Abstract
In Japan, it is possible to generate chimeric animals from specified embryos by combining animal blastocysts with human pluripotent stem (PS) cells (animal-human PS chimera). However, the production of animal-human PS chimeras has been restricted because of ethical concerns, such as the development of human-like intelligence and formation of humanized gametes in the animals, owing to the contributions of human PS cells to the brain and reproductive organs. To solve these problems, we established a novel blastocyst complementation technology that does not contribute to the gametes or the brain. First, we established GFP-expressing mouse embryonic stem cells (G-mESCs) in which the Prdm14 and Otx2 genes were knocked out and generated chimeric mice by injecting them into PDX-1-deficient blastocysts. The results showed that the G-mESCs did not contribute to the formation of gametes and the brain. Therefore, in the PDX-1-deficient mice complemented by G-mESCs without the Prdm14 and Otx2 genes, the germline was not transmitted to the next generations. This approach could address concerns regarding the development of both human gametes and a human-like brain upon mouse blastocyst complementation using human stem cells.
Keywords: blastocyst complementation, Otx2, Prdm14
Introduction
Blastocyst complementation is a biological technique used to complement genetically deficient organs or tissues by microinjecting stem cells into blastocyst-stage embryos of organogenesis-disabled mice. The first report of this technique described the complementation of T- and B-lymphocytes in Rag-deficient (Rag2−/−) mice by microinjecting intact mouse embryonic stem cells (mESCs) into blastocyst-stage embryos [4]. More recently, Kobayashi et al. (2010) succeeded in generating interspecific chimeras, namely PDX1-deficient mice with rat pancreata [13]. Additionally, rat ESCs injected into mouse blastocysts differentiated into rat spermatozoa in nude mice [11]. These studies demonstrate that blastocyst complementation is effective for generating three-dimensional and functional cells or organs in vivo. To build on this previous work, we aimed to regenerate of various organs via blastocyst complementation using human ES/iPS cells.
By injecting human pluripotent stem (PS) cells into chicken [6] and mouse blastocysts [13], groups in the US and Israel have successfully developed chimeric embryos. In Japan, it is possible to generate chimeras from specified embryos by combining animal blastocysts with human PS cells (animal-human PS chimera). However, the regulations regarding the production of animal-human PS chimeras are extremely strict, as human PS cells may form gametes or become incorporated into the brain. Therefore, the Japanese government is apprehensive that this may lead to human-like intelligence in animals or that they may form human spermatozoa and oocytes, though the study on development of animal-human PS chimera was lifted on March 1, 2019.
PRDM14 is a sequence-specific transcriptional regulator that plays key roles in promoting primordial germ cell specification and safeguarding the pluripotency of mESCs [17]. During mouse embryogenesis, the Prdm14 gene is expressed in preimplantation embryos, where its asymmetric expression promotes the allocation of cells toward various functions in the pluripotent inner cell mass [3]. PRDM14 deficiency in mice results in sterility associated with early germ cell deficiency, as cells allocated to become primordial germ cells fail to reacquire the expression of key pluripotency factors in order to undergo epigenetic reprogramming [25].
The Otx2 gene is a homeobox gene expressed in the rostral brain regions and is thought to define the anterior regions of the embryo [1]. Otx2 homozygous mutants exhibit defects of the earliest OTX2 functions in the visceral endoderm, and heterozygous mutants show defects of their cephalic neural crest cells [18]. Additionally, knockout of Otx2 resulted in the failure of the brain to form properly in the embryo [22].
Recently, gene targeting using the CRISPR/Cas9 system has become established as a means of simply editing mouse genomic DNA [5, 9]. Thus, we hypothesized that blastocyst complementation using human ES/iPS cells that lack the Prdm14 and Otx2 genes may address the concerns regarding blastocyst complementation in Japan. Therefore, we conducted experiments confirming that mESCs having CRISPR/Cas9-induced deletions of both Prdm14 and Otx2 genes do not contribute to either the gametes or the brain when used for pancreatic (PDX1-deficient) blastocyst complementation.
Materials and Methods
Culture of mESCs
Undifferentiated donor GFP-expressing mESCs (G-mESCs) were purchased from the Center for Developmental Biology, RIKEN (Kobe, Japan), and maintained on gelatin-coated dishes with feeder cells in DMEM (Cat No. 10829018, Gibco, Ireland) with 10% KnockOutTM Serum Replacement (Cat No 10828-028, Gibco), 0.1 mM 2-mercaptoethanol (Cat No. 60-24-2, Sigma-Aldrich, St. Louis, MO, USA), 0.1 mM nonessential amino acids (Cat No. 11140050, Gibco), and 1,000 units/ml of mouse leukemia inhibitory factor (Cat No. ESG1107, Chemicon, Temecula, CA, USA).
Gene targeting by CRISPR/Cas9
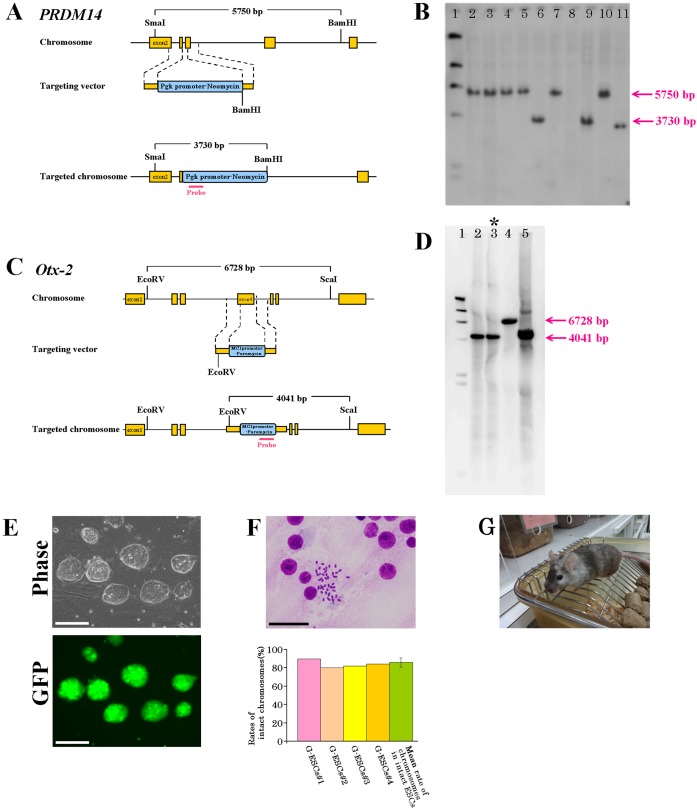
Mouse Prdm14 and Otx2 genomic clones were isolated from the C57BL/6JJcl strain mouse genomic clone library. PRDM14 CRISPR/Cas9KO plasmid (h) (Cat No. sc-404426, Santa Cruz Biotechnology, Dallas, TX, USA) was used for gene targeting. The targeting vector (Cat No. sc-404426-HDR; Santa Cruz Biotechnology) was modified as follows. Gene targeting constructs of Prdm14 were generated in a vector containing mouse Pgk promoter-neomycin, which was substituted for the exon 2 coding region of the Prdm14 gene (Fig. 1A). 5 × 106 G-mESCs were electroporated with 15 µg of linearized targeting construct in 100 µl solution V (Nucleofector Kit, Lonza, Tokyo, Japan) and one pulse of 800 V/3 µF with an Amaxa Nucleofector I (Lonza). G-mESCs were then subjected to positive selection with 250 µg/ml neomycin (Cat No. 10131035, Thermo Fisher Scientific, Waltham, MA, USA). After 7–10 days, 83 clones were isolated and screened for the presence of the targeted allele. The presence of the correct homologous recombination was confirmed by Southern blotting using a neomycin inner probe (5’-GATCGGCCATTGAACAAGAT-3’ and 5’-CTCGTCCTGCAGTTCATTCA-3’) (Fig. 1B). Additionally, PCR screening was performed using genotyping primers (5’-TCCTGCCTCGGGTTCACCTA-3’ [forward], 5’-CCAACCTCAGGTCGTCCTCC-3’ [reverse], and 5’-CCTTGCTCTGGTCAACCAGGTT-3’ [Neo]) to exclude heterozygote clones. After the establishment of PRDM14-deficient G-mESCs, gene targeting of Otx2 was also performed by CRISPR/Cas9. An OTX2 CRISPR/Cas9KO plasmid (Cat No. sc-422085, Santa Cruz Biotechnology) was used for gene targeting. The targeting vector (Cat No. sc-422085-HDR, Santa Cruz Biotechnology) was modified as follows. Gene targeting constructs of Otx2 were generated in the vector containing Mc1 promoter-puromycin, which was substituted for the exon 4 coding region of the Otx2 gene (Fig. 1C). 5 × 106 G-mESCs without Prdm14 gene were electroporated with 15 µg of linearized targeting construct under the same conditions as used previously. G-mESCs were then subjected to positive selection with 2 µg/ml puromycin (Cat No. 10131035, Thermo Fisher Scientific, Waltham, MA, USA). After 7–10 days, 51 clones were isolated and screened for the presence of the targeted allele. After targeting of the Otx2 gene, the occurrence of the correct homologous recombination was confirmed by Southern blotting using a puromycin inner probe (5’-ACAGATGGAAGGCCTCCTG-3’ and 5’-GCTCGTAGAAGGGGAGGTTG-3’) (Fig. 1D). Additionally, PCR screening was performed using genotyping primers (5’- ATCTGCAACTCCTTTAAAAG-3’ [forward], 5’-AATGCTCTGTGGCACTCGGC-3’ [reverse], and 5’- CCGGCTGGATGATCCTCCAG-3’ [Puro]) to exclude heterozygote clones. Then, Prdm14 and Otx2 gene double-knockout G-mESCs were established (Fig. 1E). The number of chromosomes was determined in 50 cells from each of these original G-mESCs, in accordance with the protocol of Sugawara et al. (2006) [20]. The results indicated that 80–90% of the G-mESCs had the correct number (Fig. 1F). Therefore, these G-mESCs were used to generate chimeras (Fig. 1G), which were Pdx1−/− mice complemented by G-mESCs without the Prdm14 and Otx2 genes (PPO mice).
Fig. 1.
Targeting of Prdm14 and Otx2 genes. A. Schematic representation of the Prdm14 gene and targeting strategy. The red-colored bar indicates the position of the probe for genomic Southern blot analysis using SmaI and BamHI digested samples (mutated allele, 3,730 bp; wild-type allele, 5,750 bp). B. Genomic DNA from ES cells was digested with SmaI and BamHI and subjected to hybridization with the probe. Lane 1, markers; Lanes 2–5, wild type; Lanes 6, gene-targeted ES clones; Lane 7, wild type; Lane 8, none; Lane 9, gene-targeted ES clones; Lane 10, wild type; Lane 11, gene-targeted ES clones. C. Schematic representation of the Otx2 gene and secondary targeting strategy. The red-colored bar indicates the position of the probe for genomic Southern blot analysis using EcoRV and ScaI digested samples (mutated allele, 4,041 bp; wild-type allele, 6,728 bp). D. Genomic DNA from ES cells was digested with EcoRV and ScaI and subjected to hybridization with the probe. Lane 1, markers; Lanes 2 and 3, gene-targeted ES clones; Lane 4, wild type; Lane 5, gene-targeted ES clone. E. Establishment of ES cells without the Prdm14 and Otx2 genes. Scale bars represent 200 µm. F. Chromosome banding of the ES cells without the Prdm14 and Otx2 genes. Scale bars represent 50 µm. G. Chimeric mice generated from the ES cells without the Prdm14 and Otx2 genes.
Embryo culture and manipulation to prepare Pdx1−/− blastocyst complementation
Pdx1+/− mice, which were developed by Hashimoto et al. [7], were backcrossed onto the IQI/Jic strain (CIEA, Kawasaki, Japan) for more than 5 generations. Then, the preparation of Pdx1 heterozygous intercrossing embryos was carried out in accordance with published protocols [16]. In brief, embryos for injection were collected upon crossing Pdx1+/− male mice and superovulated Pdx1+/− female mice. Their embryos were cultured with potassium simplex optimized medium (KSOM; ARK Resource Co., Ltd., Kumamoto, Japan) until the blastocyst stage. For micromanipulation, G-mESCs were trypsinized and suspended in G-mESCs culture medium. Approximately 10 G-mESCs were introduced into blastocyst cavities near the ICM. After blastocyst injection, mouse blastocysts were transferred into the uteri of pseudopregnant recipient ICR mice at an embryonic age of 2.5 days.
Animal husbandry
All the mice were provided with exclusive feed for laboratory animals (CA-1, CLEA, Tokyo, Japan) and tap water ad libitum. After weaning, two to three mice were kept in an open cage. The animal room was maintained at 24 ± 2°C with 55 ± 10% relative humidity and 12 h of artificial lighting from 08:00 to 20:00 and was kept under specific pathogen-free conditions, which were the same as in a previous study [8]. The glucose tolerance test was performed when mice were 6 weeks of age. The Animal Committee of the Central Institute for Experimental Animals approved this study (Permit No. 14050A).
Genotyping
The Pdx1 mutation and wild type were distinguished by genotyping PCR using mouse tail to exclude the wild type because the genotyping PCR was unable to distinguish between the Pdx1−/− and Pdx1+/− mice by the contribution of the donor G-mESCs to the tail of chimeric mouse. The genotyping PCR was performed in accordance with a previous report [7]. After selecting the mutant mice, hematopoietic cells were taken from their orbital sinus, and GFP-negative cells were sorted. Then, genotyping PCR was performed to identify the Pdx1−/− mice using the sorted GFP-negative cells.
Glucose tolerance test
All the mice were fasted for at least 16 h before the study. Then, they were challenged with an oral glucose dose of 1.0 mg/g body weight. Blood samples were taken from the retro-orbital sinus using a heparinized capillary tube at 0, 5, 15, 30, 60, 90, and 120 min after glucose administration, and blood glucose concentrations were measured using an automatic blood glucose meter (Arkray Inc., Kyoto, Japan). Blood samples at 0, 15, and 30 min after glucose administration were collected and centrifuged in heparinized tubes, and the plasma was stored at −20°C. A commercially available ELISA kit (Cat No. MS302; Morinaga Institute of Biological Science, Inc., Yokohama, Japan) was used to assay plasma insulin. Pdx1+/− and IQI/Jic strain mice were used for comparisons with PPO mice. IQI/Jic strain mice were selected as the control because the genetic background of both the Pdx1+/− mice and blastocysts to generate PPO mice was the IQI/Jic strain.
Fluorescence and phase contrast microscopy
A microscopy (VB-6010, Keyence, Osaka, Japan) and GFP filter (OP-42313, Keyence) were used to observe mouse internal organs under fluorescence and phase-contrast conditions (phase).
Immunohistochemistry
Sections fixed in 10% buffered formalin and embedded in paraffin were mounted on silane-coated glass slides and immunostained by using a Leica Bond-Max automatic immunostainer (Leica Biosystems, Mount Waverley, VIC, Australia). Paraffin sections were dewaxed in a Bond Dewax solution and rehydrated in alcohol and Bond Wash solution (Leica Biosystems). Detection was performed using a Bond Polymer Refine Detection system. Then, the sections were counterstained with hematoxylin. Immunohistochemical analysis was performed with polyclonal rabbit anti-GFP (Product code ab290, Abcam plc, Cambridge, UK).
Reproduction and germline transmission
To confirm germline transmission, the chimeras were mated to intact IQI/Jic strain mice. GFP expression of the infants was detected with a microscopy (VB-6010, Keyence) and GFP filter (OP-42313, Keyence).
Results
Reproductive rates of chimeric mice
To generate chimeric mice, G-mESCs without the Prdm14 and Otx2 genes were injected into blastocysts obtained by crossing Pdx1+/− male mice with Pdx1+/− female mice. We genotyped GFP-negative peripheral blood molecular cells from 7-week-old infants. The Pdx1−/− genotype was present in 20 of the 77 infants (26.0%, 12 males and 8 females) complemented by G-mESCs without the Prdm14 and Otx2 genes (Table 1). On the other hand, the Pdx1−/− genotype was present in 22 of the 74 infants (29.7%, 13 males and 9 females) complemented by G-mESCs with Prdm14 and Otx2 genes (Table 1). Fifteen of the PPO mice and 13 of the Pdx1−/− mice complemented by G-mESCs with Prdm14 and Otx2 genes (control) were used for analysis (Table 2). The 0% chimeras were excluded from further study. The remaining male control mice (including some >50% chimeras) were used for preliminary investigation.
Table 1. Results of chimera mice by utilization of G-mESCs without Prdm14 and Otx2 genes.
| Genotypingof recipient | UsedG-mESCs | No. of embryostransferred | No. of chimeras | Genotype of chimerasa | No. of chimerism (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −/− | +/− | +/+ | ||||||||||
| Male | Female | Male | Female | Male | Female | >50% | <50% | 100% | ||||
| Pdx1+/–× Pdx1+/– | Prdm14–/–,Otx2–/– | 220 | 77 (35.0%) | 12 | 8 | 22 | 12 | 15 | 8 | 17 (22.1%) | 52 (67.5%) | 8 (10.4%) |
| Pdx1+/–× Pdx1+/– | Prdm14+/+,Otx2+/+(intact cells) | 189 | 74 (39.1%) | 13 | 9 | 26 | 8 | 13 | 5 | 18 (24.3%) | 46 (62.2%) | 10 (13.5%) |
Table 2. Number of chimerism in Pdx1−/− mice complemented by G-ESCs without Prdm14 and Otx2 genes.
| Genotyping of recipient | Used G-mESCs | Gender | No. of chimerism (%) | |||
|---|---|---|---|---|---|---|
| 0% | >50% | <50% | 100% | |||
| Pdx1−/− | Prdm14−/−, Otx2−/− | Males | 3 | 3 | 5 | 1 |
| Females | 2 | 4 | 2 | 0 | ||
| Pdx1−/− | Prdm14+/+, Otx2+/+ (intact cells) | Males | 2 | 3 | valign="top"7 | 1 |
| Females | 2 | 5 | 2 | 0 | ||
Characteristics of PPO mice
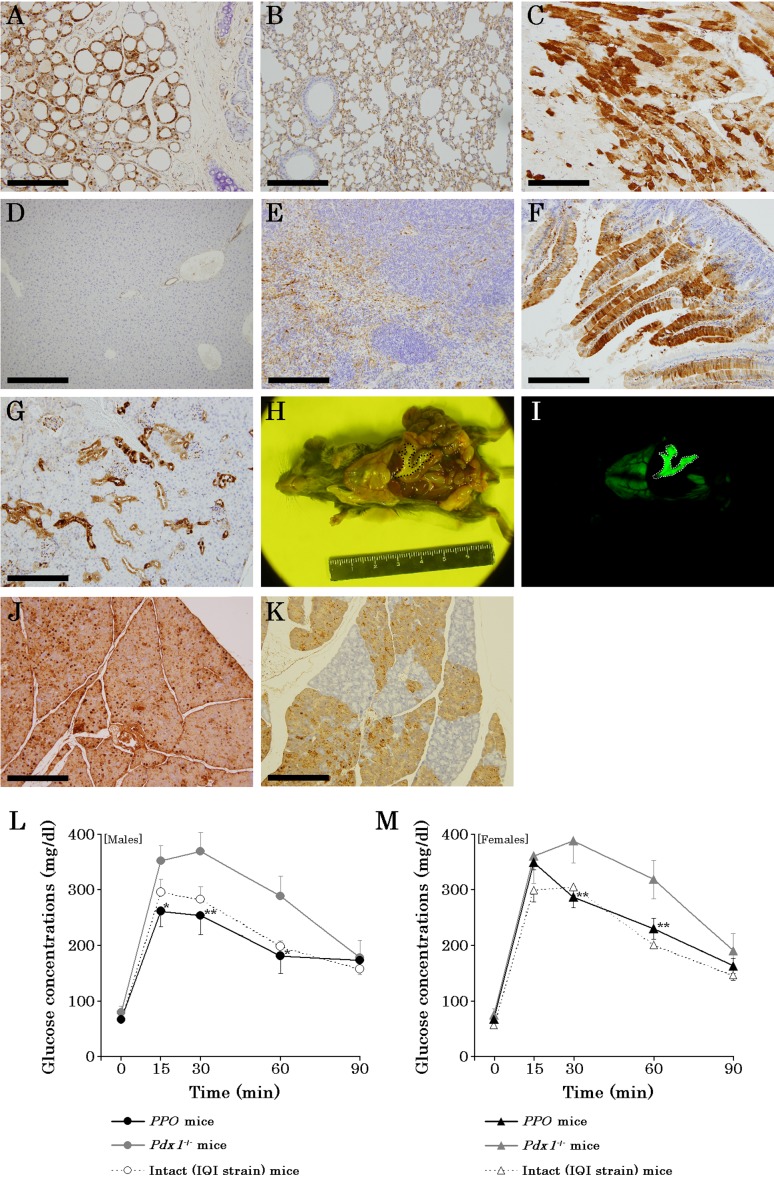
In immunohistochemical analysis, GFPs were detected from to the thyroid gland (Fig. 2A), lung (Fig. 2B), and heart (Fig. 2C) in PPO mice. Additionally, the G-mESCs contributed to the liver (Fig. 2D), spleen (Fig. 2E), intestines (Fig. 2F), and kidney (Fig. 2G) in PPO mice, although they were not detected under a fluorescent microscope (Fig. 2I). Upon observation under a fluorescent microscope, G-mESCs contributed to the pancreas highly in the abdominal cavity of the PPO mice (Figs. 2H and I) and immunohistochemistry revealed high expressions of GFP in the pancreas of PPO mice (Fig. 2J). On the other hand, the pancreatic reconstitution of Pdx1−/+ mice complemented by G-mESCs without the Prdm14 and Otx2 genes showed mosaicism (Fig. 2K). Additionally, impaired glucose tolerance of the Pdx1−/− mice without the Prdm14 and Otx2 genes was improved by blastocyst complementation, although the male Pdx1−/− mice showed impaired glucose tolerance (Fig. 2L). Glucose tolerance test of female Pdx1−/− mice also was same result (Fig. 2M).
Fig. 2.
Characteristics of PPO mice. A–G. GFP immunohistochemistry in PPO mice; thyroid gland (A), lung (B), heart (C), liver (D), spleen (E), intestines (F), and kidney (G). H. Observations of the pancreas in a male PPO mouse under phase. The area enclosed by the dotted line indicates the pancreas reconstituted by G-mESCs. I. Observations of the pancreas in a male PPO mouse under GFP fluorescence. The area enclosed by the dotted line indicates the pancreas reconstituted by G-mESCs. J. GFP immunohistochemistry of the pancreas in PPO mice. K. GFP immunohistochemistry of the pancreas in Pdx1+/− mice complemented by ES cells without the Prdm14 and Otx2 genes. L. Oral glucose tolerance test in male PPO mice at 10 weeks of age. M. Oral glucose tolerance test in female PPO mice at 10 weeks of age. Data are presented as the mean ± SE. *P<0.05, **P<0.01 (Student’s t-test) compared with value for Pdx1+/− and IQI/Jic strain mice. Black circles or trigonal marks represent PPO mice. Gray circles or trigonal marks represent Pdx1+/−. Open circles or trigonal marks represent intact mice (IQI/Jic strain). Scale bars represent the following distances: A, 100 µm; B, C, E–G, J, and K: 200 µm; and D, 400 µm.
Evaluations of the contributions of G-mESCs without the Prdm14 and Otx2 genes to gametes and the brain in chimeric mice
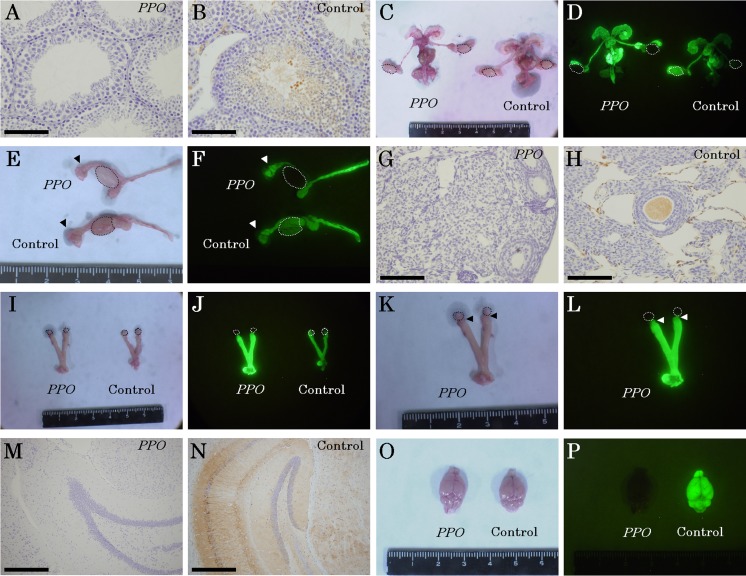
G-mESCs without the Prdm14 and Otx2 genes did not contribute to spermatozoa and the testes in Pdx1−/− mice (Figs. 3A and B), although they did contribute to other reproductive organs (Figs. 3C–F). G-mESCs without the Prdm14 and Otx2 genes did not contribute to oocytes, ovirio cells, and ovaries in the Pdx1−/− mice without the Prdm14 and Otx2 genes (Figs. 3G and H), although they did contribute to oviducts and uterus (Figs. 3I–L), which was similar to the case in the control mice. Additionally, G-mESCs without the Prdm14 and Otx2 genes did not contribute to the brain (Figs. 3M–P).
Fig. 3.
Contributions of the G-ESCs without the Prdm14 and Otx2 genes to gametes and the brain. A. GFP immunohistochemistry of spermatozoa and testes in PPO mice. B. GFP immunohistochemistry of spermatozoa and testes in control mice. C. Observations of male reproductive organs under phase. Areas enclosed by dotted lines indicate the testes. There was no macroscopic difference between the PPO and control testis. D. Observations of male reproductive organs under GFP fluorescence. Areas enclosed by dotted lines indicate the testes. The GFP was not detected in the testis of PPO mice. E. Magnification of testes and epididymis in a PPO mouse under phase. Areas enclosed by dotted lines indicate testes of PPO and control mice. Arrows indicate the epididymides. F. Magnification of testis and epididymis in PPO mouse under GFP fluorescence. Areas enclosed by the dotted lines show testis in PPO and control. Arrows show the epididymis. G. GFP immunohistochemistry of the ovary in PPO mice. H. GFP immunohistochemistry of the ovary in control mice. I. Observations of female reproductive organs under phase. Areas enclosed by dotted lines indicate the ovaries. There was no macroscopic difference between the PPO and control ovary. J. Observations of female reproductive organs under GFP fluorescence. Areas enclosed by dotted lines indicate the ovaries. The GFP was not detected in the ovary of PPO mice. K. Magnification of female reproductive organs in the PPO mouse under phase. Areas enclosed by dotted lines indicate the ovaries in the PPO mouse. Arrows indicate the oviducts. L. Magnification of female reproductive organs in the PPO mouse under GFP fluorescence. Areas enclosed by dotted lines indicate the ovaries in the PPO mouse. Arrows indicate the oviducts. M. GFP immunohistochemistry of the brain in PPO mice. N. GFP immunohistochemistry of the brain in control mice. O. Observations of the brain in PPO mice and control mice under phase. There was no macroscopic difference between the PPO and control brain. P. Observations of the brain in PPO mice and control mice under GFP fluorescence. The GFP was not detected in the brain of PPO mice. Scale bars represent the following distances: A, B, G, and H, 100 µm; M and N, 400 µm.
Reproduction and germline transmission in PPO mice
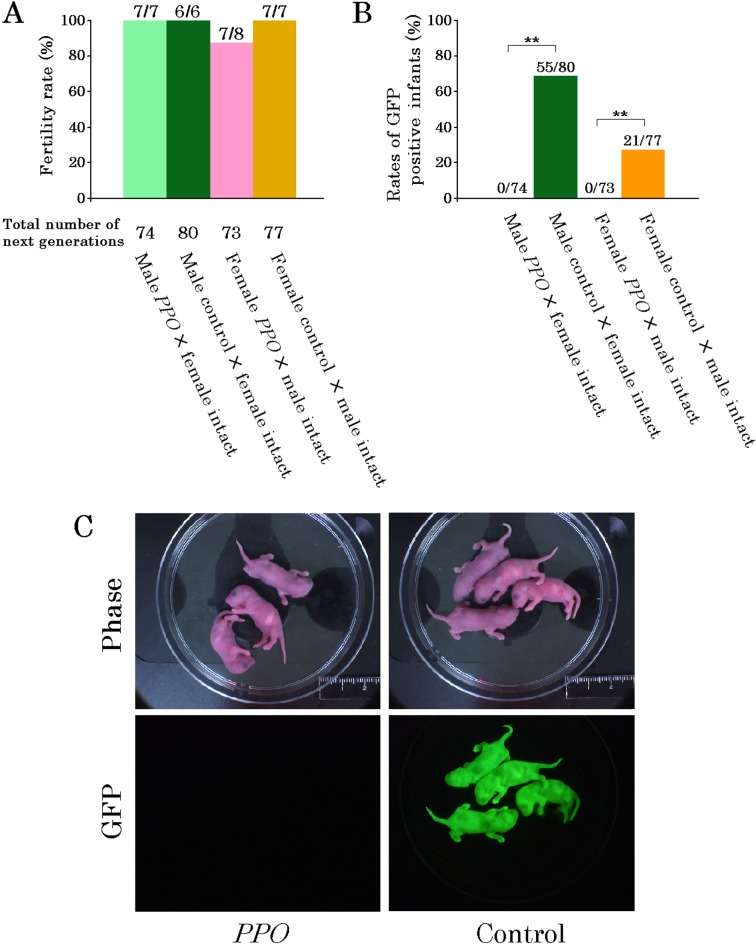
The fertility rates (Fig. 4A) and the number of infants produced by the male and female PPO mice did not differ from those of the control mice. Of the 80 male and 77 female infants produced by the control mice, the numbers of male and female GFP-positive infants were 55 (68.7%) and 21 (27.2%), respectively (Fig. 4B). On the other hand, of the 74 male and 73 female infants produced by the PPO mice (Fig. 4B), none of the infants showed GFP fluorescence (Fig. 4C).
Fig. 4.
Reproduction and germline transmission in PPO mice. A. Fertility rates and number of infants. B. Rates of GFP-positive infants from PPO and control mice. The rates were analyzed by χ2-test. **Significant difference of P<0.01. C. Confirmation of germline transmission.
Discussion
Studies examining blastocyst complementation using human PS cells are promoted by the Nakauchi group [13]. In recent years, Kobayashi et al. (2015) successfully limited and controlled PS cells for endoderm organs, especially the pancreas, via the inducible expression of the Milx1 gene, a transcription factor that induces differentiation of animal PS cells into the endoderm [14]. In the present study, we established a novel blastocyst complementation technique utilizing human PS cells as donor cells.
There is a concern in Japan that blastocyst complementation using human PS cells as donor cells may contribute to the central nervous systems of experimental animals, specifically that the contribution of human stem cells to the brain may lead to the generation of mice with human-like intelligence. Although this may seem unlikely, the possibility cannot be completely ruled out; therefore, it is necessary to address this problem. In a study conducted by Tian et al. (2002) [22], Otx2-deficient mouse embryos did not develop heads. From this finding, we hypothesized that Otx2-deficient mESCs would be unlikely to contribute to the brain in chimeric mice. Indeed, our results showed that G-mESCs without the Prdm14 and Otx2 genes did not contribute to the brains of chimeric mice, which addresses one of the concerns surrounding the use of human stem cells as donor cells for blastocyst complementation.
A second concern regarding blastocyst complementation using human stem cells as donor cells is that these cells may contribute to the gametes, leading to the generation of humanized gametes and potentially a new form of life. Yamaji et al. (2008) showed that the Prdm14 gene was associated with the development of germ cells in mice and that Prdm14-deficient mice displayed a lack of germ cells [25]. In the present study, the gametes, testes, and ovaries of PPO mice were also formed by recipient cells. In the previous study by Yamaji et al. (2008), the testes and ovaries in Prdm14-deficient mice were shown to exhibit atrophy [25]. Therefore, Prdm14-deficient ES cells may be unlikely to contribute to the gametes, testes, or ovaries. In addition, female chimeras were considered to be theoretically unable to achieve germline transmission [2] because the karyotype of the ES cells used (EGR-G101) was XY. However, our female chimeras derived from XY ES cells achieved germline transmission in similar to previous study [15], as did the male and female chimeras derived from G-mESCs with Prdm14 and Otx2 genes obtained in our study. On the other hand, the G-mESCs without the Prdm14 and Otx2 genes successfully interrupted germline transmission from the male and female chimeras. These results address the second concern about blastocyst complementation using human stem cells as donor cells. Thus, we hypothesized that the utilization of Prdm14-deficient ES cells could lead to the development of blastocyst complementation technology that does not contribute to gametes.
In the present study, CRISPR/Cas9 was used to delete the Prdm14 and Otx2 genes from a mouse model. CRISPR/Cas9 cut genomic DNA homozygosity [19, 21], as did the normal gene targeting methods [8]. However, homologous recombination via CRISPR/Cas9 was more efficient than that achieved using normal gene targeting methods [10]. Therefore, the CRISPR/Cas9 gene knockout method is suitable for use in blastocyst complementation technology, as the phenotype of the ES cells was expressed in the chimeric mice. Additionally, the pancreata of Pdx1−/− mice were complemented by G-mESCs without the Prdm14 and Otx2 genes. These findings revealed that PDX-1-deficient blastocyst complementation improved the impaired glucose tolerance, which is in agreement with the results of previous studies indicating that mouse blastocyst complementation using rat iPS cells improved diabetes in Pdx1−/− mice [13] and that transplantation of rat pancreata via rat blastocyst complementation using mouse iPS cells improved streptozotocin-induced diabetes in mice [24]. Therefore, we believe that blastocyst complementation using ES cells without the Prdm14 and Otx2 genes may offer an effective approach for studies using human stem cells [12, 23], ES cells, iPS cells, and other types of cells.
Acknowledgments
We thank Emika Sugiura and Yuki Yoshimura for help with the animal experiments. This study was supported by a Grant-in-Aid for Challenging Exploratory Research (No. 15K14374) and a Grant-in-Aid for Scientific Research (C) (No. 18K06044) to H.H. from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
References
- 1.Bally-Cuif L., Boncinelli E.1997. Transcription factors and head formation in vertebrates. BioEssays 19: 127–135. doi: 10.1002/bies.950190207 [DOI] [PubMed] [Google Scholar]
- 2.Bronson S.K., Smithies O., Mascarello J.T.1995. High incidence of XXY and XYY males among the offspring of female chimeras from embryonic stem cells. Proc. Natl. Acad. Sci. USA 92: 3120–3123. doi: 10.1073/pnas.92.8.3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burton A., Muller J., Tu S., Padilla-Longoria P., Guccione E., Torres-Padilla M.E.2013. Single-cell profiling of epigenetic modifiers identifies PRDM14 as an inducer of cell fate in the mammalian embryo. Cell Rep. 5: 687–701. doi: 10.1016/j.celrep.2013.09.044 [DOI] [PubMed] [Google Scholar]
- 4.Chen J., Lansford R., Stewart V., Young F., Alt F.W.1993. RAG-2-deficient blastocyst complementation: an assay of gene function in lymphocyte development. Proc. Natl. Acad. Sci. USA 90: 4528–4532. doi: 10.1073/pnas.90.10.4528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F.2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. doi: 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldstein R.S., Drukker M., Reubinoff B.E., Benvenisty N.2002. Integration and differentiation of human embryonic stem cells transplanted to the chick embryo. Dev. Dyn. 225: 80–86. doi: 10.1002/dvdy.10108 [DOI] [PubMed] [Google Scholar]
- 7.Hashimoto H., Kamisako T., Kagawa T., Haraguchi S., Yagoto M., Takahashi R., Kawai K., Suemizu H.2015. Expression of pancreatic and duodenal homeobox1 (PDX1) protein in the interior and exterior regions of the intestine, revealed by development and analysis of Pdx1 knockout mice. Lab. Anim. Res. 31: 93–98. doi: 10.5625/lar.2015.31.2.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hashimoto H., Yamamoto M., Sugiura E., Abe H., Kagawa T., Goto M., Takahashi R.I., Akimoto T., Suemizu H.2018. Adiponectin deficiency-induced diabetes increases TNFα and FFA via downregulation of PPARα. J. Vet. Med. Sci. 80: 662–666. doi: 10.1292/jvms.17-0641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horii T., Morita S., Kimura M., Kobayashi R., Tamura D., Takahashi R.U., Kimura H., Suetake I., Ohata H., Okamoto K., Tajima S., Ochiya T., Abe Y., Hatada I.2013. Genome engineering of mammalian haploid embryonic stem cells using the Cas9/RNA system. PeerJ 1: e230. doi: 10.7717/peerj.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horii T., Hatada I.2016. Challenges to increasing targeting efficiency in genome engineering. J. Reprod. Dev. 62: 7–9. doi: 10.1262/jrd.2015-151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isotani A., Hatayama H., Kaseda K., Ikawa M., Okabe M.2011. Formation of a thymus from rat ES cells in xenogeneic nude mouse↔rat ES chimeras. Genes Cells 16: 397–405. doi: 10.1111/j.1365-2443.2011.01495.x [DOI] [PubMed] [Google Scholar]
- 12.James D., Noggle S.A., Swigut T., Brivanlou A.H.2006. Contribution of human embryonic stem cells to mouse blastocysts. Dev. Biol. 295: 90–102. doi: 10.1016/j.ydbio.2006.03.026 [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi T., Yamaguchi T., Hamanaka S., Kato-Itoh M., Yamazaki Y., Ibata M., Sato H., Lee Y.S., Usui J., Knisely A.S., Hirabayashi M., Nakauchi H.2010. Generation of rat pancreas in mouse by interspecific blastocyst injection of pluripotent stem cells. Cell 142: 787–799. doi: 10.1016/j.cell.2010.07.039 [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi T., Kato-Itoh M., Nakauchi H.2015. Targeted organ generation using Mixl1-inducible mouse pluripotent stem cells in blastocyst complementation. Stem Cells Dev. 24: 182–189. doi: 10.1089/scd.2014.0270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuno J., Poueymirou W.T., Gong G., Siao C.J., Clarke G., Esau L., Kojak N., Posca J., Atanasio A., Strein J., Yancopoulos G.D., Lai K.M., DeChiara T.M., Frendewey D., Auerbach W., Valenzuela D.M.2015. Generation of fertile and fecund F0 XY female mice from XY ES cells. Transgenic Res. 24: 19–29. doi: 10.1007/s11248-014-9815-y [DOI] [PubMed] [Google Scholar]
- 16.Nagy A., Gertsenstein M., Vintersten K., Behringer R.2003. Manipulating the Mouse Embryo: A Laboratory Manual, Third Edition (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press). [Google Scholar]
- 17.Nakaki F., Saitou M.2014. PRDM14: a unique regulator for pluripotency and epigenetic reprogramming. Trends Biochem. Sci. 39: 289–298. doi: 10.1016/j.tibs.2014.04.003 [DOI] [PubMed] [Google Scholar]
- 18.Sakurai Y., Kurokawa D., Kiyonari H., Kajikawa E., Suda Y., Aizawa S.2010. Otx2 and Otx1 protect diencephalon and mesencephalon from caudalization into metencephalon during early brain regionalization. Dev. Biol. 347: 392–403. doi: 10.1016/j.ydbio.2010.08.028 [DOI] [PubMed] [Google Scholar]
- 19.Singh P., Schimenti J.C., Bolcun-Filas E.2015. A mouse geneticist’s practical guide to CRISPR applications. Genetics 199: 1–15. doi: 10.1534/genetics.114.169771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugawara A., Goto K., Sotomaru Y., Sofuni T., Ito T.2006. Current status of chromosomal abnormalities in mouse embryonic stem cell lines used in Japan. Comp. Med. 56: 31–34. [PubMed] [Google Scholar]
- 21.Tang C.C., Shan L.P., Wang W.M., Lu G., Tare R.S., Lee K.K.H.2017. Generation of a Bag1 homozygous knockout mouse embryonic stem cell line using CRISPR/Cas9. Stem Cell Res. (Amst.) 21: 29–31. doi: 10.1016/j.scr.2017.03.016 [DOI] [PubMed] [Google Scholar]
- 22.Tian E., Kimura C., Takeda N., Aizawa S., Matsuo I.2002. Otx2 is required to respond to signals from anterior neural ridge for forebrain specification. Dev. Biol. 242: 204–223. doi: 10.1006/dbio.2001.0531 [DOI] [PubMed] [Google Scholar]
- 23.Wang X., Li T., Cui T., Yu D., Liu C., Jiang L., Feng G., Wang L., Fu R., Zhang X., Hao J., Wang Y., Wang L., Zhou Q., Li W., Hu B.2018. Human embryonic stem cells contribute to embryonic and extraembryonic lineages in mouse embryos upon inhibition of apoptosis. Cell Res. 28: 126–129. doi: 10.1038/cr.2017.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamaguchi T., Sato H., Kato-Itoh M., Goto T., Hara H., Sanbo M., Mizuno N., Kobayashi T., Yanagida A., Umino A., Ota Y., Hamanaka S., Masaki H., Rashid S.T., Hirabayashi M., Nakauchi H.2017. Interspecies organogenesis generates autologous functional islets. Nature 542: 191–196. doi: 10.1038/nature21070 [DOI] [PubMed] [Google Scholar]
- 25.Yamaji M., Seki Y., Kurimoto K., Yabuta Y., Yuasa M., Shigeta M., Yamanaka K., Ohinata Y., Saitou M.2008. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat. Genet. 40: 1016–1022. doi: 10.1038/ng.186 [DOI] [PubMed] [Google Scholar]