Abstract
Fabry disease is a rare X-linked lysosomal storage disorder resulting from deficient activity of α-galactosidase A, leading to the accumulation of glycosphingolipids such as globotriaosylsphingosine (lyso-Gb3). The gastrointestinal symptoms of this disease may be disabling, and the life expectancy of affected patients is shortened by kidney and heart disease. Our hypothesis was that lyso-Gb3 may modify the gut microbiota. The impact of a clinically relevant concentration of lyso-Gb3 on mono- or multispecies bacterial biofilms were evaluated. A complex bacterial community from the simulated transverse colon microbiota was studied using quantitative PCR to estimate different bacterial group concentrations and a HPLC was used to estimate short-chain fatty acids concentrations. We found that lyso-Gb3 increased the biofilm-forming capacity of several individual bacteria, including Bacteroides fragilis and significantly increased the growth of B. fragilis in a multispecies biofilm. Lyso-Gb3 also modified the bacterial composition of the human colon microbiota suspension, increasing bacterial counts of B. fragilis, among others. Finally, lyso-Gb3 modified the formation of short-chain fatty acids, leading to a striking decrease in butyrate concentration. Lyso-Gb3 modifies the biology of gut bacteria, favoring the production of biofilms and altering the composition and short-chain fatty-acid profile of the gut microbiota.
Subject terms: Digestive signs and symptoms, Metabolic disorders
Introduction
Fabry disease is a rare X-linked lysosomal storage disorder caused by deficient activity of α-galactosidase A. This abnormality leads to lysosomal and extralysosomal accumulation of its substrate, globotriaosylceramide (Gb3), as well as other glycosphingolipids, such as globotriaosylsphingosine (lyso-Gb3), in a variety of cell types and plasma1,2. Classical Fabry disease first manifests in childhood, but more limited symptoms are observed in late-onset Fabry disease3,4. The symptoms are more severe in males but are also present in females5. Childhood disease is characterized by neuropathic pain, gastrointestinal symptoms, angiokeratoma, and hypohidrosis followed by development of proteinuric nephropathy, leading to end-stage renal disease requiring dialysis at a mean age of 40 years, left ventricular hypertrophy, arrhythmia, and stroke1,6. Current specific therapy includes replacement of the missing enzyme through biweekly parenteral administration of agalsidase and oral therapy with the chaperone migalastat4,7.
Most patients with Fabry disease report gastrointestinal symptoms such as abdominal pain, diarrhea, constipation, nausea, vomiting, and early satiety. These symptoms may be severe and negatively impact quality of life and body weight, potentially leading patients to undergo unnecessary surgical interventions8–11. While agalsidase therapy may improve gastrointestinal symptoms, this not always is the case, and there is a pressing need to better understand the pathogenic mechanisms of these symptoms4,5,7,12. Currently, there are two dominant hypotheses as to the mechanisms of the gastrointestinal symptoms reported in Fabry disease: dysfunction of autonomic neurons controlling gut motility13 on the one hand and vascular dysfunction and/or ischemia due to intestinal smooth-muscle or endothelial cell injury14 on the other. These mechanisms lead to a rapid gut transit time, impaired peristalsis, gastroparesis and intestinal stasis, bacterial overgrowth, and nutrient malabsorption15. Thus, gut-bacterial dysbiosis is thought to contribute to the gastrointestinal symptoms associated with Fabry disease, though until now it was thought to be secondary to stasis and dysmotility15. An altered gut microbiota may contribute to the pathogenesis and symptoms of both gastrointestinal and systemic diseases16–18. Indeed, gut biofilm–forming bacteria have been implicated in gastrointestinal disease19. Additionally, an altered microbiota may release uremic toxins or their precursors, which accelerate the progression of chronic kidney disease and cardiovascular disease, both key consequences of Fabry disease20–22, or may impair the release of protective molecules that modulate the inflammatory and immune responses, among others23.
We hypothesized that the metabolic derangement that takes place in Fabry disease may directly modify the biology of gut bacteria. Globotriaosylsphingosine (lyso-Gb3) is a deacylated form of Gb3 considered to be a diagnostic marker for Fabry disease24–26. Plasma lyso-Gb3 levels may increase up to several hundred-fold over normal control values, as compared to the 2-fold increase seen in serum Gb3. Lyso-Gb3 is more hydrosoluble than Gb3, is not trapped inside lipoproteins, and has been reported to contribute to the pathogenesis of kidney, vascular, and neuronal injury24,27–30. A dramatic increase in lyso-Gb3 concentrations was noted at tissue level in the liver and intestine of Fabry mice, clearly exceeding plasma levels24. These findings may suggest the existence of a “secret road”31: where lyso-Gb3 is secreted from the body via bile. Consequently, lyso-Gb3 may influence the gut microbiota and contribute to gastrointestinal symptoms or other manifestations of Fabry disease. Therefore, we aimed to evaluate the impact of lyso-Gb3 on intestinal bacteria in increasingly complex in vitro models. These included biofilm development in individual bacterial species; four-species biofilm, and a complex transverse colon microbiota pool sample from a dynamic human gut simulator. The results suggest that lysoGb3 may directly modify the microbiota composition as well as its secreted metabolites, potentially leading to systemic effects.
Methods
Bacteria
Five collection and 10 clinical strains were used. The collection strains supplied by American Type Culture Collection (ATCC) (Manassas, Virginia, USA) were Bacteroides fragilis ATCC 25285, Clostridium perfringens ATCC 13124, Enterococcus faecalis ATCC 29212, Escherichia coli ATCC 25922, and Klebsiella pneumoniae ATCC 23357. Furthermore, two strains of each species isolated from patient samples were used (Table 1). All patient strains were isolated and identified in the clinical microbiology department of the University Hospital Fundación Jiménez Díaz of Madrid (Spain). All strains were stocked frozen at −80 °C until the experiments were performed.
Table 1.
Origin of clinical bacterial strains used in this study.
| Species | Name | Sample | Gender | Age (years) |
|---|---|---|---|---|
| E. faecalis | Ef1 | Wound exudate | Female | 32 |
| Ef2 | Double-J catheter | Female | 85 | |
| E. coli | Ec1 | Ulcer | Female | 82 |
| Ec2 | Urine | Female | 8 | |
| K. pneumoniae | Kp1 | Urine | Female | 60 |
| Kp2 | Urine | Female | 59 | |
| B. fragilis | Bf1 | Drainage liquid | Female | 50 |
| Bf2 | Wound exudate | Male | 55 | |
| C. perfringens | Cp1 | Skin exudate | Female | 54 |
| Cp2 | Bile | Male | 68 |
Monospecies biofilm formation
Biofilm studies were performed in 96-well plates (Thermo Fisher Scientific, Waltham, Massachusetts, USA). To do this, a final concentration of 500 nM of lyso-Gb3 (Sigma-Aldrich, St. Louis Missouri, USA) was added to 106 colony-forming units (CFU)/mL of each strain inoculated in 200 µL tryptic soy broth (TSB; BD, Franklin Lakes, New Jersey, USA) supplemented with 1% glucose (Sigma-Aldrich), and the bacteria were incubated at 37 °C in 5% CO2 atmosphere for 24 h32. This lyso-Gb3 concentration was chosen since it is clinically relevant: in human and murine Fabry disease, plasma lyso-Gb3 may reach values of 400–600 nM, and in mice, these values were shown to be even higher in the liver and duodenum (10,900 and 4,100 nmol/g, respectively)24. After incubation and medium removal, samples were washed three times with 200 µL sterile 0.9% NaCl (B. Braun, Melsungen, Germany). Then, 190 µL of TSB+ 1% glucose plus 10 µL of alamarBlue® (BIORAD, California, USA) were added and incubated at 37 °C for 60 min33. After incubation, fluorescence was measured at 560 nm excitation wavelength and 590 nm emission wavelength to estimate the bacterial concentration in the biofilm33. All experiments were performed in triplicate (n = 24 per strain, 8 wells per replica).
Multispecies biofilm formation
Four representative strains of the previously assayed bacterial species were chosen and mixed (106 CFU/mL of E. coli ATCC 25922 and K. pneumoniae ATCC 23357, and 108 CFU/mL of B. fragilis ATCC 25285 and C. perfringens ATCC 13124) and incubated for 24 h in TSB+ 1% glucose with or without 500 nM lyso-Gb3 in anaerobic conditions. After incubation, each well was washed twice with 0.9% NaCl and sonicated for 5 min using an Ultrasons-H 3000840 low-power bath sonicator (J. P. Selecta, Barcelona, Spain) at 22 °C34. The concentration of bacteria in the biofilm was then estimated by applying the drop plate method35. E. coli ATCC 25922 and K. pneumoniae ATCC 23357 were quantified on chromID® CPS® Elite agar (Biomeriéux, Marcy-l'Étoile, France) in aerobic conditions, B. fragilis was quantified on Schaedler agar supplemented with neomycin and vancomycin (Biomeriéux), and counts of C. perfringens were conducted in 5% lamb’s blood agar supplemented with colistin and nalidixic acid (Biomeriéux) in anaerobic conditions. The experiment was carried out in 5 wells of 96-well plates in a volume of 200 μL/well and was repeated five times (n = 25 per species).
Multispecies biofilms were analyzed using a Leica DM IRB confocal laser-scanning microscope (Wetzlar, Lahn-Dil, Germany)36 in hydrophobic uncoated sterile 2-by-4–wells plates (ibidi GmbH, Munich, Bavaria, Germany) after staining with Live/Dead BactLight© stain (Thermo Fisher) according to manufacturer instructions.
Human gut microbiota
Dynamic multistage gut simulators are relevant for in vitro microbial ecological studies since they allow differentiation of colon region-specific populations originated from human stool samples37. We investigated the impact of lyso-Gb3 on the gut microbiota obtained from the simulated transverse colon suspension. A 50-mL colon-microbiota sample was centrifuged (10,000 × g for 10 min) and the pellet was covered with glycerol, snap-frozen in liquid nitrogen and stored at −80 °C until the experiment was performed. For experiments, the microbiota sample was suspended in 50 mL of a previously described growth medium37 supplemented with 2 g/L dehydrated purified ox-bile (Sigma-Aldrich) and buffered to pH 6.5–7.0 using a carbonate-phosphate buffer (9.240 g/L NaHCO3, 7.125 g/L Na2HPO4•12H2O, 0.470 g/L NaCl, 0.450 g/L KCl, 0.070 g/L CaCl2•12H2O, and 0.1 g/L MgCl2•6H2O) following the procedures of Durand et al.38. The suspension obtained was used to inoculate (1%) fresh buffered growth medium and incubated at 37 °C in the presence or absence of 500 nM lyso-Gb3 in an anaerobic chamber with 90% N2, 5% CO2, and 5% H2 atmosphere (Bactron II, Sheldon Manufacturing, Sunnyvale, California, USA). After incubation for 24 h, samples were centrifuged at 13,000 × g for 5 min, and the supernatant and pellet were stored at −20 °C for further analyses. All experiments were performed in triplicate (n = 3).
DNA extraction and purification
Microbial DNA was extracted as described by Moles et al.39. Briefly, the pellet from the colon microbiota culture was resuspended in 500 μL of 200 mM Tris–HCl pH 7.5, 0.5% SDS, 25 mM EDTA, 250 mM NaCl, and 3 M sodium acetate, and then incubated with 20 mg/mL lysozyme and 10 mg/mL RNAase (Sigma-Aldrich). Bacterial lysis was completed by mixing with glass beads. DNA was extracted with phenol/chloroform/isoamyl-alcohol, precipitated by adding 0.6 volumes of isopropanol and then resuspended in DNase, RNase free water (Sigma-Aldrich). The DNA yield was measured using a NanoDropH ND-1000 UV spectrophotometer (Thermo Fisher).
Quantitative PCR (qPCR)
Quantitative microbiological analysis of samples was carried out in qPCR experiments analyzed using SYBR® green methodology in a ViiA7 Real-Time PCR System (Life Technologies, Carlsbad, CA, USA). Primers, amplicon size, and annealing temperature for Akkermansia, Bacteroides, Bifidobacterium, Enterobacteriaceae, Faecalibacterium, Lactobacillus, Enterococcus, Prevotella, Roseburia, Blautia coccoides-Eubacterium rectale Cluster XIVa, Ruminococcus Cluster IV, and Clostridium leptum subgroup specific cluster IV have been described previously40. For the analysis of B. fragilis and Bilophila we used the primers and PCR conditions described by Sjögren et al.41 and Baldwin et al.42, respectively. DNA from E. coli DH5α, L. plantarum IFPL935, Enterococcus faecalis IFPL 382, Bifidobacterium breve 29M2, and B. fragilis DSM2151 were used to quantify total bacteria and Enterobacteriaceae, Lactobacillus, Enterococcus, Bifidobacterium, and Bacteroides and B. fragilis, respectively. For all other groups analyzed, samples were quantified using standards derived from targeted cloned genes using the pGEM-T cloning vector system kit (Promega, Madison, Wisconsin, USA) as described previously37.
Short-chain fatty-acid (SCFA) determination
The supernatant from the colon microbiota culture was filtered and 0.2 μL were injected into an HPLC system (Jasco, Tokyo, Japan) equipped with a UV975 detector and automatic injector37. SCFA were separated using a Rezex ROA Organic Acids column (300 × 7.8 mm) (Phenomenex, Macclesfield, UK) thermostated at 50 °C following the method described by Sanz et al.43. The mobile phase had a linear gradient of 0.005 M sulphuric acid in HPLC grade water, and the flow rate was 0.6 mL/min. The elution profile was monitored at 210 nm and peak identification was carried out by comparison between retention times and standards. ChromNAV data system software (Jasco) was used for data acquisition and processing. Calibration curves of acetic, butyric, formic, lactic and succinic acid were built up in the range concentration of 1 to 100 mM.
Statistical analysis
Statistical analyses were performed using Stata Statistical Software, Release 11 (StataCorp, College Station, Texas, USA). Prior to performing statistical calculations, the normality of each series of data was checked with the Shapiro-Wilk test. Normally distributed data were presented as mean and standard deviation and compared using the unilateral Student’s t-test. When the distribution was not normal, data were represented as median and interquartile range, and the one-sided Wilcoxon signed-rank test was applied. A level of statistical significance of P < 0.05 was considered significant.
Results
Impact of lyso-Gb3 on monospecies biofilm formation
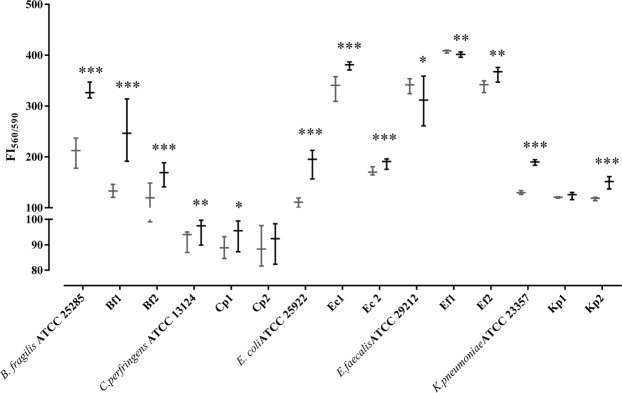
Microbial biofilms containing potential pathogens are regarded as a tipping point between healthy and diseased states in the gut mucosa44. Thus, we first explored the impact of lyso-Gb3 on biofilm formation by different individual strains, and these results are represented in Fig. 1. All strains except two (clinical strains K. pneumoniae Kp1 and C. perfringens Cp2) significantly modified their biofilm formation in the presence of lyso-Gb3. Most strains significantly increased biofilm formation in the presence of lyso-Gb3, and only two (E. faecalis ATCC 29212 and Ef1) significantly but marginally decreased their biofilm formation. Thus, the most consistent results were obtained for E. coli and B. fragilis, in which all three strains tested significantly increased the biofilm formation for each species.
Figure 1.
Impact of lyso-Gb3 on monospecies biofilm formation. Fluorescence intensity (FI) (x1000) of each bacterial-strain biofilm in presence (black) or absence (gray) of lyso-Gb3. The following represent strains derived from clinical isolates (Table 1): E. faecalis (Ef1, Ef2), E. coli (Ec1, Ec2), K. pneumoniae (Kp1, Kp2), B. fragilis (Bf1, Bf2), and C. perfringens (Cp1, Cp2). The whiskers represent the interquartile range. *p-value < 0.05, **p-value < 0.01, and ***p-value < 0.001 for Wilcoxon signed-rank test.
Impact of lyso-Gb3 on multispecies biofilm formation
We then studied the impact of lyso-Gb3 on the formation of polymicrobial biofilms containing potential pathogens. The results of multispecies (E. coli ATCC 25922, K. pneumoniae ATCC 23357, B. fragilis ATCC 25285, C. perfringens ATCC 13124) biofilm formation are shown in Fig. 2, and a representative three-dimensional representation is shown in Fig. 3. Only B. fragilis ATCC 25285 numbers significantly increased in presence of lyso-Gb3 in a multispecies biofilm model (p = 0.0236). This result is consistent with the monospecies biofilm studies.
Figure 2.
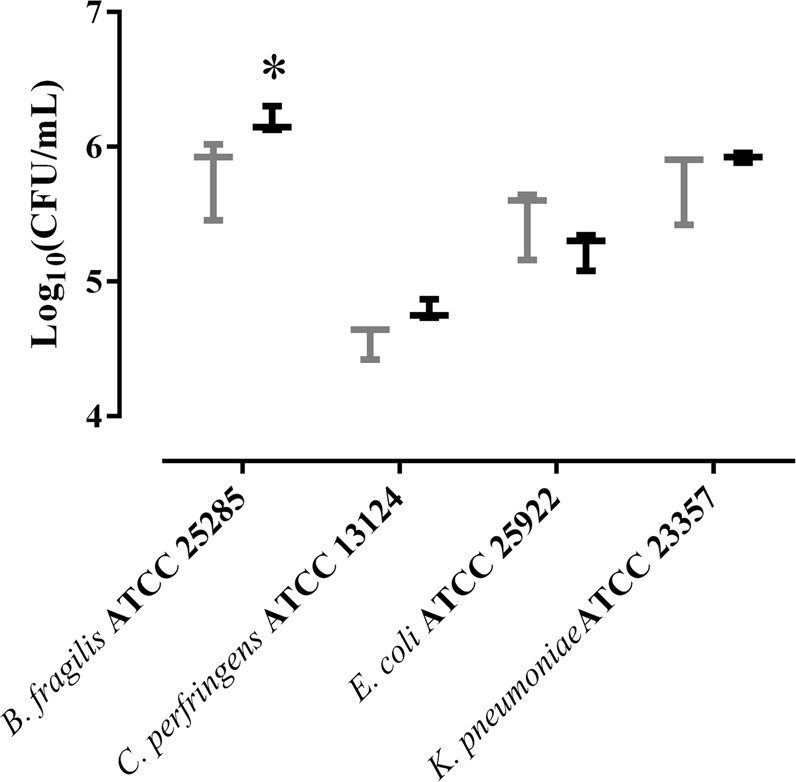
Impact of lyso-Gb3 on multispecies biofilm formation. Bacterial concentration (log CFU/mL) of each bacterial strain in a multispecies biofilm in the presence (black) or absence (gray) of lyso-Gb3. The whiskers represent the interquartile range. *p-value < 0.05 for Wilcoxon signed-rank test.
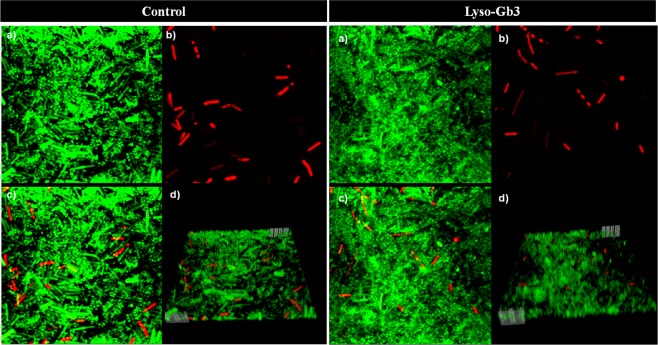
Figure 3.
Representative confocal photograph of a multispecies biofilm in the presence or absence of lyso-Gb3 using Live/Dead BactLightTM. (a) Live bacteria in green. (b) Dead bacteria in red. (c) Superposition of live and dead bacteria. (d) Three-dimensional representation of each biofilm.
Impact of lyso-Gb3 on complex bacterial communities
Since the bacterial populations of the gut microbiota are even more complex, we explored the impact of lyso-Gb3 on the composition and metabolite output of the simulated human transverse colon microbiota. Samples were analyzed for 16S rRNA qPCR quantification of bacteria (pellet) and for SCFA formation (supernatant). The targeted bacterial groups represent the predominant Gram-positive bacteria belonging to clostridial clusters XIVa and IV (Firmicutes) and Gram-negative bacteria related to Bacteroidetes. Other genera such as Lactobacillus, Bifidobacterium and Akkermansia are commonly health-related bacteria. Specific quantification of Bilophila was targeted for its correlation with intestinal discomfort45,46. The qPCR assay showed that lyso-Gb3 modified several groups of bacteria, as shown in Table 2. Most bacterial groups evaluated significantly modified their concentration in presence of 500 nM lyso-Gb3 with respect to controls, the exceptions being Bilophila, Faecalibacterium, and Ruminococcus. Those bacterial groups with significantly higher concentrations under lyso-Gb3 were Enterobacteriaceae, Enterococcus, and Prevotella. The largest effect of lyso-Gb3 to increase bacterial counts was observed for B. fragilis, as with single bacterial species and multispecies biofilm experiments. By contrast, the concentration of Akkermansia, Bacteroides, Bifidobacterium, C. leptum, B. coccoides-E. rectale, and Lactobacillus was significantly lower under lyso-Gb3 than in controls.
Table 2.
Impact of lyso-Gb3 on bacterial counts in a transverse colon microbiota sample.
| Bacterial group | Baseline | Control 24 h | 500 nM lyso-Gb3 24 h | p-value* |
|---|---|---|---|---|
| Akkermansia | 6.74 ± 0.06 | 6.22 ± 0.10 | 5.91 ± 0.10 | <0.001 |
| Bacteroides | 6.50 ± 0.22 | 7.62 ± 0.13 | 6.73 ± 0.10 | <0.001 |
| B. fragilis | 3.78 ± 0.02 | 1.55 ± 0.35 | 5.09 ± 0.18 | <0.001 |
| Bifidobacterium | 4.40 ± 0.06 | 4.09 ± 0.05 | 3.92 ± 0.10 | 0.003 |
| Bilophila | 6.75 ± 0.04 | 7.94 ± 0.09 | 7.96 ± 0.07 | 0.328 |
| Clostridium leptum | 5.26 ± 0.05 | 5.20 ± 0.16 | 4.85 ± 0.05 | 0.001 |
| Blautia coccoides-Eubacterium rectale | 6.80 ± 0.06 | 7.07 ± 0.02 | 6.81 ± 0.12 | 0.002 |
| Enterobacteriaceae | 6.76 ± 0.07 | 8.70 ± 0.08 | 8.95 ± 0.14 | 0.002 |
| Enterococcus | 6.19 ± 0.05 | 7.74 ± 0.17 | 8.06 ± 0.07 | 0.002 |
| Faecalibacterium | 7.88 ± 0.10 | 7.48 ± 0.06 | 7.53 ± 0.16 | 0.276 |
| Lactobacillus | 5.42 ± 0.10 | 4.18 ± 0.05 | 3.91 ± 0.06 | <0.001 |
| Prevotella | 2.79 ± 0.28 | 3.70 ± 0.09 | 4.34 ± 0.32 | 0.002 |
| Roseburia | 3.72 ± 0.15 | 3.11 ± 0.08 | 3.33 ± 0.16 | 0.006 |
| Ruminococcus | 3.98 ± 0.07 | 3.09 ± 0.14 | 2.94 ± 0.18 | 0.074 |
Mean ± SD of quantitative PCR counts (log copy number/mL) for the different microbial groups analyzed.
*p-values for Student’s t-test between control 24 h and 500 nM lyso-Gb3 24 h.
SCFA determination showed that lyso-Gb3 could also modify SCFA production by the gut microbiota, as shown in Table 3. The most remarkable effect observed was the significantly lower formation of butyric acid in lyso-Gb3-exposed samples than in controls. In addition, an unknown acidic compound (peak retention time 33.4) was only observed in the samples incubated with lyso-Gb3 (results not shown).
Table 3.
Impact of lyso-Gb3 on short chain fatty acid (SCFA) concentration in a transverse colon microbiota sample.
| SCFA | Baseline | Control 24 h | 500 nM lyso-Gb3 24 h | p-value* |
|---|---|---|---|---|
| Acetic acid | 1.21 ± 0.08 | 29.55 ± 2.11 | 31.28 ± 2.03 | 0.101 |
| Butyric acid | ND | 1.48 ± 0.56 | 0.68 ± 0.20 | 0.008 |
| Formic acid | 9.01 ± 0.24 | 18.46 ± 1.59 | 15.96 ± 0.11 | 0.006 |
| Lactic acid | ND | 7.21 ± 0.86 | 7.29 ± 0.42 | 0.428 |
| Succinic acid | 12.25 ± 0.7 | 14.1 ± 0.80 | 15.36 ± 0.20 | 0.006 |
SCFA concentrations in mM; expressed as mean ± SD.
*p-values for Student’s t-test between control 24 h and 500 nM lyso-Gb3 24 h. ND: not detected.
Discussion
The pathophysiology of gastrointestinal symptoms in Fabry disease is complex and multifactorial, though the fact that these symptoms stem from Gb3 and lyso-Gb3 accumulation within intestinal tissues is now widely accepted15. Here, we have shown that lyso-Gb3 may directly modify microbiota composition, potentially contributing to the gastrointestinal and systemic symptoms of Fabry disease.
The gut microbiota is a collection of archaea, bacteria, and eukarya that have evolved over thousands of years to form a symbiotic relationship with human hosts47. These microbial populations influence metabolic, immune and defense systems in the intestine and consequently, human health17,48–50. The bacterial phyla representatives of the human gut microbiota are Bacteroidetes, Firmicutes, Actinobacteria, Proteobacteria and Verrucomicrobia; additionally, the genera Faecalibacterium, Bifidobacterium, Roseburia, Ruminococcus, Bacteroides, Prevotella, Akkermansia and Oscillospira represent common core bacteria in the Western adult population51. These gut bacteria are present in both planktonic and biofilms states which may be associated with luminal material or mucosal surfaces44,52–56, such as gut biofilms commonly associated with disease44,57. According to our results, the presence of lyso-Gb3 at clinically relevant concentrations significantly favored biofilm development by B. fragilis and E. coli and by some strains of C. perfringens, E. faecalis, and K. pneumoniae. Since gut biofilms are composed of multiple microbial species17,18,53,58,59, the most realistic in vitro approach would be to simultaneously develop a multispecies biofilm using several intestinal bacterial species. Indeed, the only species significantly favored by lyso-Gb3 when this approach was taken was B. fragilis.
The dynamic and complex interactions of colonic bacteria have been nearly reproduced in vitro by using laboratory simulators of the human gut microbiome37,60,61. The transverse-colonic compartment is considered to hold most of the representative colonic bacterial groups37,61, so we adopted this approach to increase the complexity of our studies. In view of our results, all evaluated groups of bacterial underwent modified growth to a greater or lesser degree in the presence of lyso-Gb3. The most striking modification was observed in B. fragilis, as the concentration of this bacteria strain increased almost 1.5 log-fold during 24 h incubation in the presence of lyso-Gb3, whereas no growth was observed in the control. This positive impact reflected the biofilm study results. B. fragilis is a two-faced gut symbiotic bacteria62. On the one hand, lipopolysaccharide A and other polysaccharides from B. fragilis stimulate the development of regulatory T cells which, in turn, switch off inflammatory T cells, thus offering protection from local or systemic inflammatory processes59,63. On the other hand, B. fragilis strains may release an enterotoxin or B. fragilis toxin (BFT), which is associated with diarrhea in young animals64, children64,65, and adults66. BFT has been linked to colorectal adenoma, polyps, and cancer in experimental animals and humans62,67–71. Additionally, B. fragilis biofilm development has been associated with inflammatory bowel diseases52,67. Thus, changes in B. fragilis biology induced by lyso-Gb3 could potentially be linked to the gut inflammation and colonic polyps described in Fabry disease72,73.
SCFA are fermentation products of bacterial microbiota58. Acetic, propionic and butyric acids can serve as an energy source to human intestine epithelium17. The most striking impact of lyso-Gb3 observed in this study was to decrease the butyrate formation by almost 50%. This may be caused by the decrease in Firmicutes (including C. leptum and B. coccoides-E. rectale groups) counts, as Firmicutes are the main producers of butyrate17,58,74, via cross-feeding of acetate and lactate74. However, we cannot exclude a more direct impact of lyso-Gb3 on butyrate metabolism. Butyrate has several beneficial effects, including protection against colorectal cancer, chronic kidney disease, and left ventricular hypertrophy, the latter two being hallmarks of Fabry disease18,20,75. Butyrate also inhibits histone deacetylases (HDACs) and has anti-inflammatory properties23,76. Chronic low-level inflammation is thought to contribute to Fabry-disease severity by enhancing the activity of upstream enzymes, such as Gb3 synthase, which increases the availability of accumulated metabolites such as Gb377. HDAC inhibitors modify the epigenetic regulation of gene transcription and, as butyrate itself, have been beneficial in kidney disease20,78–81.
Gastrointestinal symptoms may severely compromise the quality of life of Fabry patients and lead them to undergo unnecessary surgical interventions15. While bacterial overgrowth in Fabry disease was observed in one 40-year-old patient and has been cited as a potential contributor to gastrointestinal symptoms ever since, we found no reports in which gut microbiota composition was assessed using modern techniques, and available data rely on non-specific breath tests14,82. Thus, one limitation of the present study is the lack of comparative clinical information on gut microbiota between Fabry patients and healthy controls. Given that Fabry is a rare disease, a multicenter collaborative study would be needed to address this. One of the strengths of this research is the fact that we studied a clinically relevant lyso-Gb3 concentration and observed consistent results across three different independent experimental settings for B. fragilis. The negative impact of lyso-Gb3 observed on butyrate, a SCFA with beneficial properties for cardiac hypertrophy and chronic kidney disease, lends further potential clinical relevance to our findings. In this regard, the hypothesis raised in the present study regarding a microbiota connection between glycolipid accumulation and potential local or systemic consequences are very much in line with recent developments in the interaction between gut microbiota and disease48,76,83–85. This experimental work has several limitations. First, only one clinically relevant lyso-Gb3 concentration was tested in several experimental conditions. Second, we cannot exclude that other glycosphingolipids associated to this Fabry disease such as Gb3 or lactosylceramide24, which could theoretically be present in bile31 or present within sloughing enterocytes24,86, also modulate the microbiota. However, Gb3 deposits have not been observed in enterocytes, likely due to the short half-life of these cells, since longer lived cells (neurons, podocytes, cardiomyocytes) are the ones with the largest burden of deposits87.
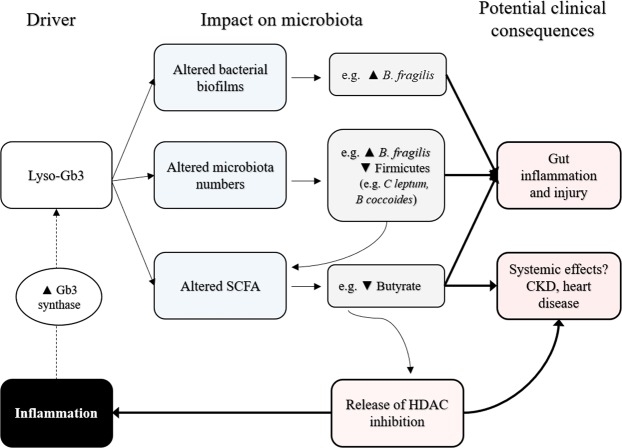
In conclusion, globotriaosylsphingosine (lyso-Gb3) may modify the growth and biofilm-forming capacity of intestinal bacteria as well as the SCFA formation pattern of healthy gut microbiota. To our knowledge, this is the first study to reveal a possible direct relationship between metabolites accumulated in Fabry disease, such as lyso-Gb3, and the gut microbiota, potentially causing an impact on the gastrointestinal and even systemic symptoms of Fabry disease, opening a whole new field of Fabry research (Fig. 4). However, more in-depth microbiologic studies are necessary to understand the molecular mechanisms linking lyso-Gb3 to bacterial metabolism, and further work exploring the in vivo clinical and therapeutic consequences of this observation would be required.
Figure 4.
Summary of research findings and working hypothesis for possible clinical implications. At clinically relevant concentrations, lyso-Gb3 modified the biofilm-forming properties and counts of diverse bacteria. Most notably, lyso-Gb3 consistently increased B. fragilis biofilm formation or counts in three different, independent experimental settings. Additionally, it led to decreased production of butyrate and an altered pattern of other short-chain fatty acids (SCFA). Experimental data from the literature have linked low butyrate levels to gastrointestinal manifestations and kidney and heart disease, which are also found in Fabry disease. Thus, butyrate has been reported to have anti-inflammatory properties attributable to its ability to inhibit histone deacetylases (HDACs)23,79–81. In this regard, we hypothesize that lyso-Gb3-induced lowering of butyrate levels may release the inhibition of butyrate on HDACs and favor systemic inflammation. Inflammation, in turn, is known to increase the activity of Gb3 synthase77, thus increasing the synthesis of metabolites such as Gb3 that accumulate in Fabry disease and potentially increasing the severity of kidney and heart disease.
Acknowledgements
JJAC is funded by an FPI grant from Spanish Ministry of Economics and Competitiveness (BES-2014-069007). The authors were further supported by FIS PI16/02057, PI18/01366, ISCIII-RETIC REDinREN RD016/0009 Fondos FEDER, Sociedad Española de Nefrología, Comunidad de Madrid B2017/BMD-3686 CIFRA2-CM, Miguel Servet MS14/00133 to MDSN and AGL2016-75951-R. The authors thank Esteban-Fernando Sáez-Martínez for his valuable technical assistance with qPCR and HPLC. We want to acknowledge Dr. Oliver Shaw for his help with the English language and María del Mar González García-Parreño for her help with the use of the confocal laser-scanning microscope. Parts of this work were previously presented at the 21st, 22nd, and 23rd Spanish Congress of Clinical Microbiology and Infectious Diseases, the 29th European Congress of Clinical Microbiology and Infectious Diseases, the 6th Update on Fabry Disease, and 15th Annual Word Symposium of Lysosomal Diseases Research & Conference.
Author Contributions
Credit for authorship should be based on: [1] substantial contributions to research design, or the acquisition, analysis or interpretation of data; [2] drafting the paper or revising it critically; [3] approval of the submitted and final versions. John-Jairo Aguilera-Correa: 1, 2, 3. Patricia Madrazo-Clemente: 1, 3. M. Carmen Martínez-Cuesta: 2, 3. Carmen Peláez: 2, 3. Alberto Ortiz: 2, 3. María Dolores Sánchez-Niño: 2, 3. Jaime Esteban: 2, 3. and Teresa Requena: 2, 3. All authors have read and approved this final submitted manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
John-Jairo Aguilera-Correa, María Dolores Sánchez-Niño, Jaime Esteban and Teresa Requena contributed equally.
References
- 1.Germain DP. Fabry disease. Orphanet journal of rare diseases. 2010;5:30. doi: 10.1186/1750-1172-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ortiz A, Sanchez-Nino MD. Diagnosis and treatment of Fabry disease. Medicina clinica. 2017;148:132–138. doi: 10.1016/j.medcli.2016.09.047. [DOI] [PubMed] [Google Scholar]
- 3.Germain Dominique P., Brand Eva, Burlina Alessandro, Cecchi Franco, Garman Scott C., Kempf Judy, Laney Dawn A., Linhart Aleš, Maródi László, Nicholls Kathy, Ortiz Alberto, Pieruzzi Federico, Shankar Suma P., Waldek Stephen, Wanner Christoph, Jovanovic Ana. Phenotypic characteristics of the p.Asn215Ser (p.N215S) GLA mutation in male and female patients with Fabry disease: A multicenter Fabry Registry study. Molecular Genetics & Genomic Medicine. 2018;6(4):492–503. doi: 10.1002/mgg3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ortiz A, et al. Fabry disease revisited: Management and treatment recommendations for adult patients. Molecular genetics and metabolism. 2018;123:416–427. doi: 10.1016/j.ymgme.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 5.Wilcox WR, et al. Improvement of Fabry Disease-Related Gastrointestinal Symptoms in a Significant Proportion of Female Patients Treated with Agalsidase Beta: Data from the Fabry Registry. JIMD reports. 2018;38:45–51. doi: 10.1007/8904_2017_28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilcox WR, et al. Females with Fabry disease frequently have major organ involvement: lessons from the Fabry Registry. Molecular genetics and metabolism. 2008;93:112–128. doi: 10.1016/j.ymgme.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 7.Wanner C, et al. European expert consensus statement on therapeutic goals in Fabry disease. Molecular genetics and metabolism. 2018;124:189–203. doi: 10.1016/j.ymgme.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Hoffmann B, Schwarz M, Mehta A, Keshav S, Fabry Outcome Survey European I. Gastrointestinal symptoms in 342 patients with Fabry disease: prevalence and response to enzyme replacement therapy. Clinical gastroenterology and hepatology: the official clinical practice journal of the American Gastroenterological Association. 2007;5:1447–1453. doi: 10.1016/j.cgh.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Keshav, S. In Fabry Disease: Perspectives from 5 Years of FOS (eds Mehta, A., Beck, M. & Sunder-Plassmann, G.) (2006). [PubMed]
- 10.Zar-Kessler C, Karaa A, Sims KB, Clarke V, Kuo B. Understanding the gastrointestinal manifestations of Fabry disease: promoting prompt diagnosis. Therapeutic advances in gastroenterology. 2016;9:626–634. doi: 10.1177/1756283X16642936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Politei J, et al. Chronic intestinal pseudo-obstruction. Did you search for lysosomal storage diseases? Molecular genetics and metabolism reports. 2017;11:8–11. doi: 10.1016/j.ymgmr.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Germain Dominique P., Arad Michael, Burlina Alessandro, Elliott Perry M., Falissard Bruno, Feldt-Rasmussen Ulla, Hilz Max J., Hughes Derralynn A., Ortiz Alberto, Wanner Christoph, Weidemann Frank, Spada Marco. The effect of enzyme replacement therapy on clinical outcomes in female patients with Fabry disease – A systematic literature review by a European panel of experts. Molecular Genetics and Metabolism. 2019;126(3):224–235. doi: 10.1016/j.ymgme.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 13.Cable WJ, Kolodny EH, Adams RD. Fabry disease: impaired autonomic function. Neurology. 1982;32:498–502. doi: 10.1212/wnl.32.5.498. [DOI] [PubMed] [Google Scholar]
- 14.O’Brien BD, et al. Pathophysiologic and ultrastructural basis for intestinal symptoms in Fabry’s disease. Gastroenterology. 1982;82:957–962. [PubMed] [Google Scholar]
- 15.Hilz MJ, et al. Non-specific gastrointestinal features: Could it be Fabry disease? Digestive and liver disease: official journal of the Italian Society of Gastroenterology and the Italian Association for the Study of the Liver. 2018;50:429–437. doi: 10.1016/j.dld.2018.02.011. [DOI] [PubMed] [Google Scholar]
- 16.Jackson MA, et al. Gut microbiota associations with common diseases and prescription medications in a population-based cohort. Nature communications. 2018;9:2655. doi: 10.1038/s41467-018-05184-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang B, Yao M, Lv L, Ling Z, Li L. The Human Microbiota in Health and Disease. Engineering. 2017;3:71–82. doi: 10.1016/J.ENG.2017.01.008. [DOI] [Google Scholar]
- 18.Feng Q, Chen WD, Wang YD. Gut Microbiota: An Integral Moderator in Health and Disease. Frontiers in microbiology. 2018;9:151. doi: 10.3389/fmicb.2018.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Srivastava A, Gupta J, Kumar S, Kumar A. Gut biofilm forming bacteria in inflammatory bowel disease. Microbial pathogenesis. 2017;112:5–14. doi: 10.1016/j.micpath.2017.09.041. [DOI] [PubMed] [Google Scholar]
- 20.Castillo-Rodriguez Esmeralda, Fernandez-Prado Raul, Esteras Raquel, Perez-Gomez Maria, Gracia-Iguacel Carolina, Fernandez-Fernandez Beatriz, Kanbay Mehmet, Tejedor Alberto, Lazaro Alberto, Ruiz-Ortega Marta, Gonzalez-Parra Emilio, Sanz Ana, Ortiz Alberto, Sanchez-Niño Maria. Impact of Altered Intestinal Microbiota on Chronic Kidney Disease Progression. Toxins. 2018;10(7):300. doi: 10.3390/toxins10070300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandez-Prado, R. et al. Nutrients Turned into Toxins: Microbiota Modulation of Nutrient Properties in Chronic Kidney Disease. Nutrients9, 10.3390/nu9050489 (2017). [DOI] [PMC free article] [PubMed]
- 22.Mafra D, Fouque D. Gut microbiota and inflammation in chronic kidney disease patients. Clinical kidney journal. 2015;8:332–334. doi: 10.1093/ckj/sfv026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aerts JM, et al. Elevated globotriaosylsphingosine is a hallmark of Fabry disease. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2812–2817. doi: 10.1073/pnas.0712309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao HC, et al. Plasma globotriaosylsphingosine (lysoGb3) could be a biomarker for Fabry disease with a Chinese hotspot late-onset mutation (IVS4+919G>A) Clinica chimica acta; international journal of clinical chemistry. 2013;426:114–120. doi: 10.1016/j.cca.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 26.Nowak A, Mechtler TP, Desnick RJ, Kasper DC. Plasma LysoGb3: A useful biomarker for the diagnosis and treatment of Fabry disease heterozygotes. Molecular genetics and metabolism. 2017;120:57–61. doi: 10.1016/j.ymgme.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 27.Choi L, et al. The Fabry disease-associated lipid Lyso-Gb3 enhances voltage-gated calcium currents in sensory neurons and causes pain. Neuroscience letters. 2015;594:163–168. doi: 10.1016/j.neulet.2015.01.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez-Nino MD, et al. Lyso-Gb3 activates Notch1 in human podocytes. Human molecular genetics. 2015;24:5720–5732. doi: 10.1093/hmg/ddv291. [DOI] [PubMed] [Google Scholar]
- 29.Sanchez-Nino MD, et al. Globotriaosylsphingosine actions on human glomerular podocytes: implications for Fabry nephropathy. Nephrology, dialysis, transplantation: official publication of the European Dialysis and Transplant Association - European Renal Association. 2011;26:1797–1802. doi: 10.1093/ndt/gfq306. [DOI] [PubMed] [Google Scholar]
- 30.Weidemann F, et al. Fibrosis: a key feature of Fabry disease with potential therapeutic implications. Orphanet journal of rare diseases. 2013;8:116. doi: 10.1186/1750-1172-8-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duro Giovanni, Zizzo Carmela, Cammarata Giuseppe, Burlina Alessandro, Burlina Alberto, Polo Giulia, Scalia Simone, Oliveri Roberta, Sciarrino Serafina, Francofonte Daniele, Alessandro Riccardo, Pisani Antonio, Palladino Giuseppe, Napoletano Rosa, Tenuta Maurizio, Masarone Daniele, Limongelli Giuseppe, Riccio Eleonora, Frustaci Andrea, Chimenti Cristina, Ferri Claudio, Pieruzzi Federico, Pieroni Maurizio, Spada Marco, Castana Cinzia, Caserta Marina, Monte Ines, Rodolico Margherita, Feriozzi Sandro, Battaglia Yuri, Amico Luisa, Losi Maria, Autore Camillo, Lombardi Marco, Zoccali Carmine, Testa Alessandra, Postorino Maurizio, Mignani Renzo, Zachara Elisabetta, Giordano Antonello, Colomba Paolo. Mutations in the GLA Gene and LysoGb3: Is It Really Anderson-Fabry Disease? International Journal of Molecular Sciences. 2018;19(12):3726. doi: 10.3390/ijms19123726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stepanovic S, et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS: acta pathologica, microbiologica, et immunologica Scandinavica. 2007;115:891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]
- 33.Pettit RK, Weber CA, Pettit GR. Application of a high throughput Alamar blue biofilm susceptibility assay to Staphylococcus aureus biofilms. Annals of clinical microbiology and antimicrobials. 2009;8:28. doi: 10.1186/1476-0711-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esteban J, et al. Evaluation of quantitative analysis of cultures from sonicated retrieved orthopedic implants in diagnosis of orthopedic infection. Journal of clinical microbiology. 2008;46:488–492. doi: 10.1128/JCM.01762-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herigstad B, Hamilton M, Heersink J. How to optimize the drop plate method for enumerating bacteria. Journal of microbiological methods. 2001;44:121–129. doi: 10.1016/S0167-7012(00)00241-4. [DOI] [PubMed] [Google Scholar]
- 36.Munoz-Egea MC, Garcia-Pedrazuela M, Mahillo-Fernandez I, Esteban J. Effect of Antibiotics and Antibiofilm Agents in the Ultrastructure and Development of Biofilms Developed by Nonpigmented Rapidly Growing Mycobacteria. Microbial drug resistance. 2016;22:1–6. doi: 10.1089/mdr.2015.0124. [DOI] [PubMed] [Google Scholar]
- 37.Barroso, E., Cueva, C., Peláez, C., Martínez-Cuesta, M. C. & Requena, T. Development of human colonic microbiota in the computer-controlled dynamic SIMulator of the GastroIntestinal tract SIMGI. Vol. 61 (2015).
- 38.Durand M, Dumay C, Beaumatin P, Morel MT. Use of the rumen simulation technique (RUSITEC) to compare microbial digestion of various by-products. Animal Feed Science and Technology. 1988;21:197–204. doi: 10.1016/0377-8401(88)90101-0. [DOI] [Google Scholar]
- 39.Moles L, et al. Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PloS one. 2013;8:e66986. doi: 10.1371/journal.pone.0066986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lozano-Ojalvo D, et al. Egg white peptide-based immunotherapy enhances vitamin A metabolism and induces RORγt+ regulatory T cells. Journal of Functional Foods. 2019;52:204–211. doi: 10.1016/j.jff.2018.11.012. [DOI] [Google Scholar]
- 41.Sjogren YM, Jenmalm MC, Bottcher MF, Bjorksten B, Sverremark-Ekstrom E. Altered early infant gut microbiota in children developing allergy up to 5 years of age. Clinical and experimental allergy: journal of the British Society for Allergy and Clinical Immunology. 2009;39:518–526. doi: 10.1111/j.1365-2222.2008.03156.x. [DOI] [PubMed] [Google Scholar]
- 42.Baldwin J, et al. Table grape consumption reduces adiposity and markers of hepatic lipogenesis and alters gut microbiota in butter fat-fed mice. The Journal of nutritional biochemistry. 2016;27:123–135. doi: 10.1016/j.jnutbio.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanz ML, et al. In vitro investigation into the potential prebiotic activity of honey oligosaccharides. Journal of agricultural and food chemistry. 2005;53:2914–2921. doi: 10.1021/jf0500684. [DOI] [PubMed] [Google Scholar]
- 44.Tytgat HLP, Nobrega FL, van der Oost J, de Vos WM. Bowel Biofilms: Tipping Points between a Healthy and Compromised Gut? Trends in microbiology. 2019;27:17–25. doi: 10.1016/j.tim.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 45.Manichanh C, et al. Anal gas evacuation and colonic microbiota in patients with flatulence: effect of diet. Gut. 2014;63:401–408. doi: 10.1136/gutjnl-2012-303013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vandeputte D, et al. Prebiotic inulin-type fructans induce specific changes in the human gut microbiota. Gut. 2017;66:1968–1974. doi: 10.1136/gutjnl-2016-313271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moeller AH, et al. Cospeciation of gut microbiota with hominids. Science. 2016;353:380–382. doi: 10.1126/science.aaf3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kundu P, Blacher E, Elinav E, Pettersson S. Our Gut Microbiome: The Evolving Inner Self. Cell. 2017;171:1481–1493. doi: 10.1016/j.cell.2017.11.024. [DOI] [PubMed] [Google Scholar]
- 49.Requena T, Martinez-Cuesta MC, Pelaez C. Diet and microbiota linked in health and disease. Food & function. 2018;9:688–704. doi: 10.1039/c7fo01820g. [DOI] [PubMed] [Google Scholar]
- 50.Rojo D, et al. Exploring the human microbiome from multiple perspectives: factors altering its composition and function. FEMS microbiology reviews. 2017;41:453–478. doi: 10.1093/femsre/fuw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shetty SA, Hugenholtz F, Lahti L, Smidt H, de Vos WM. Intestinal microbiome landscaping: insight in community assemblage and implications for microbial modulation strategies. FEMS microbiology reviews. 2017;41:182–199. doi: 10.1093/femsre/fuw045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Vos WM. Microbial biofilms and the human intestinal microbiome. NPJ biofilms and microbiomes. 2015;1:15005. doi: 10.1038/npjbiofilms.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Donelli G, Vuotto C, Cardines R, Mastrantonio P. Biofilm-growing intestinal anaerobic bacteria. FEMS immunology and medical microbiology. 2012;65:318–325. doi: 10.1111/j.1574-695X.2012.00962.x. [DOI] [PubMed] [Google Scholar]
- 54.Macfarlane S, Dillon JF. Microbial biofilms in the human gastrointestinal tract. Journal of Applied Microbiology. 2007;102:1187–1196. doi: 10.1111/j.1365-2672.2007.03287.x. [DOI] [PubMed] [Google Scholar]
- 55.Macfarlane S, Macfarlane GT. Composition and metabolic activities of bacterial biofilms colonizing food residues in the human gut. Applied and environmental microbiology. 2006;72:6204–6211. doi: 10.1128/AEM.00754-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Probert HM, Gibson GR. Bacterial biofilms in the human gastrointestinal tract. Current issues in intestinal microbiology. 2002;3:23–27. [PubMed] [Google Scholar]
- 57.Buret AG, Motta JP, Allain T, Ferraz J, Wallace JL. Pathobiont release from dysbiotic gut microbiota biofilms in intestinal inflammatory diseases: a role for iron? Journal of biomedical science. 2019;26:1. doi: 10.1186/s12929-018-0495-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thursby E, Juge N. Introduction to the human gut microbiota. The Biochemical journal. 2017;474:1823–1836. doi: 10.1042/BCJ20160510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blumberg R, Powrie F. Microbiota, disease, and back to health: a metastable journey. Science translational medicine. 2012;4:137rv137. doi: 10.1126/scitranslmed.3004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.In The Impact of Food Bioactives on Health: in vitro and ex vivo models (eds. Verhoeckx, K. et al.) (2015). [PubMed]
- 61.Barroso E, et al. Effect of lactulose-derived oligosaccharides on intestinal microbiota during the shift between media with different energy contents. Food research international. 2016;89:302–308. doi: 10.1016/j.foodres.2016.08.025. [DOI] [PubMed] [Google Scholar]
- 62.Sears C. L. Enterotoxigenic Bacteroides fragilis: a Rogue among Symbiotes. Clinical Microbiology Reviews. 2009;22(2):349–369. doi: 10.1128/CMR.00053-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ackerman J. The ultimate social network. Scientific American. 2012;306:36–43. doi: 10.1038/scientificamerican0612-36. [DOI] [PubMed] [Google Scholar]
- 64.Bressane MA, Durigon LE, Avila-Campos MJ. Prevalence of the Bacteroides fragilis Group and Enterotoxigenic Bacteroides fragilis in Immunodeficient Children. Anaerobe. 2001;7:277–281. doi: 10.1006/anae.2001.0401. [DOI] [Google Scholar]
- 65.Durmaz B, Dalgalar M, Durmaz R. Prevalence of enterotoxigenic Bacteroides fragilis in patients with diarrhea: a controlled study. Anaerobe. 2005;11:318–321. doi: 10.1016/j.anaerobe.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 66.Cohen SH, et al. Prevalence of enterotoxigenic Bacteroides fragilis in hospital-acquired diarrhea. Diagnostic microbiology and infectious disease. 2006;55:251–254. doi: 10.1016/j.diagmicrobio.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 67.Bird S, Hadjimichael E, Mehta A, Ramaswami U, Hughes D. Fabry disease and incidence of cancer. Orphanet journal of rare diseases. 2017;12:150. doi: 10.1186/s13023-017-0701-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dejea CM, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 2018;359:592–597. doi: 10.1126/science.aah3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Purcell RV, et al. Colonization with enterotoxigenic Bacteroides fragilis is associated with early-stage colorectal neoplasia. PloS one. 2017;12:e0171602. doi: 10.1371/journal.pone.0171602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Toprak NU, et al. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2006;12:782–786. doi: 10.1111/j.1469-0691.2006.01494.x. [DOI] [PubMed] [Google Scholar]
- 71.Wu S, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nature medicine. 2009;15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jack CI, Morris AI, Nasmyth DG, Carroll N. Colonic involvement in Fabry’s disease. Postgraduate medical journal. 1991;67:584–585. doi: 10.1136/pgmj.67.788.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kleinert J, et al. Anderson–Fabry disease: a case-finding study among male kidney transplant recipients in Austria. Transplant International. 2009;22:287–292. doi: 10.1111/j.1432-2277.2008.00791.x. [DOI] [PubMed] [Google Scholar]
- 74.Flint HJ, Duncan SH, Scott KP, Louis P. Interactions and competition within the microbial community of the human colon: links between diet and health. Environmental microbiology. 2007;9:1101–1111. doi: 10.1111/j.1462-2920.2007.01281.x. [DOI] [PubMed] [Google Scholar]
- 75.Patel BM. Sodium Butyrate Controls Cardiac Hypertrophy in Experimental Models of Rats. Cardiovascular toxicology. 2018;18:1–8. doi: 10.1007/s12012-017-9406-2. [DOI] [PubMed] [Google Scholar]
- 76.Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell. 2016;165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 77.Stricklett PK, Hughes AK, Ergonul Z, Kohan DE. Molecular basis for up-regulation by inflammatory cytokines of Shiga toxin 1 cytotoxicity and globotriaosylceramide expression. The Journal of infectious diseases. 2002;186:976–982. doi: 10.1086/344053. [DOI] [PubMed] [Google Scholar]
- 78.Fontecha-Barriuso M, et al. Targeting epigenetic DNA and histone modifications to treat kidney disease. Nephrology, dialysis, transplantation: official publication of the European Dialysis and Transplant Association - European Renal Association. 2018;33:1875–1886. doi: 10.1093/ndt/gfy009. [DOI] [PubMed] [Google Scholar]
- 79.Gonzalez A, et al. Sodium butyrate ameliorates insulin resistance and renal failure in CKD rats by modulating intestinal permeability and mucin expression. Nephrology, dialysis, transplantation: official publication of the European Dialysis and Transplant Association - European Renal Association. 2019;34:783–794. doi: 10.1093/ndt/gfy238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Inoue K, et al. Podocyte histone deacetylase activity regulates murine and human glomerular diseases. The Journal of clinical investigation. 2019;129:1295–1313. doi: 10.1172/JCI124030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zheng Y, Zhang Z, Zhang N. Protective Effects of Butyrate on Renal Ischemia-Reperfusion Injury in Rats. Urologia internationalis. 2019;102:348–355. doi: 10.1159/000497476. [DOI] [PubMed] [Google Scholar]
- 82.Franceschi F, Zampetti A, Gigante G, Gasbarrini A. Helicobacter pylori and small intestinal bacterial overgrowth affect gastrointestinal symptoms in Fabry’s disease. Digestive and liver disease: official journal of the Italian Society of Gastroenterology and the Italian Association for the Study of the Liver. 2015;47:618–619. doi: 10.1016/j.dld.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 83.Han B, et al. Microbial Genetic Composition Tunes Host Longevity. Cell. 2017;169:1249–1262 e1213. doi: 10.1016/j.cell.2017.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thaiss CA, et al. Microbiota Diurnal Rhythmicity Programs Host Transcriptome Oscillations. Cell. 2016;167:1495–1510 e1412. doi: 10.1016/j.cell.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 85.Thion MS, et al. Microbiome Influences Prenatal and Adult Microglia in a Sex-Specific Manner. Cell. 2018;172:500–516 e516. doi: 10.1016/j.cell.2017.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Williams JM, et al. Epithelial cell shedding and barrier function: a matter of life and death at the small intestinal villus tip. Veterinary pathology. 2015;52:445–455. doi: 10.1177/0300985814559404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Masotti M, et al. Altered globotriaosylceramide accumulation and mucosal neuronal fiber density in the colon of the Fabry disease mouse model. Neurogastroenterology and motility: the official journal of the European Gastrointestinal Motility Society. 2019;31:e13529. doi: 10.1111/nmo.13529. [DOI] [PubMed] [Google Scholar]