Abstract
BACKGROUND
Colon cancer is among the most commonly diagnosed cancers in the United States with an estimated 97220 new cases expected by the end of 2018. It affects 1.2 million people around the world and is responsible for about 0.6 million deaths every year. Despite decline in overall incidence and mortality over the past 30 years, there continues to be an alarming rise in early-onset colon cancer cases (< 50 years). Patients are often diagnosed at late stages of the disease and tend to have poor survival. We previously showed that the WNT “gatekeeper” gene, secreted frizzled-related protein 4 (SFRP4), is over-expressed in early-onset colon cancer. SFRP4 is speculated to play an essential role in cancer by inhibiting the epithelial mesenchymal transition (EMT).
AIM
To investigate the correlation between SFRP4 expression and EMT-linked genes in colon cancer and how it affects patient survival.
METHODS
SFRP4 expression relative to that of EMT-linked genes and survival analysis were performed using the University of California Santa Cruz Cancer Browser interface.
RESULTS
SFRP4 was found to be co-expressed with the EMT-linked markers CDH2, FN1, VIM, TWIST1, TWIST2, SNAI1, SNAI2, ZEB1, ZEB2, POSTN, MMP2, MMP7, MMP9, and COL1A1. SFRP4 expression negatively correlated with the EMT-linked suppressors CLDN4, CLDN7, TJP3, MUC1, and CDH1. The expression of SFRP4 and the EMT-linked markers was higher in mesenchymal-like samples compared to epithelial-like samples which potentially implicates SFRP4-EMT mechanism in colon cancer. Additionally, patients overexpressing SFRP4 presented with poor overall survival (P = 0.0293).
CONCLUSION
Considering the implication of SFRP4 in early-onset colon cancer, particularly in the context of EMT, tumor metastasis, and invasion, and the effect of increased expression on colon cancer patient survival, SFRP4 might be a potential biomarker for early-onset colon cancer that could be targeted for diagnosis and/or disease therapy.
Keywords: Secreted frizzled-related protein 4, Epithelial mesenchymal transition, Colon cancer, Survival
Core tip: We have previously shown that secreted frizzled-related protein 4 (SFRP4) is over-expressed in colon cancer, especially in patients younger than 50 years. As these early-onset colon cancers tend to be more aggressive and negatively affecting patients’ survival, we sought to evaluate whether SFRP4 is co-expressed with epithelial-mesenchymal transition-linked genes that play key roles in cancer progression. Our results using a large colon adenocarcinoma-The Cancer Genome Atlas cohort revealed that SFRP4 is over-expressed in colon cancer patients together with epithelial-mesenchymal transition-linked genes. Moreover, colon cancer patients with high expression levels of SFRP4 showed significantly poorer survival relative to colon cancer patients with lower SFRP4 expression.
INTRODUCTION
Colon cancer is an important contributor to worldwide cancer morbidity and mortality, affecting an estimated 1.2 million individuals and responsible for 0.6 million deaths every year[1]. In the United States, colon cancer is among the most commonly diagnosed cancer with an estimated 97220 new cases expected by the end of 2018 and an overall life time risk of about 4.49% for men and 4.15% for women[2,3]. Despite the decline in overall incidence and mortality of colon cancer over the past 30 years[4], there continues to be an alarming rise in the number of early-onset colon cancer cases in individuals younger than 50 years[5]. Using the Surveillance Epidemiology and End Results Program Cancer Registries database, we previously demonstrated that this rise is most significant within the age group 40-44[6]. About 70%-80% of early-onset colon cancer cases are sporadic, and these patients have aggressive features, are often diagnosed at a more advanced stage, and tend to have poor survival[5-11].
In 2016, we showed that the secreted frizzled-related protein 4 (SFRP4) gene was uniquely over-expressed in early-onset colon cancer patients[12]. This is a protein coding gene and a member of the SFRP family. The cysteine-rich domain of SFRP4 is homologous to the putative WNT-binding site of frizzled proteins, making SFRP4, like other SFRPs, a modulator of WNT signaling[13,14]. As a WNT inhibitor, SFRP4 prevents cell growth and proliferation and has been implicated in Pyle disease[15]. SFRP4 is expressed in mouse embryonic mesenchyme, cardiovascular system, and musculoskeletal system[16]. Reports show that SFRP4 plays an essential role in ovarian cancer pathogenicity by inhibiting epithelial-mesenchymal transition (EMT) and cell migration, while promoting cell adhesion[13].
Although EMT is a typical process in embryonic development, heart valve formation, and other developmental processes, its role in promoting cancer metastasis is well documented[17,18]. EMT provides a molecular environment that allows epithelial cells to lose polarity and cell-to-cell adhesion and gain migratory and invasive properties to become mesenchymal cells[17,18]. The relationship between SFRP4 and EMT-linked genes in colon cancer pathogenesis and the implication of SFRP4 overexpression in patient survival has not been investigated previously. In this study, we demonstrate a correlation between SFRP4 gene expression and the expression of some key EMT-linked genes in colon cancer and show that the overexpression of SFRP4 associates with poor survival in colon cancer patients.
MATERIALS AND METHODS
Ethics statement
This study does not involve human subjects. It utilizes the publicly available de-identified colon adenocarcinoma (COAD) data set from The Cancer Genome Atlas (TCGA).
TCGA program
A landmark cancer genomics program molecularly characterized over 20000 primary cancer and matched normal samples spanning 33 cancer types. TCGA was formed in 2005 when the U.S. National Cancer and National Human Genome Research Institutes teamed together to support the launch of the project to map comprehensively various cancer genomic changes (https://www.cancer.gov/about-nci/organization/ ccg/research/structural-genomics/tcga/history/timeline). This joint effort brought together researchers from diverse disciplines and multiple institutions. Over the next dozen years, TCGA generated over 2.5 petabytes of genomic, epigenomic, transcriptomic, and proteomic data. The data, which has already led to improvements in our ability to diagnose, treat, and prevent cancer, will remain publicly available for anyone in the research community to use (https:// www.cancer.gov /about-nci/organization/ccg/research/structural-genomics/tcga).
As of March 2019, the TCGA data portal contains 365463 pieces of genomic and clinical data for 45 projects; 68 primary sites; 33549 cases; 22872 genes, and 3142246 mutations. The TCGA-COAD project contains 461 cases including two primary sites, colon and rectosigmoid junction, and the following disease types: Adenomas and adenocarcinomas, complex epithelial neoplasms; cystic, mucinous, and serous neoplasms, epithelial neoplasms, and no other specified carcinomas.
Data set
A total of 286 of 462 “Primary Tumor” samples from TCGA-COAD cohort was used in this study for gene expression and Kaplan-Meier survival analyses[19]. Table 1 summarizes the gender, age, and tumor stage of samples.
Table 1.
Gender, age and tumor stage of The Cancer Genome Atlas-colon adenocarcinoma dataset used for analysis
| Percentage | |
| Gender | |
| Female | 46.8 |
| Male | 52.4 |
| Age of initial diagnosis | |
| Average | 66.9 ± 13 |
| Minimum | 31 |
| maximum | 90 |
| Tumor stage | |
| Stage I | 16.2 |
| Stage IA | 0.2 |
| Stage II | 6.5 |
| Stage IIA | 29.9 |
| Stage IIB | 2.2 |
| Stage IIC | 0.2 |
| Stage III | 4.3 |
| Stage IIIA | 1.7 |
| Stage IIIB | 12.8 |
| Stage IIIC | 8.9 |
| Stage IV | 10.0 |
| Stage IVA | 3.7 |
| Stage IVB | 0.4 |
| Not reported | 2.4 |
| Discrepant | 0.6 |
Gene expression
SFRP4 gene expression relative to the EMT genes, Fibronectin (FN1), Vimentin (VIM), Zinc Finger E-Box Binding Homeobox 1 (ZEB1), ZEB2, Twist Family BHLH Transcription Factor 1 (TWIST1), TWIST2, Snail Family Transcriptional Repressor 1 (SNAI1), SNAI2, N-cadherin (CDH2), Claudin 4 (CLDN4), CLDN7, Tight Junction Protein 3 (TJP3), Mucin 1 (MUC1), and E-Cadherin (CDH1), was performed using the TCGA-COAD dataset as previously described[20]. Pearson’s and Spearman’s rank coefficients and two-dimensional correlation scatter plots were generated using the University of California Santa Cruz Cancer Browser Interface (https://xena browser.net)[21].
Survival analysis
The TCGA-COAD “Primary tumor” dataset was used to perform and visualize Kaplan-Meier overall survival analysis using the University of California Santa Cruz Xena Browser for cancer genomics (https://xenabrowser.net/datapages/?Cohort-TCGA%20Colon%20Cancer%20(COAD))[22,23]. P values were obtained using the Kaplan-Meier Estimator.
RESULTS
Co-expression of SFRP4 with EMT genes
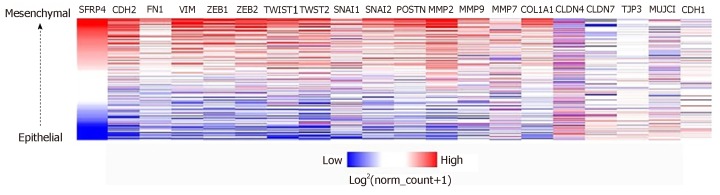
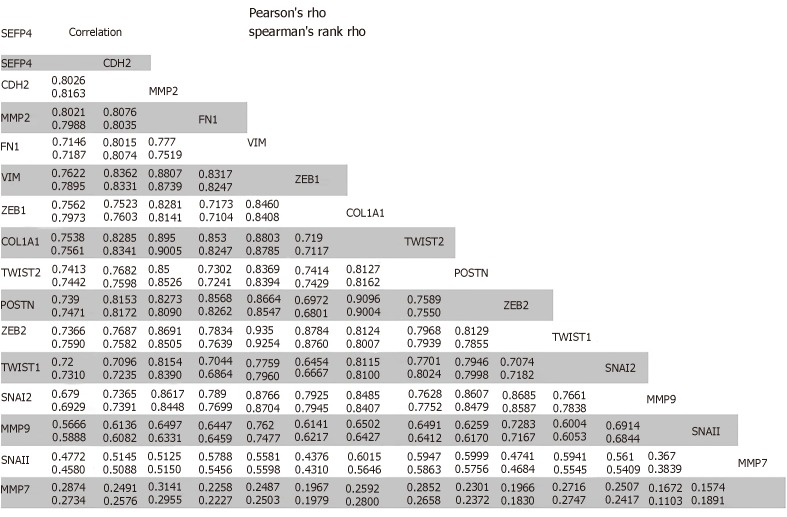
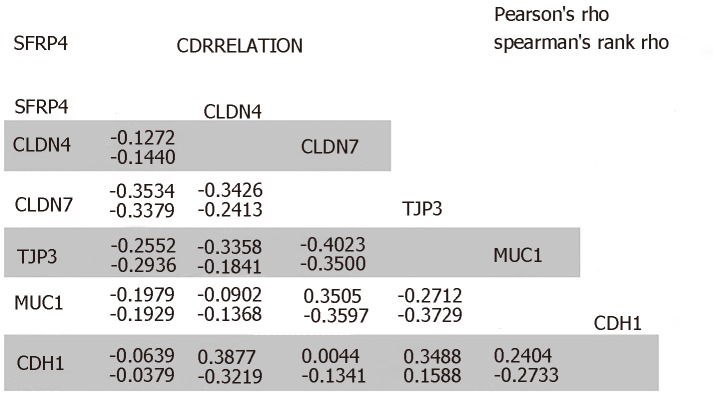
The role of SFRP4 in epithelial-mesenchymal transition was investigated in colon cancer by comparing its expression to that of some key EMT-linked genes using 286 colon cancer patients. We observed co-expression and a positive correlation by Pearson’s and Spearman’s rank coefficient analysis of SFRP4 with the EMT markers CDH2, FN1, VIM, ZEB1, ZEB2, TWIST1, TWIST2, SNAI1, SNAI2, Periostin (POSTN), Matrix Metallopeptidase 2 (MMP2), MMP9, MMP7, and Collagen 1 (COL1A1) (Figure 1 and Figure 2). A higher expression of SFRP4 and the EMT-linked markers was observed in samples that were more mesenchymal than epithelial. A negative correlation was observed between SFRP4 and CLDN4, CLDN7, TJP3, MUC1, and CDH1 (Figure 1 and Figure 3). Based on the Pearson’s and Spearman’s rank coefficients, SFRP4 shows the strongest correlation with CDH2 followed by MMP2, and the least correlation with CLDN7 (Figure 2 and Figure 3). These data suggest a role for SFRP4 in the EMT molecular pathway.
Figure 1.
SFRP4 co-expresses with epithelial mesenchymal transition-linked genes in colon cancer. SFRP4 and the epithelial mesenchymal transition-linked genes have higher expression in samples that are more mesenchymal (red) compared to the more epithelial samples with lower expression (blue).
Figure 2.
Epithelial mesenchymal transition-linked genes with positive correlation with SFRP4 expression.
Figure 3.
Epithelial mesenchymal transition-linked genes with negative correlation SFRP4 express.
Colon cancer patients over-expressing SFRP4 have poor overall survival
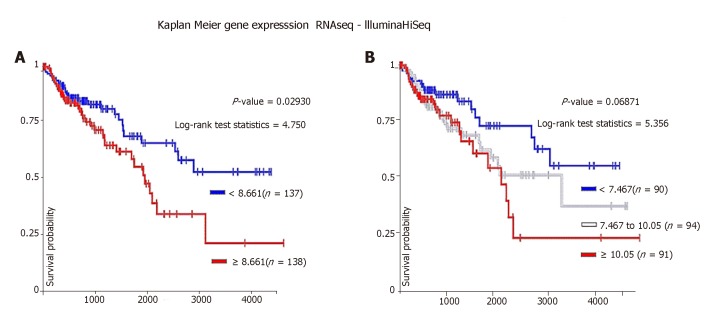
We analyzed the effect of SFRP4 overexpression on colon cancer patient survival. Our data showed that colon cancer patients with higher SFRP4 expression (≥ 8.661) had poor overall survival compared to patients with lower SFRP4 expression (< 8.661) (Figure 4A). A similar trend was observed when patient cohort was divided into three groups. Patients with intermediate levels of SFRP4 expression (7.467-10.05) showed survival curve that was between that of patients with higher (≥ 10.05) and lower (< 7.467) expression (Figure 4B). These data demonstrate that SFRP4 has an impact on colon cancer patient survival.
Figure 4.
Overall survival of colon cancer patients with low, intermediate or high SFRP4 expression. A: Patients expressing higher levels of SFRP4 (red line) had poor survival compared to those expressing lower SFRP4 levels (blue line); (B) When a three group analysis was performed, patients with higher SFRP4 expression (red line) presented with poor overall survival compared to patients with intermediate (white lines) or low expression (blue line).
DISCUSSION
Very little is known about the role of SFRP4 in cancer pathogenesis, let alone its mechanism of action. Most of the speculations on SFRP4 mechanism appear to focus on its role as a WNT signaling modulator, through which it suppresses epithelial mesenchymal transition, a mechanism that is implicated in cancer metastasis and invasion[13,17,18]. In ovarian cancer, SFRP4 regulates metastasis by inhibiting EMT-linked genes, cell proliferation, and migration, while promoting apoptosis and upregulating cell adhesion molecules such as CDH1[13,24]. A Study by Muley et al[25] showed that SFRP4 can restrict ovarian serous cystadenocarcinoma growth in mice by interfering with endothelial cell function, suggesting SFRP4 as a potent angiogenesis inhibitor. While these observations indicate that SFRP4 represses cancer pathogenesis, this is in contrast to our previous observation of SFRP4 upregulation in early-onset colon cancer[12] or its strong correlation with epithelial mesenchymal-linked genes observed in this study.
Our previous report on the overexpression of SFRP4 in early-onset colon cancer informed us of the potential role of SFRP4 in colon cancer particularly with respect to age of onset[12]. Our data showed that SFRP4 co-expresses and correlates positively with the EMT-linked genes CDH2, FN1, VIM, SNAI1, SNAI2, ZEB1, ZEB2, TWIST1, POSTN, COL1A1, MMP2, MMP7, and MMP9, many of which promote epithelial mesenchymal transition by repressing proteins that maintain tissue integrity such as CDH1[26,27]. Some of the hallmarks of EMT are cell separation and invasion, characterized by loss of cell-cell adhesion and gain of migratory and invasive properties[18]. Given the role of SFRP4 as an EMT inhibitor, overexpression in early-onset colon cancer might be a response mechanism to serve as a check on EMT in the wake of increase expression of EMT genes.
However, overexpression of SFRP4 may not always correlate with EMT suppression. There is accumulating evidence that SFRP family proteins may also have oncogenic potential in certain cancer types[28,29]. While the binding and sequestering of WNT ligand by SFRPs inhibit WNT signaling[30-33], binding may also allow SFRPs to act as carriers for WNT proteins in a diffusion gradient, thus expanding the range and activity of WNT signaling. This has been reported in Xenopus where SFRP3 and Crescent were shown potentially to activate canonical WNT signaling by promoting the diffusion of WNT8 and WNT11[34]. There have also been reports on the dimerization of the N-terminal cysteine-rich domain of SFRPs and Frizzled proteins, hence suggesting SFRPs direct activation of WNT signaling by mimicking the WNT ligand[35-37]. Additionally, it has been suggested that by antagonizing each other, the SFRPs indirectly activate WNT signaling, as seen in rat renal organogenesis where SFRP2 was reported to inhibit SFRP1 activities[38]. Also, studies have shown that WNT regulation could be concentration dependent with low SFRP concentrations activating WNT signaling and high concentrations inhibiting it[39].
Consistent with the concept of SFRPs functioning as oncogenes, SFRP1 has been reported to be over-expressed in basal-like breast cancers and implicated in brain-specific metastases, while SFRP2 induces mammary tumor formation in canines by blocking apoptosis[40,41]. Both SFRP1 and SFRP2 promote cellular invasion, proliferation, and in vivo tumor growth in renal cancer[42,43], and SFRP1 in gastric cancer. SFRP2 enhances WNT3A signaling in HEK293 and HSG cells[44] and activates WNT signaling through direct interaction with FZ4/7 in the mouse[45]. Interestingly, overexpression of SFRP4 has also been observed in stomach cancer where it associates with poor patient outcomes and interacts strongly with EMT-linked genes in gliomas[29]. Taken together, these observations suggest that besides being a WNT signaling inhibitor, the SFRP family members, including SFRP4, also have oncogenic potentials, and their mechanism of action might be context dependent or cancer type dependent.
Consistent with the important role MMP2 plays in the breakdown of the extracellular matrix and the EMT process, particularly in cell invasion and metastasis, SFRP4 expression was strongly correlated with MMP2 . In addition to CDH1, SFRP4 was negatively correlated with many other cell adhesion molecules. Given the role these genes play as gatekeepers of EMT and metastasis, it follows that SFRP4 is upregulated with EMT following downregulation of cell-adhesion molecules. Overexpression of SFRP4 is also consistent with overexpression of other EMT regulators we previously reported in early-onset colon cancer, such as cartilage oligomeric matrix protein, (COMP) which catalyzes collagen fibrillation to maintain the extracellular matrix[20,46].
Overexpression of SFRP4 in colon cancer patients resulted in poor survival that correlated with the survival analysis observed for colon cancer patients overexpressing COMP[20]. Given the implication of WNT signaling in many developmental and disease pathways[47-49] and the knowledge that SFRP4 inhibits WNT signaling, it is possible that early deaths might be due to secondary complications following overexpression of SFRP4, which interferes with the normal functions of neighboring healthy cells. However, it is also possible that early deaths are due primarily to disease independent of SFRP4, whose overexpression might only be a consequence of the disease.
Although high SFRP expression in colon cancer was observed in this study, others have suggested that SFPR4 expression in tumor may be contributed by the stroma fraction. This was documented in studies by Vincent et al[29] where a strong correlation between SFRP4 and the stroma was observed in 14 cancer types, including colorectal. Nevertheless, considering that the stroma constitutes an integral part of the tumor microenvironment, its expression of SFRP4 will undoubtedly constitute an important disease mechanism for potential targeted therapy.
In summary, we had previous shown that SFRP4 was over-expressed in early-onset colon cancer[12] and report here SFRP4 expression was correlated with EMT-linked genes. These findings highlight SFRP4 as an important contributor to the pathogenesis of colon cancer, particularly early-onset colon cancer, and therefore as a potential biomarker for the early diagnosis of colon cancer. Considering that SFRP4 modulates WNT signaling during cell proliferation, migration, and EMT, which are all hallmarks of cancer pathogenesis. Overexpression of SFRP4 in early-onset colon cancer and co-expression with EMT-linked genes provide an important first step towards understanding its molecular mechanism in young patients, particularly in the context of cancer metastasis and cell invasion.
ARTICLE HIGHLIGHTS
Research background
Colon cancer affects 1.2 million people around the world and causes about 0.6 million deaths annually. Although the overall incidence and mortality is decreasing, there has been an alarming increase in early-onset cases (< 50 years). These younger patients are often diagnosed at late stages with more aggressive tumors and tend to have poorer survival.
Research motivation
We previously showed that secreted frizzled-related protein 4 (SFRP4), speculated to play an essential role in cancer by inhibiting epithelial mesenchymal transition (EMT), is over-expressed in younger patients.
Research objectives
The aim of our study was to investigate whether there is a correlation between SFRP4 expression and EMT-linked genes in colon cancer and whether SFRP4 over-expression affects patient survival using a larger cohort of patients.
Research methods
Correlation between SFRP4 expression and EMT-linked genes along with survival analysis were performed using the University of California Santa Cruz Cancer browser interface using publicly available The Cancer Genome Atlas-colon adenocarcinoma cohort of colon cancer cases.
Research results
SFRP4 is co-expressed with EMT-linked markers, such as CDH2, FN1, VIM, TWIST1, TWIST2, SNAI1, SNAI2, ZEB1, ZEB2, POSTN, MMP2, MMP7, MMP9, and COL1A1, in colon cancer patients. SFRP4 expression negatively correlated with the EMT-linked suppressors CLDN4, CLDN7, TJP3, MUC1, and CDH1. Mesenchymal-like samples showed higher expression of SFRP4 and EMT-linked markers relative to epithelial-like samples. This implicates SFRP4 in colon cancer EMT mechanism. Additionally, patients with colon tumors over-expressing SFRP4 presented with significantly poorer overall survival.
Research conclusions
Results of this study suggest a potential role of SFRP4 in colon cancer aggressiveness, disease progression, and poorer patient survival. Although the role of SFRPs including SFRP4 as WNT signaling inhibitors is well documented in the literature, data presented in this manuscript suggest that the SFRP family proteins might also activate WNT signaling and promote cell proliferation, therefore demonstrating its oncogenic potential and suggesting its mechanism of action might be context dependent.
Research perspectives
More extensive and detailed mechanistic in vitro and in vivo studies will have to be performed to confirm these initial observations in colon cancer patients.
ACKNOWLEDGEMENTS
We would like to thank the patients without whom this work would not be possible.
Footnotes
Institutional review board statement: This study does not involve human subjects. It utilizes the publicly available de-identified colon adenocarcinoma data set from The Cancer Genome Atlas.
Conflict-of-interest statement: All authors declare no conflict of interest.
Manuscript source: Unsolicited manuscript
Peer-review started: February 15, 2019
First decision: March 14, 2019
Article in press: May 23, 2019
Specialty type: Oncology
Country of origin: United States
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Tarnawski AS, Kang GH S-Editor: Wang JL L-Editor: Filipodia E-Editor: Zhou BX
Contributor Information
Landry E Nfonsam, Department of Genetics, Children’s Hospital of Eastern Ontario, Ottawa, Ontario K1H 8L1, Canada.
Jana Jandova, Department of Surgery, University of Arizona, Tucson, AZ 85724, United States.
Hunter C Jecius, Department of Surgery, University of Arizona, Tucson, AZ 85724, United States.
Pamela N Omesiete, Department of Surgery, University of Arizona, Tucson, AZ 85724, United States.
Valentine N Nfonsam, Department of Surgery, University of Arizona, Tucson, AZ 85724, United States. vnfonsam@surgery.arizona.edu.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.American Cancer Society. Key Statistics for Colorectal Cancer 2018. Available from: https://www.cancer.org/cancer/colon-rectal-cancer/about/key-statistics.html.
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 5.Kirzin S, Marisa L, Guimbaud R, De Reynies A, Legrain M, Laurent-Puig P, Cordelier P, Pradère B, Bonnet D, Meggetto F, Portier G, Brousset P, Selves J. Sporadic early-onset colorectal cancer is a specific sub-type of cancer: a morphological, molecular and genetics study. PLoS One. 2014;9:e103159. doi: 10.1371/journal.pone.0103159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis DM, Marcet JE, Frattini JC, Prather AD, Mateka JJ, Nfonsam VN. Is it time to lower the recommended screening age for colorectal cancer? J Am Coll Surg. 2011;213:352–361. doi: 10.1016/j.jamcollsurg.2011.04.033. [DOI] [PubMed] [Google Scholar]
- 7.Bacani J, Zwingerman R, Di Nicola N, Spencer S, Wegrynowski T, Mitchell K, Hay K, Redston M, Holowaty E, Huntsman D, Pollett A, Riddell R, Gallinger S. Tumor microsatellite instability in early onset gastric cancer. J Mol Diagn. 2005;7:465–477. doi: 10.1016/S1525-1578(10)60577-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang DT, Pai RK, Rybicki LA, Dimaio MA, Limaye M, Jayachandran P, Koong AC, Kunz PA, Fisher GA, Ford JM, Welton M, Shelton A, Ma L, Arber DA, Pai RK. Clinicopathologic and molecular features of sporadic early-onset colorectal adenocarcinoma: an adenocarcinoma with frequent signet ring cell differentiation, rectal and sigmoid involvement, and adverse morphologic features. Mod Pathol. 2012;25:1128–1139. doi: 10.1038/modpathol.2012.61. [DOI] [PubMed] [Google Scholar]
- 9.Giráldez MD, Balaguer F, Bujanda L, Cuatrecasas M, Muñoz J, Alonso-Espinaco V, Larzabal M, Petit A, Gonzalo V, Ocaña T, Moreira L, Enríquez-Navascués JM, Boland CR, Goel A, Castells A, Castellví-Bel S. MSH6 and MUTYH deficiency is a frequent event in early-onset colorectal cancer. Clin Cancer Res. 2010;16:5402–5413. doi: 10.1158/1078-0432.CCR-10-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myers EA, Feingold DL, Forde KA, Arnell T, Jang JH, Whelan RL. Colorectal cancer in patients under 50 years of age: a retrospective analysis of two institutions' experience. World J Gastroenterol. 2013;19:5651–5657. doi: 10.3748/wjg.v19.i34.5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zahir MN, Azhar EM, Rafiq S, Ghias K, Shabbir-Moosajee M. Clinical features and outcome of sporadic colorectal carcinoma in young patients: a cross-sectional analysis from a developing country. ISRN Oncol. 2014;2014:461570. doi: 10.1155/2014/461570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jandova J, Xu W, Nfonsam V. Sporadic early-onset colon cancer expresses unique molecular features. J Surg Res. 2016;204:251–260. doi: 10.1016/j.jss.2016.04.068. [DOI] [PubMed] [Google Scholar]
- 13.Ford CE, Jary E, Ma SS, Nixdorf S, Heinzelmann-Schwarz VA, Ward RL. The Wnt gatekeeper SFRP4 modulates EMT, cell migration and downstream Wnt signalling in serous ovarian cancer cells. PLoS One. 2013;8:e54362. doi: 10.1371/journal.pone.0054362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J Cell Sci. 2003;116:2627–2634. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- 15.Kiper POS, Saito H, Gori F, Unger S, Hesse E, Yamana K, Kiviranta R, Solban N, Liu J, Brommage R, Boduroglu K, Bonafé L, Campos-Xavier B, Dikoglu E, Eastell R, Gossiel F, Harshman K, Nishimura G, Girisha KM, Stevenson BJ, Takita H, Rivolta C, Superti-Furga A, Baron R. Cortical-Bone Fragility--Insights from sFRP4 Deficiency in Pyle's Disease. N Engl J Med. 2016;374:2553–2562. doi: 10.1056/NEJMoa1509342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finger JH, Smith CM, Hayamizu TF, McCright IJ, Xu J, Law M, Shaw DR, Baldarelli RM, Beal JS, Blodgett O, Campbell JW, Corbani LE, Lewis JR, Forthofer KL, Frost PJ, Giannatto SC, Hutchins LN, Miers DB, Motenko H, Stone KR, Eppig JT, Kadin JA, Richardson JE, Ringwald M. The mouse Gene Expression Database (GXD): 2017 update. Nucleic Acids Res. 2017;45:D730–D736. doi: 10.1093/nar/gkw1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 18.Hay ED. An overview of epithelio-mesenchymal transformation. Acta Anat (Basel) 1995;154:8–20. doi: 10.1159/000147748. [DOI] [PubMed] [Google Scholar]
- 19.Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nfonsam VN, Nfonsam LE, Chen D, Omesiete PN, Cruz A, Runyan RB, Jandova J. COMP Gene Coexpresses With EMT Genes and Is Associated With Poor Survival in Colon Cancer Patients. J Surg Res. 2019;233:297–303. doi: 10.1016/j.jss.2018.08.021. [DOI] [PubMed] [Google Scholar]
- 21.Sanborn JZ, Benz SC, Craft B, Szeto C, Kober KM, Meyer L, Vaske CJ, Goldman M, Smith KE, Kuhn RM, Karolchik D, Kent WJ, Stuart JM, Haussler D, Zhu J. The UCSC Cancer Genomics Browser: update 2011. Nucleic Acids Res. 2011;39:D951–D959. doi: 10.1093/nar/gkq1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldman M, Craft B, Kamath A, Brooks AN, Zhu J, Haussler D. he UCSC Xena Platform for cancer genomics data visualization and interpretation. 2018 Preprint. Available from: bioRxiv:326470. [Google Scholar]
- 24.Maganga R, Giles N, Adcroft K, Unni A, Keeney D, Wood F, Fear M, Dharmarajan A. Secreted Frizzled related protein-4 (sFRP4) promotes epidermal differentiation and apoptosis. Biochem Biophys Res Commun. 2008;377:606–611. doi: 10.1016/j.bbrc.2008.10.050. [DOI] [PubMed] [Google Scholar]
- 25.Muley A, Majumder S, Kolluru GK, Parkinson S, Viola H, Hool L, Arfuso F, Ganss R, Dharmarajan A, Chatterjee S. Secreted frizzled-related protein 4: an angiogenesis inhibitor. Am J Pathol. 2010;176:1505–1516. doi: 10.2353/ajpath.2010.090465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eastham AM, Spencer H, Soncin F, Ritson S, Merry CL, Stern PL, Ward CM. Epithelial-mesenchymal transition events during human embryonic stem cell differentiation. Cancer Res. 2007;67:11254–11262. doi: 10.1158/0008-5472.CAN-07-2253. [DOI] [PubMed] [Google Scholar]
- 27.Guarino M, Rubino B, Ballabio G. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 2007;39:305–318. doi: 10.1080/00313020701329914. [DOI] [PubMed] [Google Scholar]
- 28.Vincent KM, Postovit LM. Matricellular proteins in cancer: a focus on secreted Frizzled-related proteins. J Cell Commun Signal. 2018;12:103–112. doi: 10.1007/s12079-017-0398-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vincent KM, Postovit LM. A pan-cancer analysis of secreted Frizzled-related proteins: re-examining their proposed tumour suppressive function. Sci Rep. 2017;7:42719. doi: 10.1038/srep42719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finch PW, He X, Kelley MJ, Uren A, Schaudies RP, Popescu NC, Rudikoff S, Aaronson SA, Varmus HE, Rubin JS. Purification and molecular cloning of a secreted, Frizzled-related antagonist of Wnt action. Proc Natl Acad Sci U S A. 1997;94:6770–6775. doi: 10.1073/pnas.94.13.6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leyns L, Bouwmeester T, Kim SH, Piccolo S, De Robertis EM. Frzb-1 is a secreted antagonist of Wnt signaling expressed in the Spemann organizer. Cell. 1997;88:747–756. doi: 10.1016/s0092-8674(00)81921-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin K, Wang S, Julius MA, Kitajewski J, Moos M, Jr, Luyten FP. The cysteine-rich frizzled domain of Frzb-1 is required and sufficient for modulation of Wnt signaling. Proc Natl Acad Sci U S A. 1997;94:11196–11200. doi: 10.1073/pnas.94.21.11196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang S, Krinks M, Lin K, Luyten FP, Moos M., Jr Frzb, a secreted protein expressed in the Spemann organizer, binds and inhibits Wnt-8. Cell. 1997;88:757–766. doi: 10.1016/s0092-8674(00)81922-4. [DOI] [PubMed] [Google Scholar]
- 34.Mii Y, Taira M. Secreted Frizzled-related proteins enhance the diffusion of Wnt ligands and expand their signalling range. Development. 2009;136:4083–4088. doi: 10.1242/dev.032524. [DOI] [PubMed] [Google Scholar]
- 35.Bafico A, Gazit A, Pramila T, Finch PW, Yaniv A, Aaronson SA. Interaction of frizzled related protein (FRP) with Wnt ligands and the frizzled receptor suggests alternative mechanisms for FRP inhibition of Wnt signaling. J Biol Chem. 1999;274:16180–16187. doi: 10.1074/jbc.274.23.16180. [DOI] [PubMed] [Google Scholar]
- 36.Dann CE, Hsieh JC, Rattner A, Sharma D, Nathans J, Leahy DJ. Insights into Wnt binding and signalling from the structures of two Frizzled cysteine-rich domains. Nature. 2001;412:86–90. doi: 10.1038/35083601. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez J, Esteve P, Weinl C, Ruiz JM, Fermin Y, Trousse F, Dwivedy A, Holt C, Bovolenta P. SFRP1 regulates the growth of retinal ganglion cell axons through the Fz2 receptor. Nat Neurosci. 2005;8:1301–1309. doi: 10.1038/nn1547. [DOI] [PubMed] [Google Scholar]
- 38.Yoshino K, Rubin JS, Higinbotham KG, Uren A, Anest V, Plisov SY, Perantoni AO. Secreted Frizzled-related proteins can regulate metanephric development. Mech Dev. 2001;102:45–55. doi: 10.1016/s0925-4773(01)00282-9. [DOI] [PubMed] [Google Scholar]
- 39.Bhat RA, Stauffer B, Komm BS, Bodine PV. Structure-function analysis of secreted frizzled-related protein-1 for its Wnt antagonist function. J Cell Biochem. 2007;102:1519–1528. doi: 10.1002/jcb.21372. [DOI] [PubMed] [Google Scholar]
- 40.Lee JL, Lin CT, Chueh LL, Chang CJ. Autocrine/paracrine secreted Frizzled-related protein 2 induces cellular resistance to apoptosis: a possible mechanism of mammary tumorigenesis. J Biol Chem. 2004;279:14602–14609. doi: 10.1074/jbc.M309008200. [DOI] [PubMed] [Google Scholar]
- 41.Smid M, Wang Y, Zhang Y, Sieuwerts AM, Yu J, Klijn JG, Foekens JA, Martens JW. Subtypes of breast cancer show preferential site of relapse. Cancer Res. 2008;68:3108–3114. doi: 10.1158/0008-5472.CAN-07-5644. [DOI] [PubMed] [Google Scholar]
- 42.Saini S, Liu J, Yamamura S, Majid S, Kawakami K, Hirata H, Dahiya R. Functional significance of secreted Frizzled-related protein 1 in metastatic renal cell carcinomas. Cancer Res. 2009;69:6815–6822. doi: 10.1158/0008-5472.CAN-09-1254. [DOI] [PubMed] [Google Scholar]
- 43.Yamamura S, Kawakami K, Hirata H, Ueno K, Saini S, Majid S, Dahiya R. Oncogenic functions of secreted Frizzled-related protein 2 in human renal cancer. Mol Cancer Ther. 2010;9:1680–1687. doi: 10.1158/1535-7163.MCT-10-0012. [DOI] [PubMed] [Google Scholar]
- 44.von Marschall Z, Fisher LW. Secreted Frizzled-related protein-2 (sFRP2) augments canonical Wnt3a-induced signaling. Biochem Biophys Res Commun. 2010;400:299–304. doi: 10.1016/j.bbrc.2010.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kress E, Rezza A, Nadjar J, Samarut J, Plateroti M. The frizzled-related sFRP2 gene is a target of thyroid hormone receptor alpha1 and activates beta-catenin signaling in mouse intestine. J Biol Chem. 2009;284:1234–1241. doi: 10.1074/jbc.M806548200. [DOI] [PubMed] [Google Scholar]
- 46.Halász K, Kassner A, Mörgelin M, Heinegård D. COMP acts as a catalyst in collagen fibrillogenesis. J Biol Chem. 2007;282:31166–31173. doi: 10.1074/jbc.M705735200. [DOI] [PubMed] [Google Scholar]
- 47.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 48.Steinhart Z, Angers S. Wnt signaling in development and tissue homeostasis. Development. 2018:145. doi: 10.1242/dev.146589. [DOI] [PubMed] [Google Scholar]
- 49.Yang Y. Wnt signaling in development and disease. Cell Biosci. 2012;2:14. doi: 10.1186/2045-3701-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]