SUMMARY
Facultative heterochromatin regulates gene expression, but its assembly is poorly understood. Previously, we identified facultative heterochromatin islands in the fission yeast genome and found that RNA elimination machinery promotes island assembly at meiotic genes. Here, we report that Taz1, a component of the telomere protection complex Shelterin, is required to assemble heterochromatin islands at regions corresponding to late replication origins that are sites of double-strand break formation during meiosis. The loss of Taz1 or other Shelterin subunits, including Ccq1 that interacts with Clr4/Suv39h, abolishes heterochromatin at late origins and causes derepression of associated genes. Moreover, the late origin regulator Rif1 affects heterochromatin at Taz1-dependent islands and subtelomeric regions. We explore the connection between facultative heterochromatin and replication control, and show that heterochromatin machinery affects replication timing. These analyses reveal the role of Shelterin in facultative heterochromatin assembly at late origins, which has important implications for genome stability and gene regulation.
Graphical Abstract
Eukaryotic genomes contain facultative heterochromatin domains that coordinate diverse genomic functions. Zofall et al. show that components of the telomere protection complex Shelterin assemble facultative heterochromatin at internal chromosomal sites containing late replication origins. This work reveals functional connections between facultative heterochromatin, replication control, and gene regulation.
INTRODUCTION
Eukaryotic genomes feature two major classes of chromatin structure, namely euchromatin and heterochromatin. Whereas euchromatin domains are “open” and transcriptionally poised, heterochromatic regions are “closed” and less accessible to transcriptional machinery. In addition to controlling gene expression, heterochromatin promotes genome stability by preventing recombination and by ensuring proper segregation of chromosomes (Grewal and Jia, 2007). The organization of the genome within the nuclear space is also affected by heterochromatin (Mizuguchi et al., 2014; Towbin et al., 2013). Heterochromatin is further divided into two subcategories: constitutive and facultative. Constitutive heterochromatin, for example at centromeres and telomeres, targets chromosomal regions containing repetitive DNA elements and is stably maintained throughout the cell cycle. In contrast, facultative heterochromatin often targets genes and is subjected to regulated assembly and disassembly in response to environmental or developmental cues (Trojer and Reinberg, 2007).
Heterochromatin assembly pathways are highly conserved from the fission yeast Schizosaccharomyces pombe (S. pombe) to mammals. Histone methyltransferases and deacetylases play a critical role in the assembly of heterochromatin. Clr4/Suv39h family of methyltransferases methylates histone H3 lysine 9, a hallmark of heterochromatin domains across diverse species (Jenuwein and Allis, 2001). Methylated H3K9 (H3K9me) serves as a binding site for HP1 chromodomain proteins to assemble heterochromatin (Jenuwein and Allis, 2001). Moreover, Clr4/Suv39h binds to H3K9me via its chromodomain, a feature important for spreading and epigenetic inheritance of heterochromatin (Zhang et al., 2008). Spreading of H3K9me and HP1 proteins allows effectors that interact with heterochromatin components to act across large chromosomal domains, to promote gene silencing and to maintain genome stability (Grewal and Jia, 2007).
The S. pombe genome contains constitutive heterochromatin domains at the mating type (mat) locus, centromeres and telomeres, which are coated with H3K9me and chromodomain proteins, such as Chp2, Swi6 and Chp1 (Cam et al., 2005; Sadaie et al., 2004). Heterochromatin nucleation involves RNAi machinery and DNA binding factors implicated in the recruitment of the methyltransferase Clr4 and histone deacetylases including Clr3, Clr6, and Sir2 (Grewal and Jia, 2007). A number of effectors associate with heterochromatin, including the multicomponent silencing complex SHREC (Snf2/HDAC repressor complex) containing the HDAC Clr3 (Motamedi et al., 2008; Sugiyama et al., 2007) and the RNAi-associated RITS (RNA induced transcriptional silencing) complex (Verdel et al., 2004). Surprisingly, Swi6/HP1 also interacts with Epe1 that counteracts heterochromatic silencing. Loss of Epe1 stabilizes heterochromatin and enhances the local enrichment of heterochromatin modifications (Ayoub et al., 2003; Trewick et al., 2007; Zofall and Grewal, 2006).
Discrete blocks of facultative heterochromatin islands that are modified in response to nutritional signals are also found across the S. pombe genome (Zofall et al., 2012). In particular, nitrogen starvation triggers sexual differentiation and induces disassembly of a majority of islands, several of which map to meiotic genes that are silenced during vegetative growth (Zofall et al., 2012). The assembly of heterochromatin islands at meiotic genes involves RNA elimination factors including the RNA binding protein Mmi1 (Tashiro et al., 2013; Zofall et al., 2012), which binds to transcripts containing a DSR (“determinant of selective removal”) element (Harigaya et al., 2006). Mmi1 interacts with a conserved protein Erh1 to form a stoichiometric complex called EMC (Erh1-Mmi1 complex)(Sugiyama et al., 2016), which recruits the RNA processing complex MTREC (Mtl1-Red1 core) required for the assembly of heterochromatin islands at meiotic genes (Lee et al., 2013). Importantly, not all heterochromatin islands are lost in the absence of RNA elimination machinery, particularly those lacking DSR motifs. For these “non-DSR” MTREC-independent islands, the mechanism of heterochromatin assembly has remained unclear.
Here we performed a genetic screen to identify factors required for silencing and heterochromatin formation at a non-DSR, MTREC-independent island. We identified Taz1, a counterpart of human TRF1 and TRF2 and a component of the Shelterin complex involved in telomere protection (Cooper et al., 1997; Miyoshi et al., 2008). Taz1 localizes to several non-DSR islands and its loss abolishes H3K9me and causes derepression of associated loci. Strikingly, Taz1-dependent islands contain late firing replication origins that are affected by the loss of Taz1. Other Shelterin components are also required for repression and heterochromatin assembly at Taz1-dependent islands. Moreover, we find that heterochromatin assembly factors regulate replication timing at late origins, and describe a role for Shelterin components in the assembly of facultative heterochromatin islands harboring late-firing replication origins.
RESULTS
Genetic screen for factors affecting non-DSR heterochromatin islands
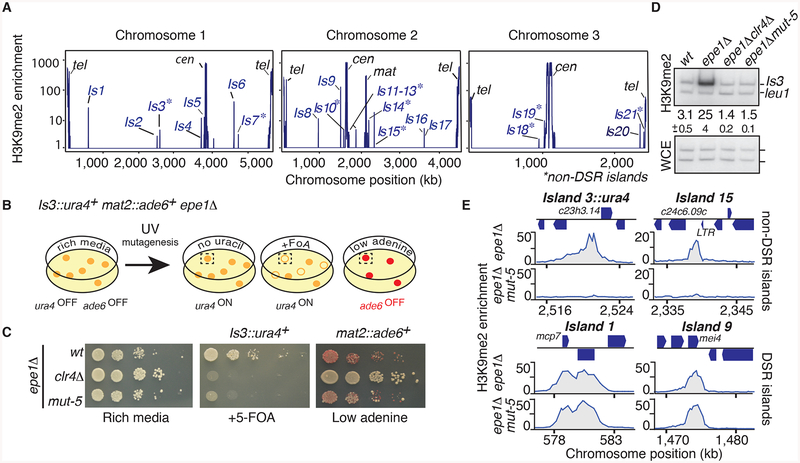
Two subcategories of facultative heterochromatin islands are found in the S. pombe genome: DSR islands that require RNA processing factors targeting DSR-containing transcripts, and non-DSR islands for which the assembly mechanism remains unknown (Figure 1A)(Zofall et al., 2012). Similar to DSR islands, non-DSR islands are remodeled in response to nutritional signals (e.g. islands 3, 7, 13, 14, 15, 19 but not island 21) (Figure S1A). To further investigate non-DSR islands, we used strain containing ura4+ reporter inserted adjacent to the SPAC23H3.14 locus at heterochromatin island 3 (Is3::ura4+) and screened for mutants that caused derepression of ura4+ (Figure 1B). The anti-silencing factor Epe1 was deleted to reinforce heterochromatic silencing of Is3::ura4+, and an ade6+ marker was inserted within the heterochromatic silent mating type locus (mat2::ade6+). By selecting colonies specifically defective in the silencing of Is3::ura4+, but not of mat2::ade6+, we could distinguish mutants specific to heterochromatin assembly at the SPAC23H3.14 locus from mutants defective in core assembly factors (such as Clr4).
Figure 1. Genetic screen for mutations that specifically compromise silencing at a non-DSR heterochromatin island.
(A) Genome-wide distribution of non-DSR and DSR/MTREC-dependent heterochromatin islands. Peaks of H3K9me2 along S. pombe chromosomes are shown. Asterisks denote non-DSR islands. Data from (Zofall et al., 2012) was used for this analysis. (B) The strategy used to isolate mutants specifically defective in silencing at island 3. (C) Serial dilution analyses to assay expression of Is3::ura4+ or mat2::ade6+. Wild type and mutant strains were spotted on the indicated medium. Red coloration on adenine-limited media indicates ade6+ repression while white color indicates expression. (D) Heterochromatin loss at island 3 in mut-5. H3K9me2 enrichment was determined by duplex PCR of DNA isolated from immunoprecipitated chromatin or from whole cell extracts (WCE). Enrichments were calculated as the ratio of band intensities observed in the ChIP sample PCR normalized to the ratio in the input (WCE) PCR. Average enrichments (upper number) and standard deviation of three measurements (lower number) are indicated. (E) H3K9me2 distribution at individual loci in the indicated strains, as determined by ChIP-chip. Note that mut-5 cells lack heterochromatin at non-DSR islands (3 and 15), whereas heterochromatin is unaffected at DSR-dependent islands (1 and 9). Chromosome positions in (A) and (E) correspond to the Sanger Center S. pombe database 2007 assembly. See also Figure S1.
We identified mutants defective in the silencing of Is3::ura4+ by replica plating colonies onto medium lacking uracil as well as onto counter-selective FOA medium. Simultaneously, we also scored mutants for the expression of mat2::ade6+. Red colonies on low adenine medium indicated that heterochromatic silencing was maintained, whereas white colony color indicated the loss of mat2::ade6+ silencing (Figure 1B and 1C). Several mutants specifically defective in Is3::ura4+ silencing (e.g. Ura+ and red color) were subjected to successive genetic backcrosses to confirm the silencing defects and to eliminate mutations that did not co-segregate with the silencing phenotype. The mut-5 mutant conferred the strongest silencing defect and was selected for further characterization.
mut-5 affects the assembly of several non-DSR heterochromatin islands
We next investigated whether changes in heterochromatin assembly accompanied the defective silencing of Is3::ura4+ in mut-5. Interestingly, we found that mut-5 alleviated the silencing of Is3::ura4+ similar to clr4Δ (Figure 1C). More importantly, the loss of silencing was linked to defective heterochromatin assembly. In particular, H3K9me levels at island 3 in mut-5 epe1Δ were decreased to levels comparable to clr4Δ epe1Δ (Figure 1D). In contrast, mut-5 caused little or no change in H3K9me at the mat locus or at centromeric repeats (Figure S1B).
We wondered whether mut-5 similarly affects the assembly of other islands. To test this, we performed genome-wide ChIP-chip analyses of H3K9me. Remarkably, H3K9me in mut-5 epe1Δ cells was dramatically reduced at several non-DSR heterochromatin islands. In particular, we detected a severe reduction of H3K9me2 at non-DSR islands 3, 7, 13, 14, 15 and 19 (Figures 1E and S1C). Importantly, mut-5 did not affect the assembly of DSR heterochromatin islands coating the meiotic genes mcp7 (island 1) or mei4 (island 9) (Figure 1E). We conclude that mut-5 is a component of a pathway involved in facultative heterochromatin assembly.
mut-5 contains a mutation in the telomere-binding protein Taz1
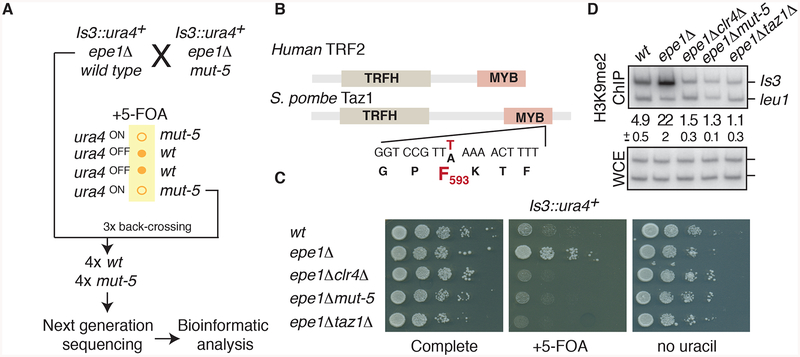
To identify the genetic locus harboring the mut-5 mutation, we first performed three backcrosses of mut-5 with the non-mutagenized parental strain. We identified the mutant phenotype in tetrad analyses by assaying for silencing of Is3::ura4+ (Figure 2A). Wild type and mut-5 segregants from the final cross were sequenced using the Illumina platform and nucleotide variants were identified using the MAQ (Mapping and Assembly with Qualities) software package (Li et al., 2008). We discovered only one substitution mutation (A-to-T) uniquely present in all mut-5 samples but absent from wild type. This mutation mapped to the taz1 gene that encodes the S. pombe ortholog of the human telomeric proteins hTRF1 and hTRF2 (Cooper et al., 1997), which are critical components of the telomere end-capping complex (de Lange, 2005). Conventional DNA sequencing confirmed the presence of the mutation within the taz1 gene specifically in segregants carrying the mut-5 allelic phenotype. The mut-5 mutation is a substitution of leucine 593 with phenylalanine (L593F) in the DNA-binding MYB domain of Taz1 (Figure 2B). The mutation did not affect the steady-state levels of the Taz1 protein (Figure S2A), however, we detected severe defects in the localization of Taz1L593F to telomeres as compared to wild type Taz1 (Figures S2B and S2C).
Figure 2. The mut-5 phenotype is associated with a mutation in the DNA binding domain of the telomere-associated protein Taz1.
(A) Identification of the mut-5 mutation by genome sequencing. The mut-5 phenotype was tracked through a series of backcrosses by assaying for FOA sensitivity of mut-5 Is3::ura4+ cells. (B) The domain structure of Taz1 and its human homologue is indicated together with the sequence substitution found in mut-5 cells. (C and D) The loss of island 3 silencing in mut-5 is phenocopied by taz1Δ. (C) Serial dilutions of the indicated strains on complete media, counter-selective (+FOA) media, and media lacking uracil. (D) ChIP of H3K9me2 at island 3 in the indicated strains. Quantitative duplex PCR was used to compare the relative enrichments of island 3 DNA in H3K9me2-immunoprecipitated and input samples. The average enrichments and standard deviations of three measurements are indicated between the panels. See also Figure S2.
Loss of Taz1 affects silencing and H3K9me at a heterochromatin island
We next tested whether the heterochromatin island assembly defects in mut-5 are indeed functionally linked to the mutation in the taz1 gene. We observed a loss of silencing at island 3 in taz1Δ that correlated with severe defects in H3K9me, similar to our results with mut-5 (Figure 2C and 2D). We conclude that Taz1 is required for heterochromatin formation and silencing at island 3. Taken together, our results suggest an important function for Taz1 in the assembly of facultative heterochromatin.
Taz1 localizes to several non-DSR heterochromatin islands
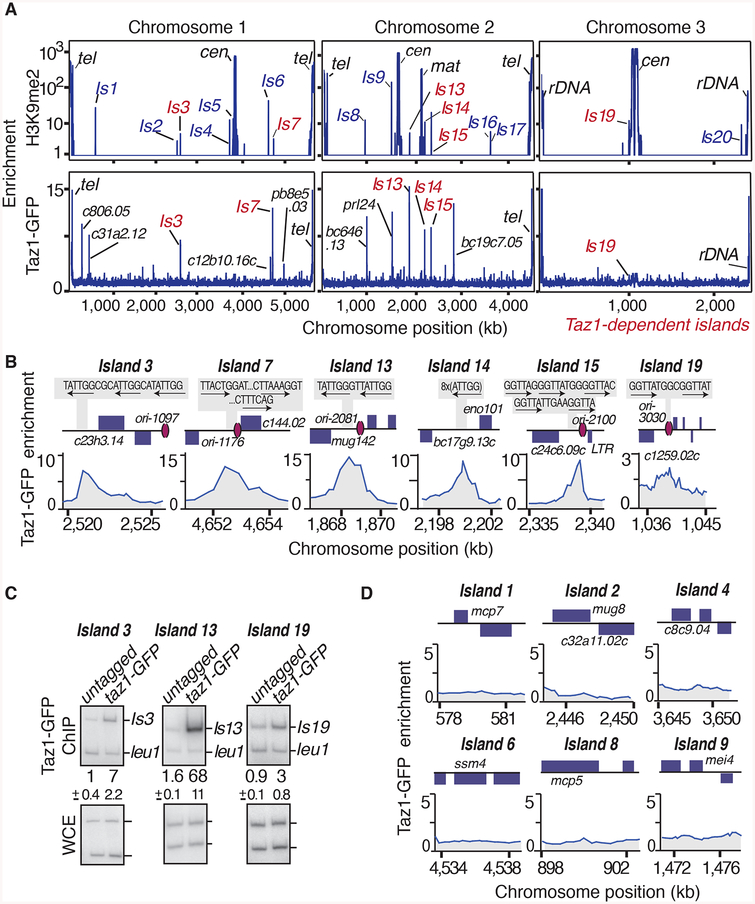
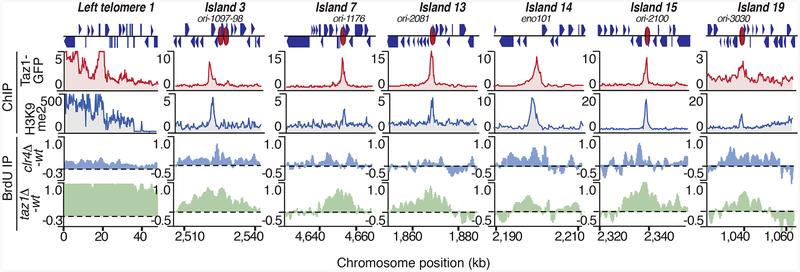
We wondered whether Taz1 physically localizes to target loci and directly assembles heterochromatin islands. ChIP-chip using either a GFP antibody targeting GFP-tagged Taz1 (Taz1-GFP) or a polyclonal antibody raised against recombinant Taz1 (Figure S3A) revealed Taz1 binding at specific sites in the genome (Figures 3A and S3A). In addition to high levels of Taz1 bound to telomeres, we detected several additional Taz1 peaks distributed across chromosome arm regions (Figures 3A and S3A), consistent with previous work (Tazumi et al., 2012). The mutation in the DNA binding domain of Taz1 (Taz1L593F) severely affected its localization on chromosome arms (Figure S3A). Remarkably, Taz1 was preferentially enriched at all non-DSR islands that showed severe defects in H3K9me2 in mut-5 (islands 3, 7, 13, 14, 15 and 19; Figure 3A and 3B). Careful examination revealed that islands 3, 13, 14, 15 and 19 contain canonical Taz1 DNA-binding sequences, whereas island 7 contains a putative variant sequence (Figure 3B). The DNA binding domain is essential for the localization of Taz1 to these heterochromatin islands (Figure S3B). We confirmed Taz1 enrichment at selected islands by conventional ChIP (Figure 3C), but could not detect Taz1 at DSR heterochromatin islands, consistent with a specific requirement for Taz1 in the assembly of Taz1-bound islands (Figure 3A and 3D).
Figure 3. Taz1 associates with Taz1-dependent heterochromatin islands.
(A) Distribution of H3K9me2 and Taz1-GFP along S. pombe chromosomes as determined by ChIP-chip. Islands affected in mut-5 cells are labeled in red. H3K9me2 data from (Zofall et al., 2012) was used. (B, C) Taz1 preferentially localizes to non-DSR islands containing telomeric repeat sequences. Note that island 7 contains a putative variant binding sequence. (B) Taz1-GFP distribution at non-DSR islands as determined by ChIP-chip. ORFs, the positions of Taz1 binding sites and late replicating origins are indicated. (C) Conventional ChIP of Taz1-GFP at the indicated islands. The average enrichments and standard deviations of three measurements are indicated. (D) Taz1-GFP distribution at DSR islands is plotted. See also Figure S3.
We noted several additional Taz1 peaks on chromosome arms at sites with no detectable H3K9me enrichment (Figure 3A). This suggests that Taz1 binding alone is not sufficient to establish detectable levels of H3K9me, and that Taz1 likely collaborates with other factors to assemble heterochromatin islands. Moreover, facultative heterochromatin assembly at these sites might be influenced by growth conditions or cell cycle stage.
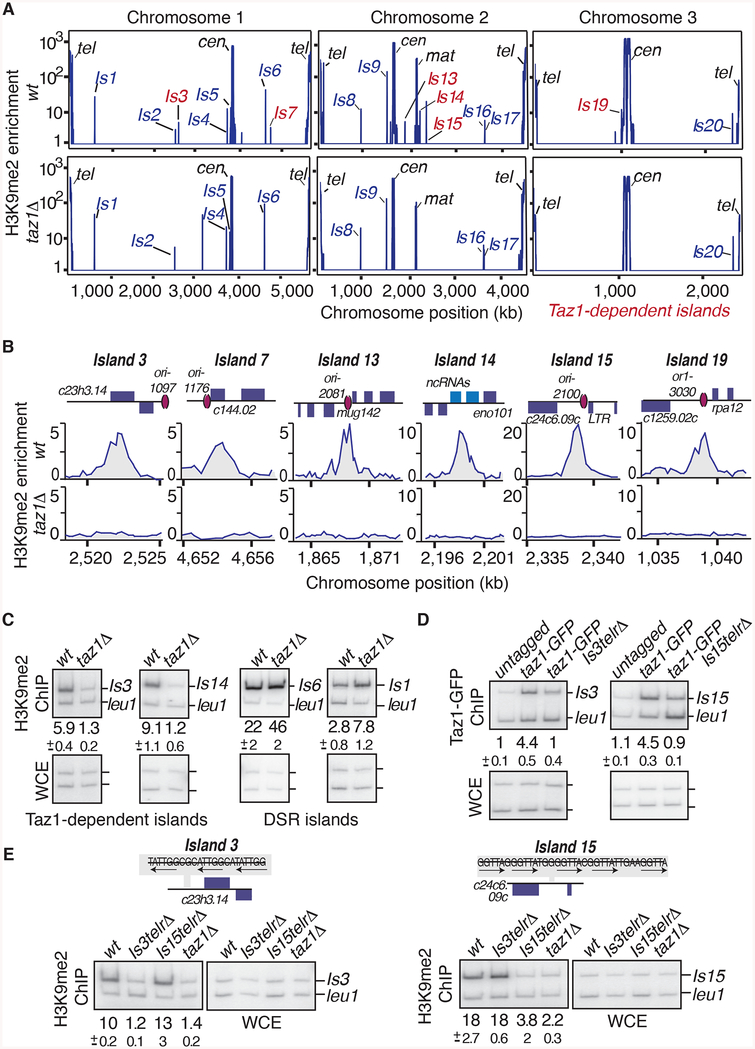
taz1Δ affects the assembly of Taz1-bound heterochromatin islands
To further investigate the role of Taz1, we compared the genome-wide distribution of H3K9me in wild type and taz1Δ. Remarkably, we observed severe defects in H3K9me at all Taz1-bound heterochromatin islands in taz1Δ (Figure 4A and 4B). This change was not due to general changes in heterochromatin assembly, as there were no defects in the assembly of DSR-containing heterochromatin islands in taz1Δ (Figures 4C and S4). In fact, H3K9me enrichment at certain DSR islands was slightly higher in taz1Δ compared to wild type (Figure 4C). Subtelomeric heterochromatin was only partially affected (data not shown), but the effects of taz1Δ are likely masked at subtelomeres by other mechanisms acting in parallel, such as RNAi (Kanoh et al., 2005). These results showing defects in H3K9me at all Taz1-bound islands in taz1Δ indicate a direct role for this protein in facultative heterochromatin assembly.
Figure 4. Taz1 localization to Taz1-dependent islands is required for heterochromatin assembly.
(A) Genome wide distribution of H3K9me2 in wild type and taz1Δ cells as determined by ChIP-chip. Taz1-bound islands are labeled in red. (B) H3K9me2 distribution at Taz1-bound islands in wild type and taz1Δ cells. ORF positions are indicated above the panels and late replication origins are marked by red ovals. Wild type H3K9me2 microarray data used in A and B are from (Zofall et al., 2012). (C) Conventional ChIP at selected islands. Duplex PCR was performed using DNA isolated from H3K9me2 immunoprecipitated fractions and input DNA from whole cell extracts. The average relative enrichments and standard deviations of replicates are shown. (D and E) Internal telomere repeat-like sequences are required for the assembly of non-DSR islands 3 and 15. (D) Conventional ChIP of Taz1-GFP in wild type and strains containing deletions of the Taz1 binding sites at island 3 (Is3telrΔ or island 15 (Is15telrΔ). (E) Conventional ChIP of H3K9me2 at islands 3 and 15 in the indicated strains. The deleted Taz1 binding motifs are indicated. The entire deleted sequence for each island can be found in Table S3. See also Figure S4.
Telomere repeat-like elements recruit Taz1 to assemble H3K9me islands
To test whether Taz1 plays a direct role in assembling heterochromatin islands, we generated strains carrying a deletion of the Taz1 binding sequences at either island 3 or island 15 (is3telrΔand is15telrΔ; Figure 4D). Remarkably, Taz1 binding and H3K9me were abolished specifically at the altered islands (Figure 4D and 4E). Importantly, in both the is3telrΔ and is15telrΔstrains, H3K9me remained unaffected at the unaltered island (Figure 4D and 4E). This result is consistent with the fact that mut-5 cells, which carry a mutation in the DNA-binding domain of Taz1, also showed loss of Taz1 binding and H3K9me (Figures S1C, S3A and S3B) and support the importance of Taz1 binding to islands for heterochromatin assembly. We conclude that the binding of Taz1 to the telomeric repeat-like elements within non-DSR islands is directly required for the formation of H3K9me at these loci.
Taz1 promotes heterochromatin-mediated gene repression
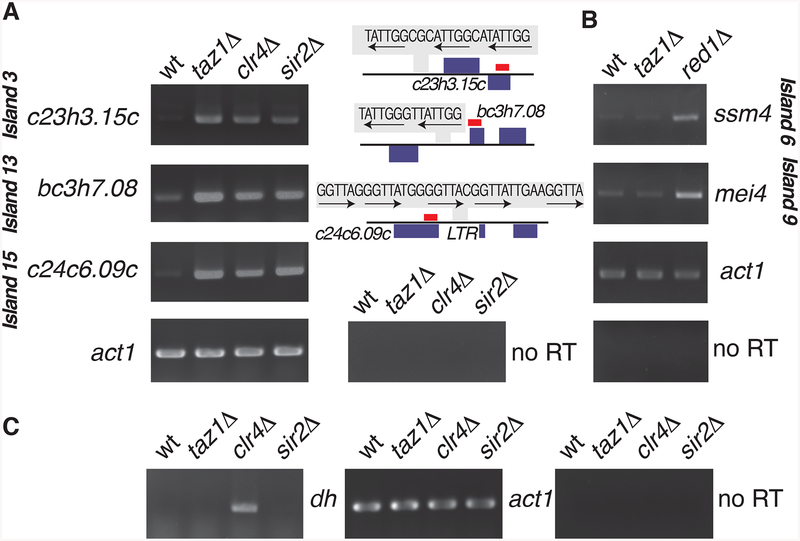
Taz1-dependent heterochromatin islands coat genes that are repressed during normal growth conditions, but expressed under stress or during meiotic induction (Chen et al., 2003; Mata et al., 2002). To determine whether Taz1-dependent heterochromatin assembly affects the expression of genes at heterochromatin islands, we compared the expression levels of loci in taz1Δ and in cells lacking the Clr4 H3K9 methyltransferase or Sir2 histone deacetylase that is also required for the assembly of heterochromatin islands (Zofall et al., 2012). Remarkably, the loss of Taz1, Clr4 or Sir2 resulted in the derepression of genes targeted by Taz1-dependent islands (Figure 5A). In contrast, we detected no change in gene expression at DSR islands in taz1Δ. As a control, we confirmed that expression of the DSR island meiotic genes ssm4 and mei4 were upregulated in a strain defective in the RNA elimination factor Red1, but not in taz1Δ (Figure 5B). Also, we observed no major defects in silencing at centromeres in taz1Δ (Figure 5C), consistent with the finding that the Taz1 mutant alleles do not affect heterochromatin formation at these loci (Figure S1B)(Tadeo et al., 2013). These results demonstrate that Taz1-dependent assembly of facultative heterochromatin islands is functionally important for the regulation of gene expression.
Figure 5. Taz1 and heterochromatin factors affect gene repression at Taz1-dependent islands.
(A) mRNA levels at the indicated Taz1-dependent islands were determined by strand specific RT-PCR. The locations of the Taz1 binding sequences and the RT-PCR amplified cDNA fragments (red bars) are indicated. (B and C) Taz1 is not required for gene repression at Taz1-independent heterochromatin sites. (B) mRNA levels at DSR heterochromatin islands in wild type, taz1Δ, and red1Δ. Note the lack of derepression at DSR-containing islands 6 and 9 targeting mei4 and ssm4 loci in taz1Δ. (C) Heterochromatin-mediated repression of pericentromeric dg/dh repeats is preserved in taz1Δ. RT-PCR of dh repeat RNAs. act1 mRNA is used as a loading control.
Taz1-dependent heterochromatin islands contain late replication origins
Taz1 plays a critical role in delayed replication initiation from certain late-firing replication origins distributed across the S. pombe genome and within subtelomeric regions (Tazumi et al., 2012). At these Taz1-affected origins, the conserved protein Rif1 directly modulates pre-replication complexes and inhibits premature activation of late origins (Hayano et al., 2012). Remarkably, Taz1-dependent facultative heterochromatin islands map to regions containing late replication origins (Figures 3B and 4B; Table S1). In particular, we found that islands 3, 7, 13, 15 and 19 contain previously annotated late firing replication origins (Hayashi et al., 2007). Notably, island 14 lacks an annotated replication origin. However, Taz1 localizes to island 14 where it is required for heterochromatin assembly (Figures 3A, 3B and 4A–C), and its loss affects the timing of replication (see below). The association of Taz1-dependent islands with late replication origins suggested the interesting possibility that heterochromatin assembly and replication timing programs might be connected.
Silencing factors affect replication timing at heterochromatin islands
We hypothesized that heterochromatin factors could affect the firing of late origins associated with Taz1-dependent islands. To test this, we analyzed the incorporation patterns of 5-bromo-2 deoxyuridine (BrdU) into DNA during replication. Cells blocked at the G2/M boundary were released and allowed to synchronously grow in medium containing BrdU, and then were arrested upon entry into early S-phase by hydroxyurea (HU). In HU-treated wild type cells, the early origins, but not the late origins, are replicated (Hayashi et al., 2007; Heichinger et al., 2006). We monitored the firing of replication origins by ChIP-chip analysis of immunoprecipitated BrdU-labeled DNA, and validated this approach by comparing the replication timing within subtelomeric regions in wild type and taz1Δ (Figures 6 and S5A). In wild type, Taz1 prevents early firing of origins at telomeres, causing cells arrested in early S-phase to show low incorporation of BrdU (Tazumi et al., 2012). However, taz1Δ showed improper early firing of origins in subtelomeric regions (Figure 6 and S5A) and early firing of specific late origins associated with prominent Taz1 peaks on chromosomal arms (Figure S5B). Importantly, early firing of late origins occurred at all Taz1-dependent heterochromatin islands (Figures 6 and S5A), including islands 3, 7, 13, 15 and 19 that contain annotated late origins. Notably, early replication was also detected at island 14 despite the lack of an annotated late origin (Figure 6).
Figure 6. Heterochromatin factors affect the timing of replication of late origins at heterochromatin islands.
Taz1 and heterochromatin factor Clr4 delay replication at telomere (1st panel from left), and Taz1-dependent islands. ChIP-chip analysis of BrdU incorporation in early S phase cells is shown. Cells carrying the cdc25–22 allele were arrested at the G2/M boundary, supplemented with HU and BrdU and released from the block by a temperature shift. The BrdU-labeled DNA was immunoprecipitated from early S-phase cells and quantified using microarray analyses. The difference between mutant and wild type cells is plotted. The distribution of Taz1-GFP and H3K9me2 is also shown. The positions of ORFs and replication origins (red ovals) are indicated above the charts. H3K9me2 microarray data from (Zofall et al., 2012) was used. See also Figure S5.
We next tested if deletion of the Taz1 binding sequences that abolishes heterochromatin affects replication timing at a specific island. Indeed, we found that the deletion of the Taz1 binding sequence at island 3 caused the loss of Taz1 binding and early firing of the adjacent late origins (Figure S5C), whereas other origins were not affected (Figure S5C). These results suggest that Taz1 acts in cis to control replication timing at island 3, consistent with previous findings that binding of Taz1 in the vicinity of late origins is sufficient to impact replication initiation (Tazumi et al., 2012). Indeed, the localized impact of Taz1 is strongly supported by statistical analysis of BrdU incorporation at individual origins. A clear effect of enhanced firing of Taz1-bound late origins was observed in taz1Δ compared to wild type (Figures S5D and S5E).
Early firing of the late origins at all Taz1-dependent heterochromatin islands prompted us to ask whether heterochromatin machinery also affects replication timing. The loss of Clr4 caused early firing of origins at subtelomeric regions (Figures 6 and S5A), but the effects were much weaker as compared to taz1Δ (Figure 6). Early firing of late origins in clr4Δ became obvious in a time course analysis (Figure S5A). Late origins at Taz1-dependent islands also fired earlier in clr4Δ than in wild type (Figures 6 and S5A). Additional statistical analyses confirmed that Clr4 preferentially affects replication timing at late origins including the Taz1-associated origins (Figure S5D and S5E). We found that clr4Δ preferentially affects Taz1-bound origins versus unbound origins (Figure S5E). Significantly, the loss of Clr4 or Taz1 had a stronger impact on late origins associated with Taz1-bound heterochromatin islands than other late origins (Figure S5F). It is notable that clr4Δ also selectively affected certain late origins not associated with H3K9me (Figure S5G), suggesting that the effects of Clr4 on late origins extend beyond non-DSR heterochromatin islands. However, we cannot rule out the possibility that Clr4 is required for heterochromatin assembly, either transiently or under specific growth conditions, at these late origins and that H3K9me is just not detectable at these sites under our current experimental conditions.
Rif1 and Shelterin components affect H3K9me and repression at Taz1-dependent islands
Taz1 is a component of the telomere capping complex Shelterin (Miyoshi et al., 2008), which protects chromosome ends from the DNA damage response, controls telomerase activity and promotes silencing (de Lange, 2005). In S. pombe, Shelterin comprises Pot1, Rap1, Tpz1, Poz1 and Ccq1 along with Taz1 (Miyoshi et al., 2008). Rap1, Tpz1 and Poz1 connect Taz1, which binds double-stranded DNA, to Pot1, a single-stranded telomeric DNA binding protein (Baumann and Cech, 2001). On the other hand, Ccq1 interacts with the HDAC Clr3 (Sugiyama et al., 2007), which is involved in heterochromatin silencing, and plays an important role in maintaining telomeres (Khair et al., 2010; Miyoshi et al., 2008; Tomita and Cooper, 2008).
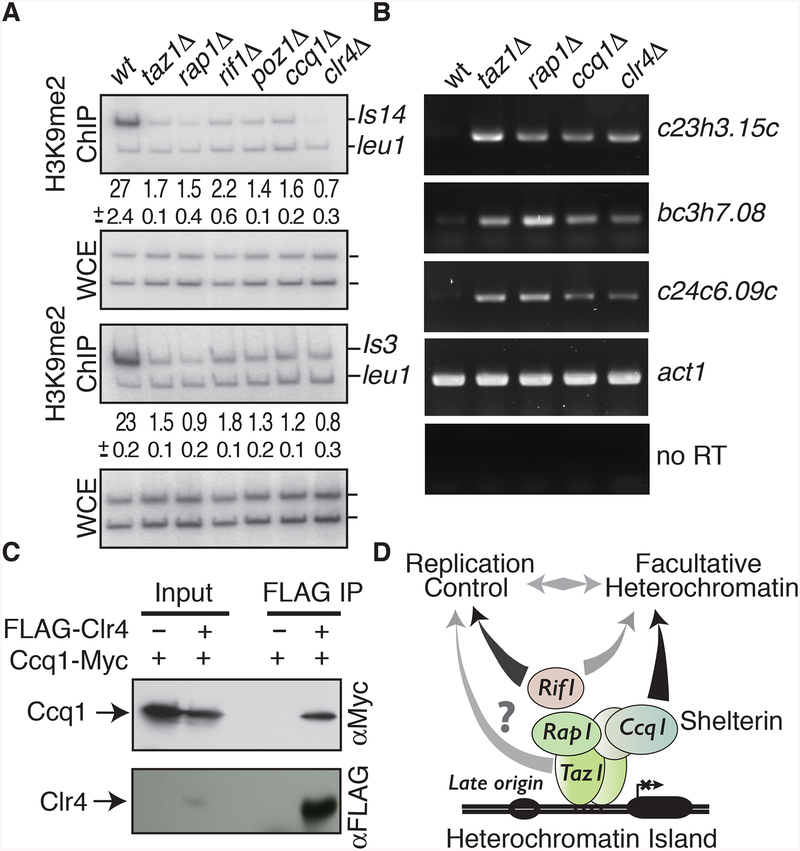
We considered that other Shelterin components could also act at Taz1-bound internal chromosomal sites. Indeed, we observed severe defects in H3K9me at Taz1-dependent islands in cells lacking Rap1, Poz1 or Ccq1 (Figure 7A). Furthermore, we found that Shelterin components are required for gene repression at these heterochromatin islands (Figure 7B). The level of derepression observed in the Shelterin mutants was comparable to clr4Δ (Figure 7B). Thus, other Shelterin components play a role in the assembly of Taz1-dependent heterochromatin islands.
Figure 7. Shelterin components and Rif1 are required for H3K9me at Taz1-dependent islands.
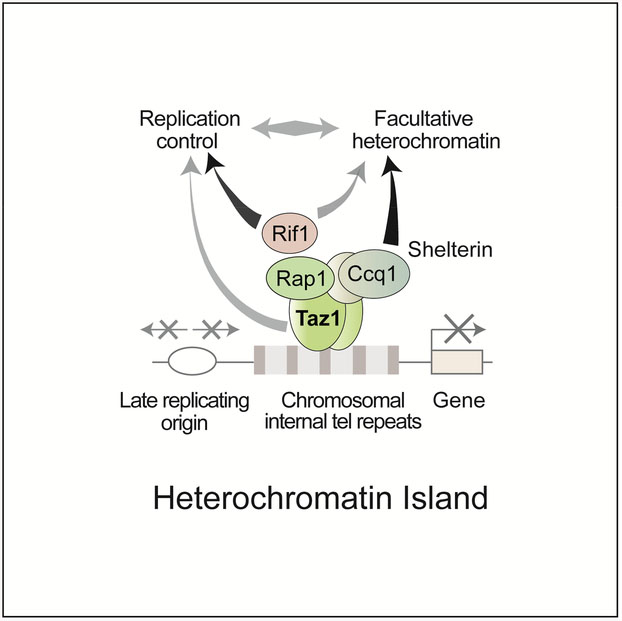
(A) Conventional ChIP combined with duplex PCR was used to assay H3K9me2 at islands 3 and 14. The average enrichments and standard deviations are indicated. (B) Shelterin components affect gene repression at Taz1 targeted islands. RT-PCR was used to examine mRNA levels at Taz1-dependent heterochromatin islands in the indicated strains. act1 mRNA was used as a loading control. (C) The Shelterin component Ccq1 associates with the Clr4 H3K9 methyltransferase, as determined by immunoprecipitation and Western blot analysis. The FLAG-tagged Clr4 co-purifying fraction was probed for Ccq1 and Clr4. (D) Model showing the involvement of Shelterin components and Rif1 in the assembly of facultative heterochromatin islands targeting late origins. Taz1 bound to DNA and other Shelterin subunits, including Ccq1, assemble facultative heterochromatin islands to regulate gene expression and control replication timing. The control of replication timing at Taz1-dependent islands also involves Rif1 (Hayano et al., 2012; Tazumi et al., 2012). Taz1 might also cooperate with additional factors, such as HDAC, to directly affect replication initiation. See also Figure S6.
We also tested whether Rif1, which associates with Taz1 and affects late origins (Hayano et al., 2012; Kanoh and Ishikawa, 2001; Tazumi et al., 2012), could also impact the assembly of heterochromatin islands. Indeed, we found that Rif1 localization could be detected at a subset of islands associated with late replication origins such as island 3 (not shown) and island 14 (Figure S6A). Moreover, we found that deletion of Rif1 affected H3K9me at Taz1-dependent, non-DSR islands (Figures 7A, S6B; Table S1). In contrast, Rif1 was found to be dispensable for H3K9me DSR-containing heterochromatin islands such as island 6 targeting ssm4 meiotic gene (Figure S6A). Interestingly, subtelomeric regions, which contain late-firing origins and show preferential enrichment of Rif1, displayed a marked decrease in H3K9me distribution in rif1Δ (Figure S6C).
Ccq1 associates with Clr4 methyltransferase
Given our results described above, we asked whether Rif1 or Shelterin components physically associate with Clr4. We did not detect an interaction between Clr4 and Taz1, Rap1 or Rif1 (Figure S6D), but we did observe a specific enrichment of Ccq1 in the protein fraction that co-purified with FLAG-tagged Clr4 (Figure 7C). We also found that Ccq1 associates with Raf1 (Figure S6D), a subunit of the Clr4 complex, ClrC (Hong et al., 2005; Horn et al., 2005; Li et al., 2005; Thon et al., 2005). Since Ccq1 was previously reported to interact with the HDAC Clr3 involved in heterochromatic silencing (Miyoshi et al., 2008; Sugiyama et al., 2007), we wondered whether Clr3 mediates the interaction between Ccq1 and ClrC. However, Ccq1 could be readily purified with Raf1 in cells lacking Clr3 (Figure S6D). Thus, in addition to its previously demonstrated role in loading the HDAC Clr3, Ccq1 likely mediates Clr4-mediated H3K9me to promote assembly of Taz1-dependent heterochromatin islands.
DISCUSSION
Heterochromatin impacts gene expression, genome organization, chromosome segregation and genome stability (Grewal and Jia, 2007). S. pombe heterochromatin domains defined by methylation of histone H3K9 (Cam et al., 2005) include small blocks of facultative heterochromatin islands that are modified in response to changes in growth conditions (Zofall et al., 2012). RNA processing factors are implicated in facultative heterochromatin formation at several meiotic genes (Tashiro et al., 2013; Zofall et al., 2012). However, the mechanisms that assemble islands elsewhere in the genome remained elusive. Here we identified Taz1, which is related to human TRF1 and TRF2 (Cooper et al., 1997), as a factor critical for the assembly of heterochromatin islands harboring late replication origins. Our results suggest that, in addition to telomere protection, Shelterin plays an important role in assembling heterochromatin islands at internal chromosomal sites. These findings have significant implications for both the maintenance of genome stability and the regulation of gene expression.
Taz1-Shelterin functions in facultative heterochromatin assembly
We describe an important role for the conserved protein Taz1 in facultative heterochromatin assembly. The taz1 mutant allele (taz1L593F) identified in our screen, as well as taz1Δ, abolish H3K9me at non-DSR heterochromatin islands associated with late replication origins. We show that Taz1 localizes to these heterochromatin islands, and that the mutation in the DNA-binding domain of Taz1 impairs its localization as well as the assembly of islands. More importantly, the deletion of Taz1 binding sequences at islands 3 or 15 abolished Taz1 binding and heterochromatin assembly specifically at these loci. Taken together, our findings provide strong support for a direct and localized role for Taz1 at specific loci.
We find that Shelterin components including Ccq1 are also required for the assembly of Taz1-dependent heterochromatin islands. Interestingly, Ccq1 associates with Clr4, but Taz1 does not. Ccq1 also associates with the HDAC Clr3, which is implicated in heterochromatic silencing (Miyoshi et al., 2008; Motamedi et al., 2008; Sugiyama et al., 2007). Ccq1 might therefore serve to connect Shelterin with chromatin modifiers involved in heterochromatic silencing. In this model (Figure 7D), Taz1 recruits Shelterin components, which in turn facilitate heterochromatin assembly via Ccq1.
Taz1 localizes to several sites in the genome that show no detectable H3K9me. Therefore, Taz1 alone might not be sufficient to assemble heterochromatin, which likely involves other factors. The possibility that Taz1 collaborates with RNA-based mechanisms to induce H3K9me is supported by our earlier finding that the loss of the RNA processing factor Mlo3 causes an increase in H3K9me at Taz1-dependent islands (e.g. island 3) (Reyes-Turcu et al., 2011). In addition, it is conceivable that facultative heterochromatin assembly at Taz1-bound sites is a regulated process that is influenced by growth conditions and/or the cell cycle and that H3K9me is not detected at these loci under our experimental conditions.
Heterochromatin silencing factors and late replication origins
Control of DNA replication origin firing is critical for coordinating the genome-wide replication program in eukaryotes (Aladjem, 2007). Taz1 controls replication firing at several late origins (Tazumi et al., 2012), including origins associated with Taz1-dependent heterochromatin islands (Figure 6), suggesting that late firing of replication origins may be necessary for the proper maintenance of heterochromatin (Gilbert, 2002; Groth et al., 2007). Regulation of origin activity might coordinate chromatin-modifying enzymes to ensure faithful propagation of heterochromatin during replication. Consistent with this, we found a link between early firing of late origins in rif1Δ (Hayano et al., 2012) and defects in heterochromatin assembly at telomeres and Taz1-dependent islands (Figure 7A and S6). On the other hand, the loss of Taz1 or Clr4, which affect the assembly of heterochromatin islands at late origins, resulted in early firing of these origins. Importantly, the loss of Taz1 or Clr4 did not indiscriminately affect replication timing of all late origins (Figure S5E). The effect on origins is specific, as deletion of the Taz1-binding sequence at an island containing late origins caused the loss of heterochromatin and resulted in early firing of the late origins only at the altered locus. Thus, these changes in replication reflect direct effects of these factors rather than indirect effects from a general change in cellular physiology.
In comparison to Taz1, the significantly weaker effects of heterochromatin factors on late origins suggest that Taz1 exploits additional mechanisms to control replication. Taz1 collaborates with Rif1 (Kanoh and Ishikawa, 2001), which controls origin activation by influencing the assembly of the pre-replication complex (Hayano et al., 2012; Hiraga et al., 2014; Mattarocci et al., 2014). Indeed, Rif1 affects replication timing at late origins associated with Taz1-dependent heterochromatin islands (Table S1) (Hayano et al., 2012; Tazumi et al., 2012). It is also possible that Taz1 and its associated factors target additional silencing effectors (such as HDACs), which can be recruited independently of H3K9me and HP1 through site-specific DNA-binding proteins, thereby masking Clr4 function at late origins. In S. cerevisiae, deletion of the histone deacetylase Rpd3, related to Clr6 in S. pombe, causes early firing of a subset of late origins (Vogelauer et al., 2002). Further studies will address the interplay between Taz1 and other factors in the control of replication at heterochromatin islands.
How might heterochromatin factors affect late origins? Heterochromatin at non-DSR islands might serve as a platform for factors involved in replication control and/or assemble chromatin structure unfavorable for pre-RC assembly, as suggested in S. cerevisiae (Crampton et al., 2008). Defects in heterochromatin factors cause increased turnover of histones, which correlates with the opening of chromatin (Aygun et al., 2013). In contrast, the loss of Epe1 stabilizes heterochromatin modifications including at Taz1-dependent islands (Isaac et al., 2007; Zofall et al., 2012) and correlates with suppression of histone turnover (Aygun et al., 2013). Interestingly, the loss of Clr4 impacts the three-dimensional organization of the genome (Mizuguchi et al., 2014), which was recently proposed to affect the positioning of late origins in the nucleus (Pichugina et al., 2016). Therefore, Clr4 could also impact replication, at least in part, by altering genome organization. Indeed, replication foci believed to contain clusters of replication origins are disrupted in clr4Δ (Kaykov and Nurse, 2015), and a link between specific nuclear positioning of late origins and replication timing has been reported in S. cerevisiae (Heun et al., 2001). Consistent with global changes in genome architecture, loss of Clr4 affects certain late origins not associated with heterochromatin islands. However, we have observed new peaks of H3K9me that map to late replication origins in cells cultured under a specific growth condition (our unpublished data). This suggests that facultative heterochromatin islands target additional origins, but these islands evade detection under normal growth conditions.
The connection between Shelterin, facultative heterochromatin and late origins has important biological implications. Changes in environmental conditions that modify heterochromatin islands might modulate replication initiation and its associated DNA metabolic processes. For example, local changes in origin efficiencies affect the distribution of meiotic double strand break (DSB) formation sites (Wu and Nurse, 2014). We note that all Taz1-dependent heterochromatin islands map to sites of meiotic DSBs, as determined by mapping of Rec12 (Figure S6E)(Cromie et al., 2007). Suppressing early firing of late origins might therefore prevent inappropriate formation of DSBs and genome instability. Indeed, fragile sites are often associated with late replication timing (Ozeri-Galai et al., 2014). Since origins fire stochastically, DSBs are expected to occur at low frequency under specific growth conditions (i.e., conditions that induce untimely expression of meiotic genes in vegetative cells). However, DSBs are readily detected upon meiotic induction, correlating with the loss of heterochromatin islands (Zofall et al., 2012). Heterochromatin machinery also suppresses inappropriate recombination (Peng and Karpen, 2008) and may collaborate with Shelterin components at internal chromosomal sites to protect the integrity of inherently unstable genomic regions.
Facultative heterochromatin islands and gene regulation
The repression of gene expression by Taz1-dependent facultative heterochromatin islands might be necessary for coordination of origin firing with transcription by RNA polymerase II, as is suggested to occur at centromeres (Li et al., 2011; Zaratiegui et al., 2011). However, in other parts of the genome replication origins are frequently found near active genes (Hayashi et al., 2007; Heichinger et al., 2006). Heterochromatin islands contain genes that are repressed during normal growth conditions, but induced under stress or during meiosis (Chen et al., 2003; Mata et al., 2002). We propose that the association of facultative heterochromatin with late origins might be critical for differential regulation of gene expression under different growth conditions. Changes in heterochromatin at these loci might be physiologically important for reprogramming gene expression patterns in response to environmental and developmental signals. Late replication at origins may contribute to repression of genes either by promoting proper maintenance of chromatin modifications or by regulating the binding of trans-acting factors required for gene transcription. Shelterin- and heterochromatin-mediated control of late origins and gene expression might therefore be coupled processes. Further work is needed to understand connections between late origins and regulation of gene expression.
In metazoans, the DNA replication program is modulated as a function of development, and mutations in replication factors cause developmental abnormalities (Aladjem, 2007; Nordman and Orr-Weaver, 2012). Whether changes in replication timing affect gene regulation in the vicinity of origins is unclear. Similar to fission yeast homologs, mammalian Shelterin subunits bind to discrete extra-telomeric sites throughout the genome, suggesting additional functions (Bosco and de Lange, 2012; Mao et al., 2007; Yang et al., 2011). In this respect, our finding that Taz1 and its interacting factors play a role in facultative heterochromatin assembly at late origins begins to shed light on extra-telomeric roles for Shelterin. Shelterin might team up with heterochromatin factors to control gene expression and other DNA metabolic processes critical for genome stability. Since heterochromatin assembly pathways involving Clr4/Suv39h are highly conserved from S. pombe to mammals, studies of Shelterin functions at heterochromatin islands will likely yield insights applicable to higher eukaryotes.
EXPERIMENTAL PROCEDURES
Strains, media and growth conditions
Strains are listed in Table S2. Standard techniques were used to culture, sporulate, cross and genetically manipulate S. pombe. Gene deletion and epitope-tagged strains were generated using PCR-module based techniques (Bahler et al., 1998). Further details including construction of strains with deleted Taz1 binding sequences at island 3 or island 15 can be found in the Supplementary Experimental Procedures and Table S3.
Genetic screen for mutants affecting non-DSR heterochromatin islands
Exponentially growing cells were plated on rich media and irradiated with UV (200 J/m2) using a UV Crosslinker (Fisher Scientific). Mutant colonies were screened for the loss of silencing of the ura4+ reporter adjacent to island 3 in an epe1Δ background (Isaac et al., 2007), and for the continued repression of mat2::ade6+. Clones unable to silence Is3::ura4+ but able to maintain ade6+ repression were isolated and backcrossed three times to a non-mutagenized strain. Genomic DNA isolated from four wild type and four mutant segregants from the final cross was sequenced using the MiSeq platform (Illumina). Sequence deviations present in all mutants and absent from all wild type colonies were identified using MAQ software. Co-segregation of the identified mutation with the silencing defect was further verified by conventional sequencing of additional clones from the final backcross.
ChIP and ChIP-chip
ChIP and microarray analyses of immunoprecipitated chromatin were performed as described previously (Cam et al., 2005). Paraformaldehyde cross-linked chromatin was sheared to 500–1000 bp and immunoprecipitated with 1–3 μg of antibodies raised against green fluorescent protein (Abcam Ab290), lysine-9 dimethylated histone H3 (Abcam, Ab1220 or Ab115159), FLAG (Sigma-Aldrich, A2220), MYC (Santa Cruz Biotechnology, c-Myc A14) or recombinant Taz1 protein (kindly provided by Julia Cooper). Immunoprecipitated and whole cell extract DNA (WCE) were analyzed by multiplex PCR. The ratio of PCR product intensities of target to control (leu1) locus obtained from immunoprecipitated samples was normalized to the ratio of PCR products obtained from whole cell extract DNA. The oligonucleotides used for multiplex PCR are listed in Table S4. For microarray analyses, immunoprecipitated and whole cell extract DNA were amplified by random priming, labeled with Cy5 (IP DNA) or Cy3 (WCE), and hybridized to a custom designed Agilent 4×44K S. pombe 60mer array described previously (Cam et al., 2005). Enrichments were calculated as the ratio of normalized Cy5 (ChIP) to Cy3 (WCE) signal. For additional information see the Supplementary Experimental Procedures.
RT-PCR
Total RNA for reverse-transcribed PCR was phenol-extracted from exponentially growing cells and contaminating DNA was removed by treatment with RNase-free DNase I (Promega). The One Step RT-PCR kit (Qiagen) was used to quantitatively generate cDNAs that were detected using agarose gels containing ethidium bromide. The oligonucleotides used for RT-PCR are listed in Table S4.
Co-immunoprecipitation and Western blot analysis
Cells expressing FLAG-tagged Clr4 or Raf1 were disrupted by grinding flash-frozen cells in dry ice and were extracted with HC buffer (150mM HEPES-KOH pH 7.6, 100mM KCl, 20% glycerol, 1mM EDTA, 0.01%NP40) supplemented by Complete Protease inhibitors (Roche). Extracts were cleared by centrifugation at 4,000×g and used to perform immunoprecipitations with anti-FLAG M2 agarose (Sigma-Aldrich). FLAG peptide eluted protein fractions were analyzed by Western blot for the presence of Myc- or GFP-tagged (Rif1, Ccq1 and Rap1) proteins, or were probed with anti-Taz1 antibody.
Replication assay
The replication assay was performed as previously described (Hayashi et al., 2007). cdc25–22 cells carrying the herpes simplex virus thymidine kinase expression module Pnmt1-TK and the human nucleoside transporter module Padh1-hENT were grown in EMM minimal media and arrested at the G2/M block by raising the temperature to 37°C for 4 hours 15 minutes. Cells were released from the block by a shift to 26°C in growth medium supplemented with 5-bromo-2′-deoxyuridine and hydroxyurea. Cells corresponding to maximal septation were collected and treated with 0.1% sodium azide, and then broken with glass beads. The DNA was phenol-extracted, sonicated and immunoprecipitated with mouse anti-BrdU antibody (BD Pharmingen) immobilized on Dynal anti-mouse IgG magnetic beads (Invitrogen). Input and precipitated DNA were purified and prepared for microarray analysis as described for the ChIP procedure.
Supplementary Material
Highlights.
Taz1-Shelterin assembles facultative heterochromatin at chromosomal internal sites
Taz1-dependent facultative heterochromatin domains harbor late replication origins
Loss of heterochromatin assembly factors affects replication at late origins
Shelterin promotes heterochromatin–mediated repression of genes near late origins
ACKNOWLEDGEMENTS
We thank Amikam Cohen, Hisao Masukata, Fuyuki Ishikawa, Gerry Smith and Julia Cooper for sharing strains and methods used in this study, and Julia Cooper for providing anti-Taz1 antibody. We also thank Ke Zhang and Chanan Rubin for their help with some experiments, Jemima Barrowman for editing the manuscript, and members of the Grewal lab for helpful suggestions. This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBER
All genome-wide datasets can be accessed at the NCBI GEO under the accession code GSE78823.
SUPPLEMENTAL INFORMATION
Supplemental information includes 6 Figures, 4 Tables and Supplemental Experimental Procedures and can be found with this article online.
REFERENCES
- Aladjem MI (2007). Replication in context: dynamic regulation of DNA replication patterns in metazoans. Nat. Rev. Genet 8, 588–600. [DOI] [PubMed] [Google Scholar]
- Aygun O, Mehta S, and Grewal SI (2013). HDAC-mediated suppression of histone turnover promotes epigenetic stability of heterochromatin. Nat. Struct. Mol. Biol 20, 547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub N, Noma K, Isaac S, Kahan T, Grewal SIS, and Cohen A (2003). A novel jmjC domain protein modulates heterochromatization in fission yeast. Mol. Cell. Biol 23, 4356–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A 3rd, Steever AB, Wach A, Philippsen P, and Pringle JR (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951. [DOI] [PubMed] [Google Scholar]
- Baumann P, and Cech TR (2001). Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292, 1171–1175. [DOI] [PubMed] [Google Scholar]
- Bosco N, and de Lange T (2012). A TRF1-controlled common fragile site containing interstitial telomeric sequences. Chromosoma 121, 465–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, Fitzgerald PC, and Grewal SIS (2005). Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet 37, 809–819. [DOI] [PubMed] [Google Scholar]
- Chen D, Toone WM, Mata J, Lyne R, Burns G, Kivinen K, Brazma A, Jones N, and Bahler J (2003). Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell 14, 214–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JP, Nimmo ER, Allshire RC, and Cech TR (1997). Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385, 744–747. [DOI] [PubMed] [Google Scholar]
- Crampton A, Chang F, Pappas DL Jr., Frisch RL, and Weinreich M (2008). An ARS element inhibits DNA replication through a SIR2-dependent mechanism. Mol. Cell 30, 156–166. [DOI] [PubMed] [Google Scholar]
- Cromie GA, Hyppa RW, Cam HP, Farah JA, Grewal SI, and Smith GR (2007). A discrete class of intergenic DNA dictates meiotic DNA break hotspots in fission yeast. PLoS Genet. 3, 1496–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T (2005). Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19, 2100–2110. [DOI] [PubMed] [Google Scholar]
- Gilbert DM (2002). Replication timing and transcriptional control: beyond cause and effect. Curr. Opin. Cell Biol 14, 377–383. [DOI] [PubMed] [Google Scholar]
- Grewal SIS, and Jia S (2007). Heterochromatin revisited. Nat. Rev. Genet 8, 35–46. [DOI] [PubMed] [Google Scholar]
- Groth A, Rocha W, Verreault A, and Almouzni G (2007). Chromatin challenges during DNA replication and repair. Cell 128, 721–733. [DOI] [PubMed] [Google Scholar]
- Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, Tsutsumi C, Chikashige Y, Hiraoka Y, Yamashita A, and Yamamoto M (2006). Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 442, 45–50. [DOI] [PubMed] [Google Scholar]
- Hayano M, Kanoh Y, Matsumoto S, Renard-Guillet C, Shirahige K, and Masai H (2012). Rif1 is a global regulator of timing of replication origin firing in fission yeast. Genes Dev. 26, 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Katou Y, Itoh T, Tazumi A, Yamada Y, Takahashi T, Nakagawa T, Shirahige K, and Masukata H (2007). Genome-wide localization of pre-RC sites and identification of replication origins in fission yeast. EMBO J. 26, 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heichinger C, Penkett CJ, Bahler J, and Nurse P (2006). Genome-wide characterization of fission yeast DNA replication origins. EMBO J. 25, 5171–5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun P, Laroche T, Raghuraman MK, and Gasser SM (2001). The positioning and dynamics of origins of replication in the budding yeast nucleus. J. Cell Biol 152, 385–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S-I, Alvino GM, Chang F, Lian H-Y, Sridhar A, Kubota T, Brewer BJ, Weinreich M, Raghuraman MK, and Donaldson AD (2014). Rif1 controls DNA replication by directing Protein Phosphatase 1 to reverse Cdc7-mediated phosphorylation of the MCM complex. Genes Dev. 28, 372–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong EJ, Villen J, Gerace EL, Gygi SP, and Moazed D (2005). A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3-K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA Biol. 2, 106–111. [DOI] [PubMed] [Google Scholar]
- Horn PJ, Bastie J-N, and Peterson CL (2005). A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev. 19, 1705–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac S, Walfridsson J, Zohar T, Lazar D, Kahan T, Ekwall K, and Cohen A (2007). Interaction of Epe1 with the heterochromatin assembly pathway in Schizosaccharomyces pombe. Genetics 175, 1549–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, and Allis CD (2001). Translating the histone code. Science 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Kanoh J, and Ishikawa F (2001). spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr. Biol 11, 1624–1630. [DOI] [PubMed] [Google Scholar]
- Kanoh J, Sadaie M, Urano T, and Ishikawa F (2005). Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr. Biol 15, 1808–1819. [DOI] [PubMed] [Google Scholar]
- Kaykov A, and Nurse P (2015). The spatial and temporal organization of origin firing during the S-phase of fission yeast. Genome Res. 25, 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khair L, Subramanian L, Moser BA, and Nakamura TM (2010). Roles of heterochromatin and telomere proteins in regulation of fission yeast telomere recombination and telomerase recruitment. J. Biol. Chem 285, 5327–5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NN, Chalamcharla VR, Reyes-Turcu F, Mehta S, Zofall M, Balachandran V, Dhakshnamoorthy J, Taneja N, Yamanaka S, Zhou M, et al. (2013). Mtr4-like protein coordinates nuclear RNA processing for heterochromatin assembly and for telomere maintenance. Cell 155, 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Goto DB, Zaratiegui M, Tang X, Martienssen RA, and Cande WZ (2005). Two novel proteins, Dos1 and Dos2, interact with Rik1 to regulate heterochromatic RNA interference and histone modification. Curr. Biol 15, 1448–1457. [DOI] [PubMed] [Google Scholar]
- Li F, Martienssen R, and Cande WZ (2011). Coordination of DNA replication and histone modification by the Rik1-Dos2 complex. Nature 475, 244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, and Durbin R (2008). Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18, 1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Seluanov A, Jiang Y, and Gorbunova V (2007). TRF2 is required for repair of nontelomeric DNA double-strand breaks by homologous recombination. Proc. Natl. Acad. Sci. U. S. A 104, 13068–13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Lyne R, Burns G, and Bahler J (2002). The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet 32, 143–147. [DOI] [PubMed] [Google Scholar]
- Mattarocci S, Shyian M, Lemmens L, Damay P, Altintas DM, Shi T, Bartholomew CR, Thomä NH, Hardy CFJ, and Shore D (2014). Rif1 controls DNA replication timing in yeast through the PP1 phosphatase Glc7. Cell Rep 7, 62–69. [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Kanoh J, Saito M, and Ishikawa F (2008). Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science 320, 1341–1344. [DOI] [PubMed] [Google Scholar]
- Mizuguchi T, Fudenberg G, Mehta S, Belton J-M, Taneja N, Folco HD, FitzGerald P, Dekker J, Mirny L, Barrowman J, et al. (2014). Cohesin-dependent globules and heterochromatin shape 3D genome architecture in S. pombe. Nature 516, 432–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi MR, Hong E-JE, Li X, Gerber S, Denison C, Gygi SP, and Moazed D (2008). HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol. Cell 32, 778–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordman J, and Orr-Weaver TL (2012). Regulation of DNA replication during development. Development 139, 455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozeri-Galai E, Tur-Sinai M, Bester AC, and Kerem B (2014). Interplay between genetic and epigenetic factors governs common fragile site instability in cancer. Cell. Mol. Life Sci 71, 4495–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, and Karpen GH (2008). Epigenetic regulation of heterochromatic DNA stability. Curr. Opin. Genet. Dev 18, 204–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichugina T, Sugawara T, Kaykov A, Schierding W, Masuda K, Uewaki J, Grand RS, Allison JR, Martienssen RA, Nurse P, et al. (2016). A diffusion model for the coordination of DNA replication in Schizosaccharomyces pombe. Sci. Rep 6, 18757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Zhang K, Zofall M, Chen E, and Grewal SIS (2011). Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin. Nat. Struct. Mol. Biol 18, 1132–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie M, Iida T, Urano T, and Nakayama J (2004). A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J. 23, 3825–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Cam HP, Sugiyama R, Noma K. i., Zofall M, Kobayashi R, and Grewal SIS (2007). SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 128, 491–504. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Thillainadesan G, Chalamcharla VR, Meng Z, Balachandran V, Dhakshnamoorthy J, Zhou M, and Grewal SI (2016). Enhancer of rudimentary cooperates with conserved RNA-processing factors to promote meiotic mRNA decay and facultative heterochromatin assembly. Mol. Cell 61, 747–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadeo X, Wang J, Kallgren SP, Liu J, Reddy BD, Qiao F, and Jia S (2013). Elimination of shelterin components bypasses RNAi for pericentric heterochromatin assembly. Genes Dev. 27, 2489–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro S, Asano T, Kanoh J, and Ishikawa F (2013). Transcription-induced chromatin association of RNA surveillance factors mediates facultative heterochromatin formation in fission yeast. Genes Cells 18, 327–329. [DOI] [PubMed] [Google Scholar]
- Tazumi A, Fukuura M, Nakato R, Kishimoto A, Takenaka T, Ogawa S, Song J-H, Takahashi TS, Nakagawa T, Shirahige K, et al. (2012). Telomere-binding protein Taz1 controls global replication timing through its localization near late replication origins in fission yeast. Genes Dev. 26, 2050–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon G, Hansen KR, Altes SP, Sidhu D, Singh G, Verhein-Hansen J, Bonaduce MJ, and Klar AJ (2005). The Clr7 and Clr8 directionality factors and the Pcu4 cullin mediate heterochromatin formation in the fission yeast Schizosaccharomyces pombe. Genetics 171, 1583–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, and Cooper JP (2008). Fission yeast Ccq1 is telomerase recruiter and local checkpoint controller. Genes Dev. 22, 3461–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin BD, Gonzalez-Sandoval A, and Gasser SM (2013). Mechanisms of heterochromatin subnuclear localization. Trends Biochem. Sci 38, 356–363. [DOI] [PubMed] [Google Scholar]
- Trewick SC, Minc E, Antonelli R, Urano T, and Allshire RC (2007). The JmjC domain protein Epe1 prevents unregulated assembly and disassembly of heterochromatin. EMBO J. 26, 4670–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P, and Reinberg D (2007). Facultative heterochromatin: is there a distinctive molecular signature? Mol. Cell 28, 1–13. [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi SP, Grewal SIS, and Moazed D (2004). RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303, 672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M, Rubbi L, Lucas I, Brewer BJ, and Grunstein M (2002). Histone acetylation regulates the time of replication origin firing. Mol. Cell 10, 1223–1233. [DOI] [PubMed] [Google Scholar]
- Wu P-YJ, and Nurse P (2014). Replication origin selection regulates the distribution of meiotic recombination. Mol. Cell 53, 655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Xiong Y, Kim H, He Q, Li Y, Chen R, and Songyang Z (2011). Human telomeric proteins occupy selective interstitial sites. Cell Res. 21, 1013–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaratiegui M, Castel SE, Irvine DV, Kloc A, Ren J, Li F, de Castro E, Marín L, Chang A-Y, Goto D, et al. (2011). RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II. Nature 479, 135–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Mosch K, Fischle W, and Grewal SIS (2008). Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat. Struct. Mol. Biol 15, 381–388. [DOI] [PubMed] [Google Scholar]
- Zofall M, and Grewal SIS (2006). Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol. Cell 22, 681–692. [DOI] [PubMed] [Google Scholar]
- Zofall M, Yamanaka S, Reyes-Turcu FE, Zhang K, Rubin C, and Grewal SIS (2012). RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science 335, 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.