The advent of single-cell RNA sequencing (scRNA-seq) has offered great opportunities for studying cardiac pathology at single cell resolution. To date, however, scRNA-seq of adult cardiomyocytes (CMs) remains limited owing to technical difficulties in single CM isolation. Adult CMs are large (~125×25μm), rod-shaped cells whose structure precludes isolation by fluorescence-activated cell sorting (FACS) or commercial single-cell microfluidic platforms1. Here, we report on successful isolation of viable single CMs through use of large particle FACS (LP-FACS), designed for the isolation of large objects, including worm/fly embryos and cell clusters. Our study demonstrates that LP-FACS enables rapid, high-throughput isolation of CMs.
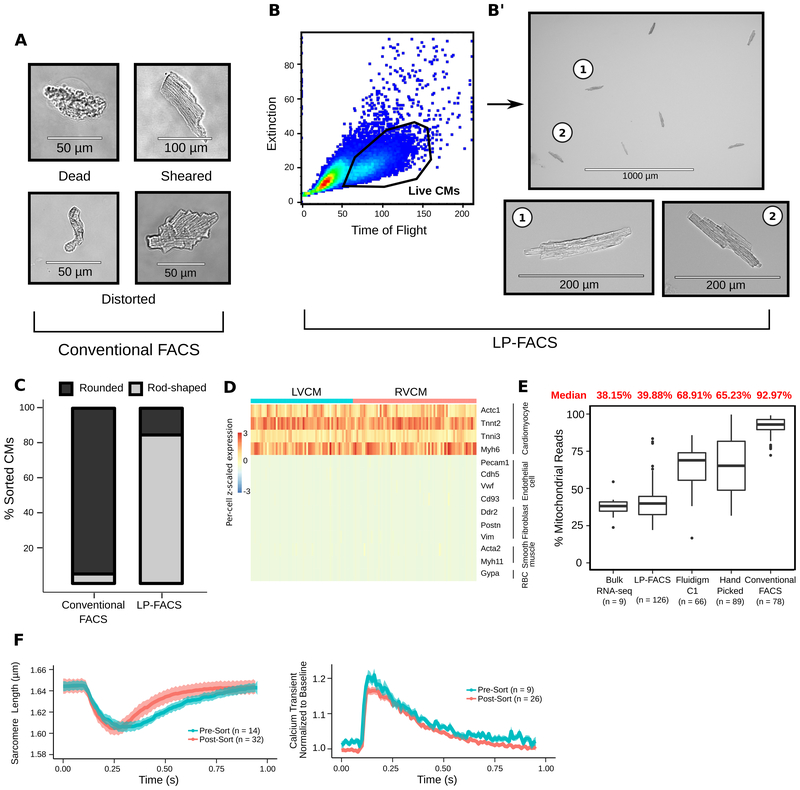
To generate viable CMs, we performed Langendorff perfusion of cannulated 12-week-old mouse heart with a standard protocol. We first attempted to isolate CMs with conventional FACS, using a commercial cell sorter with a 130 μm microfluidic channel. Live and dead CMs were identified by DAPI staining. Despite selecting DAPI-negative CMs, the vast majority of isolated cells were dead or had distorted structures (A), suggesting that the small flow size (relative to CM size) leads to terminal damage of live CMs.
We next attempted to isolate CMs through LP-FACS, using a channel size of 500 μm (COPAS-FP-500, Union Biometrica). Notably, live, rod-shaped myocytes could be easily separated from other cells with the time-of-flight (measuring axial length) and optical extinction (measuring optical density) parameters (B). When sorted, this population was enriched with healthy-appearing myocytes with intact sarcomeres (B’). Unlike conventional FACS, where only ~5% of DAPI-negative cells were rod-shaped, ~85% of the LP-FACS-isolated CMs were rod-shaped (C). Area measurement demonstrates that LP-FACS isolates CMs across the spectrum of normal size ranges (bioRxiv: https://doi.org/10.1101/654954). Additionally, LP-FACS could separate CM populations based on endogenous fluorescence (bioRxiv: https://doi.org/10.1101/654954). These support the methodological superiority of LP-FACS for CM sorting.
To test whether isolated CMs can be used for transcriptomic assays, we isolated RNA from bulk-sorted CMs (500–1500 cells) and assessed RNA quality using the Advanced Analytical Fragment Analyzer system (bioRxiv: https://doi.org/10.1101/654954. Samples presented with RNA quality number of 10.0, indicating maximal RNA quality.
We subsequently performed a proof-of-concept scRNA-seq experiment with LP-FACS-isolated adult CMs. The resulting cells selectively expressed CM markers (D). We assessed the quality of our sequenced CM libraries by analyzing the percentage of reads originating from mitochondrial transcripts. In a damaged cell, cytoplasmic RNA leaks out of the cell, while RNAs in the mitochondria are retained. Thus, a high percentage of reads mapped to mitochondrial genes indicates likely cell damage. In LP-FACS-isolated CMs, mitochondrial transcripts accounted for ~40% of reads, consistent with bulk studies2 (E). However, mitochondrial transcripts from previous single-cell studies3–5 showed abnormally high percentages of sequenced reads. These high percentages indicate likely cellular damage/rupture, supporting the use of LP-FACS to generate improved quality CM scRNA-seq libraries.
Furthermore, we detected ~3700 genes at a subsampled read depth of 200,000 reads, comparable to or superior than other CM scRNA-seq datasets (bioRxiv: https://doi.org/10.1101/654954). We successfully recovered rod-shaped CMs fixed using 80% ethanol through LP-FACS and performed scRNA-seq (bioRxiv: https://doi.org/10.1101/654954). This can be useful in experiments where immediate sorting is infeasible.
We next investigated the functional properties of LP-FACS-isolated CMs using Ionoptix (1Hz/20V/15ms). Post-sort CMs maintained sarcomeric shortening and calcium transients comparable to pre-sorted CMs (F). In particular, several important parameters, including fractional shortening, change in calcium transient, and calcium transient decay τ among others, were equivalent pre- and post-sort (bioRxiv: https://doi.org/10.1101/654954). These support maintenance of contractile function in LP-FACS-isolated CMs. While differences in CM relaxation kinetics were detected, we believe that minor technical differences contributed to this, and further optimization of sorting conditions, such as applying strict selection criteria of pre-sorted CMs, will eliminate the functional discrepancies.
Together, our data demonstrate the methodological superiority of LP-FACS for isolating single adult CMs, compared to existing methods. To our knowledge, LP-FACS is the only approach that enables both generation of high-quality scRNA-seq libraries and allows for isolation of cells for functional analysis. It is important to note that successful use of LP-FACS requires a protocol for dissociating hearts into a single cell suspension. Future improvements in dissociation methods, particularly for human tissues, will facilitate improved recovery of single CMs via LP-FACS. We envision this technology will enable researchers to integrate transcriptomic and functional data for a range of cardiac disease models at the single cell level.
(A) CM images obtained from conventional FACS, representing the vast majority of sorted cells. (B) CM isolation through LP-FACS. (C) % rod-shaped CMs from conventional FACS vs LP-FACS. 200–400 CMs were analyzed per sorting approach. (D) Heatmap of genes representing cardiac cell types. LVCM, left ventricular CM; RVCM, right ventricular CM. (E) Comparison of % mitochondrial reads resulting from methods indicated. (F) Sarcomere shortening (left) and calcium transient (right) traces for pre- and post-sorted CMs. Dark lines/shaded areas represent mean/standard error, respectively.
ACKNOWLEDGEMENTS
We thank Dr. Deborah Andrew for allowing us to use COPAS instrument. We thank Kevin Mangs, Julia Thompson, and Mike Fazzio for technical and scientific support regarding the use of the COPAS-FP. We thank Grace K Mueller for helpful feedback on experiments. This work was supported by grants from NICHD/NIH, AHA, and MSCRF.
Footnotes
DATA AND MATERIALS DISCLOSURE
All animal studies were performed in accordance with institutional guidelines/regulations of the ACUC at Johns Hopkins University. Supplementary data are at https://doi.org/10.1101/654954 and GEO (GSE133640).
REFERENCES
- 1.Ackers-johnson M, Lek W, Tan W & Foo RS Following hearts, one cell at a time: recent applications of single-cell RNA sequencing to the understanding of heart disease. Nat. Commun 8–11 (2018). doi: 10.1038/s41467-018-06894-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chevalier M et al. Transcriptomic analyses of murine ventricular cardiomyocytes. Sci. Data 1–9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeLaughter DM et al. Single-Cell Resolution of Temporal Gene Expression during Heart Development. Dev. Cell 39, 480–490 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nomura S et al. Cardiomyocyte gene programs encoding morphological and functional signatures in cardiac hypertrophy and failure. Nat. Commun 1–17 (2018). doi: 10.1038/s41467-018-06639-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gladka MM et al. Single-Cell Sequencing of the Healthy and Diseased Heart Reveals Ckap4 as a New Modulator of Fibroblasts Activation. Circulation CIRCULATIONAHA.117.030742 (2018). doi: 10.1161/CIRCULATIONAHA.117.030742 [DOI] [PubMed] [Google Scholar]