Abstract
The incidence of diabetes and related complications like nephropathy is growing rapidly and has become a major health care issue. Changes in the environment and nutritional habits have been implicated as major players. Furthermore, it is becoming increasingly clear that epigenetic factors may modulate the connections between genes and the environment. While diabetes in itself is treatable to a large extent, it is still associated with significantly increased risk for complications including chronic kidney and cardio-vascular diseases. Current treatments have added preventative approaches so as to avoid future diabetic complications. Unfortunately, diabetic patients are often plagued with the continued development of various complications even after achieving glucose control. This has been suggested to be attributable to a mysterious phenomenon termed ‘metabolic memory’ of the prior glycemic state. Recent studies have suggested that epigenetic changes to chromatin can affect gene expression in response to various stimuli, and changes in key biochemical pathways and epigenetic histone and DNA methylation patterns in chromatin have been observed in a diabetic milieu. These accumulating data suggest that metabolic or hyperglycemic memory may be due to epigenetic changes in specific target tissues altering gene expression without changing the genetic code itself. While the genetics of diabetes has long been the focus of scientific research, much less is known about the role of epigenetics and the related molecular pathways that might affect the development of diabetes and the associated complications. Further studies of epigenetic mechanisms are therefore timely and could provide valuable new insights into the pathology of diabetic complications and also uncover much needed new therapeutic targets.
It is well known that both type 1 and type 2 diabetes mellitus are associated with significantly accelerated rates of macro- and microvascular diseases such as cardiovascular complications and nephropathy. Even when blood glucose levels are under control, diabetic patients may still develop chronic kidney disease (CKD) leading to renal failure. Most of the treatment options for renal dysfunction rely on the use of limited available medications known to delay the progression toward renal failure with hopes of avoiding dialysis and/or kidney transplantation.
Genetic factors and key gene mutations have been implicated in the pathogenesis of diabetes. However, increasing evidence suggests that complex interactions between genes and the environment and related epigenetic factors may play major roles in many common human diseases such as diabetes and its complications. Chromatin provides a crucial interface between genetics and environment, and the epigenetic DNA methylation as well as posttranscriptional modifications (PTMs) of histone tails in chromatin can regulate gene transcription. Although numerous studies have identified key biochemical pathways triggered by hyperglycemia and diabetes in target cells related to inflammation and diabetic complications [1], the exact molecular mechanisms are still not very clear. Exciting recent research has implicated a role for epigenetic regulation in this context. Such investigations can provide new insights into the pathology of diabetes and its complications and lead to the development of sorely needed new treatment options.
Evidence for Metabolic Memory from Clinical Trials and Experimental Models
Evidence shows that some diabetic patients experience a continued development of diabetic complications even after achieving glucose control, suggesting a metabolic memory of prior glycemic exposure. Recent studies have suggested that this may be attributed to epigenetic changes in target cells without alterations in gene coding sequences. Several clinical trials have indicated that early intensive glycemic control leads to sustained beneficial effects. The Diabetes Control and Complications Trial (DCCT) demonstrated that type 1 diabetes mellitus patients under intensive insulin therapy had delayed progression of nephropathy, retinopathy, and neuropathy as compared to patients under conventional therapy [2]. The Epidemiology of Diabetic Complications and Interventions (EDIC) trial continued long-term follow-up of the DCCT and placed all participants on intensive glycemic control [3]. The EDIC trial has demonstrated that patients originally in the intensive treatment group during the DCCT and continued on intensive therapy for the EDIC trial continued to have significantly slower progression of key diabetic microvascular complications relative to patients who were in the conventional treatment group during DCCT. Better outcomes for macrovascular complications have been noted as well [4]. Clinical trials with T2D patients have also shown similar protection [5].
These major clinical trial findings demonstrate the importance of early metabolic control to reduce long-term complications and confirm that hyperglycemia can have long-lasting detrimental consequences. The mechanisms responsible for these persistent effects of the prior hyperglycemic state are still not well understood, and this phenomenon, termed ‘metabolic memory’ [4] has been a major challenge in the treatment of diabetic complications.
In addition, several experimental models have been used to study the molecular mechanisms of ‘metabolic memory’ Early studies in dogs found that retinal complications persisted even after reversing hyperglycemia [6]. In rats, islet transplantation after 12 weeks of diabetes could not reverse the progression of retinopathy as compared to islet transplantation after 6 weeks of diabetes [7]. Studies in streptozotocin-induced diabetic rats showed that early glycemic control has protective effects in the retina, and also demonstrated a link with histone acetylation [8].
Endothelial cells cultured in high glucose (HG) showed sustained increases in key profibrotic and extracellular matrix proteins even after normalization of glucose [9]. More recently, short-term HG culture of endothelial cells displayed sustained activation of inflammatory genes and oxidant stress even after glucose normalization [10, 11]. Additional reports demonstrated the persistence of oxidant stress for up to one week after glucose normalization, which could be blocked by antioxidants or NADPH oxidase inhibitors [12]. In another model of metabolic memory, vascular smooth muscle cells (VSMC) derived from T2D obese db/db mice displayed sustained increases in NF-κB activation, inflammatory gene expression, migration, and oxidant stress as well as increased monocyte adhesion relative to control nondiabetic db/+ VSMC even after in vitro culture [13].
These data suggest a metabolic memory of vascular dysfunction arising from prior hyperglycemia, and further emphasize the importance of strict glycemic control to reduce the progression of diabetic complications. They also suggest a role for oxidant stress in perpetuating this metabolic memory [12]. Hyperglycemia and oxidant stress can also increase the accumulation of advanced glycation end products (AGEs) further perpetuating local inflammation and oxidant stress to promote long-term damage to end organs, including the kidney. Thus AGEs, acting through receptors such as RAGE, could also contribute to hyperglycemic memory. However, the nuclear mechanisms responsible for the sustained ‘memory’ over time through multiple cell divisions at the transcriptional and epigenetic level are not fully clear and have evolved as an active area of research.
Epigenetic Regulation of Gene Expression
The basic subunit of chromatin, nucleosome, consists of DNA wrapped around a histone octamer comprised of two copies of histone H2A, H2B, H3, and H4 [14]. Transcriptional activation or repression is a dynamic process that relies on the accessibility of DNA to various transcription factors, coactivators/corepressors, and the recruitment of protein complexes that alter chromatin structure via enzymatic modifications of histone tails and nucleosome remodeling. Epigenetic changes to chromatin form an added layer of gene regulation that can be altered without altering the DNA code itself. Numerous combinations of epigenetic modifications allow for flexibility of the chromatin and transcriptional outcomes depending on the needs of the cell [15].
DNA methylation is a well-characterized epigenetic mark in the context of tumor suppressor genes and cancer [16]. However, much less is known about DNA methylation in diabetes, although a role for DNA methylation in the regulation of various genes associated with insulin expression and secretion has been implicated [17]. DNA methylation has also been shown to play a role in kidney disease. One recent study showed altered DNA methylation at key gene promoters in T1D patients with diabetic nephropathy relative to those without [18]. Changes in blood homocysteine levels affecting methyl transfer reactions by inhibiting DNA methyltransferases have also been seen in patients with CKD [19].
Histone posttranslational modifications (PTMs) such as lysine acetylation, phosphorylation, and methylation can greatly influence gene regulation. Histone acetyltransferases (HATs) and histone deacetylases (HDACs) are relatively nonselective regulators of histone lysine acetylation which can occur quite rapidly [20]. Histone methylation on the other hand is considered to be more long lasting. Histone lysine methylation is mediated by histone methyl-transferases (HMTs), and is more complex since there are several HMTs and lysine residues that can be mono-, di-, or tri-methylated. HMTs are generally more specific, usually methylating only one specific lysine residue. Typically, histone H3 lysine 4 methylation (H3K4me) is associated with gene activation, while histone H3 lysine 9 methylation (H3K9me) and H3K27me3 are associated with gene repression [21]. Furthermore, histone methylation can also be dynamically regulated through the coordinated action of HMTs and lysine demethylases (KDMs). Numerous KDMs have been identified with varying specificities [22]. Overall, gene activation or repression depends on the modification, the specific residue as well as dynamic cooperation between various chromatin factors.
Although histone lysine methylation is reversible, it remains one of the more stable epigenetic modifications with some histone lysine methylation states maintaining very low turnover rates, and hence could be key factors in metabolic memory. Histone modifications are now widely accepted to play a role in epigenetics; however, there are questions as to what role they specifically play. Histone modifications are closely linked with DNA methylation, and together they can initiate or maintain transcriptional memory [23].
Epigenetic Mechanisms in Diabetic Complications and Metabolic Memory
Numerous studies have shown epigenetic changes induced by diabetic conditions. Both in vitro and in vivo studies with diabetic stimuli like HG or environmental changes have demonstrated alterations in histone lysine acetylation and methylation patterns influenced by recruitment of HATs/HDACs or HMTs [24, 25]. In addition, recent evidence shows that HG- and TGF-β1-induced expression of fibrotic genes in renal mesangial cells was associated with changes in key histone PTMs at these gene promoters [26]. There is much interest in the biological roles of DNA methylation, histone PTMs and HMTs in metabolic memory and diabetic complication such as CKD and nephropathy that seem to persist even after treatment with drugs or glycemic control.
Emerging research has provided new insights suggesting that chromatin-based changes to histone methylation patterns may be responsible for metabolic memory. VSMC from diabetic db/db mice cultured in vitro for several passages continued to exhibit increased inflammatory gene expression as compared to VSMC from nondiabetic db/+ mice [13, 27]. This corresponded to decreased H3K9me3-repressive marks at the promoters of key inflammatory genes and was associated with decreased protein levels of Suv39h1, a known HMT mediating H3K9me3. Overexpression of Suv39h1 in the diabetic db/db VSMC partially reversed the diabetic phenotype. db/db VSMC also exhibited increased TNF-α-induced inflammatory gene expression with corresponding sustained decreases in promoter H3K9me3 and Suv39h1 occupancy. Human VSMC treated with HG demonstrated similar changes in lysine methylation [27].
Interestingly, a recent study reported elevated levels of miR-125b in diabetic db/db VSMC. MicroRNA-125b could target Suv39h1 leading to decreased H3K9me3 and a corresponding increase in inflammatory gene expression as well as enhanced VSMC monocyte binding [28]. These results indicate a sustained loss of chromatin-repressive mechanisms in the diabetic state in part through reduction in repressive marks due to the prior hyperglycemic environment and thus may play a role in metabolic memory.
Short-term hyperglycemic conditions were also found to induce long-term changes in chromatin modifications. Endothelial cells exposed to HG for 16 h and later cultured for several days in normal glucose demonstrated a sustained increase in the expression of NF-kB p65 subunit which correlated with increased promoter H3K4me1 marks and occupancy of the HMT Set7 known to regulate this mark [10, 11]. These epigenetic changes could be prevented by blocking components of the mitochondrial electron transport chain [11]. HG could also promote epigenetic changes at the promoters of fibrotic genes in renal mesangial cells [26]. Collectively, these results indicate that prior exposure to hyperglycemia can induce epigenetic changes in target cells and alter chromatin structure resulting in long-lasting repercussions for gene expression levels associated with the pathology of diabetic micro- and macrovascular complications.
Conclusions
Epigenetic modifications have been found to play an important role in altered gene expression patterns associated with various diseases [29]. Clinical and experimental studies have clearly demonstrated the deleterious effects of hyperglycemia and the importance of maintaining good glucose control to prevent the onset or severity of diabetic complications like diabetic nephropathy. Hyperglycemia can induce epigenetic changes at genes associated with diabetic complications. DNA methylation, key histone modifications and their associated chromatin modifiers have been implicated (fig. 1), and epigenetic regulation of key inflammatory and fibrotic genes has been documented in vascular and renal cells as well as in metabolic memory. In addition to hyperglycemia, diabetes is also associated with genetic, nutritional and environmental risk factors. Each of these risk factors could in itself induce epigenetic changes to the chromatin structure ultimately altering gene expression patterns in conjunction with elevated glucose in various target tissues affected by diabetic complications. How these epigenetic changes are transmitted through multiple cell cycles is still under investigation [30].
Fig. 1.
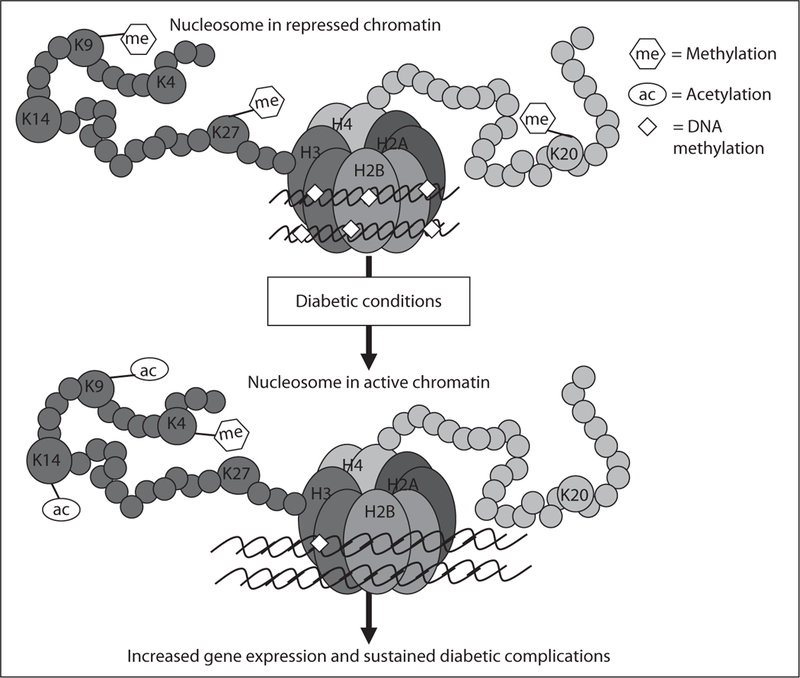
Model for epigenetic regulation of pathologic gene expression associated with diabetic complications and a potential role in metabolic memory. Posttranslational modifications of N-terminal histone tails in nucleosomes of chromatin play essential roles in gene regulation and are regulated by various chromatin modifiers. As examples, promoter trimethylation of histone H3 at lysine 9 (K9) or K27 is generally associated with repression, while methylation of histone H3 at lysine 4 or acetylation at H3 lysine 9/14 are usually associated with gene activation. In addition, promoter DNA methylation may also be involved as a key repressive mark. In the proposed model, diabetic conditions such as hyperglycemia lead to a loss of repressive modifications that normally maintain strict control of gene expression, while activating histone modifications may be increased or DNA methylation may be reduced at promoters, thus leading to relaxation or opening of the chromatin structure around key pathologic genes resulting in increased transcription. Various combinations of these chromatin modifications and others not shown here are likely to be involved. These epigenetic marks can be maintained through cell division via as yet unclear mechanisms. The occurrence and persistence of these epigenetic changes might explain altered gene expression and the metabolic memory phenomenon responsible for the sustained and persistent development of diabetic complications even after glucose normalization.
The rates of diabetes and associated complications are increasing rapidly. Given the widespread prevalence of diabetic nephropathy, CKD and the rapid transition to ESRD, further exploration into the epigenetic causes and related treatment options is imperative. Epigenetic drugs such as inhibitors of DNA methylation, HATs and HDACs, and some KDMs are already being evaluated for cancer and other diseases [16]. In addition, recent and ongoing studies investigating epigenomic DNA and histone methylation patterns are beginning to reveal key aspects of chromatin structure, gene expression and epigenetic information under diabetic states. Such epigenomic profiling approaches will be greatly aided by the recent availability of high throughput next generation sequencing technologies. No doubt will we see a lot of research activity in this field in the upcoming years that hopefully can lead to a greater understanding of the role of epigenetics in diabetes and to the identification of sorely needed new therapeutic strategies for diabetic complications and CKD.
Acknowledgments
The authors thank Dr. Marpadga Reddy for help with the manuscript, and also gratefully acknowledge funding from the National Institutes of Health (NIDDK and NHLBI) and the Juvenile Diabetes Research Foundation.
References
- 1.Brownlee M: Biochemistry and molecular cell biology of diabetic complications. Natur 2001;414:813–820. [DOI] [PubMed] [Google Scholar]
- 2.Writing Team DCCT/EDIC Research Group Effect of intensive therapy on the microvas- cular complications of type 1 diabetes mellitus. JAMA 2002;287:2563–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.EDIC Study: Sustained effect of intensive treatment of type 1 diabetes mellitus on development and progression of diabetic nephropathy: the Epidemiology of Diabetes Interventions and Complications (EDIC) study. JAMA 2003;290:2159–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nathan DM, Cleary PA, Backlund JY, et al. : Intensive diabetes treatment and cardiovascular disease in patients with type 1 diabetes N Engl J Med 2005;353:2643–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schernthaner G: Diabetes and cardiovascula disease: is intensive glucose control beneficial or deadly? Lessons from ACCORD, ADVANCE, VADT, UKPDS, PROactive, and NICE-SUGAR. Wien Med Wochenschr 2010;160:8–19. [DOI] [PubMed] [Google Scholar]
- 6.Engerman RL, Kern TS: Progression of incipient diabetic retinopathy during good glycemic control. Diabetes 1987;36:808–812. [DOI] [PubMed] [Google Scholar]
- 7.Hammes HP, Klinzing I, Wiegand S, Bretzel RG, Cohen AM, Federlin K: Islet transplantation inhibits diabetic retinopathy in the sucrose-fed diabetic Cohen rat. Invest Ophthalmol Vis Sci 1993;34:2092–2096. [PubMed] [Google Scholar]
- 8.Zhong Q, Kowluru RA: Role of histone acetylation in the development of diabetic retinopathy and the metabolic memory phenomenon. J Cell Biochem 2010;110: 1306–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roy S, Sala R, Cagliero E, Lorenzi M: Overexpression of fibronectin induced by diabetes or high glucose: phenomenon with a memory. Proc Natl Acad Sci U S A 1990;87 404–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brasacchio D, Okabe J, Tikellis C, et al. : Hyperglycemia induces a dynamic coopera- tivity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail. Diabetes 2009;58:1229–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Osta A, Brasacchio D, Yao D, et al. : Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med 2008;205:2409–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ihnat MA, Thorpe JE, Ceriello A: Hypothesis: the ‘metabolic memory’, the new challenge of diabetes. Diabet Med 2007;24: 582–586. [DOI] [PubMed] [Google Scholar]
- 13.Li SL, Reddy MA, Cai Q, et al. : Enhanced proatherogenic responses in macrophages and vascular smooth muscle cells derived from diabetic db/db mice. Diabetes 2006;55: 2611–2619. [DOI] [PubMed] [Google Scholar]
- 14.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ: Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997;389:251–260. [DOI] [PubMed] [Google Scholar]
- 15.Strahl BD, Allis CD: The language of covalent histone modifications. Nature 2000;403:41–45. [DOI] [PubMed] [Google Scholar]
- 16.Sharma S, Kelly TK, Jones PA: Epigenetics in cancer. Carcinogenesis 2010;31:27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ling C, Groop L: Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes 2009;58:2718–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bell CG, Teschendorff AE, Rakyan VK, Maxwell AP, Beck S, Savage DA: Genome-wide DNA methylation analysis for diabetic nephropathy in type 1 diabetes mellitus. BMC Med Genomics 2010;3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ekstrom TJ, Stenvinkel P: The epigenetic conductor: a genomic orchestrator in chronic kidney disease complications? J Nephrol 2009;22:442–449. [PubMed] [Google Scholar]
- 20.Roth SY, Denu JM, Allis CD: Histone acetyl-transferases. Annu Rev Biochem 2001;70: 81–120. [DOI] [PubMed] [Google Scholar]
- 21.Martin C, Zhang Y: The diverse functions of histone lysine methylation. Nat Rev Mol Cel Biol 2005;6:838–849. [DOI] [PubMed] [Google Scholar]
- 22.Shi Y, Whetstine JR: Dynamic regulation of histone lysine methylation by demethylases. Mol Cell 2007;25:1–14. [DOI] [PubMed] [Google Scholar]
- 23.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A: An operational definition of epigenetics. Genes Dev 2009;23:781–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Villeneuve LM, Natarajan R: The role of epigenetics in the pathology of diabetic complications. Am J Physiol 2010;299:F14–F25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tonna S, El-Osta A, Cooper ME, Tikellis C: Metabolic memory and diabetic nephropathy: potential role for epigenetic mechanisms. Nature Rev Nephrol 2010;6:332–341. [DOI] [PubMed] [Google Scholar]
- 26.Sun G, Reddy MA, Yuan H, Lanting L, Kato M, Natarajan R: Epigenetic histone methylation modulates fibrotic gene expression. J Am Soc Nephrol 2010;21:2069–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Villeneuve LM, Reddy MA, Lanting LL, Wang M, Meng L, Natarajan R: Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc Natl Acad Sci U S A 2008;105: 9047–9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Villeneuve LM, Kato M, Reddy MA, Wang M, Lanting L, Natarajan R: Enhanced levels of microRNA-125b in vascular smooth muscle cells of diabetic db/db mice lead to increased inflammatory gene expression by targeting the histone methyltransferase Suv39h1. Diabetes 2010;59:2904–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu L, Li Y, Tollefsbol TO: Gene-environment interactions and epigenetic basis of human diseases. Curr Issues Mol Biol 2008;10:25–36. [PMC free article] [PubMed] [Google Scholar]
- 30.Margueron R, Reinberg D: Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet 2010;11: 285–296. [DOI] [PMC free article] [PubMed] [Google Scholar]


