Abstract
The Pyrrolysyl-tRNA synthetase (PylRS) and its cognate tRNAPyl are extensively used to add non-canonical amino acids (ncAAs) to the genetic code of bacterial and eukaryotic cells. However, new ncAAs often require a cumbersome de novo engineering process to generate an appropriate PylRS/tRNAPyl pair. We here report a strategy to predict a PylRS variant with novel properties. The designed polyspecific PylRS variant HpRS catalyzes the aminoacylation of 31 structurally diverse ncAAs bearing clickable, fluorinated, fluorescent, and for the first time biotinylated entities. Moreover, we demonstrated a site-specific and copper-free conjugation strategy of a nanobody by the incorporation of biotin. The design of polyspecific PylRS variants offers an attractive alternative to existing screening approaches and provides insights into the complex PylRS-substrate interactions.
Subject terms: tRNAs, Protein design
Introduction
Site-specific incorporation of non-canonical amino acids (ncAAs) is a powerful tool to implement novel functions into the proteome1–5. The central element of a selective orthogonal ncAA incorporation system is an aminoacyl-tRNA synthetase/tRNA (aaRS/tRNA) pair that does not cross-react with the original aaRSs, tRNAs, or canonical amino acids of the host cell6. The pyrrolysyl-tRNA synthetase/tRNAPyl pair (PylRS/tRNAPyl) has become a popular choice for the genetic code expansion7,8. This success is mainly attributed to the substrate flexibility of the active site9,10, the orthogonality in prokaryotic and eukaryotic cells11, and the inherent suppression of the amber codon8. Although over 150 ncAAs were genetically encoded during the past decade, these structures were mainly based on few core motifs5. Several structural functionalities still remain elusive, e.g., biotinylated ncAAs. The main factor limiting further expansion of the genetic code is the engineering of suitable aaRS/tRNA pairs. This process requires the construction of large mutant libraries from which all non-functional and non-orthogonal aaRS variants are eliminated through alternating rounds of positive and negative selection2. In most cases, selection screens are carried out as dead-and-alive assays on agar plates2,12,13. However, the selection conditions of this approach are hardly tunable, and therefore a beneficial aaRS might be accidentally eliminated1,6,14.
Screens based on fluorescence-activated cell sorting (FACS) provide a more sensitive and quantitative read-out and have been successfully applied to evolve the aaRS specificity15–18. However, the reported experimental setups involve several negative and positive selection rounds, resulting in a cumbersome process17.
Extensive efforts have been made to create a vast number of aaRS in the last years. These synthetases are often selected and used for the incorporation of a particular ncAA, although many engineered synthetases are polyspecific and aminoacylate a range of different ncAAs19,20. Hence, the aaRS-substrate information is not fully exploited and may be used to incorporate novel ncAAs, for which the de novo selection process had failed.
The current study investigated, whether the structural features of different PylRS variants can be recombined to a new synthetase exhibiting inherited and novel properties. Therefore, we created a mutant library based on reported PylRS variants and applied a sensitive selection screen. The identified PylRS-ncAA pairs provided insights for the prediction of the “highly polyspecific pyrrolysyl-tRNA synthetase” (HpRS). HpRS showed a tailored substrate scope and genetically incorporated for the first time biotin lysine. Furthermore, we demonstrate three different conjugation strategies of a GFP-nanobody using HpRS.
Results and Discussion
Library design
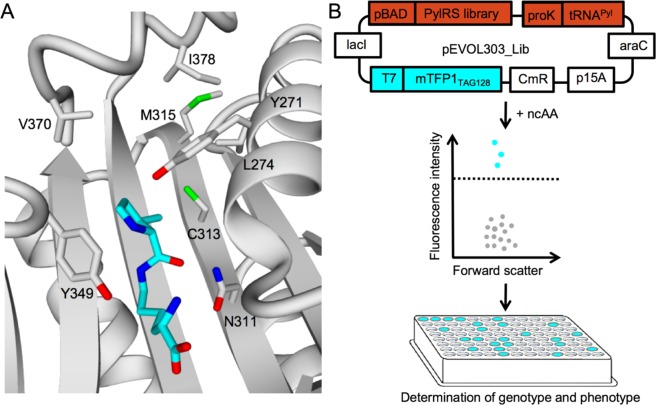
First, we designed a mutant library based on existing PylRS variants and a homology model of the Methanosarcina barkeri (M. barkeri) PylRS (Fig. 1A). The mutation Y349F enhanced the aminoacylation efficiency of PylRS and was included in the mutant library by default10. We maintained the orthogonality of the active site by retaining position N311 to eliminate a negative selection21,22. Based on these structural considerations, we selected the six amino acids Y271, L274, C313, M315, V370, and I378 to create a mutant PylRS library (Supplementary Table 1). In particular, smaller residues were inserted at these positions to increase the volume of the pocket and to allow the aminoacylation of larger ncAAs.
Figure 1.
(A) Homology model of PylRS in complex with pyrrolysine (cyan) in the binding pocket. (B) Cells transformed with the plasmid pEVOL303_Lib are screened for ncAA incorporation by mTFP1-based fluorescence intensity and forward scatter using FACS. Fluorescent cells (cyan) exceeding a set threshold are sorted separately from nonfluorescent cells (grey) for sequencing on a 96-well plate. pEVOL303_Lib encodes for the PylRS library, tRNAPyl and, mTFP1TAG128. p15A origin of replication (p15A), araC repressor gene (araC), chloramphenicol acetyltransferase marker (CmR), lac repressor (lacI), T7 promoter (T7), proK promoter (proK), araBAD promoter (pBAD).
Screening of PylRS variants
Dead-and-alive assays are the preferred method for screening large quantities of variants in a short time as demonstrated on existing aaRS/tRNA pairs2,13. Nevertheless, this screening method suffers from a harsh and hardly tunable selection pressure, causing slow cell growth and low survival rates17. Consequently, even beneficial variants might be unintentionally excluded from the library. Unlike dead-and-alive assays, fluorescence-activated cell sorting (FACS) provides a tunable sorting stringency over a broad dynamic range for the ncAA incorporation15,17,23.
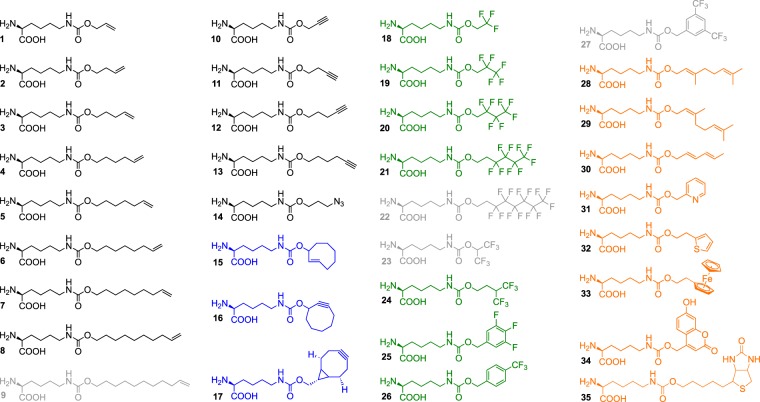
To set up a fast FACS screen, the PylRS library, tRNAPyl and the cyan monomeric teal fluorescent protein 1 (mTFP1) were expressed from a single plasmid (Fig. 1B). The mTFP1 contained an amber codon at position 128 (mTFP1TAG128) and allowed to quantify the incorporation efficiency of a ncAA according to the fluorescence intensity. E. coli cells transformed with pEVOL303_Lib were grown both in the absence and presence of ncAAs 1–23 (Fig. 2).
Figure 2.
Chemical structures of analyzed ncAAs. Class I: linear alkenes, alkynes, and azide (black). Class II: cyclic alkenes and alkynes (blue). Class III: fluorinated side chains (green). Class IV: diverse functionalities (orange). Grey ncAA structures were rejected.
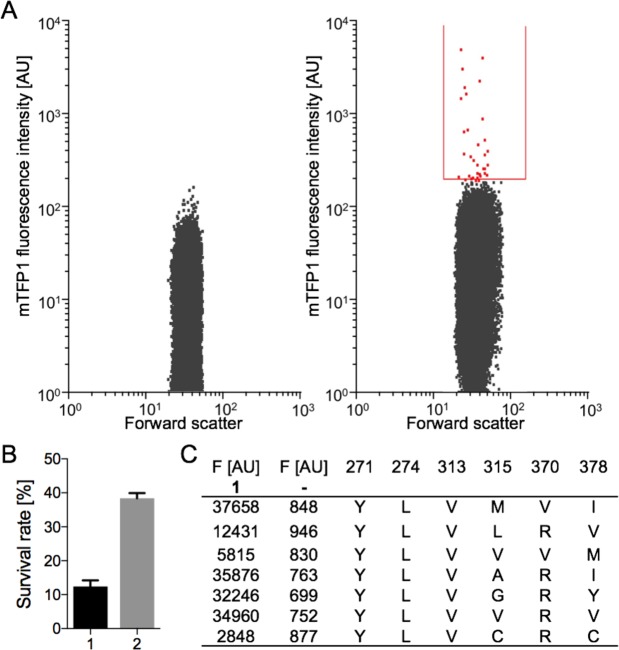
Then, the forward scatter and fluorescent signals of each cell were analysed by FACS. In the presence of ncAA 1, several cells depicted significant fluorescent signals (Fig. 3A, right plot) and negligible fluorescence in the absence of ncAAs indicates the orthogonality of the mutant library (Fig. 3A, left plot). Notably, the fluorescent cells appeared to be a heterogeneous population, suggesting that multiple PylRS variants enabled the aminoacylation of ncAA 1. All fluorescent cells exceeding a set threshold were sorted on a 96-well plate supplied with conditioned medium, recovered overnight, and the PylRS variants were sequenced. The medium contained cell-secreted growth factors that improved the survival rate of single cells three-fold compared to the untreated lysogeny broth (LB) medium (Fig. 3B). In the case of ncAA 1, seven different PylRS variants facilitated its incorporation (Fig. 3C). Next, expression of mTFP1TAG128 was repeated in the presence of the sequenced PylRS/tRNAPyl pairs and ncAA 1 to analyze the incorporation efficiency and orthogonality of the different variants (Fig. 3C).
Figure 3.
Identification of PylRS variants for the incorporation of ncAA 1. (A) (Left plot) Fluorescence intensity and forward scatter of E. coli cells harboring pEVOL303_Lib in the absence of ncAAs analyzed by FACS. (Right plot) Cells harboring pEVOL303_Lib were grown in the presence of 1. Fluorescent cells (red) within the sort gate (red line) were separated from nonfluorescent cells. (B) Survival rate of cells after FACS in LB medium (1) and conditioned medium (2). Results were obtained from five independent experiments; standard deviation is indicated by error bars. (C) Fluorescence intensities of mTFP1TAG128 expressed with 1 or without – in the present of seven different PylRS variants (mutations are shown).
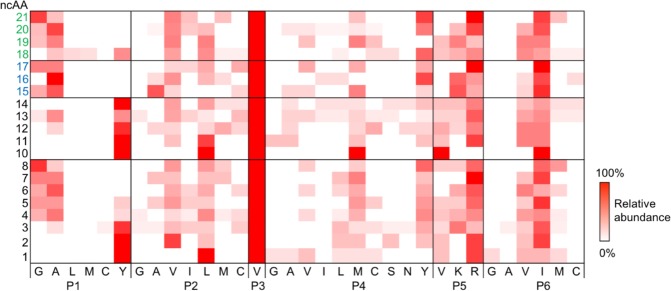
Overall, the screen identified 151 PylRS variants (110 unique and 41 redundant) that incorporated 20 out of the 23 initially tested ncAAs. The incorporation of ncAAs 9, 22, and 23 were not detected by FACS. Seventeen PylRS variants accepted multiple ncAAs. Individual ncAAs were aminoacylated by up to 16 different PylRS variants (Supplementary Table 3). The amino acid abundancies of the six mutated positions of all sorted PylRS variants are summarized for each ncAA in a heat map (Fig. 4). We divided the tested ncAAs 1–23 into three structural classes to elucidate key mutations in the binding pocket. Class I ncAAs 1–8 (alkenes) and 10–14 (alkynes) bear a linear aliphatic tail. Like a molecular ruler, these ncAAs allow to assess the maximum carbon chain length accepted by our PylRS library. Class II ncAAs 15–17 are cyclic and can be used in strain-promoted cycloadditions24,25. Highly fluorinated side chains of class III 18–23 might single out PylRS variants that accept bulky ncAAs, since the volume of a CF3 substituent (39.8 Å3) is about twice as big as a methyl-group (21.6 Å3)26.
Figure 4.
Heat map of the analyzed ncAAs 1–23 versus the six variable positions of the PylRS library (P1 = Y271, P2 = L274, P3 = C313, P4 = M315, P5 = V370, P6 = I378). Amino acids are represented by a one-letter code; the color intensity indicates the relative abundance of a mutation. Mutations not present in any of the sequenced PylRS variants have been omitted form the map. The ncAAs 9, 22, and 23 were not incorporated.
Pocket-substrate relationship
As expected, the 20 incorporated ncAAs required diverse structural adaptions of PylRS (Fig. 4). Y271 (P1) defines the bottom of the binding pocket; therefore, this position strongly varied with the ncAA size. While tyrosine was highly conserved at P1 for the short ncAAs 1, 2, 10, 11, and 14 alanine was favored by the larger ncAAs 4–7, 13, 15–17, 19, and 20. Aminoacylation of the longest ncAAs 8 and 21 was preferred by glycine at P1 of the active site. We observed a similar trend for the position L274 (P2), which constitutes the rear end of the binding pocket. While leucine was favored by the small ncAAs 1, 10, and 11, the shorter valine occurred in high abundance for larger ncAAs. Interestingly, the mutation C313V (P3), was strictly conserved throughout all sequenced variants. M315 (P4) differed widely for small ncAAs; however, tyrosine was more abundant among the larger ncAAs 8, 16, 20, and 21. V370 (P5) and I378 (P6) were not influenced by the size or chemistry of the incorporated ncAAs. Arginine and isoleucine respectively appeared with the highest frequency at these positions.
Design of a polyspecific PylRS variant
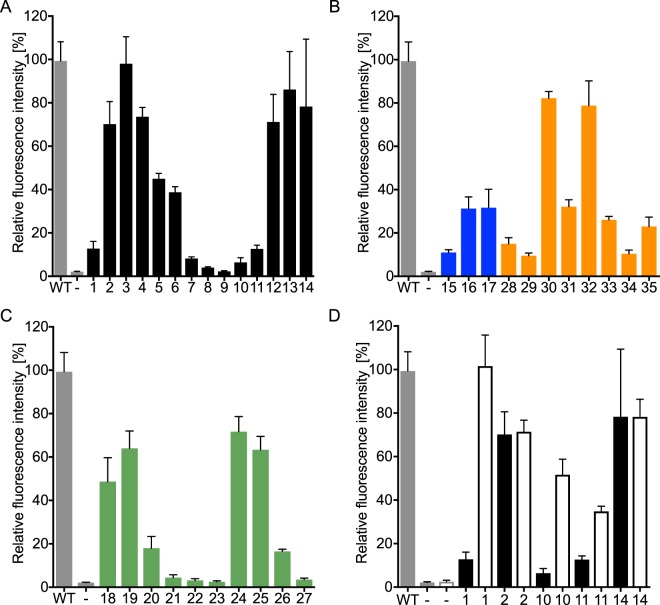
We combined potential key mutations Y271A, L274V, C313V, M315Y, Y349F, and V370R to design a highly polyspecific PylRS (HpRS) for the incorporation of large ncAAs. To test the substrate scope, the HpRS/tRNAPyl pair and mTFP1TAG128 were expressed with 1–35 in E. coli. The incorporation of the ncAAs 1–8, 10–21, 24–26, 28–35 was detected by fluorescence signal (Fig. 5) and additionally confirmed by SDS-PAGE (Supplementary Fig. 1) and ESI-TOF (Supplementary Table 4, Supplementary Fig. 2). Further, we used Alexa Fluor 532 streptavidin conjugate to confirm the incorporation of ncAA 35 in mTFP1TAG128 (Supplementary Fig. 3).
Figure 5.
HpRS-dependent incorporation of ncAAs 1–35 in mTFP1TAG128. Fluorescence intensity of mTFP1 is normalized against wild-type (WT) mTFP1 (grey bar), while expression of mTFP1TAG128 in the absence of ncAAs (−, grey bar) served as a negative control. Error bars indicate the standard deviation of three independent experiments. (A) Substrate profiles of HpRS for class I (black): ncAAs 1–14. (B) Class II (blue): ncAAs 15–17 and class IV (orange): ncAAs 28–35. (C) Class III (green): ncAAs 18–27. (D) Comparison of the aminoacylation efficiencies of HpRS (solid) and the PylRS variant (C313V, Y349F) (no fill) for ncAAs 1, 2, 10, 11, and 14.
Intriguingly, HpRS tolerated a broad range of lysine carbamates with diverse features, including aliphatic chains, fluorinated residues, cyclic structures, and a biotinylated ncAA. As predicted, HpRS was highly capable of aminoacylation of ncAAs with long side chains 3-6; yet, the smallest tested ncAAs were only moderately accepted (Fig. 5). Therefore, the mutations C313V and Y349F were combined based on the heat map to generate a PylRS particular suited for small ncAAs. Indeed, this variant showed a significant higher aminoacylation efficiency of the small ncAAs 1, 10, and 11 than HpRS (Fig. 6D).
Figure 6.
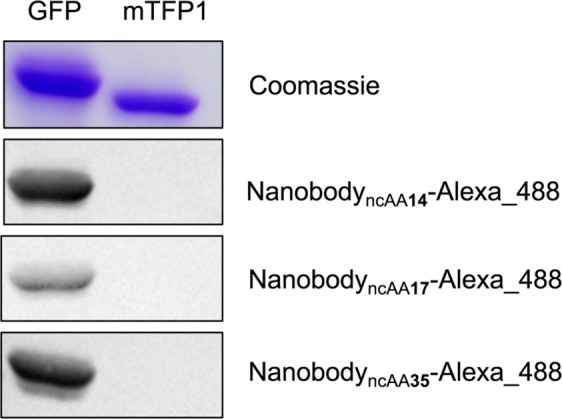
Coomassie stained SDS gel of cell lysates containing GFP or mTFP1 and western blot on the same cell lysates with an anti-GFP nanobody bearing ncAA 14, 17 or 35 conjugated to Alexa Fluor 488.
The polyspecific PylRS containing the mutations Y271A and Y349F has been reported for 15–1724,27. To compare HpRS with PylRS (Y271A, Y349F), both synthetases were co-expressed with mTFP1TAG128 in the presence of 15–17 and the fluorescence signals were analysed. The ncAAs were incorporated in mTFP1TAG128 in equivalent yields by both synthetases (Supplementary Fig. 4).
Labeling of nanobodies
Immunoassays such as flow cytometry or western blot usually require commercially available antibodies-conjugates. In contrast, nanobodies derived from Camelid antibodies, offer nowadays an attractive alternative28, as they are highly stable, small and can be expressed in heterologous systems such as E. coli in high yields29. The developed HpRS/tRNAPyl pair was employed to incorporate ncAA 14, 17, 35 in an anti-GFP nanobody, which exhibits a specific binding activity against the green fluorescence protein (GFP)30, at the exposed position K44 with expression yields of 8 mg/L, 4 mg/L, 4 mg/L in E. coli culture, respectively. The ncAA incorporation into anti-GFP nanobodyTAG44 was confirmed by SDS-PAGE (Supplementary Fig. 5) and LC-MS analysis. To conjugate the anti-GFP nanobodyncAA14, the applied azide was linked to an Alexa Fluor 488 alkyne via copper-catalyzed azide-alkyne “click” reaction. The bicyclononyne-lysine (BCN), ncAA 17, in the anti-GFP nanobodyncAA17 formed a strain-promoted [3 + 2] azide-alkyne cycloaddition with an Alexa Fluor 488 azide. The biotin containing anti-GFP nanobodyncAA35 bound successfully an Alexa Fluor 488 marked streptavidin.
A western blot of cell lysates containing GFP or mTFP1, as a negative control, demonstrated the specific detection of GFP by the established anti-GFP nanobody-conjugates (Fig. 6). Notably, the most efficient coupling reaction between anti-GFP nanobody and fluorescence-dye seemed to be the copper-catalyzed “click” reaction and the biotin-streptavidin interaction, thus resulting in the strongest western blot signal (Fig. 6).
In summary, we demonstrated that the PylRS-substrate interaction can be used to recombine key mutation to generate new PylRS variants. This seems at first glance a limiting engineering approach, however as a proof of concept the designed HpRS exhibit a remarkable substrate scope and facilitates the incorporation of a biotin bearing ncAA. A prerequisite for this method is the generation of a diverse pool of beneficial PylRS variants that was achieved by a fast and mild FACS setup. PylRS diversity for a particular ncAA provided insights into the complex PylRS-substrate interaction and allowed the prediction of polyspecific variants and can now be used as an example for others to avoid the cumbersome engineering process.
Moreover, HpRS was used to insert 14, 17 or 35 in an anti-GFP nanobody. The three different chemical handles were exploited to produce fluorescent nanobody-conjugates. In particular, the streptavidin-biotin interaction allows efficient labeling of the anti-GFP nanobody without a catalyst. Therefore, the incorporation of 35 might be a good starting point to simplify techniques such as western blotting, ELISA, flow cytometry or antibody labeling in general.
Methods
Strains
All cloning steps were performed in TOP10 E. coli (Thermo Fisher Scientific). The FACS and protein expression experiments were carried out with BL21 (DE3) (Lucigen, Middleton, WI).
Medium
A conditioned medium was used to improve the survival of sorted cells after FACS. To prepare the conditioned medium, BL21 (DE3) cultures were grown in LB (OD600~1.0), centrifuged at 8000 × g for 20 min at 4 °C and the supernatant was sterilized by passing through a 0.22 μm cellulose filter (Millipore, Bedford, MA).
Non-canonical amino acids (ncAA)
ncAA 1 (Sigma-Aldrich), ncAA 15–17 (Sirius Fine Chemicals, Bremen, Germany), and ncAAs 24–27, 30–33, as well as 35 (SUNGYOUNG Chemical Limited, Shanghai, China) were purchased from commercial sources. ncAA 34 was synthesized according to the procedure published by Luo et al.31 ncAA 2–14, 18–23, 28, and 29 were synthesized according to a modified procedure from Li et al.32 (Supplementary Information, S2).
Plasmid construction
The plasmid pEVOL303 was constructed by ligating the plasmid pEVOL33 with the commercially available pET303/CT-His (Thermo Fisher Scientific) using Gibson cloning34. Prior to ligation, pEVOL was digested with the restriction enzymes SacI and PciI. The required region of pET303 was amplified using the primers pET303_f and pET303_r (Supplementary Table 2). The resulting plasmid, pEVOL303, carried a p15A origin, a chloramphenicol resistance gene, a Methanosarcina barkeri PylRS gene downstream of a pBAD promoter, and the corresponding tRNA gene controlled by a proK promoter. The fluorescent protein gene with an N-terminal 6xHis-SUMO-tag, either wild-type mTFP1 or mTFP1TAG128 (bearing an amber codon at position 128), was cloned downstream of a T7 promoter. The amber codon was inserted using the primers mTFP_128_f and mTFP_128_r (Supplementary Table 2). The GFP-nanobody was encoded on the plasmid pOPINE30. The amber codon was inserted at position 44 using the primers GFP_Nano_f and GFP_Nano_r (Supplementary Table 2).
PylRS library construction
Methanosarcina barkeri
PylRS containing the single mutation Y349F served as the starting point for our mutant library. Six positions in PylRS, Y271, L274, C313, M315, V370, and I378, were selected for our library based on a homology model derived from the crystal structure of PylRS (PDB 4Q6G and 4CS3) (Fig. 1A,B)35,36. A pre-defined set of amino acids was inserted into the six positions, resulting in 95,040 mutants. To ensure an equal distribution of all the mutants, the library was assembled non-codon redundant and codon usage optimized by the company, Life Technologies (Thermo Fisher Scientific) and introduced to pEVOL303 via the restriction sites SalI and BglII. The resulting plasmid, pEVOL303_Lib, was transformed into BL21 (DE3) cells by applying the standard electroporation protocol. The transformation was determined with 1 × 107 CFU per preparation.
Sample preparation for FACS
BL21 (DE3) cells transformed with pEVOL303_Lib were inoculated in 50 mL LB medium supplemented with 25 µg/mL chloramphenicol (Cm), and grown until an OD600 of 0.7. Then, 450 µL of the cells was transferred into a 2 mL reaction vessel and induced with 50 µL induction medium (10 mM ncAA, 10 mM IPTG, 1% w/v arabinose) and incubated for 16 h at 37 °C and 700 rpm. The cells were diluted to a final concentration of 1 × 107 cell/mL and washed twice with an M9 minimal medium supplemented with 25 µg/mL chloramphenicol. In the beginning of each FACS experiment, the negative control (cell growth in the absence of ncAA) was screened first to ensure the discrimination of canonical amino acids by the PylRS mutant library. Cell sorting was performed by detecting the fluorescence of mTFP1 with a BD Influx (BD Biosciences) operated with filter-sterilized BD FACS Flow Sheath Fluid, a 457 nm laser for excitation, and a 480/40 bandpass filter. The selected operation mode was 1.0 Drop Single. The selection threshold (gate) was adjusted based on the fluorescent signal from the first 1 × 105 cells of each sample. Cells within the gate were sorted separately on a 96-well plate (Sigma-Aldrich) supplied with the conditioned medium containing 25 µg/mL chloramphenicol. The growth of the sorted cells was continued overnight at 37 °C and 300 rpm. Each culture was sequenced using the primers Lib_seq_f and Lib_seq_r (Supplementary Table 2). pEVOL303_Lib containing the sequenced PylRS variants were retransformed into new BL21 (DE3) cells, grown and induced with the induction medium at OD600 of 0.7. Correspondingly, PylRS, tRNAPyl, and mTFP1TAG128 were expressed. Both the incorporation of the ncAAs and the orthogonality of the PylRS variant were confirmed by fluorescence measurement of mTFP1 (excitation 462 nm, emission 492 nm) in a black, 96-well plate (Thermo Fisher Scientific) with an Infinite M1000 plate reader (Tecan, Zurich, Switzerland).
ncAA incorporation
A single BL21 (DE3) colony containing pEVOL303 encoding for HpRS, PylRS (Y271A and Y349F) or PylRS (C313V and Y349F) was selected from the LB/Cm agar plate and inoculated into 50 mL LB/Cm media, then incubated and shaken overnight at 37 °C. The resulting culture was diluted (1:100) with fresh LB/Cm media and grown to an OD600 of 1.5. The mTFP1 expression was triggered by adding 1 mM ncAA, 1 mM IPTG, and 0.1% w/v arabinose and cell growth continued at 37 °C and 700 rpm for 16 h. Fluorescence was measured correspondingly. For SDS-PAGE, the sample was heated at 75 °C for 20 min and cleared by centrifugation. For protein purification, cell pellet was suspended in lysis buffer (100 mM Tris pH 7.5, 500 mM NaCl, 20 mM imidazole, 10%, v/v glycerol), incubated at 75 °C for 20 min and cleared by centrifugation
The supernatant was loaded onto a HisTrap HP Ni–NTA column (GE Healthcare). Protein eluted by increasing the concentration of imidazole from 20 mM to 500 mM in the lysis buffer over 10 column volumes. SUMO protease was added to the eluted protein, dialyzed overnight at 4 °C against ddH2O and passed through a HisTrap HP Ni–NTA to remove the remaining N-terminal 6xHis- as well as the SUMO-tag.
The HpRS/tRNAPyl pair was employed to incorporate the respective ncAAs at position K44 of the reported anti-GFP nanobody30. BL21(DE3) were grown at 37 °C to OD600 0.7 in 500 mL LB medium. Protein synthesis was induced by 0.5 mM IPTG, 0.1% w/v arabinose, and 1 mM ncAA. Propagation was carried out at 20 °C for 20 h. Cells were harvested by centrifugation, lysed by sonication and cleared by centrifugation. The cell lysate was applied onto a HisTrap HP Ni–NTA column and eluted in 50 mM Tris-HCl pH 7.5, 300 mM NaCl, 1 mM TCEP, 500 mM Imidazole. The eluted protein was concentrated and loaded onto a HiLoad Superdex 200 (16/60) size-exclusion column (GE Healthcare) equilibrated with 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM TCEP.
Conjugation of nanobodies
500 nM purified GFP-nanobody containing 14, 17 or 35 was labeled with either Alexa 488 alkyne (40 µM, 1 h) using the Click-iT Protein Reaction Buffer Kit, Alexa 488 azide (40 µM, 6 h) or Alexa 488 streptavidin (5 µM, 1 h) at 37 °C (Thermo Fisher Scientific).
Western blot
E. coli cell lysate containing GFP or mTFP1 was electrophoretically fractionated and electroblotted onto 0.45 μm Immobilon-P polyvinylidene difluoride (PVDF) membranes (Merck, Darmstadt, Germany). After electroblotting, the membranes were washed three times for 5 min with TBS buffer and blocked in TBST (10 mM Tris, 0.9%, w/v NaCl, 0.05%, v/v, Tween-20, pH 7.4) with 1% (w/v) casein for 3 h at room temperature. The blots were probed and incubated with the conjugated GFP-nanobodies (1 µg/mL) at room temperature for 60 min. The washing steps were performed with TBST. Fluorescence was detected in an iBright FL1000 imaging system (Invitrogen).
Mass spectrometry
Pure mTFP1 and GFP-nanobodies were analysed by mass spectrometry (maXis HD ESI-TOF, Bruker). The sample was injected into a high-performance liquid chromatography (Agilent Technologies, C4 column, column volume of 5 mL) and separation was performed at a constant flow rate of 0.5 µL/min and a gradient of 80% acetonitrile and 0.1% formic acid for 8 min. Fractions were recorded according to the standard procedure.
Homology model of the Methanosarcina barkeri PylRS in complex with pyrrolysine
Homology models were generated using the YASARA Structure, Version 14.7.1737. The catalytic domain of Methanosarcina barkeri PylRS served as the template for YASARA’s homology modeling macro, using the conservative “slow” protocol with the following parameter settings: number of PSI-BLAST iterations – 10; maximum allowed BLAST E-value to consider template – 0.5; maximum oligomerization state – 2; maximum number of alignment variations per template – 4; maximum number of conformations tried per loop – 200; and maximum number of residues added to the termini – 20. The resulting homology model was based on two structures of PylRS variants, PDB 4Q6G and 4CS3; both sequences had a 98% homology with the template35,36. The YASARA algorithm performed the secondary structure prediction, loop construction, and amino-acid rotamer selection, followed by a steepest-descent energy minimization.
Supplementary information
Engineering a Polyspecific Pyrrolysyl-tRNA Synthetase by a High Throughput FACS Screen
Acknowledgements
The research reported in this publication was supported by funding from King Abdullah University of Science and Technology (KAUST). We thank the SFB749/A10 (M.G.) for financial support. We are grateful to Prof. Peter G. Schultz (The Scripps Research Institute, La Jolla, CA) for kindly providing the original pEVOL-PylRS plasmid.
Author Contributions
J.E. and M.R. conceived and supervised the study; A.H. and R.K. designed the experiments; A.H., R.K., A.A., D.R. and X.L. performed experiments, A.H., R.K., S.G. and M.G. analyzed data; A.H. and R.K. wrote the manuscript; A.H., R.K., M.G., M.R. and J.E. made manuscript revisions.
Competing Interests
The authors declare the following competing financial interest(s): Patent filed with the United States Patent and Trademark Office (USPTO) on the May 08, 2017, and assigned serial number 62/502, 860.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Adrian Hohl and Ram Karan contributed equally.
Contributor Information
Ram Karan, Email: ram.karan@kaust.edu.sa.
Magnus Rueping, Email: magnus.rueping@kaust.edu.sa.
Jörg Eppinger, Email: jorg.eppinger@kaust.edu.sa.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-48357-0.
References
- 1.Neumann H, Peak-Chew SY, Chin JW. Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat. Chem. Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 2.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 3.Lang K, et al. Genetic encoding of bicyclononynes and trans-cyclooctenes for site-specific protein labeling in vitro and in live mammalian cells via rapid fluorogenic Diels-Alder reactions. J. Am. Chem. Soc. 2012;134:10317–10320. doi: 10.1021/ja302832g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plass T, et al. Amino acids for Diels-Alder reactions in living cells. Angew. Chem. Int. Ed. Engl. 2012;51:4166–4170. doi: 10.1002/anie.201108231. [DOI] [PubMed] [Google Scholar]
- 5.Dumas A, Lercher L, Spicer CD, Davis BG. Designing logical codon reassignment - Expanding the chemistry in biology. Chem. Sci. 2015;6:50–69. doi: 10.1039/c4sc01534g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 7.Fekner T, Chan MK. The pyrrolysine translational machinery as a genetic-code expansion tool. Curr. Opin. Chem. Biol. 2011;15:387–391. doi: 10.1016/j.cbpa.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wan W, Tharp JM, Liu WR. Pyrrolysyl-tRNA synthetase: an ordinary enzyme but an outstanding genetic code expansion tool. Biochim. Biophys. Acta. 2014;1844:1059–1070. doi: 10.1016/j.bbapap.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Polycarpo CR, et al. Pyrrolysine analogues as substrates for pyrrolysyl-tRNA synthetase. FEBS Lett. 2006;580:6695–6700. doi: 10.1016/j.febslet.2006.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yanagisawa T, et al. Multistep engineering of pyrrolysyl-tRNA synthetase to genetically encode N(epsilon)-(o-azidobenzyloxycarbonyl) lysine for site-specific protein modification. Chem. Biol. 2008;15:1187–1197. doi: 10.1016/j.chembiol.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Hancock SM, Uprety R, Deiters A, Chin JW. Expanding the genetic code of yeast for incorporation of diverse unnatural amino acids via a pyrrolysyl-tRNA synthetase/tRNA pair. J. Am. Chem. Soc. 2010;132:14819–14824. doi: 10.1021/ja104609m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie J, Schultz PG. A chemical toolkit for proteins–an expanded genetic code. Nat. Rev. Mol. Cell. Biol. 2006;7:775–782. doi: 10.1038/nrm2005. [DOI] [PubMed] [Google Scholar]
- 13.Voloshchuk N, Montclare JK. Incorporation of unnatural amino acids for synthetic biology. Mol. Biosyst. 2010;6:65–80. doi: 10.1039/b909200p. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Zhang Z, Brock A, Schultz PG. Addition of the keto functional group to the genetic code of Escherichia coli. Proc. Natl. Acad. Sci. 2003;100:56–61. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santoro SW, Wang L, Herberich B, King DS, Schultz PG. An efficient system for the evolution of aminoacyl-tRNA synthetase specificity. Nat. Biotechnol. 2002;20:1044–1048. doi: 10.1038/nbt742. [DOI] [PubMed] [Google Scholar]
- 16.Link AJ, et al. Discovery of aminoacyl-tRNA synthetase activity through cell-surface display of noncanonical amino acids. Proc. Natl. Acad. Sci. 2006;103:10180–10185. doi: 10.1073/pnas.0601167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhn SM, Rubini M, Fuhrmann M, Theobald I, Skerra A. Engineering of an orthogonal aminoacyl-tRNA synthetase for efficient incorporation of the non-natural amino acid O-methyl-L-tyrosine using fluorescence-based bacterial cell sorting. J. Mol. Biol. 2010;404:70–87. doi: 10.1016/j.jmb.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Owens AE, Grasso KT, Ziegler CA, Fasan R. Two-tier screening platform for directed evolution of aminoacyl-tRNA synthetases with enhanced stop codon suppression efficiency. Chembiochem. 2017;18:1109–1116. doi: 10.1002/cbic.201700039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyake-Stoner SJ, et al. Generating permissive site-specific unnatural aminoacyl-tRNA synthetases. Biochemistry. 2010;49:1667–1677. doi: 10.1021/bi901947r. [DOI] [PubMed] [Google Scholar]
- 20.Cooley RB, Karplus PA, Mehl RA. Gleaning unexpected fruits from hard-won synthetases: probing principles of permissivity in non-canonical amino acid-tRNA synthetases. Chembiochem. 2014;15:1810–1819. doi: 10.1002/cbic.201402180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kavran JM, et al. Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation. Proc. Natl. Acad. Sci. 2007;104:11268–11273. doi: 10.1073/pnas.0704769104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang YS, et al. The de novo engineering of pyrrolysyl-tRNA synthetase for genetic incorporation of L-phenylalanine and its derivatives. Mol. Biosyst. 2011;7:714–717. doi: 10.1039/c0mb00217h. [DOI] [PubMed] [Google Scholar]
- 23.Yoo TH, Tirrell DA. High-throughput screening for methionyl-tRNA synthetases that enable residue-specific incorporation of noncanonical amino acids into recombinant proteins in bacterial cells. Angew. Chem. Int. Ed. Engl. 2007;46:5340–5343. doi: 10.1002/anie.200700779. [DOI] [PubMed] [Google Scholar]
- 24.Nikic I, et al. Minimal tags for rapid dual-color live-cell labeling and super-resolution microscopy. Angew. Chem. Int. Ed. Engl. 2014;53:2245–2249. doi: 10.1002/anie.201309847. [DOI] [PubMed] [Google Scholar]
- 25.Lang K, Chin JW. Cellular incorporation of unnatural amino acids and bioorthogonal labeling of proteins. Chem. Rev. 2014;114:4764–4806. doi: 10.1021/cr400355w. [DOI] [PubMed] [Google Scholar]
- 26.Muller K, Faeh C, Diederich F. Fluorine in pharmaceuticals: looking beyond intuition. Science. 2007;317:1881–1886. doi: 10.1126/science.1131943. [DOI] [PubMed] [Google Scholar]
- 27.Plass T, Milles S, Koehler C, Schultz C, Lemke EA. Genetically encoded copper-free click chemistry. Angew. Chem. Int. Ed. Engl. 2011;50:3878–3881. doi: 10.1002/anie.201008178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Meyer T, Muyldermans S, Depicker A. Nanobody-based products as research and diagnostic tools. Trends. Biotechnol. 2014;32:263–270. doi: 10.1016/j.tibtech.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 29.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 30.Kubala MH, Kovtun O, Alexandrov K, Collins BM. Structural and thermodynamic analysis of the GFP:GFP-nanobody complex. Protein Sci. 2010;19:2389–2401. doi: 10.1002/pro.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo J, et al. Genetically encoded optochemical probes for simultaneous fluorescence reporting and light activation of protein function with two-photon excitation. J. Am. Chem. Soc. 2014;136:15551–15558. doi: 10.1021/ja5055862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li HM, Chen CY, Padros JB. Highly efficient carbamate formation from alcohols and hindered amino acids or esters using N,N′-Disuccinimidyl Carbonate (DSC) Synlett. 2011;2011:1454–1458. doi: 10.1055/s-0030-1260584. [DOI] [Google Scholar]
- 33.Young TS, Ahmad I, Yin JA, Schultz PG. An enhanced system for unnatural amino acid mutagenesis in E. coli. J. Mol. Biol. 2010;395:361–374. doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 34.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 35.Guo LT, et al. Polyspecific pyrrolysyl-tRNA synthetases from directed evolution. Proc. Natl. Acad. Sci. 2014;111:16724–16729. doi: 10.1073/pnas.1419737111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt MJ, Weber A, Pott M, Welte W, Summerer D. Structural basis of furan-amino acid recognition by a polyspecific aminoacyl-tRNA-synthetase and its genetic encoding in human cells. Chembiochem. 2014;15:1755–1760. doi: 10.1002/cbic.201402006. [DOI] [PubMed] [Google Scholar]
- 37.Krieger E, Vriend G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015;36:996–1007. doi: 10.1002/jcc.23899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Engineering a Polyspecific Pyrrolysyl-tRNA Synthetase by a High Throughput FACS Screen