Abstract
Cellular membranes play host to a wide variety of morphologically and chemically complex processes. Although model membranes, like liposomes, are already widely used to reconstitute and study these processes, better tools are needed for making model bilayers that faithfully mimic cellular membranes. Existing methods for fabricating cell-sized (μm) or organelle-sized (tens to hundreds of nanometers) lipid vesicles have distinctly different requirements. Of particular note for biology, it remains challenging for any technique to efficiently encapsulate fragile cargo molecules or to generate liposomes with stable, asymmetric lipid leaflets within the bilayer. Here a tunable microfluidic device and protocol for fabricating liposomes with desired diameters ranging from ≈10 μm to ≈100 nm are described. Lipid vesicle size is templated by the simple inclusion of a polycarbonate filter within the microfluidic system and tuned with flow rate. It is shown that the vesicles made with this device are stable, unilamellar, lipid asymmetric, and capable of supporting transmembrane protein assembly, peripheral membrane protein binding, as well as soluble cargo encapsulation (including designer nanocages for biotechnology applications). These fabricated vesicles provide a new platform for studying the biophysically rich processes found within lipid–lipid and lipid–protein systems typically associated with cellular membranes.
Keywords: cross-flow emulsification, cryogenic electron microscopy (cryoEM), liposomes, microfluidics, phase transfer
1. Introduction
Filled with an aqueous solvent and bounded by a fluid lipid bilayer, liposomes are popular mimetics for studying biological membranes and membrane-associated biochemical activities. Giant unilamellar vesicles (GUVs, ˃1 μm) are cell-sized liposomes. Among other processes, GUVs are routinely used for studying in vitro protein synthesis,[1] actin polymerization,[2] and curvature-dependent protein–lipid dynamics.[3] Large unilamellar vesicles (LUVs, ˂1 μm) are organelle-sized liposomes. LUVs have also been used extensively to study the molecular structures and functions of membrane-binding proteins, including pore-forming toxins,[4] large GTPases of the dynamin family,[5] endosomal sorting complexes required for transport (ESCRT) protein complexes,[6] and bin-amphiphysin-rvs[7] domain proteins among many others. Despite their utility, however, liposome manufacturing protocols and the lipid membrane properties that result from each method vary widely.[8] To address these shortcomings, we sought to develop a simple and tunable microfluidic device for generating LUVs or GUVs with defined lumenal contents and asymmetric lipid-leaflet compositions. Such a device would enable researchers to reconstitute complex cellular phenomena in vitro for detailed characterizations.
A number of innovative microfluidic strategies have been developed for producing GUVs including droplet phase transfer (PT),[9–11] double emulsion templating,[12] and pulsed jetting.[13,14] These techniques allow for controlled encapsulation of molecular cargo while also conferring the ability to create symmetric and asymmetric lipid bilayers.[15] One of the challenges associated with the PT technique is with controlling initial emulsion size without impacting the integrity of encapsulated molecules or increasing processing times. A popular technique for producing emulsions is based on hand extrusion, which suffers from poor reproducibility due to variations in extrusion pressure and passage number,[16] while techniques such as vortexing and sonication may impact the integrity of encapsulated molecules.[17,18] Double emulsion templating overcomes the aforementioned challenges associated with controlling droplet size at the expense of added complexity. This approach requires access to microfabrication equipment and clean room environments, where final liposome size depends on the starting microfluidic droplet size, and this necessitates fabricating channels that are roughly the same size as the desired vesicles.[12] However, fabrication of micro (˃1 μm) or nanoscale (˂1 μm) channels typically requires access to clean room facilities and fabrication expertise. Microchannels are also prone to clogging and require filtered samples and, in many cases, chemical functionalization of the channel to promote stable droplet formation. Fabrication and operation of smaller channels (˂1 μm) require even finer control over device processing and fluidic control parameters. While it is possible to template the size of GUVs using glass capillaries, as demonstrated in the pulsed jetting technique, other challenges are introduced. Pulsed jetting requires precise alignment of instrumentation, nozzle micromanipulation and control over operating parameters such as temperature and fluid properties.[8]
Fewer microfluidic strategies exist for the formation of LUVs. While phase transfer has been adopted for LUV formation,[16,19] the most popular approach is based on hydrodynamic flow focusing where liposomes form spontaneously at the interface of an aqueous/alcohol stream.[20,21] While a powerful technique for the formation of LUVs, questions however remain over the impact of organic solvent on the lipid membrane and the functionality of encapsulated molecules.[8] Whether stable GUVs can be synthesized using this approach remains an open question. As it stands, LUV generation using traditional microfabrication techniques remains a challenge, and no technique currently exists that can be tuned to generate either GUVs or LUVs using the same methodology.
Here, we report a microfluidic device and protocol for the fabrication of either cell-sized GUVs or organelle-sized LUVs. This microfluidic approach combines a y-mixer design with cross-flow filter emulsification to rapidly produce micro- or nanoscale lipid-stabilized droplets that are subsequently converted to liposomes by the phase-transfer method.[9] Torque-balance modeling of droplet formation suggested that final liposome size depends on the polycarbonate filter pore size, allowing us to create ≈8 μm liposomes (GUVs) using a 5 μm polycarbonate filter and ≈300 nm liposomes (LUVs) using a 100 nm polycarbonate filter. Our experiments confirm that liposome size can be controlled by substituting polycarbonate filters with different pore sizes. By utilizing a number of chemical, biological, and microscopy assays, we show that at both scales the majority of generated liposomes are unilamellar and that only a small subset possesses any solvent contamination as detected by electron cryo-microscopy. Furthermore, we verify the ability of LUVs created with a complex phospholipid composition to bind to, and be remodeled by, the ESCRT-III protein charged multivesicular body protein 1B (CHMP1B). Finally, we demonstrate that, at either scale, liposomes made with our device can be used to encapsulate and protect both organic and synthetic macromolecular cargos under a variety of solution conditions.
2. Results and Discussion
2.1. Templating Vesicle Size
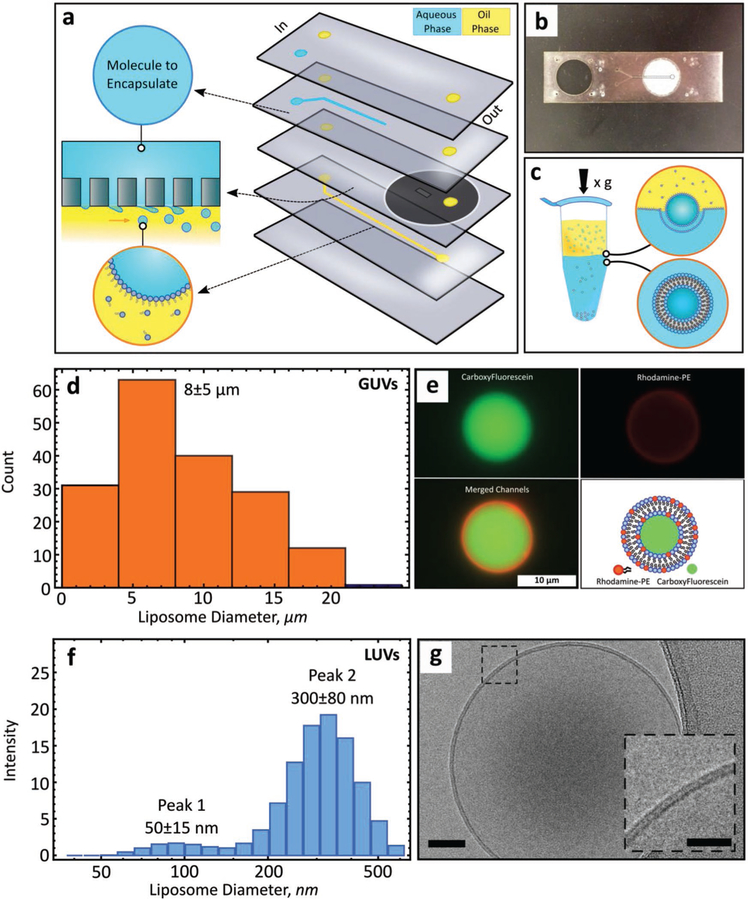
We tested whether embedding polycarbonate filters with defined pore sizes within a microfluidic device could be used to create liposomes at either the micro- or nanoscale using the PT method.[9,22] In this approach, a dispersed phase (solution to be encapsulated) is driven through a rigid filter into a shearing continuous phase (immiscible with the dispersed phase) under cross-flow emulsification conditions leading to the formation of droplets (Figure 1a). Droplet size primarily depends on the membrane pore size and can be tuned by adjusting the wall shear stress (which is a function of cross-flow velocity and fluid properties).[23] However, beyond a certain cross-flow velocity droplet size reaches a plateau.[24,25] Our microfluidic device takes advantage of this saturation regime to produce droplets of consistent size while also significantly reducing reagent consumption (requiring only hundreds of microliters as compared to tens of milliliters for traditional systems[26]). The device consists of two input channels and an off-the-shelf hydrophobic polycarbonate filter (Figure 1b). A hydrophobic membrane is used to promote stable droplet generation.[23] The geometry of the oil input channel (width: 250 μm, height: 31 μm) results in laminar flow with high shear stress throughout. Each polycarbonate layer, along with the polycarbonate filter, is added sequentially (Figure S1, Supporting Information) and permanently bonded through thermal fusion for a maximum bond strength of 4.5 MPa (Figure S2, Supporting Information).
Figure 1.
A microfluidic device for generating GUVs or LUVs in two steps. a) Schematic of the different layers used to create the final microfluidic device, with an illustration of the droplet formation process used to create micro- or nanoscale droplets. Macromolecular targets of interest are pressure driven into the first input channel (blue). Oil solvents saturated with lipids are pressure driven into the second input channel (yellow) forming water-in-oil emulsions. A polycarbonate membrane separates the two channels. b) An image of a single microfluidic device. The outlet channel has been outlined to help with visualization. c) Phase transfer of lipid-stabilized microscale or nanoscale droplets through a lipid-rich interface to form GUVS or LUVS. d) Size distribution of GUVs formed with a 5 μm pore membrane (n = 176) (mean ± s.d.). Measured by optical and fluorescent microscopy. e) Representative image of a GUV-encapsulating carboxyfluorescein (CF). Top left, CF alone. Top right, rhodamine-PE alone (0.5% mol). Bottom left, channel colocalization. Bottom right, schematic of the main features of the GUV. f) Size distribution of LUVs formed with a 100 nm pore membrane (number of independent runs, n = 3) (mean ± s.d.). Measured by DLS. g) CryoEM microscopic image of a small unilamellar liposome. Scale bar, 50 nm. Inset scale bar, 25 nm.
The existence of a droplet-size invariant regime was first established theoretically by applying a torque-balance model at the surface of the filter pore.[23] Depending on the mode of deformation, we identified droplet-size invariant regions for 5 μm and 100 nm pore polycarbonate filters (Figure S3a,b, Supporting Information). Theoretical modeling, based on the geometric constraints (Table S1, Supporting Information) used in this work, predicted low droplet diameter variation with a continuous phase flow rate set to 80 μL min−1 or greater. The existence of this region was confirmed experimentally after the transformation of droplets into liposomes using the PT method (Figure 1c), which has been shown to preserve initial droplet size upon centrifugation.[9,27] Dynamic light scattering (DLS) confirmed droplet-size invariance for a 100 nm filter pore across a variety of flow rates (80 to 230 μL min−1) (Figure S3c, Supporting Information). We estimated the shear stress within this region to be between 1500 and 4000 Pa.
Integration of a 5 μm polycarbonate filter into the microfluidic device leads to the formation of cell-sized lipid vesicles (GUVs) (8 ± 5 μm) (mean ± standard deviation (s.d.), n = 176) (Figure 1d). To visualize these vesicles, carboxyfluorescein (CF) was encapsulated into the lumen of GUVs composed of a simple lipid mixture (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC)/1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS)/cholesterol) with the addition of trace amounts of rhodamine-labeled 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (PE) for lipid bilayer visualization (Figure 1e). The size and distribution of GUVs generated here are, on average, smaller and more uniform than GUVs prepared by a similar microfluidic approach[10] and do not require detergent stabilization.[27] Our technique also enables rapid encapsulation of material within the lumen. While it may be possible to create GUVs of larger size, as shown in other works utilizing the phase-transfer protocol, the device reported here is optimized for smaller diameter GUVs (˂ 10 μm). Required modifications may include channel enlargement and larger filter pore size.
In addition, by substituting a 5 μm filter for a 100 nm filter we are able to generate organelle-sized lipid vesicles (LUVs) of 300 ± 80 nm (mean ± s.d., n = 9; Figure 1f). The presence of a minor peak (≈50 nm) may be attributed to the formation of satellite lipid particles. These particles could form as a result of either a collapsing fluid neck formed in the wake of larger liposomes traversing the two immiscible fluids[28,29] or through partial droplet coalescence (at the oil/water interface) in the capture vial, leading to the formation of secondary droplets.[30,31] Cryogenic electron microscopy (cryoEM) enabled direct visualization of LUVs and confirmed the formation of vesicles with intact, unilamellar lipid bilayers (Figure 1g). Compared to spontaneous vesiculation[16] or droplet extrusion,[16] the technique developed here confers improved control over lipid-leaflet placement, lamellarity,[32] size, and distribution. The final production rate of liposomes is on the order of a hundred or more liposomes per microliter, with a typical 100 μL liposome-loaded solution requiring ≈ 13 min to prepare (multiple batches can be prepared in parallel). While droplet microfluidics can be used to produce dramatically higher rates of liposomes, it is possible to increase the number of liposomes in solution by using concentration columns, by employing a sugar density gradient, by increasing the surface area of the buffer/oil interface in the vial, and/or by increasing the lipid concentration.[11]
2.2. Lamellarity, Permeability, and Solvent Contamination
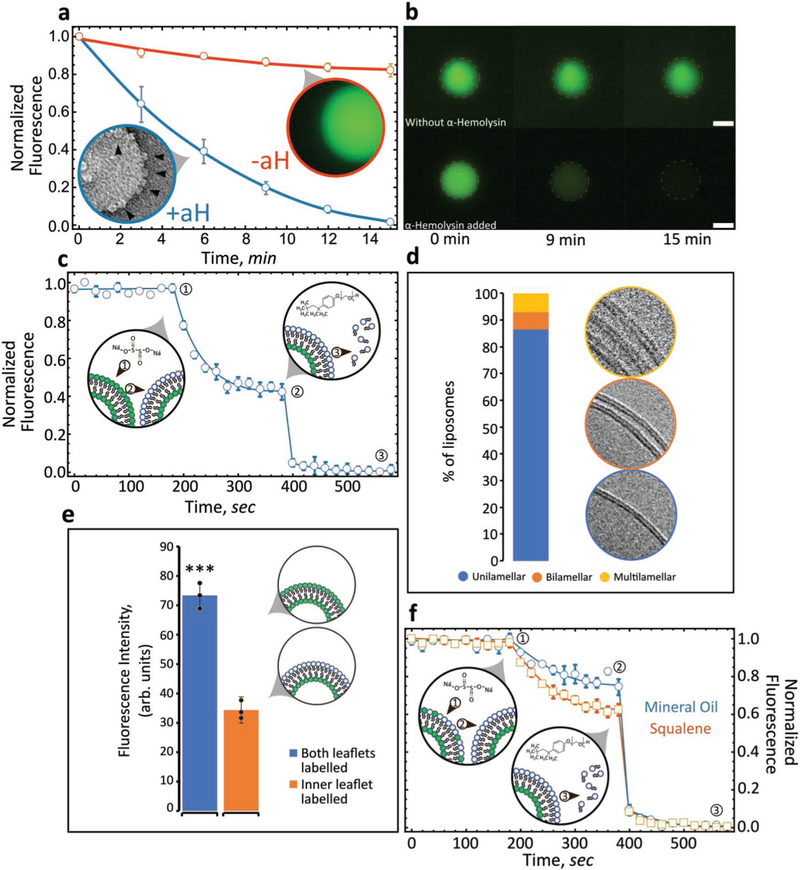
We investigated GUV lamellarity through encapsulation of the dye, carboxyfluorescein. Alpha-hemolysin (aH), a membrane-binding and pore-forming toxin, was added to the outside of liposomes. In a series of steps, monomers of aH adsorb on the lipid bilayer, followed by assembly into a heptameric pore that spans the lipid bilayer.[33,34] We observed that aH nanopores successfully assembled within the lipid bilayer and lead to the diffusion of fluorescent molecules out of the vesicle lumen (Figure 2a). Diffusion of carboxyfluorescein was tracked for 15 min with quantifiable differences between photobleached liposomes (Figure 2a,−aH) and those exposed to alpha-hemolysin (Figure 2a,+aH). Functional alpha-hemolysin insertion demonstrates three things: 1) the lipid bilayer is capable of supporting membrane insertion and pore formation; 2) the lipid bilayer is unilamellar; and 3) the bilayer remains stable following aH pore formation (Figure 2b).
Figure 2.
Lipid-leaflet lamellarity and content asymmetry of GUVs and LUVs. a) GUVs loaded with carboxyfluorescein were monitored over time by fluorescence microscopy. GUVs exposed to alpha-hemolysin (blue, +aH) were compared with control liposomes (red, −aH) and the flux of fluorescent molecules from within the lumen monitored over time. Inset: electron microscopic image of a liposome studded with aH pores (n = 5) (error bars are standard error of the mean (s.e.m.)). Data sets were min–max normalized, with +aH compared to that of −aH. b) Images of GUVs exposed (lower panels) or not exposed (upper panels) to alpha-hemolysin over time (n = 5). Scale bar, 10 μm. c) Fluorescence intensity and quenching of NBD-PC (both leaflets: POPC/POPS/cholesterol/NBD-PC) (n = 3, error bars are s.e.m.). Arrow number indicates as follows: 1) addition of quencher, 2) fluorescence quenching, 3) addition of Triton X-100 followed by bilayer solubilization and complete quenching. Data sets were min–max normalized and averaged. d) Liposome lamellarity visualized by cryoEM (lipid composition: POPC/POPS/cholesterol; n = 199). e) Fluorescence intensity of liposomes synthesized with NBD-PC present either in both leaflets or a single leaflet (n = 3). Unpaired t-test, ***p ˂ 0.001. f) Fluorescence intensity quenching of NBD-PC deposited exclusively in the inner leaflet. Inner leaflet, POPC/NBD-PC. Outer leaflet, POPS (n = 3, error bars are s.e.m.). Comparing solvents for generating asymmetric liposomes: mineral oil versus squalene. Data sets were min–max normalized.
We probed the lamellarity of nanoscale vesicles utilizing two distinctly different assays: fluorescence quenching and cryoEM. First, we utilized sodium hydrosulfite to quench fluorescence from trace 1-palmitoyl-2-{6-[(7-nitro-2–1,3-benzoxadiazol-4-yl) amino]hexanoyl}-sn-glycero-3-phosphocholine (NBD-PC) lipids. In the vicinity of the fluorophore (NBD), sodium hydrosulfite quenches the fluorescence by reducing the dye. A perfectly symmetric lipid bilayer will result in the reduction of 50% of the signal if the fluorophore (NBD-PC) is evenly distributed between the two leaflets upon exposure to the quenching agent.[9] We generated symmetric vesicles composed of POPC/POPS/cholesterol/NBD-PC (44.8:36.9:17.7:0.5 mol%) and measured NBD-PC fluorescence. For liposomes fabricated with NBD-PC in both the inner and outer leaflets, addition of the quencher reduced total fluorescence by 47% (Figure 2c), suggesting that the dye was evenly distributed in both leaflets and confirming that the majority of liposomes formed by our protocol are unilamellar. To test this notion further, we utilized cryoEM to directly visualize the lamellarity of a population of LUVs. Electron microscopy of vitrified liposomes revealed that ≈85% of LUVs prepared with this technique are unilamellar with a small fraction of multilamellar (MLV) and multivesicular (MVL) liposomes (Figure 2d; Figures S4 and S5, Supporting Information). Significantly, our approach is free of sucrose- or other sugar-containing solutions allowing for cryoEM investigation of lamellarity. Liposomes are fabricated under physiologically relevant conditions without the need for density gradients. Sucrose and glucose are routinely utilized for liposome formation and isolation; however, high concentrations of sugars are detrimental for cryoEM analysis because they lead to significant reductions in contrast.[35]
Next, we generated LUVs with asymmetric lipid leaflets. The inner leaflet comprises POPC/NBD-PC and derives from the first lipid monolayer that stabilizes the droplets during stage one. The outer leaflet comprises POPS and derives from the second monolayer that stabilizes the aqueous solution/oil interface in the vial. We confirmed the total fluorescence of fluorescent lipid headgroups (NBD-PC) present in the two leaflets through fluorescence microscopy. Quantifying the total fluorescence per lipid concentration when only a single leaflet is labeled with NBD compared to both leaflets should reveal approximately a twofold difference. As expected, total fluorescence was 53.2% greater when NBD-labeled PC (0.5 mol%) was included in both stages compared with just the first stage (Figure 2e). A fluorescent quenching assay was performed to quantify the degree of asymmetry with an inner leaflet composed of POPC/NBD-PC and the outer of POPS. Since the lipid bilayer is impermeable to sodium hydrosulfite, a completely asymmetric bilayer should maintain 100% fluorescence upon addition of the quencher. Unexpectedly, we found that oil properties play a role in the degree of asymmetry. Liposomes formed with mineral oil as the solvent, by contrast with squalene, show the greatest level of stable asymmetry, with 79% of the fluorescent intensity protected from the quencher (Figure 2f) (n = 3). Imperfect asymmetry is most likely due to the spontaneous movement of lipids from the inner leaflet to the outer leaflet due to either trading of lipids during phase transfer or flip-flop following fabrication.[10] Our findings agree with and reinforce recent publications[14,36] which demonstrate that solvent type plays a role in the final mechanical and functional properties of lipid membranes.
2.3. Characterizing Vesicle Utility
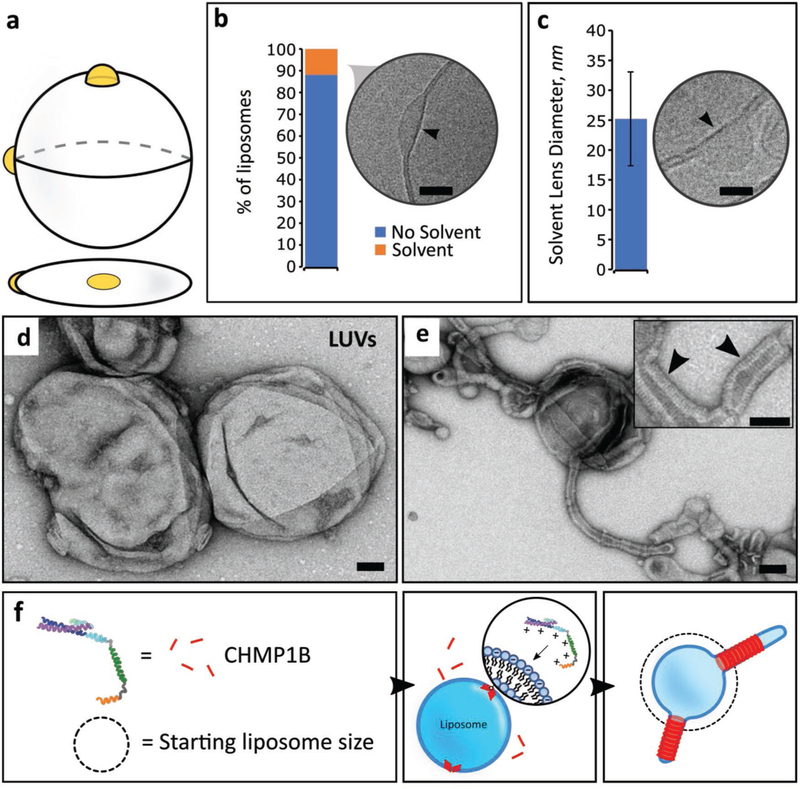
Here, we address several outstanding questions regarding the nature of solvent contamination and its impact on the quality and functionality of the lipid membrane. CryoEM imaging enabled us to quantify residual solvent inclusion within lipid bilayers formed by the PT method (Figure 3a). We observed that visible oil lenses within lipid bilayers occur in ≈12% of the total liposome population (Figure 3b). When present, solvent lenses average 25 ± 8 nm (mean ± s.d., n = 55) in diameter (Figure 3c). While large oil inclusions are straightforward to characterize (Figure S6, Supporting Information), we cannot rule out the presence of trace amounts of oil within the lipid bilayer. Interestingly, the observed mineral oil collection in lenses is reminiscent of the fatty acid lenses that form in the endoplasmic reticulum during lipid droplet biogenesis.[37] Our methodology can potentially be used to reconstitute and study this phenomenon.
Figure 3.
Characterizing oil defects and lipid bilayer properties. a) Illustration of oil accumulation as lenses within the lipid bilayer. b) Quantification of cryoEM microscopic images showing oil lenses within the bilayer (n = 1812) (lipid composition, POPC/POPS/cholesterol). Arrow points to an oil lens located on the outer edge of the liposome. Scale bar, 10 nm. c) Oil lens size diameters as observed by cryoEM (n = 55) (lipid composition, POPC/POPS/cholesterol). Arrow indicates typical morphology of solvent lenses. Scale bar, 10 nm. Error bars are s.d. d) TEM microscopic image of liposomes (control), before addition of CHMP1B (lipid composition: POPC/POPS/cholesterol). Scale bar, 50 nm. e) After addition of CHM1B to liposomes, CHMP1B accumulates on the lipid bilayer and subsequently deforms it into lipid tubules. Inset, protein striations (black arrowheads) are clearly visible under negative stain EM. Scale bar, 50 nm. f) Cartoon illustration of CHMP1B-mediated vesicle remodeling.
We assayed whether vesicles generated using our device are suitable substrates for peripheral membrane-binding proteins. CHMP1B is a human ESCRT-III protein implicated in membrane deformation processes such as recycling tubule biogenesis from endosomes.[6,38,39] CHMP1B accumulation on the surface of a lipid membrane requires the presence of negatively charged lipid headgroups, like POPS. In agreement with previous observations,[6] initially spherical liposomes (Figure 3d) are rapidly deformed by electrostatic binding of CHMP1B onto the positive (exterior) surface of liposomes consisting of POPC/POPS/cholesterol (Figure 3e). CHMP1B quickly saturates the liposomal surface, inducing strong positive curvature along the tubule axis. The outer protein shell is clearly visible under transmission electron microscopy (TEM; Figure 3e) demonstrating that CHMP1B is able to efficiently bind, stabilize and deform these vesicles (Figure 3f). This overlap of the behavior we observed for CHMP1B with previously published data reinforces the idea that the GUVs formed by our microfluidic device are biologically relevant.
2.4. Encapsulation of Synthetic and Organic Macromolecules
Liposomes have been used extensively as cargo carriers because the inner lumen of the liposome is protected from the exterior environment by the lipid bilayer. By including different cargoes in the first stage of emulsion generation, we encapsulated two cargoes with different properties. An advantage of our method is the ability to create GUVs and LUVs incorporating diverse solution conditions, e.g., high ionic strength buffers (˃ 350 × 10−3 m NaCl), for encapsulation of target proteins that would otherwise polymerize or assemble under physiological conditions.
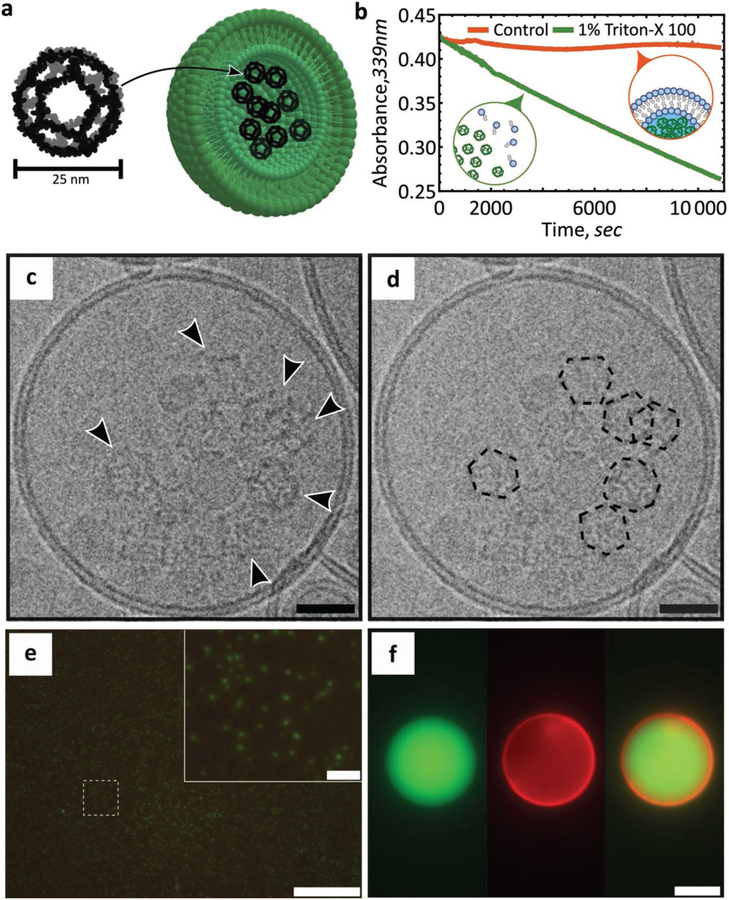
For encapsulation, we first loaded LUVs with computationally designed, self-assembling 25 nm dodecahedral nanocages (Figure 4a).[40] These recently reported[40] nanocages hold great promise for drug delivery and synthetic biology applications, including as cargo carriers,[41] as imaging probes,[42] and as scaffolds for vaccine design.[43] Initially, we purified and visualized 25 nm diameter aldolase nanocages[40,44] by negative stain TEM (Figure S7, Supporting Information). We then demonstrated nanocage encapsulation within vesicles manufactured in our microfluidic device by three methods. First, we observe that aldolase activity is sequestered within liposomes, but can be subsequently released upon the addition of a detergent (Figure 4b). Addition of Triton-X100 disrupts the lipid bilayer, releasing internal cargo into the solution, and this enables us to quantify aldolase activity over time. Second, TEM imaging revealed controlled nanocage release from detergent-exposed liposomes (Figures S8 and S9a, Supporting Information). Third, direct visualization by cryoEM demonstrated that the protein nanocages inside the synthesized liposomes matched the expected 25 nm size and assembled architecture of the designed structure (Figure 4c,d; Figure S9b, Supporting Information).[40] In addition to these dodecahedral nanocage assemblies, we also encapsulated green fluorescent protein (GFP) within both GUVs and LUVs, as determined by fluorescence microscopy (Figure 4e,f). Developing efficient means by which biological macromolecules can be encapsulated and protected while maintaining their structure and function will prove vital for future applications.
Figure 4.
Cargo encapsulation within LUVs or GUVs. a) Cartoon representation of the designed dodecahedral nanocage structure and encapsulation within a liposome. Lipid bilayer not to scale. Adapted from.[40] b) Protection and release assay of designer nanocages. Aldolase nanocages are protected within liposomes (orange) and are exposed following solubilization of the liposomes upon the addition of 1% Triton X-100 (green) (lipid composition: POPC/POPS/cholesterol; n = 3). Data sets were scaled for direct comparisons. c,d) CryoEM microscopic image of a nanocage-loaded liposome (lipid composition: POPC/POPS/cholesterol). Arrows indicate nanocages. Scale bar, 25 nm. Nanocages are outlined to aid with visualization. e) Fluorescence microscopic image of GFP encapsulated within LUVs (lipid composition: POPC/POPS/cholesterol). Scale bar 10 μm; inset scale bar, 1 μm. f) Fluorescence microscopic image of GFP encapsulated within GUVs (lipid composition: POPC/POPS/cholesterol/rhodamine-PE). Scale bar, 10 μm.
3. Conclusion
We have developed a simple, inexpensive, and modular microfluidic approach for the formation of size-controlled, lipid-content controlled, lumen-content controlled, and asymmetric liposomes that comprise, on average, a single phospholipid bilayer. Utilizing cross-flow emulsification theory, modified for the geometric properties of microfluidic channels, we identified and experimentally verified the existence of droplet-size flow invariant regions. When operating within this regime, use of the appropriate polycarbonate filter leads to the creation of either nanoscale or microscale vesicles. CryoEM studies, enabled by sugar-free solutions, revealed the extent and size of oil inclusions within liposomes. We show that our vesicles are able to support a variety of complex phenomena such as nanotubule formation and lipid bilayer remodeling by an ESCRT-III protein. The lipid vesicles created here proved remarkably stable to a host of different chemical assays, demonstrating effective encapsulation and protection of different cargoes.
4. Experimental Section
Materials:
Polycarbonate, clear, laser markable film (SD8B94) at a thickness of 50 μm was purchased from SABIC. Polycarbonate track etched, hydrophobic (≈78°–88° contact angle), 0.1 and 5 μm polycarbonate membranes were purchased from Sterlitech Corporation (PCTF0113100, PCTF5013100). Tygon 0.02″ × 0.06″ tubing was purchased from Cole-Parmer. UV-curable adhesive (3106) was purchased from Loctite. Nanosep 10 kDa MF centrifugal devices (spin columns) were obtained from VWR. Wellplates (384 well, black bottom, polystyrene) were purchased from Corning. Axygen 2 mL microtubes (MCT-200-L-C) were purchased from Axygen, Inc. Continuous carbon film grids (Formvar/Carbon Film (FCF-200-Cu)), Quantifoil holey carbon grids (2 μm hole size, 2 μm spacing, and 200 mesh) and ultrathin carbon supported by a lacey carbon film on a 400 mesh copper grid (# 0 1824) were obtained from Ted Pella, Inc. Slide-A Lyzer MINI Dialysis Devices (10k, 88 401) were purchased from Thermo Scientific.
Nanoemulsion and LUV Formation:
Lipid handling and preparation procedures followed those of previously published protocols.[45,46] Phospholipids (POPC/POPS/cholesterol (45.3:36.9:17.7 mol%)) (unless otherwise stated, this lipid ratio was used for all formulations and is the standard lipid mixture) stored in chloroform were dispersed in mineral oil at a final lipid concentration of 5 × 10−3 m (lipid/oil ratio of 0.003). Glass vials were placed into an oven overnight to evaporate any chloroform. The oil–lipid mixture was further diluted to 2 × 10−3 m using mineral oil. A 1 mL syringe loaded with 1× phosphate buffered saline (PBS) was loaded into a syringe pump (KD Scientific 200) and driven at a constant volumetric flow rate of 4 μL min−1 through the upper channel. Another syringe pump (KD Scientific 220) was loaded with a 3 mL syringe. 130 μL of emulsion was removed from the outlet of the device and added to a 2 mL microtube. Emulsions were placed into a fridge at 4 °C for 20 min to allow the lipids to equilibrate at the droplet interface. Concurrently, 130 μL of 1× PBS was added to the bottom of a 2 mL microtube. Subsequently, 170 μL of the oil–lipid mixture was placed on top of the buffer and allowed to equilibrate for 20 min at room temperature. Finally, emulsions were distributed between vials and centrifuged (4 °C for 10 min, 20 000 × g).
GUV Formation and Carboxyfluorescein Encapsulation:
Following the same protocol as outlined for LUVs, microemulsions were synthesized by keeping the dispersed phase constant at 5 μL min−1, and the oil flow rate constant at 120 μL min−1. Following incubation at room temperature, emulsions were added to the capture vial and centrifuged (10 min at 9000 × g). Carboxyfluorescein (350 × 10−3 m NaCl, 50 × 10−3 m Tris-HCl, pH 7.0, 5% (w/v) glycerol and 5 × 10−3 m β-mercaptoethanol) was encapsulated within GUVs at a final concentration of 50 × 10−6 m.
Liposome-Leaflet Asymmetry Assay:
The inner lipid leaflet was composed of POPC/NBD-PC (99.5:0.5 mol%). The outer leaflet was composed exclusively of POPS (100 mol%). In this assay, droplets were made with a stabilizing monolayer of POPC/NBD-PC (99.5:0.5 mol%). The second interface (in the vial) was created with an entirely different lipid composition, consisting of POPS (100 mol%). Emulsion preparation and collection followed previously described steps except for the addition of NBD-PC to the emulsion formation phase. Unlabeled POPS oil–lipid mixture was diluted with olive oil (1:1). 170 μL of this mixture was placed on top of 130 μL of 1× PBS buffer in a 2 mL microtube and allowed to equilibrate for 20 min at room temperature. Emulsions were centrifuged at 300 × g for 10 min at room temperature. Following previously published protocols,[47] 10 μL of unconcentrated liposome solution and 10 μL of buffer were added to one well, and 20 μL of buffer was added to another. After setting the baseline, 0.5 μL of 1 m sodium hydrosulfite prepared in 1× PBS was added to the sample followed by the addition of 2 μL of 10% Triton X-100.
Aldolase Enzyme Activity Assay:
The 2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolase activity of the I3–01 nanocage domain was monitored using an L-lactic acid dehydrogenase (LDH)-coupled assay.[44,48] 10 μL of nanocage-encapsulated liposomes was mixed with 90 μL of 1× PBS, 0.1 × 10−3 m nicotinamide adenine dinucleotide (NADH), 0.11 U μL−1 1 LDH, 1 × 10−3 m KDPG, and either including or omitting 1% Triton X-100. Loss of absorbance at 339 nm owing to oxidation of NADH was monitored using a Synergy Neo2 microplate reader. At least three replicates of the aldolase activity assay were performed.
CryoEM Imaging:
Liposomes loaded with 1× PBS were prepared as described above, and 3 mL of the sample was concentrated to 100 μL. For nanocage-encapsulated liposomes, 200 μg mL−1 of nanocages was loaded into liposomes and concentrated from 1 mL to 100 μL. For electron cryo-microscopy, 3.5 μL of these concentrated samples was applied to either glow-discharged Quantifoil holey carbon grids (2 μm hole size, 2–4 μm spacing, and 200 mesh) or ultrathin carbon film on holey carbon grids (400 mesh), blotted (6.5–8 s, −1 mm offset) and plunge-frozen in liquid ethane using a Vitrobot Mark I (FEI). Electron cryo-microscopic images were collected following low-dose procedures at liquid nitrogen temperature on a Tecnai TF20 operating at 200 kV using a Gatan 626 side-entry cryo-holder. Movies were recorded using a K2 Summit direct detector (Gatan, Pleasanton, CA) in either counting or super-resolution mode at a corrected magnification of 41 911×, corresponding to a physical pixel size of 1.193 Å, and at dose rates of ≈7 e− pixel−1 s−1 at the specimen. SerialEM[49] was used to facilitate low-dose imaging and semi-automated data collection, and each movie was recorded as a stack of 40 subframes, each of which was accumulated for 0.2 s, totaling ≈39 e− Å−2 at the specimen. Frames were aligned and summed by using MotionCor2.[50]
Statistical Analysis:
Unless otherwise stated, data were reported as mean + standard error of the mean. Each experiment was performed at least three times (n ≧ 3). Where applicable, the normalization procedure is described within the figure caption. Comparison of independent data sets was performed via the unpaired t-test. Distribution normality was checked with quantile–quantile plots with 95% confidence level and the Shapiro–Wilks test. Variance was quantified with the F-test. Significance was defined as *p ˂ 0.05, **p ˂ 0.01, and ***p ˂ 0.001. Statistical analysis was performed with R (RStudio).
Supplementary Material
Acknowledgements
The authors thank David Belnap at the University of Utah Electron Microscopy Core Laboratory for help with cryoEM image acquisition and processing, Joerg Votteler for guidance on aldolase assays, as well as Marc Porter and Hamid Ghandehari for access to the Malvern Zetasizer. The authors also thank Wes Sundquist for numerous discussions and critical reading of the manuscript. A.F. acknowledges funding by a Faculty Scholar grant from the Howard Hughes Medical Institute, the Searle Scholars Program, NIH grant 1DP2GM110772-01, NIH grant 1R01GM127673-01, the American Asthma Foundation, and the Chan Zuckerberg Biohub. J.M. and A.F. acknowledge funding by the National Institutes of Health (P50 GM082545 and R01 GM127673-01). V.R., J.M., and A.F. designed the experiments. V.R. and J.M. performed the experiments. V.R. analyzed the data. B.K.G. contributed to the device design, fabrication and characterization. V.R., J.M., and A.F. wrote the paper. All authors had direct input on experimental results and the preparation of the manuscript.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Valentin Romanov, Department of Mechanical Engineering, University of Utah, Salt Lake City, UT 84112, USA.
John McCullough, Department of Biochemistry, University of Utah, Salt Lake City, UT 84112, USA.
Bruce K. Gale, Department of Mechanical Engineering, University of Utah, Salt Lake City, UT 84112, USA
Adam Frost, Department of Biochemistry, University of Utah, Salt Lake City, UT 84112, USA; Department of Biochemistry and Biophysics, University of California, San Francisco, CA 94158, USA; California Institute for Quantitative Biomedical Research, San Francisco, CA 94158, USA; Chan Zuckerberg Biohub, San Francisco, CA 94158, USA.
References
- [1].Fujii S, Matsuura T, Sunami T, Kazuta Y, Yomo T, Proc. Natl. Acad. Sci. USA 2013, 110, 16796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Pontani L-L, van der Gucht J, Salbreux G, Heuvingh J, Joanny J-F, Sykes C, Biophys. J 2009, 96, 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Sorre B, Callan-Jones A, Manneville J-B, Nassoy P, Joanny J-F, Prost J, Goud B, Bassereau P, Proc. Natl. Acad. Sci. USA 2009, 106, 5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Anderluh G, Serra MD, Viero G, Guella G, Maček P, Menestrina G, J. Biol. Chem 2003, 278, 45216. [DOI] [PubMed] [Google Scholar]
- [5].Sweitzer SM, Hinshaw JE, Cell 1998, 93, 1021. [DOI] [PubMed] [Google Scholar]
- [6].McCullough J, Clippinger AK, Talledge N, Skowyra ML, Saunders MG, Naismith TV, Colf LA, Afonine P, Arthur C, Sundquist WI, Hanson PI, Frost A, Science 2015, 350, 1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Frost A, Unger VM, De Camilli P, Cell 2009, 137, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].van Swaay D, deMello A, Lab Chip 2013, 13, 752. [DOI] [PubMed] [Google Scholar]
- [9].Pautot S, Frisken BJ, Weitz DA, Langmuir 2003, 19, 2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hu PC, Li S, Malmstadt N, ACS Appl. Mater. Interfaces 2011, 3, 1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chiba M, Miyazaki M, Ishiwata S, Biophys. J 2014, 107, 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Deshpande S, Caspi Y, Meijering AEC, Dekker C, Nat. Commun 2016, 7, 10447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Stachowiak JC, Richmond DL, Li TH, Liu AP, Parekh SH, Fletcher DA, Proc. Natl. Acad. Sci. USA 2008, 105, 4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kamiya K, Kawano R, Osaki T, Akiyoshi K, Takeuchi S, Nat. Chem 2016, 8, 881. [DOI] [PubMed] [Google Scholar]
- [15].Lu L, Schertzer JW, Chiarot PR, Lab Chip 2015, 15, 3591. [DOI] [PubMed] [Google Scholar]
- [16].Whittenton J, Harendra S, Pitchumani R, Mohanty K, Vipulanandan C, Thevananther S, Langmuir 2008, 24, 8533. [DOI] [PubMed] [Google Scholar]
- [17].Wang W, Int. J. Pharm 1999, 185, 129. [DOI] [PubMed] [Google Scholar]
- [18].Bekard IB, Asimakis P, Bertolini J, Dunstan DE, Biopolymers 2011, 95, 733. [DOI] [PubMed] [Google Scholar]
- [19].de Matos MBC, Miranda BS, Nuari YR, Storm G, Leneweit G, Schiffelers RM, Kok RJ, J. Drug Targeting 2019, 27, 681. [DOI] [PubMed] [Google Scholar]
- [20].Jahn A, Vreeland WN, Gaitan M, Locascio LE, J. Am. Chem. Soc 2004, 126, 2674. [DOI] [PubMed] [Google Scholar]
- [21].Carugo D, Bottaro E, Owen J, Stride E, Nastruzzi C, Sci. Rep 2016, 6, 25876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhang L, Hu J, Lu Z, J. Colloid Interface Sci 1997, 190, 76. [DOI] [PubMed] [Google Scholar]
- [23].Peng SJ, Williams RA, Chem. Eng. Res. Des 1998, 76, 894. [Google Scholar]
- [24].De Luca G, Di Maio FP, Di Renzo A, Drioli E, Chem. Eng. Process 2008, 47, 1150. [Google Scholar]
- [25].Katoh R, Asano Y, Furuya A, Sotoyama K, Tomita M, J. Membr. Sci 1996, 113, 131. [Google Scholar]
- [26].Vladisavljevic GT, Schubert H, Desalination 2002, 144, 167. [Google Scholar]
- [27].Nishimura K, Suzuki H, Toyota T, Yomo T, J. Colloid Interface Sci 2012, 376, 119. [DOI] [PubMed] [Google Scholar]
- [28].Singh KK, Bart H-J, Ind. Eng. Chem. Res 2015, 54, 9478. [Google Scholar]
- [29].Bonhomme R, Magnaudet J, Duval F, Piar B, J. Fluid Mech 2012, 707, 405. [Google Scholar]
- [30].Charles GE, Mason SG, J. Colloid Sci 1960, 15, 105. [Google Scholar]
- [31].Blanchette F, Bigioni TP, Nat. Phys 2006, 2, 254. [Google Scholar]
- [32].Pautot S, Frisken BJ, Cheng J-X, Xie XS, Weitz DA, Langmuir 2003, 19, 10281. [Google Scholar]
- [33].Bhakdi S, Weller U, Walev I, Martin E, Jonas D, Palmer M, Med. Microbiol. Immunol 1993, 182, 167. [DOI] [PubMed] [Google Scholar]
- [34].Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE, Science 1996, 274, 1859. [DOI] [PubMed] [Google Scholar]
- [35].Matias VRF, Al-Amoudi A, Dubochet J, Beveridge TJ, J. Bacteriol 2003, 185, 6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Campillo C, Sens P, Köster D, Pontani L-L, Lévy D, Bassereau P, Nassoy P, Sykes C, Biophys. J 2013, 104, 1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Robenek H, Hofnagel O, Buers I, Robenek MJ, Troyer D, Severs NJ, J. Cell Sci 2006, 119, 4215. [DOI] [PubMed] [Google Scholar]
- [38].Dobro MJ, Samson RY, Yu Z, McCullough J, Ding HJ, Chong PL-G, Bell SD, Jensen GJ, Mol. Biol. Cell 2013, 24, 2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Reid E, Connell J, Edwards TL, Duley S, Brown SE, Sanderson CM, Hum. Mol. Genet 2005, 14, 19. [DOI] [PubMed] [Google Scholar]
- [40].Hsia Y, Bale JB, Gonen S, Shi D, Sheffler W, Fong KK, Nattermann U, Xu C, Huang P-S, Ravichandran R, Yi S, Davis TN, Gonen T, King NP, Baker D, Nature 2016, 535, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Schoonen L, van Hest JCM, Nanoscale 2014, 6, 7124. [DOI] [PubMed] [Google Scholar]
- [42].Lin X, Xie J, Niu G, Zhang F, Gao H, Yang M, Quan Q, Aronova MA, Zhang G, Lee S, Leapman R, Chen X, Nano Lett 2011, 11, 814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].He D, Marles-Wright J, New Biotechnol 2015, 32, 651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Votteler J, Ogohara C, Yi S, Hsia Y, Nattermann U, Belnap DM, King NP, Sundquist WI, Nature 2016, 540, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Noireaux V, Libchaber A, Proc. Natl. Acad. Sci. USA 2004, 101, 17669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Fujii S, Matsuura T, Sunami T, Nishikawa T, Kazuta Y, Yomo T, Nat. Protoc 2014, 9, 1578. [DOI] [PubMed] [Google Scholar]
- [47].McIntyre JC, Sleight RG, Biochemistry 1991, 30, 11819. [DOI] [PubMed] [Google Scholar]
- [48].Griffiths JS, Wymer NJ, Njolito E, Niranjanakumari S, Fierke CA, Toone EJ, Bioorg. Med. Chem 2002, 10, 545. [DOI] [PubMed] [Google Scholar]
- [49].Mastronarde DN, J. Struct. Biol 2005, 152, 36. [DOI] [PubMed] [Google Scholar]
- [50].Zheng S, Palovcak E, Armache J-P, Cheng Y, Agard D, Nat. Methods 2016, 14, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.