Abstract
Prenatal genomic evaluation of the fetus is available at decreasing cost and with a faster turnaround time. However, fetal genotype-phenotype correlations are in their infancy. By comparison, pediatric and adult genotype-phenotype databases are well-established and publicly accessible. A similar system for fetal genomics is lacking. When a fetal anomaly is identified by ultrasound imaging, a genetic diagnosis provides important information. However, fetal prognostic counseling is problematic if the only available information is based on outcomes following postnatal diagnoses. The same conditions identified prenatally may have more benign or more deleterious outcomes. Also, the condition may evolve over the pregnancy itself. As genomic testing increasingly examines fetal DNA at a single nucleotide level, the concomitant in utero phenotype deserves equal attention. Often the reports of fetal phenotype are limited. Among sonologists, an increased awareness of attaining and communicating detailed fetal phenotypes is needed. The interpretation of expanded prenatal sequencing is reliant on deeper fetal phenotyping. The information gained significantly impacts clinical care and understanding of fetal development. This case series highlights: the broad spectrum of fetal phenotypes for known genetic conditions, phenotype progression during pregnancy, and the need to supplement systematic imaging with descriptive details when assessing fetuses with malformations.
Introduction
Prenatal evaluation of the fetus is rapidly changing. The quantity and detail of genetic information available in the prenatal period is unprecedented. Prenatal imaging resolution continues to improve and alternative modalities including MRI and 3D and 4D ultrasound are entering the prenatal testing space. When an ultrasound abnormality is identified, chromosome microarray (CMA) is now the first-line test1. Following a normal CMA, specific gene panels and sequencing, both exome and whole genome, are often considered given data showing an improved diagnostic yield in anomalous fetuses2,3. The combined advances in both genetic investigations and imaging are uncovering fetal phenotypes atypical from those classically associated with a newborn, child or adult with the same genetic condition. This is epitomized by fetal Noonan syndrome where pleural effusions and polyhydramnios in the second trimester fetus, and not the typical pulmonic stenosis, are common4,5. As such reports grow, and because these syndromes are by nature rare, the necessity for careful and systematic fetal phenotype-genotype determination becomes critically important.
From the laboratory standpoint, incorporation of standardized nomenclature such as Human Phenotype Ontology (HPO) in conjunction with sequencing technologies provides a resource for systematic assessment of genotype-phenotype correlations. However, as HPO is designed for pediatric and adult phenotypes, many key prenatal characteristics, such as amniotic fluid abnormalities and single umbilical artery, are absent6. Furthermore, clinicians often include only minimal ultrasound information on laboratory requisitions limiting detailed and thorough classification of primary and secondary anomalies. Though retrospective review of imaging studies may add further detail, if the original pictures are not obtained with an eye to dysmorphia and minor anomalies, the complete presentation of the fetus with a primary anomaly may be lost.
We sought to highlight cases from our own institutions to emphasize these facets of the growing field of fetal genotype-phenotype correlations, including the need for detailed and documented imaging, conveyance of this information to the laboratory, and a commitment to documentation of evolving imaging findings as the pregnancy progresses, including fetal loss and new third trimester imaging findings.
Methods
Two centers (Brigham and Women’s Hospital and University of North Carolina at Chapel Hill) providing prenatal diagnostic testing were asked to contribute cases. The criteria for inclusion were genetic testing by chromosome microarray (CMA), directed gene panel or exome sequencing AND ultrasound findings which did not suggest the genetic diagnosis. The choice of genetic diagnostic study performed was at the discretion of each center. All studies were performed in CLIA-certified laboratories, including confirmation of exome sequencing results when obtained in a research protocol. Ultrasound assessment of the fetuses occurred before the genetic result. Abnormalities on the ultrasound were classified as structural anomalies (causing mortality or requiring surgical correction), minor anomalies, and the traditional soft marker variants associated with aneuploidy. The information supplied to the testing laboratory was reviewed. Given the years over which these cases were collected and the more recent progression to prenatal exome sequencing, cases were not excluded if the genetic testing was performed in the neonate or stillbirth provided the ultrasound findings had prompted the genetic study, (often antepartum studies had been declined). These cases are included as the genetic testing was obtained from cord blood by prior consent or as part of a research initiative for rapid neonatal exome sequencing. Newborn outcomes were obtained from medical record review.
As this is a series of case reports, institutional review board approval was not needed (Brigham and Women’s Hospital) or was included within the consents for exome sequencing research (University of North Carolina at Chapel Hill).
Results
Case 1
A 44 yo G5 P3013 at 37 0/7 weeks presented for consultation regarding a right-sided pleural effusion. The only other finding was mild polyhydramnios (deepest vertical pocket 8 cm). A normal cardiac exam was obtained, and the estimated fetal weight was within normal limits (3680 g; 85%). Biophysical profile was 8/8. The prenatal course had been uncomplicated except for gestational diabetes (GDM), well-controlled with insulin. Cell-free DNA for the major autosomal and sex aneuploidies was unremarkable; fetal survey at 18 weeks demonstrated normal fetal biometry and anatomy. Due to GDM, frequent ultrasounds (29 0/7, 32 0/7, 33 6/7, 34 5/7 weeks) were obtained for routine fetal surveillance. All ultrasounds confirmed a structurally-normal fetus with reassuring fetal surveillance, > 95% estimated fetal weight with high normal amniotic fluid or mild polyhydramnios.
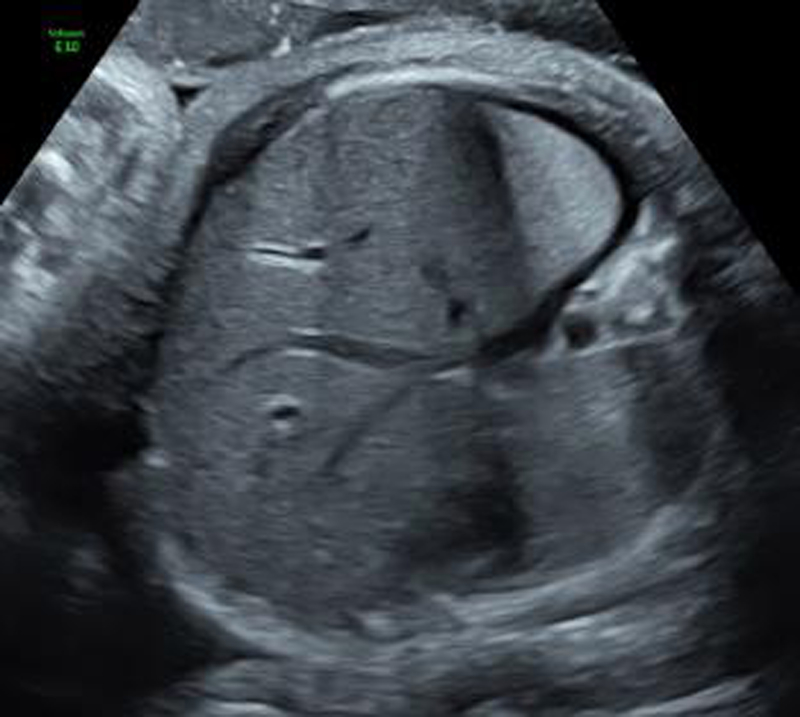
At 36 5/7 weeks, the US was unchanged except for a trace right-sided pleural effusion. Two days later (37 0/7 weeks), a follow-up ultrasound demonstrated an increased right-sided pleural effusion (now moderate) and a new trace left-sided pleural effusion (Figure 1). No other changes were noted; there was mild polyhydramnios and neither skin edema or ascites were noted. Given these findings, the patient was induced with subsequent delivery of a male infant, 3280 g at 37 1/7 weeks, Apgars 8/8. Initial newborn exam was normal with exception of mildly increased work of breathing and the infant was transferred to the Neonatal Intensive Care Unit for management of the pleural effusions. Initial chest x-ray revealed bilateral pleural effusions, right greater than left. Over the subsequent 2 weeks, due to imaging findings and continued oxygen requirement, the infant had serial right pleurocenteses which were suggestive of chylothorax (triglycerides 188 mg/dL, total protein 3.6 g/dL (blood 5.2 g/dL), albumin 2.8 g/dL (blood 3.8 g/dL), LDH 188 U/L (blood 359 U/L)).
Figure 1.
View of the fetal thorax at 36 weeks (Case 1) demonstrating bilateral pleural effusions, right greater than left.
Chromosome microarray on cord blood obtained at delivery was ordered for the indications of “mild right pleural effusion, trace left pleural effusion, mild polyhydramnios”. Seven days later, CMA revealed a pathogenic 2.8 Mb loss involving 69 genes at the site of the classic velocardiofacial deletion (also known as DiGeorge or 22q11.2 deletion syndrome).
Given this result, calcium testing was initiated in the neonate and was notable for hypocalcemia, which was corrected with IV calcium gluconate supplementation over the next 2 days. Neonatal echocardiogram was notable only for a dysplastic pulmonary valve with elongated leaflets and normal valvular function without obstruction. An EEG obtained for two episodes of abnormal movements was normal for age. The newborn screen returned remarkable for severe combined immunodeficiency (SCID) initially attributed to an ill infant, although was likely ultimately reflective of the immune dysfunction observed in DiGeorge syndrome.
Case 2
A 33 yo G2P1001 was a late entry to care at 20 weeks gestation with an US revealing anhydramnios and bilateral echogenic kidneys with multiple small cysts. Given these findings, autosomal recessive polycystic kidney disease was suspected; however, PKHD1 gene testing for carrier status in the mother was unremarkable. Care was transferred at 24 5/7 weeks and bilateral echogenic kidneys of normal size with multiple small cysts and anhydramnios was confirmed on ultrasound. Subjectively the fetal lungs appeared small without measurement and the cardiac image filled the chest. There were additional notes of possible bilateral club feet and non-visualization of the stomach and bladder. The imaging was limited by the anhydramnios, especially the face and detailed cardiac exam; given this, other potential anomalies could not be excluded. Fetal percutaneous umbilical blood sampling was technically not attainable and chorionic villus sampling was declined. Estimated fetal weight was 26% (702 g +/− 119 g). By 29 weeks, in addition to the above findings, fetal growth restriction was noted (1105 g, 2%). At 34 3/7 weeks, she presented with 24 hours of no fetal movements and was found to have a fetal demise.
She delivered a 1565 g infant. Autopsy was restricted to a gross examination only (by patient request) and cord blood was obtained for genetic testing with placental tissue for backup. Gross fetal examination was notable for ambiguous genitalia, small kidneys for gestational age by palpation, cleft lip and palate, a flat nasal bridge, and imperforate anus.
CMA ordered with the noted indication of “bilateral multicystic dysplastic kidneys; anhydramnios” revealed a pathogenic 3.0 Mb deletion on chromosome 22, containing 73 genes at the classic site of the velocardiofacial deletion (DiGeorge or 22q11.2 deletion syndrome). Maternal studies were unremarkable. The father has not been able to obtain testing.
Case 3
A 32 yo G1P0 at 17 3/7 weeks presented for her fetal survey. Her medical and surgical histories were unremarkable. Family history was notable only for a 2nd degree relative with a congenital cardiac anomaly. Initial ultrasound at 8 weeks showed a fetal heart rate of 80; fetal nuchal lucency at 11 weeks was within normal limits (1.5 mm) and fetal heart rate was normal. Screening for Down syndrome was low-risk by combined first trimester testing (1 in 6,500). Maternal genetic carrier screening for 39 conditions was unremarkable. The fetal survey was notable for bilateral echogenic kidneys of normal size, mildly dilated renal pelvises bilaterally (3 mm right and 4 mm left), and 3 cortical cysts in the left kidney, one in the right kidney. The remainder of the fetal survey was normal, with normal fluid and biometry. One week later, ultrasound performed at the time of amniocentesis was unchanged. The patient elected to terminate the pregnancy at 20 weeks by dilation and evacuation (D & E). Pathology confirmed the gestational age, fetal sex, and unilateral renal dysgenesis with disorganized glomerulus formation and scattered cysts. A simple cortical cyst was found in the contralateral kidney. The bladder was fully distended with slight distension of ureters bilaterally. No other anomalies were noted but the fetal exam was limited due to structural disruption at the time of the D & E procedure.
CMA from the amniocentesis was sent with an indication of “echogenic kidneys”. The analysis revealed a 1.39 Mb deletion at 17q12, which included two genes of note-- HNF1B and LHX1. Mutations and deletions in HNF1B (also referred to as TCF2) have been found in patients with renal cysts and diabetes syndrome7–9. Deletion of LHX1 is associated with additional features including cognitive impairment, seizures, and structural brain abnormalities10. The deletion was considered to have clinical significance. Parental CMAs were normal.
Case 4
A 35 yo G1P0 was noted on her fetal survey at 19 5/7 weeks to have an absent nasal bone, mildly echogenic bowel, and abnormal cardiac images. Her medical and surgical histories were unremarkable. Family history was notable for a maternal uncle with intellectual disability. Her gynecologic history was notable for prior dermoid removal and endometriosis. The pregnancy was spontaneous following an infertility evaluation. Initial imaging at 7 weeks noted a gestational age consistent with her LMP. At 13 4/7 weeks the nuchal lucency was 2.4 mm, within normal limits for gestational age. Cell-free DNA screening for aneuploidy (21, 18,13, X, Y) was unremarkable. Maternal carrier screening for the common 23 cystic fibrosis gene mutations was unremarkable and fragile X screening was negative.
At 18 6/7 weeks, the fetal survey was notable for absent nasal bone, increased nuchal fold (6–7 mm), a cardiac bradyarrhythmia (heart rate 70) with an active fetus, echogenic focus and prominent aorta with abnormal arch views and normal 4-chamber view. The remaining major structures were normal. Amniotic fluid was normal, and biometry was consistent with gestational age. Fetal echo was notable for fetal heart rate of 65 with periods of normal sinus rhythm, mild pulmonic stenosis, a dilated pulmonary artery, and a moderately dilated ascending aorta. The findings were consistent with a mild biventricular outflow obstruction in the setting of bradyarrhythmia. These findings were unchanged one week later. SSa and SSb antibody screening was negative.
At 23 weeks, a normal fetal heart rate was noted. Fetal growth was adequate and there were no other documented changes. The following 9 ultrasound evaluations were stable with a normal fetal heart rate, absent nasal bone, and generous nuchal fold. Fetal growth and amniotic fluid were appropriate throughout the pregnancy. At 39 weeks, new-onset moderate oligohydramnios was noted. Induction was recommended and initiated, resulting in a forceps-assisted vaginal delivery of a female infant, Apgars 8/9, weight 3350 g. The infant did well and was discharged with the patient on postpartum day two.
A CMA initiated at 17 5/7 weeks for the noted indication of “fetal echogenic bowel and absent nasal bridge” was unremarkable. CMV PCR of the amniotic fluid was unremarkable and anti-SSa and SSb studies were negative. Exome sequencing was performed on a research basis at 7 days of age following new-onset seizures and noted facial dysmorphia. The results were positive for a pathogenic variant within the PACS1 gene, c.607C>T in exon 4. The variant was determined to be de novo based on parental studies and was pathogenic based on a literature review of reported cases with neonatal-onset seizures, dysmorphia, cardiac anomalies and developmental disability11. In vivo and in vitro analyses of this PACS1 variant supported a pathogenic classification; functional studies in zebrafish supported a dominant–negative function of this gene in relation to craniofacial phenotypes11.
Case 5
A 30 yo G4P2012 presented at 20 6/7 weeks as a referral for suspected fetal anomalies. Her medical, surgical and obstetric histories were unremarkable, and the pregnancy had been uncomplicated. Bilateral renal agenesis, anhydramnios and a complex congenital heart defect with a significant pericardial effusion, suspected hypoplastic left heart and AV canal defect with a pericardial effusion were noted. Fetal biometry was appropriate for gestational age. Subsequent ultrasounds were unchanged. The patient delivered at term and desired palliative care. The neonate passed away shortly after delivery and postnatal exam was notable for skin sloughing on the scalp consistent with aplasia cutis and nail dysplasia. Newborn weight was 2860 g. No further external findings were noted. Autopsy was declined.
Chromosome microarray was ordered with a noted indication of multiple malformations and was normal. The family was enrolled in the trio exome sequencing research study and a de novo variant (c. 86A>G; p. Asn29Ser) in KCTD1 was noted12. Heterozygous pathogenic variants in the KCTD1 gene are associated with autosomal dominant scalp-ear-nipple (SEN) syndrome13. SEN syndrome is characterized by anomalies of the scalp, breast, and external ears, but may include other less frequently observed anomalies of the nails, teeth and kidney13. KCTD1 N29S is a rare missense variant observed in 2 of 60,706 unselected individuals that changes a single well-conserved amino acid from an asparagine to a serine in the “bric-a-brac, tram track, and broad complex” (BTB) domain of the KCTD1 gene. Although this variant has not been previously reported as pathogenic, several nearby missense changes within the same BTB domain (including the adjacent residue) are associated with scalp-ear-nipple syndrome13. The KCTD1 N29S variant is therefore considered likely pathogenic. Renal/urinary tract and cardiac abnormalities are infrequently reported in the albeit few reported cases of postnatally diagnosed SEN making this prenatal presentation, with severely disrupted renal and cardiac systems during embryonic development, implicating the KCTD1 gene as critical in human development.
Discussion
This initial series of select cases highlights a growing knowledge of fetal dysmorphology due to genetic causes. These cases underscore the discordance of the fetal phenotype from the traditional features associated with the genetic disorder as detected in the newborn / child. While 22q11.2 deletion syndrome is known for its wide spectrum of presentation, the first two cases highlight how these fetal findings can present with quite different phenotypic presentations. Additionally, these cases reveal the difficulties faced when formulating even short-term outcomes as fetal demise is not commonly associated with 22q11.2 deletion syndrome. In the newborn case, as 22q11.2 deletion syndrome was not suspected based on the fetal presentation, the delayed diagnosis led this neonate to develop hypocalcemia. These two cases emphasize the necessity for consistent and detailed ultrasound reporting, and neonatal / childhood follow-up.
Cases 3– 5 highlight how additional considerations emerge as more extensive sequencing is paired with detailed fetal phenotyping. Case 3 denotes a clinical situation in which sequencing expanded the counseling from isolated renal system abnormalities to concerns for a broader syndrome that included developmental delay. In case 4, sequential ultrasound surveillance provided the natural history of a fetus later found to have a PACS1 mutation. Resolution of initial bradycardia was falsely reassuring as to the health of this fetus, although an absent nasal bone was noted throughout gestation. Lastly, case 5 is an example in which the fetal phenotype of a syndrome, in this case scalp-ear-nipple (SEN), presented with lethal renal anomalies further refining the embryologic impact of the altered genes.
Experience from pediatric dysmorphology should inform ultrasound evaluations for fetal phenotyping. The goals of pediatric dysmorphology are a systematic attainment of a diagnosis, utilization of diagnostic information for prognostication, attaining recurrence information and, lastly, often ending a diagnostic odyssey for the parents14,15. Several dysmorphology forms are published, but a single format is not followed by all. Instead, an approach of assessment and documentation at two levels of examination is common - first delineation of major systems by positive, negative or uninterpretable findings and, secondarily, a deliberate evaluation for minor findings. These minor findings themselves alone may not be of significance but can help inform a total phenotype16.
Shifting workflow from imaging of a major anomaly and offering diagnostic testing to comprehensive fetal phenotype assessment takes time to assimilate into practice. Extending dysmorphology to antenatal imaging is challenging on several fronts. With increasing access to genomic technologies, the genetic education of providers ordering these tests is more diverse. Secondly, subtle features and minor anomalies require time and attention to specific and detailed images, even with the best resolution imaging technologies. Lastly, communication between the ordering provider, the sonologist, and the molecular laboratory is not routine. When a genetic test is ordered, a simultaneous commitment to documenting the totality of the fetal image and communicating this information to the molecular laboratory should be made. While gestalt plays an important role in both fetal and pediatric dysmorphology, an emphasis on a systematic approach and not rapid diagnosis should be emphasized.16 Ultrasound documentation protocols by body system and inclusive of amniotic fluid and fetal weight are encouraged17,18,19. A second analysis focused on subtle, minor findings also should be systematic, referencing normal fetal measurements as needed20, 21–24 (Table 1). Minor features entered verbatim can now be retrieved by artificial intelligence software searches of electronic medical records. Lastly, the ability to prognosticate fetal outcome is supplemented by serial imaging.
Table 1.
Examples of current public-facing databases for genotype-phenotype exploration (fetal findings uncommon) (modified from Köhler et al., 2017, Table 1)25.
| Name | URL |
|---|---|
| PhenomeCentral | phenomecentral.org |
| DDD (Deciphering Developmental Disorders) | www.ddduk.org |
| DECIPHER (DatabasE of genomiC varIation and Phenotype in Humans using Ensembl Resources) | decipher.sanger.ac.uk |
| ECARUCA (European Cytogeneticists Association Register of Unbalanced Chromosome Aberrations) | http://umcecaruca01.extern.umcn.nl:8080/ecaruca/ecaruca.jsp |
| The 100,000 Genomes Project | https://www.genomicsengland.co.uk/ |
| Geno2MP (Exome sequencing data linked to phenotypic information from a wide variety of Mendelian gene discovery projects) | http://geno2mp.gs.washington.edu |
| NIH UDP (Undiagnosed Diseases Program) | available via phenomecentral.org |
| NIH UDN (Undiagnosed Diseases Network) | available via phenomecentral.org |
| HDG (Human Disease Gene Website series) | www.humandiseasegenes.com |
| Phenopolis (An open platform for harmonization and analysis of sequencing and phenotype data) | https://phenopolis.github.io |
| GenomeConnect (Patient portal developed by ClinGen) | www.genomeconnect.org |
| FORGE Canada & Care4Rare Consortium | available via phenomecentral.org |
| RD-Connect | platform.rd-connect.eu |
| Genesis | https://thegenesisprojectfoundation.org |
Our series benefited from genetic testing on cord blood or fetopsy specimens when in utero sampling was not obtained. Both participating sites had access to exome sequencing on a research basis and each center worked closely with their ultrasound, molecular diagnostics and neonatology teams for a comprehensive case description. Despite such coordinated care, the indication for the molecular cytogenetic studies was often truncated to one anomaly or the descriptor, multiple anomalies.
The limitation of a small sample size is inherent in a case series. As an initial commentary, however, these cases underscore potential areas of future research including the biology behind the discordance of fetal and newborn / childhood phenotypes, and the challenges to dysmorphology-based imaging. As a limited series, the counseling burden associated with variants of unknown significance was not addressed but is an additional area for ongoing assessment.2,3,12. Curation of fetal genetic variants will depend on close communication between the clinicians and testing laboratories to assist with not only the understanding of the known pathogenic variants but also those with less robust information.
As genetic testing shifts to assessing expanded genomic regions, deeper fetal phenotyping is needed. Resources brought to this initiative should include transmission of fetal phenotype- genotype information to publicly-available databases (Table 2). Furthermore, findings need to be translated into an expanded ontology format, inclusive of uniquely prenatal characteristics, that will promote utilization in bioinformatic algorithms.
Table 2.
An approach to ultrasound dysmorphology after one or more congenital anomalies is identified. Open response would allow searching by artificial intelligence within electronic medical records (modified from Philip et al., 2010)20.
| Fetal category | Subsection | Drop down | Comments |
|---|---|---|---|
| Cranium | |||
| Skull shape | Normal / Abnormal | ||
| Frontal | |||
| Hairline | Normal / Abnormal | ||
| Orbit size | Normal / Abnormal | ||
| Orbital distance | Normal / Abnormal | ||
| Mouth / lips | Normal / Abnormal | ||
| Profile | |||
| Forehead | Normal / Abnormal | ||
| Nasal bridge | Normal / Abnormal | ||
| Philtrum | Normal / Abnormal | ||
| Chin alignment | Normal / Abnormal | ||
| Ear placement | Normal / Abnormal | ||
| Anterior | |||
| Effusions including ascites | Yes / No | ||
| Situs | Normal / Abnormal | ||
| Genitalia | Male / Female | ||
| Extremities – contractures, abnormal positioning, webbing | Normal / Abnormal | ||
| Extremities – number of digits, digital posturing, syndactyly | Normal / Abnormal | ||
| Tone | Normal / Abnormal | ||
| Movement | Normal / Abnormal |
As genetic technologies advance, this is an opportune time to enlist clinicians in the first step of annotating fetal phenotype-genotype correlations. Clinicians should be recognized for their role as contributors to defining an evolving number of genetic presentations in the fetus. Well-curated fetal phenotype-genotype correlations will advance patient care and increase understanding of fetal development. Close collaboration between sonologists, laboratory geneticists, dysmorphologists and a publicly-available database with prenatal genotype-phenotype information are critical to achieving these goals.
Bulleted statements.
What is already known: Prenatal diagnosis has expanded beyond karyotype and chromosomal microarray to include next generation sequencing techniques that provide variant level information and interpretation that requires integration with detailed fetal phenotypes.
What does the study add: Fetal genotype-phenotype correlations are broad and disparate from the pediatric presentation, emphasizing the necessity of detailed prenatal imaging, documentation, and communication of imaging findings with sequencing facilities where variant analysis is performed to expand our understanding of fetal development and prenatal presentations of genetic conditions.
Funding
Dr. Gray was supported by the BIRCWH career development award K12 HD051959 from the National Institutes of Health (NIH) to Harvard Medical School. Dr. Vora was supported by the K23 HD088742 career development award from the NIH National Institute of Child Health and Human Development.
Footnotes
Conflicts of interest
Dr. Gray reports consulting for Quest Diagnostics and Illumina. The other authors report no conflicts of interest.
Data sharing
Data sharing not applicable – no new data generated.
References
- 1.Committee on Genetics and the Society for Maternal-Fetal Medicine. Committee Opinion No.682: Microarrays and Next-Generation Sequencing Technology: The Use of Advanced Genetic Diagnostic Tools in Obstetrics and Gynecology. Obstet Gynecol. 2016 Dec;128(6):e262–8. [DOI] [PubMed] [Google Scholar]
- 2.Lord J, McMullan DJ, Eberhardt RY, Rinck G, Hamilton SJ, Quinlan-Jones E, et al. Prenatal exome sequencing analysis in fetal structural anomalies detected by ultrasonography (PAGE): a cohort study. Lancet. 2019 Feb 23;393(10173):747–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petrovski S, Aggarwal V, Giordano JL, Stosic M, Wou K, Bier L, et al. Whole-exome sequencing in the evaluation of fetal structural anomalies: a prospective cohort study. Lancet. 2019 Feb 23;393(10173):758–67. [DOI] [PubMed] [Google Scholar]
- 4.Benacerraf BR, Greene MF, Holmes LB. The prenatal sonographic features of Noonan’s syndrome. J Ultrasound Med. 1989 Feb;8(2):59–63. [DOI] [PubMed] [Google Scholar]
- 5.Nisbet DL, Griffin DR, Chitty LS. Prenatal features of Noonan syndrome. Prenat Diagn. 1999 Jul;19(7):642–7. [DOI] [PubMed] [Google Scholar]
- 6.Köhler S, Doelken SC, Mungall CJ, Bauer S, Firth HV, Bailleul-Forestier I, et al. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014 Jan;42(Database issue):D966–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bingham C, Bulman MP, Ellard S, Allen LI, Lipkin GW, Hoff WG, et al. Mutations in the hepatocyte nuclear factor-1beta gene are associated with familial hypoplastic glomerulocystic kidney disease. Am J Hum Genet. 2001 Jan;68(1):219–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verhave JC, Bech AP, Wetzels JFM, Nijenhuis T. Hepatocyte Nuclear Factor 1β-Associated Kidney Disease: More than Renal Cysts and Diabetes. J Am Soc Nephrol. 2016 Feb;27(2):345–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duval H, Michel-Calemard L, Gonzales M, Loget P, Beneteau C, Buenerd A, et al. Fetal anomalies associated with HNF1B mutations: report of 20 autopsy cases. Prenat Diagn. 2016 Aug;36(8):744–51. [DOI] [PubMed] [Google Scholar]
- 10.George AM, Love DR, Hayes I, Tsang B. Recurrent Transmission of a 17q12 Microdeletion and a Variable Clinical Spectrum. Mol Syndromol. 2012 Jan;2(2):72–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schuurs-Hoeijmakers JHM, Oh EC, Vissers LELM, Swinkels MEM, Gilissen C, Willemsen MA, et al. Recurrent de novo mutations in PACS1 cause defective cranial-neural-crest migration and define a recognizable intellectual-disability syndrome. Am J Hum Genet. 2012 Dec 7;91(6):1122–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vora NL, Powell B, Brandt A, Strande N, Hardisty E, Gilmore K, et al. Prenatal exome sequencing in anomalous fetuses: new opportunities and challenges. Genet Med. 2017 Nov;19(11):1207–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marneros AG, Beck AE, Turner EH, McMillin MJ, Edwards MJ, Field M, et al. Mutations in KCTD1 cause scalp-ear-nipple syndrome. Am J Hum Genet. 2013 Apr 4;92(4):621–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donnenfeld AE. Fetal dysmorphology. Ultrasound Obstet Gynecol. 1997 Feb;9(2):73–5. [DOI] [PubMed] [Google Scholar]
- 15.Accardo JA. 50 Years Ago in The Journal of Pediatrics: Dysmorphology (Teratology). J Pediatr. 2016 Dec;179:67. [DOI] [PubMed] [Google Scholar]
- 16.Chitayat D, Babul-Hirji R. Genetic counselling in prenatally diagnosed non-chromosomal fetal abnormalities. Curr Opin Obstet Gynecol. 2000 Apr;12(2):77–80. [DOI] [PubMed] [Google Scholar]
- 17.AIUM-ACR-ACOG-SMFM-SRU Practice Parameter for the Performance of Standard Diagnostic Obstetric Ultrasound Examinations. J Ultrasound Med. 2018 Nov;37(11):E13–24. [DOI] [PubMed] [Google Scholar]
- 18.Monk D, Moore GE. Intrauterine growth restriction--genetic causes and consequences. Semin Fetal Neonatal Med. 2004 Oct;9(5):371–8. [DOI] [PubMed] [Google Scholar]
- 19.Sagi-Dain L, Peleg A, Sagi S. Risk for chromosomal aberrations in apparently isolated intrauterine growth restriction: A systematic review. Prenat Diagn. 2017 Nov;37(11):1061–6. [DOI] [PubMed] [Google Scholar]
- 20.Philip N, Quarello E, Gorincour G, Sigaudy S. [Fetal dysmorphology: a practical approach in utero]. Gynecol Obstet Fertil. 2010 Nov;38(11):677–85. [DOI] [PubMed] [Google Scholar]
- 21.Goldstein I, Reiss A, Rajamim B-S, Tamir A. Nomogram of maxillary bone length in normal pregnancies. J Ultrasound Med. 2005 Sep;24(9):1229–33. [DOI] [PubMed] [Google Scholar]
- 22.Goldstein I, Tamir A, Itskovitz-Eldor J, Zimmer EZ. Growth of the fetal nose width and nostril distance in normal pregnancies. Ultrasound Obstet Gynecol. 1997 Jan;9(1):35–8. [DOI] [PubMed] [Google Scholar]
- 23.Goldstein I, Tamir A, Weiner Z, Jakobi P. Dimensions of the fetal facial profile in normal pregnancy. Ultrasound Obstet Gynecol. 2010 Feb;35(2):191–4. [DOI] [PubMed] [Google Scholar]
- 24.Goldstein I, Tamir A, Zimmer EZ, Itskovitz-Eldor J. Growth of the fetal orbit and lens in normal pregnancies. Ultrasound Obstet Gynecol. 1998 Sep;12(3):175–9. [DOI] [PubMed] [Google Scholar]
- 25.Köhler S, Vasilevsky NA, Engelstad M, Foster E, McMurry J, Aymé S, et al. The Human Phenotype Ontology in 2017. Nucleic Acids Res. 2017 Jan 4;45(D1):D865–76. [DOI] [PMC free article] [PubMed] [Google Scholar]