Abstract
Three mitochondrial metabolic pathways are required for efficient energy production in eukaryotic cells: the electron transfer chain (ETC), fatty acid β-oxidation (FAO), and the tricarboxylic acid cycle. The ETC is organized into inner mitochondrial membrane supercomplexes that promote substrate channeling and catalytic efficiency. Although previous studies have suggested functional interaction between FAO and the ETC, their physical interaction has never been demonstrated. In this study, using blue native gel and two-dimensional electrophoreses, nano-LC-MS/MS, immunogold EM, and stimulated emission depletion microscopy, we show that FAO enzymes physically interact with ETC supercomplexes at two points. We found that the FAO trifunctional protein (TFP) interacts with the NADH-binding domain of complex I of the ETC, whereas the electron transfer enzyme flavoprotein dehydrogenase interacts with ETC complex III. Moreover, the FAO enzyme very-long-chain acyl-CoA dehydrogenase physically interacted with TFP, thereby creating a multifunctional energy protein complex. These findings provide a first view of an integrated molecular architecture for the major energy-generating pathways in mitochondria that ensures the safe transfer of unstable reducing equivalents from FAO to the ETC. They also offer insight into clinical ramifications for individuals with genetic defects in these pathways.
Keywords: fatty acid oxidation, mitochondrial respiratory chain complex, mitochondrial metabolism, protein structure, proteomics, electron microscopy (EM), primary metabolism, stimulated emission depletion microscopy (STED), supercomplex, trifunctional protein (TFP)
Introduction
Mitochondrial fatty acid β-oxidation (FAO),2 the electron transport chain (ETC), and the tricarboxylic acid (TCA) cycle are interrelated pathways involved in cellular bioenergetics in eukaryotes (1). FAO is composed of two major components, the transport of substrates into mitochondria through carnitine palmitoyltransferases I and II (CPTI and CPTII) linked by a carnitine-acylcarnitine translocase, and a catabolic β-oxidation enzymatic spiral, which for long-chain substrates are composed of very-long-chain–CoA dehydrogenase (VLCAD) and the mitochondrial trifunctional protein (TFP) (2, 3). TFP is an α2β2 tetramer containing the activities enoyl–CoA hydratase, 3-hydroxyacyl-CoA dehydrogenase (LCHAD, both a part of the HADHA subunit), and 3-ketoacyl-CoA thiolase (HADHB) (4). VLCAD is a homodimer containing a FAD cofactor that is the first step in the mitochondrial matrix for oxidation of the acyl-CoA substrate (5). Reducing equivalents from VLCAD in the form of FAD (FADH2) are transferred, through a series of redox reactions involving electron transfer flavoprotein (ETF) and electron flavoprotein dehydrogenase (ETFDH), to coenzyme Q (QH2) and then into ETC complex III. NADH generated from LCHAD is utilized as substrate by complex I of the ETC. The acetyl-CoA end product of FAO can enter the TCA cycle, where additional NADH and FADH2 are produced. The FAO-generated NADH and QH2 are potentially exposed to oxidation in the reactive environment of the mitochondrial matrix and rely on safe transfer of electron-reducing equivalents from NADH and QH2 to ETC to generate ATP. Any reduction in FAO–ETC linkage will lead to inhibition of ATP generation from FAO. Thus, a suitable physical interaction is required to ensure the safe transfer of electron equivalents from FAO to ETC. The mechanism of this transfer to ETC has not been elucidated.
We have previously purified a sub-mitochondrial particle by sucrose gradient centrifugation from digitonin-treated rat heart mitochondria (3). The supercomplex fractions (first four) contain the enzymatic activities of FAO for long-chain substrates and deliver reducing equivalents to the ETC with high catalytic efficiency. However, those enzyme activities do not exist in all other lower molecular fractions (3). Our findings of a highly-efficient functional interaction between FAO and ETC in supercomplexes suggested that ETFDH should physically interact with ETC complex III at its Q reduction site, whereas TFP should interact with the NADH-binding domain of complex I, allowing for channeling of reducing equivalents to the appropriate ETC components and potentially effectively preventing reducing equivalents from being nonspecifically oxidized. A number of older publications have reported functional studies consistent with interactions of various components of FAO and ETC, but the structural basis of this phenomenon remains unknown (6–8). It is now clear that the enzymes of ETC are arranged in situ as macromolecular supercomplexes containing complexes I, III, and IV, with different stoichiometries that increase catalytic efficiency and protein stability (9–13). However, a direct physical relationship between FAO and ETC has not been demonstrated. Here, we report evidence using multiple modalities, including functional assays, direct visualization, and proteomic techniques, that the enzymes of FAO physically interact with those of ETC. These results provide the first view of an integrated molecular architecture for the major energy-generating pathways in the mitochondria. Our findings also offer significant insight into clinical ramifications and diagnostic possibilities in patients with genetic energy metabolism disorders.
Results
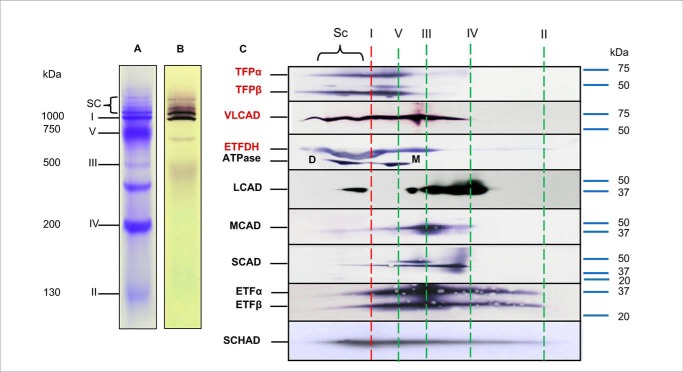
To examine FAO–ETC protein interactions, digitonin-solubilized mitochondria from rat heart mitochondria were separated using blue native-PAGE (BN-PAGE) in the first dimension. The first-dimension separation showed a typical pattern of ETC complexes and supercomplexes with complex I activity identified in all supercomplex bands (Fig. 1, A and B) (14). Western blotting was performed after second-dimension separation on a denaturing gel using a variety of antibodies (Fig. 1C). Notably, VLCAD, HADHA (TFPα), HADHB (TFPβ), and ETFDH, all involved in long-chain FAO, were mainly found to localize in the same high-molecular-mass region of the gel as the ETC supercomplexes, rather than at their native molecular mass. This suggests that the long-chain FAO and ETC proteins were separating together as a macromolecular complex in BNGE. ETFα, ETFβ, and short-chain 3-hydroxyacyl-CoA dehydrogenase (SCHAD) were widely distributed across the BN-polyacrylamide gel. Complex V serves as the lower molecular boundary between the supercomplexes
Figure 1.
Interaction of FAO enzymes with ETC supercomplexes. Mouse heart mitochondria were treated with digitonin and analyzed using BN and a second dimension of SDS-PAGE. A, Coomassie Blue-stained BN-PAGE shows multiple ET-related bands: supercomplexes (SCs); complex I (∼1,000 kDa); complex V (∼750 kDa); complex III (∼500 kDa); complex IV (∼200 kDa); and complex II (∼130 kDa). B, in-gel enzyme activity stain shows that both the isolated complex I and the SC bands have complex I activity. C, two-dimensional separation of mitochondrial proteins. BN-PAGE was followed by SDS-PAGE and then Western blotting with antibodies against FAO enzymes, including TFP, VLCAD, ETFDH, LCAD, MACD, SCAD, ETF, and SCHAD. Anti-ATPase antibody was used as a marker to reference the locations of the other protein. The ATPase dimer is labeled as D, and the monomer as M, which serves as the low molecular mass boundary of the SCs. The position of molecular mass markers on the full gel is shown to the right of C. Long-chain FAO proteins are seen to distribute in the high-molecular-mass portion of the BN gel rather than in the region corresponding to each isolated protein. The vertical red dashed lines indicate the locations of individual ETC complexes on BN-PAGE as shown in A.
In contrast, a relatively small proportion of long-chain, medium-chain, and short-chain acyl-CoA dehydrogenases (LCAD, MCAD, and SCAD, respectively) are localized in the high-molecular-mass region, with most migrating as smaller molecular mass species, suggesting only a relatively weak interaction with the larger molecular mass supercomplexes (Fig. 1C).
Digitonin-treated mitochondria were subsequently separated by sucrose gradient centrifugation, divided into 12 fractions, and then analyzed by BN-PAGE. Coomassie Blue protein staining and in-gel complex I activity identified the densest first four fractions as containing ETC supercomplexes (refer to Fig. 5A). MCAD activity localized to lower molecular mass fractions of a sucrose gradient, consistent with being present as an isolated protein (Fig. 2). This is in contrast to our previous findings that the VLCAD and TFP migrated at the top of the gradient in the same fractions as ETC SCs (3).
Figure 5.
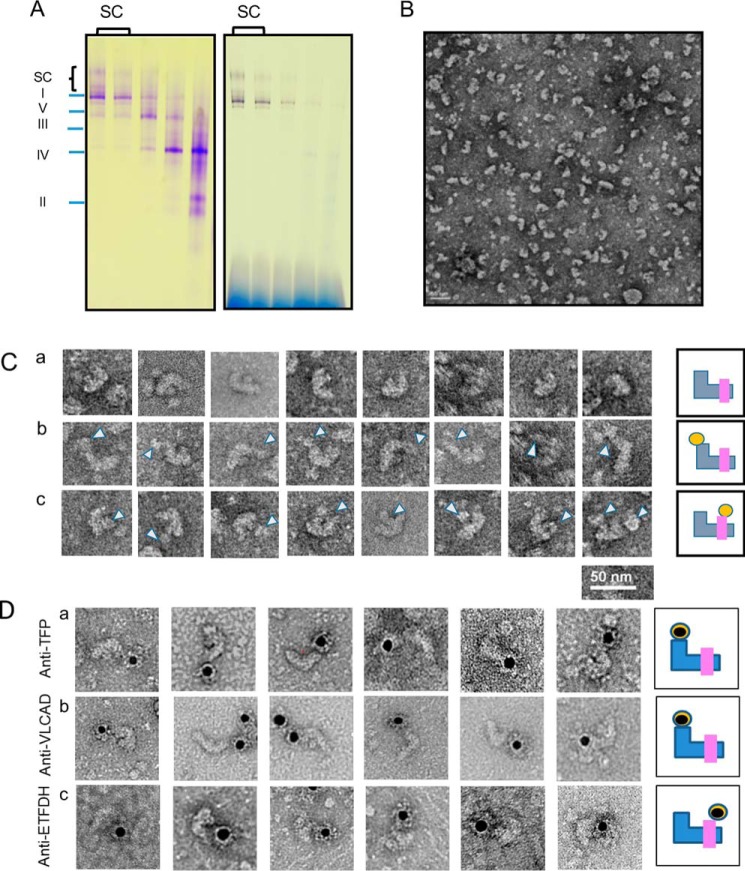
EM visualization of rat heart mitochondria FAO–ETC protein complexes. A, FAO–digitonin-permeabilized rat heart mitochondria were separated by a sucrose gradient centrifugation, and the gradient fractions were analyzed by BN-PAGE. The left panel shows a protein stain of the gel, and the right panel shows a complex I activity stain. The complex I and supercomplex fractions of the sucrose gradient were combined for EM analysis. B, EM grid with negative staining shows that there are variable forms of SC particles. C, panel a, particles are the main supercomplex form identified, representing one molecule each of complexes I, III, and IV. Panel b, additional particle binds on top of the short arm of complex I in some particles (as indicated by arrowheads). Panel c, extra particle also is seen binding to the top of complex III. In the final image in this row, both extra masses are seen simultaneously with the short (matrix) arm of complex I and the top of complex III (as indicated by arrowheads). Schematics to the right of each image stylize the three types of association with blue representing complex I; pink representing complex III; and orange representing complex IV. D, gold-labeled antisera as indicated to the left of the figure were incubated with supercomplexes on EM grids and then imaged. The black dots are the gold particles, and a summary of each set of binding experiments is cartooned to the right of the images.
Figure 2.
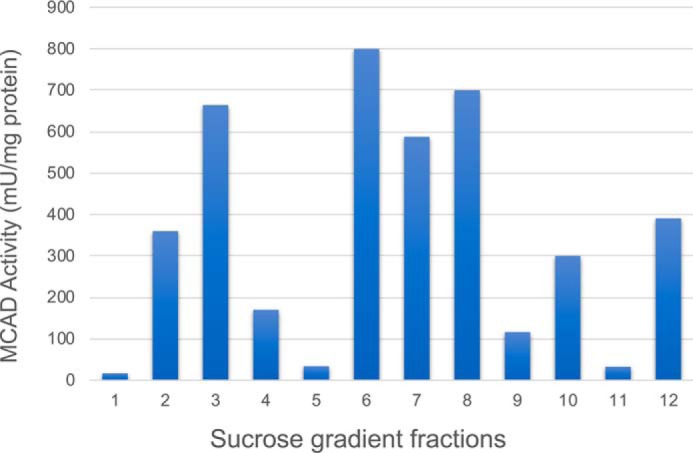
MCAD activity in solubilized mitochondria after sucrose density centrifugation. The lowest numbered fractions are the densest. Digitonin-treated rat heart mitochondria were separated by sucrose gradient centrifugation and divided into 12 fractions. SCs were mainly distributed in the first four (highest density) fractions (see Fig. 5, A and B). The graph shows the distribution of oxidation of C8CoA (MCAD substrate). The distribution of MCAD activity is highest in the middle gradient fractions, representing isolated homotetramers. However, a significant amount of activity still associates in the highest molecular mass fractions (lowest gradient numbers) containing the ETC supercomplexes. In contrast, most VLCAD activity has previously been shown to associate with the high-molecular mass fractions (3).
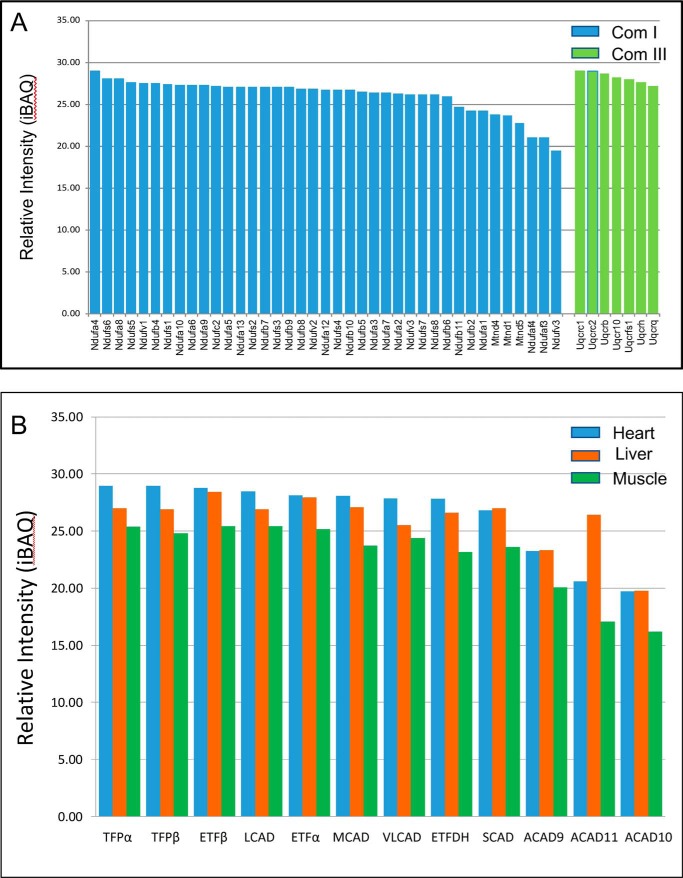
To demonstrate direct contact of FAO and ETC proteins, isolated mouse heart mitochondria supercomplexes were analyzed by mass spectrometry (MS). Approximately 39 complex I subunits, 7 complex III subunits, and 12 FAO enzymes were identified. The abundance of FAO enzymes was similar to that of complex I and III subunits (Fig. 3, A and B, and Tables S1–S3), suggesting equal contributions of each to a potential energy complex. MCAD and SCAD were also found in high-molecular-mass fractions from mouse heart mitochondria, but in lower abundance than the long-chain–specific enzymes, consistent with Western blot findings.
Figure 3.
Fatty acid oxidation proteins are associated with those of ETC. WT mouse heart mitochondria were solubilized and separated on a sucrose gradient, and high-molecular mass fractions containing the ETC supercomplexes were collected and analyzed for proteins by MS. A, MS identified 41 complex I subunits, the most abundant of which were NDUFA4, NDUFS6, and NDUFA8. None of the NADH-binding domain subunits were identified. Seven complex III subunits were also seen. B, MS analysis of the high-molecular mass sucrose gradient fractions of solubilized mitochondria from three tissues also identified 12 FAO enzymes. The full list of identified proteins can be found in Tables S1–S4.
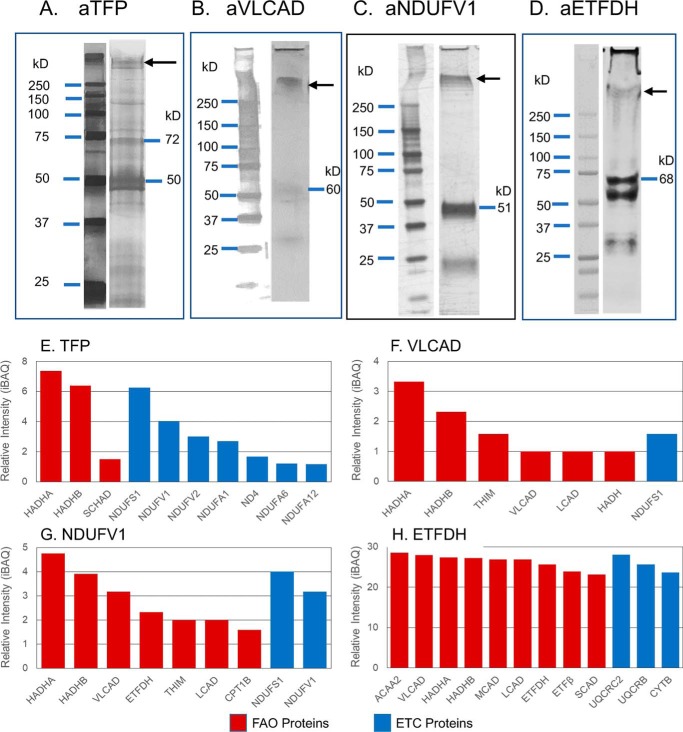
To characterize the physical relationship of these proteins, mitochondria next were treated with ethylene glycol bis-succinimidylsuccinate (EGS), a protein cross-linker, and the extract was immunoprecipitated with a variety of antibodies against FAO and ETC proteins. Cross-linked complexes identified by Western blotting (Fig. 4, A–D) were subjected to immunoprecipitation, and the captured proteins were identified through MS (Fig. 4, E–H). Use of nonspecific primary antibodies did not identify a specific pattern of protein precipitation. Treatment with antiserum against TFP co-precipitated seven complex I subunits in high abundance along with VLCAD and the expected TFP subunits (Fig. 4E). Among the seven complex I subunits, the most notable are NDUFS1 (NADH-ubiquinone oxidoreductase 75-kDa subunit, mitochondrial), NDUFV1 (NADH-ubiquinone oxidoreductase flavoprotein 1, 52-kDa subunit, mitochondrial), and NDUFV2 (NADH-ubiquinone oxidoreductase flavoprotein 2, 24-kDa subunit, mitochondrial). These are the components that form the NADH-binding domain of complex I (10, 15). These findings support that TFP interacts with the NADH-binding domain of complex I, as well as with VLCAD. Precipitation with anti-VLCAD antibody identified both subunits of TFPα and TFPβ, but only one of the ETC complex I NDUFS1, an NADH-binding domain subunit of complex I, suggesting that TFP is closer to VLCAD and complex I than VLCAD is to complex I (Fig. 4F). Treatment with antibody to NDUFV1 precipitated both TFP subunits and, to a lesser extent, VLCAD, further supporting that TFP is the main FAO binding partner of complex I (Fig. 4G). Treatment with anti-ETFDH antibody co-precipitated the core II subunit of complex III (cytochrome b-c1 complex subunit 2; gene UQCRC2) with greatest efficiency, along with the long-chain FAO proteins (Fig. 4H).
Figure 4.
Mass spectrometry analysis of proteins isolated by co-immunoprecipitation from mouse heart mitochondria treated with protein cross-linking agents. Rat heart mitochondria were incubated with the chemical cross-linker EGS, incubated with an anti-TFP, anti-VLCAD, anti-ETFDH, or anti-ETFDH antibody. The cross-linked products were co-immunoprecipitated using protein A–Sepharose 4B, followed by SDS-PAGE separation, in-gel protein digest, and MS analysis. A–D, Western blottings of the SDS-polyacrylamide gels with the indicated antisera identified larger molecular mass bands consistent with other proteins cross-linked to the primary antigen as seen to the right of A–D. The region of the SDS-polyacrylamide gel corresponding to that indicated by the arrows was cut from the gel and subjected to MS analysis. E–H, proteins identified by MS analysis of high-molecular mass bands from SDS-PAGE of cross-linked samples. Red bars represent FAO proteins, and blue bars represent ETC proteins. E, co-immunoprecipitation with anti-TFP antibody identified three complex I NADH-binding domain subunits. F, treatment with anti-VLCAD antibody also precipitated HADHA and HADHB subunits and ETC complex I subunit, NDUFS1, one of the NADH-binding domain subunits seen to bind to TFP. G, precipitation with an anti-NDUFV1 antibody precipitated HADHA and HADHB and NDUFS1 subunit of complex I. H, treatment with anti-ETFDH antibody precipitated one complex III subunit, core II, plus numerous membrane and matrix-associated FAO proteins (HADHA, HADHB, and VLCAD), along with other acyl-CoA dehydrogenases and ETF subunits.
Immunoelectron microscopy (IEM) is a standard technique used to visualize ETC supercomplexes (10, 15). To demonstrate the physical approximation of FAO proteins to ETC supercomplexes, digitonin-treated rat heart mitochondrial supercomplexes were separated with sucrose gradient centrifugation, and high-molecular-mass fractions were applied to EM grids (Fig. 5, A and B). Fig. 5A shows that the high-molecular-mass portion of the gradient was enriched for ETC SCs. EM grids with purified unlabeled ETC supercomplexes (Fig. 5B) were shown to have additional structures attached to the matrix arm of complex I and the top of complex III (Fig. 5C, panels b and c). Supercomplexes were treated with gold bead–conjugated secondary antibodies after treatment with TFP, VLCAD, or ETFDH primary antibodies (Fig. 5D). Approximately 70–80% of gold bead–labeled anti-TFP and anti-VLCAD antiserum were found bound to a protein particle located immediately adjacent to the matrix arm of complex I containing the NADH-binding domain (Fig. 5D, panels a and b). In contrast, ∼60% of gold bead–conjugated ETFDH antibodies were found predominantly bound to a protein particle located near the Q-reduction site of complex III (Fig. 5D, panel c).
The physical distance between FAO and ETC proteins was estimated directly with two-color stimulated emission depletion (STED) imaging, which is capable of identifying protein–protein co-localization at a resolution of 40 nm (Fig. 6) (16). Five antibody pairs were selected (Table S4): HADHA/NDUFS1; HADHA/VLCAD; core II/ETFDH; NDUFS1/VDAC1 (an outer mitochondrial membrane voltage-dependent anion channel); and core II/NDUFS1. NDUFS1/VCAC 1 essentially serves as a negative control with a molecular distance of >30–40 nm between them, whereas core II/NDUFS1 are known to be 15–20 nm distant from each other in ETC supercomplexes (17, 18). The fluorescent Alexa Fluor® 555 (pseudocolored green)- and Alexa Fluor® 647 (pseudocolored red)-labeled secondary antibody (Table S4) images were merged such that the generated yellow color represents the degree of co-localization of the individually selected protein pair. Fig. 6, A–E, shows a single z-stack image obtained through STED imaging of each antibody pair. Three-dimensional reconstructions of each interaction can be viewed in Movies S1–S5. STED imaging revealed that HADHA and NDUFS1 (Fig. 6A) have the highest contact among the four pairs of antibodies as observed through their co-localization on the three-dimensional movies. The HADHA/VLCAD antibody pair (Fig. 6B) produced the next highest co-localization, followed by complex III's core II and ETFDH (Fig. 6C), which shows slightly more co-localization than core II/NDUFS1. The negative control pair, VDAC1 and NDUFS1, showed no co-localization (Fig. 6D). These results indicate that the FAO–ETC interactions are all at physical distances similar to or closer than previously described distances between supercomplex components.
Figure 6.
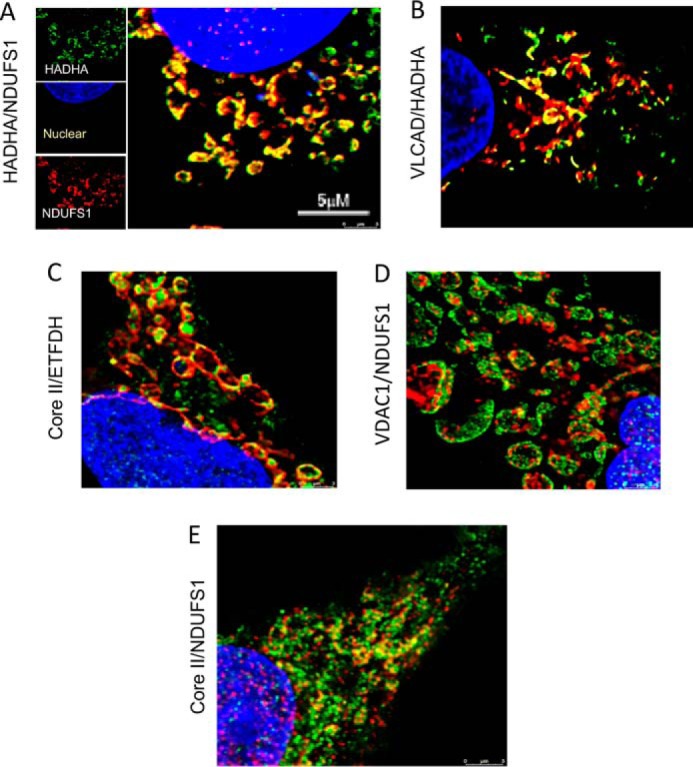
STED imaging of HEPG2 cells. Cells grown in monolayers were separately decorated with different pairs of primary antibodies with either a red or green fluorometric label. The target protein pairs of antibodies include rabbit anti-HADHA/mouse anti-NDUFS1 (A), rabbit anti-HADHA/mouse anti-VLCAD (B), and rabbit anti-core II/mouse anti-ETFDH (C). As negative control, cells were treated with the antibody pair rabbit anti-VDAC1/mouse anti-NDUFS1 (D). Antibodies to core II and NDUFS1 served as an internal collaboration showing results for a known molecular distance of 15–20 nm (E). Nuclei were stained with DAPI. A, three images on the left show the individual color signals for anti-HADHA (green), NDUFS1 (green), and DAPI (blue). The large image is the merged image with overlapping signals shown as yellow. B–E, each panel shows only the merged image of each pair of antibody pairs.
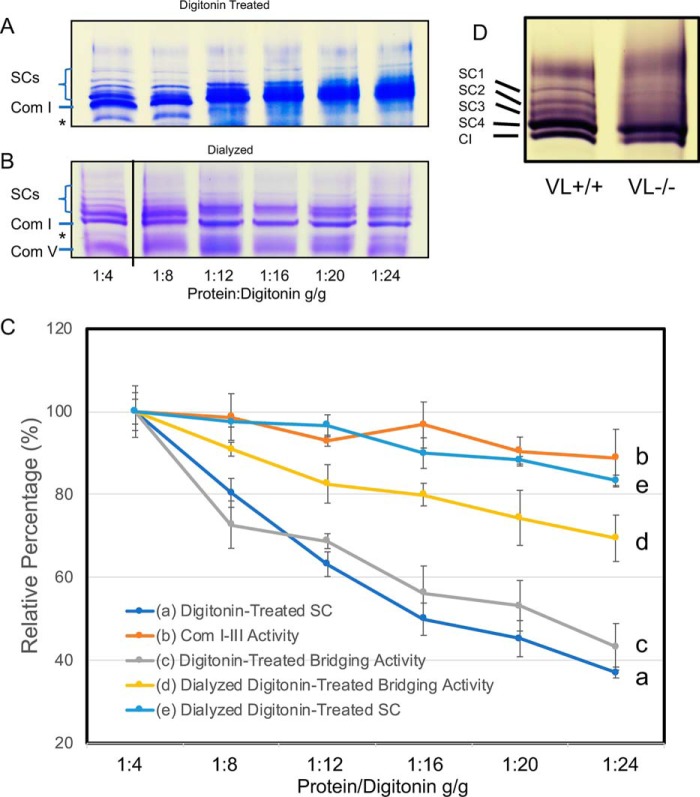
The physiologic relevance of the interaction between ETC and FAO was explored through additional functional assays. Isolated mitochondria were treated with increasing ratios of mitochondrial protein/digitonin (1:4, 1:8, 1:12, 1:16, 1:20 to 1:24 (mg/mg)) followed by BN-PAGE analysis. All supercomplex protein density levels and FAO–ETC bridging activities were compared with that of samples treated with a ratio of 1:4. Supercomplex bands on BNGE reduced ∼40% with increasing ratios of digitonin/protein (Fig. 7, A and C, line a) indicating loss of integrity of the supercomplexes. FAO–ETC bridging activity was reduced in parallel to ∼42% of control under the same conditions (Fig. 7C, line c). Dialysis of the samples prior to BNGE to remove digitonin partially restored the presence of supercomplexes (Fig. 7, B and C, line e). Notably, ETC-linked complex I–III activity was preserved at the highest digitonin concentration (Fig. 7C, line b). These results suggest that the interaction of FAO with ETC is weaker than the interaction between ETC complexes in supercomplexes. Following dialysis of the samples to remove digitonin, supercomplexes were partially restored (Fig. 7, B and C, line e) with a recovery of 45% as determined by BNGE. Similarly, bridging activity was partially restored after dialysis (Fig. 6C, line d). In total, these results indicate that the FAO enzymes physically and functionally interact with the ETC supercomplexes, albeit more weakly than the interaction between ETC complexes in supercomplexes.
Figure 7.
Functional studies of rat mitochondrial FAO–ETC complexes. A, BNGE of mitochondrial samples treated with increasing concentrations of digitonin (as per the x axis of C). Com I, ETC complex I. Only the top portion of the gel containing the SCs and complex I is shown. The band immediately below Com I is nonspecific and present in all samples regardless of treatment. B, samples as in A were dialyzed to remove the digitonin and then analyzed by BNGE. Samples are arranged as in A. Note that the 1:4 dilution slot was electronically spliced to be next to the remaining slots on the same gel. C, functional assays of rat mitochondrial samples treated with digitonin. Yellow (line d): linked ETC complex I to III assay. Gray (line c): linked FAO–ETC assay of dialyzed digitonin-treated samples. Orange (line b): linked enzymatic assays of rat mitochondrial samples. Dark blue (line a): densitometric tracing of the supercomplex bands on the blue native gel shown in A; and light blue (line e): samples from a dialyzed to remove digitonin. The y axis shows the percent of each respective measurement compared with the 1:4 sample. The x axis shows the protein/digitonin ratio. D, complex I activity gel stain of BNGE of digitonin-treated heart mitochondria isolated from wild type (WT) and VLCAD-deficient mice (VL−). Supercomplex 1–3 bands are missing in the VLCAD-deficient mitochondria. SC1–4, supercomplex bands. CI, ETC complex I.
Finally, VLCAD knockout and WT mouse mitochondria were analyzed by BN-PAGE. ETC supercomplexes SC1–3 were clearly disrupted in mitochondria from VLCAD-deficient mice (Fig. 7D). In WT animals, complex I activity in these bands contained ∼13% of total complex I activity measured by the in-gel activity stain. However, these same three supercomplex bands contained only 2.9% of the total complex I activity in VLCAD-deficient mitochondria.
Discussion
Respiratory chain supercomplexes have only relatively recently been recognized as the in situ physiologic configuration of the ETC, physically linking ETC complexes into a catalytically more efficient unit. They contain varying combinations of the individual ETC complexes with the most frequent documented being complex I/IIIn, complex I/IIIn/IVn, and complex III/IVn (10, 19–22).
Our BN-2D Western blotting (Fig. 1) and MS analysis of mouse heart mitochondrial supercomplexes (Figs. 1 and 3 and Tables S1–S3) indicate that fatty acid oxidation enzymes interact with respiratory chain proteins in supercomplexes, especially TFP, VLCAD, and ETFDH.
We have previously demonstrated that all of the enzymatic functions for complete oxidation of palmitoyl-CoA to acetyl-CoA co-separate on sucrose density gradients with ETC supercomplexes (the highest molecular mass fractions), and reducing equivalents generated by this reaction could be delivered to ETC cytochrome c at rates that are compatible with metabolic channeling (3). In contrast, in this study we demonstrate that octanoyl-CoA oxidation, representing MCAD, separates in lower molecular mass fractions of a sucrose gradient (Fig. 2). These findings provide evidence that FAO and ETC functionally interact in a macromolecular complex in such a way as to promote functional efficiency.
Complementary cross-linking immunoprecipitation, STED, and immunoelectron microscopy studies clearly show a specific interaction between FAO and ETC proteins, with VLCAD and TFP being close to complex I, and ETFDH being close to core II of complex III, containing the Q-reduction site (23). Such interactions would effectively ensure the transfer of reducing equivalents from FAO to ETC. Functional studies confirmed that the FAO–ETC interaction can be disrupted more readily than complex I and III interactions, and that it can be partially restored in vitro by dialyzing digitonin from mitochondrial preparations. Note that the gold bead–labeled ETFDH signal is less consistently oriented relative to complex III than is TFP and complex I. This is probably due to lack of membranes on the electron microscope grid, allowing some freedom of movement of the two relative to each other. This is supported by previous structural studies of respiratory chain complexes (24). Finally, disruption of ETC supercomplexes in mitochondria isolated from VLCAD-deficient hearts clearly demonstrates the importance of this protein to the integrity of the mitochondrial protein architecture.
Crystal structures of VLCAD (5), TFP (25), ETFDH (24), and the ETC supercomplexes (17) have all previously been described. Examination of the enzymatic domains of TFP shows that the LCHAD and 3-enoyl–CoA hydratase domains are situated next to each other on either end of the molecule with enoyl-CoA thiolase domains between them. This structure would accommodate the binding of VLCAD to the hydratase domain and the LCHAD domain to the NADH-inding domain of complex I, bringing VLCAD in approximation to complex I. These findings are consistent with our results. ETFDH is an integral membrane protein, with two highly hydrophobic peptide segments located on the surface facing the membrane. The ubiquinone-binding pocket is located at the hydrophobic surface. QH2 generated by ETFDH should be released into the CoA pool in the inner mitochondrial membrane. Core II is a subunit of complex III containing CoQ, located on the matrix side of the mitochondrial inner membrane. Thus, ETFDH and Com III are predicted to contact each other, presumably in close approximation to the core II CoQ-binding subunit, again consistent with our results.
Increasing concentrations of digitonin disrupted the ETC–FAO protein complexes, thereby disrupting the physical and functional interaction between the two sets of proteins and corresponding to a decrease in electron transfer activity from FAO to ETC. Supercomplexes and electron transfer from FAO to ETC were recovered by dialysis to remove digitonin from samples. Disruption of ETC supercomplexes in mitochondria isolated from VLCAD-deficient hearts clearly demonstrates the importance of this protein to the integrity of the mitochondrial protein architecture.
In total, our results allow for the development of a model of the interactions between FAO and ETC that supports the channeling of reducing equivalents from FAO to ETC. It is likely that the FAO substrate-shuttling system, composed of carnitine palmitoyltransferases I and II and the carnitine–acylcarnitine translocase, is likely to interact with the membrane-associated VLCAD through carnitine palmitoyltransferase II, although this interaction may be fluid. TFP is linked both with the complex I NADH-binding domain and VLCAD, likely on opposite sides of the TFP molecule, whereas ETFDH interacts with complex III at its matrix side. FAO matrix enzymes, LCAD, MCAD, SCAD, and SCHAD, are mainly localized to the matrix but may interact transiently with the complex.
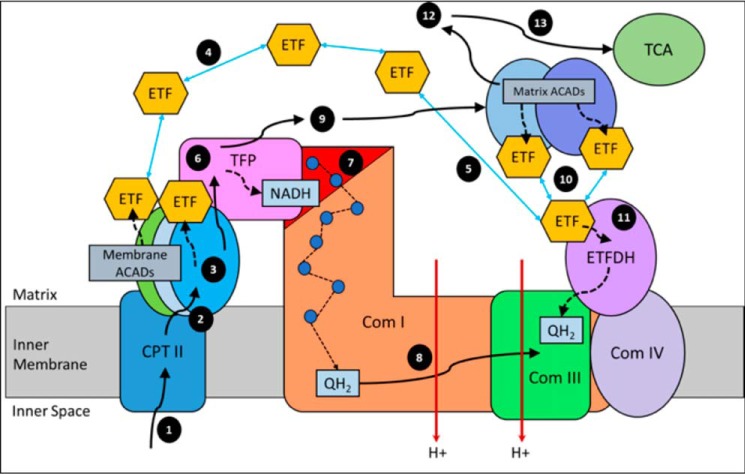
The model in Fig. 8 depicts the path of oxidation of long-chain fatty acids. Step numbers refer to those indicated in the figure. For steps 1–3, long-chain acyl-CoA substrates are transferred into mitochondria as acylcarnitines, which cross from the intermembrane space into VLCAD through CPTII in the inner membrane. VLCAD then accepts and catalyzes the released long-chain acyl-CoA substrate to its enoyl–CoA product with reduction of ETF. The protein complex promotes metabolite channeling for all these reactions. For steps 4 and 5, reduced ETF is released from VLCAD into the mitochondrial matrix, where it is free to find its redox partner, ETFDH, and shuttle its reducing equivalents (QH2) to ETC complex III. Alternatively, for high catalytic efficiency of transfer of electrons from FAO to ETC, the ETF may remain associated with the macromolecular FAO–ETC complex and instead slide down the membrane-associated proteins to more efficiently contact ETFDH. For steps 6 and 7, long-chain enoyl–CoAs, from VLCAD, channel directly to TFP where the next three reactions in the cycle occur, producing one molecule of acetyl-CoA and an acyl-CoA substrate that is two carbons shorter. As the acyl-CoAs become shorter, they become more hydrophilic, allowing them to be released into the matrix. NADH generated by the LCHAD reaction of TFP can directly channel to the NADH-binding domain of complex I. For step 8, in complex I NADH is oxidized through iron–sulfur clusters to generate QH2 at the Q-binding domain, which is then transported through the membrane electron channel of complex I to complex III. For steps 9 and 10, medium- and short-chain acyl-CoA substrates produced by TFP are transferred to MCAD and SCAD in the matrix or in a more weakly-associating peripheral domain of the multifunctional FAO–ETC complex. ETF is once again reduced and released to ETFDH, as in step 5. The remaining FAO reactions are catalyzed by monofunctional enzymes that are likely also weakly associated at the periphery of the complex. For step 11, ETFDH oxidizes reduced ETF by reducing CoQ to QH2, which is then channeled to ETC complex III. Finally, the acetyl-CoA generated by FAO is free to enter the TCA cycle or to be utilized for ketone body production (steps 12 and 13).
Figure 8.
Working model of fatty acid oxidation and respiratory chain protein interactions. See text for description.
Of note, our experiments suggest that the enzymes of medium and short-chain FAO are more weakly associated with the FAO–ETC protein complex than the high-molecular-mass mitochondrial fraction containing the long-chain FAO and ETC enzymes that can catalyze the complete oxidation of palmitoyl-CoA to acetyl-CoA with very-high efficiency (3). Because VLCAD and TFP, but not MCAD, SCAD, and SCHAD, are associated with the inner mitochondrial membrane, we surmise that the interaction of the medium- and short-chain FAO enzymes with the long-chain enzymes is weaker than that of VLCAD and TFP with complex I, and thus they are prone to loss with attempted separation and purification.
FAO relies on the safe transfer of the electron carriers NADH and QH2 to the ETC and efficient generation of ATP. The interaction between TFP/complex I and ETFDH/complex III can ensure safe transfer of electron equivalents to ETC and ultimate ATP generation. The physical relationship between FAO and ETC is of great potential relevance to clinical disorders involving these two pathways. Collectively, ∼1:4,000 people are affected with genetic defects of the respiratory chain or fatty acid oxidation, which share many clinical similarities and biochemical findings in patients. Although these similarities have been vaguely attributed to cellular energy deficit, our findings suggest a more direct explanation. In our model, deficiency of FAO proteins could disrupt the structure of the macromolecular enzyme complex and lead to secondary dysfunction of ETC as was seen in mitochondria from VLCAD-deficient mice. Resultant exacerbation of the ATP deficiency related to decreased reducing equivalents from FAO fueling oxidative phosphorylation and oxidative stress related to escape of damaging reactive oxygen species from ETC supercomplexes could expand the phenotype of FAO deficiencies, especially lactic acidosis. Similarly, secondary disruption of FAO enzymes in the context of an ETC deficiency could explain the hypoglycemia seen in some patients. Finally, the concept of synergistic heterozygosity has been used to explain an observed phenomenon of a clinically relevant reduction in energy production in patients with evidence of partial defects in multiple steps in energy pathways (26, 27). This study offers a possible structural basis for this phenomenon, as reduction in subunits of a multifunctional protein complex (or the present of structurally abnormal subunits) could lead to destabilization of the complex with resultant decrease in the efficiency of energy production. Additional experiments in model cellular and animal systems will be necessary to explore this hypothesis.
Experimental procedures
Institutional animal care and use committee assurance
All animal studies were reviewed and approved by the University of Pittsburgh Institutional Animal Care and Use Committee.
Mitochondria preparation
Mitochondria were prepared from freshly isolated mouse/rat heart, muscle, and liver and stored at −80 °C as described previously (3).
BNGE, 2D electrophoresis, and Western blotting
Mitochondrial samples were prepared and subjected to nondenaturing electrophoresis on blue native gels as described previously (3). A constant amount of protein was loaded across each gel. The complex I in-gel activity stain was performed as described previously (28). For second-dimensional separation, a strip of the native gel corresponding to a single sample well was rotated 90° and placed on a 12% SDS-polyacrylamide gel. This gel was subjected to electrophoresis as a second-dimensional gel as described previously (29). Following electrophoresis, the gel was visualized by Western blotting as described previously (3).
Sucrose gradient centrifugation
Mitochondrial samples were prepared and separated by sucrose gradient electrophoresis as described previously (3). The supercomplex fractions were combined and used for electron microscopic (EM) analysis.
Cross-linking and co-immunoprecipitation
Rat heart mitochondria were prepared as above and then resuspended in 1 ml of 10 mm phosphate buffer, pH 7.5, for 10 min. A 100 mm stock solution EGS (Thermo Fisher Scientific) was made with dimethyl sulfoxide (DMSO). This solution was added to 1 ml of mitochondrial suspension (∼3 mg/ml protein) to yield a final concentration of 1.5 mm, after which it was incubated with shaking at room temperature for 1 h. The reaction was terminated by adding 1/10 (v/v) of 50 mm Tris-HCl. Antibodies against TFP, ETFDH, VLCAD, and NDUFV1 at 1:1000 dilution were separately added to the sample and incubated at room temperature for 1 h. Protein A–coupled Sepharose beads were washed with PBS, and 0.3 ml were added to the sample. The sample was shaken for 30 min at room temperature. Protein A–Sepharose 4B beads were precipitated through brief a centrifugation and at least five washes with 900 mm NaCl in 20 mm Tris-HCl, pH 8.0, 0.5 mm EDTA, 0.5% Nonidet P-40. To the precipitated sample, 100 μl of SDS-sample buffer was added, and the mixture was heated at 90 °C for 4 min, after which the beads were removed through centrifugation. The supernatant was then subjected to SDS-PAGE and visualized with silver staining. Western blotting was performed on parallel gels to identify the cross-linked products containing FAO proteins. Protein bands of interest were excised from a silver-stained gel and sent for MS analysis.
Protein component analysis by nano-LC-MS/MS and database analysis
The bands corresponding to four respiratory SCs bands and isolated complex I on a blue native- or SDS-polyacrylamide gel were excised and analyzed by MS. Proteins were identified and quantified by a nano-LC-MS/MS (nanoscale-LC tandem MS) label-free proteomics approach, essentially as described previously (30). MaxQuant software version 1.5.2.8 was applied for protein identification and label-free quantification by means of peptide peak areas. The MS raw files were searched against a mouse database consisting of 16,658 Mus musculus sequences downloaded in 2014 from UniProt (http://www.uniprot.org/).3 Carbamidomethylation of cysteines was set as a fixed modification, whereas methionine oxidation and protein N-terminal acetylation were set as dynamic modifications. The false discovery rate (FDR) was assessed by searching against a reverse decoy database, and FDRs of protein and peptide identification were both set to 0.01.
IEM
Primary antibodies to various FAO proteins (TFP, ETFDH, and VLCAD) and secondary antibodies conjugated with 10 nm colloidal gold beads (Millipore Sigma) were used to perform IEM as described previously (31–33). The rat heart/liver mitochondrial supercomplexes were purified as above. At a concentration of 0.4 mg/ml, the sample was dialyzed against a buffer containing 30 mm HEPES, pH 7.4, 75 mm KCl, 2% sucrose, 5% glycerol, 0.1% digitonin. Primary rabbit anti-TFP/VLCAD/ETFDH polyclonal of IgG at 1000-fold dilution was added to the sample, which was then incubated at 4 °C for 1 h. A solution diluted 50-fold of 10-nm gold particles conjugated to goat anti-rabbit IgG was then added to the reaction mixture and incubated at 4 °C for 1 h. The resulting sample was directly applied to freshly glow-discharged carbon-coated 400-mesh copper grids and negatively stained with 2% solution of uranyl acetate. The samples were imaged on an FEI TF20 transmission electron microscope (Thermo Fisher Scientific Materials and Structural Analysis Division, Hillsboro, OR) operating at 200 kV. Micrographs were collected on an UltraScan 4000 CCD camera (Gatan Inc., Pleasanton, CA), 4k × 4k pixels, at a nominal microscope magnification of ×50,000 corresponding to a calibrated pixel size of 2 Å at the sample.
STED microscopy
STED imaging was performed largely as described previously (34–36).
HepG2 cells seeding
HepG2 cells were seeded at 70,000 cells/well on coated coverslips in 1 ml of Dulbecco's modified Eagle's medium in a 12-well plate and were grown at 37 °C overnight to ensure a monolayer. Each slide cover/well was treated with 300 μl of poly-l-lysine at room temperature for 5 min prior to seeding with cells. The solution was then taken out from the wells and dried at room temperature for 15 min.
Cell fixing
To fix the cells, monolayers were first washed three times with phosphate-buffered saline (PBS) at room temperature. They were then incubated with 300 μl of 2% paraformaldehyde at room temperature for 30 min, followed by another three PBS washes.
Cell immunostaining
Immunostaining began with an incubation in permeabilization buffer (0.1% TritonTM X-100 in PBS) at room temperature for 15 min, after which the cells were rehydrated with three PBS washes. The cells were blocked with 5% donkey serum in PBB (0.5% BSA + PBS) for 45 min at room temperature. Cells were washed five times with PBB, and subsequently treated with primary antibody pairs diluted in PBB (varying from 1:50 to 1:300). The primary antibody pairs can be found in Table S4. The treated cells were incubated at 4 °C overnight, after which the cells were washed five times with PBB at room temperature. The rest of the immunostaining was completed in a darkroom at room temperature. The treated cells were incubated for 1 h with secondary antibodies (Table S5) diluted in 1:1,000 PBB. This was followed by five PBB washes. NucBlueTM fixed cell ReadyProbesTM reagent (Thermo Fisher Scientific, Waltham, MA), containing 4′,6-diamidino-2-phenylindole (DAPI) dye, was used to stain the cells (500 μl/well) for 1 min and washed five times with PBB at room temperature. A drop of ProLongTM diamond antifade mountant (Thermo Fisher Scientific) was added onto microscopic slides, and the seeded coverslips were inverted and placed on the microscope slide. The slides were dried overnight at 4 °C. Cell imaging was completed through STED on a Leica TCS SP8 STED ×3 nanoscope (Leica Biosystems, Buffalo Grove, IL) at the Center for Biologic Imaging, University of Pittsburgh. Samples were imaged with a 100 × 1.4 NA Leica STED objective.
FAO–ETC bridging activity assay
This reaction reflects the functional interaction of FAO and ETC, starting with the VLCAD FAO long-chain-substrate palmitoyl-CoA, and electron transfer across TFP, complex I, and complex III, and measuring cytochrome c reduction as the reaction end point as described previously (3).
Treatment of rat heart mitochondria with variable concentrations of digitonin
Rat heart mitochondria were treated with digitonin as for BNGE, with the exception of use of variable concentrations of digitonin/protein ratios: 1:4, 1:8, 1:12. 1:16. 1:20, and 1:24 (protein/digitonin mg/mg). The mixture was incubated on ice for 20 min and centrifuged at high speed, and the supernatant was either directly analyzed by BNGE or partially dialyzed against 30 mm HEPES, pH 7.2, 150 mm potassium acetate, 10% glycerol, and 0.06% digitonin for 5 h at 4 °C prior to analysis (37). The density of the supercomplex bands on BNGE images was analyzed using ImageJ software (National Institutes of Health). Means and averages of 3–5 samples were calculated and compared by a Student's t test.
Author contributions
Y. W., J. F. C., S. W., and J. V. conceptualization; Y. W. and J. V. data curation; Y. W. and J. V. formal analysis; Y. W., J. P., N. G., A. M. M., J. F. C., M. W., S. P. M., S. B., H. A., C. S. C., and M. J. C. investigation; Y. W., C. S. C., and S. W. methodology; Y. W., J. P., N. G., A. M. M., J. F. C., S. W., and J. V. writing-review and editing; J. F. C., C. S. C., and S. W. supervision; J. V. resources; J. V. funding acquisition; J. V. writing-original draft; J. V. project administration.
Supplementary Material
This work was supported by National Institutes of Health Grant 1S10OD025009-01 (to J. F. C.) and in part by National Institutes of Health Grant S10OD021540 (to S. W.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1–S4 and Movies S1–S5.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- FAO
- fatty acid β-oxidation
- TCA
- tricarboxylic acid
- SC
- supercomplex
- DAPI
- 4′,6-diamidino-2-phenylindole
- ETC
- electron transfer chain
- Com
- complex
- TFP
- trifunctional protein
- VLCAD
- enzyme very-long-chain acyl-CoA dehydrogenase
- IEM
- immunogold EM
- SCHAD
- short-chain 3-hydroxyacyl-CoA dehydrogenase
- EGS
- ethylene glycol bis(succinimidyl succinate)
- SCAD
- short-chain acyl-CoA dehydrogenase
- LCAD
- long-chain acyl-CoA dehydrogenase
- MCAD
- medium-chain acyl-CoA dehydrogenase
- ETFDH
- electron flavoprotein dehydrogenase
- QH2
- coenzyme Q
- STED
- stimulated emission depletion
- FDR
- false discovery rate
- BNGE
- blue native polyacrylamide gel electrophoresis
- CPT
- carnitine palmitoyltransferase.
References
- 1. Vockley J., Bennett M. J., and Gillingham M. B. (2016) in The Online Metabolic and Molecular Bases of Inherited Disease (Beaudet A. L., Vogelstein B., Kinzler K. W., Antonarakis S. E., Ballabio A., Gibson K. M., and Mitchell G., eds) McGraw-Hill Co., Inc., New York [Google Scholar]
- 2. Longo N., Frigeni M., and Pasquali M. (2016) Carnitine transport and fatty acid oxidation. Biochim. Biophys. Acta 1863, 2422–2435 10.1016/j.bbamcr.2016.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang Y., Mohsen A. W., Mihalik S. J., Goetzman E. S., and Vockley J. (2010) Evidence for physical association of mitochondrial fatty acid oxidation and oxidative phosphorylation complexes. J. Biol. Chem. 285, 29834–29841 10.1074/jbc.M110.139493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liang K., Li N., Wang X., Dai J., Liu P., Wang C., Chen X. W., Gao N., and Xiao J. (2018) Cryo-EM structure of human mitochondrial trifunctional protein. Proc. Natl. Acad. Sci. U.S.A. 115, 7039–7044 10.1073/pnas.1801252115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McAndrew R. P., Wang Y., Mohsen A. W., He M., Vockley J., and Kim J. J. (2008) Structural basis for substrate fatty acyl chain specificity: crystal structure of human very-long-chain acyl-CoA dehydrogenase. J. Biol. Chem. 283, 9435–9443 10.1074/jbc.M709135200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Parker A., and Engel P. C. (2000) Preliminary evidence for the existence of specific functional assemblies between enzymes of the β-oxidation pathway and the respiratory chain. Biochem. J. 345, 429–435 10.1042/bj3450429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sumegi B., and Srere P. A. (1984) Binding of the enzymes of fatty acid β-oxidation and some related enzymes to pig heart inner mitochondrial membrane. J. Biol. Chem. 259, 8748–8752 [PubMed] [Google Scholar]
- 8. Sumegi B., Porpaczy Z., and Alkonyi I. (1991) Kinetic advantage of the interaction between the fatty acid β-oxidation enzymes and the complexes of the respiratory chain. Biochim. Biophys. Acta 1081, 121–128 10.1016/0005-2760(91)90016-B [DOI] [PubMed] [Google Scholar]
- 9. Acín-Pérez R., Fernández-Silva P., Peleato M. L., Pérez-Martos A., and Enriquez J. A. (2008) Respiratory active mitochondrial supercomplexes. Mol. Cell 32, 529–539 10.1016/j.molcel.2008.10.021 [DOI] [PubMed] [Google Scholar]
- 10. Dudkina N. V., Kudryashev M., Stahlberg H., and Boekema E. J. (2011) Interaction of complexes I, III, and IV within the bovine respirasome by single particle cryoelectron tomography. Proc. Natl. Acad. Sci. U.S.A. 108, 15196–15200 10.1073/pnas.1107819108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Althoff T., Mills D. J., Popot J. L., and Kühlbrandt W. (2011) Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1. EMBO J. 30, 4652–4664 10.1038/emboj.2011.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schägger H., de Coo R., Bauer M. F., Hofmann S., Godinot C., and Brandt U. (2004) Significance of respirasomes for the assembly/stability of human respiratory chain complex I. J. Biol. Chem. 279, 36349–36353 10.1074/jbc.M404033200 [DOI] [PubMed] [Google Scholar]
- 13. Acín-Pérez R., Bayona-Bafaluy M. P., Fernández-Silva P., Moreno-Loshuertos R., Pérez-Martos A., Bruno C., Moraes C. T., and Enríquez J. A. (2004) Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol. Cell 13, 805–815 10.1016/S1097-2765(04)00124-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jha P., Wang X., and Auwerx J. (2016) Analysis of mitochondrial respiratory chain supercomplexes using blue native polyacrylamide gel electrophoresis (BN-PAGE). Curr. Protoc. Mouse Biol. 6, 1–14 10.1002/9780470942390.mo150182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schäfer E., Seelert H., Reifschneider N. H., Krause F., Dencher N. A., and Vonck J. (2006) Architecture of active mammalian respiratory chain supercomplexes. J. Biol. Chem. 281, 15370–15375 10.1074/jbc.M513525200 [DOI] [PubMed] [Google Scholar]
- 16. Neumann D., Bückers J., Kastrup L., Hell S. W., and Jakobs S. (2010) Two-color STED microscopy reveals different degrees of colocalization between hexokinase-I and the three human VDAC isoforms. PMC Biophys. 3, 4 10.1186/1757-5036-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kühlbrandt W. (2015) Structure and function of mitochondrial membrane protein complexes. BMC Biol. 13, 89 10.1186/s12915-015-0201-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lobo-Jarne T., and Ugalde C. (2018) Respiratory chain supercomplexes: structures, function and biogenesis. Semin. Cell Dev. Biol. 76, 179–190 10.1016/j.semcdb.2017.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Berry E. A., and Trumpower B. L. (1985) Isolation of ubiquinol oxidase from Paracoccus denitrificans and resolution into cytochrome bc1 and cytochrome c-aa3 complexes. J. Biol. Chem. 260, 2458–2467 [PubMed] [Google Scholar]
- 20. Stroh A., Anderka O., Pfeiffer K., Yagi T., Finel M., Ludwig B., and Schägger H. (2004) Assembly of respiratory complexes I, III, and IV into NADH oxidase supercomplex stabilizes complex I in Paracoccus denitrificans. J. Biol. Chem. 279, 5000–5007 10.1074/jbc.M309505200 [DOI] [PubMed] [Google Scholar]
- 21. Genova M. L., and Lenaz G. (2014) Functional role of mitochondrial respiratory supercomplexes. Biochim. Biophys. Acta 1837, 427–443 10.1016/j.bbabio.2013.11.002 [DOI] [PubMed] [Google Scholar]
- 22. Guo R., Zong S., Wu M., Gu J., and Yang M. (2017) Architecture of human mitochondrial respiratory megacomplex I2III2IV2. Cell 170, 1247–1257.e12 10.1016/j.cell.2017.07.050 [DOI] [PubMed] [Google Scholar]
- 23. Zhang Z., Huang L., Shulmeister V. M., Chi Y. I., Kim K. K., Hung L. W., Crofts A. R., Berry E. A., and Kim S. H. (1998) Electron transfer by domain movement in cytochrome bc1. Nature 392, 677–684 10.1038/33612 [DOI] [PubMed] [Google Scholar]
- 24. Zhang J., Frerman F. E., and Kim J. J. (2006) Structure of electron transfer flavoprotein–ubiquinone oxidoreductase and electron transfer to the mitochondrial ubiquinone pool. Proc. Natl. Acad. Sci. U.S.A. 103, 16212–16217 10.1073/pnas.0604567103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xia C., Fu Z., Battaile K. P., and Kim J. P. (2019) Crystal structure of human mitochondrial trifunctional protein, a fatty acid cytochrome b oxidation metabolon. Proc. Natl. Acad. Sci. U.S.A. 116, 6069–6074 10.1073/pnas.1816317116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vockley J., Rinaldo P., Bennett M. J., Matern D., and Vladutiu G. D. (2000) Synergistic heterozygosity: disease resulting from multiple partial defects in one or more metabolic pathways. Mol. Genet. Metab. 71, 10–18 10.1006/mgme.2000.3066 [DOI] [PubMed] [Google Scholar]
- 27. Schuler A. M., Gower B. A., Matern D., Rinaldo P., Vockley J., and Wood P. A. (2005) Synergistic heterozygosity in mice with inherited enzyme deficiencies of mitochondrial fatty acid β-oxidation. Mol. Genet. Metab. 85, 7–11 10.1016/j.ymgme.2004.09.006 [DOI] [PubMed] [Google Scholar]
- 28. Nijtmans L. G., Henderson N. S., and Holt I. J. (2002) Blue native electrophoresis to study mitochondrial and other protein complexes. Methods 26, 327–334 10.1016/S1046-2023(02)00038-5 [DOI] [PubMed] [Google Scholar]
- 29. Reifschneider N. H., Goto S., Nakamoto H., Takahashi R., Sugawa M., Dencher N. A., and Krause F. (2006) Defining the mitochondrial proteomes from five rat organs in a physiologically significant context using 2D blue-native/SDS-PAGE. J. Proteome Res. 5, 1117–1132 10.1021/pr0504440 [DOI] [PubMed] [Google Scholar]
- 30. Sahebekhtiari N., Thomsen M. M., Sloth J. J., Stenbroen V., Zeviani M., Gregersen N., Viscomi C., and Palmfeldt J. (2016) Quantitative proteomics suggests metabolic reprogramming during ETHE1 deficiency. Proteomics 16, 1166–1176 10.1002/pmic.201500336 [DOI] [PubMed] [Google Scholar]
- 31. Shingler K. L., Cifuente J. O., Ashley R. E., Makhov A. M., Conway J. F., and Hafenstein S. (2015) The enterovirus 71 procapsid binds neutralizing antibodies and rescues virus infection in vitro. J. Virol. 89, 1900–1908 10.1128/JVI.03098-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hua J., Huet A., Lopez C. A., Toropova K., Pope W. H., Duda R. L., Hendrix R. W., and Conway J. F. (2017) Capsids and genomes of jumbo-sized bacteriophages reveal the evolutionary reach of the HK97 fold. MBio 8, e01579 10.1128/mBio.01579-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Conway J. F., Wikoff W. R., Cheng N., Duda R. L., Hendrix R. W., Johnson J. E., and Steven A. C. (2001) Virus maturation involving large subunit rotations and local refolding. Science 292, 744–748 10.1126/science.1058069 [DOI] [PubMed] [Google Scholar]
- 34. Saydmohammed M., Yagi H., Calderon M., Clark M. J., Feinstein T., Sun M., Stolz D. B., Watkins S. C., Amack J. D., Lo C. W., and Tsang M. (2018) Vertebrate myosin 1d regulates left-right organizer morphogenesis and laterality. Nat. Commun. 9, 3381 10.1038/s41467-018-05866-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tao J., Bulgari D., Berkhoudt D. A., Calderon M. J., Watkins S. C., Fonseca Velez H. J., Sabeva N., Deitcher D. L., and Levitan E. S. (2019) Drosophila Ptp4E regulates vesicular packaging for monoamine-neuropeptide co-transmission. J. Cell Sci. 132, jcs224568 10.1242/jcs.224568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Van Laar V. S., Arnold B., Howlett E. H., Calderon M. J., St Croix C. M., Greenamyre J. T., Sanders L. H., and Berman S. B. (2018) Evidence for compartmentalized axonal mitochondrial biogenesis: mitochondrial DNA replication Increases in distal axons as an early response to Parkinson's disease-relevant stress. J. Neurosci. 38, 7505–7515 10.1523/JNEUROSCI.0541-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bazán S., Mileykovskaya E., Mallampalli V. K., Heacock P., Sparagna G. C., and Dowhan W. (2013) Cardiolipin-dependent reconstitution of respiratory supercomplexes from purified Saccharomyces cerevisiae complexes III and IV. J. Biol. Chem. 288, 401–411 10.1074/jbc.M112.425876 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.