Abstract
The genetically diverse Orthocoronavirinae (CoV) family is prone to cross species transmission and disease emergence in both humans and livestock. Viruses similar to known epidemic strains circulating in wild and domestic animals further increase the probability of emergence in the future. Currently, there are no approved therapeutics for any human CoV presenting a clear unmet medical need. Remdesivir (RDV, GS-5734) is a monophosphoramidate prodrug of an adenosine analog with potent activity against an array of RNA virus families including Filoviridae, Paramyxoviridae, Pneumoviridae, and Orthocoronavirinae, through the targeting of the viral RNA dependent RNA polymerase (RdRp). We developed multiple assays to further define the breadth of RDV antiviral activity against the CoV family. Here, we show potent antiviral activity of RDV against endemic human CoVs OC43 (HCoV-OC43) and 229E (HCoV-229E) with submicromolar EC50 values. Of known CoVs, the members of the deltacoronavirus genus have the most divergent RdRp as compared to SARS- and MERS-CoV and both avian and porcine members harbor a native residue in the RdRp that confers resistance in beta-CoVs. Nevertheless, RDV is highly efficacious against porcine deltacoronavirus (PDCoV). These data further extend the known breadth and antiviral activity of RDV to include both contemporary human and highly divergent zoonotic CoV and potentially enhance our ability to fight future emerging CoV.
Keywords: Coronavirus, Emerging viruses, Broad-spectrum antivirals, GS-5743, Remdesivir
Abbreviations: CoV, Orthocoronavirinae; HCoV-OC43, Human coronavirus OC43; HCoV-229E, Human coronavirus 229E; PDCoV, Porcine deltacoronavirus; Remdesivir (GS-5734), RDV; RdRp, RNA dependent RNA polymerase
Highlights
-
•
In vitro antiviral assays were developed for human CoV OC43 and 229E and the zoonotic PDCoV.
-
•
The nucleoside analog RDV inhibited HCoV-OC43 and 229E as well as deltacoronavirus member PDCoV.
-
•
RDV has broad-spectrum antiviral activity against CoV and should be evaluated for future emerging CoV.
1. Introduction
The genetically diverse Orthocoronavirinae (CoV) family is divided into four genera (alpha, beta, gamma, and deltacoronavirus) and thus far human CoV are limited to the alpha and beta genera. Human CoVs OC43, 229E, NL63 and HKU1 cause 10% of all upper and lower respiratory tract infections, which typically present with common-cold like symptoms but can cause more severe disease in young children as well as people with underlying respiratory conditions (i.e. asthma, COPD) and the elderly (Dijkman et al., 2012; Falsey et al., 2002). In children, severe respiratory tract CoV infections require hospitalization in about 10% of cases and have been associated with febrile seizure in those less than 1 year old (Carman et al., 2018; Heimdal et al., 2019). CoV infection can also be severe in the elderly requiring hospitalization and can even cause acute respiratory distress syndrome (ARDS) (Falsey et al., 2002; Vassilara et al., 2018). Zoonotic CoVs have a natural predilection for emergence into new host species giving rise to new diseases most recently exemplified in humans by severe acute respiratory syndrome coronavirus (SARS-CoV), and Middle East respiratory syndrome coronavirus (MERS-CoV) (de Wit et al., 2016). Interestingly, all known human CoVs are thought to have emerged as zoonoses from wild or domestic animals (Hu et al., 2015a; Huynh et al., 2012; Menachery et al., 2016; Vijgen et al., 2005). This emergence paradigm is not unique to human CoVs. Novel animal CoVs like porcine epidemic diarrhea virus (PEDV), porcine delta coronavirus (PDCoV) and swine acute diarrhea syndrome coronavirus (SADS-CoV) have recently emerged causing the deaths of millions of piglets and billions of dollars in agricultural losses (Hu et al., 2015b; Huang et al., 2013; Zhou et al., 2018). While chloroquine, ribavirin, lopinavir and interferons have all been tested against multiple CoV in vitro, currently, there are no approved therapeutics for any human CoV (Chan et al., 2013; de Wilde et al., 2014; Shen et al., 2016). To address an unmet medical need for the treatment of current human CoV infections and to maximize pandemic preparedness, broad spectrum antiviral therapies are needed that are effective against current and future emerging CoV given the numerous examples of novel CoV emergence.
Remdesivir (RDV, GS-5734) is a monophosphoramidate prodrug of an adenosine analog with demonstrated antiviral activity against an array of RNA virus families including Filoviridae, Paramyxoviridae, Pneumoviridae, and CoV (Lo et al., 2017; Sheahan et al., 2017; Warren et al., 2016). The antiviral mechanism for RDV has been demonstrated to be through delayed chain termination of nascent viral RNA of for Ebola virus, Nipah virus and respiratory syncytial virus (Jordan et al., 2018; Lo et al., 2017; Tchesnokov et al., 2018, 2019; Warren et al., 2016). We previously reported the antiviral activity of RDV against a genetically diverse panel of human endemic, emerging and zoonotic CoV including HCoV-NL63 (alpha 1b), mouse hepatitis virus (MHV, beta 2a), SARS-CoV and related Bat CoVs WIV1 and SHC014 (beta 2b), as well as MERS-CoV and related Bat CoV HKU5 (beta 2c) (Agostini et al., 2018; Sheahan et al., 2017). Upon passage of MHV in the presence of RDV, resistance mutations arise in the RNA dependent RNA polymerase (RdRp) that confer resistance (i.e. up to a 5-fold shift in EC50) demonstrating that the RdRp is a target of RDV antiviral activity (Agostini et al., 2018). The CoV RdRp is highly conserved within genogroups (i.e. beta 2b) but amino acid identity between groups varies from 70 to 90% (Sheahan et al., 2017). Here, we expand upon our earlier work to better understand the spectrum of RDV efficacy among human and zoonotic CoV. We show that RDV inhibits endemic human CoVs 229E and OC43 as well as a member of the deltacoronavirus genus, PDCoV, which have the most divergent RdRp of known CoV as compared to SARS- and MERS-CoV. These data further illuminate the breadth and antiviral activity of RDV against the CoV family and suggest RDV as a potential antiviral for current endemic and epidemic CoV as well as future emerging CoV.
2. Materials and methods
2.1. Viruses and cells
Human colorectal carcinoma (HCT-8, CCL-244) cells were purchased from American Type Culture Collection (ATCC, Manassas, VA) and maintained in RPMI-1640 (ThermoFisher Scientific), 10% fetal bovine serum (FBS, Hyclone, ThermoFisher Scientific) and antibiotic/antimycotic (anti/anti, Gibco, ThermoFisher Scientific). Human hepatoma (Huh7) cells were kindly provided by Dr. Mark Heise at UNC Chapel Hill. Huh7 cells were grown in Dulbecco's Modified Eagle's Medium (DMEM, Gibco, ThermoFisher Scientific), 10% FBS (Hyclone) and anti/anti (Gibco, ThermoFisher Scientific). Porcine kidney (LLC-PK1) cells were purchased from the UNC Tissue Culture Facility and maintained in DMEM, 5% Fetal Clone 2 (Hyclone, ThermoFisher Scientific), non-essential amino acids (NEAA, Gibco, ThermoFisher Scientific), 10 mM HEPES (Gibco, ThermoFisher Scientific), anti/anti (Gibco, ThermoFisher Scientific). Human lung fibroblast (MRC5) cells were purchased from ATCC (CCL-171) and maintained in MEM (Gibco), 10% FBS (Hyclone, ThermoFisher Scientific) and anti/anti (Gibco, ThermoFisher Scientific). The VR-1558 strain of HCoV-OC43 was purchased from ATCC, passaged once on HCT-8 cells and amplified once on Huh7 cells to create a working stock. The VR-740 strain of HCoV-229E was purchased from ATCC, passaged once on MRC5 cells and amplified once on Huh7 cells to create a working stock. Porcine deltacoronavirus (PDCoV) strain OH-FD22 LLCPK P5 was kindly provided by Dr. Linda Saif at Ohio State University. PDCoV virus stock was created through passage on LLC-PK1 cells in Optimem (Gibco, ThermoFisher Scientific), NEAA (Gibco, ThermoFisher Scientific), 10 mM HEPES (Gibco, ThermoFisher Scientific), anti/anti (Gibco), 0.3% tryptose phosphate broth and 0.0025% pancreatin (Sigma-Aldrich, St. Louis, MO).
2.2. Remdesivir (RDV)
RDV was synthesized at Gilead Sciences Inc. (Siegel et al., 2017) and its chemical identity and purity were determined by nuclear magnetic resonance, high-resolution mass spectrometry, and high-performance liquid chromatography (HPLC) analysis. RDV was made available to the University of North Carolina at Chapel Hill (UNC) under a material transfer agreement with Gilead Sciences. RDV was solubilized in 100% DMSO for in vitro studies.
2.3. HCoV-OC43 antiviral focus forming assay in Huh7 cells
Poly-L Lysine (Gibco, ThermoFisher Scientific) coated 96-well plates were seeded with 2E+05 Huh7 cells/well. The following day, cells were infected with HCoV-OC43 (MOI = 0.025) diluted in “infection medium” (DMEM, 5% FBS, anti/anti, all ThermoFisher Scientific) in the presence of DMSO or a dose response of RDV for 1 h at 32 °C. After 1 h, cells were overlaid with Optimem (Gibco, ThermoFisher Scientific), 1% carboxymethyl cellulose, 2% FBS, NEAA, anti/anti that contained DMSO or the dose response of RDV. Final DMSO concentration was held at 0.05% in all conditions. Four to eight replicate wells were allotted to each condition. Non-infected cells treated with DMSO were also included as a “100% inhibition” control. After 72 h treatment at 32 °C, cells were fixed in 10% buffered formalin (ThermoFisher Scientific), permeabilized with 0.1% Triton X-100 (Sigma-Aldrich, St. Louis, MO), blocked in PBS 1% BSA/0.2% skim milk and the endogenous peroxidase was quenched with 3% hydrogen peroxide (ThermoFisher Scientific). OC43 antigen was stained with antibody (primary: mouse anti-OC43 nucleoprotein antibody (Millipore MAB9013), secondary: goat anti-mouse HRP antibody (KPL 474–1806)) and visualized with DAB reagent (ThermoFisher Scientific). Infected cell foci were visualized and quantitated via CTL ImmunoSpot ELIspot reader (CTL, Cleveland, OH, USA). The effective concentration that reaches 50% decrease in viral replication was defined as the EC50 value. The EC50 value was calculated from a dose-response curve using 4-parameter (variable slope) equation (Equation (1)) in GraphPad Prism 8 (GraphPad).
| Y = 100/(1 + 10^((LogEC50-X)*HillSlope)) | (1) |
Where Y represents % of inhibition and X represents RDV concentration. Mock infected cells were used as control for 100% inhibition and cells treated with vehicle alone was used as control for 0% inhibition.
2.4. HCoV-OC43 antiviral fluorescence in situ hybridization (FISH) imaging assay
In a 12-well plate, calf skin collagen (Sigma-Aldrich, St. Louis, MO) coated glass cover slips (18 mm round #1, Neuvitro, ThermoFisher Scientific) were seeded with 2E+05 Huh7 cells/well 24 h prior to infection. Cells were infected with HCoV-OC43 (MOI = 0.025) diluted in “infection medium” containing either 0.25, 0.1, 0.025 or 0.01 μM RDV or DMSO vehicle control for 1 h at 32 °C. After 1 h, monolayers were washed and infection medium containing the previously described DMSO or RDV was added. After 6 days at 32 °C, coverslips were fixed with 3.7% formaldehyde in PBS for 10 min, washed with PBS and stored in 70% ethanol until staining. 48 unique oligonucleotide probes against ORF1a coupled with Quasar 570 were fabricated by LGC Biosearch Technologies. Fixed coverslips were FISH stained and nuclei were counterstained with Hoechst 33258 (ThermoFisher Scientific) according to the protocol provided by LGC. Coverslips were imaged on a Keyence BZX-700 automated microscope. Images were acquired with the same settings and cell quantitation was performed using the Keyence Hybrid Cell Count analysis package (Matsuda et al., 2018).
2.5. HCoV-OC43 genomic and subgenomic qRT-PCR assay
12-well plates of Huh7 cells were prepared without coverslips, infected and treated similarly to those described for FISH. After 6 days, total RNA was isolated via Qiagen RNeasy kit, quantified by NanoDrop (Thermo) and stored at −80 °C until analysis. Previously published TaqMan style “PrimeTime” qRT-PCR assays were generated by Integrated DNA technologies (IDT) to measure genomic RNA (Orf1b, All 5′-3′, Forward GTGGATTCTGCTCAAG, Probe (5′ 6-FAM/ZEN/3′ IBFQ) ATTCACAGACTGCAGAAACAGCGCATTCTGTA Reverse AATACCTTTCTTGGCTCGAGTAAT) and subgenomic RNA (Nucleocapsid, All 5′-3′, Forward CGATGAGGCTATTCCGACTAGGT, Probe (5′ 6-FAM/ZEN/3′ IBFQ) TCCGCCTGGCACGGTACTCCCT, Reverse CTTCCTGAGCCTTCAATATAGTAACC) (Loens et al., 2012; Walsh et al., 2013). Single step qRT-PCR was performed using Fast Virus 1-Step Master Mix (ThermoFisher Scientific) and associated program in a Roche Light Cycler qPCR machine. Each reaction contained 100 ng of total RNA for experimental samples. Copy number/reaction was generated using an HCoV-OC43 RNA standard curve.
2.6. Cytopathic effect based HCoV-229E antiviral assay
The day prior to infection, black-walled clear-bottom 96-well plates (Corning #3904) were seeded with 2E+05 Huh7 cells/well. Cells were infected with HCoV-229E (MOI = 0.15) diluted in infection medium (DMEM, 5% FBS, anti/anti) in the presence of DMSO or a dose response of RDV and incubated 32 °C for 6 days. Eight replicate wells were allotted to each concentration condition. DMSO was held constant in all conditions at 0.05%. Non-infected cells treated with DMSO were also included as a “100% inhibition” control. At the termination of the assay, cytopathic effect (CPE) was measured in a luminometer (Promega Glomax, Promega, Madison, WI) by CellTiter-Glo Assay (Promega, Madison, WI).
2.7. Porcine deltacoronavirus (PDCoV) antiviral assay in LLCKP1 and Huh7 cells
The day prior to infection, black-walled clear-bottom 96-well plates (Corning, Kennebunk, ME) were seeded with 2E+05 LLC-PK1 cells/well. Cells were infected with PDCoV (MOI = 0.025) diluted in “infection medium” (DMEM, HEPES, NEAA, anti/anti, 0.3% tryptose phosphate broth and 0.0025% pancreatin) in the presence of RDV (10–0.0015 μM) or DMSO for 3 days at 37 °C. Eight replicate wells were allotted to each concentration condition. Final DMSO concentration was held constant at 0.05% in all conditions. Non-infected cells treated with DMSO were also included as a “100% inhibition” control. At the termination of the assay, CPE was measured in a luminometer (Promega Glomax) by CellTiter-Glo Assay (Promega). We performed similar antiviral assays with HCoV-229E in LLC-PK1 cells with the following changes: cells were infected with HCoV-229E at an MOI of 0.15 in the presence of (10–0.0015 μM) or DMSO diluted in PDCoV infection medium for 6 days at 32 °C. For Huh7-based PDCoV assay, conditions were similar to the LLC-PK1-based assay but with the following changes: TPCK Trypsin at 1 μg/mL was used rather than pancreatin, and cells were infected with PDCoV at a MOI of 0.0006 (titer was determined on Huh7 cells). We performed similar antiviral assays with HCoV-229E in Huh7 cells using the same media conditions used for PDCoV with the following changes: cells were infected with HCoV-229E at a MOI of 0.15 for 6 days at 32 °C.
2.8. Assessing cytotoxicity with RDV treatment
To assess RDV cytotoxicity in Huh7 and LLC-PK1 cells, cells were seeded and treated with DMSO or RDV (10–0.0015 μM) diluted in the appropriate infection medium and concurrent with each antiviral assay described above. At the termination of each assay, cytoxicity was measured via CellTiter-Glo assay (Promega) read on a Promega luminometer. CC50 value was defined by the concentration required to reach 50% cell death and determined by fitting a dose-response to Equation (1), where the EC50 value is replaced by the CC50 value, using GraphPad Prism 8. Cell-free samples were used as 100% cytotoxic controls and vehicle-only cells were used as 0% cytotoxic controls.
2.9. Coronavirus phylogenetic analysis
Multiple sequence alignments and phylogenetic trees were constructed using the Geneious Tree Builder in Geneious 9.1.3 (http://www.geneious.com). Tree visualization, customization and sequence identity heat map creation was performed in Evolview (www.evolgenius.info/evolview). The accession numbers utilized for phylogenetic analysis were: alphacoronavirus (group 1a) PEDV (porcine epidemic diarrhea virus, NC_003436); alphacoronavirus (group 1b) HCoV-229E (JX503060), HCoV-NL63 (JX504050); betacoronavirus (group 2a) HCoV-OC43 (AY903460), HCoV-HKU1 (DQ415904), MHV (mouse hepatitis virus, AY910861.1); betacoronavirus (group 2b) SARS-CoV (AY278741), BtCoV-SHC014 (bat coronavirus SHC014, KC881005), BtCoV-WIV1 (bat coronavirus WIV1, KF367457), BtCoV-HKU3 (bat coronavirus HKU3, DQ22305); betacoronavirus (group 2c) BtCoV-HKU5 (bat coronavirus HKU5, NC_009020), MERS-CoV (JX869059); betacoronavirus (group 2d) HKU9 (bat coronavirus HKU9, EF065516); gammacoronavirus (group 3) AIBV (avian infectious bronchitis virus, NC_001451); deltacoronavirus (group 4) PDCoV (KR265863), HKU12 (thrush coronavirus, FJ376621), HKU13 (munia coronavirus, FJ376622), HKU16 (white-eye coronavirus, JQ065044), HKU17 (sparrow coronavirus JQ065045), HKU18 (magpie robin coronavirus, JQ065046), HKU19 (night heron coronavirus, JQ065047), HKU20 (wigeon coronavirus, JQ065048), HKU21 (common moorhen coronavirus, JQ065049).
3. Results
3.1. Remdesivir (RDV) is a potent antiviral against human coronavirus OC43
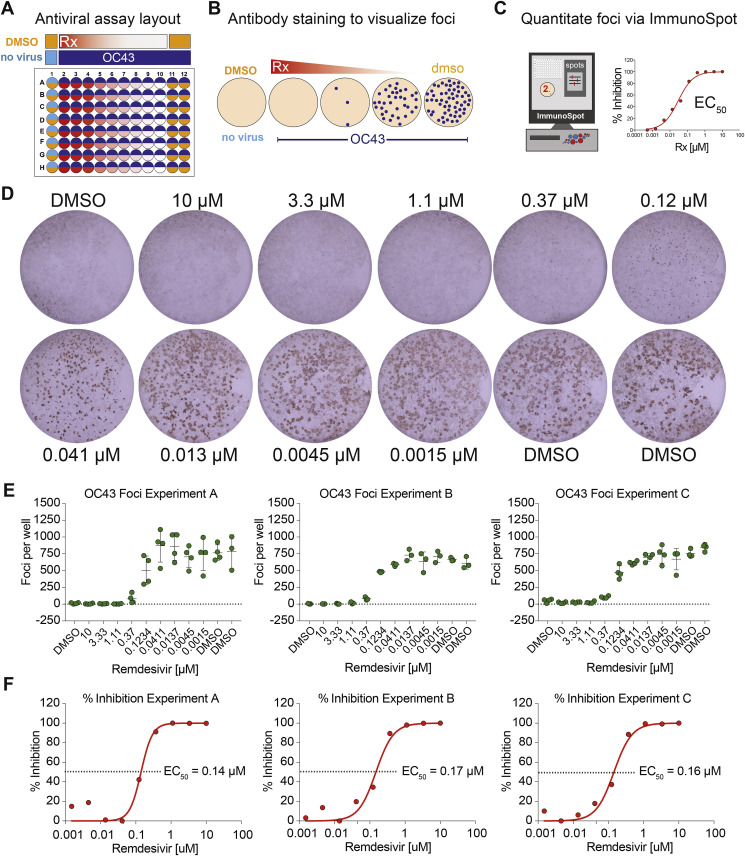
The EC50 values of RDV have previously been reported for MERS-CoV [0.03 μM in Calu-3 cells; 0.074 μM in primary human airway epithelial cells (HAE)], SARS-CoV (0.069 μM in HAE), and MHV (0.03 μM in DBT cells) (Agostini et al., 2018; Sheahan et al., 2017). We sought to determine the EC50 values of RDV for HCoV-OC43 and HCoV-229E, which typically cause upper respiratory infection in children but can cause more severe lower respiratory infection in adults with underlying respiratory conditions (i.e. asthma, COPD) and the elderly (Falsey et al., 2002). Since HCoV-OC43 does not cause an overt cytopathic effect (CPE) in Huh7 cells, we established a focus forming reduction assay (FFRA) for HCoV-OC43 (Fig. 1 A) in 96-well plates based on nucleocapsid antigen staining (Fig. 1B) and quantification via Elispot reader for increased throughput (Fig. 1C). Among the numerous viral genomic and subgenomic messenger RNAs and protein products generated during CoV replication, the nucleocapsid viral messenger RNA and protein are the most abundant. We consistently observed an RDV dose-dependent reduction in HCoV-OC43 antigen foci (Fig. 1D and E). Similarly, the EC50 (0.14, 0.17, 0.16 μM, mean = 0.15 μM, standard deviation = 0.015) was highly consistent from experiment to experiment (Fig. 1F). Importantly, the CC50 obtained in Huh7 cells was >10 μM. Thus, for this assay system, the selectivity index (SI = CC50/EC50) was >66. This assay is driven by the detection nucleocapsid, the most abundant viral protein during CoV replication. Thus, the dynamic range of detection is maximized due to our antibody and viral antigen pairing, which may have been notably lower had we chosen to measure the expression of a viral protein with lower expression (e.g. non-structural protein 2) (Neuman et al., 2008). Conversely, due to the high abundance of nucleocapsid mRNA, RDV may cause reductions in viral RNAs that are not detectable in this assay until a certain threshold is achieved that results in significant diminishment of nucleocapsid protein production.
Fig. 1.
HCoV-OC43 Focus forming antiviral assay. A) HCoV-OC43 antiviral assay plate layout in Huh7 cells treated with DMSO or a dose response of RDV. B) A dose dependent reduction in viral foci is visualized through antibody staining. C) The numbers of foci are quantitated on an ImmunoSpot Elispot reader to generate % inhibition values. D) Example foci from a complete dose response of RDV. E) The quantitated number of spots per well for three independent experiments (A, B, C). Each dot represents the data from one well in a multiwell plate. The line is drawn at the mean and error bars represent the standard deviation. F) EC50 values were generated through graphing the percent inhibition from the above data in Graphpad Prism 8.
3.2. RNA FISH and qRT-PCR demonstrate RDV diminishes HCoV-OC43 viral RNA
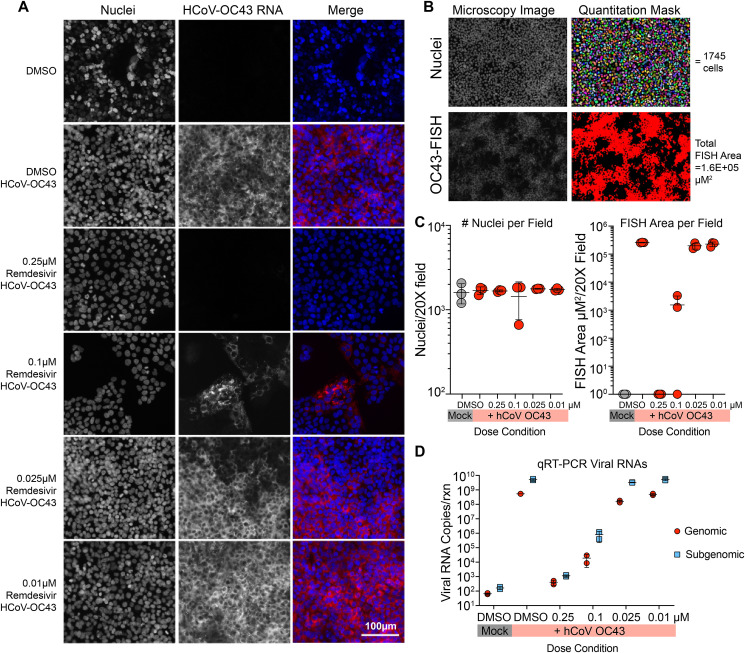
We aimed to corroborate the protein-based FFRA data for HCoV-OC43 with multiple viral RNA-based techniques. First, we employed an RNA fluorescence in situ hybridization (FISH) microscopy-based technique. The HCoV-OC43 ORF1a region of the genomic RNA was stained with 48 unique oligonucleotide probes conjugated to a Quasar 570 fluorophore. With RNA FISH visualized through fluorescence microscopy, we observed an RDV dose-dependent reduction in HCoV-OC43 genomic RNA (Fig. 2 A). To quantify this trend, cell number in three random microscopy fields per drug condition was determined by automated enumeration of Hoechst stained nuclei along with the total HCoV-OC43 FISH area/field (Fig. 2B). The number of nuclei per random field were consistent among drug conditions (Fig. 2C). Similarly, we observed more than a 5-log difference in HCoV-OC43 FISH area comparing mock and infected cells in the absence of RDV. At the highest dose of RDV (0.25 μM), HCoV-OC43 FISH signal was not detected but as RDV dose was diminished, virus replication gradually increased to levels similar to that in untreated cells (Fig. 2C). Similar data were generated from concurrent and equivalent studies by quantitative reverse transcriptase PCR (qRT-PCR) measuring both genomic (ORF1b) and subgenomic (nucleocapsid) viral RNA species. Due to the increased sensitivity of PCR-based assays over microscopy, the dynamic range of the qRT-PCR assay was more than 6 logs when comparing mock to infected replicates (Fig. 2D). Similar to FISH, levels of HCoV-OC43 genomic and subgenomic RNA treated with 0.25 μM RDV were similar to that of mock infected and increased in an RDV dose dependent manner. The resultant EC50 (0.02 μM) generated from the genomic RNA assay was 7.5-fold lower than that obtained by FFRA but this is not unexpected given the increased sensitivity of PCR-based assays. It is important to note that our qPCR assay may be underestimating the potency of RDV. The antiviral effect of RDV has been reported to be through delayed chain termination (Jordan et al., 2018; Lo et al., 2017; Tchesnokov et al., 2018, 2019; Warren et al., 2016). Thus, our assay should detect both functional and non-functional truncated defective viral RNAs as long as the target sequence is present.
Fig. 2.
RNA FISH and qRT-PCR demonstrate RDV diminishes HCoV-OC43 viral RNA. (A) Photomicrographs of HCoV-OC43 genomic RNA fluorescence in situ hybridization (FISH). Huh7 infected with HCoV-OC43 at an MOI of 0.025 treated with dilutions of RDV or DMSO. Cell nuclei were counterstained with Hoechst. The bar is 100 μM. Representative pictures from two independent experiments are shown. (B) Quantitation of RNA FISH signal through the creation of signal area masks for nuclei and HCoV-OC43 FISH signals. (C) Enumeration of nuclei and HCoV-OC43 RNA FISH area in three random fields per condition. (D) Confirmation of HCoV-OC43 FISH data with qRT-PCR for viral genomic (ORF1b) and subgenomic (nucleocapsid). Parallel plates were infected and treated similarly to those in A.
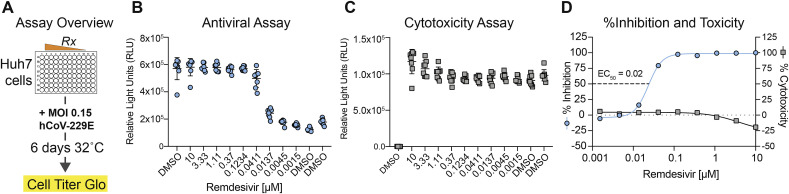
3.3. Potent antiviral activity of RDV against human coronavirus 229E
Unlike HCoV-OC43, HCoV-229E infection of Huh7 causes CPE. Thus, we established a CPE and CellTiter-Glo-based antiviral assay for HCoV-229E (Fig. 3 A). An RDV dose-dependent reduction in HCoV-229E replication was observed (Fig. 3B) without drug-induced cytotoxicity (Fig. 3C). Over five independent experiments, we obtained an average EC50 of 0.024 ± 0.018 μM (mean±standard deviation) (Fig. 3D). Importantly, since cytotoxicity (CC50 > 10 μM) was not observed in Huh7 cells across the dose range measured in the assay (10 μM–0.0015 μM) (Fig. 3B), the SI for this assay was >400.
Fig. 3.
HCoV 229E antiviral assay. A) The antiviral activity of RDV against HCoV-229E was measured in a cytopathic effect-based assay in Huh7 cells by CellTiter-Glo assay. Each dot represents the data from one well in a 96-well plate. The line is at the mean and error bars represent the standard deviation. B) Cytotoxicity of RDV was measured via CellTiter-Glo assay. C) Percent inhibition and percent cytotoxicity from data in panel A and panel B to determine EC50 and CC50. Five independent studies were performed (average EC50 = 0.024 μM). Representative data from a single experiment is shown.
3.4. Porcine deltacoronavirus is susceptible to the antiviral activity of RDV
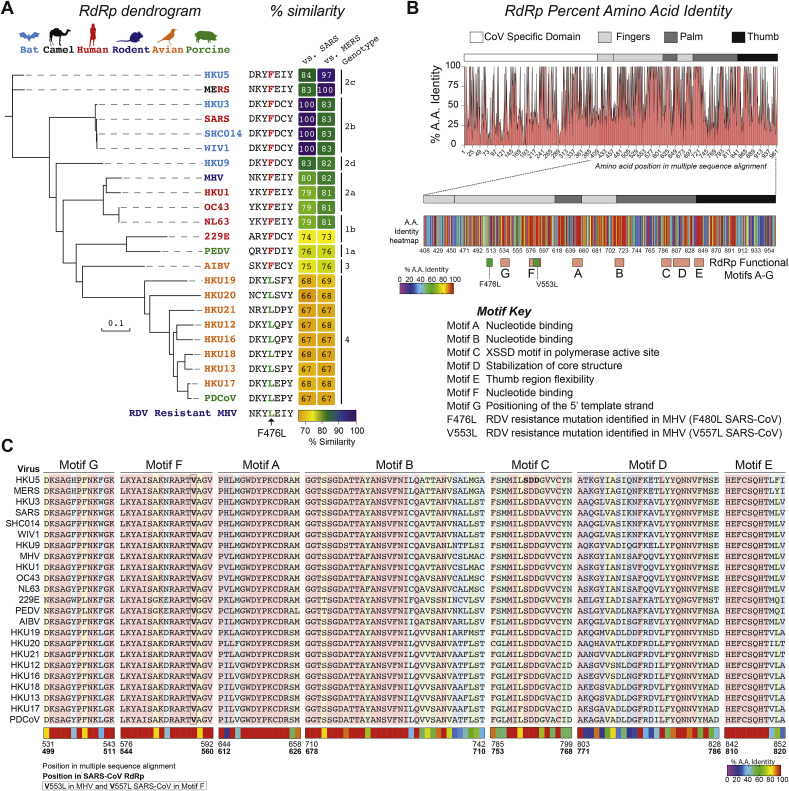
The genetically diverse CoV family infects a wide variety of avian and mammalian hosts. Of the four CoV genera, the deltacoronavirus have the most divergent RNA dependent RNA polymerase (RdRp) as compared to SARS- and MERS-CoV (67–69% amino acid similarity to SARS- or MERS-CoV) (Fig. 4 A). Interestingly, deltacoronavirus RdRp naturally harbor a leucine at residue 483 which is associated with partial resistance (i.e. up to a 5-fold shift in EC50) to RDV in MHV (F476L) and SARS-CoV (F480L) at the homologous positions (Fig. 4A). We mapped the percent amino acid identity for the CoVs described in Fig. 4A to determine if variation was localized to one specific region or functional domain (Fig. 4B). While there is variation across the entire protein, there are regions of concentrated heterogeneity in between motifs B (nucleotide binding) and C (SDD motif in the active site) and in the C-terminal region of the thumb domain (Fig. 4B). Importantly, most of the RdRp functional domains (A-G) as described by Xu et al., in 2003 for SARS-CoV (Fig. 4C) are highly conserved (i.e. 100% identity) (Xu et al., 2003).
Fig. 4.
Variation in CoV RdRp and susceptibility to RDV. (A) Dendrogram showing genetic relatedness of representative human and zoonotic CoV RdRp proteins. Font color of virus name corresponds to natural host for virus shown above. Amino acid sequence alignment shows 483L RDV resistance mutation is naturally occurring in deltacoronavirus (genotype 4). The percent similarity of each RdRp as compared to SARS-CoV and MERS-CoV is indicated in the heat map. (B) Variation in CoV specific, fingers, palm and thumb domains of RdRp shown in % amino acid identity plot (top) and in the more detailed heat map showing % identity per residue within the RdRp functional domains (A–G) and RDV resistance mutations identified in MHV (F476L, V553L) mapped. (C) Amino acid multiple sequence alignments for each RdRp functional motif showing high conservation. Amino acid positions are noted in the multiple sequence alignment which corresponds to positions above as well as in SARS-CoV RdRp (bold).
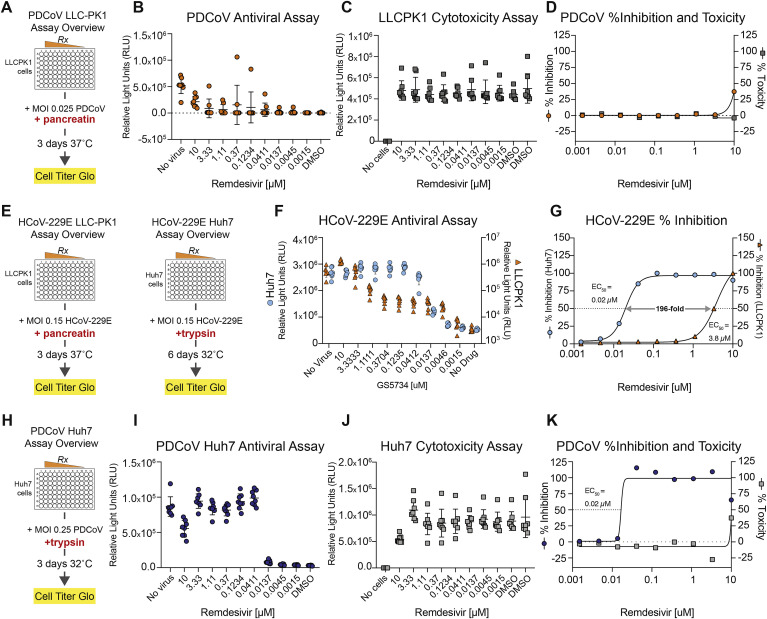
Given the divergence of the deltacoronavirus RdRp and the naturally occurring putative resistance mutation, we sought to determine the susceptibility of members of the deltacoronavirus genus to the antiviral effect of RDV. Using porcine deltacoronavirus (PDCoV) as a model, we first established an antiviral assay in the porcine kidney epithelial cell line, LLC-PK1, which support robust PDCoV replication (Fig. 5 A). Enteric CoV (i.e. PDCoV, PEDV) require the addition of digestive enzymes (i.e. trypsin, pancreatin) to culture medium for efficient replication and CPE (Beall et al., 2016; Hu et al., 2015b). In LLC-PK1 cells with serum-free medium and pancreatin, RDV did not diminish PDCoV replication greater than 50% thus the EC50 could not be determined (Fig. 5B–D). To ascertain whether PDCoV was naturally resistant to RDV or if the LLC-PK1 cell harbored an unknown defect in a cellular process required for antiviral activity (i.e. nucleotide uptake, metabolism, etc.), we performed similar antiviral assays with HCoV-229E in LLC-PK1 cells (Fig. 5E). Similar to PDCoV, RDV was not potent against HCoV-229E in LLC-PK1 cells (EC50 = 3.8 μM, Fig. 5F and G) yet the antiviral activity of RDV against HCoV-229E in Huh7 cells in PDCoV assay medium (i.e. serum free medium with trypsin) were similar to those in Fig. 3 (EC50 = 0.02 μM, Fig. 5G). These data suggest LLC-PK1 cells are deficient in a cellular process required for the antiviral activity of RDV. Importantly, in Huh7 cells cultured in TPCK trypsin-containing and serum-free media, (Fig. 5H), PDCoV replication was dose-dependently reduced (Fig. 5I) with an EC50 value of 0.02 μM (Fig. J and K, a CC50 value > 10 μM and a SI > 500). All together with previous publications, these data demonstrate that a panel of CoV representing family-wide genetic diversity in the RdRp are susceptible to the inhibition by RDV.
Fig. 5.
Porcine deltacoronavirus is susceptible to the antiviral activity of RDV. A) Overview of the CPE-based antiviral assay for PDCoV in LLC-PK1 cells. (B) Representative relative light units (RLU) raw data from one of four independent PDCoV assays in LLC-PK1 cells. C) Example cytoxicity data (RLU) measured via CellTiter-Glo for RDV in LLC-PK1 cells. (D) RDV EC50 and CC50 curves for PDCoV in LLC-PK1 cells. (E) Overview of RDV CPE-based antiviral assays in either LLC-PK1 or Huh7 cells for HCoV-229E using PDCoV media conditions. (F) RLU raw data for HCoV-229E in LLC-PK1 or Huh7 cells using media formulations for PDCoV. (G) EC50 curves for raw data shown in F. (H) Overview of RDV CPE-based antiviral assay for PDCoV in Huh7 cells. (I) Representative RLU raw data one of five independent PDCoV assays in Huh7 cells. (J) Example cytoxicity data (RLU) measured via CellTiter-Glo for RDV in Huh7 cells for assay described in “H”. (K) Representative RDV EC50 and CC50 curves for PDCoV in Huh7 cells. For B, C, F, I and J, each dot represents the data from one well in a 96-well plate. The line is at the mean and error bars represent the standard deviation.
4. Discussion
Effective broad-spectrum therapies are needed for the emerging viral threats of today, like Ebola and MERS-CoV, as well as those that have yet to emerge. There are multiple examples of novel CoV emergence including all six human CoV, which are thought to have emerged as zoonoses (Hu et al., 2015a; Huynh et al., 2012; Menachery et al., 2016; Vijgen et al., 2005). Unlike SARS- and MERS-CoV, which are known to cause more sporadic outbreaks, the other four human CoV are endemic causing annual widespread morbidity in infants and the elderly, potentially requiring hospitalization. (Carman et al., 2018; Falsey et al., 2002; Varghese et al., 2018). Although rare, endemic human CoV like HCoV-229E and HCoV-OC43 can also cause severe respiratory disease (pneumonia, ARDS, etc.) in subsets of patients, with presentation similar to SARS- and MERS-CoV (Falsey et al., 2002; Patrick et al., 2006; Vassilara et al., 2018). Thus, the capacity for human CoV to cause severe disease is not unique to emerging viruses like SARS- and MERS-CoV. While there are no CoV specific antivirals approved for human use, many approved therapies for other indications have been evaluated against CoV in hopes of repurposing. The antimalarial, chloroquine, reportedly has moderate antiviral activity in vitro against SARS-CoV (IC50 = 3 μM), MERS-CoV (IC50 = 3–4 μM), HCoV-OC43 (IC50 = 0.3 μM) and 229E (IC50 = 3.3 μM) but failed to exert an antiviral effect against SARS-CoV in mice (Barnard et al., 2006; de Wilde et al., 2014; Shen et al., 2016). Inhibition of viral protease has also been evaluated with lopinavir, a protease inhibitor designed for human immunodeficiency virus, which like chloroquine exerts a moderate antiviral effect on CoV replication (EC50 values: MERS-CoV 8 μM, SARS-CoV 17.1 μM, HCoV-229E 6.6 μM) (de Wilde et al., 2014). RDV is a broad-spectrum antiviral drug with potent in vitro antiviral activity and in vivo therapeutic efficacy against CoV (Agostini et al., 2018; Murphy et al., 2018; Sheahan et al., 2017). However, the full breadth of activity against CoV has yet to be determined. Previously, we demonstrated antiviral activity of RDV against SARS-CoV and related zoonotic bat-CoV (beta 2b), MERS-CoV, and related bat-CoV (beta 2c), HCoV-NL63 (alpha 1b), and MHV (beta 2a) (Agostini et al., 2018; Sheahan et al., 2017). In addition, the parental nucleoside (GS-441524) of RDV (i.e. the prodrug of GS-5734) has a reported EC50 of 0.7 μM against feline infectious peritonitis virus (FIP, alpha 1a CoV) and could reverse lethal disease in experimental infectious of cats (Murphy et al., 2018). Here, we show that RDV has potent antiviral activity against HCoV-OC43, HCoV-229E, and PDCoV which are in subgenera beta 2a, alpha 1b, and delta 4, respectively. While we have not yet tested a representative from alpha 1a (e.g. PEDV) or gamma (e.g. avian infectious bronchitis virus, AIBV), CoV members with similar RdRps, like HCoV-229E, have been evaluated. The other outstanding subgenera yet to be evaluated is beta 2d, which is currently comprised of only bat-CoV (Lau et al., 2010). Culture systems for beta 2d viruses are currently not available. Thus, with this report, we demonstrate potent antiviral activity of RDV against representatives capturing near family-wide CoV RdRp diversity including deltacoronavirus which have the most divergent RdRp of known CoV as compared to SARS- and MERS-CoV. These data suggest that future emerging CoV may be similarly susceptible to the inhibition by RDV.
The CoV RNA replication complex is comprised of multiple viral proteins one of which, the viral RdRp, generates nascent genomic and subgenomic viral RNAs. While the CoV RdRp is relatively conserved, the amino acid percent similarity can vary from 67 to 100%. For antiviral drugs that interact and interfere with RdRp function, variation in amino acid sequence and resultant protein structure can have profound effects on susceptibility. Lo et al. recently reported a correlation between nucleotide interaction motifs “A” and “B” in the RdRp and susceptibility to the antiviral activity of RDV (Lo et al., 2017). Motifs A and B were highly conserved within virus families and generally correlated with RDV antiviral activity but for members of Flaviviridae (i.e. West Nile virus EC50 = >30 μM, yellow fever virus EC50 = 11 μM, tick borne encephalitis virus EC50 = 2.1 μM, hepatitis C virus EC50 = 4.1 μM), the potency of RDV was mixed even though motifs A and B were for the most part highly conserved. Thus, other factors in the RdRp structure or function likely mediate this discrepancy. Mutations (F476L and V553L), identified through passage of MHV in the presence of RDV, provide partial resistance to RDV antiviral activity (i.e. 2.4-fold and 5-fold shift in EC50, respectively). Interestingly, though MHV and SARS-CoV RdRp are 80% identical, resistance is transferable to SARS-CoV via reverse genetic introduction of homologous mutations (F480L and V557L) (Agostini et al., 2018). Here, we show that PDCoV, a member of the deltacoronavirus genus, is exquisitely sensitive to RDV despite naturally harboring a one of the two RdRp resistance mutations identified in MHV. Without a reverse genetic system for PDCoV, it is not possible to definitively determine if this homologous mutation (483L) alters the antiviral effect of RDV on PDCoV. In the very recent publication describing the SARS-CoV RdRp structure, the authors suggest the F480L mutation, located in the fingers domain contacting motif B, impacts active site dynamics related to catalysis (Kirchdoerfer and Ward, 2019). Interestingly, Kirchdorfer and Ward suggest the V557L (V553L in MHV) resistance mutation in the SARS-CoV RdRp increases stringency of base pairing thereby increasing polymerase fidelity allowing for better exclusion of RDV from the active site (Kirchdoerfer and Ward, 2019). Future studies will focus on passage of PDCoV for RDV resistance to determine if genetically divergent CoV take similar or different paths towards acquiring RDV resistance.
Broad-spectrum approaches maximize the utility of antivirals with proven efficacy against current epidemic strains as well as zoonotic viruses that will likely seed future emergence. Here, we further describe the breadth of RDV antiviral activity against both human and zoonotic CoV. We found that RDV had decreased potency against 229E and PDCoV in the porcine kidney cell line, LLC-PK1, although these viruses were similarly sensitive in the human hepatoma cell, Huh7. Although different from RDV, normal uptake and metabolism has been reported for the nucleoside analog lamivudine (i.e. 3 TC) in LLC-PK1 (Leung and Bendayan, 2001). Interestingly, uptake and the antiviral activity of ribavirin, another nucleoside analog, was reported to vary significantly among human liver cell lines, which was directly correlated with antiviral activity against poliovirus indicating that variation in small molecule uptake/transport can impact antiviral efficacy (Ibarra and Pfeiffer, 2009). While RDV does have altered metabolism in mice due to the circulating serum carboxylesterase 1c (Ces1c) that is absent in humans (Sheahan et al., 2017), it is potently antiviral against MHV in mouse DBT cells (Agostini et al., 2018). Therefore, the Ces1c metabolism issue with RDV is specific to the mouse in vivo system. Although it is likely that LLC-PK1 cells are deficient in a cellular process required for the antiviral activity of RDV, without further testing in other porcine cell lines, it is not possible to ascertain whether this is cell line specific or species specific or to speculate about the potential efficacy against PDCoV in pigs. Here, we have focused our efforts on developing in vitro assays for human and zoonotic CoV but animal models exist for HCoV-229E and OC43 as well as PDCoV within which in vivo efficacy could be evaluated (Jung et al., 2015; Lassnig et al., 2005; St-Jean et al., 2004). Given that we have demonstrated antiviral activity against a panel of CoVs with the great RdRp diversity, RDV may too be efficacious against future emerging CoV as long as they fall within the spectrum of known genetic diversity. To prepare for the potential of drug resistance to single therapies, multiple novel broad-spectrum antiviral strategies are needed. Moreover, with desired safety profiles, combination broad-spectrum antiviral therapies with disparate mechanisms of action may be more effective at treating emerging viral syndromes in parallel prior to etiological agent identification. If available, new antiviral therapies would markedly expand the current treatment options available to clinicians. The current front-line agents treating emerging viral epidemics (i.e. ribavirin, interferon, lopinavir/ritonavir, and corticosteroids) often do not improve outcomes over supportive care (Zumla et al., 2016). Therefore, our studies provide further evidence of the antiviral activity of RDV against endemic, epidemic, and zoonotic CoV, which may be useful in treating patients with severe endemic CoV infections, MERS-CoV patients or emerging CoV diseases in the future provided that the drug safety profile is proven to be compatible with its use in the affected patient populations.
Acknowledgements.
We would like to acknowledge the following funding sources, Antiviral Drug Discovery and Development Center (5U19AI109680) and a partnership grant from the National Institutes of Health (NIH), United States (5R01AI132178). KD was supported by a fellowship from the NIH National Institute of Allergy and Infectious Diseases virology training grant (T32 AI007419).
References
- Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R., Ray A.S., Cihlar T., Siegel D., Mackman R.L., Clarke M.O., Baric R.S., Denison M.R. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9 doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard D.L., Day C.W., Bailey K., Heiner M., Montgomery R., Lauridsen L., Chan P.K., Sidwell R.W. Evaluation of immunomodulators, interferons and known in vitro SARS-coV inhibitors for inhibition of SARS-coV replication in BALB/c mice. Antivir. Chem. Chemother. 2006;17:275–284. doi: 10.1177/095632020601700505. [DOI] [PubMed] [Google Scholar]
- Beall A., Yount B., Lin C.M., Hou Y., Wang Q., Saif L., Baric R. Characterization of a pathogenic full-length cDNA Clone and transmission model for porcine epidemic diarrhea virus strain PC22A. mBio. 2016;7 doi: 10.1128/mBio.01451-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman K.B., Calik M., Karal Y., Isikay S., Kocak O., Ozcelik A., Yazar A.S., Nuhoglu C., Sag C., Kilic O., Dinleyici M., Lacinel Gurlevik S., Yimenicioglu S., Ekici A., Perk P., Tosun A., Isik I., Yarar C., Arslantas D., Dinleyici E.C., E.S.G Viral etiological causes of febrile seizures for respiratory pathogens (EFES Study) Hum. Vaccines Immunother. 2018:1–7. doi: 10.1080/21645515.2018.1526588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Chan K.H., Kao R.Y., To K.K., Zheng B.J., Li C.P., Li P.T., Dai J., Mok F.K., Chen H., Hayden F.G., Yuen K.Y. Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J. Infect. 2013;67:606–616. doi: 10.1016/j.jinf.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Jochmans D., Posthuma C.C., Zevenhoven-Dobbe J.C., van Nieuwkoop S., Bestebroer T.M., van den Hoogen B.G., Neyts J., Snijder E.J. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 2014;58:4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkman R., Jebbink M.F., Gaunt E., Rossen J.W., Templeton K.E., Kuijpers T.W., van der Hoek L. The dominance of human coronavirus OC43 and NL63 infections in infants. J. Clin. Virol. 2012;53:135–139. doi: 10.1016/j.jcv.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsey A.R., Walsh E.E., Hayden F.G. Rhinovirus and coronavirus infection-associated hospitalizations among older adults. J. Infect. Dis. 2002;185:1338–1341. doi: 10.1086/339881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimdal I., Moe N., Krokstad S., Christensen A., Skanke L.H., Nordbo S.A., Dollner H. Human coronavirus in hospitalized children with respiratory tract infections: a nine-year-long, population-based study from Norway. J. Infect. Dis. 2019 Apr 8;219(8):1198–1206. doi: 10.1093/infdis/jiy646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Ge X., Wang L.F., Shi Z. Bat origin of human coronaviruses. Virol. J. 2015;12:221. doi: 10.1186/s12985-015-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Jung K., Vlasova A.N., Chepngeno J., Lu Z., Wang Q., Saif L.J. Isolation and characterization of porcine deltacoronavirus from pigs with diarrhea in the United States. J. Clin. Microbiol. 2015;53:1537–1548. doi: 10.1128/JCM.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.W., Dickerman A.W., Pineyro P., Li L., Fang L., Kiehne R., Opriessnig T., Meng X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio. 2013;4 doi: 10.1128/mBio.00737-13. e00737-00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh J., Li S., Yount B., Smith A., Sturges L., Olsen J.C., Nagel J., Johnson J.B., Agnihothram S., Gates J.E., Frieman M.B., Baric R.S., Donaldson E.F. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J. Virol. 2012;86:12816–12825. doi: 10.1128/JVI.00906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra K.D., Pfeiffer J.K. Reduced ribavirin antiviral efficacy via nucleoside transporter-mediated drug resistance. J. Virol. 2009;83:4538–4547. doi: 10.1128/JVI.02280-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P.C., Liu C., Raynaud P., Lo M.K., Spiropoulou C.F., Symons J.A., Beigelman L., Deval J. Initiation, extension, and termination of RNA synthesis by a paramyxovirus polymerase. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Hu H., Eyerly B., Lu Z., Chepngeno J., Saif L.J. Pathogenicity of 2 porcine deltacoronavirus strains in gnotobiotic pigs. Emerg. Infect. Dis. 2015;21:650–654. doi: 10.3201/eid2104.141859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassnig C., Sanchez C.M., Egerbacher M., Walter I., Majer S., Kolbe T., Pallares P., Enjuanes L., Muller M. Development of a transgenic mouse model susceptible to human coronavirus 229E. Proc. Natl. Acad. Sci. U. S. A. 2005;102:8275–8280. doi: 10.1073/pnas.0408589102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Poon R.W., Wong B.H., Wang M., Huang Y., Xu H., Guo R., Li K.S., Gao K., Chan K.H., Zheng B.J., Woo P.C., Yuen K.Y. Coexistence of different genotypes in the same bat and serological characterization of Rousettus bat coronavirus HKU9 belonging to a novel Betacoronavirus subgroup. J. Virol. 2010;84:11385–11394. doi: 10.1128/JVI.01121-10. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Leung S., Bendayan R. Uptake properties of lamivudine (3TC) by a continuous renal epithelial cell line. Can. J. Physiol. Pharmacol. 2001;79:59–66. [PubMed] [Google Scholar]
- Lo M.K., Jordan R., Arvey A., Sudhamsu J., Shrivastava-Ranjan P., Hotard A.L., Flint M., McMullan L.K., Siegel D., Clarke M.O., Mackman R.L., Hui H.C., Perron M., Ray A.S., Cihlar T., Nichol S.T., Spiropoulou C.F. GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci. Rep. 2017;7:43395. doi: 10.1038/srep43395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loens K., van Loon A.M., Coenjaerts F., van Aarle Y., Goossens H., Wallace P., Claas E.J., Ieven M., Group G.S. Performance of different mono- and multiplex nucleic acid amplification tests on a multipathogen external quality assessment panel. J. Clin. Microbiol. 2012;50:977–987. doi: 10.1128/JCM.00200-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M., Tsurusaki S., Miyata N., Saijou E., Okochi H., Miyajima A., Tanaka M. Oncostatin M causes liver fibrosis by regulating cooperation between hepatic stellate cells and macrophages in mice. Hepatology. 2018;67:296–312. doi: 10.1002/hep.29421. [DOI] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Sims A.C., Debbink K., Agnihothram S.S., Gralinski L.E., Graham R.L., Scobey T., Plante J.A., Royal S.R., Swanstrom J., Sheahan T.P., Pickles R.J., Corti D., Randell S.H., Lanzavecchia A., Marasco W.A., Baric R.S. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. U. S. A. 2016 Mar 15;113(11):3048–3053. doi: 10.1073/pnas.1517719113. Epub 2016 Mar 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy B.G., Perron M., Murakami E., Bauer K., Park Y., Eckstrand C., Liepnieks M., Pedersen N.C. The nucleoside analog GS-441524 strongly inhibits feline infectious peritonitis (FIP) virus in tissue culture and experimental cat infection studies. Vet. Microbiol. 2018;219:226–233. doi: 10.1016/j.vetmic.2018.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Joseph J.S., Saikatendu K.S., Serrano P., Chatterjee A., Johnson M.A., Liao L., Klaus J.P., Yates J.R., 3rd, Wuthrich K., Stevens R.C., Buchmeier M.J., Kuhn P. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J. Virol. 2008;82:5279–5294. doi: 10.1128/JVI.02631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick D.M., Petric M., Skowronski D.M., Guasparini R., Booth T.F., Krajden M., McGeer P., Bastien N., Gustafson L., Dubord J., Macdonald D., David S.T., Srour L.F., Parker R., Andonov A., Isaac-Renton J., Loewen N., McNabb G., McNabb A., Goh S.H., Henwick S., Astell C., Guo J.P., Drebot M., Tellier R., Plummer F., Brunham R.C. An outbreak of human coronavirus OC43 infection and serological cross-reactivity with SARS coronavirus. Can. J. Infect Dis. Med. Microbiol. 2006;17:330–336. doi: 10.1155/2006/152612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B., Leist S.R., Pyrc K., Feng J.Y., Trantcheva I., Bannister R., Park Y., Babusis D., Clarke M.O., Mackman R.L., Spahn J.E., Palmiotti C.A., Siegel D., Ray A.S., Cihlar T., Jordan R., Denison M.R., Baric R.S. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Yang Y., Ye F., Liu G., Desforges M., Talbot P.J., Tan W. Safe and sensitive antiviral screening platform based on recombinant human coronavirus OC43 expressing the luciferase reporter gene. Antimicrob. Agents Chemother. 2016;60:5492–5503. doi: 10.1128/AAC.00814-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel D., Hui H.C., Doerffler E., Clarke M.O., Chun K., Zhang L., Neville S., Carra E., Lew W., Ross B., Wang Q., Wolfe L., Jordan R., Soloveva V., Knox J., Perry J., Perron M., Stray K.M., Barauskas O., Feng J.Y., Xu Y., Lee G., Rheingold A.L., Ray A.S., Bannister R., Strickley R., Swaminathan S., Lee W.A., Bavari S., Cihlar T., Lo M.K., Warren T.K., Mackman R.L. Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f][triazin-4-amino] adenine C-nucleoside (GS-5734) for the treatment of Ebola and emerging viruses. J. Med. Chem. 2017;60:1648–1661. doi: 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- St-Jean J.R., Jacomy H., Desforges M., Vabret A., Freymuth F., Talbot P.J. Human respiratory coronavirus OC43: genetic stability and neuroinvasion. J. Virol. 2004;78:8824–8834. doi: 10.1128/JVI.78.16.8824-8834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchesnokov E.P., Feng J.Y., Porter D.P., Gotte M. Mechanism of inhibition of Ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses. 2019;11 doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchesnokov E.P., Raeisimakiani P., Ngure M., Marchant D., Gotte M. Recombinant RNA-dependent RNA polymerase complex of Ebola virus. Sci. Rep. 2018;8:3970. doi: 10.1038/s41598-018-22328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese L., Zachariah P., Vargas C., LaRussa P., Demmer R.T., Furuya Y.E., Whittier S., Reed C., Stockwell M.S., Saiman L. Epidemiology and clinical features of human coronaviruses in the pediatric population. J. Pediatr. Infect. Dis. Soc. 2018;7:151–158. doi: 10.1093/jpids/pix027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilara F., Spyridaki A., Pothitos G., Deliveliotou A., Papadopoulos A. A rare case of human coronavirus 229E associated with acute respiratory distress syndrome in a healthy adult. Case Rep. Infect. Dis. 2018;2018:6796839. doi: 10.1155/2018/6796839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Moes E., Thoelen I., Wollants E., Lemey P., Vandamme A.M., Van Ranst M. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J. Virol. 2005;79:1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh E.E., Shin J.H., Falsey A.R. Clinical impact of human coronaviruses 229E and OC43 infection in diverse adult populations. J. Infect. Dis. 2013;208:1634–1642. doi: 10.1093/infdis/jit393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C., Larson N., Strickley R., Wells J., Stuthman K.S., Van Tongeren S.A., Garza N.L., Donnelly G., Shurtleff A.C., Retterer C.J., Gharaibeh D., Zamani R., Kenny T., Eaton B.P., Grimes E., Welch L.S., Gomba L., Wilhelmsen C.L., Nichols D.K., Nuss J.E., Nagle E.R., Kugelman J.R., Palacios G., Doerffler E., Neville S., Carra E., Clarke M.O., Zhang L., Lew W., Ross B., Wang Q., Chun K., Wolfe L., Babusis D., Park Y., Stray K.M., Trancheva I., Feng J.Y., Barauskas O., Xu Y., Wong P., Braun M.R., Flint M., McMullan L.K., Chen S.S., Fearns R., Swaminathan S., Mayers D.L., Spiropoulou C.F., Lee W.A., Nichol S.T., Cihlar T., Bavari S. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Liu Y., Weiss S., Arnold E., Sarafianos S.G., Ding J. Molecular model of SARS coronavirus polymerase: implications for biochemical functions and drug design. Nucleic Acids Res. 2003;31:7117–7130. doi: 10.1093/nar/gkg916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Fan H., Lan T., Yang X.L., Shi W.F., Zhang W., Zhu Y., Zhang Y.W., Xie Q.M., Mani S., Zheng X.S., Li B., Li J.M., Guo H., Pei G.Q., An X.P., Chen J.W., Zhou L., Mai K.J., Wu Z.X., Li D., Anderson D.E., Zhang L.B., Li S.Y., Mi Z.Q., He T.T., Cong F., Guo P.J., Huang R., Luo Y., Liu X.L., Chen J., Huang Y., Sun Q., Zhang X.L., Wang Y.Y., Xing S.Z., Chen Y.S., Sun Y., Li J., Daszak P., Wang L.F., Shi Z.L., Tong Y.G., Ma J.Y. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.Y. Coronaviruses - drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]