Abstract
Endothelial lipase (EL) is a strong determinant of structural and functional properties of high-density lipoprotein (HDL). We examined whether the antioxidative capacity of HDL is affected by EL. EL-modified HDL (EL-HDL) and control EV-HDL were generated by incubation of HDL with EL- overexpressing or control HepG2 cells. As determined by native gradient gel electrophoresis, electron microscopy, and small-angle X-ray scattering EL-HDL is smaller than EV-HDL. Mass spectrometry revealed an enrichment of EL-HDL with lipolytic products and depletion of phospholipids and triacylglycerol. Kinetics of conjugated diene formation and HPLC-based malondialdehyde quantification revealed that EL-HDL exhibited a significantly higher resistance to copper ion-induced oxidation and a significantly higher capacity to protect low-density lipoprotein (LDL) from copper ion-induced oxidation when compared to EV-HDL. Depletion of the lipolytic products from EL-HDL abolished the capacity of EL-HDL to protect LDL from copper ion-induced oxidation, which could be partially restored by lysophosphatidylcholine enrichment. Proteomics of HDL incubated with oxidized LDL revealed significantly higher levels of methionine 136 sulfoxide in EL-HDL compared to EV-HDL. Chloramine T (oxidizes methionines and modifies free thiols), diminished the difference between EL-HDL and EV-HDL regarding the capacity to protect LDL from oxidation. In absence of LDL small EV-HDL and EL-HDL exhibited higher resistance to copper ion-induced oxidation when compared to respective large particles. In conclusion, the augmented antioxidative capacity of EL-HDL is primarily determined by the enrichment of HDL with EL-generated lipolytic products and to a lesser extent by the decreased HDL particle size and the increased activity of chloramine T-sensitive mechanisms.
Keywords: Oxidation, LDL, HDL, Proteomics, Mass spectrometry
1. Introduction
Numerous studies have demonstrated the relationship between structural and compositional features and antioxidative capacity of high-density lipoprotein (HDL) [1,2]. The well-established capacity of HDL to inactivate lipid hydroperoxides (LOOH) and protect low-density lipoprotein (LDL) from oxidation is largely determined by the HDL content of apolipoprotein A-I (apoA-I) and the oxidative status of apoA-I methionine (Met) residues [3]. ApoA-I mediates both the removal of LOOH molecules from LDL as well as the reduction of HDL-associated LOOH to redox-inactive lipid hydroxides [4]. Other HDL-associated apolipoproteins and enzymes such as paraoxonase 1 (PON1), platelet-activated factor-acetylhydrolase (PAF-AH), and lecithin:cholesterol acyltransferase (LCAT) also contribute to the antioxidative capacity of HDL [1,5].
Besides HDL proteins, HDL lipids also shape the antioxidative capacity of HDL [6]. This is primarily due to the impact of lipids on the fluidity of the HDL surface and on the size of the HDL core. For example, enrichment of HDL with ceramide, sphingomyelin or cholesterol decreases the antioxidative capacity of HDL. This is thought to be due to a decreased HDL surface fluidity and in consequence reduced transfer of LOOH from LDL to HDL [7]. The antioxidative capacity of HDL is also affected by alterations in triacylglycerol (TAG) and cholesteryl ester (CE) levels in the HDL core, whereby an increased TAG content diminishes apoA-I mediated inactivation of LOOH [8,9].
Endothelial lipase (EL) is a strong negative regulator of HDL plasma levels [10] and a potent modulator of HDL lipid composition [11,12]. EL belongs to the TAG lipase family and is mainly synthesized and secreted by vascular endothelial cells [13,14]. By its pronounced phospholipase activity EL depletes HDL phospholipids (PL) thereby producing lysophospholipids (LysoPL) and fatty acids (FA) [15]. EL-mediated depletion of HDL-PL alters structural and functional properties of HDL [11,16]. However, the role of EL in atherosclerosis still remains inconclusive: in one study EL promoted the progression of atherosclerosis in apolipoprotein E (apoE)-deficient mice [17], whereas in another study EL had no impact on atherosclerosis development in apoE- or LDL receptor-deficient mice [18]. Similarly inconclusive data have been published on the impact of EL on the antioxidative capacity of HDL: augmented antioxidative capacity of HDL isolated from EL deficient mice was observed in one [19] but not in another study [20].
The overall antioxidative capacity of HDL as well as the relative contribution of HDL-associated proteins and lipids to the antioxidative capacity of HDL are largely determined by the composition and size of HDL particles [1]. Considering this point as well as the pronounced EL-mediated alterations in HDL structure and composition [11,12,16], we hypothesized that EL may alter the antioxidative capacity of HDL. Therefore, the present study has been designed to investigate the relationship between the composition, structure and antioxidative capacity of in vitro generated EL-modified HDL (EL-HDL).
2. Materials and methods
2.1. Cell culture
HepG2 cells (ATCC®, HB-8065™) were cultured in DMEM supplemented with 2 mmol/L glutamine, 1% PS (100 U/mL penicillin, 100 μg/mL streptomycin) and 10% fetal calf serum (FCS).
2.2. Human plasma
Normolipidemic plasma of 18 healthy donors (whose identity was not disclosed to the researcher) was purchased from the blood bank of the Medical University of Graz. The mean donor age was 44.8 ± 11.6 years, with a range of 24–65, and 3 out of 18 donors were female. The regular written informed consent for blood donation covers the donation of residual material for research purposes. The ethics committee of the Medical University of Graz has no objections to the use of the anonymized material for research purposes.
2.3. Isolation of HDL from human plasma
HDL preparation from human plasma was performed by a one-step density gradient ultracentrifugation method using long centrifuge tubes (16 × 76 mm; Beckman), as described [21]. Briefly, the density-adjusted plasma (1.24 g/mL with potassium bromide) was layered underneath a potassium bromide-density solution (1.063 g/mL). Samples were centrifuged at 330,000 ×g for 6 h (centrifuge: Beckman Optima L-80 ultracentrifuge, rotor: Sorvall T-1270). Subsequently, the collected HDL was concentrated by Viva Spin Tubes (Sartorius, Vienna, Austria), desalted by gel filtration on Sephadex PD-10 columns (GE Healthcare, Munich, Germany) and stored at −80 °C for further experiments.
2.4. Preparation of EL-HDL and control EV-HDL
HepG2 cells (2 × 106) were plated onto 60 mm dishes and incubated under standard conditions as described in Section 3.1. After 24 h, cells were washed once with DMEM without FCS and infected with multiplicity of infection (MOI) 10 of adenovirus encoding human EL (EL-Ad) or empty adenovirus containing no recombinant cDNA (EV-Ad) [22] in DMEM without FCS for 2 h. After removal of infection media, cells were incubated with fresh DMEM containing 10% FCS for 28 h. Thereafter, cells were washed once with DMEM without FCS and each plate was incubated under cell culture conditions with 2 mg HDL protein, re-suspended in 1.8 mL DMEM without FCS, in the absence or presence of 4% (final concentration) of non-esterified fatty acid (NEFA)- free bovine serum albumin (BSA) (Sigma-Aldrich, Vienna, Austria) for 16 h. After incubation, the media were collected and spun at 1100 ×g for 3 min to remove cellular debris. EL-HDL and EV-HDL were isolated from media by density gradient ultracentrifugation, desalted and purified by fast protein liquid chromatography (FPLC) as described in Section 3.9.
2.5. Lysophosphatidylcholine (LPC) enrichment of HDL
Enrichment of HDL with LPC was performed as described in our previous study [23]. Required amounts of LPC, which were stored dissolved in chloroform/methanol at −20 °C under argon atmosphere, were dried under a stream of nitrogen and re-dissolved in PBS. The lipolytic products-depleted EL-HDL and EV-HDL (1 mg/mL) (generated in the presence of 4% BSA) were incubated with 0.6 mmol/L LPC 16:0 or 18:1 at 37 °C for 2 h. Unbound LPC were removed by gel filtration. The efficiency of LPC enrichment was examined by mass spectrometry as described in Section 3.6.
2.6. Non-denaturing gradient-gel electrophoresis
Non-denaturing gradient-gel electrophoresis of HDL and FPLC fractions was performed as previously described [16]. Briefly, aliquots of HDL (10 μg) and FPLC fractions (15 μL) were electrophoresed on 4–16% non-denaturing polyacrylamide gels upon dilution with native sample buffer (LifeTechnologies, Vienna, Austria) at 125 V for 4 h at room temperature. Gels were stained with Sudan black (Sigma-Aldrich, Vienna, Austria) or fixed with 10% sulfosalicylic acid for 30 min and then stained with Coomassie Brillant Blue G250 or used for Western blotting analysis. The high molecular weight marker NativeMark (LifeTechnologies, Vienna, Austria) was used as standard.
2.7. Targeted lipidomic analysis
Three hundred μg HDL protein were transferred to 2 mL Safe-Lock PP-tubes and extracted according to Matyash et al. [24]. In brief, samples were homogenized using two 6 mm steal beads on a Mixer Mill (Retsch, Haan, GER; 2x15sec, frequency 30/s) in 700 μL MTBE/MeOH (3/1, v/v) containing 500 pmol butylated hydroxytoluene, 1% acetic acid, and 100 pmol of internal standards (ISTD, 17:0–17:0 PC, 12:0–12:0 PC, 12:0–12:0 PE, 17:0–17:0 PA, 17:0 FA, d18:1/17:0 Cer, 14:0–14:0 DG, 17:0 LPC, 17:0–17:0–17:0 TG, 15:0–15:0–15:0 TG, Avanti Polar Lipids) per sample. Total lipid extraction was performed under constant shaking for 60 min at RT. After addition of 140 μL dH2O and further incubation for 30 min at RT, samples were centrifuged at 1000 ×g for 15 min to establish phase separation. Five hundred μL of the upper, organic phase were collected and dried under a stream of nitrogen. Lipids were resolved in 250 μL 2-propanol/methanol/water (7/2.5/1, v/v/v) for UPLC-QqQ analysis. Chromatographic separation was modified according to Knittelfelder et al. [25] using an AQUITY-UPLC system (Waters Corporation), equipped with a Kinetex EVOC18 column (2.1 × 50 mm, 1.7 μm; Phenomenex) starting a 25 min gradient with 100% solvent A (MeOH/H2O, 1/1, v/v; 10 mM ammonium acetate, 0.1% formic acid). A EVOQ Elite™ triple quadrupole mass spectrometer (Bruker) equipped with an ESI source was used for detection. Lipid species were analyzed by selected reaction monitoring (PC: [MH] + to m/z 184, 25 eV, PE: [MH] + to -m/z 141, 20 eV, PI: [MH] to corresponding [FA]-, 50 eV, LPC: [MH] + to m/z 184, 22 eV, LPE: [MH] + to -m/z 141, 17 eV, Cer: [MH] + to m/z 264, 22 eV, TG: [MNH4] + to corresponding [DG-H2O]+, 22 eV, DG: [MNH4] + to [RCOO+58]+, 15 eV, CE: [MNH4] + to m/z 369, FC: [M-H2O]+, 0 eV, FA: [M-H]-, 0 eV, SM: [MH] + to m/z 184, 23 eV). Data acquisition was done by MS Workstation (Bruker). Data were normalized for recovery, extraction-, and ionization efficacy by calculating analyte/ISTD ratios (AU) and expressed as AU/mg protein.
2.8. Small-angle X-ray scattering (SAXS)
SAXS data for suspensions of HDL were recorded on an in-house SAXS instrument (SAXSess mc2, Anton Paar, Graz, Austria) equipped with a Kratky camera, a sealed X-ray tube source and a two-dimensional Princeton Instruments PI•SCX:4300 (Roper Scientific) CCD detector. The scattering patterns were measured with a 90-min exposure time (540 frames, each 10 s) for several solute concentrations in the range from 0.5 to 1.0 mg/mL. Radiation damage was excluded based on a comparison of individual frames of the 90-min exposures, with no detectable changes. A range of momentum transfer of 0.012 < s < 0.63 Å−1 was covered (s = 4π sin(θ)/λ, with 2θ scattering angle and λ = 1.5 Å X-ray wavelength). All SAXS data were analyzed with the package ATSAS (version 2.5). The data were processed with the SAXSQuant software (version 3.9) and desmeared using the programs GNOM and GIFT [26]. The forward scattering, I(0), the radius of gyration, Rg, the maximum dimension, Dmax, and the inter-atomic distance distribution functions, (P(R)), were computed with the program GNOM and GIFT. The masses of the solutes were evaluated by comparison of the forward scattering intensity with human serum albumin reference solution (molecular mass 69 kDa) and using the Porod's law. To generate ab initio shape models, a total number of 50 models were calculated using the program DAMMIF [27] and aligned, and averaged using the program DAMCLUST.
2.9. Negative stain electron microscopy for HDL visualization
Glow discharged carbon coated copper grids were used. The grids were placed on 100 μL of HDL suspension (0.8 mg/mL HDL protein) on parafilm for 1 min. The specimens were washed three times with 200 μL water. Samples were blotted with filter paper and placed on a drop of 2% aqueous uranyl acetate solution for 1 min, blotted with filter paper and air dried at room temperature [28]. Specimens were examined with an FEI Tecnai G 2 equipped with an ultrascan 1000 CCD camera (Gatan).
2.10. FPLC of native (nHDL) and modified HDL
FPLC was performed as described in our previous report [29]. Briefly, an ÄKTA pure FPLC System (GE Healthcare, Munich, Germany) equipped with a Superdex 200 Increase 10/300 column (GE Healthcare, Munich, Germany) was used with PBS as mobile phase. After loading, HDL samples were separated with a constant flow of 0.5 mL/min, and 0.5 mL fractionation between of 9 to 13.5 mL elution volume were collected. The software UNICORN (GE Healthcare, Munich, Germany) was used for analysis.
2.11. SDS-PAGE and Western blotting
Preparation of heparin media and analyses of EL overexpression were performed as described [22]. HDL-associated proteins (10 μg) and cell lysates (30 μg) were separated by 12% SDS-PAGE at 175 V for 90 min. Separated proteins were transferred to polyvinylidene difluoride (PVDF) membranes (Carl Roth, Karlsruhe, Germany) with blotting buffer (Tris, glycine, EDTA, sodium azide) at 150 mA for 90 min. Membranes were blocked at RT in 10% skim milk for 2 h followed by overnight incubation at 4 °C with antibodies specific for: human PON1 (Abcam, ab24261, Cambridge, UK), human apoA-I (Novus biological, NB100–65491, Littleton, CO, USA, or Academy biomedical, 11A-G2b, Houston, TX, USA), human LCAT (Novus biological, Littleton, CO, USA), human PAF-AH (Cayman chemical, 160603, Ann Arbor, MI, USA), α-tubulin (Cell signaling technology, 11H10, Leiden, Netherlands) and albumin (Abcam, ab 83465, Cambridge, UK). After washing and incubation with the appropriate secondary antibody (Dako, Vienna, Austria), protein signals were visualized by incubation with Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific, Schwerte, Germany) using the ChemiDoc system (Bio-Rad Laboratories, Vienna, Austria).
2.12. LDL preparation and oxidation
Human LDL was isolated from plasma as described previously [30]. LDL was dialyzed against PBS (pH 7.4) without EDTA and diluted to a final concentration of 0.3 mg apoB/mL. Oxidation was performed by addition of a CuCl2 solution giving a final concentration of 10 μmol/L. After 4 h oxidation was stopped with EDTA (200 μmol/L) and the LDL was dialyzed against PBS (GIBCO, Life technologies, pH 7.40).
2.13. Kinetics of lipoprotein oxidation (conjugated dienes formation)
Copper ion-induced formation of conjugated dienes was examined as described [31]. Briefly, CuCl2 (2 μmol/L) was added to the indicated concentrations of lipoproteins and the formation of conjugated dienes was continuously monitored at 234 nm by a spectrophotometer (Hitachi U-2000) at 37 °C for 100 min. The lag phase was determined with a simple sigmoidal model. The mathematical properties of the model were determined using the software package MATHEMATICA version 11 (Wolfram Research Inc., Champaign, IL, USA) and implemented in a Microsoft EXCEL sheet for convenient usage.
2.14. Malondialdehyde (MDA) analysis
MDA content was measured in the reaction mixtures described in Section 2.13. at termination of the oxidation (120 min), using a HPLC method described by Pilz et al. [32] after derivatization with 2,4-dinitrophenylhydrazine (DNPH). The slight modifications of the method have been reported [33]. Briefly, for alkaline hydrolysis of protein-bound MDA, 25 μL of 6 mol/L sodium hydroxide was added to 0.125 mL of the samples (1.5 mL Eppendorf tubes) and incubated at 60 °C for 30 min. The hydrolyzed sample was deproteinized with 62.5 μL 35% (v/v) perchloric acid. The supernatant (125 μL) obtained after centrifugation (14,000 g; 2 min) was mixed with 12.5 μL DNPH solution and incubated for 10 min. This reaction mixture, diluted derivatized standard solutions (0.625 nmol/mL – 10 nmol/mL) and reagent blanks were injected into the HPLC system (injection volume: 40 μL). The MDA standard was prepared by dissolving 25 μL 1,1,3,3-tetramethoxypropane (TMP) in 100 mL bidistilled H2O (stock solution: 1 mmol/L). The hydrolysis was performed with 200 μL TMP stock solution in 10 mL 1% sulfuric acid and incubation for 2 h at RT [34].The resulting MDA standard of 20 nmol/mL was further diluted with 1% sulfuric acid to the final concentrations. The DNPH derivaives (hydrazones) were isocratically separated on a 5-μm ODS hypersil column (150 × 4.6 mm) guarded by a 5-μm ODS hypersil column (10 × 4.6 mm; Uniguard holder) with a mobile phase consisting of a 0.2% (v/v) acetic acid solution (bidistilled water) containing 50% acetonitrile (v/v) at a flow rate of 0.8 mL/min. The HPLC separations were performed with an L-2200 autosampler (injection volume: 40 μL), a L-2130 HTA pump and a L-2450 diode array detector (all: VWR Hitachi Vienna; Austria). Detector signals (absorbance at 310 nm) were recorded and the program EZchrom Elite (VWR) was used for data requisition and analysis.
2.15. Oxidation of HDL Met residues
Blocking of Met residues was performed as described previously [3]. Shortly, FPLC-purified EV-HDL and EL-HDL (1.5 mg protein/mL) were incubated at 4 °C for 1 h with 2 mmol/L chloramine T (Sigma Aldrich, Vienna, Austria), which oxidizes protein Met residues into corresponding sulfoxides [35] as well as modifies free SH groups [36]. Chloramine T was removed by gel filtration on Sephadex PD-10 columns (GE Healthcare, Munich, Germany).
2.16. Analyses of apo A-I Met residues by proteomics
CuCl2-oxidized LDL (50 μg) was incubated with EV-HDL or EL-HDL (both 50 μg, three biological replicates each) for 30, 75 and 120 min at 37 °C, followed by termination of the reaction with acetone and detection of oxidized apoA-I Met residues by proteomics. For tryptic digest, 50 μg of HDL protein was precipitated with 4 volumes of acetone at −20 °C overnight, solubilized in 50 μL 100 mmol/L ammonium bicarbonate, reduced with 50 μL of 10 mmol/L DTT for 20 min under shaking at 550 rpm at 56 °C and alkylated with 50 μL of 60 mmol/L iodoacetamide under shaking at 550 rpm at RT for 15 min. Protein was digested by adding 1 μg modified trypsin (Promega) and shaking overnight at 550 rpm at 37 °C. The resulting peptide solution was acidified by adding 3 μL of 5% formic acid. Thereafter, 250 ng of the digest was injected and concentrated on the enrichment column (C18, 5 μm, 100 Å, 5 × 0.3 mm) for 2 min using 0.1% formic acid as isocratic solvent at 5 μL/min flow rate. The column was then switched in the nanoflow circuit and the sample was loaded on the nanocolumn (Acclaim PepMap RSLC nanocolumn; C18, 2 μm, 100 Å, 500 × 0.075 mm), at a flow rate of 250 nL/min at 60 °C and separated using the following gradient: solvent A: water, 0.1% formic acid; solvent B: acetonitrile, 0.1% formic acid; 0–2 min: 4% B; 2–90 min: 4–25% B; 90–95 min: 25–95% B, 96–110 min: 95% B; 110–110.1 min: 4% B; 110.1–125 min: 4% B. The sample was ionized in the nanospray source equipped with stainless steel emitters (Thermo Fisher Scientific, Vienna, Austria) and analyzed in a Thermo Orbitrap velos Pro mass spectrometer in positive ion mode by alternating full scan MS (m/z 300 to 2000, 60,000 resolution) in the ICR cell and MS/MS by CID of the 20 most intense peaks in the ion trap with dynamic exclusion enabled. The LC-MS/MS data were analyzed by searching the human Swiss-Prot protein database containing all common contaminants with Proteome Discoverer 1.4 (Thermo Fisher Scientific) and Mascot 2.4.1 (MatrixScience, London, UK). Carbamidomethylation of cysteine residues was entered as fixed and methionine oxidation as well as cysteine sulfonation as variable modification. Detailed search criteria were used as follows: trypsin; max. Missed cleavage sites: 2; search mode: MS/MS ion search with decoy database search included (false discovery rate < 1%); precursor mass tolerance ± 10 ppm; product mass tolerance ± 0.7 Da. Label free quantitation of precursor ions was done with Proteome Discoverer 1.4 (Thermo Fisher Scientific) to determine peptide intensities. ApoA-I peptides with detected oxidative modifications as well as their unmodified counterparts were relatively quantified and normalized on apoA-I total protein intensity, which was calculated by summing up all individual apoA-I peptide intensities for each sample. Overall 51 different peptides of apoA-I were detected. Values at time point zero were subtracted.
2.17. Statistical analyses
Data are represented as the means + standard error of mean (S.E.M.). Statistical significance between two groups was determined by two-tailed unpaired t-test followed by Bonferroni correction for multiple testing. Comparisons among multiple groups were performed using one-way analysis of variance (ANOVA), with the Bonferroni post hoc test to determine p values. Statistical analyses were performed using Graph Pad Prism 5.0. software (GraphPad Software, La Jolla, CA, USA). Statistically significant differences between groups are indicated by P-values of < 0.05 (* or #), < 0.01 (** or ##), or < 0.001 (*** or ###).
3. Results
3.1. EL alters structure and size of HDL
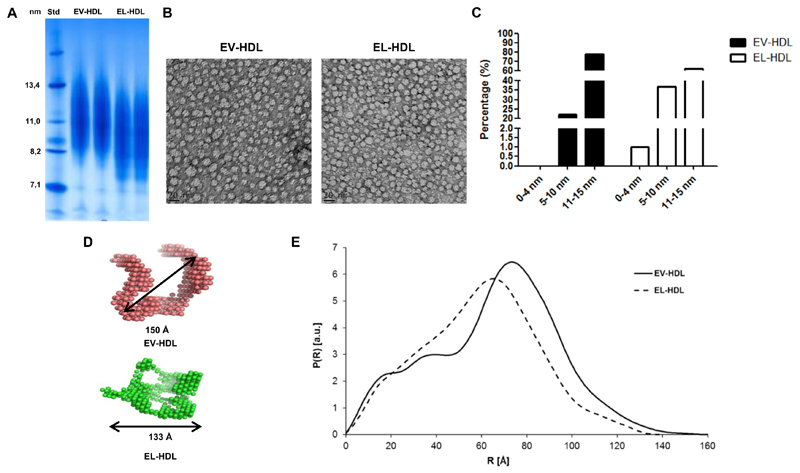
To study the impact of EL on the antioxidative capacity of HDL, we prepared EL-modified HDL and control HDL by incubating isolated HDL with EL- and EV-overexpressing HepG2 cells (Supplementary Fig. S1), followed by re-isolation of EL-HDL and EV-HDL by ultracentrifugation and subsequent purification by FPLC. As revealed by non-denaturing gradient-gel electrophoresis and subsequent protein staining, EL-HDL particles were smaller compared to EV-HDL (Fig. 1A). This result was further confirmed by electron microscopy, showing greater abundance of small (0 nm–10 nm), and lower abundance of large (10 nm–15 nm) particles in the population of EL-HDL compared to EV-HDL particles (Fig. 1B, C). Impact of EL on the size and structure of HDL was assessed by SAXS analysis which showed a mean diameter of 133 Ǻ for EL-HDL, compared to 150 Ǻ for EV-HDL (Fig. 1D). Furthermore, SAXS analysis revealed an increased distance distribution P(R) between 20 and 40 Ǻ for EL-HDL, indicating decreased lipid content. In addition, EL-HDL exhibited decreased maximal spatial dimension, exemplified by reduced density distribution R > 80 Ǻ and maximal diameter (Dmax), respectively (Fig. 1E).
Fig. 1. EL alters HDL size and structure.
A) EV-HDL and EL-HDL were generated by incubation of human HDL with EV or EL overexpressing cells under cell culture conditions for 16 h. Thereafter, HDL was purified by ultracentrifugation and FPLC. 10 μg isolated HDL protein were electrophoresed on 4–20% non-denaturing polyacrylamide gels followed by Coomassie Brillant Blue staining. Protein size annotations refer to protein marker bands on the gel. B) Electron micrographs of EV-HDL and EL-HDL. Particle window size was 20 nm. C) Calculated particle size distribution. The number of analyzed particles was 1999 for EV-HDL and 1336 for EL-HDL. The data show abundance of sized EV-HDL and EL-HDL particles as percentage of the total number of analyzed particles. D) Ab initio shape models and E) pairwise distance distribution function of modified HDLs were generated by SAXS analysis and subsequent modeling.
3.2. EL-HDL exhibits altered lipid composition
Considering the established relationship between HDL lipid composition and the antioxidative capacity of HDL, we examined lipid composition of purified EV-HDL and EL-HDL. As revealed by MS analyses (Table 1), the levels of phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), triacylglycerol (TAG), and diacylglycerol (DAG) were significantly decreased, whereas the levels of lysophosphatidylcholine (LPC), lysophosphatidylethanolamine (LPE), FA, and ceramide (Cer) were significantly increased in EL-HDL when compared to EV-HDL. The levels of free cholesterol (FC) showed a trend to be higher in EL-HDL compared to EV-HDL, however, the p-values were no longer significant after Bonferroni correction for multiple comparison. Cholesterol ester (CE) and sphingomyelin (SM) content were not significantly different between EL-HDL and EV-HDL.
Table 1. MS analyses of EV-HDL and EL-HDL lipids.
| AU/mg HDL protein | EV-HDL | EL-HDL | p-value |
|---|---|---|---|
| PC | 461.7 ± 19.39 | 172.1 ± 5.73 | 0.0001 |
| PE | 18.01 ± 0.35 | 3.53 ± 0.22 | < 0.0001 |
| PI | 156.0 ± 6.25 | 4.01 ± 0.13 | < 0.0001 |
| TAG | 51.67 ± 1.79 | 19.52 ± 1.91 | 0.0003 |
| DAG | 45.74 ± 1.52 | 11.88 ± 0.42 | < 0.0001 |
| LPC | 41.37 ± 0.68 | 108.2 ± 3.80 | < 0.0001 |
| LPE | 0.16 ± 0.004 | 0.19 ± 0.003 | 0.0033 |
| FA | 223.3 ± 6.78 | 1834 ± 222.1 | 0.0019 |
| FC | 0.95 ± 0.05 | 1.32 ± 0.05 | 0.0067 |
| CE | 1036 ± 16.06 | 1051 ± 102.0 | 0.8936 |
| Cer | 4.065 ± 0.10 | 5.229 ± 0.14 | 0.0025 |
| SM | 67.89 ± 2.24 | 75.17 ± 4.58 | 0.2265 |
Lipids from EV-HDL and EL-HDL (300 μg protein) purified by ultracentrifugation followed by FPLC were extracted and analyzed by MS. Results are mean ± SEM of 3 independent modifications of human HDL. The differences between EV-HDL and EL-HDL were analyzed by two-tailed unpaired t-test between EV- and EL-HDL. P-values of < 0.004 are considered significant after a Bonferroni correction for multiple testing. PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; TAG, triacylglycerol; DAG, diacylglycerol; LPC, lysophosphatidylcholine, LPE, lysophosphatidylethanolamine, FC, free cholesterol, CE, cholesteryl ester; Cer, Ceramide; SM, Sphingomyelin; MS, mass spectrometry; SEM, standard error of mean;
3.3. EL modification reduces HDL-associated enzymes
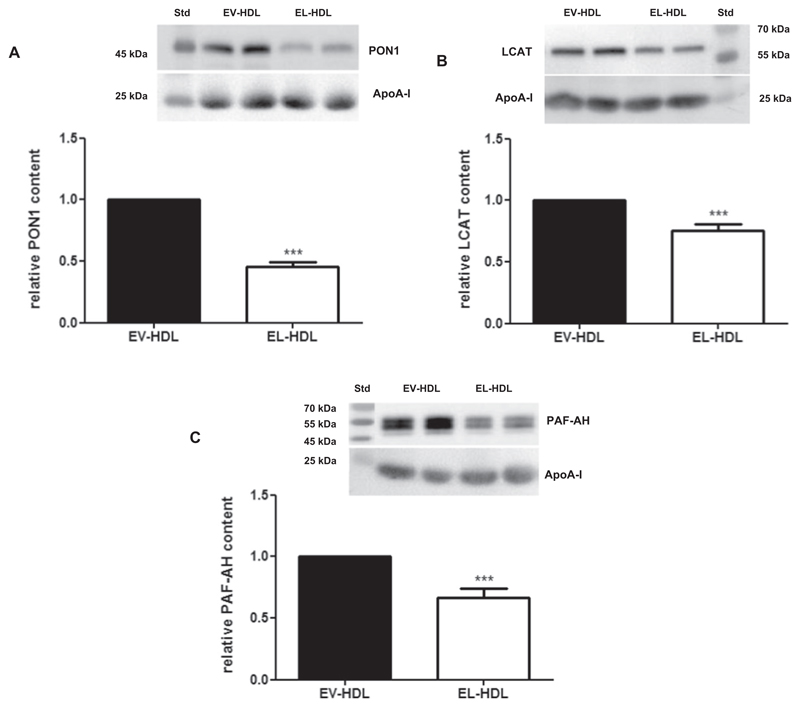
As revealed by Western blotting analyses, the abundance of HDL-associated enzymes, known to contribute to the antioxidative capacity of HDL, such as PON1 (Fig. 2A), LCAT (Fig. 2B) and PAF-AH (Fig. 2C),was significantly lower in EL-HDL when compared to EV-HDL.
Fig. 2. Abundance of PON1, LCAT and PAF-AH is decreased in EL-HDL.
HDL proteins (10 μg/lane) were separated by 12% SDS-PAGE followed by Western blotting analyses of A) PON1, B) LCAT, and C) PAF-AH. Results show representative Western blots and corresponding densitometric analyses. Densitometric values of PON1, LCAT and PAF-AH were normalized to the corresponding apoA-I signals. Results obtained by densitometry are means + SEM of 4 independent modifications of human HDL, each loaded in duplicates and analyzed by two-tailed unpaired t-test. ***P < 0.001.
3.4. EL-HDL exhibits increased antioxidative capacity
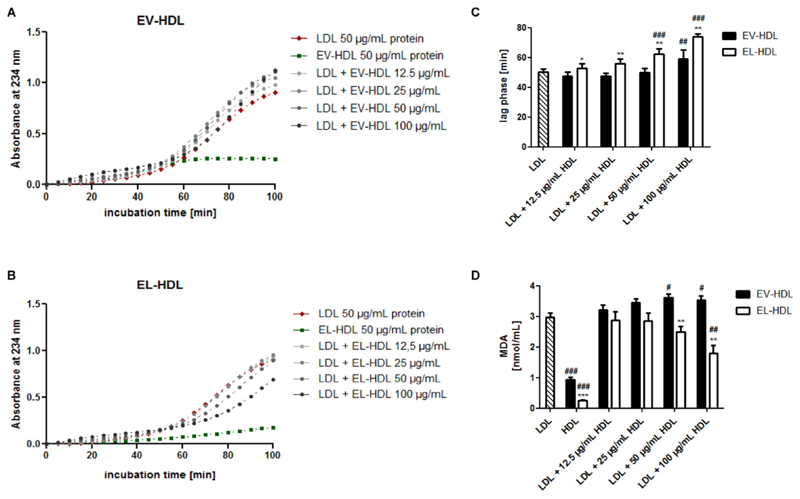
Next, we studied the capacity of EL-HDL to protect LDL from copper ion-induced oxidation as well as its resistance to copper ion-induced oxidation in the absence of LDL. Importantly, EL-HDL dose dependently attenuated LDL oxidation as illustrated with the longer lag phases (significant at the two highest EL-HDL concentrations) and lower MDA concentrations (significant at the highest EL-HDL concentrations) in the presence of EL-HDL compared to LDL alone (Fig. 3C, D). Oppositely, EV-HDL promoted LDL oxidation as exemplified by unaltered lag phases (except in the presence of highest EV-HDL concentration) and significantly higher MDA levels (at the two highest EV-HDL concentrations) compared to LDL alone (Fig. 3C, D). Although the lag phase for the cooper ion-induced oxidation of EV-HDL and EL-HDL in the absence of LDL could not be calculated, the traces in Fig. 3A and B clearly show a slower formation of conjugated dienes in EL-HDL compared to EV-HDL. In line, in the absence of LDL, the MDA levels were significantly lower in EL-HDL compared to EV-HDL (Fig. 3D). Together, EL-modification of HDL increases the resistance of HDL to copper ion-induced oxidation and the capacity of HDL to protect LDL from copper ion-induced oxidation.
Fig. 3. EL increases the antioxidative capacity of HDL.
Human LDL, EV-HDL and EL-HDL were incubated either individually or in the indicated combinations with 2 μmol/L CuCl2 at 37 °C followed by reaction termination after 120 min. The formation of conjugated dienes was monitored at 234 nm. Representative kinetic graphs of lipoprotein incubation with CuCl2 are shown in A) and B). C) The lag phase was determined with a simple sigmoid model. D) MDA levels were measured in the reaction mixture obtained after 120 min by HPLC. Results are means + SEM of 4 independent modifications of human HDL. The differences between EV-HDL and EL-HDL (with or without LDL) were analyzed by two-tailed unpaired t-test followed by Bonferroni correction for multiple comparison (*P < 0.05, **P < 0.01, ***P < 0.001) and between EV-HDL or EL-HDL (with or without LDL) and LDL alone by one-way ANOVA followed by Bonferroni post hoc test (#P < 0.05, ##P < 0.01, ###P < 0.001).
3.5. Depletion of lipolytic products abolishes capacity of EL-HDL to protect LDL from copper ion-induced oxidation
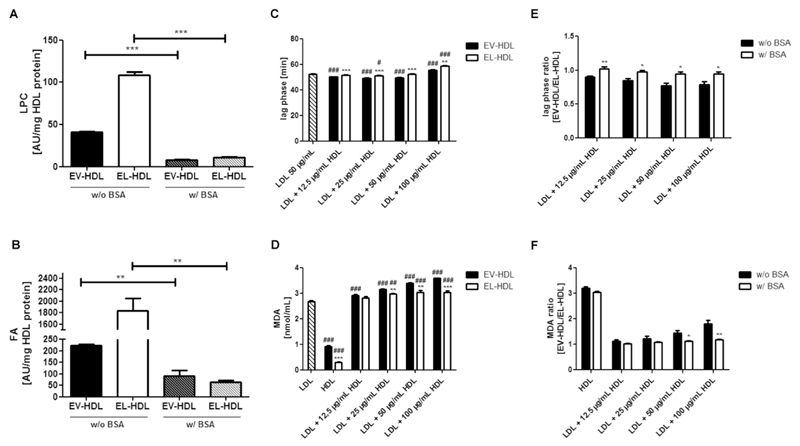
Because the action of EL on HDL markedly increases the content of lipolytic products in HDL (Table 1), we hypothesized that the accumulated lipolytic products might be responsible for the augmented antioxidative capacity of EL-HDL. To test this hypothesis, we depleted lipolytic products in EL-HDL and EV-HDL by performing HDL modification in the presence of 4% BSA followed by purification of HDL by ultracentrifugation and FPLC. As revealed by Western blotting using an antibody which detects both human albumin and BSA, purified EV-HDL and EL-HDL were free of plasma albumin and BSA Supplementary Fig. S2). This procedure markedly decreased the LPC (Fig. 4A), FA (Fig. 4B), and LPE (Supplementary Fig. S3F), without significantly affecting the content of other lipids (Supplementary Fig. S3) in EV-HDL and EL-HDL. Importantly, depletion of lipolytic products abolished the capacity of EL-HDL to protect LDL from copper ion-induced oxidation; the lag phases were shorter or similar compared to that of LDL alone (except in the presence of 100 μg/mL EL-HDL) (Fig. 4C) and the MDA levels were significantly higher compared to LDL alone (Fig. 4D). This finding is in sharp contrast to the results with EL-HDL prepared in the absence of BSA, thus containing high levels of lipolytic products (compare Fig. 4C, D with Fig. 3C, D). In contrast to EL-HDL, the capacity of EV-HDL to protect LDL from copper ion-induced oxidation was not affected by the depletion of lipolytic products; the duration of lag phases and MDA levels were similar as observed for EV-HDL which was prepared in the absence of BSA (compare Fig. 4C, D and Fig. 3C, D). In line with the presented results, depletion of lipolytic products diminished the difference between EV-HDL and EL-HDL regarding the duration of lag phase and MDA levels. This is illustrated by higher lag phase ratios (significant at 12.5 and 50 μg/mL HDL) as well as lower MDA ratios (at the highest HDL concentrations) of EV-HDL and EL-HDL prepared in the presence (w/ BSA) compared to the ratios of EV-HDL and EL-HDL prepared in the absence of BSA (w/o BSA) (Fig. 4E, F). Furthermore, the resistance of EL-HDL to copper ion-induced oxidation in the absence of LDL was not significantly altered upon depletion of lipolytic products. The amounts of generated MDA and the difference in MDA content between EV-HDL and EL-HDL (Fig. 4D) were similar to what we found with EV-HDL and EL-HDL prepared in the absence of BSA (Fig. 3D). Together, depletion of lipolytic products abolished the capacity of EL-HDL to protect LDL from copper ion-induced oxidation without affecting the resistance of EL-HDL to copper ion-induced oxidation.
Fig. 4. Role of lipolytic products in the antioxidative capacity of EL-HDL.
EV-HDL and EL-HDL were prepared in the absence (w/o BSA) or presence (w/ BSA) of 4% BSA followed by ultracentrifugation and FPLC. (A) LPC and (B) FA contents were determined by MS. LDL, EV-HDL and EL-HDL (the latter two prepared in the presence of 4% BSA) were incubated either individually or in the indicated combinations with 2 μmol/L CuCl2 at 37 °C followed by reaction termination after 120 min. (C) The formation of conjugated dienes was monitored at 234 nm. (D) MDA levels were measured in the reaction mixture obtained after 120 min by HPLC. (E) Lag phase and (F) MDA ratios of EV-HDL and EL-HDL prepared w/o or w/ 4% BSA. Results are means + SEM of 4 independent modifications of human HDL. The indicated differences in (A) and (B), as well as between EV-HDL and EL-HDL (with or without LDL) (C, D), and between w/o BSA and w/ BSA (E,F) were analyzed by two-tailed unpaired t-test followed by Bonferroni correction for multiple comparison (*P < 0.05, **P < 0.01, ***P < 0.001). The differences between EV-HDL or EL-HDL (with or without LDL) and LDL (C, D) were analyzed by one-way ANOVA followed by Bonferroni post hoc test (#P < 0.05, ##P < 0.01, ###P < 0.001). w/o, without; w/, with;
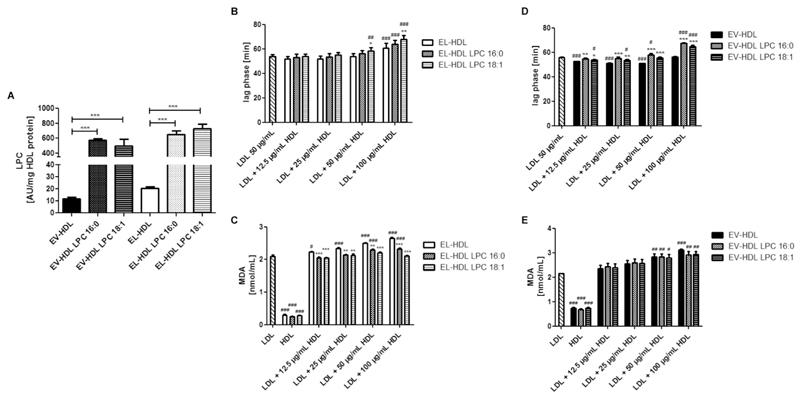
3.6. LPC enrichment partially restores the capacity of lipolytic products-depleted EL-HDL to protect LDL from copper ion-induced oxidation
To substantiate the role of lipolytic products in the antioxidative capacity of EL-HDL, we enriched the lipolytic products-depleted EL-HDL (and EV-HDL) with LPC 16:0 or LPC 18:1 (Fig. 5A). Enrichment of the lipolytic products-depleted EL-HDL with LPC 18:1 markedly improved the capacity of that HDL to protect LDL from oxidation. This is exemplified by significantly longer lag phases at the two highest HDL concentrations and significantly lower MDA concentrations at all tested LPC-enriched HDL concentrations when compared to the lipolytic products-depleted EL-HDL (Fig. 5B, C). Enrichment of the lipolytic products-depleted EL-HDL with LPC 16:0 did not affect the lag phase duration (Fig. 5B), but significantly attenuated MDA formation when compared to the lipolytic products-depleted EL-HDL (Fig. 5C). The resistance of EL-HDL to copper ion-induced oxidation (in the absence of LDL) was not significantly affected by LPC enrichment (Fig. 5C). Enrichment of the lipolytic products-depleted EV-HDL with LPC 16:0 or LPC 18:1 significantly prolonged the lag phase (Fig. 5D) but had no impact on the rate of MDA formation (Fig. 5E). Together, LPC enrichment partially restores the diminished capacity of lipolytic products-depleted EL-HDL to protect LDL from copper ion-induced oxidation.
Fig. 5. Impact of LPC enrichment on the antioxidative capacity of EL-HDL.
(A) LPC content of EV-HDL and EL-HDL enriched or not with LPC 16:0 or 18:1 determined by MS. LDL, EV-HDL and EL-HDL (the latter two enriched or not with LPC) were incubated either individually or in the indicated combinations with 2 μmol/L CuCl2 at 37 °C followed by reaction termination after 120 min. (B,D) The formation of conjugated dienes was monitored at 234 nm. (C, E) MDA levels were measured after 120 min by HPLC. Results are means ± SEM of 4 independent modifications of human HDL. The differences in (A) were analyzed by two-tailed unpaired t-test followed by Bonferroni correction for multiple comparison. The differences between non-enriched and LPC-enriched EV-HDL or EL-HDL (with or without LDL) (B-E) (*P < 0.05, **P < 0.01, ***P < 0.001) as well as the differences between EV-HDL or EL-HDL (with or without LPC enrichment, with or without LDL) and LDL (B-E) (#P < 0.05, ##P < 0.01, ###P < 0.001) were analyzed by one-way ANOVA followed by Bonferroni post hoc test.
3.7. Role of apoA-I Met residues and chloramine T-sensitive antioxidative activities
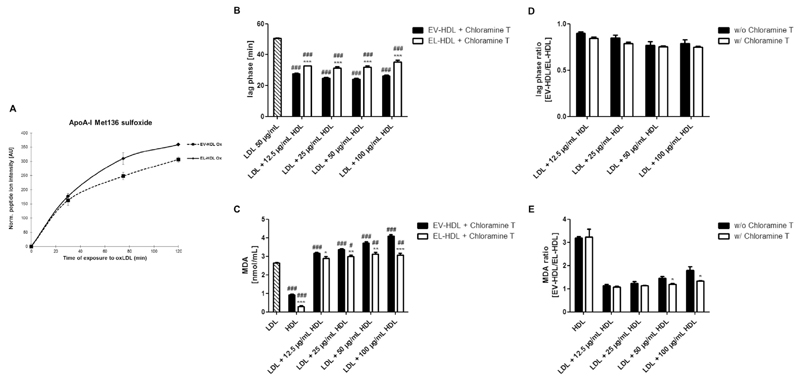
Previous studies have shown that HDL size and structure affect conformation of HDL-associated proteins, resulting in altered accessibility and in turn altered antioxidative activity of Met residues and free-SH groups [1,3]. We therefore investigated the contribution of apoA-I Met residues to the antioxidative capacity of EL-HDL. For this purpose, purified EV-HDL and EL-HDL were incubated in the absence or presence of copper oxidized LDL at 37 °C for up to 120 min. We detected higher levels of apoA-I Met136 sulfoxides (contained in the peptide WQEEM (136)ELYR) in EL-HDL compared to EV-HDL (P = 0.028), however, upon Bonferroni correction for multiple comparison this difference did not remain statistically significant (Fig. 6A). The levels of other apoA-I Met sulfoxides were not significantly different (before and after Bonferroni correction) or not sufficiently covered in EV-HDL and EL-HDL (not shown). Incubation of EV-HDL and EL-HDL in the absence of oxidized LDL resulted in very low and similar levels of apoA-I sulfoxides in both HDL types (not shown).
Fig. 6. Role of apoA-I Met residues and chloramine T-sensitive antioxidative activities.
(A) CuCl2-oxidized LDL (50 μg) was incubated with EV-HDL or EL-HDL (50 μg) for 30, 75 and 120 min at 37 °C. ApoA-I Met sulfoxides were measured by proteomics. Human LDL, chloramine T-treated EV-HDL and EL-HDL were incubated either individually or in the indicated combinations with 2 μmol/L CuCl2 at 37 °C followed by reaction termination after 120 min. The formation of conjugated dienes was monitored at 234 nm. B) The lag phase was determined with a simple sigmoid model. C) MDA levels were measured in the reaction mixture obtained after 120 min by HPLC. Results in (A) are mean ± SEM of 3 independent modifications of human HDL and the differences between EV-HDL and EL-HDL at each indicated time-point were analyzed by two-tailed paired t-test followed by Bonferroni correction for multiple comparison. Results in B-E are means + SEM of 4 independent modifications of human HDL. The differences between EV-HDL and EL-HDL (with or without LDL) (B, C) and between w/o and w/ chloramine T (D, E) were analyzed by two-tailed unpaired t-test followed by Bonferroni correction for multiple comparison (*P < 0.05, **P < 0.01, ***P < 0.001). The differences between EV-HDL or EL-HDL (with or without LDL) and LDL (B, C) were analyzed by one-way ANOVA followed by Bonferroni post hoc test (#P < 0.05, ##P < 0.01, ###P < 0.001).
To address the role of Met residues (and free SH groups) in the antioxidative capacity of EL-HDL, we examined the antioxidative capacity of chloramine T (oxidizes Met residues to the respective sulfoxides and modifies free thiols)-treated EV-HDL and EL-HDL [3,36]. As revealed by proteomics, 85–90% of apoA-I Met residues in EV-HDL and EL-HDL were oxidized following chloramine T treatment. Chloramine T abolished the capacity of both EV-HDL and EL-HDL to protect LDL from oxidation exemplified by significantly shorter lag phases and significantly higher MDA levels in the presence of EV-HDL or EL-HDL compared to LDL alone (Fig. 6B, C). Chloramine T did not alter the difference between EV-HDL and EL-HDL in terms of the duration of the lag phase (compare Fig. 6B and Fig. 3C) as illustrated through the similar lag phase ratios (Fig. 6D). In contrast, however, chloramine T-treatment of HDL diminished the difference in MDA levels between EV-HDL and EL-HDL (compare Fig. 6C with Fig. 3D), resulting in a lower MDA ratios of chloramine T-treated compared to chloramine T-untreated EV-HDL and EL-HDL, which upon Bonferroni correction for multiple comparison did not remain significant (Fig. 6E). The oxidation of EV-HDL and EL-HDL in the absence of LDL was not affected by chloramine T as the MDA levels were similar in chloramine T-treated compared to untreated EV-HDL and EL-HDL (see also Fig. 6C and 3D). This is further exemplified by the similar MDA ratio of chloramine T-treated versus untreated EV-HDL to EL-HDL, respectively (Fig. 6E). From these results we concluded that the chloramine T-sensitive anti-oxidative mechanisms contribute to the capacity of EL-HDL to protect LDL from copper ion-induced oxidation without affecting the resistance of EL-HDL to copper ion-induced oxidation in the absence of LDL.
3.8. Antioxidative capacity – role of HDL particle size
Considering established inverse relationship between HDL size and its antioxidative capacity [37], the augmented antioxidative capacity of EL-HDL may (at least in part) be due to its smaller size. Accordingly, the smallest particles may exhibit the highest antioxidative capacity. We therefore prepared different sized fractions of EL-HDL and EV-HDL by FPLC (Suppl. Fig. S4A) and tested their capacity to protect LDL from copper ion-induced oxidation as well as their sole effect on resistance to copper ion-induced oxidation. We observed a trend of a negative relationship between EL-HDL particle size and the capacity to protect LDL from copper ion-induced oxidation (large versus small particle size - Suppl. Fig. S4B, C). However, the opposite was found for EV-HDL (large versus small particle size - Suppl. Fig. S4D, E). Interestingly, both small EV-HDL and small EL-HDL particles exhibited significantly higher resistance to copper ion-induced oxidation in the absence of LDL, when compared to respective large particles (Suppl. Fig. S4C, E). The increased antioxidative capacity of small compared to large EV-HDL and EL-HDL particles was accompanied by significantly lower PI, TAG, FC, Cer, and SM contents in small EV-HDL (Suppl. Fig. S5) as well significantly lower TAG, FC, and Cer contents in small EL-HDL (Suppl. Fig. S6) compared to the respective large HDL particles. Interestingly, LPC content was similar in the different sized fractions of EV-HDL and EL-HDL (Suppl. Fig. S5 and S6).
3.9. Comparison of structural and functional features of native HDL (nHDL) with EV-HDL and EL-HDL
To examine whether and how modified HDL differ from native nHDL, we compared structural and functional features of EV-HDL and EL-HDL with nHDL isolated from human plasma by ultracentrifugation and further purified by FPLC. As revealed by non-denaturing gradient-gel electrophoresis nHDL particles were larger compared to EV-HDL and EL-HDL (Supplementary Fig. S7A). SAXS-derived low-resolution models showed a mean diameter of 160 Ǻ for nHDL compared to 150 Ǻ for EV-HDL and 133 Ǻ for EL-HDL (Supplementary Fig. S7B). Furthermore, the SAXS data indicate that processing of nHDL to EV-HDL and even more to EL-HDL leads to a compaction of the particles and a decrease in the lipid content. This is exemplified by the lower distance distribution of nHDL in the region between 20 and 40 Ǻ compared to EV-HDL and EL-HDL, and a shift of the maximum from 90 Ǻ (nHDL) to 73 Ǻ (EV-HDL) and 65 Ǻ (EL-HDL) (Suppl. Fig. S7C). In line with SAXS data, the MS analyses revealed significantly higher PC, PE, PI, TAG, DAG, and SM levels in nHDL compared to EV-HDL and EL-HDL (Supplementary Fig. S8). Furthermore, LPC and LPE were similar in nHDL and EV-HDL and significantly lower compared to EL-HDL. The tested HDL contained similar levels of CE and Cer. Interestingly, the FC content of nHDL was significantly higher compared to EV-HDL but similar to that of EL-HDL. Moreover, while the content of LCAT and PAF-AH was higher in nHDL compared to EV-HDL, the content of PON1 was comparable in nHDL and EV-HDL (Supplementary Fig. S9). Despite structural and compositional differences between nHDL and EV-HDL, the resistance of nHDL to copper ion-induced oxidation and the capacity of nHDL to protect LDL from copper ion-induced oxidation were not significantly different from that of EV-HDL (Supplementary Fig. S10).
4. Discussion
Due to the fact that the capacity of HDL to protect LDL from oxidation is determined by structural features and composition of HDL [1], we hypothesized that EL (by altering structure and composition) [11,16,38] would markedly affect the antioxidative capacity of HDL. To test our hypothesis we modified HDL by incubation with EL-overexpressing cells and examined the antioxidative capacity of EL-modified HDL by monitoring its resistance to copper ion-induced oxidation as well as its capacity to attenuate copper ion-induced LDL oxidation.
In the present study, in vitro EL-modification markedly augmented the antioxidative capacity of HDL. This is in contrast to the observed increased antioxidative capacity of HDL isolated from EL-deficient mice [19]. It remains to be determined whether species-specific differences or differences between in vivo and in vitro models may explain the increasing effect of EL-overexpression and EL deficiency on HDL antioxidative capacity. However, one should bear in mind that the augmenting effect of EL deficiency on HDL antioxidative capacity was observed in one [19] but not another study [20].
In contrast to EL-modified HDL, both nHDL and EV-HDL promoted copper-ion induced LDL oxidation, highlighting the impact of EL-modification on the antioxidative capacity of HDL under the experimental conditions applied in the present study. Furthermore, the comparable magnitude of the copper-ion induced oxidation of LDL in the presence of nHDL or EV-HDL, excludes that the prooxidative feature of EV-HDL is due to the incubation of HDL with control HepG2 cells.
Previous studies have shown that small, dense HDL particles exhibit higher antioxidative capacity than larger, less dense particles [1]. Several mechanisms and molecular determinants including enrichment in LCAT, PAF-AH, and PON1, as well as the relative enrichment in apoA-I, altered apoA-I conformation, or altered lipid composition and packing of surface lipids were proposed to explain better antioxidative properties of small HDL particles [39–42]. Also the significantly higher resistance to copper ion-induced oxidation of small compared to large EL-HDL particles, suggests that the smaller size of EL-HDL contributes, at least in part, to its augmented antioxidative capacity when compared to EV-HDL. The lower content of LCAT, PAF-AH, and PON1 in EL-HDL argues against a significant contribution of these enzymes to the increased antioxidative capacity of EL-HDL. Furthermore, the lower content of LCAT and PAF-AH in EV-HDL compared to nHDL most probably reflects loss of these enzymes during incubation with HepG2 cells and/or during subsequent ultracentrifugation and FPLC purification.
A striking compositional feature of EL-HDL was its enrichment with LysoPL and FA. The depletion of lipolytic products from EL-HDL attenuated its capacity to protect LDL from copper ion-induced oxidation without affecting the resistance of EL-HDL to copper ion-induced oxidation in the absence of LDL. Conversely, the replenishment of the lipolytic products-depleted EL-HDL with LPC 16:0 or 18:1 improved but not completely restored the capacity of that HDL to protect LDL from oxidation. Although LPC 16:0 and 18:1 belong to the most abundant LPC species in EL-HDL [15], the replenishment of lipolytic products-depleted EL-HDL with single LPC species can hardly compensate for the BSA-mediated loss of a naturally occurring combination of LysoPL and FA found in EL-HDL [15]. Accordingly, only enrichment of HDL with a natural combination of lipolytic products as generated by the action of EL on HDL would probably fully restore the capacity of lipolytic products-depleted EL-HDL to protect LDL from oxidation.
Nevertheless, our results strongly indicate that the accumulation of EL-generated lipolytic products underlie the augmented capacity of EL-HDL to protect LDL from oxidation. It is likely that the presence of lipolytic products in EL-HDL increases the surface fluidity of HDL, thus facilitating the interaction of EL-HDL with oxidized LDL. The more profound restoration of EL-HDL's protective capacity upon enrichment with LPC 18:1, compared to LPC 16:0, likely reflects the superior effect of unsaturated compared to saturated LPC on the surface fluidity of HDL. Increased surface fluidity in turn may facilitate the transfer of LOOH from oxidized LDL to EL-HDL followed by conversion of LOOH to LOH by Met residues of apoA-I. Indeed, a previous study showed that LPC enrichment increases HDL surface fluidity [43] resulting in a decreased packing of surface lipids. This was accompanied by an augmented capacity of HDL to incorporate oxidation products including LOOH. Others, however, have shown that in vitro LPC enrichment reduced apoA-I content and the antioxidative capacity of HDL [44]. This suggests that the impact of LPC enrichment on the structural and functional properties of HDL is dependent on the structure and composition of HDL prior to enrichment. More specifically, the decreased content of phospholipids and lipolytic products in HDL is likely a prerequisite for the augmentation of HDL's antioxidative capacity by LPC enrichment. In line, in the present study LPC enrichment more profoundly augmented antioxidative capacity of the lipolytic products depleted EL-HDL compared to EV-HDL.
It is well documented that the antioxidative capacity of HDL is regulated by the abundance and conformation of apoA-I, whereby inactivation of LOOH by HDL is driven by apoA-I Met residues [3]. Incubation of EV-HDL and EL-HDL with oxLDL resulted in a trend of a higher oxidation rate of apoA-I Met136 in EL-HDL compared to EV-HDL, suggesting a more pronounced antioxidative activity of Met136 in apoA-I of EL-HDL. It is conceivable that the altered conformation of apoA-I, as a consequence of the EL-induced reduction in HDL particle size underlies the augmented antioxidative activity of apoA-I Met136 of EL-HDL. Additionally, an altered lipid content and composition of EL-HDL may facilitate the exposure of Met136 to the aqueous phase resulting in its augmented antioxidative capacity. In the present study, chloramine T profoundly attenuated the capacity of both EV-HDL and EL-HDL to protect LDL from oxidation. These findings indicate that the chloramine T-sensitive amino acids, primarily Met residues of apoA-I, but also free SH groups in other HDL associated proteins [1,3], are important for the capacity of both EV-HDL and EL-HDL to protect LDL from oxidation. The fact that chloramine T diminished the difference between EL-HDL and EV-HDL regarding the capacity to protect LDL from oxidation, strongly suggests that chloramine T-sensitive mechanisms contribute to the better antioxidative capacity of EL-HDL. Interestingly, chloramine T did not affect the resistance of EV-HDL and EL-HDL to copper ion-induced oxidation in the absence of LDL, suggesting that different mechanisms are responsible for preventing HDL autooxidation and HDL-mediated protection of LDL from oxidation.
Enrichment of HDL surface lipids with FC, SM or Cer negatively affects the transfer of LOOH from LDL to HDL for subsequent reduction to inactive LOH by Met residues of apoA-I [1]. Accordingly, a higher content of FC and Cer in EL-HDL, is in contrast with the observed increased antioxidative capacity of EL-HDL. However, it is likely that accumulated lipolytic products by increasing the fluidity of the EL-HDL surface can counteract the augmented rigidity caused by FC and Cer. Indeed, higher resistance to copper ion-induced oxidation of small EV-HDL and small EL-HDL particles was accompanied by lower FC and Cer levels when compared to respective large HDL particles. Importantly, the small and large EV-HDL and EL-HDL particles did not differ in the abundance of LPC. This strongly suggests that the EL-generated lipolytic products enriched in EL-HDL may outperform the impact of FC and Cer on the antioxidative capacity of HDL. The content of several lipid species (including phospholipids, TAG, DAG, FC, and SM) was lower in EV-HDL compared to nHDL, indicating either a transfer of lipids to or degradation by HepG2 cells during modification. Interestingly, the FC levels of EL-HDL and nHDL were similar but significantly higher compared to EV-HDL (Suppl. Fig. S8). This finding possibly reflects a compensation of FC loss by an increased cholesterol efflux from EL-overexpressing HepG2 cells during in vitro HDL modification, similarly to results described in a previous study in EL-overexpressing macrophages [45].
The smaller size of EL-HDL compared to EV-HDL in consequence leads to higher EL-HDL particle numbers in antioxidative assays normalized to protein. Accordingly, the increased antioxidative capacity of EL-HDL observed in the present study may be a consequence of these higher EL-HDL particle numbers. However, the fact that lipolytic product depletion decreased both the antioxidative capacity (Fig. 4) and size of EL-HDL (Suppl. Fig. S11) strongly indicates that enrichment with lipolytic products and not HDL particle numbers is the reason for the increase in antioxidative capacity of EL-HDL. Nevertheless, further experiments are required to elucidate the relative contribution of HDL particle numbers (size) to the increased antioxidative capacity of EL-HDL.
Considering results obtained by structural modeling of lipoprotein particles [42], one can speculate that the pronounced EL-induced perturbations in HDL size and composition promote the redistribution of CE to the HDL surface, leading to disturbed packing of surface lipids, which in turn may promote an efficient insertion of LOOHs (despite increased FC and Cer). Additionally, a recent study demonstrated a negative correlation between the TAG content of HDL and the HDL antioxidative capacity [46]. Accordingly, the decreased TAG levels might also contribute to the increased antioxidative activity of EL-HDL. Because the content of the minor PL species phosphatidylserine, which is a known enhancer of HDL functionality [37], could not be determined due to technical limitation, its contribution to antioxidative capacity of EL-HDL could not be addressed.
We have recently shown that EL diminishes cholesterol efflux capacity of HDL, which is the first step of reverse cholesterol transport, the best-studied atheroprotective activity of HDL [16]. This, together with EL-mediated augmentation of HDL's antioxidative capacity, which is also an important atheroprotective feature of HDL, may help understanding inconclusive data on the role of EL in atherosclerosis.
This study is not without limitations. First, the relative contribution of HDL particle concentration to the increased antioxidative capacity of EL-HDL could not be assessed. Our efforts to determine HDL particle concentrations in EV-HDL and EL-HDL preparations by nuclear magnetic resonance (NMR) spectroscopy failed. This is due to the fact that NMR spectroscopy allows quantification of HDL particles only in a physiological environment, full serum or plasma, but not in suspensions containing only HDL. Despite optimization of HDL purification procedure by FPLC, the albumin/BSA-free HDL was obtained in only 60–70% of preparations. Only the fractions which were, as revealed by Western blotting, free of albumin/BSA were used for antioxidative assays, whereas the preparations which after FPLC still contained residual albumin were used for SAXS or electron microscopy. Accordingly, there is some albumin and overlap of HDL with albumin in the preparations shown in Fig. 1A, Suppl. Fig. S7, or S11 but not in Suppl. Fig. S2 and S4, the latter two being representative of the preparations free of albumin/BSA used for functional assays. We cannot exclude that our albumin/BSA-free preparations also contain albumin/BSA, however, at concentrations which are below the detection limit of the applied Western blotting analyses. Furthermore, HDL was modified in the absence of serum and accordingly, in the presence of sub-physiological levels of HDL remodeling proteins which in vivo, together with EL, determine structural and functional properties of HDL [47,48]. Additionally, we tested the impact of HDL on copper-ion induced oxidation of LDL which is only one out of several known mechanisms of LDL oxidation [49]. Accordingly, our results have limited capacity to predict impact of EL on the capacity of HDL to protect LDL from oxidation in vivo.
5. Conclusions
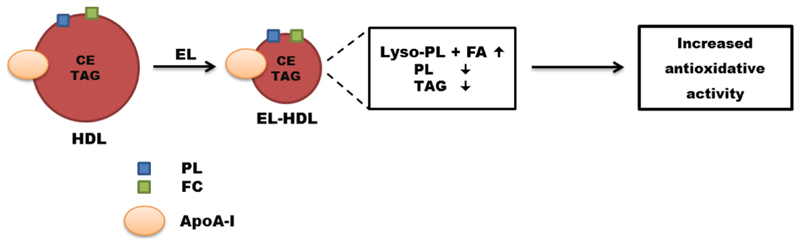
Here, we presented evidence that in vitro EL modification of HDL generates small HDL particles with altered composition and augmented antioxidative capacity. The augmented antioxidative capacity of EL-modified HDL is primarily due to the enrichment of HDL with EL-generated lipolytic products and to a lesser extent due to decreased HDL particle size and increased activity of chloramine T-sensitive mechanisms (Fig. 7).
Fig. 7. Consequences of EL action on HDL.
In vitro EL action on HDL generates small HDL particles with altered lipid and protein composition and increased antioxidative activity.
Supplementary Material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbalip.2019.06.011.
Acknowledgements
This work was supported by the Austrian Science Fund FWF (P27166-B23 to SF, P28854 and I3792 to TM, and W1226 DK-MCD to DK and TM). TM was supported by the President's International Fellowship Initiative of CAS (No. 2015VBB045), the National Natural Science Foundation of China (No. 31450110423), the Austrian Research Promotion Agency (FFG: 864690, 870,454), the Integrative Metabolism Research Center Graz, the Austrian infrastructure program 2016/2017, BioTechMed-Graz, the Styrian government (Zukunftsfonds), and the OMICS center Graz. The funders had no roles in the study design, collection, analysis and interpretation of data, writing report or submission of the article.
The authors thank Margarete Lechleitner, Dominique Pernitsch, Therese Macher, and Gerd Kager for the expert technical assistance.
Abbreviations
- LDL
low-density lipoprotein
- HDL
high-density lipoprotein
- apoA-I
apolipoprotein A-I
- PAF-AH
platelet-activating factor-acetyl hydrolase
- LCAT
lecithin:cholesterol acyltransferase
- PON1
paraoxonase 1
- HDL-C
HDL cholesterol
- EL
endothelial lipase
- HSPG
heparin sulfate proteoglycan
- PL
phospholipids
- LysoPL
lysophospholipids
- FFA
free fatty acids
- FCS
fetal calf serum
- NEFA
non-esterified fatty acids
- BSA
bovine serum albumin
- FPLC
fast protein liquid chromatography
- HPLC
high performance liquid chromatography
- SAXS
small-angle x-ray scattering
- PC
mass spectrometry (MS), phosphatidylcholine
- PE
phosphatidylehtanolamine
- PI
phosphatidylinositol
- TAG
triacylglycerol
- DAG
diacylglycerol
- LPC
lysophosphatidylcholine
- LPE
lysophosphatidylethanolamine
- FC
free cholesterol
- CE
cholesteryl ester
- Cer
ceramide
- SM
sphingomyelin
- MDA
malondialdehyde
- Met
methionine
- EV
empty virus
Footnotes
Transparency document
The Transparency document associated with this article can be found, in online version.
Declaration of Competing Interest
The authors declare no conflicts of interest.
References
- [1].Karlsson H, Kontush A, James RW. Functionality of HDL: antioxidation and detoxifying effects. Handb Exp Pharmacol. 2015;224:207–228. doi: 10.1007/978-3-319-09665-0_5. [DOI] [PubMed] [Google Scholar]
- [2].Soran H, Schofield JD, Durrington PN. Antioxidant properties of HDL. Front Pharmacol. 2015;6:222. doi: 10.3389/fphar.2015.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zerrad-Saadi A, Therond P, Chantepie S, Couturier M, Rye KA, Chapman MJ, Kontush A. HDL3-mediated inactivation of LDL-associated phospholipid hydroperoxides is determined by the redox status of apolipoprotein A-I and HDL particle surface lipid rigidity: relevance to inflammation and atherogenesis. Arterioscler Thromb Vasc Biol. 2009;29:2169–2175. doi: 10.1161/ATVBAHA.109.194555. [DOI] [PubMed] [Google Scholar]
- [4].Garner B, Witting PK, Waldeck AR, Christison JK, Raftery M, Stocker R. Oxidation of high density lipoproteins. I. Formation of methionine sulfoxide in apolipoproteins AI and AII is an early event that accompanies lipid peroxidation and can be enhanced by alpha-tocopherol. J Biol Chem. 1998;273:6080–6087. doi: 10.1074/jbc.273.11.6080. [DOI] [PubMed] [Google Scholar]
- [5].Gugliucci A, Menini T. Paraoxonase 1 and HDL maturation. Clin Chim Acta. 2015;439:5–13. doi: 10.1016/j.cca.2014.09.016. [DOI] [PubMed] [Google Scholar]
- [6].Kontush A, Lhomme M, Chapman MJ. Unraveling the complexities of the HDL lipidome. J Lipid Res. 2013;54:2950–2963. doi: 10.1194/jlr.R036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Vila A, Korytowski W, Girotti AW. Dissemination of peroxidative stress via intermembrane transfer of lipid hydroperoxides: model studies with cholesterol hydroperoxides. Arch Biochem Biophys. 2000;380:208–218. doi: 10.1006/abbi.2000.1928. [DOI] [PubMed] [Google Scholar]
- [8].Sparks DL, Davidson WS, Lund-Katz S, Phillips MC. Effects of the neutral lipid content of high density lipoprotein on apolipoprotein A-I structure and particle stability. J Biol Chem. 1995;270:26910–26917. doi: 10.1074/jbc.270.45.26910. [DOI] [PubMed] [Google Scholar]
- [9].Curtiss LK, Bonnet DJ, Rye KA. The conformation of apolipoprotein A-I in high-density lipoproteins is influenced by core lipid composition and particle size: a surface plasmon resonance study. Biochemistry. 2000;39:5712–5721. doi: 10.1021/bi992902m. [DOI] [PubMed] [Google Scholar]
- [10].Ishida T, Choi S, Kundu RK, Hirata K, Rubin EM, Cooper AD, Quertermous T. Endothelial lipase is a major determinant of HDL level. J Clin Invest. 2003;111:347–355. doi: 10.1172/JCI16306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gauster M, Oskolkova OV, Innerlohinger J, Glatter O, Knipping G, Frank S. Endothelial lipase-modified high-density lipoprotein exhibits diminished ability to mediate SR-BI (scavenger receptor B type I)-dependent free-cholesterol efflux. Biochem J. 2004;382:75–82. doi: 10.1042/BJ20031882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nijstad N, Wiersma H, Gautier T, van der Giet M, Maugeais C, Tietge UJ. Scavenger receptor BI-mediated selective uptake is required for the remodeling of high density lipoprotein by endothelial lipase. J Biol Chem. 2009;284:6093–6100. doi: 10.1074/jbc.M807683200. [DOI] [PubMed] [Google Scholar]
- [13].Jaye M, Lynch KJ, Krawiec J, Marchadier D, Maugeais C, Doan K, South V, Amin D, Perrone M, Rader DJ. A novel endothelial-derived lipase that modulates HDL metabolism. Nat Genet. 1999;21:424–428. doi: 10.1038/7766. [DOI] [PubMed] [Google Scholar]
- [14].Hirata K, Dichek HL, Cioffi JA, Choi SY, Leeper NJ, Quintana L, Kronmal GS, Cooper AD, Quertermous T. Cloning of a unique lipase from endothelial cells extends the lipase gene family. J Biol Chem. 1999;274:14170–14175. doi: 10.1074/jbc.274.20.14170. [DOI] [PubMed] [Google Scholar]
- [15].Gauster M, Rechberger G, Sovic A, Horl G, Steyrer E, Sattler W, Frank S. Endothelial lipase releases saturated and unsaturated fatty acids of high density lipoprotein phosphatidylcholine. J Lipid Res. 2005;46:1517–1525. doi: 10.1194/jlr.M500054-JLR200. [DOI] [PubMed] [Google Scholar]
- [16].Schilcher I, Kern S, Hrzenjak A, Eichmann TO, Stojakovic T, Scharnagl H, Duta-Mare M, Kratky D, Marsche G, Frank S. Impact of endothelial lipase on cholesterol efflux capacity of serum and high-density lipoprotein. Sci Rep. 2017;7 doi: 10.1038/s41598-017-12882-7. 12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ishida T, Choi SY, Kundu RK, Spin J, Yamashita T, Hirata K, Kojima Y, Yokoyama M, Cooper AD, Quertermous T. Endothelial lipase modulates susceptibility to atherosclerosis in apolipoprotein-E-deficient mice. J Biol Chem. 2004;279:45085–45092. doi: 10.1074/jbc.M406360200. [DOI] [PubMed] [Google Scholar]
- [18].Ko KW, Paul A, Ma K, Li L, Chan L. Endothelial lipase modulates HDL but has no effect on atherosclerosis development in apoE−/− and LDLR−/− mice. J Lipid Res. 2005;46:2586–2594. doi: 10.1194/jlr.M500366-JLR200. [DOI] [PubMed] [Google Scholar]
- [19].Escola-Gil JC, Chen X, Julve J, Quesada H, Santos D, Metso J, Tous M, Jauhiainen M, Blanco-Vaca F. Hepatic lipase- and endothelial lipase-deficiency in mice promotes macrophage-to-feces RCT and HDL antioxidant properties. Biochim Biophys Acta. 2013;1831:691–697. doi: 10.1016/j.bbalip.2013.01.003. [DOI] [PubMed] [Google Scholar]
- [20].Hara T, Ishida T, Kojima Y, Tanaka H, Yasuda T, Shinohara M, Toh R, Hirata K. Targeted deletion of endothelial lipase increases HDL particles with anti-inflammatory properties both in vitro and in vivo. J Lipid Res. 2011;52:57–67. doi: 10.1194/jlr.M008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Trieb M, Horvath A, Birner-Gruenberger R, Spindelboeck W, Stadlbauer V, Taschler U, Curcic S, Stauber RE, Holzer M, Pasterk L, Heinemann A, et al. Liver disease alters high-density lipoprotein composition, metabolism and function. Biochim Biophys Acta. 2016;1861:630–638. doi: 10.1016/j.bbalip.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gauster M, Hrzenjak A, Schick K, Frank S. Endothelial lipase is inactivated upon cleavage by the members of the proprotein convertase family. J Lipid Res. 2005;46:977–987. doi: 10.1194/jlr.M400500-JLR200. [DOI] [PubMed] [Google Scholar]
- [23].Curcic S, Holzer M, Pasterk L, Knuplez E, Eichmann TO, Frank S, Zimmermann R, Schicho R, Heinemann A, Marsche G. Secretory phospholipase A2 modified HDL rapidly and potently suppresses platelet activation. Sci Rep. 2017;7 doi: 10.1038/s41598-017-08136-1. 8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res. 2008;49:1137–1146. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Knittelfelder OL, Weberhofer BP, Eichmann TO, Kohlwein SD, Rechberger GN. A versatile ultra-high performance LC-MS method for lipid profiling. J Chromatogr B Analyt Technol Biomed Life Sci. 2014;951–952:119–128. doi: 10.1016/j.jchromb.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Jansson J, Schillen K, Nilsson M, Soderman O, Fritz G, Bergmann A, Glatter O. Small-angle X-ray scattering, light scattering, and NMR study of PEO-PPO-PEO triblock copolymer/cationic surfactant complexes in aqueous solution. J Phys Chem B. 2005;B109:7073–7083. doi: 10.1021/jp0468354. [DOI] [PubMed] [Google Scholar]
- [27].Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J Appl Crystallogr. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Didichenko SA, Navdaev AV, Cukier AM, Gille A, Schuetz P, Spycher MO, Therond P, Chapman MJ, Kontush A, Wright SD. Enhanced HDL functionality in small HDL species produced upon remodeling of HDL by reconstituted HDL, CSL112: effects on cholesterol efflux, anti-inflammatory and antioxidative activity. Circ Res. 2016;119:751–763. doi: 10.1161/CIRCRESAHA.116.308685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Holzer M, Kern S, Birner-Grunberger R, Curcic S, Heinemann A, Marsche G. Refined purification strategy for reliable proteomic profiling of HDL2/3: impact on proteomic complexity. Sci Rep. 2016;6 doi: 10.1038/srep38533. 38533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jurgens G, Ashy A, Esterbauer H. Detection of new epitopes formed upon oxidation of low-density lipoprotein, lipoprotein (a) and very-low-density lipoprotein. Use of an antiserum against 4-hydroxynonenal-modified low-density lipoprotein. Biochem J. 1990;265:605–608. doi: 10.1042/bj2650605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Esterbauer H, Gebicki J, Puhl H, Jurgens G. The role of lipid peroxidation and antioxidants in oxidative modification of LDL. Free Radic Biol Med. 1992;13:341–390. doi: 10.1016/0891-5849(92)90181-f. [DOI] [PubMed] [Google Scholar]
- [32].Pilz J, Meineke I, Gleiter CH. Measurement of free and bound malondialdehyde in plasma by high-performance liquid chromatography as the 2,4-dinitrophenylhydrazine derivative. J Chromatogr B Biomed Sci Appl. 2000;742:315–325. doi: 10.1016/s0378-4347(00)00174-2. [DOI] [PubMed] [Google Scholar]
- [33].Horl G, Froehlich H, Ferstl U, Ledinski G, Binder J, Cvirn G, Stojakovic T, Trauner M, Koidl C, Tafeit E, Amrein K, et al. Simvastatin efficiently lowers small LDL-IgG immune complex levels: a therapeutic quality beyond the lipid-lowering effect. PLoS One. 2016;11:e0148210. doi: 10.1371/journal.pone.0148210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Esterbauer H, Lang J, Zadravec S, Slater TF. Detection of malonaldehyde by high-performance liquid chromatography. Methods Enzymol. 1984;105:319–328. doi: 10.1016/s0076-6879(84)05041-2. [DOI] [PubMed] [Google Scholar]
- [35].Garner B, Waldeck AR, Witting PK, Rye KA, Stocker R. Oxidation of high density lipoproteins. II. Evidence for direct reduction of lipid hydroperoxides by methionine residues of apolipoproteins AI and AII. J Biol Chem. 1998;273:6088–6095. doi: 10.1074/jbc.273.11.6088. [DOI] [PubMed] [Google Scholar]
- [36].Oda T, Tokushige M. Chemical modification of tryptophanase by chloramine T: a possible involvement of the methionine residue in enzyme activity. J Biochem. 1988;104:178–183. doi: 10.1093/oxfordjournals.jbchem.a122437. [DOI] [PubMed] [Google Scholar]
- [37].Camont L, Lhomme M, Rached F, Le Goff W, Negre-Salvayre A, Salvayre R, Calzada C, Lagarde M, Chapman MJ, Kontush A. Small, dense high-density lipoprotein-3 particles are enriched in negatively charged phospholipids: relevance to cellular cholesterol efflux, antioxidative, antithrombotic, anti-inflammatory, and antiapoptotic functionalities. Arterioscler Thromb Vasc Biol. 2013;33:2715–2723. doi: 10.1161/ATVBAHA.113.301468. [DOI] [PubMed] [Google Scholar]
- [38].Riederer M, Kofeler H, Lechleitner M, Tritscher M, Frank S. Impact of endothelial lipase on cellular lipid composition. Biochim Biophys Acta. 2012;1821:1003–1011. doi: 10.1016/j.bbalip.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kontush A, Chantepie S, Chapman MJ. Small, dense HDL particles exert potent protection of atherogenic LDL against oxidative stress. Arterioscler Thromb Vasc Biol. 2003;23:1881–1888. doi: 10.1161/01.ATV.0000091338.93223.E8. [DOI] [PubMed] [Google Scholar]
- [40].Kontush A, Chapman MJ. Antiatherogenic small, dense HDL—guardian angel of the arterial wall? Nat Clin Pract Cardiovasc Med. 2006;3:144–153. doi: 10.1038/ncpcardio0500. [DOI] [PubMed] [Google Scholar]
- [41].Kontush A, Therond P, Zerrad A, Couturier M, Negre-Salvayre A, de Souza JA, Chantepie S, Chapman MJ. Preferential sphingosine-1-phosphate enrichment and sphingomyelin depletion are key features of small dense HDL3 particles: relevance to antiapoptotic and antioxidative activities. Arterioscler Thromb Vasc Biol. 2007;27:1843–1849. doi: 10.1161/ATVBAHA.107.145672. [DOI] [PubMed] [Google Scholar]
- [42].Kumpula LS, Kumpula JM, Taskinen MR, Jauhiainen M, Kaski K, Ala-Korpela M. Reconsideration of hydrophobic lipid distributions in lipoprotein particles. Chem Phys Lipids. 2008;155:57–62. doi: 10.1016/j.chemphyslip.2008.06.003. [DOI] [PubMed] [Google Scholar]
- [43].Seu KJ, Cambrea LR, Everly RM, Hovis JS. Influence of lipid chemistry on membrane fluidity: tail and headgroup interactions. Biophys J. 2006;91:3727–3735. doi: 10.1529/biophysj.106.084590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Rached F, Lhomme M, Camont L, Gomes F, Dauteuille C, Robillard P, Santos RD, Lesnik P, Serrano CV, Jr, Chapman MJ, Kontush A. Defective functionality of small, dense HDL3 subpopulations in ST segment elevation myocardial infarction: relevance of enrichment in lysophosphatidylcholine, phosphatidic acid and serum amyloid A. Biochim Biophys Acta. 2015;1851:1254–1261. doi: 10.1016/j.bbalip.2015.05.007. [DOI] [PubMed] [Google Scholar]
- [45].Qiu G, Hill JS. Endothelial lipase promotes apolipoprotein AI-mediated cholesterol efflux in THP-1 macrophages. Arterioscler Thromb Vasc Biol. 2009;29:84–91. doi: 10.1161/ATVBAHA.108.176487. [DOI] [PubMed] [Google Scholar]
- [46].Persegol L, Darabi M, Dauteuille C, Lhomme M, Chantepie S, Rye KA, Therond P, Chapman MJ, Salvayre R, Negre-Salvayre A, Lesnik P, et al. Small dense HDLs display potent vasorelaxing activity, reflecting their elevated content of sphingosine-1-phosphate. J Lipid Res. 2018;59:25–34. doi: 10.1194/jlr.M076927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Tall AR. Plasma high density lipoproteins: therapeutic targeting and links to atherogenic inflammation. Atherosclerosis. 2018;276:39–43. doi: 10.1016/j.atherosclerosis.2018.07.004. [DOI] [PubMed] [Google Scholar]
- [48].Tani M, Horvath KV, Lamarche B, Couture P, Burnett JR, Schaefer EJ, Asztalos BF. High-density lipoprotein subpopulation profiles in lipoprotein lipase and hepatic lipase deficiency. Atherosclerosis. 2016;253:7–14. doi: 10.1016/j.atherosclerosis.2016.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Brites F, Martin M, Guillas I, Kontush A. Antioxidative activity of high-density lipoprotein (HDL): mechanistic insights into potential clinical benefit. BBA Clin. 2017;8:66–77. doi: 10.1016/j.bbacli.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.