Abstract
Mitochondrial lipid overload in skeletal muscle contributes to insulin resistance, and strategies limiting this lipid pressure improve glucose homeostasis; however, comprehensive cellular adaptations that occur in response to such an intervention have not been reported. Herein, mice with skeletal muscle-specific deletion of carnitine palmitoyltransferase 1b (Cpt1bM−/−), which limits mitochondrial lipid entry, were fed a moderate fat (25%) diet, and samples were subjected to a multimodal analysis merging transcriptomics, proteomics, and nontargeted metabolomics to characterize the coordinated multilevel cellular responses that occur when mitochondrial lipid burden is mitigated. Limiting mitochondrial fat entry predictably improves glucose homeostasis; however, remodeling of glucose metabolism pathways pales compared with adaptations in amino acid and lipid metabolism pathways, shifts in nucleotide metabolites, and biogenesis of mitochondria and peroxisomes. Despite impaired fat utilization, Cpt1bM−/− mice have increased acetyl-CoA (14-fold) and NADH (2-fold), indicating metabolic shifts yield sufficient precursors to meet energy demand; however, this does not translate to enhance energy status as Cpt1bM−/− mice have low ATP and high AMP levels, signifying energy deficit. Comparative analysis of transcriptomic data with disease-associated gene-sets not only predicted reduced risk of glucose metabolism disorders but was also consistent with lower risk for hepatic steatosis, cardiac hypertrophy, and premature death. Collectively, these results suggest induction of metabolic inefficiency under conditions of energy surfeit likely contributes to improvements in metabolic health when mitochondrial lipid burden is mitigated. Moreover, the breadth of disease states to which mechanisms induced by muscle-specific Cpt1b inhibition may mediate health benefits could be more extensive than previously predicted.
Keywords: skeletal muscle metabolism, fatty acid metabolism, lipid oxidation, glucose metabolism, mitochondria, peroxisome, genomics, metabolomics, proteomics, systems biology
Introduction
In the absence of increased energy demand, heightened lipid pressure in a hyperlipidemic environment can overwhelm mitochondrial β-oxidative demand, resulting in greater incomplete fatty acid oxidation (1–3). This excess mitochondrial lipid burden appears to be a key contributor toward insulin resistance, whereas strategies that limit mitochondrial fatty acid entry improve glucose homeostasis and mitigate lipid-induced insulin resistance (3–9). Comprehensive examination of models that limit skeletal muscle mitochondrial lipid overload is likely to facilitate identification of novel mechanisms through which alleviating the mitochondrial lipid burden improves whole-body glucose homeostasis. One such model that links improved glucose homeostasis with reduced mitochondrial fatty acid entry is the skeletal muscle-specific, Cpt1b-deficient (Cpt1bM−/−) mouse (8, 9).
Carnitine palmitoyltransferase 1 (Cpt1) is localized to the outer mitochondrial membrane and is the rate-limiting enzyme controlling mitochondrial lipid entry. Cpt1 facilitates mitochondrial lipid entry by converting acyl-CoAs to acylcarnitines, which then enter the mitochondrion by passing through the transmembrane-spanning carnitine–acylcarnitine translocase. Once within the mitochondrial matrix, acylcarnitines are converted back into acyl-CoAs, which then enter the β-oxidative pathway for use as metabolic fuel. Given its critical role in this process, it is not surprising that inhibition of Cpt1 activity greatly limits mitochondrial fatty acid oxidation (9–12). Three isoforms of Cpt1 have been discovered (Cpt1a, Cpt1b, and Cpt1c) with Cpt1b being predominant in skeletal muscle. Studies by Mynatt and co-workers (8, 9) showed Cpt1bM−/− mice have reduced mitochondrial lipid oxidation and excess intramuscular lipids (triglycerides, diglycerides, and ceramides); however, these mice exhibit improved glucose homeostasis and were resistant to diet-induced obesity. These results support the concept that mitochondrial lipid overload contributes to glucose intolerance, and strategies that limit mitochondrial fat entry in skeletal muscle can limit lipid-induced insulin resistance. As such, Cpt1bM−/− mice provide a valuable experimental model that can be used to investigate adaptations that improve metabolic health when skeletal muscle mitochondrial lipid overload is mitigated. Currently, comprehensive effects of muscle-specific Cpt1b deficiency are unknown; therefore, the purpose of this study is to address this gap by employing an integrative systems biology approach to assess global molecular differences in skeletal muscle from Cpt1bfl/fl versus Cpt1bM−/− mice. Specifically, we analyzed skeletal muscle (mixed gastrocnemius) samples through datasets derived from transcriptomic, proteomic, and metabolomic platforms and interrogated the unique and common responses among the different methodologies. This integrative approach revealed substantial remodeling of substrate metabolism pathways that are consistent with our previous findings and provides insight into potential mechanisms that could contribute to the beneficial phenotypes that result when mitochondrial lipid entry is limited specifically in skeletal muscle.
Results
Global changes in gene, protein, and metabolite levels
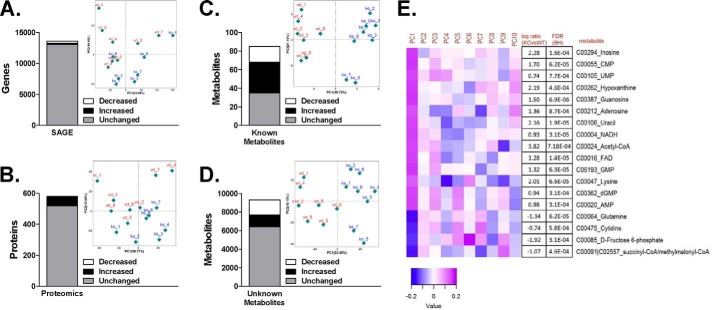
Transcriptome analysis via SAGE5 detected 26,639 genes, of which 13,602 were deemed as reliably identified (gene count >2 in at least one sample). 539 genes (∼4% of all detected genes) displayed an absolute fold-change of ≥1.5 between genotypes, with 229 genes up-regulated and 310 genes down-regulated in Cpt1bM−/− mice (Fig. 1A). Proteomics analysis yielded identifications for 3,036 peptides and 591 proteins across all samples. After curation for high-quality peptides, the final quantitative dataset was based on 2,955 peptides and 581 proteins, with 408 proteins containing at least two unique peptides. 62 proteins (∼11% of the identified proteome) were ≥1.5-fold different between genotypes with 60/62 being significantly higher in Cpt1bM−/− mice (Fig. 1B). Nontargeted metabolomics analysis annotated 9,374 m/z features displaying chromatographic peak-like qualities. Currently, 85 of these features have been identified in our library. The remaining 9,289 features likely correspond to other unidentified polar, water-soluble metabolites, but they may also contain isotope and adduct variant metabolites. Using a p < 0.05 threshold, 59% of the identified metabolites were different between genotypes, with 33 increased and 17 decreased in Cpt1bM−/− mice (Fig. 1C). Additionally, 31% of the unidentified metabolite peaks were different as 1,245 metabolites were greater in Cpt1bM−/− mice, whereas 1,637 metabolites were lower compared with Cpt1bfl/fl littermates (Fig. 1D).
Figure 1.
Overview of comprehensive changes identified by SAGE, proteomics, and metabolomics platforms. Stacked bar charts were prepared depicting the overall changes (decreased, increased, and unchanged) in Cpt1bM−/− mice relative to Cpt1bfl/fl littermate controls identified from SAGE (A), proteomics (B), known metabolites (C), and unknown metabolites (D). A principal components analysis incorporating data from both Cpt1bfl/fl (WT; red) and Cpt1bM−/− (KO; blue) mice was performed on all 'omics datasets, and results are insets within A–D. Using the known metabolomics dataset, a heatmap was generated plotting the top hits along the first 10 principal components (PC1–10) that explain 95% of the variance in the dataset (E). Eighteen individual metabolites were identified through this analysis and are listed along with the log ratio (KO versus WT) and FDR.
Principal component analysis (PCA) was performed on SAGE, proteomics, and metabolomics datasets to detect potential outliers and to further investigate how global gene, protein, and metabolite expression could differentiate between experimental groups (Fig. 1, A–D, insets). Whereas a moderate level of group separation was noted for transcriptomic and proteomic data, PCA results from the metabolomics dataset (both known and unknown metabolites) revealed a clear separation. Additionally, 41 of the 50 identified metabolites that were significantly different between Cpt1fl/fl versus Cpt1bM−/− mice were also significantly correlated (p ≤ 0.0251 and FDR <0.1) with glucose homeostasis (Fig. S1C). To further identify metabolites that were key contributors to the top principal components explaining the difference between Cpt1bfl/fl versus Cpt1bM−/− mice, we generated a variable-loading heatmap for the first 10 principal components that explains 95% of the variance within the dataset, and we further annotated it with the log ratio and false discovery rates of the metabolites (Fig. 1E). The heatmap shows the greatest contribution from these metabolites primarily involved the first PC (PC1), with some showing increased expression in Cpt1bM−/− mice (Fig. 1E, purple) and others reduced expression (blue). It is interesting to note that 11 of the 18 metabolites identified in PC1 are nucleic acid constituents, including nucleobases (hypoxanthine (purine analogue) and uracil (pyrimidine analogue)), ribonucleosides (inosine, guanosine, adenosine, and cytidine), ribonucleotides (cytidine monophosphate (CMP), uridine monophosphate (UMP), guanosine monophosphate (GMP), and adenosine monophosphate (AMP)), and a deoxyribonucleotide (deoxyguanosine monophosphate (dGMP)). These nucleic acid constituents are key contributors to the nucleotide pool that are the building blocks for DNA and RNA; therefore, we examined the SAGE dataset for pathways involved in nucleic acid metabolism. As shown in Fig. S2, gene-set enrichment analysis (GSEA) showed that neither the purine nor the pyrimidine metabolism pathways were significantly enriched, suggesting these pathways may not be the primary impetus driving changes in these metabolites. Alternatively, nucleotides are also critical to the synthesis of key factors involved in energy metabolism pathways, including nucleoside triphosphates (ATP, GTP, CTP, and UTP), reducing equivalents (NAD and FAD), and coenzyme A (CoA). As a result, much of the analyses performed using Cpt1bM−/− mice indicates significant remodeling of energy metabolism pathways, and many of these alterations are discussed in greater detail in the context of relevant metabolic pathways below.
GSEA and ingenuity pathway analysis (IPA)
To gain understanding of the possible effects of transcriptomic changes on biological processes in Cpt1bM−/− mice, we subjected SAGE data to GSEA. Results show up-regulation of several pathways involved in lipid metabolism, amino acid metabolism, pyruvate metabolism, and mitochondrial function (Table S1) in Cpt1bM−/− mice. A parallel investigation using over-representation analysis of “biofunctions” via IPA software identified significant differences in several biofunctions, including activation of lipid metabolism, carbohydrate metabolism, carboxylic acid transport, thermoregulation, and regulation of muscle mass and body size (Table S2). Remarkably, these transcriptomic adaptations were equally consistent with models predictive of better metabolic health: for example, predicted reductions in glucose metabolism disorders, hepatic steatosis, fatigue, cardiac hypertrophy, and premature mortality (Table S2). Because GSEA and IPA results revealed that remodeling of many substrate metabolism pathways in Cpt1bM−/− mice likely predicts improved metabolic health, we mapped the global gene expression differences onto a comprehensive KEGG metabolic network map (Fig. S3), highlighting the key areas of cellular metabolism affected between the genotypes. Individual pathways of interest will be discussed in greater detail below.
IPA results for Cpt1b network and fatty acid metabolism pathways
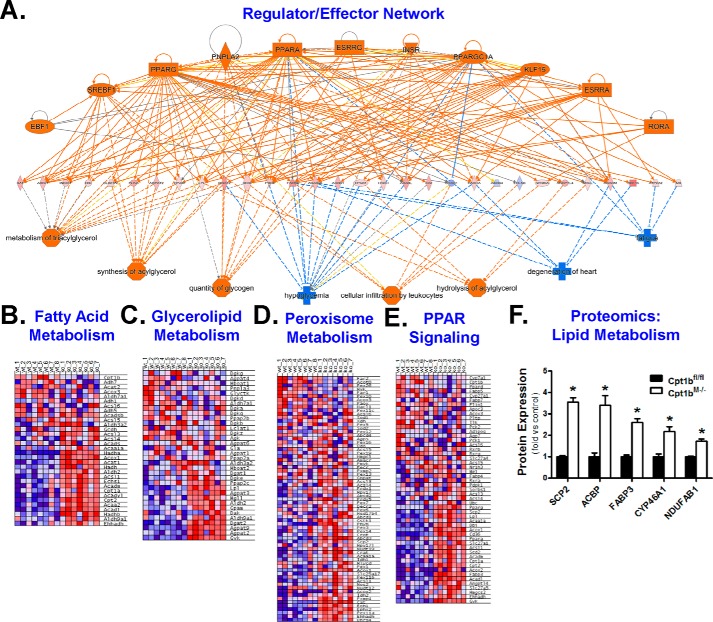
To identify possible transcriptional networks governing the observed gene expression changes, we constructed a regulator/effector network based on curated information available from the IPA Knowledge Base. The network consists of three layers with the top layer being transcription regulators, the middle layer being observed gene expression changes, and the bottom layer being predicted phenotypic effects (Fig. 2A). This network map indicated changes in genes that regulate substrate metabolism, and metabolic disease phenotypes were likely mediated by induction of overlapping transcription factor networks involving peroxisome proliferator-activated receptor (Ppar) pathways, sterol regulatory element-binding transcription factor 1 (Srebf1), retinoic acid receptor–related orphan receptor α (Rora), Krüppel-like factor 15 (Klf15), estrogen-related receptor α (Esrra), and PPARγ coactivator 1-α (Pgc-1α). Analysis of the portion of the network directly linked to Cpt1b (Fig. S4A) revealed induction of genes involved in fatty acid metabolism (Fig. 2B), glycerolipid metabolism (Fig. 2C), and peroxisomal metabolism (Fig. 2D), which were some of the most robustly up-regulated pathways noted in the GSEA (Table S1). Peroxisomal adaptations showed particular enrichment of genes involved in fatty acid oxidation (Fig. S4B). Additionally, whereas the individual PPAR isoforms did not appear to be regulated, the PPAR signaling network was identified as strongly enhanced via GSEA (Fig. 2E). Mapping the PPAR network indicated that the genes regulated in Cpt1bM−/− mice were primarily involved in lipid transport and oxidation (Fig. S4C). Proteomic analysis further supported this finding through the observed induction of several proteins involved in lipid metabolism in Cpt1bM−/− skeletal muscle (Fig. 2F).
Figure 2.
Cpt1bM−/− mice substantially remodel lipid handling pathways. IPA of SAGE data was used to develop a global regulator/effector network (A). GSEA was used to generate heatmaps showing that Cpt1bM−/− (KO; n = 7) mice exhibit up-regulation (red) of pathways involved in fatty acid metabolism (B), glycerolipid metabolism (C), peroxisome metabolism (D), and PPAR signaling (E) versus Cpt1bfl/fl (WT; n = 8) littermate controls. Peptides related to lipid metabolism were also identified in the proteomics analysis (F). *, p < 0.05. The following key for regulator/effector network is as follows: regulators (top); effectors (bottom); genes (middle); orange (activation/up-regulation); blue (inhibition/down-regulation); solid lines (direct relationship); dashed lines (indirect relationship); ellipse (transcription factor); rectangle (ligand-dependent nuclear receptor); diamond (enzyme); triangle (kinase); trapezoid (transporter); octagon (function); plus (disease).
Integrative analysis: insulin-signaling pathway, pyruvate handling, and glycolysis
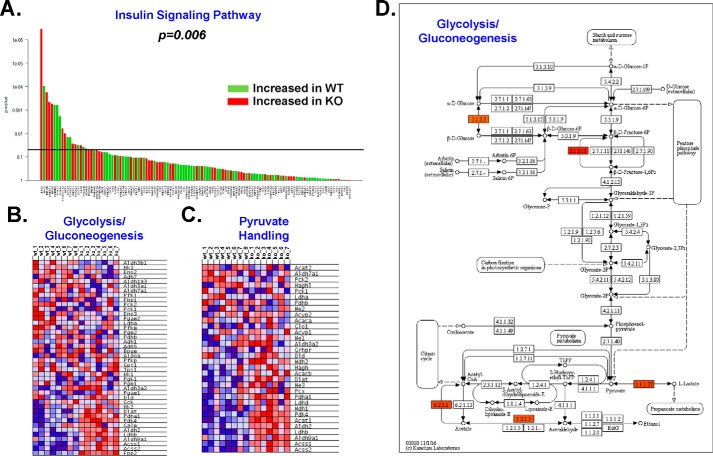
Fig. S1B shows Cpt1bM−/− mice have improved glucose homeostasis because they have lower baseline glucose levels and maintain lower blood glucose throughout a glucose tolerance test than the Cpt1bfl/fl controls, which is consistent with previous findings (8, 9, 13). However, using Akt phosphorylation as a measure of insulin sensitivity, we did not detect notable differences in insulin signaling (8, 9). In this study, globaltest analysis of 118 genes linked to the insulin-signaling cascade indicated significant alterations in this pathway (p = 0.006) within the basal (i.e. not insulin-stimulated) state (Fig. 3A). It is, however, worth noting that roughly half of the differentially regulated genes were increased, while the other half were decreased. We next investigated gene enrichment within glycolysis (Fig. 3B) and pyruvate handling (Fig. 3C) and observed significant up-regulation of genes in both pathways. Mapping changes to a KEGG pathway map suggests remodeling of the glycolytic pathway was modest and most likely driven primarily by adaptations in pyruvate handling (Fig. 3D). This coincides well with the KEGG map for the pyruvate handling pathway (Fig. S4D) and provides mechanistic insight helping to explain our previous observations showing increased pyruvate dehydrogenase activity in Cpt1bM−/− skeletal muscle (8, 9).
Figure 3.
Cpt1bM−/− mice up-regulate glucose-handling pathways. GSEA revealed significant remodeling of the insulin-signaling pathway (A). Heatmaps generated via GSEA detailed alterations in glycolysis/gluconeogenesis (B) and pyruvate handling (C) pathways. KEGG painter software was used to generate a metabolic network map showing changes in the glycolysis/gluconeogenesis pathway (D). The following key is used: red (activation/up-regulation); blue (inhibition/down-regulation).
Integrative analysis: TCA cycle
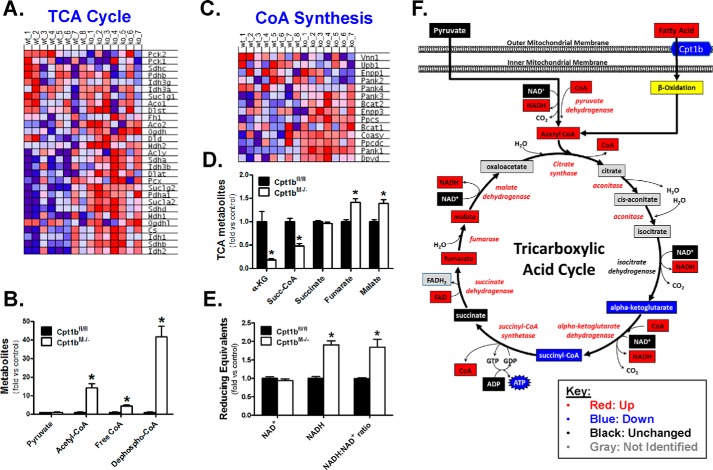
GSEA revealed genes linked to TCA cycle function were increased in Cpt1bM−/− skeletal muscle (Fig. 4A), which supports the observed heightened pyruvate oxidation in these mice (8, 9). Analysis of the identified metabolites indicated that although the pyruvate levels were not different between genotypes, acetyl-CoA levels were nearly 14-fold higher in Cpt1bM−/− skeletal muscle (Fig. 4B). Heightened acetyl-CoA production appears to be at least partially facilitated by a 4.5-fold increase in free CoA (Fig. 4B). Interestingly, enrichment of the CoA biosynthesis pathway was observed in Cpt1bM−/− mice (Fig. 4C), which included a significant increase in expression of the gene encoding the rate-limiting enzyme in CoA biosynthesis, pantothenate kinase 1 (Pank1). This was confirmed by RT-PCR (Fig. S5A), and CT values (22–24) indicate Pank1 is a fairly abundant transcript in muscle. Additionally, perhaps the most compelling evidence for enhanced CoA biosynthesis is that Cpt1bM−/− mice exhibit 41-fold greater levels of the metabolite precursor for CoA synthesis, dephospho-CoA (Fig. 4B).
Figure 4.
Effect of Cpt1b deficiency on the TCA cycle. GSEA identified up-regulation (red) of genes involved in the TCA cycle in Cpt1bM−/− (KO) mice (A). Metabolomics screening identified a high number of metabolites linked to carbon entry into the TCA cycle (B), TCA cycle intermediates (D), and reducing equivalents (E). GSEA revealed enrichment of the coenzyme A (CoA) biosynthesis pathway. Compiled results from the SAGE and metabolomics platforms are depicted in F. *, p < 0.05. The abbreviations used are as follows: α-KG, α-ketoglutarate; Succ-CoA, succinyl-CoA.
Differences in CoA and acetyl-CoA could also be impacted by a variety of other factors including but not limited to the following: changes in flux rates; acyl-CoA synthetase activity (Acss1–3, Acsf1–4, Acsm1–5, Acsl1–6, Acsbg1/2, and Fatp1–6); carnitine acyltransferase activity (CrAT, CrOT, Cpt1a, Cpt1b, Cpt1c, and Cpt2); activity of dehydrogenase enzymes with a lipoic acid moiety (pyruvate dehydrogenase, α-ketoglutarate dehydrogenase, and branched-chain alpha-keto acid dehydrogenase); acetyl-CoA acyltransferase activity (Acat1/2 and Acaa1/2); acyl-CoA thioesterase activity (Acot1–13 and Them4/5); acetoacetyl-CoA synthetase activity (Aacs); acetyl-CoA consuming reactions (CS, ACC, and HS2, etc.); as well as activity of various acetyltransferase and acyltransferase enzymes that regulate post-translational modification. Differential expression of several genes involved in these pathways was observed in the SAGE dataset, so we attempted to validate these with RT-PCR. We previously reported CrAT, CrOT, Cpt2, CS, Pdha1, Sdhb, Bckdh, and Fatp1 are significantly higher in Cpt1bM−/− skeletal muscle (8, 9), so we did not repeat these analyses. Newly identified genes involved in CoA/acetyl-CoA metabolism that were differentially regulated in the SAGE dataset included the following: Acss1, Acss2, Acsl1, Acot2, Acot8, and Acot13. Of these, only Acot2, Acot8, and Acot13 were significantly different (Fig. S5B).
Elevated acetyl-CoA provides more carbons that can enter the TCA cycle in Cpt1bM−/− mice. Although our metabolomics platform did not resolve citrate or isocitrate, earlier results show Cpt1bM−/− mice have increased complete oxidation (i.e. CO2 production) of nonlipid substrates (pyruvate and leucine) that provide acetyl-CoA precursors (8, 9), suggesting greater potential for substrate entry into the TCA cycle. Because Cpt1bM−/− mice likely have greater entry of carbons into the citrate pool, it is interesting to note that α-ketoglutarate and succinyl-CoA levels are substantially lower in these mice versus littermate controls (Fig. 4D). Alternatively, downstream of these points in the TCA cycle, succinate is similar between genotypes, whereas fumarate and malate are both significantly elevated in Cpt1bM−/− mice (Fig. 4D). Also noteworthy is the fact that NAD+ levels were similar between genotypes, and NADH levels were higher in Cpt1bM−/− mice, resulting in an increased NADH/NAD+ ratio (Fig. 4E). A visual summary depicting changes in genes and metabolites related to the TCA cycle and reducing equivalents is shown in Fig. 4F.
Integrative analysis: amino acid metabolism
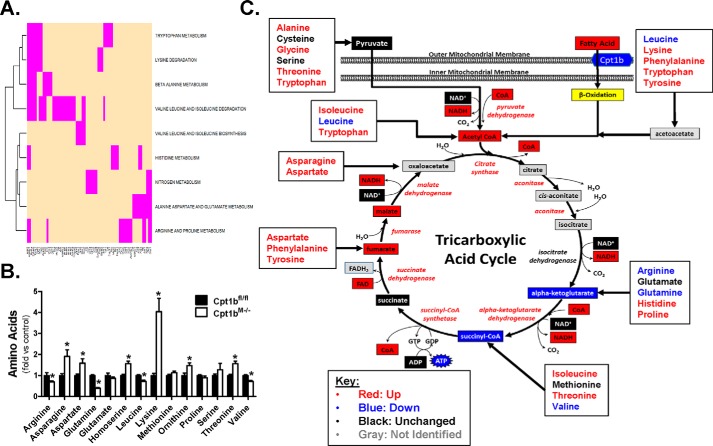
Leucine oxidation is increased in Cpt1bM−/− mice (8, 9), suggesting that when mitochondrial lipid entry is impaired, they become more reliant on amino acids as fuel. Results in Table S1 confirm substantial remodeling at the gene level of several pathways involved in amino acid metabolism, and heatmaps for each identified pathway can be found in Fig. S6. Using genes that contributed to core enrichment in all of these amino acid metabolism pathways, a heatmap was made showing commonality of all regulated genes among the amino acid metabolism pathways (Fig. 5A). As shown, little overlap between regulated genes is apparent for most of these pathways (Fig. 5A), indicating the adaptations identified by GSEA (Table S1) is due to expansive remodeling of amino acid metabolism, rather than a result of changes within a small set of genes common to all amino acid metabolism pathways. Because the genes involved in numerous amino acid metabolism pathways were regulated, the metabolomics dataset was probed to identify amino acids in skeletal muscle. Results show decreases in l-arginine, l-glutamine, l-leucine, and l-valine levels in Cpt1bM−/− mice, whereas increased levels of l-asparagine, l-aspartate, l-homoserine, l-lysine, l-ornithine, and l-threonine were identified (Fig. 5B). Although results from this nontargeted metabolomics screen did not provide a comprehensive amino acid panel, the amino acids that were identified show very similar patterns to those found in our previously targeted metabolomics analysis (9). Collectively, the metabolomics platforms from both investigations show Cpt1bM−/− mice have 13 amino acids increased in skeletal muscle, four amino acids are decreased, and three amino acids remain similar when compared with Cpt1bfl/fl controls. Because amino acids can be used as metabolic fuel, results from these datasets were used to develop an integrative map showing points of entry of carbons derived from amino acid substrates into the TCA cycle (Fig. 5C). As shown, amino acids that can be metabolized to pyruvate, fumarate, or oxaloacetate are almost universally increased. Likewise, amino acids that can be catabolized to produce acetyl-CoA are primarily increased, except leucine. Alternatively, amino acid-derived metabolites that enter the TCA cycle as either α-ketoglutarate or succinyl-CoA are not as consistently present at higher concentrations. Notably, the amino acids most commonly considered to be important in generating α-ketoglutarate are glutamate and glutamine, which are unaltered and decreased, respectively, in Cpt1bM−/− mouse muscle. Of additional interest, the most robustly up-regulated pathway identified by GSEA was leucine, isoleucine, and valine degradation (Table S1), and two of these amino acids (leucine and valine) are reduced in Cpt1bM−/− mice, which likely represent greater branched chain amino acid utilization.
Figure 5.
Substantial remodeling of amino acid metabolism pathways in Cpt1bM−/− mice. A shows a heatmap of the genes that were regulated in several pathways of amino acid metabolism, which showed core enrichment. Changes in amino acid levels were detected by metabolomics analyses (B) and incorporated into the TCA cycle map (C). *, p < 0.05.
Integrative analysis: oxidative phosphorylation
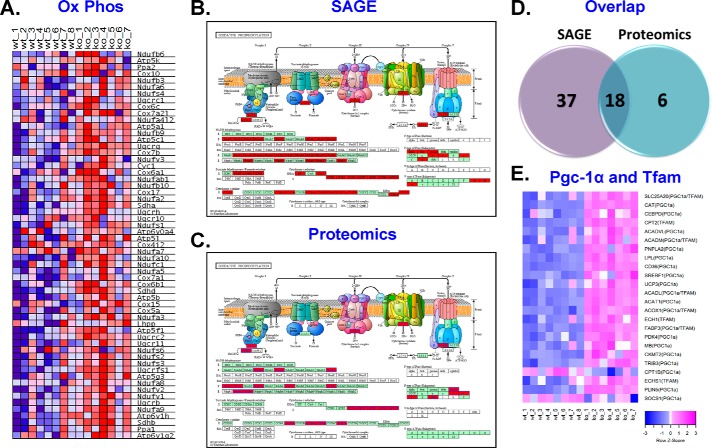
With widespread alterations in substrate metabolism pathways in Cpt1bM−/− skeletal muscle, it was important to determine whether alterations in electron transport chain (ETC) components were also present. The heatmap in Fig. 6A shows genes encoding proteins involved in oxidative phosphorylation were enriched in Cpt1bM−/− mice. Additionally, analysis of the proteomics dataset for Gene Ontology-based enrichment of cellular compartments (Fig. S7A) demonstrated clear enrichment for mitochondrial respiratory chain components (Fig. S7B). A graphical representation of specific subunits of each ETC complex showing significant differences between genotypes for SAGE (Fig. 6B) and proteomics (Fig. 6C) datasets reveals significant remodeling of the ETC in Cpt1bM−/− mice. The SAGE dataset identified 55 genes in the oxidative phosphorylation pathway to be regulated, whereas the proteomics screen identified 24 proteins that were altered in the ETC complexes (despite limited coverage of the proteome). Importantly, there appears to be reasonable agreement between SAGE and proteomics results as 18 of the regulated proteins were also increased at the gene level (Fig. 6D). All differentially expressed genes and proteins were up-regulated, which supports the concept of enhanced mitochondrial biogenesis in Cpt1bM−/− skeletal muscle. This is likely driven by activation of Pgc-1α and transcription factor A, mitochondrial (Tfam; Fig. 6E). It is also worth noting that changes in the ETC appear to be almost exclusively due to induction of nuclear-encoded subunits as none of the genes identified by the SAGE analysis were encoded by mitochondrial DNA (mtDNA), although only one protein encoded by mtDNA (ATP6) was significantly altered.
Figure 6.
Cpt1bM−/− mice exhibit signs of mitochondrial biogenesis. Cpt1bM−/− (KO) mice exhibit increased (red) enrichment of genes involved in the oxidative phosphorylation (Ox Phos) system (A). Subunits of the electron transport chain were consistently increased (red) in Cpt1bM−/− mice at the gene (B) and protein (C) levels, and the results between the SAGE and proteomics platforms were generally consistent (D). These adaptations are likely driven by induction of Pgc-1α and TFAM (E).
AMPK signaling
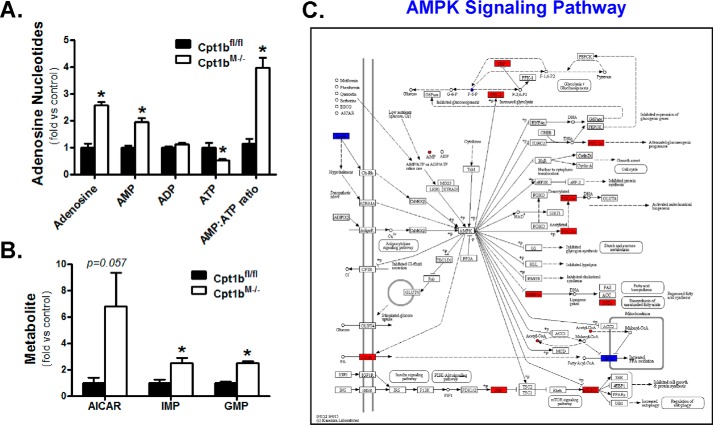
Because flux through the TCA cycle generates reducing equivalents that support energy production, the increased NADH/NAD+ ratio (Fig. 4F) suggests there are ample reducing equivalents to support ATP synthesis in Cpt1bM−/− mice. With this in mind, it is interesting that analysis of adenosine nucleotides found ATP content is actually lower in Cpt1bM−/− mouse skeletal muscle, whereas AMP levels and free adenosine are increased (Fig. 7A). The elevated AMP/ATP ratio in Cpt1bM−/− mice suggests they are in an energy-insufficient state. The 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) is a naturally occurring intermediate in the synthesis of inosine monophosphate (IMP), which can be used to produce AMP and guanosine monophosphate (GMP). As shown in Fig. 7B, IMP and GMP levels are also significantly higher in Cpt1bM−/− mice, but a >6-fold increase in the metabolic precursor, AICAR, approached statistical significance (p = 0.057). Because both AMP and AICAR are known activators of AMP-activated protein kinase (AMPK), the SAGE results were examined for evidence of activation of this pathway. Fig. 7C provides a visualization showing changes in several genes downstream from AMPK, which is consistent with activation of this energy-sensing pathway. These findings provide a mechanism that helps explain the increase in AMPK phosphorylation observed previously in Cpt1bM−/− mouse skeletal muscle (9).
Figure 7.
Energy deficit in Cpt1bM−/− mice likely activates AMPK signaling pathways. Adenosine nucleotides (A) and intermediates of nucleotide synthesis (B) are altered in Cpt1bM−/− mice. Results from the GSEA and metabolomics were combined into a KEGG network search to show regulation related to AMPK signaling (C). The following key is used: red (activation/up-regulation); blue (inhibition/down-regulation). *, p < 0.05.
Discussion
The purpose of this investigation was to integrate information yielded from three distinct 'omics technologies (SAGE, proteomics, and nontargeted metabolomics) to identify regulated metabolic systems in skeletal muscle from Cpt1bM−/− mice. We previously reported Cpt1bM−/− mice exhibit changes in substrate oxidation (8, 9); therefore, initial analyses of our 'omics datasets were targeted toward pathways involved in carbohydrate, amino acid, and lipid metabolism. Results confirmed the extensive remodeling of these pathways, with the most pronounced adaptations being observed in amino acid and lipid metabolism pathways, whereas shifts in glucose metabolism were modest. Strikingly, a biofunctional analysis, based on a large body of published gene-disease associations, indicated the adaptations were consistent with decreased predicted risk of glucose metabolism disorders, hepatic steatosis, fatigue, cardiac hypertrophy, and premature mortality, thus signifying that the breadth of disease states to which muscle-specific Cpt1b inhibition may mediate improvements in health may exceed that which had been previously believed.
Cpt1b has a critical role in facilitating mitochondrial long-chain fatty acid (LCFA) entry; thus, inhibiting this enzyme markedly reduces mitochondrial LCFA oxidation. A potentially underappreciated role of Cpt1b lies in the fact that the acylcarnitine metabolites produced via Cpt1b are also readily exported from the cell, which is particularly relevant in conditions of lipid excess (1–3). Limitations in the ability to both use lipids as fuel, as well as export excess lipid from the cell in the form of acylcarnitines, lead to increased intramuscular lipid accrual in Cpt1bM−/− mice as verified by elevated triglycerides, diglycerides, and ceramide metabolites (9). Accordingly, it is not surprising that a primary molecular response is to remodel lipid metabolism pathways. Regulator/effector network analysis of the SAGE data predicts this might occur through activation of Srebf1-, Rora-, Klf15-, Esrra-, Pgc-1α-, and/or Ppar-regulated pathways. Of note, some adaptations induced by these transcriptional regulators may actually exacerbate ectopic lipid buildup, as evidence herein shows Cpt1bM−/− mice have increased expression of genes and proteins that drive cellular lipid uptake, transport, and storage. Conversely, despite the inability of mitochondria to directly utilize LCFAs, our results show significant up-regulation of lipid catabolism pathways. Of interest, genes encoding peroxisomal proteins were identified as the second most robustly-enriched metabolic pathway via GSEA, and our previous data confirm that peroxisomal function is heightened in Cpt1bM−/− skeletal muscle (8, 9). Notably, genes involved in peroxisomal fat oxidation were particularly enriched. This is potentially important as unesterified fatty acids can enter peroxisomes; therefore, peroxisomal lipid entry does not require a carnitine shuttle system. Within their matrix, peroxisomes contain carnitine acetyltransferase (CrAT) and carnitine octanoyltransferase (CrOT), which convert chain-shortened moieties of peroxisomal oxidation into acylcarnitines. These products can then be exported into the cytosol, where they can enter the mitochondrial matrix independent of Cpt1. As a result, peroxisomes are capable of at least partially rescuing LCFA oxidation when Cpt1 is inhibited (14). With this in mind, defining whether recruitment of peroxisomes in skeletal muscle of Cpt1bM−/− mice is a critical adaptation that helps combat a potentially lipotoxic environment and/or contributes to the beneficial metabolic phenotype is an important consideration that warrants further investigation.
Cpt1b deletion limits the ability to use lipids to meet energy demand; therefore, Cpt1bM−/− mice must adapt to rely more heavily on other substrates. In this regard, earlier studies show Cpt1 inhibition increases reliance on carbohydrates (4, 5, 7, 15–21). Similarly, we found Cpt1bM−/− mice show enhanced whole-body glucose oxidation (indirect calorimetry), improved glucose tolerance, as well as increased pyruvate dehydrogenase activity and pyruvate oxidation in skeletal muscle homogenates (8, 9). It is also important to note that Cpt1 inhibition mitigates lipid-induced insulin resistance in skeletal muscle, despite the fact that such interventions promote a potentially lipotoxic environment (3, 8, 9). Results from the present analysis support these concepts and suggest that at least a portion of the adaptive increase in carbohydrate utilization in skeletal muscle of Cpt1bM−/− mice is encoded at the molecular level, as GSEA revealed enrichment of pathways involved in glycolysis, pyruvate handling, and insulin signaling. It is, however, worth mentioning that skeletal muscle is predicted to account for ∼20% of the basal metabolic rate (22), and this can increase greatly during exercise (23), emphasizing skeletal muscle has significant substrate requirements. Skeletal muscle of Cpt1bM−/− has limited ability to utilize the most prevalent substrate available (lipid); therefore, the adaptations in glucose-handling pathways actually seem relatively modest compared with what we expected. In this regard, it is important to recall that glucose is an essential substrate for brain and blood cells, and the body goes to great lengths to protect against hypoglycemia. As such, it is tempting to speculate that the relatively modest remodeling of glucose-handling pathways in skeletal muscle of Cpt1bM−/− mice is necessary to protect against the possibility of hypoglycemia. If true, this would also require adaptations in tissues other than skeletal muscle, which provides an interesting model to identify novel mechanisms of organ systems' cross-talk that regulate whole-body metabolic health that is worth future study.
Cpt1bM−/− mice also exhibit remarkable remodeling of amino acid metabolism pathways. Earlier, we reported that Cpt1bM−/− mice have heightened leucine oxidation (8, 9), suggesting adaptations occur that allow for greater protein catabolism. Findings herein strongly support this as follows: 1) pathways involved in amino acid metabolism represented 9 of the 21 significantly-enriched pathways identified via GSEA, and 2) the metabolomics screen revealed the majority of amino acids are significantly different between Cpt1bfl/fl versus Cpt1bM−/− mice. In fact, based upon the results of this study, it can be argued that adaptations in amino acid metabolism pathways actually exceed those directing carbohydrate metabolism. In this regard, futile protein cycling has been linked to increased energy expenditure (24) and could potentially contribute to the metabolically advantageous phenotype observed in Cpt1bM−/− mice; however, the importance of adaptations in amino acid metabolism pathways in this rodent model remains to be more fully understood. It is also worth noting that we previously reported that Cpt1bM−/− mice begin to exhibit lower lean mass at ∼17–20 weeks of age (8, 9). Mice in this study were 14–16 weeks of age at the time of harvest, which likely precedes significant differences in lean mass; however, the detected up-regulation of amino acid metabolism in this study likely contributes to lower lean mass in Cpt1bM−/− mice as they age.
SAGE and proteomics datasets both predicted enhanced mitochondrial biogenesis in skeletal muscle from Cpt1bM−/− mice. AMPK and Pgc-1α are activated under conditions of energy deficit and promote mitochondrial biogenesis. Cpt1bM−/− mice exhibit an increased AMP/ATP ratio (Fig. 7A), as well as activation of pAMPK and up-regulation of Pgc-1α (9); therefore, it is likely that activation of these pathways contributes to the observed mitochondrial biogenesis. Skeletal muscle from traditional models of energy-deficit (caloric restriction, prolonged fasting, and acute exercise, etc.) exhibit an increased AMP/ATP ratio, as well as decreases in NADH- and FADH2-reducing equivalents. Because mitochondrial ATP production is supported by delivery of reducing equivalents to the electron transport chain, it is surprising to note that lower ATP levels in Cpt1bM−/− mouse muscle occurs despite greater mitochondrial content and ample reducing equivalents (increased NADH levels and greater NADH/NAD+ ratio). Roughly 95% of the total cellular NADH pool is estimated to reside within mitochondria (25); thus, an apparent mismatch between NADH availability and ATP content is seemingly paradoxical. One possibility is that despite increased mitochondrial biogenesis, the relative lack of induction of genes and proteins encoded by mitochondrial DNA may have created mitochondria with an impaired ability to utilize reducing equivalents to generate the proton-motive force required to support ATP synthesis. While potentially compelling, the prospect that the newly formed mitochondria are inherently dysfunctional seems unlikely because the isolated mitochondria from Cpt1bM−/− mice exhibited similar functionality (oxygen consumption, respiratory control ratio, and uncoupled respiration) as littermate controls (9). This does not, however, rule out the possibility that other cellular alterations occur in Cpt1bM−/− mice that either 1) limit the ability to produce ATP and/or 2) increase ATP utilization. Finally, reducing equivalents are involved in a myriad of other redox reactions throughout the cell, so another possibility is that the increased NADH levels are linked to regulation of cellular redox balance; however, identifying specific pathways requires further study.
Cpt1b converts LCFA-CoAs into long-chain acylcarnitines, resulting in the liberation of CoA. As such, manipulation of Cpt1b activity has the potential to affect the cellular CoA pool. Because Cpt1b acts directly on LCFA-CoA, it stands to reason that manipulating Cpt1 activity would affect the LCFA-CoA pool; however, previous studies that have measured LCFA-CoA levels in models of variable Cpt1 activity have yielded inconsistent findings (15, 26–28). At this time, we cannot offer a resolution to this debate as targeted acyl-CoA metabolic profiling in Cpt1bM−/− mice has not been performed; however, our results clearly indicate significant differences in CoA metabolism. Substantial increases in free CoA (4.5-fold), dephospho-CoA (41-fold), enrichment of genes involved in CoA biosynthesis, and up-regulation of acyl-CoA thioesterases point toward a system that is attempting to increase CoA availability. Recent work suggests increased CoA biosynthesis occurs in systems attempting to drive LCFA oxidation (29), which is consistent with the extensive remodeling of lipid metabolism pathways in Cpt1bM−/− mice. The rise in CoA availability almost certainly contributes to the 14-fold increase in acetyl-CoA levels in Cpt1bM−/− mice. Heightened acetyl-CoA levels are usually found in states of energy surplus (30); thus, the robust increase in acetyl-CoA in Cpt1bM−/− mice signifies yet another seemingly paradoxical metabolite signature in a mouse model that otherwise exhibits a phenotype similar to models of energy deficit. With this in mind, it is important to note that whereas acetyl-CoA can enter the TCA cycle for use as metabolic fuel, it can also be used as a substrate in other reactions within the cell. Of particular relevance, surplus acetyl-CoA and NADH are known to promote protein acetylation (30). Although analysis of the SAGE data did not reveal enrichment of pathways involved in protein acetylation or deacetylation, there are observations from this study that support this possibility. First, numerous proteins in the TCA cycle and oxidative phosphorylation system are acetylated, and it has been shown that hyperacetylation of mitochondrial proteins diminishes ATP production (21–33), which resembles the energy-deficit phenotype observed in Cpt1bM−/− mice. Second, at the cellular level, acetylation of histone residues is a critical post-translational modification that regulates gene transcription within the nucleus. It has been postulated that heightened acetyl-CoA drives acetylation of histone residues to promote cell growth and survival even under states of energy deficit (30, 34), which also resembles the results from Cpt1bM−/− mice. Ultimately, however, the potential role that enhanced CoA biosynthesis and/or acetylation plays in the phenotype observed in Cpt1bM−/− mice remains speculative and warrants further study.
In summary, despite variable coverage of the proteome and identified metabolites, the combined use of SAGE, proteomics, and nontargeted metabolomics identified regulation of several pathways that were consistent across multiple platforms. Furthermore, the fact that robust differences in numerous metabolites were observed increases confidence that changes at the gene and protein levels represent functional adaptations. Importantly, the findings herein revealed that the extensive remodeling of substrate metabolism pathways in Cpt1bM−/− mice is consistent with metabolically healthy phenotypes. With this in mind, the large publicly available datasets developed from this comprehensive systems biology approach are expected to help investigators define specific mechanisms that contribute to the notable improvements in metabolic health when the mitochondrial lipid burden is mitigated in skeletal muscle.
Experimental procedures
Experimental model
For animals, the skeletal muscle-specific Cpt1b-deficient mice (Cpt1bM−/−) and littermate controls (Cpt1bfl/fl) were developed as described (8, 9). Briefly, floxed Cpt1b mice were bred to mice expressing Cre recombinase under the control of the myosin light chain 3, skeletal muscle isoform (Mlc1f promoter) (The Jackson Laboratory, Stock no. 024713) to delete Cpt1b in skeletal muscle. As validated in our previous studies (8, 9), this deletes Cpt1b expression and function specifically in skeletal muscle with no effect on cardiac muscle. Mice were group-housed at room temperature under a 12:12-h light/dark cycle and allowed ad libitum access to food and water. In an effort to be able to best compare results from this systems biology study to previous findings (8, 9, 13), mice were fed the same rodent breeder chow (Purina Rodent Chow no. 5015, Purina Mills, St. Louis, MO), which provides 20% of calories from protein, 26% from fat, and 54% from carbohydrate. At 12–14 weeks of age, body weight was measured (Fig. S1A), and a glucose tolerance test was performed as described below to confirm the Cpt1bM−/− mice used in this study exhibited the established improvement in glucose homeostasis (Fig. S1B). Tissue harvest occurred at 14–16 weeks of age, which is a time point when fat mass has consistently been shown to be lower in Cpt1bM−/− mice, yet lean mass is typically not yet different (8, 9). Following a 4-h food pull, Cpt1bfl/fl (n = 8) and Cpt1bM−/− (n = 8) male mice were euthanized by cervical dislocation, and tissues were collected, snap-frozen in liquid nitrogen, and stored at −80 °C until subsequent analyses could be performed. Mixed gastrocnemius (MG) skeletal muscle was powdered and used for all assays presented. The Pennington Biomedical Research Center has an AALAC-approved Comparative Biology Core facility and veterinary staff that continuously monitor the health of the animals via a sentinel program and daily inspection. All animal studies performed were approved by the Pennington Biomedical Research Center Institutional Animal Care and Use Committee.
Method details
Glucose tolerance test
Glucose tolerance tests were performed after a 4-h fast as described previously (9, 13). Briefly, after measuring baseline blood glucose levels via tail vein, mice received a 0.2-ml intraperitoneal injection of 20% d-glucose (40 mg of glucose per mouse), and blood glucose levels were subsequently monitored at 20, 40, and 60 min post-injection.
RNA and protein isolation
RNA and protein were extracted from 20 to 30 mg of powdered MG skeletal muscle using TRIzol (Thermo Fisher Scientific, Waltham, MA), as described previously (8, 9). Briefly, samples were homogenized in 1 ml of TRIzol and allowed to sit at room temperature for 5 min, and then 0.2 ml of chloroform was added. The samples were shaken vigorously for 15 s and allowed to sit at room temperature for 2–3 min before they were centrifuged (12,000 × g; 15 min; 4 °C) to induce phase separation. Roughly 600 μl of the upper aqueous supernatant containing RNA was transferred to a new microcentrifuge tube, whereupon 600 μl of 70% ethanol was added, and the samples were vortexed. RNA was then isolated using an RNeasy kit (Qiagen, Valencia, CA) with DNase treatment per the manufacturer's instructions. RNA content and quality (260:280 ratio range 1.9–2.1) were assessed using a Nanodrop 1000 and used for downstream SAGE studies. The lower organic phase developed following TRIzol extraction contained protein samples that were shipped to the Duke Proteomics Core Facility (Durham, NC). Following centrifugation, phase separation was performed by adding chloroform, and the aqueous layer was discarded, and protein was precipitated from the organic layer by methanol precipitation. After washing and sonication of the protein pellet, the precipitate was reconstituted in 50 μl of 0.25% Rapigest SF (Waters) in 50 mm ammonium bicarbonate, pH 8.0. Bradford assays were performed to determine protein quantity and were used for subsequent proteomics analysis.
SAGE
A primary goal of this study was to detect comprehensive changes in gene expression in skeletal muscle from Cpt1bfl/fl versus Cpt1bM−/− mice, and thus SAGE was performed by the Genomics Core at the Pennington Biomedical Research Center (Baton Rouge, LA). The SAGE analysis was performed as reported earlier (35, 36). Briefly, gene expression profiling was performed by expression tag sequencing (SAGE) on an AB SOLiD 5500XL next-generation sequencing instrument using reagent kits from the manufacturer (Applied Biosystems, Foster City, CA). Sequence reads were aligned to mouse reference RefSeq transcripts (version mm9) via SOLiDSAGE (Applied Biosystems). Only uniquely mapped sequence reads were counted to generate the expression count level for each respective RefSeq gene. Changes in certain genes detected via SAGE were confirmed by RT-PCR using described methods (8, 9) and primers shown in Table S3.
Proteomics
Another goal of this study was to detect broad-scale adaptations in proteins involved in metabolism in Cpt1bM−/− mice, with a particular interest in mitochondrial adaptations. As such, we had a proteomics analysis performed by the Duke Proteomics Core (Durham, NC). TRIzol-extracted protein (20 μg) from each sample (concentration range 1–7.5 μg/μl) was normalized to an equal volume (20 μl) to yield a 1 μg/μl sample. This sample was reduced and denatured with 10 mm DTT and heating at 80 °C for 10 min. Samples were subsequently alkylated with 20 mm iodoacetamide at room temperature in the dark for 30 min and then digested with 1:50 (w/w) trypsin/protein overnight at 37 °C. After digestion, 25 fmol/μl trypsinized yeast alcohol dehydrogenase 1 (MassPrep, Waters) was added as a surrogate standard to each sample, and samples were acidified to 1% TFA and heated to 60 °C for 2 h to hydrolyze Rapigest. After centrifugation, samples were transferred to liquid chromatography (LC) vials, and a quality control (QC) pool was prepared by mixing equal volumes of each sample. Quantitative one-dimensional LC/tandem MS (LC/MS/MS) was performed on 1 μg of the peptide digests. Individual peptides were scored using PeptideProphet algorithm (Elucidator), and the data were annotated at a <1% false discovery rate (FDR).
Quantitative one-dimensional LC/tandem MS (LC/MS/MS) was performed on 1 μg of the peptide digests per sample in singlicate. After a single column conditioning run of the QC pool, samples were analyzed in randomized order and interspersed with four additional analyses of the QC pool. Samples were analyzed using a nanoAcquity UPLC system (Waters) coupled to a Synapt G2 high definition mass spectrometry (HDMS) high-resolution accurate mass tandem mass spectrometer (Waters) via a nanoelectrospray ionization source. Briefly, samples were first trapped on a Symmetry C18 300 × 180-mm trapping column (5 μl/min at 99.9/0.1 v/v H2O/MeCN), after which the analytical separation was performed using a 1.7-μm Acquity BEH130 C18 75 × 250-mm column (Waters) using a 90-min gradient of 5–40% acetonitrile with 0.1% formic acid at a flow rate of 300 nl/min and a column temperature of 45 °C. Data collection on the Synapt G2 mass spectrometer was performed in ion-mobility–assisted data-independent acquisition (HDMSE) mode, using 0.6 s alternating cycle time between low (6 V) and high (27–50 V) collision energy (CE). Scans performed at low CE measured peptide accurate mass and intensity (abundance), whereas scans at elevated CE allowed for qualitative identification of the resulting peptide fragments via database searching. The total analysis cycle time for each sample injection was ∼2 h. Following the 21 total analyses, data were imported into Rosetta Elucidator version 3.3 (Rosetta Biosoftware, Inc.), and all LC-MS runs were aligned based on the accurate mass and retention time of detected ions (“features”) using the PeakTeller algorithm in Elucidator. Relative peptide abundance was calculated based on the area-under-the-curve of aligned features across all runs. The overall dataset had 168,700 quantified features, and 108,834 high collision energy (peptide fragment) spectra that were subjected to database searching. This MS/MS data were searched against a custom RefSeq database with Mus musculus taxonomy that contained a reversed-sequence “decoy” database for false discovery rate determination. ProteinLynx Global Server version 2.5.2 was utilized to produce fragment ion spectra and to perform the database searches for the HDMSE runs, respectively. Included in the database searches were fixed modifications on Cys (carbamidomethyl) and variable modifications on Met (oxidation) and Asn/Gln (deamidation). Mascot Distiller was used to search data-dependent acquisition data from the first QC analysis, as well as pkl files for two representative HDMSE within the Elucidator. Additional variable post-translational modifications searched with Mascot included N-terminal acetylation and phosphorylation of Ser/Thr/Tyr.
A 10-μl aliquot of each sample was injected through a Synergi 2.5-μm Hydro-RP 100, 100 × 2.00-mm LC column (Phenomenex, Torrance, CA) held at 25 °C. The mass spectrometer was run in negative ionization mode similar to a previously reported method (37). The chromatographic elution was introduced to the Exactive Plus Orbitrap mass spectrometer (Thermo Fisher Scientific) via an electrospray ionization source through a 0.1-mm internal diameter fused silica capillary tube. Instrumental parameters were as follows: 3 kV spray voltage, 10 nitrogen sheath gas flow rate (unitless), 320 °C capillary temperature, 3e6 AGC target, 140,000 resolution, and a scan window of 85–800 m/z from 0 to 9 min and 100 to 1,000 m/z from 9 to 25 min. Solvent A was composed of 97:3 water/methanol, 10 mm tributylamine, and 15 mm acetic acid. Solvent B was methanol. The gradient was as follows: 0–5 min, 0% B; 5–13 min, 20% B; 13–15.5 min, 55% B; 15.5–19 min, 95% B; and 19–25 min, 0% B. The flow rate was maintained 200 μl/min for the duration.
Metabolomics
A third goal of this study was to get a comprehensive view of global adaptations in metabolism in Cpt1bM−/− skeletal muscle, while also providing detail on metabolites involved in glucose and amino acid metabolism, as well as energy status and redox balance. To achieve this goal, nontargeted metabolomics was performed by the Biological and Small Molecule Mass Spectrometry Core at the University of Tennessee (Knoxville, TN). Metabolite extraction was performed at 4 °C unless stated otherwise. To 1.5-ml microcentrifuge tubes containing 10–15 mg of powdered mixed gastrocnemius skeletal muscle, 1.3 ml of extraction solvent was added consisting of a 40:40:20 HPLC-grade methanol, acetonitrile, and water with 0.1 m formic acid (38). Samples were thoroughly mixed by vortexing before extraction was allowed to proceed for 20 min at −20 °C. Following extraction, samples were centrifuged (5 min at 16.1 relative centrifugal force), and supernatants were transferred to new vials. Pellets were resuspended in an additional 50 μl of extraction solvent, and extraction was again allowed to proceed for 20 min at −20 °C before being centrifuged. Supernatants were again transferred to vials, and the process was repeated once more with an additional 50 μl of extraction solvent. The combined supernatants were dried under nitrogen gas and subsequently resuspended in 300 μl of sterile water. The samples were transferred to autosampler vials and immediately placed in an Ultimate 3000 RS autosampler (Dionex, Sunnyvale, CA) cooled to 4 °C. Samples were analyzed using UPLC–high-resolution mass spectrometry run in negative ionization mode similar to a previously reported method (37). Metabolite identification was performed by first converting raw spectrum files to the open-source mzML format using the Proteowizard package (39). Total ion chromatograms were automatically corrected based on retention times for each sample using MAVEN software (Princeton University) (40, 41). Metabolites were identified based on retention times, and m/z (±5 ppm) and chromatographic intensities were integrated for each sample. Unidentified spectral features were annotated using MAVEN's automated peak detection feature, selecting m/z features exhibiting chromatographic peak-like qualities. Peak detection settings were as follows: 10 ppm mass domain resolution; 10 scan time domain resolution; 11 scan extracted ion chromatogram (EIC) smoothing; 0.50 min peak grouping; 5 scan baseline smoothing; 80% chromatograph intensity dropped baseline; 1 minimum peak per group; 3 signal to baseline and signal to blank ratios; 5 scan minimum peak width; and 10,000 ion minimum peak intensity, using default peak classifier model.
Quantification and statistical analysis
Gene expression data analysis
Differential analysis of RNA read count data were performed using DESeq2 software (42), which models read counts as a negative binomial distribution and uses an empirical Bayes shrinkage-based method to estimate signal dispersion and fold-changes. Gene expression signals were logarithmically transformed (to base 2) for all downstream analyses (the lowest expression value being set to 1 for this purpose).
Pathway enrichment analysis
Pathway enrichment was conducted via three separate approaches: (a) competitive gene-scoring based on GSEA (43); (b) over-representation analysis based on a pre-filtered list of differentially expressed genes (Ingenuity Pathway Analysis (IPA), Qiagen, Redwood City, www.qiagen.com/ingenuity); and (c) enrichment analysis of pre-selected pathways via the globaltest package in R (http://bioconductor.org)6 (44). GSEA was performed by first ranking the expression of all genes in the Cpt1bfl/fl (WT) and Cpt1bM−/− (KO) groups via the signal-to-noise ratio (SNR) metric, and then employing a weighted Kolmogorov–Smirnov test to determine whether the gene SNRs deviate significantly from a uniform distribution in a priori defined gene-sets (pathways) derived from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (45) via the Molecular Signatures Database repository (MSigDb, http://software.broadinstitute.org/gsea/msigdb/6 (46, 47). Statistical significance for the observed enrichment was ascertained by permutation testing over size-matched gene-sets. Significant gene-sets were selected by control of the FDR at 25% (48). The per-sample expression profiles of genes contributing to core enrichment of the significant pathways were visualized via row-normalized blue-red heatmaps with blue representing lower levels and red representing higher gene expression levels.
IPA analysis was carried out on differentially expressed genes (with absolute fold-change ≥1.5-fold and FDR ≤10% between the two groups; therefore, genes chosen for the analysis were based on both statistical significance and effect size. Over-representation was tested on “biological function” and “upstream regulator”-based gene-sets available in the IPA Knowledge Base. In both cases, a Fisher's exact test was first performed to determine whether there was a high degree of overlap between the differentially expressed genes and the target genes ascribed to the biological function or upstream regulator entity by IPA. The overall activation or inhibition status was then inferred based on the degree of consistency (up- or down-regulation) in the expression patterns of its target genes, expressed as a signed z-score. A z-score ≥2 or ≤−2 was considered to be evidence for activation and inhibition, respectively.
In addition to the above two “competitive test” methods (49), we also queried gene expression differences via a “self-contained” test for gene-set enrichment known as the globaltest. The globaltest method is based on regression analysis of the association of a gene-set's component gene expression to a phenotype, and it tests the null hypothesis that no gene in the gene-set is associated with the phenotype (with no additional consideration for genes in other gene-sets). We applied globaltest to the same gene expression data as used for GSEA analysis, and queried KEGG-specific gene-sets obtained from the package kegg.db (50). Gene annotations were provided as Entrez IDs to match the gene-set annotations utilized by globaltest from org.Mm.eg.db (release 3.3) (51).
For visual representations of gene and metabolite expression pattern differences in pathways, we used the Search and Color Pathway tool from the KEGG Mapper suite of tools (www.genome.jp/kegg/mapper.html).6 A total of 389 genes with a nominal p value of ≤0.01 and an absolute fold-change of ≥1.5-fold between the Cpt1bfl/fl and Cpt1bM−/− groups were selected for representation on the KEGG pathway maps. Forty four differentially expressed metabolites (FDR ≤0.05) were similarly selected. Up-regulated genes and metabolites were color-coded in red and down-regulated entities were shown in blue.
Statistical analysis
Data depicted in bar graphs are presented as mean ± S.E. A two-tailed Student's t test was used to detect significant differences in metabolites, proteins, and genes tested via RT-PCR. Statistical significance for these tests was established a priori at p ≤ 0.05.
Data and software availability
The SAGE dataset has been uploaded to the Gene Expression Omnibus (GEO) website (accession number GSE107138), and Figshare was used to upload the proteomics (10.6084/m9.figshare.5695474) and metabolomics (10.6084/m9.figshare.5695444) datasets. The SAGE and proteomics datasets that were uploaded excluded an outlier (KO8) that was identified in both, while WT8 was removed as an outlier from the metabolomics dataset.
Author contributions
S. G., S. E. W., B. V., T. M. M., D. S. B., J. M. S., S. P. D., S. R. C., R. L. M., and R. C. N. data curation; S. G., J. M. S., S. P. D., and R. C. N. software; S. G., D. S. B., J. M. S., S. P. D., S. R. C., R. L. M., and R. C. N. formal analysis; S. G., J. M. S., S. P. D., R. L. M., and R. C. N. validation; S. G., S. E. W., B. V., T. M. M., D. S. B., J. M. S., S. P. D., S. R. C., R. L. M., and R. C. N. methodology; S. G., R. L. M., and R. C. N. writing-original draft; S. G., R. L. M., and R. C. N. writing-review and editing; S. E. W., R. L. M., and R. C. N. funding acquisition; S. E. W., B. V., T. M. M., R. L. M., and R. C. N. investigation; J. M. S., R. L. M., and R. C. N. supervision; R. L. M. and R. C. N. conceptualization; R. L. M. and R. C. N. resources; R. C. N. visualization; R. C. N. project administration.
Supplementary Material
Acknowledgments
We thank Jaycob Warfel, Susan Newman, Claudia Kruger, and Richard Carmouche for critical advice, reagents, and/or technical assistance. We appreciate Drs. Matthew Foster, J. Will Thompson, and Arthur Moseley from the Duke Proteomics Core Facility (Duke University School of Medicine) for performing the proteomic analysis. This work utilized PBRC core facilities (Genomics and Transgenic and Animal Phenotyping) that are supported in part by COBRE Grant 3P30-GM118430 and NORC Grant 2P30-DK072476 from the National Institutes of Health.
This work was supported in part by National Institutes of Health Grants 3P30-GM118430 (to R. C. N.) and 1R01-DK103860 (to R. C. N.), American Diabetes Association Grant 1-10-BS-129 (to R. L. M.), and National Institutes of Health Grant 1R01-DK089641 (to R. L. M.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
This article contains Figs. S1–S7 and Tables S1–S3.
The SAGE dataset has been uploaded to the Gene Expression Omnibus (GEO) website under Accession Number GSE107138.
Figshare was used to upload the proteomics under Accession Number 10.6084/m9.figshare.5695474 and metabolomics under Accession Number 10.6084/m9.figshare.5695444) datasets.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- SAGE
- serial analysis of gene expression
- AMPK
- AMP-activated protein kinase
- LCFA
- long-chain fatty acid
- MG
- mixed gastrocnemius
- AICAR
- 5-aminoimidazole-4-carboxamide ribonucleotide
- PCA
- principal component analysis
- GSEA
- gene-set enrichment analysis
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- MSigDB
- molecular signatures database
- IPA
- ingenuity pathway analysis
- QC
- quality control
- FDR
- false discovery rate
- UPLC
- ultraperformance liquid chromatography
- SNR
- signal-to-noise ratio
- TCA
- tricarboxylic acid
- ETC
- electron transport chain
- Tfam
- transcription factor A, mitochondrial
- CrAT
- carnitine acetyltransferase
- CrOT
- carnitine octanoyltransferase
- CE
- collision energy
- PPAR
- peroxisome proliferator-activated receptor
- HDMS
- high definition mass spectrometry.
References
- 1. Muoio D. M., Noland R. C., Kovalik J. P., Seiler S. E., Davies M. N., DeBalsi K. L., Ilkayeva O. R., Stevens R. D., Kheterpal I., Zhang J., Covington J. D., Bajpeyi S., Ravussin E., Kraus W., Koves T. R., and Mynatt R. L. (2012) Muscle-specific deletion of carnitine acetyltransferase compromises glucose tolerance and metabolic flexibility. Cell Metab. 15, 764–777 10.1016/j.cmet.2012.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Noland R. C., Koves T. R., Seiler S. E., Lum H., Lust R. M., Ilkayeva O., Stevens R. D., Hegardt F. G., and Muoio D. M. (2009) Carnitine insufficiency caused by aging and overnutrition compromises mitochondrial performance and metabolic control. J. Biol. Chem. 284, 22840–22852 10.1074/jbc.M109.032888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Koves T. R., Ussher J. R., Noland R. C., Slentz D., Mosedale M., Ilkayeva O., Bain J., Stevens R., Dyck J. R., Newgard C. B., Lopaschuk G. D., and Muoio D. M. (2008) Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 7, 45–56 10.1016/j.cmet.2007.10.013 [DOI] [PubMed] [Google Scholar]
- 4. Barnett M., Collier G. R., and O'Dea K. (1992) The longitudinal effect of inhibiting fatty acid oxidation in diabetic rats fed a high fat diet. Horm. Metab. Res. 24, 360–362 10.1055/s-2007-1003335 [DOI] [PubMed] [Google Scholar]
- 5. Deems R. O., Anderson R. C., and Foley J. E. (1998) Hypoglycemic effects of a novel fatty acid oxidation inhibitor in rats and monkeys. Am. J. Physiol. 274, R524–R528 10.1152/ajpregu.1998.274.2.R524 [DOI] [PubMed] [Google Scholar]
- 6. Keung W., Ussher J. R., Jaswal J. S., Raubenheimer M., Lam V. H., Wagg C. S., and Lopaschuk G. D. (2013) Inhibition of carnitine palmitoyltransferase-1 activity alleviates insulin resistance in diet-induced obese mice. Diabetes 62, 711–720 10.2337/db12-0259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Timmers S., Nabben M., Bosma M., van Bree B., Lenaers E., van Beurden D., Schaart G., Westerterp-Plantenga M. S., Langhans W., Hesselink M. K., Schrauwen-Hinderling V. B., and Schrauwen P. (2012) Augmenting muscle diacylglycerol and triacylglycerol content by blocking fatty acid oxidation does not impede insulin sensitivity. Proc. Natl. Acad. Sci. U.S.A. 109, 11711–11716 10.1073/pnas.1206868109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vandanmagsar B., Warfel J. D., Wicks S. E., Ghosh S., Salbaum J. M., Burk D., Dubuisson O. S., Mendoza T. M., Zhang J., Noland R. C., and Mynatt R. L. (2016) Impaired mitochondrial fat oxidation induces FGF21 in muscle. Cell Rep. 15, 1686–1699 10.1016/j.celrep.2016.04.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wicks S. E., Vandanmagsar B., Haynie K. R., Fuller S. E., Warfel J. D., Stephens J. M., Wang M., Han X., Zhang J., Noland R. C., and Mynatt R. L. (2015) Impaired mitochondrial fat oxidation induces adaptive remodeling of muscle metabolism. Proc. Natl. Acad. Sci. U.S.A. 112, E3300–E3309 10.1073/pnas.1418560112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chase J. F., and Tubbs P. K. (1972) Specific inhibition of mitochondrial fatty acid oxidation by 2-bromopalmitate and its coenzyme A and carnitine esters. Biochem. J. 129, 55–65 10.1042/bj1290055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kerner J., and Hoppel C. (2000) Fatty acid import into mitochondria. Biochim. Biophys. Acta 1486, 1–17 10.1016/S1388-1981(00)00044-5 [DOI] [PubMed] [Google Scholar]
- 12. Koves T. R., Noland R. C., Bates A. L., Henes S. T., Muoio D. M., and Cortright R. N. (2005) Subsarcolemmal and intermyofibrillar mitochondria play distinct roles in regulating skeletal muscle fatty acid metabolism. Am. J. Physiol. Cell Physiol 288, C1074–C1082 10.1152/ajpcell.00391.2004 [DOI] [PubMed] [Google Scholar]
- 13. Warfel J. D., Vandanmagsar B., Wicks S. E., Zhang J., Noland R. C., and Mynatt R. L. (2017) A low fat diet ameliorates pathology but retains beneficial effects associated with CPT1b knockout in skeletal muscle. PLoS ONE 12, e0188850 10.1371/journal.pone.0188850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Noland R. C., Woodlief T. L., Whitfield B. R., Manning S. M., Evans J. R., Dudek R. W., Lust R. M., and Cortright R. N. (2007) Peroxisomal-mitochondrial oxidation in a rodent model of obesity-associated insulin resistance. Am. J. Physiol. Endocrinol. Metab. 293, E986–E1001 10.1152/ajpendo.00399.2006 [DOI] [PubMed] [Google Scholar]
- 15. Dobbins R. L., Szczepaniak L. S., Bentley B., Esser V., Myhill J., and McGarry J. D. (2001) Prolonged inhibition of muscle carnitine palmitoyltransferase-1 promotes intramyocellular lipid accumulation and insulin resistance in rats. Diabetes 50, 123–130 10.2337/diabetes.50.1.123 [DOI] [PubMed] [Google Scholar]
- 16. Hübinger A., Knode O., Susanto F., Reinauer H., and Gries F. A. (1997) Effects of the carnitine-acyltransferase inhibitor etomoxir on insulin sensitivity, energy expenditure and substrate oxidation in NIDDM. Horm. Metab. Res. 29, 436–439 10.1055/s-2007-979072 [DOI] [PubMed] [Google Scholar]
- 17. Lopaschuk G. D., McNeil G. F., and McVeigh J. J. (1989) Glucose oxidation is stimulated in reperfused ischemic hearts with the carnitine palmitoyltransferase 1 inhibitor, Etomoxir. Mol. Cell. Biochem. 88, 175–179 [DOI] [PubMed] [Google Scholar]
- 18. Martin C., Odeon M., Cohen R., and Beylot M. (1991) Mechanisms of the glucose lowering effect of a carnitine palmitoyl transferase inhibitor in normal and diabetic rats. Metabolism 40, 420–427 10.1016/0026-0495(91)90154-O [DOI] [PubMed] [Google Scholar]
- 19. Ratheiser K., Schneeweiss B., Waldhäusl W., Fasching P., Korn A., Nowotny P., Rohac M., and Wolf H. P. (1991) Inhibition by etomoxir of carnitine palmitoyltransferase I reduces hepatic glucose production and plasma lipids in non-insulin-dependent diabetes mellitus. Metabolism 40, 1185–1190 10.1016/0026-0495(91)90214-H [DOI] [PubMed] [Google Scholar]
- 20. Tutwiler G. F., Brentzel H. J., and Kiorpes T. C. (1985) Inhibition of mitochondrial carnitine palmitoyl transferase A in vivo with methyl 2-tetradecylglycidate (methyl palmoxirate) and its relationship to ketonemia and glycemia. Proc. Soc. Exp. Biol. Med. 178, 288–296 10.3181/00379727-178-42012 [DOI] [PubMed] [Google Scholar]
- 21. Wolf H. P., and Engel D. W. (1985) Decrease of fatty acid oxidation, ketogenesis and gluconeogenesis in isolated perfused rat liver by phenylalkyl oxirane carboxylate (B 807–27) due to inhibition of CPT I (EC 2.3.1.21). Eur. J. Biochem. 146, 359–363 10.1111/j.1432-1033.1985.tb08661.x [DOI] [PubMed] [Google Scholar]
- 22. Wang Z., Heshka S., Gallagher D., Boozer C. N., Kotler D. P., and Heymsfield S. B. (2000) Resting energy expenditure–fat-free mass relationship: new insights provided by body composition modeling. Am. J. Physiol. Endocrinol. Metab. 279, E539–E545 10.1152/ajpendo.2000.279.3.E539 [DOI] [PubMed] [Google Scholar]
- 23. Noland R. C. (2015) Exercise and regulation of lipid metabolism. Prog. Mol. Biol. Transl. Sci. 135, 39–74 10.1016/bs.pmbts.2015.06.017 [DOI] [PubMed] [Google Scholar]
- 24. She P., Reid T. M., Bronson S. K., Vary T. C., Hajnal A., Lynch C. J., and Hutson S. M. (2007) Disruption of BCATm in mice leads to increased energy expenditure associated with the activation of a futile protein turnover cycle. Cell Metab. 6, 181–194 10.1016/j.cmet.2007.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. White A. T., and Schenk S. (2012) NAD(+)/NADH and skeletal muscle mitochondrial adaptations to exercise. Am. J. Physiol. Endocrinol. Metab. 303, E308–E321 10.1152/ajpendo.00054.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. An J., Muoio D. M., Shiota M., Fujimoto Y., Cline G. W., Shulman G. I., Koves T. R., Stevens R., Millington D., and Newgard C. B. (2004) Hepatic expression of malonyl-CoA decarboxylase reverses muscle, liver and whole-animal insulin resistance. Nat. Med. 10, 268–274 10.1038/nm995 [DOI] [PubMed] [Google Scholar]
- 27. Lopaschuk G. D., Wall S. R., Olley P. M., and Davies N. J. (1988) Etomoxir, a carnitine palmitoyltransferase I inhibitor, protects hearts from fatty acid-induced ischemic injury independent of changes in long chain acylcarnitine. Circ. Res. 63, 1036–1043 10.1161/01.RES.63.6.1036 [DOI] [PubMed] [Google Scholar]
- 28. Obici S., Feng Z., Arduini A., Conti R., and Rossetti L. (2003) Inhibition of hypothalamic carnitine palmitoyltransferase-1 decreases food intake and glucose production. Nat. Med. 9, 756–761 10.1038/nm873 [DOI] [PubMed] [Google Scholar]
- 29. Jackowski S., and Leonardi R. (2014) Deregulated coenzyme A, loss of metabolic flexibility and diabetes. Biochem. Soc. Trans. 42, 1118–1122 10.1042/BST20140156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cai L., and Tu B. P. (2011) On acetyl-CoA as a gauge of cellular metabolic state. Cold Spring Harb. Symp. Quant. Biol. 76, 195–202 10.1101/sqb.2011.76.010769 [DOI] [PubMed] [Google Scholar]
- 31. Davies M. N., Kjalarsdottir L., Thompson J. W., Dubois L. G., Stevens R. D., Ilkayeva O. R., Brosnan M. J., Rolph T. P., Grimsrud P. A., and Muoio D. M. (2016) The acetyl group buffering action of carnitine acetyltransferase offsets macronutrient-induced lysine acetylation of mitochondrial proteins. Cell Rep. 14, 243–254 10.1016/j.celrep.2015.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hirschey M. D., Shimazu T., Goetzman E., Jing E., Schwer B., Lombard D. B., Grueter C. A., Harris C., Biddinger S., Ilkayeva O. R., Stevens R. D., Li Y., Saha A. K., Ruderman N. B., Bain J. R., et al. (2010) SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464, 121–125 10.1038/nature08778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim S. C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., Grishin N. V., White M., Yang X. J., and Zhao Y. (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618 10.1016/j.molcel.2006.06.026 [DOI] [PubMed] [Google Scholar]
- 34. Vander Heiden M. G., Cantley L. C., and Thompson C. B. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Matsumura H., Krüger D. H., Kahl G., and Terauchi R. (2015) SuperSAGE as an analytical tool for host and viral gene expression. Methods Mol. Biol. 1236, 181–195 10.1007/978-1-4939-1743-3_14 [DOI] [PubMed] [Google Scholar]
- 36. Salbaum J. M., Kruger C., MacGowan J., Herion N. J., Burk D., and Kappen C. (2015) Novel mode of defective neural tube closure in the non-obese diabetic (NOD) mouse strain. Sci. Rep. 5, 16917 10.1038/srep16917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lu W., Clasquin M. F., Melamud E., Amador-Noguez D., Caudy A. A., and Rabinowitz J. D. (2010) Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal. Chem. 82, 3212–3221 10.1021/ac902837x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rabinowitz J. D., and Kimball E. (2007) Acidic acetonitrile for cellular metabolome extraction from Escherichia coli. Anal. Chem. 79, 6167–6173 10.1021/ac070470c [DOI] [PubMed] [Google Scholar]
- 39. Chambers M. C., Maclean B., Burke R., Amodei D., Ruderman D. L., Neumann S., Gatto L., Fischer B., Pratt B., Egertson J., Hoff K., Kessner D., Tasman N., Shulman N., Frewen B., et al. (2012) A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 10.1038/nbt.2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Clasquin M. F., Melamud E., and Rabinowitz J. D. (2012) LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine. Curr. Protoc. Bioinformatics Chapter 14, Unit 14.11 10.1002/0471250953.bi1411s37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Melamud E., Vastag L., and Rabinowitz J. D. (2010) Metabolomic analysis and visualization engine for LC-MS data. Anal. Chem. 82, 9818–9826 10.1021/ac1021166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Love M. I., Huber W., and Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., and Mesirov J. P. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Goeman J. J., van de Geer S. A., de Kort F., and van Houwelingen H. C. (2004) A global test for groups of genes: testing association with a clinical outcome. Bioinformatics 20, 93–99 10.1093/bioinformatics/btg382 [DOI] [PubMed] [Google Scholar]
- 45. Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., and Kanehisa M. (1999) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27, 29–34 10.1093/nar/27.1.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lek M., Karczewski K. J., Minikel E. V., Samocha K. E., Banks E., Fennell T., O'Donnell-Luria A. H., Ware J. S., Hill A. J., Cummings B. B., Tukiainen T., Birnbaum D. P., Kosmicki J. A., Duncan L. E., Estrada K., et al. (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J. P., and Tamayo P. (2015) The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell. Syst. 1, 417–425 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Reiner A., Yekutieli D., and Benjamini Y. (2003) Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19, 368–375 10.1093/bioinformatics/btf877 [DOI] [PubMed] [Google Scholar]
- 49. Goeman J. J., and Bühlmann P. (2007) Analyzing gene expression data in terms of gene sets: methodological issues. Bioinformatics 23, 980–987 10.1093/bioinformatics/btm051 [DOI] [PubMed] [Google Scholar]
- 50. Carlson M. (2016) KEGG.db: A set of annotation maps for KEGG. R package version 3.2.3 10.18129/B9.bioc.KEGG.db [DOI] [Google Scholar]
- 51. Carlson M. (2019) org.Mm.eg.db: Genome wide annotation for Mouse. R package version 3.8.2 10.18129/B9.bioc.org.Mm.eg.db [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The SAGE dataset has been uploaded to the Gene Expression Omnibus (GEO) website (accession number GSE107138), and Figshare was used to upload the proteomics (10.6084/m9.figshare.5695474) and metabolomics (10.6084/m9.figshare.5695444) datasets. The SAGE and proteomics datasets that were uploaded excluded an outlier (KO8) that was identified in both, while WT8 was removed as an outlier from the metabolomics dataset.