Abstract
Anthropogenic landscape changes such as land use change and habitat fragmentation are known to alter wildlife diversity. Since host and parasite diversities are strongly connected, landscape changes are also likely to change wildlife parasite diversity with implication for wildlife health. However, research linking anthropogenic landscape change and wildlife parasite diversity is limited, especially comparing effects of land use change and habitat fragmentation, which often cooccur but may affect parasite diversity substantially differently. Here, we assessed how anthropogenic land use change (presence of plantation, livestock foraging and human settlement) and habitat fragmentation may change the gastrointestinal parasite diversity of wild mammalian host species (n = 23) in Anamalai hills, India. We found that presence of plantations, and potentially livestock, significantly increased parasite diversity due possibly to spillover of parasites from livestock to wildlife. However, effect of habitat fragmentation on parasite diversity was not significant. Together, our results showed how human activities may increase wildlife parasite diversity within human-dominated landscape and highlighted the complex pattern of parasite diversity distribution as a result of cooccurrence of multiple anthropogenic landscape changes.
Subject terms: Ecology, Ecology
Introduction
Land use change and habitat fragmentation are two major landscape-level outcomes of human activities that significantly impact biodiversity1–3. Hence, considerable research on biodiversity change in human-dominated landscape have been conducted, which has resulted in improved understanding of how these two human impacts on landscape can impact biodiversity1,4,5. These anthropogenic factors can also modify host–parasite interactions, which, in turn, can lead to either increase or decrease in parasite diversity6–8. Understanding how these factors may influence parasite diversity is ecologically important for multiple reasons. For instance, parasites regulate host population dynamics9, alter species communities10 and constitute a significant proportion of total biomass of any ecosystem11, which is not surprising considering parasites comprise at least 40% of all animal species on earth12. Despite their ecological importance, our knowledge on parasite diversity is limited13,14, particularly in the context of increasing human impact on environment, underlining a significant research gap15,16. The gap is specifically wide for wildlife hosts and urgent research is required in the face of recent increased emergence of novel pathogens of wildlife origin7,17,18. It is, thus, crucial to answer how anthropogenic land use change and habitat fragmentation may impact parasite diversity in the wild.
Land use change can affect parasites both directly and indirectly. By altering environment (for example, through pollution), land use change may render transmission of environmentally-transmitted parasites difficult. This is particularly true for parasites that has life stages outside host body. However, land use change can indirectly impact parasite diversity by altering host diversity as it is one of the strongest predictors of parasite diversity19–22. By decreasing host diversity and abundance, land use change can deplete richness of parasites particularly those that require multiple obligatory hosts23. This is evident when many host species that are threatened in their natural habitat appear to harbour fewer parasites24. On the other hand, land use change can also increase parasite diversity in multiple ways. Land use change can increase parasite diversity by increasing host diversity. For instance, land use change such as agricultural field or land-fill can act as resource traps and amplify host diversity artificially25. Land use change can also increase parasite diversity by introducing non-native parasites such as parasites of domestic and feral animals and even from humans26.
It is also important to distinguish between different types of land use change and their effects on parasites27. One type of land use change that has not been studied well is the effect of plantation on wildlife parasites27. Plantations are usually monocultures of exotic or native plant species grown as timber or fuel wood or as cash crops and have a large and increasing footprint in wildlife habitats worldwide28. They can sometime act as refuge to wildlife but usually with a biotic homogenising effect29,30. Consequently, plantation may also increase but homogenise parasite community. Plantations are often accompanied by settlement of labourers and livestock foraging31,32. These changes within a wildlife habitat can both increase or decrease parasite diversity. Parasite diversity may decrease if wildlife hosts avoid human areas to lessen confrontation with humans and resource competition with livestock. On the other hand, generalist species may actually thrive in human settlements by utilizing novel resources33,34. Herbivore species may also prefer to stay closer to human settlements and livestock (“spatial refugia”) that may displace predators35–38. Moreover, many wildlife, over time, may actually get habituated to humans and livestock and aggregate near human-dominated landscape39,40. These aggregations may eventually increase parasite diversity by increasing contact between native host species. Such situations may also increasingly expose wildlife to humans and human-associated animals, such as livestock and commensals, increasing chance of spillover of non-native parasites to wildlife.
Habitat fragmentation may lead to higher parasite diversity because heavily fragmented habitats may disrupt wildlife dispersal and increase host diversity in smaller fragments. Such increase in host diversity in a smaller patch may alter host characteristics such as home range, abundance and intra and interspecific contacts thus increasing overlap among host species making host individuals exposed to higher parasite infections41,42. These effects are likely to be greatest in the smallest and most isolated of the fragments3,43. By disrupting host dispersal, fragmentations can also adversely affect parasite diversity. This could be especially true for parasites who require multiple host species to complete its life cycle, such as those that are transmitted trophically44. So far, many studies looked into this effect but the results have been mixed6,41,45–47.
The Anamalai (Elephant hills in Tamil) hills of southern India is a highly biodiverse rainforest habitat of Western Ghats, which holds about 30% of India’s plant and vertebrate species diversity in less than 6% of the country’s area48. It is also one of the most altered natural habitats in India and typifies different levels of land use change and habitat fragmentation rampant in Indian wildlife habitats. Large section of the habitat is highly modified due to land use change, bordered by large, relatively undisturbed tropical rainforests. The landscape is a matrix of over 40 rainforest fragments (1–2,500 ha in size),often surrounded by plantations (coffee, tea and cardamom), roads, hydroelectric dams and settlements49. Highly-modified fragments contain within them human settlements and have higher livestock pressures than other remote, less disturbed fragments. In spite of such high levels of land use change and habitat fragmentation, the Anamalai hills still harbour a large number of wildlife whose ranges often unavoidably overlap with humans and livestock50–53. In fact, large number of wildlife species are regularly observed within human-dominated habitats and this concurrence with humans often precipitates into wildlife-human conflicts49,50,54–56. It is possible that many of the wildlife are important reservoirs of multiple environmentally-transmitted parasites. In fact, recent studies have recorded important parasite groups within certain host species populations that may cause Ascariasis, Trichuriasis and Strongylodiasis in humans45,46,57,58.
To assess the effect of land use change (plantation, livestock foraging and human settlements) and habitat fragmentation on parasite diversity, we studied gastrointestinal parasites of wild mammalian hosts across rainforest fragments in Anamalai hills. We employed statistical models to test these effects. We predicted a positive impact of land use change on parasite diversity due to increased host diversity and an increased exposure of wildlife to humans and livestock. For habitat fragmentation too, we predicted an increase in parasite diversity with decrease in habitat size and increase in habitat isolation. Our alternative predictions were that land use change and habitat fragmentation could actually deplete parasite diversity by decreasing host diversity in disturbed fragments. Finally, it is also possible that land use change and habitat fragmentation may not significantly impact parasite diversity either by not impacting host community or by not spillover from non-native hosts such as livestock and humans.
Materials and Methods
Ethical statement
For this study, faecal samples were collected only noninvasively. As a result, no animal was sacrificed or harmed during sampling. Part of the sampling was done within Anamalai Tiger Reserve, which is a protected area. Hence, appropriate written permission was taken from the Tamil Nadu Forest Department (Letter Ref. No. WL 5/58890/2008, dated 2 September 2009). All methods were carried out in accordance with relevant guidelines and regulations and, all experimental protocols were approved by the Institutional Animal Ethical Committee.
Study site
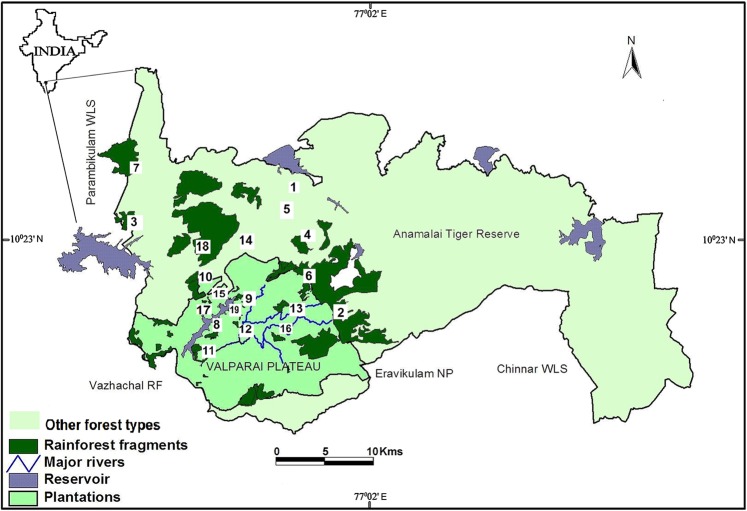
Located south of the Palghat gap (11°N) of the Western Ghats, Anamalai hills once had large tracts of tropical rainforest dotted with few tribal settlements. Between 1860 and 1930, British colonisers started clearing the rainforests extensively for cultivation of tea and coffee and developing teak and Eucalyptus plantations, particularly in the Valparai Plateau59. As a result, the Anamalais today consists of both a relatively undisturbed, large (958.59 km2; ~100,000 ha) tropical rainforest within the protected Anamalai Tiger Reserve (ATR; 10°12′–10°35′N and 76°49′–77°24′E) and about 1,000 ha highly degraded Valparai Plateau (Fig. 1). The plateau consists of many tea estates and other plantations, which are surrounded by four protected areas—ATR in Tamil Nadu state and three others in Kerala state. The major vegetation types include scrub forests in the rain-shadow areas in the eastern foothills, dry and moist deciduous forests (<800 m), mid-elevation tropical wet evergreen forest (600–1,500 m) and high-altitude shola-grassland ecosystems (>1,500 m)60. Although a large part of the tropical wet evergreen forests occurs within ATR, many of the smaller (<200 ha) fragments are found in private estates in the Valparai plateau. These small fragments are highly degraded and disturbed due to fuel-wood collection and livestock grazing. Valparai town also is a part of the plateau and around 200,000 people live across the town and plantations60. Due to the ongoing habitat fragmentation, the whole landscape is a matrix of over 40 rainforest fragments, ranging 1 ha-2,500 ha in size and often surrounded by plantations (coffee, tea and cardamom), roads, hydroelectric dams and settlements49. Based on size range (2–2,500 ha), level of perceived human disturbance and access, we selected 19 forest fragments in total for sampling —16 mid-elevation tropical rainforest and three low-elevation dry and moist deciduous forest fragments (Fig. 1).
Figure 1.
Map of Anamalai hills, Western Ghats, India with numbered study fragments. (1) Aliyar dam, (2) Akkamalai, (3) Anaikundi, (4) Andiparai, (5) Attakatty, (6) Iyerpadi (7) Karian_shola, (8) Korangumudi, (9) Monica_estate, (10) Monomboly, (11) Nirar_dam, (12) Pannimedu, (13) Puthuthottam, (14) Sethumadai (15) Shekkalmudi (16) Sirukundra (7) Uralikal, (18) Varagaliyar and (19) Varattuparai WLS: Wildlife sanctuary; RF: Reserve Forest; NP: National Park
Host sampling
Between Oct 2013 and Oct 2015, faecal samples were collected from populations of mammalian wildlife. We collected fresh faecal samples non-invasively during the day on transects (400 m-3 km in length). For large and medium herbivores and primates, we followed individuals and collected fresh faeces when animals defaecated. For elusive species such as carnivores, we identified home range based on secondary information and faecal samples were identified based on morphology and also using nearby secondary signs such as pug-marks or hoof-prints. To avoid sampling the same individual repeatedly, only one sample of a host species was collected from each spot and the sample source was either marked or removed whenever possible. However, if removal was not possible (for example, due to large quantity), then we marked. To avoid contamination from soil, samples were collected from the inside of the bolus or only top pellet was collected from a heap. We immediately fixed each sample in 10% formaldehyde solution (50 ml), labelled the containers with the information of origin (fragment name, date, time and host species) and stored them at room temperature until parasitological screening. Differences in sampling effort can confound the comparison of diversity among replicates. We accounted for differences in number of host species encountered by calculating richness estimates with the assumption that each faecal sample represents single individual. We used bootstrap, which is a resampling method for estimating the whole sampling distribution of richness by sampling with replacement from the original sample, that can offer greater precision than jackknife estimates, especially when sample sizes are small61.
Parasite sampling
Employing both the flotation and sedimentation techniques (NaNO3 solution), we screened the faecal samples for the presence of helminth eggs, larvae and protozoan cysts62. For each concentration technique, we examined two slides per sample. Slides were examined under a light microscope (400X). Eggs and cysts were first examined at 10× magnification and then their size was measured with a micrometre eyepiece (0.1 μm) at 40× magnification. To facilitate identification of parasite eggs, we often added a drop of Lugol’s iodine solution to the slides to highlight detailed structures. In addition, photographs of each parasite species have been archived and are available for examination on request. We identified parasites to the lowest possible taxonomic level using published keys63,64. Differences in sampling effort can confound the comparison of diversity among replicates. We accounted for differences in number of parasite taxon encountered by calculating richness estimates with the assumption that each faecal sample represents single host individual. We used bootstrap, which is a resampling method for estimating the whole sampling distribution of richness by sampling with replacement from the original sample. Bootstrap can offer greater precision over jackknife estimator, especially when sample sizes are small61. This method is particularly recommended for parasite richness estimation65.
Land use data
In Anamalai hills, land use change manifests in largely three forms—presence of human settlements, plantations and livestock foraging. There are only few large (>1000 ha) fragments that are legally protected and thus undisturbed. Many of the studied fragments share more than one type of land use change. For instance, some fragments with human settlements may also have livestock present. For the current study, we identified 18 fragments with land use change, out of which 15 (83.3%) had plantation, in contrast to three (16.7%). Eleven (61.1%) of the fragments have significant livestock foraging pressure, in contrast to seven (38.9%) fragments without livestock. Finally, ten (55.6%) of the fragments had human settlements within them, in contrast to eight (44.4%) without settlements.
Habitat fragmentation data
To measure effect of habitat fragmentation, we used fragment size and isolation distance between fragments. According to the equilibrium theory of island biogeography, organism dispersal probability declines as distance between islands increases, reducing rates of immigration and, in turn, reducing diversity66–68. Assuming each forest fragment as an island, their isolation was summarized with an isolation index which was calculated as the sum of the square root of the distances to the nearest equivalent (no smaller than 80% of size) or larger fragment (Dahl, 2004). Data on fragment size, distance between fragments and presence of human settlements, plantations and livestock were collected from earlier studies from Anamalai hills45,60.
Data analyses
To assess the effects of land use change and habitat fragmentations on bootstrap estimate of parasite taxon richness, we created two different linear mixed effects models69. Each model included random effects of host species and fragments to account for multiple observations within each fragment (across host species) and across fragments. In the land use model, the predictor variables (fixed effects) were presence of plantation, human settlement and livestock. The predictor variables for the habitat fragmentation model were fragment size and fragment isolation index. In both the models, we incorporated both bootstrap estimates of host species richness and host body mass as co-predictors as these were known to effect parasite richness. We retrieved host body mass data from online ecological database70. Given the potential role of host density as a covariate to parasite richness (discussed in Introduction), we also planned collection of host density data. This exercise, however, became logistically and financially impractical due to the large number of host species (>20) studied, 19 rainforest patches sampled (totaling >12,000 ha in size), often lack of visibility within the rainforest patches and limited time and resources available to us. Consequently, we dropped this covariate from our study and decided instead to investigate the specific hypotheses born out of the current study (discussed in Discussion) in future, based on a logistically manageable subset of study sites and hosts.
After fitting these models to the data, we also compared and selected the best fit model using lowest AIC value71. At the end, diagnostics were run to check distribution of the residuals for each model. This analysis was conducted in the lme4 package72. We also assessed the effects of land use change and habitat fragmentation on bootstrap estimates of host species richness using two linear models. In the land use model, the predictor variables were presence of plantation, human settlement and livestock. The predictor variables for the habitat fragmentation model were fragment size and fragment isolation index. We followed the same strategy as described above for model fitting, fitting diagnostics and model selection. Finally, we tested whether land use change homogenized the composition of the parasite community. We used a multivariate nonparametric Analysis of Variance (permAnoVa; 1,000 permutations) based on the Jaccard dissimilarity index for a matrix of parasite presence/absence. We calculated the variance of homogeneity of parasite communities within each fragment based on disturbed vs. undisturbed divisions using the betadisper function of the vegan package in R73.
Results
Sample diversity
From 19 forest fragments, we collected 4,056 mammalian faecal samples belonging to 23 mammalian wildlife species and two livestock species—domestic goats Capra aegagrus and cattle Bos taurus. Analyses were done only on wildlife samples. Number of samples varied from 41 in Uralikal to 495 in Puthuthottam (Table 1). Number of samples for each host species varied between six in otter Lutra lutra and 623 in gaur B. gaurus). In total, seven protozoa (18.42%) and 32 helminth (81.58%) species were recorded, including five trematodes, five cestodes and 20 nematodes. At least seven different parasites, belonging to different parasite groups, were recorded in ≥20 different host species—protozoa Coccidia sp. (23 hosts); cestodes Hymenolepis nana (20 hosts) and Moniezia sp. (22 hosts); and nematodes Gongylonema sp. (20 hosts), Strongyloides sp. (23 hosts), Trichuris sp. (24 hosts) and Ascaris sp. (26 hosts). On the other hand, cestode Dipylidium sp. and nematode Parascaris sp. were found only in small Indian civet Viverricula indica and Indian porcupine Hystrix indica samples, respectively.
Table 1.
Bootstrap estimate of host richness in each fragment of Anamalai hills, India.
| Fragment name | Bootstrap estimate of host species richness | SE | Sample size |
|---|---|---|---|
| Aliyar dam | 21.9 | 0.8 | 210 |
| Akkamalai | 18.9 | 0.7 | 199 |
| Anaikundi | 20.1 | 0.8 | 150 |
| Andiparai | 22.6 | 1 | 256 |
| Attakatty | 11.7 | 0.6 | 15 |
| Iyerpadi | 25.5 | 1 | 398 |
| Karian_shola | 25.3 | 0.9 | 244 |
| Korangumudi | 24.9 | 0.8 | 356 |
| Monica estate | 20.4 | 0.9 | 124 |
| Monomboly | 26.6 | 1 | 181 |
| Nirar dam | 24.2 | 0.9 | 167 |
| Pannimedu | 17.5 | 1 | 55 |
| Puthuthottam | 24.9 | 0.8 | 426 |
| Sethumadai | 18.1 | 0.9 | 65 |
| Shekkalmudi | 16.0 | 0.8 | 42 |
| Sirukundra | 17.9 | 0.8 | 127 |
| Uralikal | 11.5 | 0.5 | 41 |
| Varagaliyar | 21.6 | 1 | 162 |
| Varattuparai | 23.4 | 0.9 | 397 |
Host and parasite diversity and disturbance
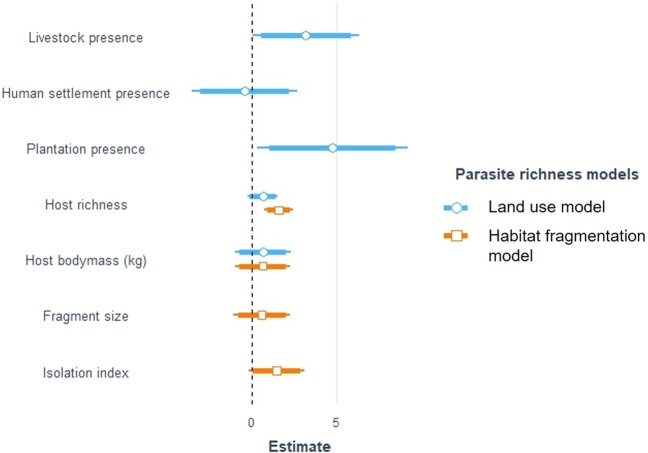
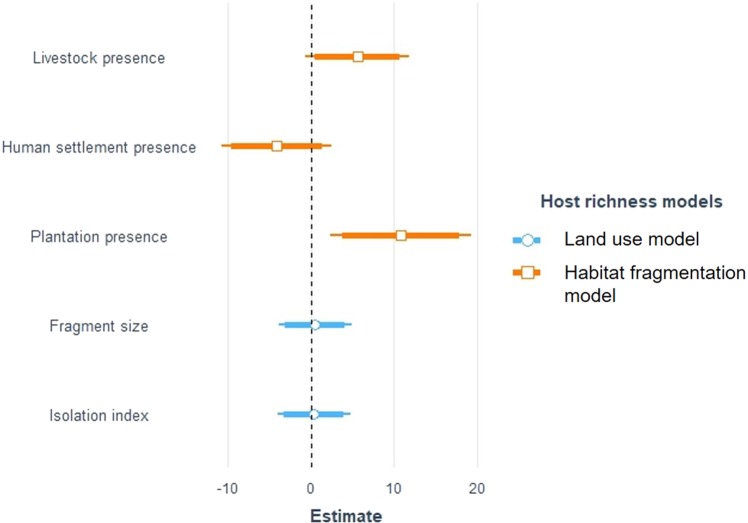
For parasite diversity analysis, the human disturbance model was the best fit (Table 2). Parasite diversity was significantly driven by presence of plantation (estimate = 4.779, CIProfile = 0.326–9.232, t = 2.103, p < 0.05). Presence of livestock had a substantial but not significant positive effect (estimate = 3.209, CIProfile = −0.052–6.366, t = 1.992, p > 0.05). Effects of settlement, host richness and host body mass on parasite richness were not significant (Fig. 2). For host diversity analysis the human disturbance model was again the best fit (Table 3). Presence of plantation was the only predictor that had a significant positive effect on host diversity (estimate = 10.798, CIProfile = 2.302–19.294, t = 2.726, p < 0.05 (Fig. 3)—almost half of all host species occur in plantations. Although presence of livestock did not have a significant effect, its wide confidence interval was mostly on the positive side suggesting potential positive impact—limited by sample size—on host richness (estimate = 5.602, CIProfile = −0.639–11.843, t = 1.925, p > 0.05). Similarly, presence of human settlement did not significantly affect host richness, however, the substantial effect was mostly on the negative side, suggesting potential negative effect on host diversity (estimate = −4.112, CIProfile = −10.717–2.492, t = −1.335, p > 0.05).
Table 2.
Comparison between two different models to explain bootstrap estimate of parasite taxon richness in Anamalai hills, India.
| Models | K | logLik | AIC | delta | weight |
|---|---|---|---|---|---|
| Plantation + Settlement + Livestock + Host richness + Host body size (Land use model) | 9 | −667.54 | 1353.07 | 0 | 0.887 |
| Fragment size + Isolation index + Host richness + Host body size (Habitat fragmentation model) | 8 | −670.59 | 1357.19 | 4.112 | 0.113 |
Figure 2.
Unstandardized effect size of predictor variables on bootstrap estimate of parasite taxon richness in rainforest fragments of Anamalai hills, India. Estimates were plotted to scale. Intercepts were omitted to avoid distortion of scale. Land use model was the best fitted model based on AIC. Confidence intervals are represented by the lines around the points— thick (α = 0.10) and thin (α = 0.05).
Table 3.
Comparison between two different models to explain bootstrap estimate of host species richness in Anamalai hills, India.
| Models | K | logLik | AIC | delta | weight |
|---|---|---|---|---|---|
| Plantation + Settlement + Livestock(Land use model) | 5 | −46.73 | 103.47 | 0 | 0.971 |
| Fragment size + Isolation index(Habitat fragmentation model) | 4 | −51.25 | 110.5 | 7.029 | 0.029 |
Figure 3.
Unstandardized effect size of predictor variables on bootstrap estimate of host species richness in rainforest fragments of Anamalai hills, India. Estimates were plotted to scale. Intercepts were omitted to avoid distortion of scale. Land use model was the best fitted model based on AIC. Confidence intervals are represented by the lines around the points— thick (α = 0.10) and thin (α = 0.05). Host sample sizes are given in Table 1.
We recorded 12 parasites (ten helminths and two protozoa) that occurred only in plantations. Six of the ten helminths were nematodes (60%), while rest were trematodes (30%) and one cestode (10%). Fragments without plantations did not harbour any parasite taxon exclusively, which means parasites in those undisturbed fragments also occured in plantations. Fragments with livestock harboured three parasite taxa (two nematodes and one cestode) exclusively relative to their undisturbed counterpart. However, only one parasite taxon (Taenia sp.) exclusively occurred in livestock disturbed fragments, while other two nematodes also occurred in the plantations. Its counterpart undisturbed fragments only harboured one taxon exclusively (Paragonimus sp.), which however also occurred in plantations. Finally, settlements harboured three nematode taxa exclusively in comparison to their undisturbed counterpart. Only one of these taxa (Uncinaria sp.) were exclusive to settlements across all fragments. Undisturbed counterpart of settlements harboured only one parasite taxon (Sarcocystis sp.) exclusively.
Parasite and host homogeneity
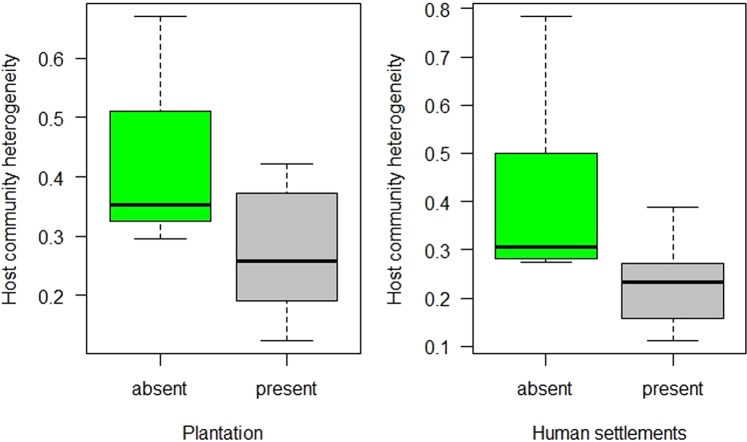
Parasite communities within disturbed forest fragments were not significantly more homogeneous than the undisturbed ones due to presence of either plantations (F = 2.58, p > 0.05), livestock (F = 0.04, p > 0.05) or settlements (F = 3.55, p > 0.05). Host communities within plantations (TukeyHSD; p < 0.05; Fig. 4a) and human settlements (TukeyHSD; p < 0.05; Fig. 4b) were, however, significantly more homogeneous than undisturbed fragments. Finally, we did not find any of the disturbance variables to significantly alter the parasite community composition between undisturbed and disturbed fragments.
Figure 4.
Host community heterogeneity between undisturbed (absent) and disturbed (present) rainforest fragments of Anamalai hills, India. Community heterogeneity is the within group dispersion values based on Jaccard distance for presence/absence data.
Discussion
Our results reveal that rainforest fragments with plantations (and potentially with livestock) in Anamalai hills harbour significantly higher parasite diversity than undisturbed fragments. Interestingly, some of the disturbed fragments (at least, fragments with plantations) also has significantly more host diversity than the undisturbed fragments, however host diversity was not found to significantly affect parasite diversity.
In Anamalai hills, plantations (coffee, tea and cardamom) had more mammalian wildlife species richness than the undisturbed fragments. This was not particularly a surprising result because studies have reported similar high richness in vertebrate species from plantation within wildlife habitats74,75. In fact, earlier studies from Western Ghats also found high vertebrate richness within or around plantations with large variations depending on plantation types, from open tea to more shaded coffee and cardamom plantations30,76,77. The reason for such increased host diversity is thought to be an increase in habitat heterogeneity within plantations. Increased habitat heterogeneity is thought to generate greater diversity of niches consequently facilitating cooccurrence of many species78,79. However, such increase in species richness is often accompanied by more generalist and wide-ranging species being more abundant within the plantations and a loss of community heterogeneity relative to undisturbed habitats80–82. We found similar loss of heterogeneity for host species in disturbed habitats with plantations and settlements (Fig. 4).
Effect of livestock presence on host species richness was positive but not statistically significant at α = 0.05. The effect, however, was significant at α = 0.10, which suggested potential, but weak effect that was reflected by the almost equal number of wildlife species recorded from these two groups of fragments (nLivestock = 20 and nUndisturbed = 22). Interaction between livestock and wildlife is complicated. For instance, while a number of studies found evidence of competitive exclusion between livestock and large herbivore83, many other recorded resource sharing between these two groups84,85. Yet still, many other studies did not find any relationship between the two86. The outcome of the interaction may depend on the ecological similarity between the two groups (Niche overlap), availability of natural resources that may vary between habitats (between low to high productivity) and also degree of behavioural habituation by the wildlife. The wildlife community that we studied was an ecologically broad one consisting of wildlife with very different ecology. Therefore, while some of the species—such as spotted deer Axis axis and sambar deer Rusa unicolor, who were found only in the undisturbed fragments—may face resource competition from livestock grazing, others (for example, small carnivores and primates) may not face any competition. In addition, many large herbivores, such as gaur B. gaurus and Indian elephants Elephas maximus indicus, who despite resource competition, may still use the disturbed fragments as corridors contributing to host richness. These processes together may explain almost similar host species richness between fragments with and without livestock grazing.
We did not find any significant effect of human settlement on host diversity but the trend is negative (Fig. 3). While human settlement may attract and facilitate generalist and weedy species with high tolerance for disturbance (for example, rodents, which were not sampled in the present study), many elusive species such as carnivores may be adversely affected and may prefer to avoid fragments with settlements87. Still, we recorded overall a large host species richness (host richnessSettlement = 19, host richnessUndisturbed = 23) from around the settlement in Anamalai hills. This could be explained by the facts that many of these settlements may attract wildlife with unintentionally supplemented resources such as planted fruit trees60. Additionally, the high level of fragmentation of the landscape meant large herbivores and carnivores may not have much choice but to disperse through human settlements54,56. We did not find evidence of habitat fragmentation (fragment size and isolation) influencing host species richness in Anamalai hills (Fig. 3). This is in line with findings from across studies that reported effects of fragmentation on species communities are often weak88. Effects of habitat fragmentation on species diversity is also highly context-specific and varies considerably between animal groups, ecosystems and kinds of human activities prevalent in the landscape88–92. In Anamalai hills, habitat fragmentation is widespread, which likely disrupts animal movement to some extent but, in the absence of hunting, perhaps not substantially. For instance, studies recorded use of certain plantation as corridors to connect with isolated undisturbed habits51,52,77. However, the adverse outcome of these movement through human-dominated habitats is the increase in wildlife-human conflict54,56.
Among the different types of land use change in Anamalai hills, plantations had the strongest positive effect on parasite diversity (Fig. 2). Increase in number of parasite taxa in modified fragments ranged between one to ten, with eight parasite taxa that were recorded exclusively in these fragments (Table 4). However, this increased richness in disturbed fragments were likely not driven by host richness as host richness had a small and statistically not significant effect on parasite richness in the disturbance model (Fig. 2). This is in contrast to the predominant patterns across most studies on parasite diversity that found host richness to be the strongest predictor of parasite richness19–22. However, there could be potential deviations from this rule, particularly due to human impacts21,93,94. For instance, many human parasites may spillover to wildlife (anthropozoonoses) as humans regularly come in contact with wildlife95–100. In addition to livestock, humans may also introduce many non-wildlife species such as feral dogs and cats into wildlife habitats and these species may share parasites with wildlife26. In such cases, parasite richness in wildlife would be more than in the undisturbed fragments. Indeed, all but one (Schistosoma sp.) of the parasites that we found exclusively in plantations also occurred in cattle (Table 4). Surprisingly, wildlife parasite taxa that were present in the livestock foraging fragments did not occur in cattle samples from the same fragments. This was also the case for the wildlife parasites that only occurred in settlement but not in undisturbed fragments. We did not find any significant effect of host body mass on parasite diversity (Fig. 2). This is in contrast to many studies that found a significant relationship between these two variables101,102. On the other hand, many other empirical studies did not find any relationship between body mass and parasite richness when accounting for host phylogenetic relationships103,104. Such contradictory results may suggest that relationship between host body mass and parasite diversity is a factor of body mass and life history traits, which vary between ecologically different groups of hosts22. Based on these observations, the broad ecological diversity among host species in the present study might have confounded this relationship.
Table 4.
Parasite taxa that were found only in disturbed or undisturbed fragments in Anamalai hills, India.
| Parasite taxa | Parasite group | Family | In livestock samplesa | Known human caseb |
|---|---|---|---|---|
| Plantation only | ||||
| Baylisascaris sp. | Nematodes | Ascaridoidea | Present | present |
| Nematodirus sp. | Nematodes | Trichostrongyloidea | Present | present |
| Enterobius sp. | Nematodes | Oxyuroidea | Present | present |
| Dictyocaulus sp. | Nematodes | Trichostrongyloidea | Absent | absent |
| Uncinaria sp. | Nematodes | Ancylostomatoidea | Absent | present |
| Schistosoma sp. | Trematodes | Schistosomatidae | Absent | present |
| Metastrongylus sp. | Nematodes | Metastrongyloidea | Absent | present |
| Clonorchis sp. | Trematodes | Opisthorchiidae | Present | present |
| Toxoplasma sp. | Apicomplexa | Present | present | |
| Isospora sp. | Apicomplexa | Present | present | |
| Paragonimus sp. | Trematodes | Paragonimidae | Present | present |
| Dipylidium sp. | Cestodes | Dilepididae | Present | present |
| Livestock presence only | ||||
| Dictyocaulus sp. | Nematodes | Trichostrongyloidea | Absent | absent |
| Taenia sp. | Cestodes | Taeniidae | Absent | present |
| Metastrongylus sp. | Nematodes | Metastrongyloidea | Absent | present |
| Undisturbed (Livestock) only | ||||
| Paragonimus | Trematodes | Paragonimidae | Present | present |
| Settlement only | ||||
| Dictyocaulus sp. | Nematodes | Trichostrongyloidea | Absent | absent |
| Uncinaria sp. | Nematodes | Ancylostomatoidea | Absent | present |
| Metastrongylus sp. | Nematodes | Metastrongyloidea | Absent | present |
| Undisturbed (Settlement) only | ||||
| Sarcocystis sp. | Apicomplexa | Present | present | |
Parasite taxa in bold font were found in the corresponding fragment group exclusively. aCurrent study; bNatural History Museum parasite database, London, UK.
Our results did not find any significant effect of habitat fragmentation on parasite richness (Fig. 2). This was expected as we did not find any effect of fragmentation on host richness either. This lack of relationship between fragmentation and parasite diversity could also be an outcome of large home ranges and low habitat specialisation of most of the host species in our study. Many of the species that we sampled were large herbivores or carnivores (e.g., Elephas maximus, Bos gaurus, Panthera tigris, Panthera pardus) with larg home range and they disperse across fragments. The level of fragment isolation (Median distance = 30.2 km) may not be a deterrent to their dispersal. Similarly, many host species in the study community, such as bonnet macaque Macaca radiata, wild pig Sus scrofa, small Indian civet Viverricula indica, are habitat generalists. These species, according to the distribution-abundance relationship hypothesis6, may be adapted to smaller, fragmented habitats. These hosts may then spread parasites across habitats, independent of the level of habitat fragmentation.
Conclusion
In the present study, we demonstrated that human-driven land use changes increased parasite diversity in a rainforest habitat and presence of potential spillover of parasites from livestock to wildlife. We also showed that the observed pattern of parasite diversity was not driven by habitat fragmentation.
One of the limitations of this study was that it could not test the effect of land use change and habitat fragmentation on the relationship between host density and parasite diversity. Host density is an important predictor of parasite diversity and in nature, host density is linked to host ecology (e.g., home range). However, land use change can unpredictably change host density, which may have a complex outcome for parasite diversity. It will thus be worthwhile in future to explore this question in the present system. Additionally, with the present evidence of potential anthropozonosis, it will be important in future to compare parasite from the present study to samples from humans, livestock and commensal animals in the fragments. Finally, as far as land use change and habitat fragmentation of wildlife habitats in India are concerned, the present study represents a case study with particular relevance for tropical rainforest habitats. However, there exists a large diversity in habitats and levels of disturbance in India. Given the increased threat to wildlife health from anthropogenic environmental change, it will thus be crucial for wildlife conservation biologists to study the patterns of parasite diversity in other types of habitats, especially those with already threatened wildlife.
Acknowledgements
D.C. gratefully acknowledges Steffen Foerster for initial discussions on model selection. D.C. also used part of Fulbright fellowship to work on the current paper, for which he gratefully acknowledges support of US-India Education Foundation (US-IEF), New Delhi. G.U. gratefully acknowledges funding awarded to him by Department of Biotechnology, Government of India (Ref. No. BTPR4503/BCE/8/899/2012 dated 09-11-2012).
Author Contributions
Sampling and data collection were done by D.M.R., S.T. and D.C. D.C. conducted data analysis and wrote the manuscript. G.U. conceived, designed and lead the study, organised funding and reviewed the manuscript.
Data Availability
The datasets generated and analysed during the current study are available from the corresponding author on request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Foley JA, et al. Global consequences of land use. science. 2005;309:570–574. doi: 10.1126/science.1111772. [DOI] [PubMed] [Google Scholar]
- 2.Goudie, A. S. Human impact on the natural environment. (John Wiley & Sons, 2018).
- 3.Haddad NM, et al. Habitat fragmentation and its lasting impact on Earth’s ecosystems. Sci. Adv. 2015;1:e1500052. doi: 10.1126/sciadv.1500052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dornelas M, et al. Assemblage time series reveal biodiversity change but not systematic loss. Science. 2014;344:296–299. doi: 10.1126/science.1248484. [DOI] [PubMed] [Google Scholar]
- 5.McGill BJ, Dornelas M, Gotelli NJ, Magurran AE. Fifteen forms of biodiversity trend in the Anthropocene. Trends Ecol. Evol. 2015;30:104–113. doi: 10.1016/j.tree.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 6.Bordes F, et al. Habitat fragmentation alters the properties of a host–parasite network: rodents and their helminths in South‐East Asia. J. Anim. Ecol. 2015;84:1253–1263. doi: 10.1111/1365-2656.12368. [DOI] [PubMed] [Google Scholar]
- 7.Keesing F, et al. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature. 2010;468:647. doi: 10.1038/nature09575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wood CL, Sandin SA, Zgliczynski B, Guerra AS, Micheli F. Fishing drives declines in fish parasite diversity and has variable effects on parasite abundance. Ecology. 2014;95:1929–1946. doi: 10.1890/13-1270.1. [DOI] [PubMed] [Google Scholar]
- 9.Anderson RM, Jackson HC, May RM, Smith AM. Population dynamics of fox rabies in Europe. Nature. 1981;289:765. doi: 10.1038/289765a0. [DOI] [PubMed] [Google Scholar]
- 10.Wood CL, et al. Parasites alter community structure. Proc. Natl. Acad. Sci. 2007;104:9335–9339. doi: 10.1073/pnas.0700062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuris AM, et al. Ecosystem energetic implications of parasite and free-living biomass in three estuaries. Nature. 2008;454:515. doi: 10.1038/nature06970. [DOI] [PubMed] [Google Scholar]
- 12.Dobson A, Lafferty KD, Kuris AM, Hechinger RF, Jetz W. Homage to Linnaeus: how many parasites? How many hosts? Proc. Natl. Acad. Sci. 2008;105:11482–11489. doi: 10.1073/pnas.0803232105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bordes F, Morand S. Parasite diversity: an overlooked metric of parasite pressures? Oikos. 2009;118:801–806. doi: 10.1111/j.1600-0706.2008.17169.x. [DOI] [Google Scholar]
- 14.Poulin R. Parasite biodiversity revisited: frontiers and constraints. Int. J. Parasitol. 2014;44:581–589. doi: 10.1016/j.ijpara.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Dunn RR, Harris NC, Colwell RK, Koh LP, Sodhi NS. The sixth mass coextinction: are most endangered species parasites and mutualists? Proc. R. Soc. Lond. B Biol. Sci. 2009;276:3037–3045. doi: 10.1098/rspb.2009.0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunn RR, Davies TJ, Harris NC, Gavin MC. Global drivers of human pathogen richness and prevalence. Proc. R. Soc. Lond. B Biol. Sci. 2010;277:2587–2595. doi: 10.1098/rspb.2010.0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones KE, et al. Global trends in emerging infectious diseases. Nature. 2008;451:990. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith KF, et al. Global rise in human infectious disease outbreaks. J. R. Soc. Interface. 2014;11:20140950. doi: 10.1098/rsif.2014.0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dallas TA, et al. Gauging support for macroecological patterns in helminth parasites. Glob. Ecol. Biogeogr. 2018;27:1437–1447. doi: 10.1111/geb.12819. [DOI] [Google Scholar]
- 20.Kamiya T, O’dwyer K, Nakagawa S, Poulin R. Host diversity drives parasite diversity: meta-analytical insights into patterns and causal mechanisms. Ecography. 2014;37:689–697. doi: 10.1111/j.1600-0587.2013.00571.x. [DOI] [Google Scholar]
- 21.Krasnov BR, Shenbrot GI, Khokhlova IS, Degen AA. Relationship between host diversity and parasite diversity: flea assemblages on small mammals. J. Biogeogr. 2004;31:1857–1866. doi: 10.1111/j.1365-2699.2004.01132.x. [DOI] [Google Scholar]
- 22.Morand S. (macro-) Evolutionary ecology of parasite diversity: From determinants of parasite species richness to host diversification. Int. J. Parasitol. Parasites Wildl. 2015;4:80–87. doi: 10.1016/j.ijppaw.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lafferty KD. Environmental parasitology: what can parasites tell us about human impacts on the environment? Parasitol. Today. 1997;13:251–255. doi: 10.1016/S0169-4758(97)01072-7. [DOI] [PubMed] [Google Scholar]
- 24.Altizer S, Nunn CL, Lindenfors P. Do threatened hosts have fewer parasites? A comparative study in primates. J. Anim. Ecol. 2007;76:304–314. doi: 10.1111/j.1365-2656.2007.01214.x. [DOI] [PubMed] [Google Scholar]
- 25.Becker Daniel J, Hall Richard J, Forbes Kristian M, Plowright Raina K. & Altizer Sonia. Anthropogenic resource subsidies and host–parasite dynamics in wildlife. Philos. Trans. R. Soc. B Biol. Sci. 2018;373:20170086. doi: 10.1098/rstb.2017.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weinstein SB, Lafferty KD. How do humans affect wildlife nematodes? Trends Parasitol. 2015;31:222–227. doi: 10.1016/j.pt.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 27.Gottdenker NL, Streicker DG, Faust CL, Carroll CR. Anthropogenic land use change and infectious diseases: a review of the evidence. Ecohealth. 2014;11:619–632. doi: 10.1007/s10393-014-0941-z. [DOI] [PubMed] [Google Scholar]
- 28.Brockerhoff EG, Jactel H, Parrotta JA, Quine CP, Sayer J. Plantation forests and biodiversity: oxymoron or opportunity? Biodivers. Conserv. 2008;17:925–951. doi: 10.1007/s10531-008-9380-x. [DOI] [Google Scholar]
- 29.Solar RRdC, et al. How pervasive is biotic homogenization in human-modified tropical forest landscapes? Ecol. Lett. 2015;18:1108–1118. doi: 10.1111/ele.12494. [DOI] [PubMed] [Google Scholar]
- 30.Bali A, Kumar A, Krishnaswamy J. The mammalian communities in coffee plantations around a protected area in the Western Ghats, India. Biol. Conserv. 2007;139:93–102. doi: 10.1016/j.biocon.2007.06.017. [DOI] [Google Scholar]
- 31.Wakker, E., Watch, S. & Rozario, J. de. Greasy palms: the social and ecological impacts of large-scale oil palm plantation development in Southeast Asia. Greasy Palms Soc. Ecol. Impacts Large-Scale Oil Palm Plant. Dev. Southeast Asia (2004).
- 32.Richardson, B. Plantation Infrastructure and Labor Mobility in Guiana and Trinidad. Migr. Dev. Implic. For 205–224 (1975).
- 33.McKinney ML, Lockwood JL. Biotic homogenization: a few winners replacing many losers in the next mass extinction. Trends Ecol. Evol. 1999;14:450–453. doi: 10.1016/S0169-5347(99)01679-1. [DOI] [PubMed] [Google Scholar]
- 34.Sih A, Ferrari MC, Harris DJ. Evolution and behavioural responses to human-induced rapid environmental change. Evol. Appl. 2011;4:367–387. doi: 10.1111/j.1752-4571.2010.00166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muhly TB, Semeniuk C, Massolo A, Hickman L, Musiani M. Human activity helps prey win the predator-prey space race. PLoS One. 2011;6:e17050. doi: 10.1371/journal.pone.0017050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berger J. Fear, human shields and the redistribution of prey and predators in protected areas. Biol. Lett. 2007;3:620–623. doi: 10.1098/rsbl.2007.0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hebblewhite M, et al. Human activity mediates a trophic cascade caused by wolves. Ecology. 2005;86:2135–2144. doi: 10.1890/04-1269. [DOI] [Google Scholar]
- 38.Ripple WJ, Beschta RL. Linking a cougar decline, trophic cascade, and catastrophic regime shift in Zion National Park. Biol. Conserv. 2006;133:397–408. doi: 10.1016/j.biocon.2006.07.002. [DOI] [Google Scholar]
- 39.Samia DS, Nakagawa S, Nomura F, Rangel TF, Blumstein DT. Increased tolerance to humans among disturbed wildlife. Nat. Commun. 2015;6:8877. doi: 10.1038/ncomms9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blumstein DT. Habituation and sensitization: new thoughts about old ideas. Anim. Behav. 2016;120:255–262. doi: 10.1016/j.anbehav.2016.05.012. [DOI] [Google Scholar]
- 41.Gillespie TR, Chapman CA. Forest fragmentation, the decline of an endangered primate, and changes in host–parasite interactions relative to an unfragmented forest. Am. J. Primatol. Off. J. Am. Soc. Primatol. 2008;70:222–230. doi: 10.1002/ajp.20475. [DOI] [PubMed] [Google Scholar]
- 42.Kowalewski MM, et al. Black and gold howler monkeys (Alouatta caraya) as sentinels of ecosystem health: patterns of zoonotic protozoa infection relative to degree of human–primate contact. Am. J. Primatol. 2011;73:75–83. doi: 10.1002/ajp.20803. [DOI] [PubMed] [Google Scholar]
- 43.McCallum H, Dobson A. Disease, habitat fragmentation and conservation. Proc. R. Soc. Lond. B Biol. Sci. 2002;269:2041–2049. doi: 10.1098/rspb.2002.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lafferty KD. The evolution of trophic transmission. Parasitol. Today. 1999;15:111–115. doi: 10.1016/S0169-4758(99)01397-6. [DOI] [PubMed] [Google Scholar]
- 45.Chakraborty D, et al. Mammalian gastrointestinal parasites in rainforest remnants of Anamalai Hills, Western Ghats, India. J. Biosci. 2015;40:399–406. doi: 10.1007/s12038-015-9517-5. [DOI] [PubMed] [Google Scholar]
- 46.Hussain S, Ram MS, Kumar A, Shivaji S, Umapathy G. Human presence increases parasitic load in endangered lion-tailed macaques (Macaca silenus) in its fragmented rainforest habitats in southern India. PLoS One. 2013;8:e63685. doi: 10.1371/journal.pone.0063685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wells K, Smales LR, Kalko EK, Pfeiffer M. Impact of rain-forest logging on helminth assemblages in small mammals (Muridae, Tupaiidae) from Borneo. J. Trop. Ecol. 2007;23:35–43. doi: 10.1017/S0266467406003804. [DOI] [Google Scholar]
- 48.Mudappa, D. & Raman, T. S. Beyond the borders: Wildlife conservation in landscapes fragmented by plantation crops in India. 21 (Nature Conservation Foundation, 2012).
- 49.Mudappa, D. & Raman, T. S. Rainforest restoration and wildlife conservation on private lands in the Western Ghats. Mak. Conserv. Work 210–240 (2007).
- 50.Singh M, et al. Distribution, population structure, and conservation of lion-tailed macaques (Macaca silenus) in the Anaimalai Hills, Western Ghats, India. Am. J. Primatol. Off. J. Am. Soc. Primatol. 2002;57:91–102. doi: 10.1002/ajp.10037. [DOI] [PubMed] [Google Scholar]
- 51.Kumar MA, Mudappa D, Raman TS. Asian elephant Elephas maximus habitat use and ranging in fragmented rainforest and plantations in the Anamalai Hills, India. Trop. Conserv. Sci. 2010;3:143–158. doi: 10.1177/194008291000300203. [DOI] [Google Scholar]
- 52.Navya R, Athreya V, Mudappa D, Raman TS. Assessing leopard occurrence in the plantation landscape of Valparai, Anamalai Hills. Curr. Sci. 2014;107:1381–1385. [Google Scholar]
- 53.Kumaraguru A, Saravanamuthu R, Brinda K, Asokan S. Prey preference of large carnivores in Anamalai Tiger Reserve, India. Eur. J. Wildl. Res. 2011;57:627–637. doi: 10.1007/s10344-010-0473-y. [DOI] [Google Scholar]
- 54.Sidhu, S., Raghunathan, G., Mudappa, D. & Raman, T. R. Conflict to Coexistence: Human-Leopard Interactions in a Plantation Landscape in Anamalai Hills, India. Conserv. Soc. 15 (2017).
- 55.Baskaran N, Kannan G, Anbarasan U, Thapa A, Sukumar R. A landscape-level assessment of Asian elephant habitat, its population and elephant–human conflict in the Anamalai hill ranges of southern Western Ghats, India. Mamm. Biol.-Z. Für Säugetierkd. 2013;78:470–481. doi: 10.1016/j.mambio.2013.04.007. [DOI] [Google Scholar]
- 56.Kumar, A. M., Malizia, N. & Koschinsky, J. Spatial Analysis of Human-Elephant Conflicts in a Fragmented Tropical Landscape. (GeoDa Center for Geospatial Analysis and Computation 2011).
- 57.Chakraborty D, Tiwari S, Reddy DM, Umapathy G. Prevalence of gastrointestinal parasites in civets of fragmented rainforest patches in Anamalai Hills, Western Ghats, India. J. Parasitol. 2016;102:463–467. doi: 10.1645/15-834. [DOI] [PubMed] [Google Scholar]
- 58.Tiwari S, Reddy DM, Pradheeps M, Sreenivasamurthy GS, Umapathy G. Prevalence and co-occurrence of gastrointestinal parasites in Nilgiri Langur (Trachypithecus johnii) of fragmented landscape in Anamalai Hills, Western Ghats, India. Curr. Sci. 2017;113:2194. doi: 10.18520/cs/v113/i11/2194-2200. [DOI] [Google Scholar]
- 59.Congreve, H. The Anamalais. (1942).
- 60.Umapathy G, Kumar A. The occurrence of arboreal mammals in the rain forest fragments in the Anamalai Hills, south India. Biol. Conserv. 2000;92:311–319. doi: 10.1016/S0006-3207(99)00097-X. [DOI] [Google Scholar]
- 61.Hellmann JJ, Fowler GW. Bias, precision, and accuracy of four measures of species richness. Ecol. Appl. 1999;9:824–834. doi: 10.1890/1051-0761(1999)009[0824:BPAAOF]2.0.CO;2. [DOI] [Google Scholar]
- 62.Gillespie TR. Noninvasive assessment of gastrointestinal parasite infections in free-ranging primates. Int. J. Primatol. 2006;27:1129. doi: 10.1007/s10764-006-9064-x. [DOI] [Google Scholar]
- 63.Sloss, M. W., Kemp, R. L. & Zajac, A. M. Veterinary clinical parasitology. Vet. Clin. Parasitol. Iowa State Univ. Press Google Sch (1994).
- 64.Foreyt, W. J. Veterinary parasitology reference manual. (John Wiley & Sons, 2013).
- 65.Poulin Robert. Comparison of Three Estimators of Species Richness in Parasite Component Communities. The Journal of Parasitology. 1998;84(3):485. doi: 10.2307/3284710. [DOI] [PubMed] [Google Scholar]
- 66.Whittaker, R. J. & José, M. F-P. Island biogeography: ecology, evolution, and conservation. Oxford University Press, (2007).
- 67.MacArthur RH, Wilson EO. An equilibrium theory of insular zoogeography. Evolution. 1963;17:373–387. doi: 10.1111/j.1558-5646.1963.tb03295.x. [DOI] [Google Scholar]
- 68.Wilson, E. O. & MacArthur, R. H. The theory of island biogeography. Princeton University Press, (1967).
- 69.Bolker BM, et al. Generalized linear mixed models: a practical guide for ecology and evolution. Trends Ecol. Evol. 2009;24:127–135. doi: 10.1016/j.tree.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 70.Jones KE, et al. PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals: Ecological Archives E090-184. Ecology. 2009;90:2648–2648. doi: 10.1890/08-1494.1. [DOI] [Google Scholar]
- 71.Aho K, Derryberry D, Peterson T. Model selection for ecologists: the worldviews of AIC and BIC. Ecology. 2014;95:631–636. doi: 10.1890/13-1452.1. [DOI] [PubMed] [Google Scholar]
- 72.Bates D, Sarkar D, Bates MD, Matrix L. The lme4 package. R Package Version. 2007;2:74. [Google Scholar]
- 73.Oksanen, J. et al. Vegan: Community Ecology Package. R package vegan, vers. 2.2-1; 2015 (2018).
- 74.Bhagwat SA, Willis KJ, Birks HJB, Whittaker RJ. Agroforestry: a refuge for tropical biodiversity? Trends Ecol. Evol. 2008;23:261–267. doi: 10.1016/j.tree.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 75.Felton A, Knight E, Wood J, Zammit C, Lindenmayer D. A meta-analysis of fauna and flora species richness and abundance in plantations and pasture lands. Biol. Conserv. 2010;143:545–554. doi: 10.1016/j.biocon.2009.11.030. [DOI] [Google Scholar]
- 76.Ranganathan J, Daniels RR, Chandran MS, Ehrlich PR, Daily GC. Sustaining biodiversity in ancient tropical countryside. Proc. Natl. Acad. Sci. 2008;105:17852–17854. doi: 10.1073/pnas.0808874105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sidhu S, Shankar Raman TR, Goodale E. Effects of plantations and home-gardens on tropical forest bird communities and mixed-species bird flocks in the southern Western Ghats. J. Bombay Nat. Hist. Soc. 2010;107:91. [Google Scholar]
- 78.MacArthur RH, MacArthur JW. On bird species diversity. Ecology. 1961;42:594–598. doi: 10.2307/1932254. [DOI] [Google Scholar]
- 79.Rosenzweig, M. L. Species diversity in space and time. (Cambridge University Press, 1995).
- 80.Harvey CA, Villalobos JAG. Agroforestry systems conserve species-rich but modified assemblages of tropical birds and bats. Biodivers. Conserv. 2007;16:2257–2292. doi: 10.1007/s10531-007-9194-2. [DOI] [Google Scholar]
- 81.Waltert M, Bobo KS, Sainge NM, Fermon H, Mühlenberg M. From forest to farmland: habitat effects on Afrotropical forest bird diversity. Ecol. Appl. 2005;15:1351–1366. doi: 10.1890/04-1002. [DOI] [Google Scholar]
- 82.Waltert M, et al. Assessing conservation values: biodiversity and endemicity in tropical land use systems. PLoS One. 2011;6:e16238. doi: 10.1371/journal.pone.0016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Madhusudan MD. Recovery of wild large herbivores following livestock decline in a tropical Indian wildlife reserve. J. Appl. Ecol. 2004;41:858–869. doi: 10.1111/j.0021-8901.2004.00950.x. [DOI] [Google Scholar]
- 84.Rannestad OT, Danielsen T, Moe SR, Stokke S. Adjacent pastoral areas support higher densities of wild ungulates during the wet season than the Lake Mburo National Park in Uganda. J. Trop. Ecol. 2006;22:675–683. doi: 10.1017/S0266467406003610. [DOI] [Google Scholar]
- 85.Sitters J, Heitkönig IM, Holmgren M, Ojwang GS. Herded cattle and wild grazers partition water but share forage resources during dry years in East African savannas. Biol. Conserv. 2009;142:738–750. doi: 10.1016/j.biocon.2008.12.001. [DOI] [Google Scholar]
- 86.Khan JA. Conservation and management of Gir lion sanctuary and national park, Gujarat, India. Biol. Conserv. 1995;73:183–188. doi: 10.1016/0006-3207(94)00107-2. [DOI] [Google Scholar]
- 87.Vanthomme H, Kolowski J, Korte L, Alonso A. Distribution of a community of mammals in relation to roads and other human disturbances in Gabon, Central Africa. Conserv. Biol. 2013;27:281–291. doi: 10.1111/cobi.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fahrig L. Habitat fragmentation: A long and tangled tale. Glob. Ecol. Biogeogr. 2019;28:33–41. doi: 10.1111/geb.12839. [DOI] [Google Scholar]
- 89.Fahrig L, et al. Is habitat fragmentation bad for biodiversity? Biol. Conserv. 2019;230:179–186. doi: 10.1016/j.biocon.2018.12.026. [DOI] [Google Scholar]
- 90.Fletcher RJ, Jr, et al. Is habitat fragmentation good for biodiversity? Biol. Conserv. 2018;226:9–15. doi: 10.1016/j.biocon.2018.07.022. [DOI] [Google Scholar]
- 91.Simmonds, J. S., van Rensburg, B. J., Tulloch, A. I. & Maron, M. Landscape-specific thresholds in the relationship between species richness and natural land cover. J. Appl. Ecol (2019).
- 92.Wintle Brendan A., Kujala Heini, Whitehead Amy, Cameron Alison, Veloz Sam, Kukkala Aija, Moilanen Atte, Gordon Ascelin, Lentini Pia E., Cadenhead Natasha C. R., Bekessy Sarah A. Global synthesis of conservation studies reveals the importance of small habitat patches for biodiversity. Proceedings of the National Academy of Sciences. 2018;116(3):909–914. doi: 10.1073/pnas.1813051115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wood, C. L. & Johnson, P. T. 2018 How does space influence the relationship between host and parasite diversity? J. Parasitol. 102, 485–495 (2016). [DOI] [PubMed]
- 94.Wood CL, et al. Human impacts decouple a fundamental ecological relationship—The positive association between host diversity and parasite diversity. Glob. Change Biol. 2018;24:3666–3679. doi: 10.1111/gcb.14159. [DOI] [PubMed] [Google Scholar]
- 95.Eley RM, Strum SC, Muchemi G, Reid GDF. Nutrition, body condition, activity patterns, and parasitism of free-ranging troops of olive baboons (Papio anubis) in Kenya. Am. J. Primatol. 1989;18:209–219. doi: 10.1002/ajp.1350180304. [DOI] [PubMed] [Google Scholar]
- 96.Gillespie TR, Chapman CA. Prediction of parasite infection dynamics in primate metapopulations based on attributes of forest fragmentation. Conserv. Biol. 2006;20:441–448. doi: 10.1111/j.1523-1739.2006.00290.x. [DOI] [PubMed] [Google Scholar]
- 97.Gillespie TR, et al. Demographic and ecological effects on patterns of parasitism in eastern chimpanzees (Pan troglodytes schweinfurthii) in Gombe National Park, Tanzania. Am. J. Phys. Anthropol. 2010;143:534–544. doi: 10.1002/ajpa.21348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Howells ME, Pruetz J, Gillespie TR. Patterns of gastro-intestinal parasites and commensals as an index of population and ecosystem health: the case of sympatric Western chimpanzees (Pan troglodytes verus) and Guinea baboons (Papio hamadryas papio) at Fongoli, Senegal. Am. J. Primatol. 2011;73:173–179. doi: 10.1002/ajp.20884. [DOI] [PubMed] [Google Scholar]
- 99.Messenger AM, Barnes AN, Gray GC. Reverse zoonotic disease transmission (zooanthroponosis): a systematic review of seldom-documented human biological threats to animals. PloS One. 2014;9:e89055. doi: 10.1371/journal.pone.0089055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sá RM, et al. Gastrointestinal symbionts of chimpanzees in Cantanhez National Park, Guinea-Bissau with respect to habitat fragmentation. Am. J. Primatol. 2013;75:1032–1041. doi: 10.1002/ajp.22170. [DOI] [PubMed] [Google Scholar]
- 101.Bordes F, Morand S, Ricardo G. Bat fly species richness in Neotropical bats: correlations with host ecology and host brain. Oecologia. 2008;158:109–116. doi: 10.1007/s00442-008-1115-x. [DOI] [PubMed] [Google Scholar]
- 102.Ezenwa VO, Price SA, Altizer S, Vitone ND, Cook KC. Host traits and parasite species richness in even and odd-toed hoofed mammals, Artiodactyla and Perissodactyla. Oikos. 2006;115:526–536. doi: 10.1111/j.2006.0030-1299.15186.x. [DOI] [Google Scholar]
- 103.Morand S, Poulin R. Density, body mass and parasite species richness of terrestrial mammals. Evol. Ecol. 1998;12:717–727. doi: 10.1023/A:1006537600093. [DOI] [Google Scholar]
- 104.Nunn CL, Altizer S, Jones KE, Sechrest W. Comparative tests of parasite species richness in primates. Am. Nat. 2003;162:597–614. doi: 10.1086/378721. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analysed during the current study are available from the corresponding author on request.