including 11 novel and 48 known lncRNAs. Of the 59 DELs, we identified five known lncRNAs by a BLAST search of the lncRNA database and NONCODE database. In particular Malat1 (Neat2) had a huge FPKM (≥50) and at least a two FC in the RAPA and STV groups (Table 1), which most likely promotes autophagy. The clusters, chromosome distribution, and proportion of lncRNAs under different conditions were analyzed and the results shown there was no chromosome preference of lncRNAs under the different conditions (Fig 3C, Rapa; Fig 3D, 3-MA and Fig 3E, STV). More than 90% DELs in Raw264.7 cells were lincRNAs, whereas intronic and anti-sense lncRNA only accounted for a minor percentage (<10%). Hence, lincRNAs were more responsive during autophagy in our research model, while intronic and anti-sense lncRNAs were less affected.
Fig 3. LncRNA expression file.
(A) The numbers of DEL under each condition. (B) Two expression clusters containing 59 DELs. (C-E) Different type and chromosome distribution of DEL in each group (C: 50 nM Rapa, D: 3 mM 3-MA, E: STV).
Table 1. Known lncRNAs from the lncRNA and NONCODE databases.
| LncRNA.Transcript.ID | FPKM | Log2FC | LncRNADB | ||||||
|---|---|---|---|---|---|---|---|---|---|
| NC | Rapa | 3-MA | STV | Chr. | Rapa | 3-MA | STV | ||
| TCONS_00039341 | 1.1168 | 3.0858 | 0.8130 | 17.3318 | 19 | 1.47 | -0.46 | 3.96 | Neat1 |
| TCONS_00024164 | 0.7419 | 1.7895 | 0.6883 | 1.2134 | 14 | 1.27 | -0.11 | 0.71 | Dleu2_hg |
| TCONS_00038768 | 95.3854 | 147.3102 | 71.0835 | 220.7682 | 19 | 0.63 | -0.42 | 1.21 | Malat1(Neat2) |
| TCONS_00031986 | 0.6253 | 0.5744 | 1.1604 | 0.0663 | 17 | -0.12 | 0.89 | -3.24 | Linc1257 |
| TCONS_00034127 | 2.9721 | 2.2811 | 3.0819 | 1.1861 | 17 | -0.38 | 0.05 | -1.33 | Beta-MHC-AS |
| TCONS_00003923 | 6.5679 | 26.4309 | 5.2733 | 16.0668 | 1 | 2.01 | -0.32 | 1.29 | Gas5 |
Bold: FPMK ≥ 5 in four groups
LncRNA targets for pathway analysis
Cis and trans targets were used as input for pathway analysis. DAVID online software was used to analyze the statistical enrichment of DEL target genes in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Based on the results of significantly DEL analysis, different autophagy-responsive pathways were found under different conditions (S2 Table). In the RAPA group, there were four lncRNA targets involved in the phosphoinositide 3-kinase (PI3K)-Akt pathway (ENSMUSG00000004056, ENSMUSG00000021457, ENSMUSG00000023067, and ENSMUSG00000025856), two involved in the mammalian target of rapamycin (mTOR) pathway (ENSMUSG00000004056 and ENSMUSG00000028278), and five involved in the tuberculosis pathway (ENSMUSG00000004056, ENSMUSG00000007891, ENSMUSG00000013160, ENSMUSG00000018819, and ENSMUSG00000021457). In the 3-MA group, ENSMUSG00000033467 was involved in the Janus kinase-signal transducers and activators of transcription signaling pathway, and in the STV group, there were four targets (ENSMUSG00000020248, ENSMUSG00000021457, ENSMUSG00000039217, and ENSMUSG00000051439) involved in the tuberculosis pathway. Autophagy relative lncRNA-targets pair’s datasets shown in Table 2.
Table 2. Autophagy-related lncRNAs.
| Treat | Transcript.id | LncRNA. Symbol | NC. FPKM | Treat.FPKM | FDR | log2FC | Targets Gene.ID | Symbol |
|---|---|---|---|---|---|---|---|---|
| Rapa | TCONS_00023913 | - | 0.0000 | 1.1339 | 1.84297E-14 | 6.39 | ENSMUSG00000098557 | Kctd12 |
| TCONS_00082080 | NONMMUT073660 | 0.0000 | 1.9297 | 1.60726E-06 | 4.82 | ENSMUSG00000063663 | Brwd3 | |
| TCONS_00055298 | NONMMUT046615 | 0.5167 | 2.0777 | 0.007054927 | 1.96 | ENSMUSG00000028278 | Rragd | |
| TCONS_00053382 | NONMMUT048051 | 0.0000 | 1.5544 | 1.03152E-05 | 4.62 | ENSMUSG00000044303 | Cdkn2a | |
| TCONS_00004955 | NONMMUT000128 | 0.8223 | 6.9005 | 1.10592E-06 | 2.99 | ENSMUSG00000025917 | Cops5 | |
| TCONS_00060740 | NONMMUT054895 | 4.6242 | 0.0000 | 0 | -7.74 | ENSMUSG00000019054 | Fis1 | |
| TCONS_00038768 | NONMMUT033610 | 95.3854 | 147.3102 | 1.00592E-06 | 0.63 | ENSMUSG00000031447 | Lamp1 | |
| 3-MA | TCONS_00014193 | NONMMUT011771 | 0.7140 | 0.0000 | 0 | -7.05 | ENSMUSG00000018659 | Pnpo |
| TCONS_00008237 | NONMMUT008160 | 1.1633 | 0.0000 | 1.16417E-08 | -5.30 | ENSMUSG00000025364 | Pa2g4 | |
| TCONS_00008237 | NONMMUT008160 | 1.1633 | 0.0000 | 1.16417E-08 | -5.30 | ENSMUSG00000025373 | Rnf41 | |
| TCONS_00060740 | NONMMUT054895 | 4.6248 | 0.0000 | 0 | -8.26 | ENSMUSG00000019054 | Fis1 | |
| TCONS_00023913 | - | 0.0000 | 1.4805 | 0 | 7.27 | ENSMUSG00000098557 | Kctd12 | |
| TCONS_00060740 | NONMMUT054895 | 4.6248 | 0.0000 | 0 | -8.26 | ENSMUSG00000004846 | Plod3 | |
| TCONS_00064926 | NONMMUT056532 | 0.7829 | 0.1901 | 0.006511504 | -1.93 | ENSMUSG00000018659 | Pnpo | |
| TCONS_00023915 | - | 0.7297 | 2.5738 | 2.9502E-07 | 1.83 | ENSMUSG00000098557 | Kctd12 | |
| TCONS_00014137 | NONMMUT010720 | 0.3265 | 1.1308 | 7.68694E-05 | 1.78 | ENSMUSG00000000753 | Serpinf1 | |
| TCONS_00004516 | LincRNA-COX2 | 1.4150 | 0.3644 | 0.002440544 | -1.87 | ENSMUSG00000032487 | Ptgs2 | |
| TCONS_00008236 | NONMMUT008160 | 0.4915 | 1.9999 | 0.000182137 | 1.98 | ENSMUSG00000025373 | Rnf41 | |
| TCONS_00008236 | NONMMUT008160 | 0.4915 | 1.9999 | 0.000182137 | 1.98 | ENSMUSG00000025364 | Pa2g4 | |
| TCONS_00004955 | NONMMUT000128 | 0.8224 | 3.0498 | 0.003987109 | 1.84 | ENSMUSG00000025917 | Cops5 | |
| TCONS_00053382 | NONMMUT048051 | 0.0000 | 3.1468 | 9.10383E-14 | 6.10 | ENSMUSG00000044303 | Cdkn2a | |
| TCONS_00038768 | NONMMUT033610 | 95.3854 | 71.0835 | 0.000455226 | -0.52 | ENSMUSG00000031447 | Lamp1 | |
| STV | TCONS_00008830 | - | 0.0000 | 1.1312 | 4.02227E-05 | 4.46 | ENSMUSG00000075000 | Nrbf2 |
| TCONS_00015161 | NONMMUT008283 | 0.0000 | 0.7484 | 2.74567E-05 | 4.52 | ENSMUSG00000020448 | Rnf185 | |
| TCONS_00061211 | - | 0.0000 | 1.4425 | 5.94488E-05 | 4.40 | ENSMUSG00000029223 | Uchl1 | |
| TCONS_00061397 | NONMMUT054532 | 0.0000 | 1.6107 | 5.94488E-05 | 4.40 | ENSMUSG00000008348 | Ubc | |
| TCONS_00010935 | NONMMUT011771 | 0.9580 | 0.0000 | 0.000655226 | -4.56 | ENSMUSG00000018659 | Pnpo | |
| TCONS_00027620 | NONMMUT023686 | 0.5352 | 3.0793 | 0.003522683 | 2.36 | ENSMUSG00000022346 | Myc | |
| TCONS_00060740 | NONMMUT054895 | 4.6195 | 0.9621 | 0.009364357 | -2.32 | ENSMUSG00000004846 | Plod3 | |
| TCONS_00060740 | NONMMUT054895 | 4.6195 | 0.9621 | 0.009364357 | -2.32 | ENSMUSG00000019054 | Fis1 | |
| TCONS_00071969 | - | 9.5469 | 1.5670 | 0.004403232 | -2.60 | ENSMUSG00000066232 | Ipo7 | |
| TCONS_00004955 | NONMMUT000128 | 0.8214 | 10.9721 | 2.65116E-06 | 3.49 | ENSMUSG00000025917 | Cops5 | |
| TCONS_00030139 | NONMMUT029200 | 4.2132 | 32.8201 | 3.72584E-05 | 2.86 | ENSMUSG00000004069 | Dnaja3 | |
| TCONS_00038768 | NONMMUT033610 | 220.7682 | 1.13152E-05 | 1.21 | ENSMUSG00000031447 | Lamp1 |
-: indicates novel lncRNAs found in this research
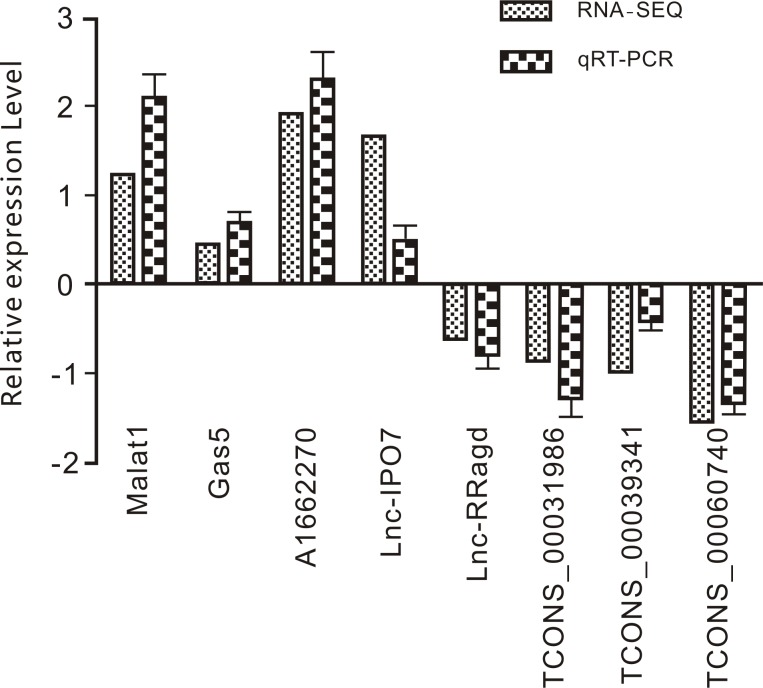
Validation of the transcription levels of eight lncRNAs
To confirm the differential expression of eight lncRNAs under Rapa condition that potentially function in autophagy, qPCR was performed (Fig 4). The lncRNAs were found to involved in autophagy pathways and were known lncRNAs from this work that included Malat1, Gas5, AI662270, Lnc-IPO7, Lnc-Rragd, TCONS-00031986, TCONS-00039341, and TCONS-00060740. Our qPCR results showed that the relative expression levels of the eight lncRNAs were consistent with those of RNA-seq.
Fig 4. RT-qPCR validation of eight LncRNA relative expression level upon 50 nM Rapa stimulation 2 hours in Raw264.7 cell lines (Mean±SD, n = 3).
Effects of Malat1 on autophagy in macrophages
Among the lncRNAs selected above, Malat1 has been the most extensively studied in many cancers and may have autophagy-promoting effects. Fluorescent in situ hybridization targeting ribonucleic acid molecules (RNA-FISH) showed that Malat1 was mainly distributed in the cytoplasm (Fig 5A). Autophagy flux check revealed that overexpression of Malat1 could promote autophagy (Fig 5B and 5E). Lentivirus-mediated Malat1 overexpression (Fig 5C) and small interfering RNA (siRNA) and antisense oligo (ASO)-mediated Malat1 knockdown (Fig 5D) were performed in Raw264.7 cells. Furthermore, western blot analysis confirmed that the overexpression of malat1 (pd-Malat1) could promote microtubule-associated protein light chain 3 (LC3) conversion and an increase in LC3 II expression. After the addition of choroquine diphosphate (CQ), the accumulation of LC3 in each group increased, but accumulation of the PD-malat1 group was the most obvious (Fig 5F and 5H). While knockdown of malat1 expression (Malat1-si), increased the expression of p62, the ratio of LC3 I/II decreased (Fig 5G and 5J). Malat1 functions as a CeRNA to regulate autophagy [38–40]. Furthermore, bioinformatics analysis of chromatin immunoprecipitation (Chip)-seq data suggested that human Malat1 could sponge many autophagy-related miRNAs, such as hsa-mir-23-3p, which had the same miRNA response elements (MREs) in mice, as determined by a BLAST search of the miRBase database of mature miRNA (S2 Fig).
Fig 5. Effects of Malat1 on autophagy in macrophages.
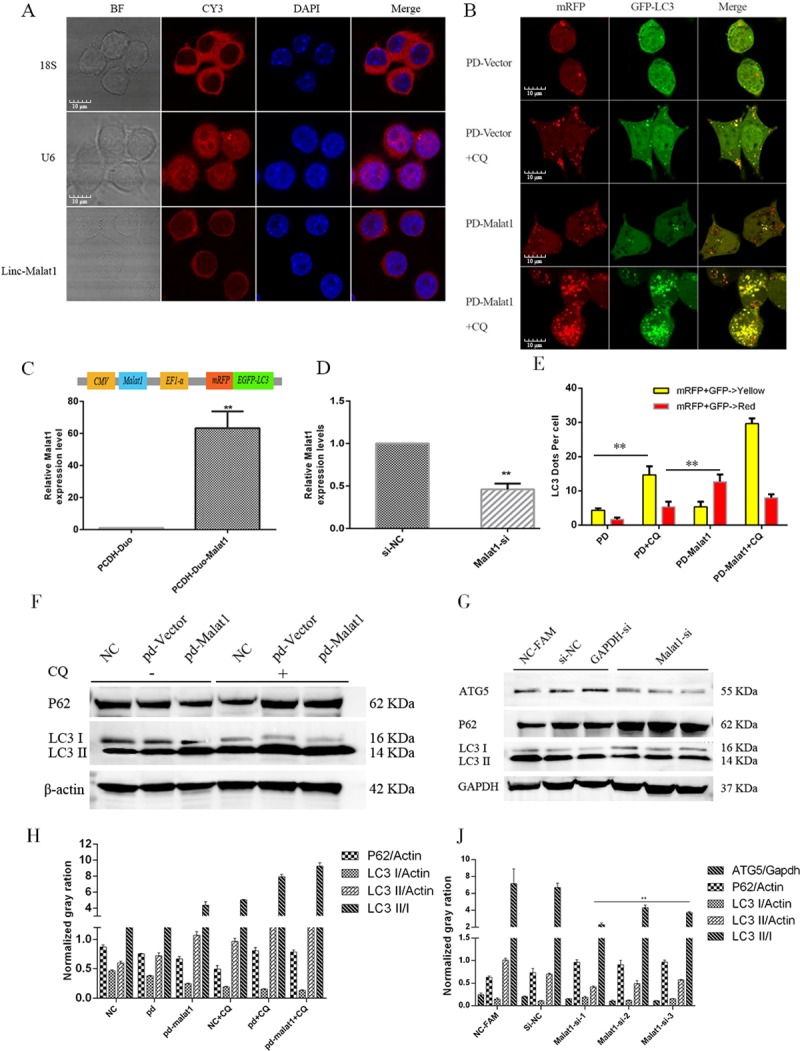
(A) RNA-FISH was performed to confirm sub cellular distribution of Malat1 in Raw264.7 cells (Scale bar = 10 μm, n = 3). (B) Confocal microscope was used to check the autophagy flux in different groups (Scale bar = 10 μm, n = 3). (C) The expression of Malat1 in Raw264.7 cells infected with PCDH-Duo-Malat1 (PD-Malat1) or vector (PCDH-Duo) Lentivirus was assessed by RT-qPCR (Mean±SD; n = 3). GAPDH was used as the endogenous control. (D) The expression of Malat1 was assessed by RT-qPCR in Raw264.7 cells transfected with Malat1-SiRNA and ASO mix (Mean±SD; n = 3). (E) Fluorescent LC3 dots per cell was quantified by Image J software, the data represent the Mean±SD of three independent experiments. (F and H) Autophagy marker P62 and LC3 were assessed by Western blot in PD-Vector or PD-Malat1 group. The expression of autophagy marker protein includes P62, LC3 was normalized against β-actin. (G and J) Autophagy marker P62, ATG5 and LC3 were assessed by Western blot in the Malat1-siRNA or Si-NC groups. The expression of autophagy marker protein includes ATG5, P62 and LC3 was normalized against GAPDH. *P<0.05 or ** P<0.01 in experiments vs. the corresponding control group.
Lamp1 is the direct target of mir-23-3p in macrophages
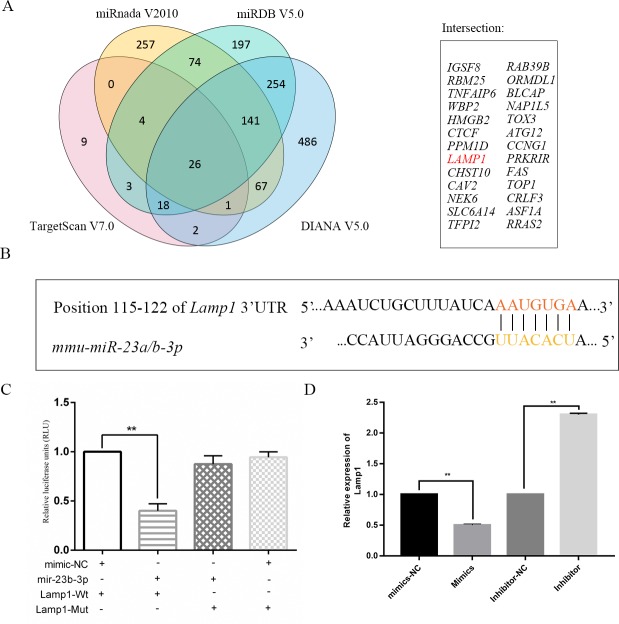
mir-23-3p was selected from the human Malat1 CeRNA network because it was confirmed to affect autophagy in previous reports. Four different miRNA TargetScan software programs with strict parameters were employed to determine the potential targets of mir-23-3p with high confidence, namely TargetScan V7.0 (Only top 100 conserved with a cumulative weighted context ++ score ≤ -0.4), Miranda v2010 (mirsvr_score ≤ -1.0), miRDB V5.0 (score ≥ 80), and DIANA V5.0 (miTG score ≥ 0.80). Consequently, we identified 26 mRNAs as potential targets of mir-23-3p (Fig 6A). The target of each miRNA was mapped to the protein-protein interaction (PPI) network. The results showed that mir-23-3p might be involved in the PI3K-Akt pathway, and co-expression of Lamp1 and Akt1 was found (S3 Fig). Lamp1 was chosen from all of the potential targets of mir-23-3p, depending on insilco analysis of candidate target genes and existing reports, as well as several lines of evidence supporting the fact that LAMP1 protein plays a key role in the fusion of autophagosome and lysosomes [41–43]. The mir-23-3p MRE was localized in 115–122 nt (Fig 6B) of Lamp1 3‘-UTR. Our dual luciferase reporter assay results suggested that mir-23-3p could directly target Lamp1 3‘-UTR (Fig 6C and S4 Fig). The mRNA expression of Lamp1 was detected by qPCR, and the results indicated that overexpression of mir-23-3p could significantly reduce the expression of Lamp1 in Raw264.7 cells. However, these effects were reversed in the inhibitor group (Fig 6D). The protein expression of LAMP1 in the cells treated with different concentrations of mir-23-3p mimics was detected by western blotting. The results showed that Lamp1 was significantly downregulated in a concentration-dependent manner (Fig 7C), and opposite results were obtained with mir-23-3p inhibitors (Fig 7E).
Fig 6. Lamp1 is a target of Mir-23b-3p in macrophage.
(A) Four different databases were used to cross-predict mir-23b-3p targets and 26 potential targets presented in Venn. (B) The bioinformatics prediction of mir-23a/b-3p MRE in Lamp1. (C) Luciferase reporter assay was performed in 293T cells co-transfected with pmirGLO-Lamp1-Wt or Mut and mimics or mimics-NC in Raw264.7 cells. (D) RT-qPCR was performed to examine the expression of Lamp1 in Raw264.7 cells transfected with mimics-NC, mimics, inhibitor-NC, or inhibitor.
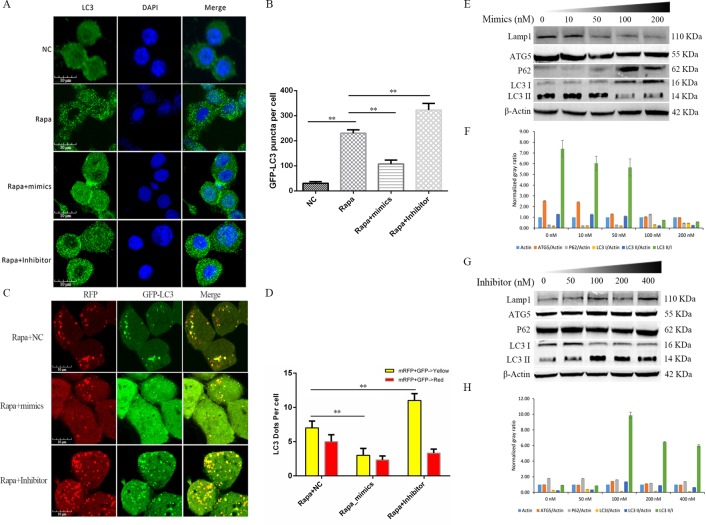
Fig 7. mir-23b-3p inhibits autophagy in macrophage.
(A) Immunofluorescence of endogenous LC3 was checked in Raw264.7 cells under different treatment conditions (Scale bar = 10 μm). (B) GFP-LC3 puncta per cell was quantified by Image J software, the data represent the Mean±SD of three independent experiments. (C) Autophagy flux analysis was conducted in Raw264.7 cells in Rapa+NC, Rapa+mimics, or Rapa + Inhibitor group to confirm mir-23-3p function (Scale bar = 10 μm). (D) Fluorescent LC3 dots per cell was quantified by Image J software, the data represent the Mean±SD of three independent experiments. (E-H) Western blot was used to detect the protein levels of Lamp1, ATG5, P62, LC3 I/II in Raw264.7 cells treated with concentration gradients of (E) mimics (0–200 nM) or (G) inhibitors (0–400 nM). Normalized gray ratio was also compared, the expression of protein was normalized against β-actin (Mean±SD; n = 3).
Mir-23-3p inhibits autophagy in macrophages
The regulatory function of mir-23-3p in autophagy has been reported, but whether it takes part in the autophagy of macrophage cells is still unknown. We first analyzed the expression of the endogenous autophagy marker LC3B in macrophages by immunofluorescence. when mir-23-3p was over-expressed (mimics) in Rapa group in macrophages could down-regulation expression of LC3B and reduced LC3B puncta. Furthermore, LC3B puncta accumulates in the RAPA group treated with mir-23b-23 inhibitor compared to the RAPA group (Fig 7A), which was verified by autophagy flux analysis. Moreover, autophagy flux results showed that mir-23-3p might regulate late autophagy due to enhanced co-localization of membrane bound red fluorescence protein (mRFP) and enhanced green fluorescent protein (EGFP) signals (Fig 7B). western blot analysis of macrophages treated with different concentrations of mir-23-3p mimics (Fig 7C and 7D) and inhibitors (Fig 7E and 7F) also showed that the endogenous autophagy markers, including autophagy related 5 (ATG5), p62, and the ratio of LC3 II/I, had a corresponding trend. These results demonstrate that mir-23-3p suppresses autophagy in macrophages.
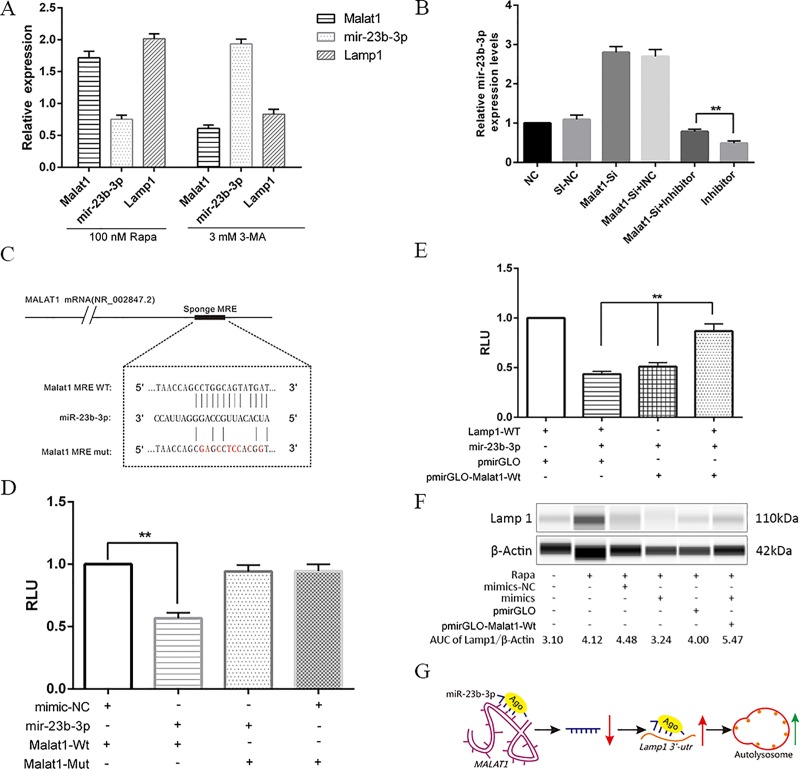
Malat1 functions as a decoy of mir-23-3p to regulate Lamp1 expression in macrophages
LncRNA Malat1 has been widely reported to regulate gene expression through the sponge mechanism. Lamp1 mediates the fusion of autophagosome with lysosomes, and is closely related to autophagy and infection immunity. Malat1 LncRNA CeRNA network analysis and mir-23-3p target analysis showed that LncRNA Malat1 may inhibit the autophagy of macrophages by release the inhibition of mir-23-3p on Lamp1 through the CeRNA mechanism. Therefore, whether malat1 promotes macrophage autophagy through CeRNA mechanism is worthy of further study. qPCR was used to analyze the differential expression of Lamp1, mir-23-3p, and Malat1 with treatment of 100 nM RAPA or 3 mM 3-MA, and the results showed that Lamp1 had the same expression trend as Malat1 (Fig 8A). Expression of mir-23-3p in the control, Malat1-si, Malat1-si+Inhibitor normal control(INC), Malat1-si+inhibitor, or inhibitor groups was detected by qPCR, and the results suggested that Malat1-siRNA had promoted the upregulation of Lamp1 (Fig 8B). To investigate whether the expression of mir-23-3p was regulated by lncRNA, Diana Tools (http://diana.imis.athena-innovation.gr/), RNA22, and Segal Lab (http://genie.weizmann.ac.il/) were jointly used to predict the lncRNA that might target mir-23-3p. Bioinformatics analysis revealed that Malat1 lncRNA contained a putative 13-mer-binding motif within mir-23-3p (Fig 8C). Malat1 was cloned downstream of the luciferase gene. The construct was named PmirGLO-Malat1-WT/Mut, and it was transfected with mir-23-3p mimics, inhibitor, or corresponding NC. A dual luciferase assay was conducted, and the results indicated that the luciferase activity reduced by ~40% when co-transfected with Malat1-Wt and mimics compared with the mimic-NC; however, suppression of luciferase activity was completely abolished in transfected luciferase reporter plasmid of PmirGLO-Malat1-Mut than in wild-type vector of pmirGLO (Fig 8D). Moreover, as expected, the rescue experiments showed that Malat1 promoted luciferase expression in a mir-23-3p-dependent manner upon transient transfection with the pmirGLO-Malat1-Wt plasmid into Raw264.7 cells (Fig 8E). We further used Simple Western technology (Protein Simple, USA) to confirm that the interaction between Malat1 and mir-23-3p could regulate the protein expression of LAMP1. The results showed that transient transfection with pmirGLO-Malat1-Wt plasmids significantly elevated Lamp1 expression compared with pmirGLO empty vector (Fig 8F). Interestingly, while knockdown of malat1 expression (si-Malat1) compared to the si-NC group downregulated the expression of Lamp1, suppression of mir-23-3p by its inhibitor increased lamp1 expression compared to the si-Malat1 with inhibitor NC group (S5 Fig). In short, autophagy stress can change the lncRNA profiles, including Malat1, which serves as a CeRNA to regulate Lamp1 expression by sponging mir-23-3p in macrophages (Fig 8G).
Fig 8. Malat1 functions as CeRNA to regulate Lamp1 expression by sponging mir-23-3p in macrophage cells.
(A) RT-qPCR was performed to evaluate the expression levels of Malat1, mir-23b-3p, and Lamp1 in Raw264.7 cells respective treatment with 50 nM Rapa 2h or 3 mM 3-MA 12h (Mean±SD; n = 3). (B) RT-qPCR was used to test mir-23b-3p expression in the different groups, including NC, Si-NC, Malat1-siRNA, Malat1-siRNA+inhibitor-NC (INC), Malat1-siRNA+inhibitor, or inhibitor (Mean±SD; n = 3). (C) The bioinformatics predicted MRE of mir-23b-3p on Malat1. (D) Luciferase reporter assay was performed in 293T cells co-transfected with pmirGLO-Malat1-Wt or Mut and mimics or mimics-NC (Mean±SD; n = 3). (E) Luciferase reporter rescue experiment was conducted to confirm co-transfected pmirGLO-Malat1-Wt, pmirGLO-Lamp1-Wt and mir-23b-3p mimics effect the relative luciferase activity in 293T cells (Mean±SD; n = 3). (F) WES was used to confirm over-expression Malat1 can promote Lamp1 expression in Raw264.7 cells. (G) Graphic summary of Malat1 function in this work.
Discussion
It has been found that 80% of ncRNAs are lncRNAs, a class of transcripts involved in gene expression and regulatory functions [44]. Unlike miRNA, which has been broadly investigated in autophagy [45], the functions of most lncRNAs and their potential roles in autophagy are still unclear. High-throughput sequencing provides a convenient and reliable approach for researching ncRNAs at the transcription level [46,47]. With the development of sequencing technology and bioinformatics, the functions of lncRNAs in many cells and species have been described in previous studies [48–51]. However, few studies have focused on lncRNAs and their functions in the autophagy of macrophages, which plays a key role in the immune system.
Cell autophagy is precisely controlled and is highly regulated by multiple signaling pathways, orchestrated by more than 30 autophagy-related proteins organized in several functional units and many other proteins that regulate ncRNAs, including miRNAs and lncRNAs. New evidence has indicated that lncRNAs are emerging as critical regulators of autophagy and the immune process at the epigenetic and transcriptional levels in mammals in response to different stressors. Macrophages are linchpins of innate immunity, responding to invading microorganisms by initiating coordinated inflammatory and antimicrobial programs. Autophagy is a powerful weapon that macrophage cells use to defend against pathogenic microorganism infection, but its regulation in macrophages by lncRNAs is poorly defined. Here, through transcriptomic, bioinformatics, and functional studies, we focused on the specific DEL malat1. Systematic information about this lncRNA was acquired and the interaction between Malat1-mir-23-3p-Lamp1 was found, suggesting that through the competitive adsorption of mir-23- 3p, Malat1 releases inhibition of Lamp1 and promotes autophagy in macrophages. These data provide a better understanding of lncRNA function in macrophages as well as a basis for further investigation into the roles and mechanisms of lncRNAs, especially Lnc-Malat1, in the pathogenesis of macrophages.
There have been few studies on the regulation of macrophage autophagy by lncRNAs. Mao et al. [52] suggested that lncRNAs may be important regulators of the lipolysaccharide-induced innate immune response in bone marrow-derived macrophages. For example, downregulation of maternally expressed 3 lncRNA eliminates mycobacteria in macrophages via autophagy [53]. Some studies have used microarrays to demonstrate that there is substantial abnormal lincRNA and mRNA expression profiles in response to pathogenic microorganisms in monocyte-derived human macrophages infected with human immunodeficiency virus or Mycobacterium tuberculosis [54]. LncRNAs are known to be involved in regulating autophagy, but sufficient data are lacking regarding their expression profile and role in autophagy in macrophages; thus, additional studies are needed to identify and clarify their roles and potential mechanisms.
In this study, RNA-Seq data were used to identify all of the potential transcripts, followed by computational analysis to determine putative lncRNAs under different autophagy stressors in Raw264.7 cells. A total of 94761 transcripts were reconstructed from the data. In comparison with 27099 transcripts (based on ESEMBL GTF file for Mouse), the number of identified transcripts, by assembled transcriptome, increased two-fold (Log2FC ≥ 2,94761) due to both novel isoforms of known genes as well as new genes. In total, 26282 of 94761 transcripts were predicted to be unknown transcripts and analyzed to identify putative lncRNA using credible computational methods. Finally, we obtained 1112 putative lncRNAs, including 831 large intergenic, 129 intronic, and 152 anti-sense lncRNA, in which 59 differentially expressed transcripts exhibited more than a 1.5-FC under different conditions. A total of 240 novel lncRNA sequences were found in macrophages in this work, and 8 were confirmed by qPCR in Rapa conditions.
The differential expression of lncRNA in cells can be induced by pathogens and may regulate the host response to pathogens [55]. A growing body of literature has reported the specific involvement of lncRNAs in the host cell response towards bacterial infections [56]. Viruses can hijack a host lncRNA to replicate [57]. Increasing evidence suggests that lncRNAs may regulate toll-like receptor signaling and innate immunity in APCs including macrophages [58]. However, the lncRNA profile in macrophages is still large unknown. In this study, we identified 271 novel lncRNAs in a macrophage autophagy model. Among them, there were 67 DELs in the RAPA treatment group, 37 of which were upregulated and 9 that were downregulated. Analysis of the lncRNA cis and trans target pathways showed that DELs acted as key regulators of pathways, such as the PI3K-Akt, mTOR and HIF-1α pathway. We also acquired a series of autophagy-related lncRNA-target interaction pairs for further studies.
One of the eight confirmed DELs, a newly identified functional lincRNA named Malat1, also known as a Neat2, has been recently studied in many fields. LncRNA Malat1 is a large, infrequently spliced ncRNA that is highly conserved in mammalian evolution and widely expressed in many tissues among primates, with a homology of up to 90% between human and Mus musculus at the 3' end of the nucleotide sequence of 5 kb, suggesting that Malat1 plays an important role in the evolution process. The full sequence of Malat1 has a 4 highly homologous region in human and Mus musculus, suggesting that these regions are most likely functional regions of Malat1[59,60]. LncRNA Malat1 plays a role in multiple physiological processes, such as alternative splicing, nuclear organization, and epigenetic modulation of gene expression, and it is also closely related to various pathological processes ranging from diabetes complications to cancer. It has been reported that Malat1 can directly interact with a variety of important transcription factors to regulate gene expression, including SRSF1-3, JUN, DND1, BAF57, HuR and YAP [60]. At the same time, it was also reported that LncRNA Malat1 expression is regulated by Ago2, but the detailed mechanism of action is unknown. Recently, LncRNA Malat1 also positively regulates cell motility via the transcriptional and/or post-transcriptional regulation of motility-related genes [61], and modulates many genes through the sponge mechanism via different miRNAs in cancer, indicating its ability to regulate gene expression in cytoplasm [62,63]. Malat1 has been extensively studied in pan-cancer and is reportedly an anti-oncogene, but its autophagy function in macrophage has rarely been reported.
Recently, existing research has specified that Malat1 regulates autophagy through the sponge of mir-23-3p, which targets HMGB2 and ATG12 in gastric cancer [64]. In these studies, Malat1 promoted autophagy in Raw264.7 cells. Malat1 responded to RAPA stimulation, and was involved in the PI3K-Akt pathway in autophagy in macrophages. Interestingly, our unpublished RNA-seq data also showed that Malat1 was significantly upregulated in macrophages infected with BacillusCalmette-Guerin (BCG) at 12 h compared to NC (data not published). To summarize, the information suggests that Malat1 may play a crucial role in macrophages. Overexpression of Mala1 promoted macrophage autophagy protein marker expression and autophagy flux, but blocked Malat1 inhibition of autophagy. These results suggest that Malat1 promotes autophagy in macrophages.
Until recently, supported lncRNAs could modulate broadly biological functions through a variety of interactions between RNA-protein, RNA-DNA, or RNA-RNA. Among them, CeRNA mechanisms were the most frequently reported. LncRNAs and miRNAs and protein complexes can associate with each other and regulate gene expression through post-transcriptional regulation. miRNAs and lncRNAs mediate dynamic regulation of gene expression, and the ternary regulatory relationship of miRNA-lncRNA-mRNA expands the central principle of genetic information and is increasingly being studied by researchers. Malat1 functions as a sponge to regulate gene expression in many studies; however, whether malat1 functions as a CeRNA and its new sponge miRNA and new targets are important research questions in macrophage.
In exploring the potential mechanism of Malat1, human ChIP-seq data from SatrBase V4 and autophagy background genes were used to construct the Malat1 autophagy CeRNA network at first. As a result, we have obtained a better understanding of the possible functioning mode of Malat1 in autophagy process. Malat1-miRNA was picked out from human autophagy CeRNA network to further analysis in study. Next, RNA22 software was used to explore its interaction in mouse Malat1 sequence.
We found that Malat1 may be as decoy of mmu-mir-23a/b-3p, and interaction between malat1 and mir-23-3p has been confirmed in gastric cancers cells through RIP experiments [39,64]. Collectively, our work focuses on discovering new targets of mir-23-3p could involve in the autophagy process in macrophages. Bioinformatics analysis was performed to explore the potential targets of mir-23-3p. The related miRNAs of mir-23-3p were predicted using miRanda, TargetScan, miRDB, and DIANA databases. The minimum free energy, seed region, and miRNA recognition elements of miRNA-mRNA duplexes were calculated to choose superior candidates [40]. We found 26 mRNA may be the targets of mir-23-3p, including Lamp1, ATG12, HMGB2, ORMDL1, and CTCF. ATG12 and HMGB2 have already been reported to be involved in autophagy. We further searched for targets of mir-23-3p through document mining, and a related PPI network was constructed by STRING (https://string-db.org/cgi/input.pl). The results showed that mir-23-3p may be involved in the PI3K-Akt pathway. Lamp1 was selected as a superior candidate according to gene function and human Malat1 CeRNA network. The dual luciferase reporter assay and western blotting were used to further verify the interaction between mir-23-3p and Lamp1. The results showed that mir-23-3p mimics significantly reduced the luciferase activity and expression of Lamp1 to different levels compared with the mimic-NC group. After confirming the interaction, we found that mir-23-3p could alternate autophagy level in macrophages. With the results of immunofluorescence, autophagy flux, and western blotting, we proved mir-23-3p inhibitor autophagy through targets Lamp1 in macrophage. Ultimately, the expression of Malat1, Lamp1, and mir-23-3p was examined through qPCR, and we found that Malat1 had the same expression trend as Lamp1 but the opposite expression trend as mir-23-3p. Based on the dual luciferase reporter assay and rescue experiment, the interaction between Malat1 and mir-23-3p was confirmed, and overexpressed malat1 promoted Lamp1 expression through WES. malat1-mir-23-3p-Lamp1 regulation axis in autophagy in macrophage was confirmed. We found malat1 responds to different autophagy stress and exhibited the capacity of sponge mir-23-3p, which can regulate a new crucial autophagy relative target Lamp1, and it took part in autophagy lysosome formation. Combined with literature analysis and pathway analysis of mir-23-3p, shown it may have the crucial role to regulate autophagy in macrophage through broadly affects some key genes, including Lamp1, ATG12, HMGB2, and AKT1/2. As far as we know, there is no report about the sponge regulation between Malat1, mir-23-3p, and Lamp1 in autophagy in macrophage currently.
The phosphatidylinositol 3-kinase/AKT/mammalian target of rapamycin (PI3K/AKT/mTOR) axis is a key pathway implicated in regulating autophagy. Studies have indicated that Malat1 can regulate autophagy through the PI3K/Akt pathway. Li X et al [65] reported that Malat1 knockdown markedly suppressed autophagy by inhibited PI3K, Akt and p70S6K phosphorylation in HUVECs cell lines. But the mechanism of 3-MA and Malat1′s regulation of PI3K/AKT pathway on the autophagy process in Raw264.7 was uncertain.
3-Methyladenine (3-MA) blocks autophagy by inhibiting PI3K, and PI3K activity is essential for nucleation and assembly of early membrane pools in autophagosome formation. Malat1 ceRNA network analysis indicated that Malat1 can bind to mir-23-3p and participate in PI3K-AKT signaling pathway to regulate macrophage autophagy. The detection of 3-MA at mRNA level inhibited the expression of Malat1, which may be due to the action of 3-MA on the early stage of autophagy, and the negative feedback mechanism after the inhibition of PI3K-AKT signaling pathway reduced the expression of Malat1, increasing the expression of mir-23-3p further inhibits the PI3K-AKT signaling pathway, but this requires further experimental evidence.
In brief, we obtained a detailed lncRNA profile and autophagy-related lncRNA-mRNA pairs with different autophagy stressors in macrophages. Our results strongly suggest that Malat1 functions as a CeRNA to regulate Lamp1 expression by sponging mir-23-3p in macrophages. In this Malat1-mir-23-3p-Lamp1 axis, Malat1 releases inhibition of Lamp1 and may promote autophagy in macrophages. These results provide a basis for further investigation of the roles and mechanisms of lncRNA, especially Malat1, in the pathogenesis of macrophages and the immune system.
Supporting information
(DOCX)
(XLSX)
(DOCX)
LncRNA and mRNA compare in RNA length (A), exon number (B) and ORF length (C).
(TIF)
(JPG)
(TIF)
(TIF)
(JPG)
(A) The Map of Double fluorescence labeling lentivirus autophagy flux detection vector and (B) Homology modeling of expression product.
(TIF)
Acknowledgments
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Abbreviations
- LncRNA
long noncoding RNA
- 3′-UTR
3′-untranslated region
- ASO
Antisense oligonucleotide
- Luc
luciferase
- RLU
Relative luciferase unit
- 3-MA
3-Methyladenine
- RAPA
Rapamycin
- STV
starvation
- DEG
differentially expressed genes
- DEL
differentially expressed LncRNAs
- LC3
Microtubule-associated protein light chain 3
Data Availability
All clean reads of this work are available from the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) database (PRJNA544884).
Funding Statement
This work was supported by grants from the National Natural Science Foundation of China (81860355), 2015 Ningxia Scientific and technological innovation leader training project (KJT2015020), First-class Discipline construction of clinical medicine of Ningxia Medical University (NXYLXUK2017A05) and Natural Science Foundation of Ningxia Province of China (2018AAC03088).
References
- 1.Fang S, Zhang L, Guo J, Niu Y, Wu Y, Li H, et al. NONCODEV5: a comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res.2018; 46(D1):D308–d314. 10.1093/nar/gkx1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dykes I, Emanueli C Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genomics Proteomics Bioinformatics.2017. 10.1016/j.gpb.2016.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Backofen R, Gorodkin J, Hofacker IL, Stadler PF Comparative RNA Genomics. Methods Mol Biol.2018; 1704:363–400. 10.1007/978-1-4939-7463-4_14 [DOI] [PubMed] [Google Scholar]
- 4.Adelman K, Egan E Non-coding RNA: More uses for genomic junk. Nature.2017; 543(7644):183–185. 10.1038/543183a [DOI] [PubMed] [Google Scholar]
- 5.Elkon R, Agami R Characterization of noncoding regulatory DNA in the human genome. Nat Biotechnol.2017; 35(8):732–746. 10.1038/nbt.3863 [DOI] [PubMed] [Google Scholar]
- 6.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet.2015; 47(3):199–208. 10.1038/ng.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmitt AM, Garcia JT, Hung T, Flynn RA, Shen Y, Qu K, et al. An inducible long noncoding RNA amplifies DNA damage signaling. Nat Genet.2016; 48(11):1370–1376. 10.1038/ng.3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mondal T, Subhash S, Vaid R, Enroth S, Uday S, Reinius B, et al. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures. Nat Commun.2015; 6:7743 10.1038/ncomms8743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L, Yang Z, Trottier J, Barbier O, Wang L Long noncoding RNA MEG3 induces cholestatic liver injury by interaction with PTBP1 to facilitate shp mRNA decay. Hepatology.2017; 65(2):604–615. 10.1002/hep.28882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell.2013; 52(1):101–112. 10.1016/j.molcel.2013.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du Z, Sun T, Hacisuleyman E, Fei T, Wang X, Brown M, et al. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat Commun.2016; 7:10982 10.1038/ncomms10982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaiti F, Fernandez-Valverde SL, Nakanishi N, Calcino AD, Yanai I, Tanurdzic M, et al. Dynamic and Widespread lncRNA Expression in a Sponge and the Origin of Animal Complexity. Mol Biol Evol.2015; 32(9):2367–2382. 10.1093/molbev/msv117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu X, Zhang Y, Li T, Ma Z, Jia H, Chen Q, et al. Long non-coding RNA Linc-RAM enhances myogenic differentiation by interacting with MyoD. Nat Commun.2017; 8:14016 10.1038/ncomms14016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernardes de Jesus B, Marinho SP, Barros S, Sousa-Franco A, Alves-Vale C, Carvalho T, et al. Silencing of the lncRNA Zeb2-NAT facilitates reprogramming of aged fibroblasts and safeguards stem cell pluripotency. Nat Commun.2018; 9(1):94 10.1038/s41467-017-01921-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Z, Carter AC, Chang HY Mechanistic insights in X-chromosome inactivation. Philos Trans R Soc Lond, B, Biol Sci.2017; 372(1733). 10.1098/rstb.2016.0356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noack F, Calegari F Epitranscriptomics: A New Regulatory Mechanism of Brain Development and Function. Front Neurosci.2018; 12:85 10.3389/fnins.2018.00085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmitt AM, Chang HY Long Noncoding RNAs in Cancer Pathways. Cancer Cell.2016; 29(4):452–463. 10.1016/j.ccell.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kugelberg E Antiviral immunity: Linking lncRNA to IFN regulation. Nat Rev Immunol.2014; 14(12):779 [Google Scholar]
- 19.Atianand MK, Cafferey DR, Fitzgerald KA Immunobiology of Long Noncoding RNAs. Annu Rev Immunol.2017. 10.1146/annurev-immunol-041015-055459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Highlighting advances in autophagy. Nat Cell Biol.2018; 20(3):223 10.1038/s41556-018-0061-z [DOI] [PubMed] [Google Scholar]
- 21.Kuballa P, Nolte WM, Castoreno AB, Xavier RJ Autophagy and the immune system. Annu Rev Immunol.2012; 30(611–646. 10.1146/annurev-immunol-020711-074948 [DOI] [PubMed] [Google Scholar]
- 22.Chen Z, Wang T, Liu Z, Zhang G, Wang J, Feng S, et al. Inhibition of Autophagy by MiR-30A Induced by Mycobacteria tuberculosis as a Possible Mechanism of Immune Escape in Human Macrophages. Jpn J Infect Dis.2015; 68(5):420–424. 10.7883/yoken.JJID.2014.466 [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Wang P, Wan L, Xu S, Pang D The emergence of noncoding RNAs as Heracles in autophagy. Autophagy.2017; 13(6):1004–1024. 10.1080/15548627.2017.1312041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frankel LB, Lubas M, Lund AH Emerging connections between RNA and autophagy. Autophagy.2017; 13(1):3–23. 10.1080/15548627.2016.1222992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang L, Wang H, Shen Q, Feng L, Jin H Long non-coding RNAs involved in autophagy regulation. Cell Death Dis.2017; 8(10):e3073 10.1038/cddis.2017.464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomson DW, Dinger ME Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet.2016; 17(5):272–283. 10.1038/nrg.2016.20 [DOI] [PubMed] [Google Scholar]
- 27.Tay Y, Rinn J, Pandolfi PP The multilayered complexity of ceRNA crosstalk and competition. Nature.2014; 505(7483):344–352. 10.1038/nature12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang K, Liu CY, Zhou LY, Wang JX, Wang M, Zhao B, et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat Commun.2015; 6:6779 10.1038/ncomms7779 [DOI] [PubMed] [Google Scholar]
- 29.Li L, Chen H, Gao Y, Wang YW, Zhang GQ, Pan SH, et al. Long noncoding RNA MALAT1 promotes aggressive pancreatic cancer proliferation and metastasis via the stimulation of autophagy. Mol Cancer Ther.2016. 10.1158/1535-7163.MCT-16-0008 [DOI] [PubMed] [Google Scholar]
- 30.Li Z, Li J, Tang N Long noncoding RNA Malat1 is a potent autophagy inducer protecting brain microvascular endothelial cells against oxygen-glucose deprivation/reoxygenation-induced injury by sponging miR-26b and upregulating ULK2 expression. Neuroscience.2017; 354:1–10. 10.1016/j.neuroscience.2017.04.017 [DOI] [PubMed] [Google Scholar]
- 31.Fu Z, Luo W, Wang J, Peng T, Sun G, Shi J, et al. Malat1 activates autophagy and promotes cell proliferation by sponging miR-101 and upregulating STMN1, RAB5A and ATG4D expression in glioma. Biochem Biophys Res Commun.2017; 492(3):480–486. 10.1016/j.bbrc.2017.08.070 [DOI] [PubMed] [Google Scholar]
- 32.Cui H, Banerjee S, Guo S, Xie N, Ge J, Jiang D, et al. Long noncoding RNA Malat1 regulates differential activation of macrophage and response to lung injury. JCI Insight.2019. 10.1172/jci.insight.124522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo H, Bu D, Sun L, Fang S, Liu Z, Zhao Y Identification and function annotation of long intervening noncoding RNAs. Brief Bioinform.2017; 18(5):789–797. 10.1093/bib/bbw046 [DOI] [PubMed] [Google Scholar]
- 34.Li J, Ma W, Zeng P, Wang J, Geng B, Yang J, et al. LncTar: a tool for predicting the RNA targets of long noncoding RNAs. Brief Bioinform.2015; 16(5):806–812. 10.1093/bib/bbu048 [DOI] [PubMed] [Google Scholar]
- 35.Bustin SA, Wittwer CT MIQE: A Step Toward More Robust and Reproducible Quantitative PCR. Clin Chem.2017; 63(9):1537–1538. 10.1373/clinchem.2016.268953 [DOI] [PubMed] [Google Scholar]
- 36.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy.2016; 12(1):1–222. PMC4835977 10.1080/15548627.2015.1100356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Storey JD, Tibshirani R Statistical significance for genomewide studies. Proc Natl Acad Sci U S A.2003; 100(16):9440–9445. 10.1073/pnas.1530509100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.YiRen H, YingCong Y, Sunwu Y, Keqin L, Xiaochun T, Senrui C, et al. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via mir-23-3p sequestration in gastric cancer. Mol Cancer.2017; 16(1):174 10.1186/s12943-017-0743-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang J, Yang Y, Fang F, Liu K MALAT1 modulates the autophagy of retinoblastoma cell through miR-124-mediated stx17 regulation. J Cell Biochem.2018; 119(5):3853–3863. 10.1002/jcb.26464 [DOI] [PubMed] [Google Scholar]
- 40.Guo D, Ma J, Yan L, Li T, Li Z, Han X, et al. Down-Regulation of Lncrna MALAT1 Attenuates Neuronal Cell Death Through Suppressing Beclin1-Dependent Autophagy by Regulating Mir-30a in Cerebral Ischemic Stroke. Cell Physiol Biochem.2017; 43(1):182–194. 10.1159/000480337 [DOI] [PubMed] [Google Scholar]
- 41.Alessandrini F, Pezzè L, Ciribilli Y LAMPs: Shedding light on cancer biology. Semin Oncol.2017; 44(4):239–253. 10.1053/j.seminoncol.2017.10.013 [DOI] [PubMed] [Google Scholar]
- 42.Cohen-Dvashi H, Israeli H, Shani O, Katz A, Diskin R Role of LAMP1 Binding and pH Sensing by the Spike Complex of Lassa Virus. J Virol.2016; 90(22):10329–10338. 10.1128/JVI.01624-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cohen-Dvashi H, Cohen N, Israeli H, Diskin R Molecular Mechanism for LAMP1 Recognition by Lassa Virus. J Virol.2015; 89(15):7584–7592. 10.1128/JVI.00651-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Redis RS, Calin GA SnapShot: Non-coding RNAs and Metabolism. Cell Metab.2017; 25(1):220–220.e221. 10.1016/j.cmet.2016.12.012 [DOI] [PubMed] [Google Scholar]
- 45.Aredia F, Scovassi AI A new function for miRNAs as regulators of autophagy. Future Med Chem.2017; 9(1):25–36. 10.4155/fmc-2016-0173 [DOI] [PubMed] [Google Scholar]
- 46.Ilott NE, Ponting CP Predicting long non-coding RNAs using RNA sequencing. Methods.2013; 63(1):50–59. 10.1016/j.ymeth.2013.03.019 [DOI] [PubMed] [Google Scholar]
- 47.Uchida S. High-Throughput Methods to Detect Long Non-Coding RNAs. High Throughput.2017; 6(3). 10.3390/ht6030012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu X, Chen S, Jia C, Xue S, Dou C, Dai Z, et al. Gene expression profile and long non-coding RNA analysis, using RNA-Seq, in chicken embryonic fibroblast cells infected by avian leukosis virus J. Arch Virol.2018; 163(3):639–647. 10.1007/s00705-017-3659-8 [DOI] [PubMed] [Google Scholar]
- 49.Golicz AA, Singh MB, Bhalla PL The Long Intergenic Noncoding RNA (LincRNA) Landscape of the Soybean Genome. Plant Physiol.2018; 176(3):2133–2147. 10.1104/pp.17.01657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chekanova JA Long non-coding RNAs and their functions in plants. Curr Opin Plant Biol.2015; 27:207–216. 10.1016/j.pbi.2015.08.003 [DOI] [PubMed] [Google Scholar]
- 51.Gao X, Ye J, Yang C, Luo L, Liu Y, Ding J, et al. RNA-seq analysis of lncRNA-controlled developmental gene expression during puberty in goat & rat. BMC Genet.2018; 19(1):19 10.1186/s12863-018-0608-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mao AP, Shen J, Zuo Z Expression and regulation of long noncoding RNAs in TLR4 signaling in mouse macrophages. BMC Genomics.2015; 16:45 10.1186/s12864-015-1270-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pawar K, Hanisch C, Palma Vera SE, Einspanier R, Sharbati S Down regulated lncRNA MEG3 eliminates mycobacteria in macrophages via autophagy. Sci Rep.2016; 6:19416 10.1038/srep19416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang X, Yang J, Wang J, Wen Q, Wang H, He J, et al. Microarray analysis of long noncoding RNA and mRNA expression profiles in human macrophages infected with Mycobacterium tuberculosis. Sci Rep.2016; 6(38963 10.1038/srep38963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duval M, Cossart P, Lebreton A. Mammalian microRNAs and long noncoding RNAs in the host-bacterial pathogen crosstalk. Semin Cell Dev Biol.2017; 65:11–19. 10.1016/j.semcdb.2016.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zur Bruegge J, Einspanier R, Sharbati S A. Long Journey Ahead: Long Non-coding RNAs in Bacterial Infections. Front Cell Infect Microbiol.2017; 7:95 10.3389/fcimb.2017.00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kotzin JJ, Mowel WK, Henao-Mejia J. Viruses hijack a host lncRNA to replicate. Science.2017; 358(6366):993–994. 10.1126/science.aar2300 [DOI] [PubMed] [Google Scholar]
- 58.Murphy MB, Medvedev AE. Long noncoding RNAs as regulators of Toll-like receptor signaling and innate immunity. J Leukoc Biol.2016; 99(6):839–850. 10.1189/jlb.2RU1215-575R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eissmann M, Gutschner T, Hammerle M, Gunther S, Caudron-Herger M, Gross M, et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol.2012; 9(8):1076–1087. 10.4161/rna.21089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma XY, Wang JH, Wang JL, Ma CX, Wang XC, Liu FS,et al. Malat1 as an evolutionarily conserved lncRNA, plays a positive role in regulating proliferation and maintaining undifferentiated status of early-stage hematopoietic cells. BMC Genomics.2015; 16:676 10.1186/s12864-015-1881-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun Q, Hao Q, Prasanth KV.Nuclear Long Noncoding RNAs: Key Regulators of Gene Expression. Trends Genet.2017. 10.1016/j.tig.2017.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou HJ, Wang LQ, Wang DB, Yu JB, Zhu Y, Xu QS, et al. Long non-coding RNA MALAT1 contributes to inflammatory response of microglia following spinal cord injury via modulating miR-199b/IKKbeta/NF-kappaB signaling pathway. Am J Physiol Cell Physiol.2018. 10.1152/ajpcell.00278.2017 [DOI] [PubMed] [Google Scholar]
- 63.Yan W, Wu Q, Yao W, Li Y, Liu Y, Yuan J, et al. MiR-503 modulates epithelial-mesenchymal transition in silica-induced pulmonary fibrosis by targeting PI3K p85 and is sponged by lncRNA MALAT1. Sci Rep.2017; 7(1):11313 10.1038/s41598-017-11904-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.An Y, Zhang Z, Shang Y, Jiang X, Dong J, Yu P, et al. mir-23-3p regulates the chemoresistance of gastric cancer cells by targeting ATG12 and HMGB2. Cell Death Dis.2015; 6:e1766 10.1038/cddis.2015.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li S, Pan X, Yang S, Ma A, Yin S, Dong Y,et al. LncRNA MALAT1 promotes autophagy in HUVECs by inhibiting the PI3K/AKT pathway.J Cell Biochem. 2019. March;120(3):4092–4101. 10.1002/jcb.27694 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(XLSX)
(DOCX)
LncRNA and mRNA compare in RNA length (A), exon number (B) and ORF length (C).
(TIF)
(JPG)
(TIF)
(TIF)
(JPG)
(A) The Map of Double fluorescence labeling lentivirus autophagy flux detection vector and (B) Homology modeling of expression product.
(TIF)
Data Availability Statement
All clean reads of this work are available from the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) database (PRJNA544884).