Abstract
Prochlorococcus is a major contributor to primary production, and globally the most abundant photosynthetic genus of picocyanobacteria because it can adapt to highly stratified low-nutrient conditions that are characteristic of the surface ocean. Here, we examine the structural adaptations of the photosynthetic thylakoid membrane that enable different Prochlorococcus ecotypes to occupy high-light (HL), low-light (LL) and nutrient-poor ecological niches. We used atomic force microscopy (AFM) to image the different photosystem I (PSI) membrane architectures of the MED4 (HL) Prochlorococcus ecotype grown under high-light and low-light conditions in addition to the MIT9313 (LL) and SS120 (LL) Prochlorococcus ecotypes grown under low-light conditions. Mass spectrometry quantified the relative abundance of PSI, photosystem II (PSII) and cytochrome b6f complexes and the various Pcb proteins in the thylakoid membrane. AFM topographs and structural modelling revealed a series of specialised PSI configurations, each adapted to the environmental niche occupied by a particular ecotype. MED4 PSI domains were loosely packed in the thylakoid membrane, whereas PSI in the LL MIT9313 is organised into a tightly-packed pseudo-hexagonal lattice that maximises harvesting and trapping of light. There are approximately equal levels of PSI and PSII in MED4 and MIT9313, but nearly two-fold more PSII than PSI in SS120, which also has a lower content of cytochrome b6f complexes. SS120 has a different tactic to cope with low-light levels, and SS120 thylakoids contained hundreds of closely packed Pcb-PSI supercomplexes that economise on the extra iron and nitrogen required to assemble PSI-only domains. Thus, the abundance and widespread distribution of Prochlorococcus reflect the strategies that various ecotypes employ for adapting to limitations in light and nutrient levels.
Introduction
By virtue of its abundance in the oceans Prochlorococcus is one of the most important photosynthetic organisms on Earth. A global abundance of 2.9 ± 0.1 × 1027 Prochlorococcus cells fix 4 gigatonnes of carbon per year1, which is comparable to the total primary productivity of the world’s croplands2. Prochlorococcus is found in the oligotrophic ocean with a distribution between approximately 45°N and 40°S and is present throughout the euphotic zone down to a depth of about 200 metres3. Prochlorococcus is also notable for its unique pigmentation, being the only type of marine phytoplankton to use divinyl derivatives of chlorophyll a and b (Chl a and Chl b), bound to Pcb proteins, to capture light energy and drive photosynthesis4. Prochlorococcus Chls exceed 50% of marine Chl mass in large expanses of the ocean3,5.
There are six major clades of Prochlorococcus that have been cultured and evolutionary diversification has been strongly tied to environmental conditions, with clades broadly classified as being either high-light (HL)-adapted or low-light (LL)-adapted ecotypes6-12. HL-adapted ecotypes such as MED4 and MIT9312 have a lower Chl b:Chl a ratio, and are typically the most abundant organisms in oligotrophic surface waters of the open ocean although they are present throughout the entire euphotic zone13-16. LL-adapted ecotypes such as NATL2A, SS120 and MIT9313 have a higher Chl b:Chl a ratio and grow optimally under much lower light intensities8,17. Elevated Chl b levels allow these strains to absorb more light in the blue region of the spectrum, which is prevalent at the lower depths in the euphotic zone18, conferring a competitive advantage in this ecological niche17.
Prochlorococcus differs from marine Synechococcus, with which it shares a relatively recent common ancestor10,12,19-21, in that its light-harvesting antenna complexes are formed from integral membrane Pcb proteins, rather than the membrane-extrinsic phycobilisome complexes found in most cyanobacteria22. Pcb proteins have six transmembrane helices, and significant homology with the Chl binding PSII subunit CP43 and the iron-limitation IsiA23,24 protein. Prochlorococcus ecotypes rely on different Pcb-PSI supercomplexes to meet their light harvesting requirements; in SS120 light-harvesting capacity is enhanced by surrounding PSI trimers with an 18-membered PcbG ring (PcbG18PSI3)25,26, similar to the IsiA-PSI supercomplex found in Synechocystis when grown under iron limited conditions27,28. In SS120 and MIT9313 PSII dimers are flanked by 8 Pcb proteins (Pcb8PSII2)24,26. The HL-adapted ecotype MED4 does not produce an 18 membered Pcb-PSI supercomplex in iron rich or depleted conditions, although it does appear to assemble a Pcb-PSII supercomplex26.
Taking into account the abundance of Prochlorococcus in the oceans, with each cell housing roughly 5 μm2 of thylakoids29, we estimate that the combined surface area of energy-absorbing Prochlorococcus membranes is 28 times the surface area of the Earth. Despite the global importance and scale of these membranes, little is known about their supramolecular organisation and how they vary between different ecotypes to allow adaptation to different light and nutrient conditions. Photosynthetic membrane organisation can be probed by atomic force microscopy (AFM)30-38. AFM of cyanobacterial thylakoids from Thermosynechococcus elongatus and Synechococcus sp. PCC 7002 revealed long-range semi-crystalline PSI-only membrane arrays, and more heterogeneous membrane domains where PSI is interspersed amongst membrane complexes such as PSII and the cytochrome b6f complex39. AFM of thylakoid membranes from Synechococcus sp. PCC 7942 showed a disordered membrane system with PSI intermixed with PSII, which was co-localised with the cytochrome b6f complex40. Here, we use a combination of AFM, mass spectrometry and pigment analysis to elucidate the organisation and composition of photosynthetic membranes from Prochlorococcus, to see how membrane architectures vary with ecotype and how they are optimised to function in their respective ecological niches in order to harvest, transfer and trap light energy.
Results
Supramolecular organisation of thylakoids from the MED4 ecotype grown under LL
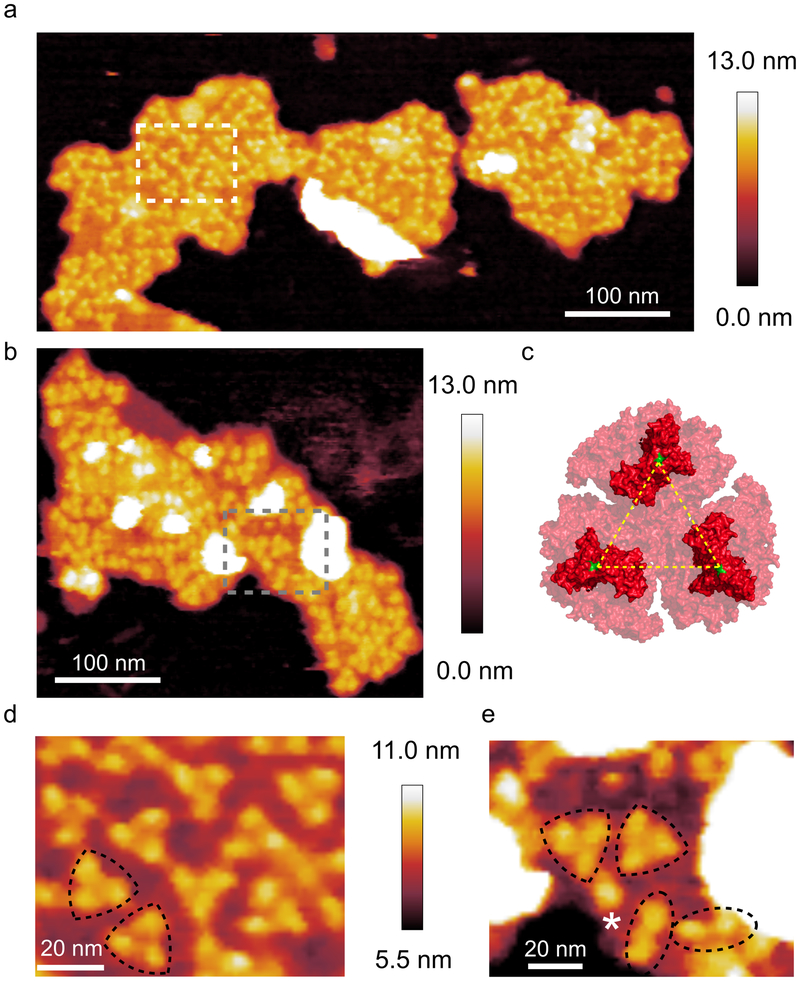
Purified thylakoid membranes were prepared from cells of the MED4 ecotype grown at 5 μmol photons m−2 s−1, by fractionation on continuous sucrose gradients containing 0.1% digitonin (Supplementary Fig. 1). AFM analysis of membrane samples collected from throughout the sucrose gradient showed multiple membrane patches housing trimeric protein complexes (Fig. 1a,b,d,e), which were reminiscent of the PSI complexes in AFM topographs of T. elongatus thylakoids39. As no crystal structure of the MED4 PSI complex is available, the structure of the T. elongatus PSI trimer (PDB ID: 1JB0) was used for reference (Fig. 1c). The trimeric features in the topographs had an average height above the mica surface and the lipid bilayer of 10.1 ± 0.6 nm and 3.4 ± 0.3 nm respectively; the average lateral distance between monomers was 10.4 ± 0.9 nm. These dimensions are consistent with the trimeric PSI structure41-43 and were assigned as such.
Fig. 1. AFM of PSI in thylakoid membrane patches from LL-grown MED4.
(a) AFM topograph of a membrane patch showing trimeric PSI complexes; the area delineated by the white box is shown in (d). (b) A second membrane patch also showing PSI, which has a more disorganised, less densely packed architecture; the area delineated by the grey box is shown in (e). (c) The crystal structure of the trimeric PSI complex from T. elongatus seen from the cytoplasmic face of the membrane (PDB ID: 1JB0). The yellow lines represent a distance of 9.1 nm measured from proline 29 (green) of the PsaC subunit. (d) Zoomed view of the area highlighted in (a) showing the trimeric PSI complexes (outlined in black) in more detail. (e) Zoomed view of the area outlined in (b). Trimeric, dimeric and monomeric PSI complexes are highlighted with black triangles, black ovals and white asterisks respectively. AFM imaging was conducted to a similar standard on a minimum of 10 independent membrane preparations from this ecotype, grown under the same light conditions. Large flat areas of membrane were selected in which protein complexes were clearly visible and they were imaged at the highest resolution.
The somewhat disorganised arrangement of trimeric PSI complexes in MED4 membrane patches (Fig. 1a,b) differs from the paracrystalline PSI organisation often found in AFM topographs of T. elongatus membranes39. However, for both paracrystalline and disorganised PSI domains, the high density of PSI packing appears to preclude the presence of other protein complexes, and there was no evidence in AFM topographs for PSII, cytochrome b6f complex or Pcb antenna complexes in these MED4 membranes. The density of PSI complexes was calculated for the membrane patches shown in Fig. 1a; for ease of comparison, the data were calculated as PSI monomer equivalents rather than whole trimers. The membrane densities were 4604 (left) and 5203 (right) PSI monomer equivalents per μm2 and for the membrane patch in Fig. 1b it was calculated to be 3102 complexes per μm2. Using a value of 96 Chl molecules per PSI monomer the density of Chl in these membrane patches was calculated as 442024, 499510 and 293236 molecules of Chl per μm2 of thylakoid membrane respectively.
Another feature of the MED4 membrane patches was the presence of dimeric and monomeric complexes (highlighted by dotted ovals and a white asterisk in Fig. 1e respectively); these complexes have been assigned as dimeric and monomeric PSI on the basis of their height and their lateral dimensions. This is consistent with membrane patches from T. elongatus39 where several membrane patches were imaged that contained monomeric, dimeric and trimeric PSI complexes. The combined ratio of PSI monomer equivalents in trimeric vs monomeric or dimeric PSI in the membrane patches in Fig 1 is 5.3.
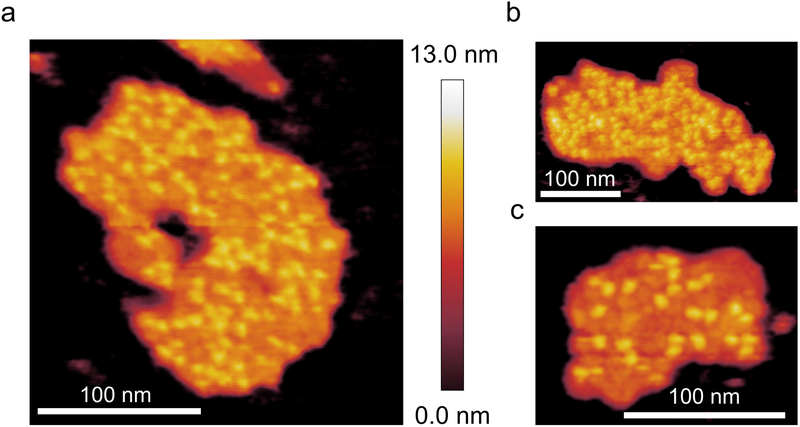
Supramolecular organisation of thylakoids from the MED4 ecotype grown under HL
In thylakoid membranes purified from MED4 cells grown in HL the PSI complexes have a disordered distribution similar to that of the thylakoid membranes purified from LL-grown cells. The density of PSI complexes in these HL-grown membrane patches is generally lower that their LL-grown counterparts and is highly variable; the PSI densities of membrane patches in Fig. 2a-c are 4283, 3742 and 2635 PSI monomer equivalents per μm2 of thylakoid membrane, respectively. Using a value of 96 Chl molecules per PSI monomer equivalent the Chl density of these patches was calculated at 411168, 359232 and 252960 Chl molecules per μm2 of thylakoid membrane, respectively, somewhat lower than for LL-grown MED4 (442024, 499510 and 293236 Chl molecules per μm2). Another difference between HL-grown and LL-grown membranes is the proportion of PSI complexes in a trimeric configuration; the combined number of PSI complexes (as monomer equivalents) forming trimers in the HL-grown membrane patches from Fig. 2a-c is 132. There are also proportionally more PSI complexes in either a dimeric or monomeric state; the combined total (monomers plus dimers) from the three membrane patches in Fig. 2 is 121 giving a ratio of trimeric to non-trimeric PSI complexes of 1.1, significantly lower than the 5.3 observed for LL-grown membranes. In summary, the effect of increasing the light used to grow MED4 from 5 to 250 μmol photons m−2 s−1, is a reduced packing density of PSI complexes, lower by approximately 17% on average, and a significantly reduced population of PSI trimers, in favour of more monomers and dimers.
Fig. 2. AFM of PSI in MED4 thylakoid membrane patches grown under HL.
(a) and (b) AFM topographs of membrane patches showing PSI complexes at a relatively high density. (c) AFM topograph of a membrane patch with a lower density of PSI complexes. AFM imaging was conducted to a similar standard on a minimum of 10 independent membrane preparations from this ecotype, grown under the same light conditions. Large flat areas of membrane were selected in which protein complexes were clearly visible and they were imaged at the highest resolution.
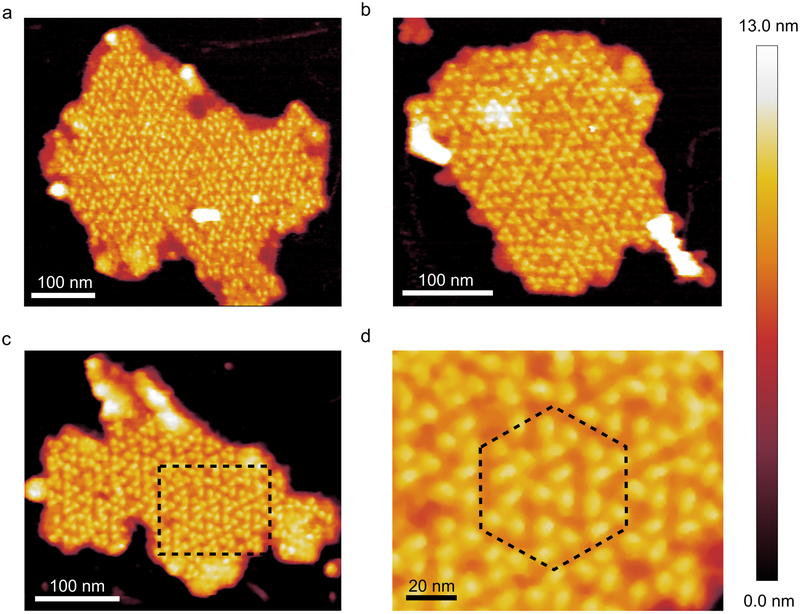
AFM of thylakoid membranes from the MIT9313 ecotype
Trimeric PSI complexes could also be imaged in thylakoid membranes from MIT9313 cells, but their organisation differed from that seen in MED4 membrane patches. Fig. 3a-d shows that MIT9313 PSI complexes were almost exclusively organised into a pseudo-hexagonal lattice (Fig. 3d), similar to the paracrystalline PSI-only domains of thylakoid membranes from T. elongatus39, an arrangement that leaves no room for PSII, cytochrome b6f complex or Pcb antenna proteins. In this LL-adapted ecotype, the tight packing of PSI complexes increases the abundance of PSI in the thylakoid membrane relative to the MED4 ecotype, with 5377, 5982 and 5391 PSI complexes per μm2 and 516258, 574302 and 517572 molecules of Chl per μm2 in Fig. 3a-c respectively. Unlike MED4, PSI complexes in the MIT9313 membranes are nearly all trimeric, with few PSI monomers and dimers; the combined ratio of trimeric to non-trimeric PSI (that is, PSI monomers and dimers) in the patches in Fig 3 is 44.5, significantly higher than MED4 membrane patches grown in either HL or LL. The average height of the PSI complexes in the MIT9313 membrane patches from the mica and bilayer surface is 10.1 ± 0.4 nm and 3.3 ± 0.4 nm respectively. The average distance between constituent monomers of the trimeric PSI complexes from the MIT9313 membrane patches was 10.2 ± 0.7 nm. These measurements are consistent with the crystal structure of the T. elongatus PSI trimer39 and almost identical to the dimensions measured for the MED4 PSI trimer.
Fig. 3. AFM of PSI in thylakoid membrane patches from MIT9313.
(a), (b) and (c) form a gallery of thylakoid membrane patches in which PSI is packed into a pseudo-hexagonal organisation; these large patches of PSI trimers do not appear to contain any other protein complexes. (d) Zoomed in view of the area highlighted in (c) with the “unit cell” of the hexagonally packed complexes outlined by the black dotted line. AFM imaging was conducted to a similar standard on a minimum of 10 independent membrane preparations from this ecotype, grown under the same light conditions. Large flat areas of membrane were selected in which protein complexes were clearly visible and they were imaged at the highest resolution.
AFM of thylakoid membranes from the SS120 ecotype
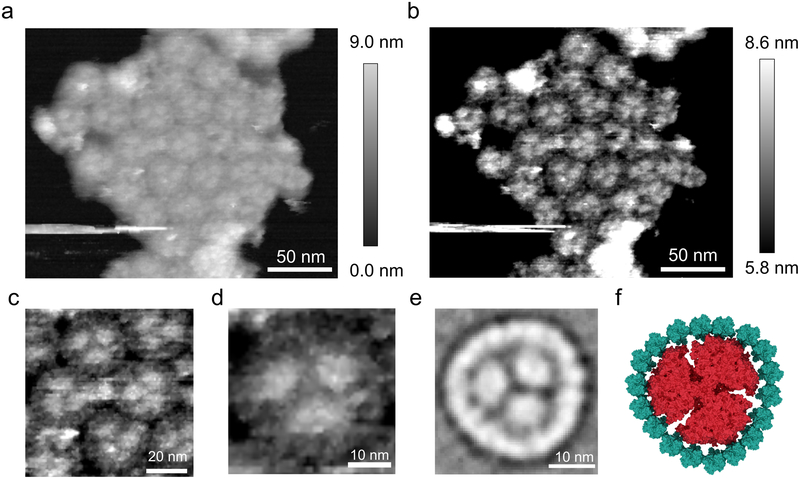
Solubilisation of membranes from the SS120 ecotype yields PSI supercomplexes, in which a PSI trimer is surrounded by an 18 membered ring of the Pcb protein25,26. However, the supramolecular arrangement of these Pcb-PSI supercomplexes was unknown, so AFM topographs were recorded for thylakoid membranes from SS120 (Fig. 4a,b), revealing several closely packed Pcb-PSI supercomplexes. The resolution is sufficient to identify individual components, including trimeric PSI cores, and each surrounding ring comprised of Pcb proteins (Fig. 4c,d). For comparison, a homologous PSI supercomplex, the IsiA-PSI supercomplex, was purified from an iron-limited Synechocystis sp. PCC 6803 culture and imaged by negative stain TEM. The projection map for the IsiA-PSI supercomplex (Fig. 4e), generated by averaging 52 particles, shows a trimeric PSI core surrounded by an 18-membered ring of the IsiA protein, a homologue of the PSII subunit CP43 and the light harvesting antenna Pcb proteins. Fig. 4f shows a model of the IsiA-PSI supercomplex constructed from the PSI crystal structure (PDB ID:1JB0) and the CP43 subunit from the PSII crystal structure (PDB ID: 3WU2)44, which shows the similarities between the Pcb-PSI supercomplexes in the AFM topographs and the IsiA-PSI structures. Furthermore, the average diameter of the putative Pcb ring in the AFM topographs was 32.8 ± 0.9 nm, consistent with the 33.0 nm diameter of the 18 membered Pcb ring determined by negative stain TEM of the isolated Pcb-PSI supercomplex25.
Fig. 4. AFM imaging of clustered Pcb-PSI supercomplexes in thylakoid membrane patches from SS120.
(a) A cluster of approximately 30 closely packed Pcb-PSI supercomplexes (b) The same membrane patch as displayed in (a) with the z-scale altered to highlight the structural elements of the Pcb-PSI supercomplex. The trimeric PSI core can clearly be seen in several of the supercomplexes with individual Pcb subunits also visible in the rings surrounding the PSI trimer. (c) Zoomed in view of (b) showing the interactions between adjacent Pcb-PSI supercomplexes in the membrane patch in (a) and (b). (d) Zoomed view of the membrane patch in (a) and (b) showing a single Pcb-PSI supercomplex; the trimeric core can be clearly identified, as can subunits within the Pcb ring. (e) Averaged projection map of the top-down view of the IsiA-PSI supercomplex purified from iron-limited Synechocystis sp. PCC 6803, homologous structure to the Pcb-PSI supercomplex, and generated by averaging projections of 52 negatively stained particles taken at room temperature by transmission electron microscopy (TEM). This averaged projection map shows the trimeric PSI complexes surrounded by an 18 membered ring of the IsiA protein, a homologue of the Pcb and CP43 proteins. (f) Model of the Pcb-PSI supercomplex based on the AFM data in (b), the PSI crystal structure (PDB ID: 1JB0) and the crystal structure of the CP43 subunit from the PSII crystal structure (PDB ID: 3WU2). AFM imaging was conducted to a similar standard on a minimum of 10 independent membrane preparations from this ecotype, grown under the same light conditions. Large flat areas of membrane were selected in which protein complexes were clearly visible and they were imaged at the highest resolution.
It was also possible to image a much larger membrane patch that contained over a hundred Pcb-PSI supercomplexes (Fig. 5a). The average height of the PSI complexes in this membrane patch was 9.6 ± 0.2 nm above the mica surface and 3.4 ± 0.2 nm above the membrane bilayer, comparable to the height of the PSI crystal structure from T. elongatus.
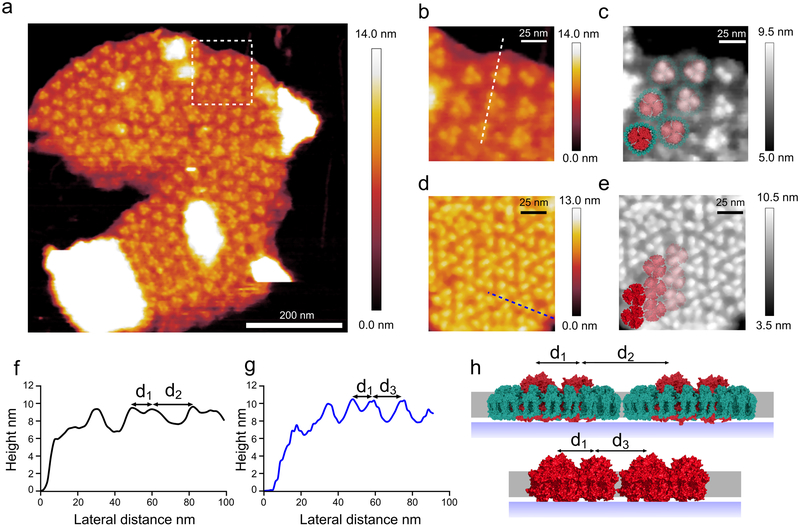
Fig. 5. Medium resolution AFM topograph of a large membrane patch from the SS120 ecotype.
(a) This image shows a membrane patch where the trimeric PSI core can be seen within hundreds of Pcb-PSI supercomplexes. (b) Magnified view of the area outlined by the white box in (a) showing the Pcb-PSI supercomplexes in more detail; the white line shows the location of the height profile in (f). (c) Grey scale of the same view as in (b) with the Pcb-PSI supercomplex model fitted to the AFM data. (d) An area of membrane from the MIT9313 membrane patch shown in Fig. 3c highlighting the difference in PSI packing between Pcb-PSI supercomplexes and “naked” PSI trimers in the thylakoid membrane; the presence of the Pcb ring in (b) leads to fewer PSI complexes per μm2 of thylakoid membrane. The blue line shows the location of the height profile in (g). (e) Grey scale of the same area as (d) with the PSI crystal structure fitted to the AFM data. (f) Height profile of dashed white line in (b) showing distances between the PSI complexes; the distance between PSI monomers in the same supercomplex (d1) is 9.3 nm and the distance between PSI monomers in adjacent supercomplexes (d2) is 20.4 nm. (g) Height profile of dashed blue line in (d) showing distances between PSI complexes in “naked” PSI trimers; the distance between constituent monomers in the PSI trimer (d1) is 10.1 nm, consistent with d1 measured in Pcb-PSI supercomplexes. The distance between PSI monomers in adjacent PSI trimers (d3) is measured at 14.0 nm, less than that measured from the Pcb-PSI supercomplexes owing to the absence of the Pcb ring. (h) A membrane model showing the distances between Pcb-PSI supercomplexes (top) and “naked” PSI trimers (bottom) with the distances measured from (f) and (g) shown. AFM imaging was conducted to a similar standard on a minimum of 10 independent membrane preparations from this ecotype, grown under the same light conditions. Large flat areas of membrane were selected in which protein complexes were clearly visible and they were imaged at the highest resolution.
The thylakoid membrane in Fig. 5 is very densely packed with Pcb-PSI supercomplexes (Fig. 5b,c), leaving no room for the other photosynthetic protein complexes such as PSII and the cytochrome b6f complex, and indicating that “PSI-only” zones are a feature of all three ecotypes. The density of Pcb-PSI supercomplexes in the membrane patch shown in Fig. 5a is 893 per μm2, equivalent to 2679 PSI complexes per μm2. The average PSI density was 5583 PSI complexes per μm2 for the other LL-adapted strain MIT9313; thus, the Pcb ring reduces the number of PSI complexes that can pack into the same area (Fig. 5d,e) and increases the distance between adjacent PSI trimers (Fig. 5f-h). The exact number of Chl pigments bound to each type of Pcb protein is unavailable; however by sequence comparison with the IsiA protein from Synechocystis it is apparent that the two proteins are almost identical24. Assuming that each Pcb protein binds 15 Chl molecules, the same number as the IsiA protein, the number of Chl molecules in the Pcb-PSI supercomplex is 55825. Using this number the density of Chl was calculated at 498294 molecules of Chl per μm2 based on the AFM data in Fig. 5. This density of Chl molecules is comparable to the MED4 ecotype but not as high as the other LL-adapted ecotype MIT9313.
Comparison of long range order of PSI complexes between Prochlorococcus ecotypes, MIT9313, MED4, SS120, and T. elongatus
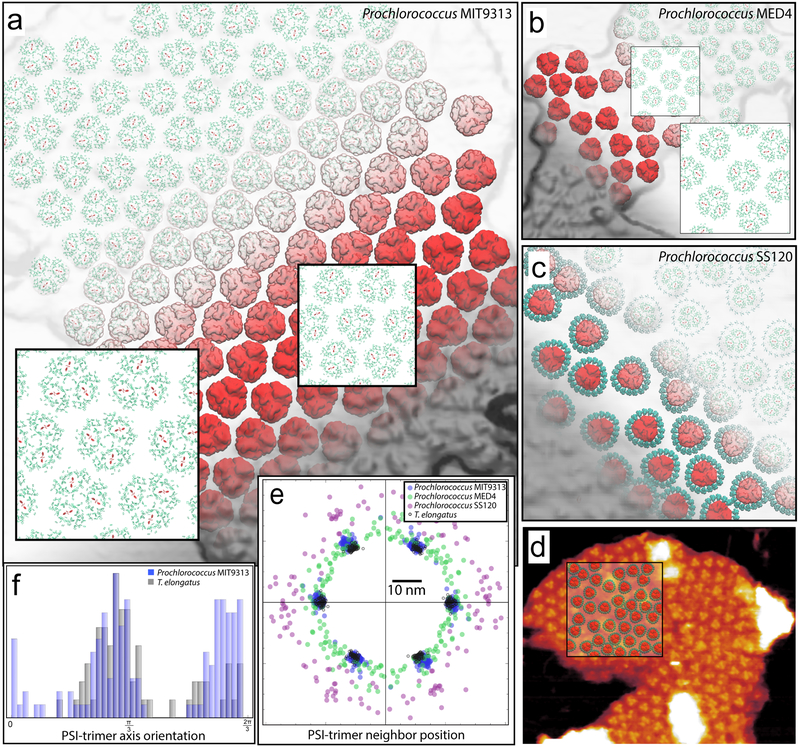
Structural models based on AFM topographs were constructed for the PSI trimer containing thylakoid domains of Prochlorococcus ecotypes, MIT9313 (Figs. 3b, 6a), MED4 (Figs. 1a, 6b), SS120 (Figs. 5a, 6c, 6d), and compared to a corresponding T. elongatus membrane model39 (Figs. 6e, 6f). The structural models reveal the packing pattern of constituent proteins, particularly the relative position and orientation of neighbouring PSI trimers, thereby permitting a comparison between the membrane architectures of different ecotypes (Fig. 6e). A near-periodic arrangement of PSI trimers, reported earlier for T. elongatus thylakoid domains39, is observed for the MIT9313 ecotype (Fig. 3b, 6a, 6e); a strong orientational correlation between neighbouring PSI trimers is also present for MIT9313, but up to an arbitrary π/3 rotation of the trimers (Fig. 6f).
Fig. 6. Structural models for the PSI trimer domains from the Prochlorococcus ecotypes MIT9313, MED4, and SS120 grown under LL.
The models for MIT9313 (a), MED4 (b), and SS120 (c) domains are arranged according to AFM topographs from Figs. 3b, 1a, and 5a, respectively. The protrusions of the PsaC-D-E subunits of PSI trimers (red), modelled according to PDB ID: 1JB041, can be seen to correspond to AFM topological features (grey). Pcb units (blue) surrounding the PSI trimers in SS120 are modelled after CP43, PDB ID: 3WU244. Constituent Chls are represented as porphyrin rings (red: PSI reaction center; green: PSI antenna; blue: Pcb (SS120 only)). The insets in (a) and (b) show typical Chl packing patterns, which for MIT9313 (a) reveal an arrangement similar to the one reported for T. elongatus39 (see (e)). The relative location of the SS120 model (c) with respect to the AFM topograph of Fig. 5a is shown in (d). Long range order of PSI trimers is shown in (e) and (f) in terms of the neighbouring trimer positions and orientations, respectively, for the aforementioned ecotypes in comparison with T. elongatus39. The x-axis in (e) for each set is chosen arbitrarily for alignment purposes. The MIT9313 trimer spacings (blue) display a near-periodic arrangement resembling that of T. elongatus (grey circles); the MED4 (green) and SS120 (purple) ecotypes do not represent a periodic arrangement pattern for constituent proteins, with the spacing between PSI trimers in the SS120 ecotype being notably larger due to the presence of surrounding Pcb units. Orientational correlations between PSI trimers are shown in (f) in terms of histograms for the angle between the symmetry axes of neighbouring trimers. Due to the C3-symmetry of the trimer, only the region (0, 2π/3) is shown. The double peak for MIT9313 in contrast with T. elongatus shows a bi-modal distribution of orientation correlations, i.e., an arbitrary π/3 rotation of PSI trimers is more predominant in MIT9313 compared with T. elongatus. MED4 and SS120 trimer orientations do not display correlated behaviour and are therefore not shown. The models presented contain: MIT9313 (a): 133 PS1 trimers with 38,304 Chls; MED4 (b): 57 PS1 trimers with 16,416 Chls; SS120 (c): 42 PS1 trimers and 728 Pcb units with 21560 Chls.
The packing patterns of MIT9313 and T. elongatus PSI domains are nearly identical (Fig. 6e), thereby implying that MIT9313 has similar inter-PSI exciton sharing properties as T. elongatus39. The model for the MED4 ecotype (Figs. 6b) represents a packing density of PSI trimers similar to that of MIT9313, but without any apparent periodicity (Fig. 6e). The presence of the surrounding Pcb units for SS120 (Fig. 6c) results in an increased trimer-trimer separation as well as a lack of periodicity (Fig. 6e).
Quantification of PSI, PSII, cytochrome b6f, ATP synthase and Pcb proteins by mass spectrometry
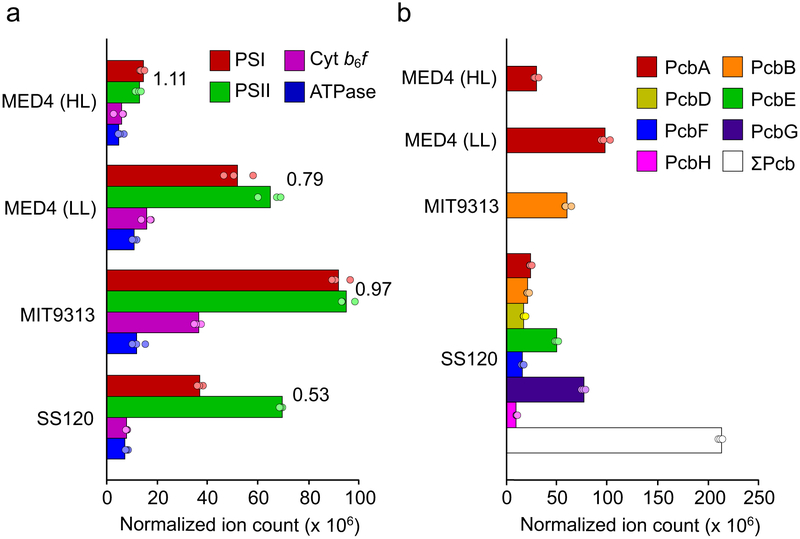
The AFM analyses presented in Fig. 1-5 show the arrangements of individual complexes, with no averaging, in membranes patches, from samples retrieved from sucrose density gradients following treatment of thylakoids with digitonin. In all cases we observe closely packed PSI trimers, in some cases with a surrounding Pcb ring, but with no PSII and cytochrome b6f complexes present. The packing density leaves no room for these complexes in the AFM topographs, yet they are required in a functioning photosynthetic cell. Thylakoid membranes adhere to the mica substrate in an orientation that displays the protruding cytoplasmic face of PSI; while this aids identification of PSI by AFM the poorly-protruding cytoplasmic faces of PSII and cytochrome b6f complexes are difficult to identify. In order to obtain an averaged view of the composition of membranes prepared from the three Prochlorococcus ecotypes, we used analysis by mass spectrometry. The number (mean ± SD) of proteins identified in three replicate analyses of each ecotype was 864 ± 5 (MED4 grown under HL), 781 ± 5 (MED4 grown under LL), 521 ± 9 (SS120) and 946 ± 11 (MIT9313). Label-free protein quantification of the complete data-set gave inter-replicate correlation coefficients of 0.993-0.999 (see Supplementary Fig. 2). Normalized ion counts for subunits PsaA and PsaB (PSI), PsbA and PsbB (PSII), PetA, PetB and PetC (cytochrome b6f) and AtpF and AtpG (ATP synthase) are shown in Supplementary Table 1 and in Fig.7a. As shown in Fig. 7a, levels of PSI in thylakoids purified from the three ecotypes are all either close to or lower than the levels of PSII, with PSI:PSII ratios (expressed as monomer equivalents) of 1.11 (MED4 grown under HL), 0.79 (MED4 grown under LL), 0.97 (MIT9313) and 0.53 (SS120). A previous quantitative proteomic analysis of MED4 grown under a 24-hour light-dark illumination regime45 revealed a PSI:PSII ratio of 0.66 while the results of another study of SS120 cells cultured under constant blue light46 gave a ratio of 1.19. Therefore, although deviation from the expected 1:1 PSI:PSII ratio for Prochlorococcus cells25 is observable, PSI is not the dominant photosystem complex in Prochlorococcus, in marked contrast with model strains such as Synechocystis sp. PCC6803 and Synechococcus sp. PCC7002.
Fig. 7. Comparison of the relative levels of PSI, PSII, cytochrome b6f, ATP synthase and Pcb proteins in Prochlorococcus marinus ecotypes MED4, MIT9313 and SS120.
Proteins extracted from thylakoid membranes were analysed by mass spectrometry (MS) and quantified by the iBAQ method (see Materials and Methods). MED4 was grown under both HL and LL. MIT9313 and SS120 were grown under LL. (a) Levels of PSI, PSII, cytochrome b6f and ATP synthase were calculated from the sum of the normalized ion counts (see Supplementary Table 1a-d) of subunits PsaA and PsaB (PSI), PsbA and PsbB (PSII), PetA, PetB and PetC (Cyt b6f), AtpF and AtpG (ATP synthase). These subunits were selected as representative of their respective protein complexes owing to their detection in all analyses; PSI:PSII ratios were calculated from the averages. (b) The normalized ion counts (see Supplementary Table 1, a-d) of the divinyl chlorophyll a/b light-harvesting protein isoforms identified. PcbA is the only Pcb isoform present in MED4. Although MIT9313 contains both pcbA and pcbB in its genome, only PcbB was identified. For the SS120 ecotype, in which 7 out of a total of 8 Pcb isoforms were identified (PcbC was not detected, as in a previous study26), the sum of all Pcb ion counts is also shown. The data are presented as the average of three repeat MS analyses of samples derived from 10 pooled cell cultures per ecotype/light level with dots representing values for individual analyses.
Fig. 7a also shows the levels of cytochrome b6f and the ATP synthase in both the MED4 grown under HL, MED4 grown under LL, MIT9313 and SS120 membranes. The lowest level of the cytochrome b6f complex was found in MED4 grown under HL, only slightly less than that of the SS120 ecotype. Cytochrome b6f was detected at significantly higher levels in MED4 grown under LL and MIT9313, approximately 3 times and 6 times that detected in the HL-grown MED4 sample, respectively. Furthermore, levels of cytochrome b6f are positively correlated with PSI (p < 0.00001) and PSII (p = 0.003, see Supplementary Fig. 3), highlighting the functional linkage between these complexes. The levels of the ATP synthase were less variable between the ecotypes with the HL-grown MED4 cells again having the lowest levels, approximately 2-fold higher in SS120 and approximately 3-fold higher in MIT9313 and LL-grown MED4.
The different ecotypes of Prochlorococcus marinus contain a variety of Pcb protein isoforms encoded within their genomes. MED4 only carries pcbA and the corresponding protein was detected in this analysis in both the HL- and LL-grown MED4 thylakoid membranes, with a three-fold higher level in LL-grown membranes (Fig. 7b). MIT9313 carries both pcbA and pcbB, but mass spectrometry analysis detected only the latter isoform and at less than the PcbA level of MED4 grown under LL. The genome of SS120 encodes eight Pcb isoforms and all except PcbC were detected, giving a combined Pcb level 7.0- 2.2- and 3.6-fold greater than that in HL- and LL-grown MED4, and MIT9313 respectively. The ratio of combined-Pcb:PSI was 2.08, 1.89, 0.66 and 5.74 in in HL-grown MED4, LL-grown MED4, MIT9313 and SS120 respectively.
Quantification of the Chl content of Prochlorococcus ecotypes MED4, MIT9313 and SS120
Chl b:a ratios in the thylakoid membranes purified from MED4 grown under HL, MED4 grown under LL, MIT9313 and SS120 were determined from reverse-phase HPLC of methanol-extracted pigments. Chl a and Chl b were separated (Supplementary Fig. 4) and collected before being buffer exchanged into 90% acetone. The amount of Chl a was calculated from the absorption at 663 nm using the molar extinction coefficient45 of 78.75 × 103 M−1.cm−1 and the amount of Chl b was calculated from the absorption at 647 nm using the molar extinction coefficient47 46.61 × 103 M−1.cm−1. The Chl b:a ratios were 0.06, 0.27, 1.52 and 2.36 for thylakoid membranes from HL-grown MED4, LL-grown MED4, MIT9313 and SS120 respectively (Supplementary Fig. 5a). In each pigment extract the combined Chl a and b concentration was calculated to be 26.6, 62.5, 115.8 and 91.4 μg Chl per mg of protein in HL-grown MED4, LL-grown MED4, MIT9313 and SS120 respectively (Supplementary Fig. 5b).
Discussion
Identification of domains of PSI complexes in Prochlorococcus ecotypes
Previous studies have shown that AFM can be used to identify photosystems in oxygenic phototrophs, and to determine their organisation in thylakoid membranes30-36,39,40. The most accurate measurement recorded by the AFM is the distance measured in the Z-plane, henceforth referred to as the height, which is typically accurate to 0.1 nm. The average heights measured for complexes in MED4 and MIT9313 membranes, and the average lateral distance between monomers, are consistent with both the crystal structure of the PSI trimer (PDB ID:1JB0) and previous measurements of PSI in thylakoid membranes39, allowing the identification of these complexes as trimeric PSI. This study39 found that the lumenal face of PSI-rich cyanobacterial thylakoids generally adsorbed to the mica substrate, which, in terms of AFM imaging, favours the marked topographic features of PSI over the low-topology PSII and cytochrome b6f complexes on the cytoplasmic face of the membrane. The same constraints apply to the topographs of Prochlorococcus membranes reported herein; thus, despite the abundance of PSII in all three ecotypes revealed by our mass spectrometry analyses, PSI-rich membrane domains feature in Fig. 1-5. The close packing and high density of PSI complexes in these domains, whether as trimers (Fig. 1, 2, 3, 6a, 6b) or in supercomplexes with a Pcb antenna ring (Fig. 4, 5, 6c), leaves no room for either PSII or cytochrome b6f complexes, which must reside in other domains of the thylakoid system. The presence of PSI-enriched areas of membrane is consistent with previous studies of thylakoid membranes from cyanobacteria and stromal lamellae in plant chloroplasts. These arrangements of PSI could optimise energy trapping and electron transport in these organisms, for example to mitigate ‘spillover’ of excitation energy when PSI and PSII are in close proximity48. The segregation of PSI from PSII in large domains is believed to be an adaptation that plants49, algae50, cyanobacteria39 and now Prochlorococcus appear to employ to ensure efficient PSII function. Another aspect of electron transport to consider is that the larger the PSI domains are, the further reduced plastocyanin molecules must diffuse from the cytochrome b6f complex in order to deliver electrons to the acceptor side of the PSI complex, which will change the rate at which linear electron flow takes place. The same is true for the diffusion of ferredoxin for the purposes of cyclic electron transport; it is possible that the size of the PSI domains in Prochlorococcus ecotypes optimises the balance between linear and cyclic electron flow to control the production of ATP and NADPH depending on the metabolic needs of the organism. For example, it has been shown in plants that differences in the size of grana, which predominantly contain PSII and the cytochrome b6f complex, can have a marked effect on cyclic and linear electron flow36. However, further elucidation of the organisation of PSII and the cytochrome b6f complex is required before conclusions can be drawn on cyclic and linear electron flows in Prochlorococcus and other cyanobacteria.
As discussed above, this Prochlorococcus study did not identify membrane domains where PSI is interspersed with other photosynthetic complexes such as PSII, possibly because the lumenal face of PSI-rich membranes tends to adsorb efficiently to the mica substrate used for AFM imaging. In contrast, previous studies did identify and image thylakoid membrane regions in T. elongatus and Synechococcus 7942 where PSI complexes co-localise with PSII and the cytochrome b6f complex39,40. Mass spectrometry analysis of thylakoids from Prochlorococcus ecotypes did however detect peptides from these two protein complexes; the PSI:PSII ratios in thylakoids from MED4 grown under HL, MED4 grown under LL, MIT9313 and SS120 cells were 1.11, 0.79, 0.97 and 0.53, respectively. The lower PSI:PSII ratio in SS120 membranes could be a consequence of the membrane area occupied by Pcb proteins, thereby decreasing the space available for PSI.
MED4
Imaging the native membrane arrangements of PSI and other complexes in Prochlorococcus ecotypes provides an opportunity to calculate membrane densities of PSI complexes and Chl pigments; in the case of the MED4 ecotype we were also able to compare the effects of growing the cells at 5 and 250 μmol photons m−2 s−1. This higher figure is lower than irradiances experienced by MED4 in surface waters15, but it is close to the limit we could achieve for laboratory-grown cultures. The averages for membrane packing density of PSI and Chl for MED4 grown under LL were 4303 and 411590 per μm2, respectively and the averages for MED4 grown under HL were 3553 and 341120 per μm2. At both light intensities the PSI domains imaged in this ecotype had a somewhat random distribution and were loosely packed in the thylakoid membrane, reminiscent of PSI organisation in Synechocystis sp. PCC 680339 and Synechococcus sp. PCC 794240. The major difference between the two light conditions is the five-fold increased prevalence of monomeric or dimeric PSI under HL growth conditions. The reasons for this are unclear, although it has been shown in thylakoid membranes from other cyanobacteria the probability that light energy absorbed by a PSI monomer within a trimeric complex has a probability of 0.35 of being trapped by a neighbouring monomer39. Adapting to HL conditions could require not only a lower packing density of PSI within the membrane, but also a switch away from a trimeric configuration. At lower light intensities there is greater need for efficient energy harvesting and very densely packed PSI complexes as well as a minimal number of monomers and dimers, as seen here for the LL-adapted MIT9313 ecotype, could confer an advantage at lower irradiances.
The MED4 strain is most abundant in surface waters of the open ocean15; closer to the surface of the ocean there is greater irradiance, so HL-adapted ecotypes such as MED4 do not need to waste metabolic resources by packing their thylakoid membranes full of Chl-containing proteins. Whilst having a greater light harvesting capacity could confer a competitive advantage, the oligotrophic waters that the MED4 strain occupies are very low in nutrients and synthesising photosynthetic reaction centres is costly to the cyanobacterium in terms of nitrogen and iron. It is likely that the markedly lower concentration of photosystems in this strain relative to the LL-adapted MIT9313 strain is a result of having to balance the need to absorb light energy with the metabolic requirements of producing photosystem complexes. The mass spectrometry data would seem to support this hypothesis as we see a large reduction in the levels of PSI, PSII, cytochrome b6f and Pcb proteins in HL-grown MED4 relative to LL-grown MED4.
The higher Chl b:a ratio measured for LL-grown MED4, and much higher value for the MIT9313 and SS120 ecotypes is in keeping with the idea that Chl b is utilised more extensively by LL-adapted strains. Having more Chl b increases spectral coverage in the blue region of the spectrum, which is the only light present deeper in the euphotic zone that these ecotypes occupy.
MIT9313
In contrast to MED4, the thylakoids of the LLIV ecotype MIT9313 contain domains that consist of densely packed PSI complexes, organised into pseudo-hexagonal arrays very similar in size and appearance to those previously imaged in T. elongatus39 and suggesting that this type of crystalline packing of PSI may be a common membrane motif across many different species of cyanobacteria. MIT9313 thylakoids (Fig. 3) had average PSI and Chl densities of 5583 and 536044 per μm2 respectively.
The structural model of the MIT9313 membrane, based on AFM data (Fig. 6a,e), showed that MIT9313 features a near-identical packing pattern to that of T. elongatus39. This previous study allowed calculation of the probability that harvested excitation energy is either trapped within a particular trimer (0.35), or that it can migrate to another trimer (0.07)39. Thus, it appears that the main point of having tightly packed PSI complexes is not to create inter-trimer energy transfer networks, but simply to achieve the highest possible density of Chl pigments in the thylakoid membranes. We speculate that these more densely packed PSI domains in MIT9313 are an adaptation to the lower light conditions and the higher availability of nutrients in the deeper euphotic zone16. With iron and nitrogen less scarce in this zone the cost of synthesising photosystems becomes less severe and the cyanobacteria are able to produce more PSI relative to their HL-adapted counterparts. By packing PSI in a pseudo-hexagonal lattice MIT9313 gains a competitive advantage in the LL environment of the lower euphotic zone.
Using cryo-EM tomography data obtained for MED4 and MIT931329 it was possible to estimate that these HL-adapted and LL-adapted ecotypes contain ~6 μm2 and ~22 μm2 of thylakoid membrane area, respectively. Considering that there are 1.8 times as many PSI complexes in MIT9313 thylakoid membranes per unit area and cells house approximately 3.7-fold more membrane, each MIT9313 cell could house as much as 6.7 times the number of PSI complexes relative to the HL-adapted MED4 ecotype. Pigment analysis showed that thylakoid membranes from MIT9313 had significantly more Chl per mg of protein than MED4 and SS120 (Supplementary Fig. 5b), supporting the hypothesis that MIT9313 has adapted to LL by maximising the number of pigments in the thylakoid membrane.
SS120
This ecotype employs a different strategy to combat the limitations of LL levels, and we found a different organisation of PSI in the LLII strain SS120; it was possible to image densely packed Pcb-PSI supercomplexes confirming the structures of isolated complexes25 and additionally showing the organisation of these supercomplexes in the native membrane environment. SS120, which has abundant Pcb proteins (Fig. 7b), contains large membrane domains comprising hundreds of trimeric PSI complexes, with each trimer surrounded by a multi-subunit ring (Fig. 4, 5); these complexes are very similar in appearance and dimensions to the Pcb-PSI supercomplex25,26 and were assigned as Pcb-PSI supercomplexes. The SS120 membrane patch measured in Fig. 5 contained 893 Pcb-PSI supercomplexes per μm2, with 2679 PSI complexes per μm 2 and 498294 Chl pigments per μm2, assuming 15 Chl molecules per Pcb protein. This 52% reduction in the levels of PSI in SS120 relative to MIT9313 is in good agreement with the mass spectrometry analysis of purified thylakoids which reveals a 59% reduction in the total number of PSI complexes. Relative to MIT9313, adaptation to LL in SS120 appears to involve the synthesis and assembly of fewer PSI reaction centres and filling the membrane area gained with Pcb light-harvesting complexes. This strategy is likely to require a lower energy input for biosynthesis, given the lower ratio of proteins to be synthesised per bound pigment (kDa/pigment) in antenna proteins generally, with respect to reaction centre complexes. For the Pcb proteins found in Prochlorococcus, assuming similarity with IsiA proteins, which bind 15 Chls, this ratio was estimated to be 2.6, whereas it is 6.5 for marine cyanobacterial phycoerythrin, and 3.7 for cyanobacterial PSI22. Thus, a combination of nitrogen and light limitation disfavours phycobilisomes and favours Pcb antenna complexes, in Prochlorococcus SS120 at least. These features of SS120 confer significant competitive advantages over other picoplankton, allowing this ecotype to thrive in low nutrient oligotrophic waters found in the open ocean5. SS120 deploys an increased level of Chl b to adapt to the LL environment. Pigment analyses of SS120 membranes show a Chl b:a ratio 39.3-fold, 8.7-fold and 1.6-fold greater than for HL-grown MED4, LL-grown MED4 and MIT9313, respectively (Supplementary Fig. 5). This increased level of Chl b gives SS120 greater access to the blue regions of the solar spectrum that represent the only light available at a depth of 200 meters18. Furthermore, The roughly double PSII:PSI ratio of the SS120 ecotype with respect to the MIT9313 ecotype may require the presence of a Pcb antenna ring to increase the functional cross section of PSI to balance the excitation of PSI with PSII, in order to ensure efficient linear electron transport. The above considerations also apply to the formation of Pcb-PSII complexes, although the membrane regions housing them were not imaged in the present study. Finally, the positive correlation between levels of PSI and cytochrome b6f complexes, and the increased proportion of PSII that emerged from mass spectrometry analysis of SS120 might reflect an adaptation to life in iron-deficient environments. Both PSI and cytochrome b6f complexes place demands on the supply of iron, whereas PSII has a lower iron requirement.
Mass spectrometry analysis of SS120 thylakoid membranes also highlighted the presence of approximately 7.0, 1.8 and 3.5 times more total Pcb proteins relative to HL-MED4, and MIT9313 membranes respectively. It has been reported that the Pcb-PSI supercomplex comprises an 18-membered ring of PcbG25,26; in agreement with this analysis, our mass spectrometry data shows that PcbG is the most abundant Pcb protein in SS120, and thus likely the identity of the Pcb subunits imaged in Fig. 4. The presence of 6 other Pcb proteins is consistent with previous analysis of the antenna complexes in SS120 cells26. It has been shown that PcbA acts as an antenna for the PSII complex and PcbG and PcbC can make up the ring surrounding trimeric PSI complexes26. Despite the presence of the pcbC gene in this ecotype the PcbC protein could not be detected. PcbC is involved in the iron starvation response in SS120, where it replaces the PcbG protein in the 18 membered ring around the PSI trimer26. However, the SS120 cells studied in the present work were not grown under iron starvation conditions, which accounts for the absence of PcbC from our mass spectrometry analysis. Although present in the cell, the functions of the other Pcb proteins, PcbB, D, E, F, H, are unknown. However, it is interesting to note that the ratio of total Pcb:PSI is 5.74; close to the ratio of 6 required for a Pcb-PSI supercomplex composed of 18 Pcb proteins and 3 PSI complexes. This similarity could imply that these other Pcb proteins are involved in forming Pcb-PSI supercomplexes, but this interpretation must be viewed with caution as it has been shown that a certain population of Pcb proteins associates with PSII in this ecotype24. It is likely that there is a population of PSI complexes that are not associated with a Pcb ring, which we were not able to image with AFM.
In conclusion, we have presented the first high resolution imaging of the photosystems in the thylakoid membranes from one HL-adapted ecotype and two LL-adapted ecotypes of the globally important cyanobacterium, Prochlorococcus. The AFM topographs were used as the basis for constructing structural models of PSI-rich domains of thylakoids from Prochlorococcus ecotypes, MED4, MIT9313 and SS120 that reveal the packing patterns of PSI complexes. Additionally we have used mass spectrometry to quantify the major photosynthetic protein complexes in each ecotype. The organisation and composition of thylakoid membranes can be related to the particular environmental niches occupied by each ecotype. Whilst this is the first step in understanding the architecture of the photosynthetic membrane system in Prochlorococcus there are still several questions that remain unanswered, including the organisation of PSII and the cytochrome b6f complexes. It has been shown that Pcb proteins act as antenna complexes for PSII but how these supercomplexes associate in the membrane environment is still unclear, as are the structures of the various Pcb proteins. With such knowledge, it would be possible to build a functional model of the complete photosynthetic membrane system in one of the most important photosynthetic organisms.
Materials and methods
Cell growth
LL-grown Prochlorococcus spp. MED4, SS120 and MIT9313 were grown at 22°C in PCR-S11 medium51 at a white light intensity of 5 μmol photons m−2 s−1. HL-grown MED4 grown at a white light intensity of 250 μmol photons m−2 s−1; a comparable light intensity to HL-grown MED4 in previous studies52. Cells were harvested in the exponential phase by centrifugation at 10000 rpm using a JA-25.50 rotor (Beckman) at 20°C for 30 minutes and flash frozen in liquid nitrogen before storing at −80°C for subsequent membrane isolation.
Crude membrane preparation
A volume of 1.0 ml of resuspended cells was added to 1.0 ml of glass beads and cells were broken by 6 rounds of bead beating for 30 seconds in a Mini bead beater (Biospec products). The cell lysate was removed from the glass beads by pipette and then layered onto a 11.5 ml sucrose step gradient composed of 9.5 ml of 30% (w/w) sucrose on a 2.0 ml 50% (w/w) sucrose cushion. The sucrose gradient was then centrifuged at 30000 rpm in an SW41 rotor (Beckman) at 4 °C for 30 minutes. The thylakoid membranes were present at the interface between the 30% and 50% (w/w) sucrose volumes, which were harvested and either immediately used for AFM analysis or were flash frozen in liquid nitrogen and then stored at −80 °C for later use
Preparation of membranes for AFM analysis
Harvested crude membranes were loaded onto 11.5 ml continuous sucrose gradients made from equal volumes of 20% and 50% (w/w) sucrose which contained 0.1% digitonin (w/w). These sucrose gradients were centrifuged at 40000 rpm in an SW41 rotor (Beckman) at 4°C for 2 hours. The thylakoid membranes were present as a green smear running roughly the length of the gradients; membranes were harvested from throughout the gradient and used for AFM analysis.
AFM imaging
Instrumentation
Membrane samples were imaged using a multimode VIII AFM with a Nanoscope 8.0 controller (Bruker Nano Surfaces Business).
Sample adsorption
Approximately 5 μl of membrane sample was pipetted onto a freshly cleaved mica substrate before 45 μl of buffer containing 10 mM HEPES pH 7.5 and 100 mM KCl was pipetted onto the mica. The membranes were incubated for 1 hour before being washed 3 times with 50 μl of the same buffer, with the final wash left on the surface. The mica disk was then mounted onto the (J-scanner) AFM scanner.
Sample imaging
Samples were imaged using Peak Force Nanomechanical Mapping™ (PF-QNM) mode under liquid using a Peak Force frequency of 2 kHz. An SNL AFM probe (Bruker Nano Surfaces Business) mounted in a MTFML fluid cell (Bruker Nano Surfaces Business) was used to image membrane samples. Once the probe had been loaded into the fluid cell the reservoirs were filled with buffer containing 10 mM HEPES pH 7.5 and 100 mM KCl and the cell was mounted onto the AFM at which point the laser was aligned with the probe. A Peak Force amplitude of 5-20 nm was used and images were taken at 256 × 256 or 512 × 512 pixels. The force imparted on the sample during imaging was varied between 5-1000 pN and image processing was performed using NanoScope Analysis 1.9 or Gwyddion v2.4753. Heights and distances are expressed as the mean ± the standard deviation.
Construction of structural models for Prochlorococcus MIT9313, MED4, SS120 ecotypes
The construction of structural models for PSI containing thylakoid membranes, based on AFM topographs, follows the protocol employed earlier for T. elongatus PSI domains39. Specifically, the crystal structure, PDB:1JB041 of PSI trimers from T. elongatus is used to match the protrusion profiles of PsaC-D-E subunits onto the topological features revealed by AFM. Mathematica54 was employed with image recognition methods to determine the position and orientation of each trimer with respect to the AFM density. The structural models were manually refined iteratively using VMD55. For ecotypes MIT9313 (Fig. 6a) and MED4 (Fig. 6b), the corresponding structural models contain only PSI-trimers, whereas for SS120 (Fig. 6c) the membrane domain contains also surrounding Pcb units modelled in terms of CP43, PDB ID: 3WU244. Even though the structural models show atomistic detail (Figs. 6a, 6b, 6c) as determined by the underlying crystal structures41,44, the resolution of the models should be considered to be limited by the native AFM resolution, namely, 9.9 Å, 14.6 Å, 18.6 Å, for MIT9313 (Fig. 6a), MED4 (Fig. 6b), and SS120 (Fig. 6c) models, respectively. Excitonic connectivity between PSI subunits in a thylakoid membrane was calculated on an effective Hamiltonian formulation39,56.
Proteomic analysis of thylakoid membranes
Thylakoid membranes (50 μg protein) from Prochlorococcus marinus ecotypes MED4, SS120 and MIT9313 were processed using a 2-D clean-up kit (GE Healthcare) to isolate the proteins from lipids and cofactors. The extracted proteins were solubilized, S-alkylated, digested and analysed by nano-flow liquid chromatography coupled to mass spectrometry as previously described57. Tryptic peptides were resolved using a 3-hour gradient and the mass spectrometer was programmed for data dependent acquisition with 10 product ion scans (centroid, resolution 15000, automatic gain control 5e4, maximum injection time 20 ms, isolation window 1.2 Th, normalized collision energy 32, intensity threshold 2.5e5) per full mass spectrometry scan (profile, resolution 60000, automatic gain control 3e6, maximum injection time 100 ms). Protein identification was carried out using MaxQuant v. 1.5.3.3058 to search the Prochlorococcus marinus reference proteome databases for ecotypes: (1) MED4 (www.uniprot.org/proteomes/UP000001026, 1924 proteins, downloaded on 17-03-16), (2) SS120 (www.uniprot.org/proteomes/UP000001420, 1881 proteins, downloaded on 17-03-16), (3) MIT9313 (www.uniprot.org/proteomes/UP000001423, 2830 proteins, downloaded on 31-10-16). Default database search parameters were used and protein quantification was enabled by selecting the iBAQ option59,60 (Intensity-Based Absolute Quantification, a widely accepted and validated label-free protein quantification method). Identification and quantitative results were further processed using Perseus software v. 1.5.3.261. Protein amounts were normalized to compensate for random variation in sample loadings and tryptic peptide spectral acquisition patterns by applying a factor derived from the ion intensity of the trypsin auto-digestion peptide VATVSLPR62 (see Supplementary Table 2) present in all analyses.
Pigment analysis
Membrane samples (140 μg protein) were pelleted by centrifugation at 270000 × g at 4 °C for 1 hour. Chls were extracted from membrane pellets by addition of 100 μl methanol and vortexing at room temperature under dim green light. The extracted pigments were separated from insoluble material by centrifugation (15,000 rpm, 4°C, 15 mins) and 80 μl of the supernatant was immediately analysed by reverse phase high performance liquid chromatography (HPLC) on an Agilent 1200 HPLC system using a Discovery® HS C18 5 μm column (column dimensions: 25 cm × 4.6 mm) pre-equilibrated in 84:9:7 acetonitrile:methanol:water (solvent A). Pigments were separated at a solvent flow rate of 1 ml min−1 using a mobile phase consisting of solvent A and solvent B (68:32 methanol:ethyl acetate) and a linear gradient from 100% solvent A to 100% solvent B over 12 minutes followed by isocratic elution with 100% solvent B for 6 minutes63. The column was re-equilibrated with 100% solvent A for 6 minutes prior to injection of the next sample. Absorbance was monitored at 653 nm and 663 nm using a diode-array detector; divinyl-chlorophyll b and divinyl-chlorophyll a eluted at ~13.8 minutes and ~15.3 minutes, respectively, as determined by their absorbance spectra.
Collected Chl a and Chl b solutions were placed in an Eppendorf concentrator plus and centrifuged under vacuum until the solvent evaporated. The solid pigments were then resuspended in 90% (v/v) acetone and the Chl a content was calculated from measuring the absorption at 664 nm using the extinction coefficient of 78.75 × 103 M−1.cm−1. The Chl b content was calculated using the absorption at 647 nm using an extinction coefficient of 46.61 × 103 M−1.cm−1 respectively45. All absorption spectra were taken using a Cary 60 (Agilent technologies).
Protein content calculation
The protein concentration of samples was calculated as previously described64.
Purification of IsiA-PSI supercomplexes
IsiA-PSI supercomplexes were purified as previously described27.
TEM imaging
A solution containing IsiA-PSI supercomplexes was pipetted (~20 μl) onto a charged carbon coated grid and incubated for 2 minutes. The sample was negatively stained with 0.75% w/v uranyl formate and imaged with a Philips CM100 microscope that was equipped with a Gatan Ultrascan 667 CCD camera. Particles were viewed with magnification of x1000- x52000. 52 particles were chosen from a field of IsiA-PSI supercomplexes observed by negative stain TEM and image processing was performed by Digital Micrograph (Gatan. Inc.) and the IMAGIC-5 image processing system
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (http://proteomecentral.proteomexchange.org) with the dataset identifier PXD013506. All other data can be obtained from the corresponding author upon request. The following figures have associated raw data: Figs. 7, Supplementary Figs. 2-5.
Supplementary Material
Acknowledgements
This work was supported by Advanced Award 338895 from the European Research Council which funded CM-C, PJJ, JWC, PQ and provided partial support for CNH. CNH and AH also gratefully acknowledges financial support from the Biotechnology and Biological Sciences Research Council (BBSRC UK), award number BB/M000265/1. CNH, MS and ZLS were supported by the Photosynthetic Antenna Research Center (PARC), an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Award Number DE-SC 0001035. MS and ZLS were also supported by the National Science Foundation (MCB1616590) and the National Institutes of Health (NIH 9P41GM104601) DJS acknowledges funding from NERC (NE/N003241/1) and The Leverhulme Trust (RPG-2014-354). MJD acknowledges support from the Biotechnology and Biological Sciences Research Council (UK) (BB/M012166/1). MPJ would like to acknowledge BBSRC Grant BB/P002005/1 and the Grantham Centres for Sustainable Futures, University of Sheffield, for G.E.M.’s studentship.
Footnotes
Competing Interest Statement
The authors declare no competing interests.
References
- 1.Flombaum P, Gallegos JL, Gordillo RA, Rincón J, Zabala LL, Jiao N, Karl DM, Li WK, Lomas MW, Veneziano D & Vera CS Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl Acad. Sci USA 110, 9824–9829 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huston MA & Wolverton S The global distribution of net primary production: resolving the paradox. Ecol. Monographs 79, 343–377 (2009). [Google Scholar]
- 3.Partensky F & Garczarek L Prochlorococcus: Advantages and limits of minimalism. Annu. Rev. Marine. Sci. 2, 305–331 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Goericke R & Repeta DJ The pigments of Prochlorococcus marinus: the presence of divinyl chlorophyll a and b in a marine procaryote. Limnol. Oceanogr. 37, 425–433 (1992). [Google Scholar]
- 5.Partensky F, Hess WR & Vaulot D Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63, 106–127 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Partensky F, Hoepffner N, Li W & Ulloa O Photoacclimation of Prochlorococcus sp. (Prochlorophyta) strains isolated from the North Atlantic and the Mediterranean Sea. Plant Physiol. 101, 285–296 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore LR, Goericke R & Chisholm SW Comparative physiology of Synechococcus and Prochlorococcus: influence of light and temperature on growth, pigments, fluorescence and absorptive properties. Marine Ecol. Progress Series 116, 259–275 (1995). [Google Scholar]
- 8.Moore LR, Goericke R & Chisholm SW Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature, 393, 464–467 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Ferris MJ & Palenik B Niche adaptation in ocean cyanobacteria. Nature 396, 226–228 (1998). [Google Scholar]
- 10.Urbach E, Scanlan DJ, Distel DL, Waterbury JB & Chisholm SW Rapid diversification of marine picophytoplankton with dissimilar light-harvesting structures inferred from sequences of Prochlorococcus and Synechococcus (Cyanobacteria). J. Mol. Evol. 46, 188–201 (1998). [DOI] [PubMed] [Google Scholar]
- 11.West NJ & Scanlan DJ Niche-partitioning of Prochlorococcus populations in a stratified water column in the Eastern North Atlantic Ocean. Appl. Environ. Microbiol. 65, 2585–2591 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rocap G, Distel DL, Waterbury JB & Chisholm SW Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl. Environ. Microbiol. 68, 1180–1191 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zwirglmaier K, Jardillier L, Ostrowski M, Mazard S, Garczarek L, Vaulot D, Not F, Massana R, Ulloa O & Scanlan DJ Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ. Microbiol. 10, 147–161 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Martiny AC, Tai APK, Veneziano D, Primeau F & Chisholm SW Taxonomic resolution, ecotypes and the biogeography of Prochlorococcus. Environ. Microbiol. 11, 823–832 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Malmstrom RR, Coe A, Kettler GC, Martiny AC, Frias-Lopez J, Zinser ER & Chisholm SW Temporal dynamics of Prochlorococcus ecotypes in the Atlantic and Pacific oceans. The ISME J. 4, 1252 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Biller SJ, Berube PM, Lindell D & Chisholm SW Prochlorococcus: the structure and function of collective diversity. Nature Rev. Microbiol 13, 13 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Moore LR, & Chisholm SW. Photophysiology of the marine cyanobacterium Prochlorococcus: ecotypic differences among cultured isolates. Limnol. Oceanogr. 44, 628–638 (1999). [Google Scholar]
- 18.Kirk JTO The nature of the underwater light field Light and Photosynthesis In Aquatic Ecosystems, Cambridge University Press; (1994) [Google Scholar]
- 19.Palenik BP & Haselkorn R. Multiple evolutionary origins of prochlorophytes, the chlorophyll b-containing prokaryotes. Nature 355, 265–267 (1992). [DOI] [PubMed] [Google Scholar]
- 20.Urbach E, Robertson DL & Chisholm SW Multiple evolutionary origins of prochlorophytes within the cyanobacterial radiation. Nature 355, 267 (1992). [DOI] [PubMed] [Google Scholar]
- 21.Scanlan DJ & West NJ Molecular ecology of the marine cyanobacterial genera Prochlorococcus and Synechococcus. FEMS Microbiol. Ecol. 40, 1–12 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Ting CS, Rocap G, King J & Chisholm SW Cyanobacterial photosynthesis in the oceans: the origins and significance of divergent light-harvesting strategies. Trends Microbiol. 10, 134–142 (2002) [DOI] [PubMed] [Google Scholar]
- 23.La Roche J, Van der Staay GWM, Partensky F, Ducret A, Aebersold R, Li R, Golden SS, Hiller RG, Wrench PM, Larkum AWD & Green BR Independent evolution of the prochlorophyte and green plant chlorophyll a/b light-harvesting proteins. Proc. Natl Acad. Sci USA, 93, 15244–15248 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen M & Bibby TS Photosynthetic apparatus of antenna-reaction centres supercomplexes in oxyphotobacteria: insight through significance of Pcb/IsiA proteins. Photosynthesis Res. 86, 165–173 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Bibby TS, Nield J, Partensky F & Barber J Oxyphotobacteria: Antenna ring around photosystem I. Nature 413, 590 (2001a). [DOI] [PubMed] [Google Scholar]
- 26.Bibby TS, Mary I, Nield J, Partensky F & Barber J Low-light-adapted Prochlorococcus species possess specific antennae for each photosystem. Nature 424, 1051 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Bibby TS, Nield J & Barber J Iron deficiency induces the formation of an antenna ring around trimeric photosystem I in cyanobacteria. Nature 412, 743 (2001b). [DOI] [PubMed] [Google Scholar]
- 28.Boekema EJ, Hifney A, Yakushevska AE, Piotrowski M, Keegstra W, Berry S, Michel KP, Pistorius EK & Kruip J A giant chlorophyll–protein complex induced by iron deficiency in cyanobacteria. Nature 412, 745 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Ting CS, Hsieh C, Sundararaman S, Mannella C & Marko M Cryo-electron tomography reveals the comparative three-dimensional architecture of Prochlorococcus, a globally important marine cyanobacterium. J. Bacteriol. 189, 4485–4493 (2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirchhoff H, Lenhert S, Büchel C, Chi L & Nield J Probing the organization of photosystem II in photosynthetic membranes by atomic force microscopy. Biochemistry 47, 431–440 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Sznee K, Dekker JP, Dame RT, van Roon H, Wuite GJL & Frese RN Jumping mode atomic force microscopy on grana membranes from spinach. J. Biol. Chem. 286, 39164–39171 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson MP, Vasilev C, Olsen JD & Hunter CN Nanodomains of cytochrome b6f and photosystem II complexes in spinach grana thylakoid membranes. Plant Cell 26, 3051–3061 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Onoa B, Schneider AR, Brooks MD, Grob P, Nogales E, Geissler PL, Niyogi KK & Bustamante C Atomic force microscopy of photosystem II and its unit cell clustering quantitatively delineate the mesoscale variability in Arabidopsis thylakoids. PloS ONE 9, p.e101470 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phuthong W, Huang Z, Wittkopp TM, Sznee K, Heinnickel ML, Dekker JP, Frese RN, Prinz FB & Grossman AR The use of contact mode atomic force microscopy in aqueous medium for structural analysis of spinach photosynthetic complexes. Plant Physiol. 169, 1318–1332 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tietz S, Puthiyaveetil S, Enlow HM, Yarbrough R, Wood M, Semchonok DA, Lowry T, Li Z, Jahns P, Boekema EJ & Lenhert S Functional implications of photosystem II crystal formation in photosynthetic membranes. J. Biol. Chem. 290, 14091–14106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wood WH, MacGregor-Chatwin C, Barnett SF, Mayneord GE, Huang X, Hobbs JK, Hunter CN & Johnson MP Dynamic thylakoid stacking regulates the balance between linear and cyclic photosynthetic electron transfer. Nature Plants 4, 116 (2018) [DOI] [PubMed] [Google Scholar]
- 37.Liu LN & Scheuring S Investigation of photosynthetic membrane structure using atomic force microscopy. Trends in Plant Science 18, 277–286 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Kumar S, Cartron ML, Mullin N, Qian P, Leggett GL, Hunter CN & Hobbs JK Direct imaging of protein organisation in an intact bacterial organelle using high-resolution Atomic Force Microscopy. ACS Nano 11, 126–133 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacGregor-Chatwin C, Sener M, Barnett SF, Hitchcock A, Barnhart-Dailey MC, Maghlaoui K, Barber J, Timlin JA, Schulten K & Hunter CN Lateral segregation of photosystem I in cyanobacterial thylakoids. The Plant Cell 29, 1119–1136 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casella S, Huang F, Mason D, Zhao GY, Johnson GN, Mullineaux CW & Liu LN Dissecting the native architecture and dynamics of cyanobacterial photosynthetic machinery. Molecular Plant 10, 1434–1448 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jordan P, Fromme P, Witt HT, Klukas O, Saenger W & Krauß N Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature 411, 909 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Malavath T, Caspy I, Netzer-El SY, Klaiman D & Nelson N Structure and function of wild-type and subunit-depleted photosystem I in Synechocystis. Biochimica et Biophysica Acta (BBA)-Bioenergetics 1859, 645–654 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Kubota-Kawai H, Mutoh R, Shinmura K, Sétif P, Nowaczyk MM, Rögner M, Ikegami T, Tanaka H & Kurisu G X-ray structure of an asymmetrical trimeric ferredoxin–photosystem I complex. Nature Plants 4, 218–224 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Umena Y, Kawakami K, Shen JR & Kamiya N Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473, 55 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Waldbauer JR, Rodrigue S, Coleman ML & Chisholm SW Transcriptome and proteome dynamics of a light-dark synchronized bacterial cell cycle. PloS ONE 7, e43432 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Domínguez-Martín MA, Gómez-Baena G, Díez J, López-Grueso MJ, Beynon RJ & García-Fernández JM Quantitative Proteomics Shows Extensive Remodeling Induced by Nitrogen Limitation in Prochlorococcus marinus SS120. MSystems 2, e00008–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeffrey ST & Humphrey GF New spectrophotometric equations for determining chlorophylls a, b, c1 and c2 in higher plants, algae and natural phytoplankton. Biochemie und Physiologie der Pflanzen 167, 191–194 (1975). [Google Scholar]
- 48.Biggins J & Bruce D Regulation of excitation energy transfer in organisms containing phycobilins. Photosynthesis research 20, 1–34 (1989). [DOI] [PubMed] [Google Scholar]
- 49.Andersson B & Anderson JM Lateral heterogeneity in the distribution of chlorophyll-protein complexes of the thylakoid membranes of spinach chloroplasts. Biochimica et Biophysica Acta (BBA)-Bioenergetics 593, 427–440 (1980). [DOI] [PubMed] [Google Scholar]
- 50.Engel BD, Schaffer M, Cuellar LK, Villa E, Plitzko JM & Baumeister W Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography. Elife 4, e04889 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rippka R, Coursin T, Hess W, Lichtlé C, Scanlan DJ, Palinska KA, Iteman I, Partensky F, Houmard J & Herdman M Prochlorococcus marinus Chisholm et al. 1992 subsp. pastoris subsp. nov. strain PCC 9511, the first axenic chlorophyll a2/b2-containing cyanobacterium (Oxyphotobacteria). International Journal of Systematic and Evolutionary Microbiology 50, 1833–1847 (2000). [DOI] [PubMed] [Google Scholar]
- 52.Bonisteel EM, Turner BE, Murphy CD, Melanson JR, Duff NM, Beardsall BD, Xu K, Campbell DA & Cockshutt AM Strain specific differences in rates of Photosystem II repair in picocyanobacteria correlate to differences in FtsH protein levels and isoform expression patterns. PloS ONE 13, e0209115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nečas D & Klapetek P Gwyddion: an open-source software for SPM data analysis. Open Physics 10, 181–188 (2012). [Google Scholar]
- 54.Wolfram Research, Inc., Mathematica, Version 11.3, Champaign, IL: (2018). [Google Scholar]
- 55.Humphrey W; Dalke A; Schulten K VMD − Visual Molecular Dynamics. J. Mol. Graphics 14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
- 56.Sener MK, Park S, Lu D, Damjanović A, Ritz T, Fromme P & Schulten K Excitation migration in trimeric cyanobacterial photosystem I. J. Chem. Phys. 120, 11183–11195 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Hitchcock A, Jackson PJ, Chidgey JW, Dickman MJ, Hunter CN & Canniffe DP Biosynthesis of chlorophyll a in a purple bacterial phototroph and assembly into a plant chlorophyll-protein complex ACS Synth. Biol 5, 948–954 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Cox J & Mann M MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W & Selbach M Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011). [DOI] [PubMed] [Google Scholar]
- 60.Fabre B, Lambour T, Bouyssié D, Menneteau T, Monsarrat B, Burlet-Schiltz O & Bousquet-Dubouch M-P Comparison of label-free quantification methods for the determination of protein complexes subunits stoichiometry. EuPA Open Proteomics 4, 82–86 (2014). [Google Scholar]
- 61.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein M, Geiger T, Mann M & Cox J The Perseus computational platform for comprehensive analysis of (prote) omics data. Nature Methods 13, 731–740 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Li L, Bebek G, Previs SF, Smith JD, Sadygov RG, McCullough AJ, Willard B & Kasumov T Proteome dynamics reveals pro-inflammatory remodeling of plasma proteome in a mouse model of NAFLD. Journal of proteome research 15, 3388–3404 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garcia‐Plazaola JI & Becerril JM A rapid high‐performance liquid chromatography method to measure lipophilic antioxidants in stressed plants: simultaneous determination of carotenoids and tocopherols. Phytochemical Analysis 10, 307–313 (1999). [Google Scholar]
- 64.Kalb VF Jr & Bernlohr RW A new spectrophotometric assay for protein in cell extracts. Analytical biochemistry 82, 362–371 (1977). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (http://proteomecentral.proteomexchange.org) with the dataset identifier PXD013506. All other data can be obtained from the corresponding author upon request. The following figures have associated raw data: Figs. 7, Supplementary Figs. 2-5.