Abstract
While density functional theory (DFT) is often an accurate and efficient methodology for evaluating molecular properties such as energies and multipole moments, this approach often yields larger errors for response properties such as the dipole polarizability (α), which describes the tendency of a molecule to form an induced dipole moment in the presence of an electric field. In this work, we provide static α tensors (and other molecular properties such as total energy components, dipole and quadrupole moments, etc.) computed using quantum chemical (QC) and DFT methodologies for all 7,211 molecules in the QM7b database. We also provide the same quantities for the 52 molecules in the AlphaML showcase database, which includes the DNA/RNA nucleobases, uncharged amino acids, several open-chain and cyclic carbohydrates, five popular pharmaceutical molecules, and 23 isomers of C8Hn. All QC calculations were performed using linear-response coupled-cluster theory including single and double excitations (LR-CCSD), a sophisticated approach for electron correlation, and the d-aug-cc-pVDZ basis set to mitigate basis set incompleteness error. DFT calculations employed the B3LYP and SCAN0 hybrid functionals, in conjunction with d-aug-cc-pVDZ (B3LYP and SCAN0) and d-aug-cc-pVTZ (B3LYP).
Subject terms: Quantum chemistry, Density functional theory, Chemical physics, Chemical physics, Electronic structure of atoms and molecules
| Design Type(s) | chemical structure classification objective • chemical reaction data analysis objective • modeling and simulation objective |
| Measurement Type(s) | chemical structure analysis |
| Technology Type(s) | ab initio quantum chemistry computational method |
| Factor Type(s) | atom |
| Sample Characteristic(s) |
Machine-accessible metadata file describing the reported data (ISA-Tab format)
Background & Summary
The molecular dipole polarizability, α, describes the tendency of a molecule to form an induced dipole moment in the presence of an external electric field. Knowledge of this fundamental response property is central to describing non-bonded interactions (such as induction and dispersion) between molecules in clusters or the condensed phase1–3, computing Raman and sum frequency generation (SFG) spectra4–7, and developing polarizable force fields8–12. When compared to other ground-state molecular properties (e.g., multipole moments), the theoretical prediction of the α tensor is considerably more difficult to obtain, as this quantity is often more sensitive to the description of the underlying molecular electronic structure. In this regard, benchmark ab initio calculations of α are quite challenging to perform, as they require a simultaneous treatment of sophisticated electron correlation effects as well as mitigation of basis set incompleteness error to ensure sufficiently accurate and converged results.
To obtain benchmark values for α in molecular systems with a sizeable HOMO-LUMO gap (i.e., systems that are well-described by a single-reference wavefunction), one can utilize quantum chemical methods such as linear-response coupled-cluster theory (LR-CC)13–15, which provides an accurate and reliable treatment of electron correlation. The downside of such wavefunction-based approaches is the large (and often prohibitive) computational cost associated with the inclusion of higher order excitations in the CC expansion. For example, LR-CC at the lowest order includes single and double excitations (LR-CCSD), and scales as O(n6), where n is a measure of the system size (i.e., the number of orbitals). This computational cost keeps increasing as higher order excitations are included, and scales as O(n8) with the inclusion of triple excitations (LR-CCSDT) and O(n10) with the further inclusion of quadruple excitations (LR-CCSDTQ). As a result of this steep rise in the cost, such calculations are computationally prohibitive, even when one is dealing with relatively small molecules containing only 10–15 heavy (non-hydrogen) atoms. In addition to the computational cost required for a wavefunction-based treatment of the electron correlation, the error introduced by the use of a finite one-electron basis set is another factor that needs to be considered when computing α. In this regard, basis set incompleteness error in the prediction of α can be more severe than the error due to the lack of higher order (e.g., beyond doubles) excitations16–19.
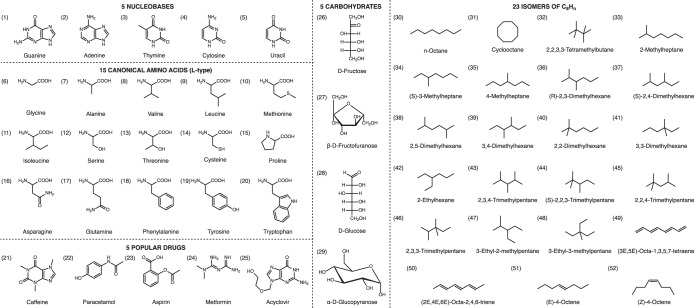
In this work, we provide static (frequency-independent) α tensors computed using LR-CCSD and hybrid density functional theory (DFT) for all molecules in the QM7b20–22 and AlphaML showcase databases23. The QM7b database20–22 has become one of the de facto standard databases for machine-learning (ML) applications in chemistry, and contains N = 7,211 small organic molecules with up to seven heavy atoms (i.e., C, N, O, S, and Cl) and varying levels of H saturation. Recently introduced by Wilkins et al.23 for testing the transferability of ML-based predictions of α, the AlphaML showcase database consists of N = 52 larger organic molecules (with up to 16 heavy atoms), and includes the DNA/RNA nucleobases, uncharged amino acids, several open-chain and cyclic carbohydrates, five popular pharmaceutical molecules, and 23 isomers of C8Hn (see Fig. 1). The diversity of structures in this combination of databases includes alkanes, alkenes, alkynes, (hetero)cycles, carbonyl and carboxyl groups, cyanides, amides, alcohols, amines, thiols, ethers, and epoxides, thereby providing a meaningful survey of α across a wide swath of chemical compound space.
Fig. 1.
Names and indices of the 52 molecules in the AlphaML showcase database. (1–5) DNA/RNA nucleobases; (6–20) uncharged canonical amino acids (L-type); (21–25) popular pharmaceutical molecules; (26–29) open-chain and cyclic carbohydrates; and (30–52) 23 isomers of C8Hn. Throughout this work, these molecules will be specified by “showcase” followed by the corresponding index (padded to four digits with leading zeros, e.g., showcase0001 to showcase0052).
Reference values for α were obtained with LR-CCSD with the doubly-augmented d-aug-cc-pVDZ basis set of Woon and Dunning19, as this method (when employed in conjunction with a sufficiently large and diffuse one-particle basis set) has been shown to yield accurate and reliable predictions for α16–18,24. The use of d-aug-cc-pVDZ greatly mitigates the basis set incompleteness error at the double-ζ level, and the validity of this basis set choice will be critically examined and discussed in more detail below. For comparative purposes, we also provide finite-field DFT values for α obtained with the popular B3LYP25,26 and SCAN027 hybrid functionals in conjunction with the d-aug-cc-pVDZ (B3LYP and SCAN0) and d-aug-cc-pVTZ (B3LYP only) basis sets. Throughout the remainder of this work, the d-aug-cc-pVXZ basis sets (with X = D and T) will be referred to as daXZ, and all LR-CCSD/daDZ calculations will simply be denoted by CCSD/daDZ unless otherwise specified.
Methods
In this section, we provide the conventions used in generating and processing the geometries of the molecules in the QM7b and AlphaML showcase databases, all relevant computational details to ensure reproducibility of the quantum mechanical data, as well as a summary of the codes employed in this work.
Molecular cartesian coordinates in the QM7b and AlphaML showcase databases
The molecular geometries for all 7,211 species in the QM7b database20–22 were obtained online via the quantum-machine.org website28. All QM7b molecular geometries were first translated to their respective center of nuclear (ionic) charge, to remove the origin-dependence of the higher-order (i.e., quadrupole) multipole moments. Using farthest-point sampling (FPS)29, all molecules were then reordered using a kernel-based similarity measure30, and relabelled accordingly from molecule0001 to molecule7211 (again padded to four digits with leading zeros). For consistency with the QM7b database, all 52 molecules in the AlphaML showcase database (see Fig. 1) were optimized with DFT using the PBE functional31 and a converged numerical atom-centered basis (i.e., tight settings with the tier-2 basis set in FHI-AIMS)32. All AlphaML showcase molecules were also translated to their respective center of nuclear (ionic) charge, and are labelled from showcase0001 to showcase0052, as depicted in Fig. 1. All 7,263 structures are available on Materials Cloud33, according to the format described below in the Data Records section.
Details of the quantum mechanical calculations
All CCSD/daDZ, B3LYP/daDZ, and B3LYP/daTZ calculations were carried out using Psi4 v1.134, while all SCAN0/daDZ calculations were performed with Q-Chem v5.035. At the CCSD/daDZ level, all α tensors, unrelaxed dipole moments, μ, and unrelaxed quadrupole moments, Q, were calculated using LR-CCSD/daDZ, with the exception of the ten largest molecules in the AlphaML showcase database (e.g., (18) Phenylalanine, (19) Tyrosine, (20) Tryptophan, (21) Caffeine, (23) Aspirin, (25) Acyclovir, (26) D-Fructose, (27) β-D-Fructofuranose, (28) D-Glucose, and (29) α-D-glucopyranose, see Fig. 1). For these molecules, the memory requirements required to solve the Λ-CC equations at the LR-CCSD/daDZ level were computationally prohibitive, and only energy calculations with CCSD/daDZ could be performed with the available computational resources. For consistency, this required the use of the orbital-unrelaxed finite-field method, in which the molecular orbitals were obtained from a field-free (unperturbed) Hartree-Fock calculation. To obtain μ and α, we computed first and second derivatives of the CCSD/daDZ energy (U) with respect to an external electric field, E, i.e., μ = ∂U/∂E and α = ∂2U/∂E2. Q values were not computed for the ten largest molecules in the AlphaML showcase database. All DFT calculations used the orbital-relaxed finite-field method, in which a self-consistent field (SCF) was obtained in the presence of each applied field, and α was computed via α = ∂μ/∂E. All other molecular properties at the DFT level (vide infra) were obtained directly from the field-free (unperturbed) calculation. All derivatives were computed numerically using two-point (for first derivatives) and three-point (for second derivatives) central difference formulae and a step size of E = 1.8897261250 × 10−5 atomic units.
For all LR-CCSD/daDZ calculations, the convergence criteria were set to their default values in Psi4, i.e., E_convergence = 1.0E-10 and D_convergence = 1.0E-10 for the energy and density during the solution of the HF equations, and E_convergence = 1.0E-08 and R_convergence = 1.0E-07 for the energy and residuals during the solution of the CCSD equations. For the ten largest molecules in the AlphaML showcase database, the finite-field CCSD/daDZ calculations were performed using the following convergence criteria in Psi4: E_convergence = 5.0E-10 and D_convergence = 5.0E-10 for the energy and density during the solution of the HF equations. Significantly tighter convergence criteria of E_convergence = 5.0E-10 and R_convergence = 5.0E-09 were employed for the energy and residuals during the solution of the CCSD equations to minimize errors in the numerical evaluation of μ and α. The frozen core (FC) approximation and scf_type = direct were used for all LR-CCSD/daDZ and CCSD/daDZ calculations. For all B3LYP/daDZ and B3LYP/daTZ calculations, the convergence criteria in Psi4 were again set to tight values to minimize numerical error in the finite-difference evaluation of α: E_convergence = 1.0E-10 and D_convergence = 1.0E-10 for the energy and density during the solution of the Kohn-Sham equations. For all the SCAN0/daDZ calculations, the convergence criteria were set to scf_convergence = 1.0E-10 and thresh = 1.0E-13 for the DIIS error and integral thresholding in Q-Chem. The Dunning-style daDZ and daTZ basis sets19 were obtained from the EMSL Basis Set Library36,37.
Data Records
In this section, we briefly describe the molecular properties that have been computed in this work, as well as the conventions used to store and retrieve the generated data. In addition to a select set of molecular properties (such as energetic components, dipole and quadrupole moments, orbital eigenvalues, etc.), the provided data will also include the full output files from all of the calculations performed herein. In what follows, we focus the discussion on α, as this molecular response property is arguably the most challenging quantity computed in this work. In particular, we provide a statistical summary of the CCSD/daDZ α data in the QM7b and AlphaML databases, as well as a comparative analysis of the different quantum mechanical methods employed in this work.
Included molecular properties and file format
To store and disseminate the data generated in this work, we have created the following four data packages: CCSD_daDZ, B3LYP_daDZ, SCAN0_daDZ, and B3LYP_daTZ. Each data package contains 7,263 standard xyz files, and has been named according to the level of theory used to generate the data contained therein. Each of the included xyz files contains the translated geometries and calculated properties for a single molecule in the QM7b and AlphaML showcase databases. As described above, the 7,211 molecules in the QM7b database are contained in xyz files labelled from molecule0001 to molecule7211, and the 52 molecules in the AlphaML showcase database are contained in xyz files labelled from showcase0001 to showcase0052 (see Fig. 1).
All computed properties (for a given molecule) are provided on the “comment line” (i.e., the second line) of the corresponding xyz file (as comma-separated values), following the order provided in Table 1 (for CCSD_daDZ) and Table 2 (for B3LYP_daDZ, SCAN0_daDZ, and B3LYP_daTZ). Common molecular properties included in all four data packages are: the isotropic polarizability (αiso),
| 1 |
anisotropic polarizability (αaniso),
| 2 |
all symmetry-unique components of the polarizability (α) tensor (i.e., αxx, αyy, αzz, αxy, αxz, αyz), all components of the dipole moment (μ) vector (i.e., μx, μy, μz), and all symmetry-unique components of the quadrupole moment (Q) vector (i.e., Qxx, Qyy, Qzz, Qxy, Qxz, Qyz). In the CCSD_daDZ data package only, the following molecular properties are also included: the Hartree-Fock total energy (), same-spin () and opposite-spin () correlation energies at the level of second-order Møller-Plesset perturbation (MP2) theory, and same-spin () and opposite-spin () correlation energies at the CCSD level. In the B3LYP_daDZ, SCAN0_daDZ, and B3LYP_daTZ data packages only, the following molecular properties are also included: the DFT total energy () and the eigenvalues corresponding to the HOMO () and LUMO (). All data described above is provided in atomic units and available for download on Materials Cloud33.
Table 1.
Calculated properties at the CCSD/daDZ level.
| No. | Property | Description |
|---|---|---|
| 01 | Tag | “Properties” string |
| 02 | α iso | isotropic polarizability (see Eq. (1)) |
| 03 | α aniso | anisotropic polarizability (see Eq. (2)) |
| 04–09 | α | polarizability tensora |
| 10–12 | μ | (unrelaxed) dipole momentb |
| 13–18 | Q | (unrelaxed) quadrupole momentc,d |
| 19 | HF total energy | |
| 20 | MP2 same-spin correlation energy | |
| 21 | MP2 opposite-spin correlation energy | |
| 22 | CCSD same-spin correlation energy | |
| 23 | CCSD opposite-spin correlation energy |
All properties are in atomic units and are provided on the “comment line” (i.e., the second line) of a standard xyz file.
a04–09: αxx, αyy, αzz, αxy, αxz, αyz.
b10–12: μx, μy, μz.
c13–18: Qxx, Qyy, Qzz, Qxy, Qxz, Qyz.
dQ values are not provided for the ten largest molecules in the AlphaML showcase database (see text for details).
Table 2.
Calculated properties at the B3LYP/daDZ, SCAN0/daDZ, and B3LYP/daTZ levels.
| No. | Properties | Description |
|---|---|---|
| 01 | Tag | “Properties” string |
| 02 | α iso | isotropic polarizability (see Eq. (1)) |
| 03 | α aniso | anisotropic polarizability (see Eq. (2)) |
| 04–09 | α | polarizability tensora |
| 10–12 | μ | dipole momentb |
| 13–18 | Q | quadrupole momentc |
| 19 | DFT total energy | |
| 20 | HOMO energy | |
| 21 | LUMO energy |
All properties are in atomic units and are provided on the “comment line” (i.e., the second line) of a standard xyz file.
a04–09: αxx, αyy, αzz, αxy, αxz, αyz.
b10–12: μx, μy, μz.
c13–18: Qxx, Qyy, Qzz, Qxy, Qxz, Qyz.
Statistical summary of the CCSD/daDZ α data
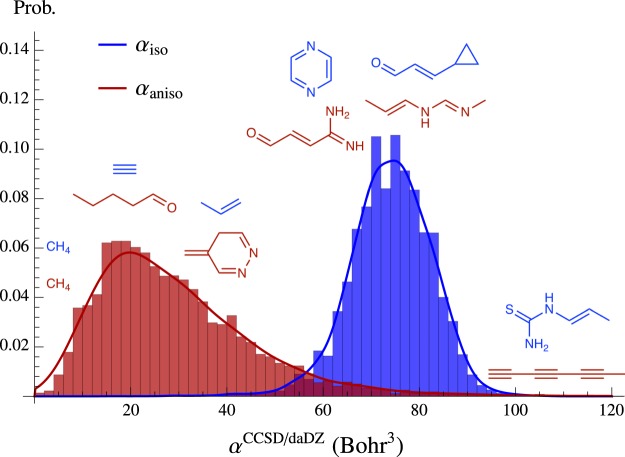
To provide an overview of the α data, Fig. 2 contains the normalized probability distributions of the CCSD/daDZ isotropic (αiso, blue) and anisotropic (αaniso, red) polarizabilities in the QM7b database. This is accompanied by Table 3, which provides a statistical analysis of all α data generated in this work. From Fig. 2 and Table 3, one can see that the CCSD/daDZ αiso values in the QM7b database have a range of 16.80–106.50 Bohr3, and are centered around a mean value of 〈α〉 = 74.07 Bohr3. With a standard deviation (σ) that is nearly two times larger than αiso, the CCSD/daDZ αaniso values in this database are characterized by a broader distribution that is significantly skewed to the right. We note in passing that the range of αaniso is larger than αiso by ≈64%, and includes minimum values that are significantly smaller (cf. Bohr3 vs. Bohr3) and maximum values that are significantly larger (cf. Bohr3 vs. Bohr3). Also depicted in Fig. 2 are the subset of molecules in the QM7b database with the smallest and largest αiso and αaniso values (as well as those molecules with intermediate αiso and αaniso values of ≈20, 40, 60, and 80 Bohr3). From these molecules, one clearly sees that αiso is an extensive quantity that grows with molecular size, and αaniso (which is a measure of the anisotropy in the α tensor, see Eq. (2)) is largest for molecules with elongated and non-spherical/asymmetric shapes.
Fig. 2.
Normalized probability distributions of the CCSD/daDZ isotropic (αiso, blue) and anisotropic (αaniso, red) polarizabilities in the QM7b database. Molecules with the smallest and largest αiso and αaniso values are depicted at the left and right of the figure, respectively. Molecules with intermediate αiso and αaniso values (≈20, 40, 60, and 80 Bohr3) are depicted in the middle of the figure. Histogram bin widths were set to 2.0 Bohr3.
Table 3.
Statistical analysis of the isotropic (αiso) and anisotropic (αaniso) polarizabilities in the QM7b and AlphaML showcase databases computed at the CCSD/daDZ, B3LYP/daDZ, SCAN0/daDZ, and B3LYP/daTZ levels.
| α | Level | 〈α〉 | σ | α min | α max |
|---|---|---|---|---|---|
| QM7b Database | |||||
| α iso | CCSD/daDZ | 74.07 | 8.47 | 16.80 | 106.50 |
| B3LYP/daDZ | 75.97 | 9.14 | 17.32 | 117.29 | |
| SCAN0/daDZ | 74.27 | 9.06 | 16.94 | 116.64 | |
| B3LYP/daTZ | 75.90 | 9.14 | 17.30 | 117.27 | |
| α aniso | CCSD/daDZ | 29.09 | 16.18 | 2.16E-4 | 147.49 |
| B3LYP/daDZ | 32.08 | 18.93 | 4.13E-4 | 181.51 | |
| SCAN0/daDZ | 31.51 | 18.58 | 3.83E-3 | 177.21 | |
| B3LYP/daTZ | 32.08 | 18.94 | 2.74E-4 | 181.74 | |
| AlphaML Showcase Database | |||||
| α iso | CCSD/daDZ | 99.59 | 20.87 | 43.94 | 156.44 |
| B3LYP/daDZ | 101.56 | 22.14 | 44.33 | 156.38 | |
| SCAN0/daDZ | 99.37 | 21.97 | 42.75 | 153.08 | |
| B3LYP/daTZ | 101.42 | 22.14 | 44.25 | 156.24 | |
| α aniso | CCSD/daDZ | 35.66 | 32.52 | 6.93 | 171.44 |
| B3LYP/daDZ | 39.19 | 40.07 | 7.52 | 229.32 | |
| SCAN0/daDZ | 38.35 | 39.91 | 7.36 | 231.07 | |
| B3LYP/daTZ | 39.18 | 40.11 | 7.56 | 229.66 | |
Statistical quantities (in Bohr3) include: 〈α〉 (mean), σ (standard deviation), αmin (minimum value), and αmax (maximum value).
The statistical summary of the α data corresponding to the 52 molecules in the AlphaML showcase database (see Table 3) also illustrates that the αiso (αaniso) distributions in this database are characterized by 〈α〉 values that are larger by ≈34% (≈23%) and σ values that are 2.5× (2.0×) larger than that found in the QM7b database. In addition, the range of αiso (αaniso) values is approximately 25% (12%) larger in the AlphaML showcase database, and does not include symmetric molecules with vanishingly small αaniso values. Taken together, these statistical measures reflect the fact that the molecules in the AlphaML showcase database, which includes the DNA/RNA nucleobases, uncharged amino acids, several open-chain and cyclic carbohydrates, five popular pharmaceutical molecules, and 23 isomers of C8Hn, are (in general) larger and more diverse than those contained in the QM7b database (see Fig. 1).
Comparative analysis of the quantum mechanical methodologies
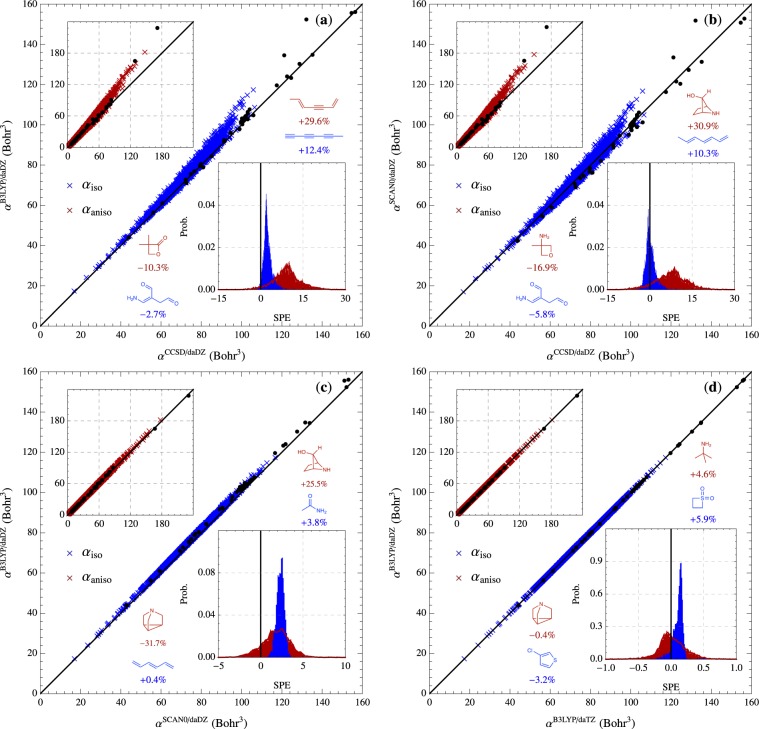
To investigate the performance of different quantum mechanical methodologies in calculating the α tensor in the QM7b and AlphaML showcase databases, a detailed statistical error analysis was carried out for the following combinations of methods (Level//Reference Level): B3LYP/daDZ//CCSD/daDZ and SCAN0/daDZ//CCSD/daDZ (to compare the electron correlation level while keeping the basis set fixed), B3LYP/daDZ//SCAN0/daDZ (to compare the exchange-correlation functional while keeping the basis set fixed), and B3LYP/daDZ//B3LYP/daTZ (to quantify the basis set incompleteness error at the B3LYP level). A summary of statistical error measures, including the mean signed error, , mean absolute error, , and root-mean-square error, , are provided in Table 4 as well as the corresponding percent errors, , , and . To visualize these differences in more detail, correlation plots (corresponding to the four method combinations above) for αiso and αaniso (as well as probability distributions of the signed percent error (SPE)) are provided in Fig. 3.
Table 4.
Comparative analysis of the isotropic (αiso) and anisotropic (αaniso) polarizabilities in the QM7b and AlphaML showcase databases computed at the CCSD/daDZ, B3LYP/daDZ, SCAN0/daDZ, and B3LYP/daTZ levels.
| α | Level | Reference Level | MSE (MSPE) | MAE (MAPE) | RMSE (RMSPE) |
|---|---|---|---|---|---|
| QM7b Database | |||||
| α iso | B3LYP/daDZ | CCSD/daDZ | 1.91 (2.52) | 1.92 (2.54) | 2.32 (2.97) |
| SCAN0/daDZ | CCSD/daDZ | 0.20 (0.21) | 0.97 (1.29) | 1.41 (1.78) | |
| B3LYP/daDZ | SCAN0/daDZ | 1.70 (2.31) | 1.70 (2.31) | 1.73 (2.36) | |
| B3LYP/daDZ | B3LYP/daTZ | 0.08 (0.11) | 0.09 (0.12) | 0.11 (0.14) | |
| α aniso | B3LYP/daDZ | CCSD/daDZ | 2.99 (9.19) | 3.03 (9.34) | 4.48 (10.4) |
| SCAN0/daDZ | CCSD/daDZ | 2.42 (7.34) | 2.59 (7.93) | 4.01 (9.25) | |
| B3LYP/daDZ | SCAN0/daDZ | 0.57 (1.78) | 0.64 (2.09) | 0.85 (2.52) | |
| B3LYP/daDZ | B3LYP/daTZ | <0.01 (0.02) | 0.05 (0.17) | 0.07 (0.25) | |
| AlphaML Showcase Database | |||||
| α iso | B3LYP/daDZ | CCSD/daDZ | 1.97 (1.83) | 2.21 (2.09) | 3.87 (3.22) |
| SCAN0/daDZ | CCSD/daDZ | −0.22 (−0.43) | 2.15 (2.08) | 3.81 (3.24) | |
| B3LYP/daDZ | SCAN0/daDZ | 2.19 (2.29) | 2.19 (2.29) | 2.34 (2.44) | |
| B3LYP/daDZ | B3LYP/daTZ | 0.13 (0.14) | 0.13 (0.14) | 0.14 (0.15) | |
| α aniso | B3LYP/daDZ | CCSD/daDZ | 3.52 (7.83) | 3.66 (8.10) | 9.87 (9.97) |
| SCAN0/daDZ | CCSD/daDZ | 2.69 (5.25) | 3.37 (6.73) | 10.0 (9.11) | |
| B3LYP/daDZ | SCAN0/daDZ | 0.84 (2.57) | 0.94 (2.65) | 1.44 (3.57) | |
| B3LYP/daDZ | B3LYP/daTZ | <0.01 (0.01) | 0.08 (0.23) | 0.11 (0.31) | |
Statistical quantities (in Bohr3) are computed with respect to the Reference Level and include: mean signed error (MSE), mean absolute error (MAE), and root-mean-square error (RMSE). Corresponding percent errors (MSPE, MAPE, RMSPE) are provided in %. When computing the MSPE, MAPE, and RMSPE values for αaniso in the QM7b database, molecules with very small αaniso values (i.e., molecule0001: methane, Bohr3 and molecule1009: neopentane, Bohr3) were excluded, as these molecules yielded large (but physically insignificant) percent errors.
Fig. 3.
Correlation plots of the isotropic (αiso, blue, main plots) and anisotropic (αaniso, red, upper left insets) polarizabilities in the QM7b and AlphaML showcase databases computed at different levels of theory. Normalized probability distributions of the signed percent errors (SPE, computed with respect to the Reference Level of theory on the x-axis) are provided in the lower right insets of each panel. Histogram bin widths were set to 0.1% for panels (a) B3LYP/daDZ vs. CCSD/daDZ, (b) SCAN0/daDZ vs. CCSD/daDZ, and (c) B3LYP/daDZ vs. SCAN0/daDZ, and 0.01% for panel (d) B3LYP/daDZ vs. B3LYP/daTZ. For completeness, αiso and αaniso values in the AlphaML showcase database are depicted as black circles in the main plots and upper left insets of each panel. Molecules corresponding to the minimum and maximum αiso and αaniso SPE values are also depicted in each panel. When computing the minimum and maximum αaniso SPE values, molecules with very small αaniso values (i.e., molecule0001: methane, Bohr3 and molecule1009: neopentane, Bohr3) were excluded, as these molecules yielded large (but physically insignificant) SPE values.
When comparing B3LYP/daDZ to the reference CCSD/daDZ level for the molecules in the QM7b database, one sees that B3LYP/daDZ yields essentially identical MSE and MAE values for αiso (i.e., 1.91 Bohr3 and 1.92 Bohr3, respectively), indicating that B3LYP/daDZ systematically overestimates αiso values by ≈2.5% (see Table 4). With an RMSE value that is ≈21% greater than the MAE, the magnitudes of the B3LYP/daDZ errors show substantial variations from molecule to molecule; this is particularly evident for the molecules with large αiso values in Fig. 3(a). When comparing SCAN0/daDZ to CCSD/daDZ, one sees that SCAN0/daDZ outperforms B3LYP/daDZ by a large margin in the prediction of αiso, yielding reductions of ≈90%, ≈50%, and ≈40% in the MSE, MAE, and RMSE values, respectively. In this regard, our finding that the SCAN0 functional provides greatly improved estimates for αiso is also consistent with the recent benchmark study by Lao et al.18 on the dipole polarizability surface of the gas-phase water molecule. With an MSE value that is nearly 5× smaller than the MAE, it is also worth noting that SCAN0/daDZ αiso values only have a slightly positive systematic error. From a quick glance at Fig. 3(b), it is clear that SCAN0/daDZ (like B3LYP/daDZ) also has more difficulties when treating molecules with large αiso values; this is indicative of the challenges that one faces when computing α, a response property which becomes substantially more non-additive as the size and complexity of the molecules increase. When comparing B3LYP/daDZ to SCAN0/daDZ, one obtains nearly identical MSE, MAE, and RMSE values, which indicates that: (i) B3LYP/daDZ systematically overestimates αiso with respect to SCAN0/daDZ, and (ii) the magnitudes of the B3LYP/daDZ errors do not show substantial molecule-to-molecule variations. Both of these findings are confirmed in Fig. 3(c), where one sees that: (i) the SPE distribution is centered around 2.3% (and not zero), and (ii) there is clearly a very strong linear correlation between the B3LYP/daDZ and SCAN0/daDZ αiso values that does not deteriorate with molecular size and complexity. When comparing B3LYP/daDZ to B3LYP/daTZ, one finds that the increase from double- to triple-ζ in the underlying basis set does not lead to significantly different αiso values. This finding is consistent with the (in general) rapid convergence of DFT with respect to the occupied space and the relatively weak dependence of DFT on the virtual/unoccupied space.
From a quick glance at Table 4, one also sees that both B3LYP and SCAN0 (when compared to the reference CCSD/daDZ level) yield larger errors when predicting αaniso than αiso. When comparing B3LYP/daDZ to CCSD/daDZ, for example, the MSPE, MAPE, and RMSPE values increased from 2.52%, 2.54%, and 2.97% for αiso to 9.19%, 9.34%, and 10.4% for αaniso; a similar increase was observed when comparing SCAN0/daDZ to CCSD/daDZ. These findings demonstrate that the tensorial properties of α (which govern αaniso) are more difficult to predict than the average of the diagonal elements (i.e., αiso values). When compared to CCSD/daDZ, SCAN0/daDZ is no longer performing substantially better than B3LYP/daDZ and now exhibits a (nearly) systematic overestimation of αaniso. By looking at the upper left insets in Fig. 3(a,b), one again sees that the errors made by B3LYP and SCAN0 increase for molecules with larger αaniso values; this is indicative of the increasing importance of including electron correlation effects when predicting αaniso for molecules that are larger in size and potentially more anisotropic. Among the DFT functionals at the daDZ level, one sees that B3LYP/daDZ overestimates αaniso with respect to SCAN0/daDZ in most cases, and that B3LYP/daDZ and SCAN0/daDZ are now in better agreement with each other than with CCSD/daDZ. When comparing B3LYP/daDZ and B3LYP/daTZ, the B3LYP functional again shows rapid convergence with respect to the underlying basis set in the prediction of αaniso.
When performing a similar analysis for the molecules in the AlphaML showcase database, most of the findings described above for the QM7b database still hold. One interesting distinction is the finding that SCAN0/daDZ no longer outperforms B3LYP/daDZ when predicting αiso values for the larger and more complex molecules contained in the AlphaML showcase database; in the same breath, we note that SCAN0/daDZ still maintains a relatively small MSE value, which is indicative of an error profile that is more random (and less systematic) than B3LYP/daDZ (see Fig. 3(a,b)).
Technical Validation
In this section, we explore the validity and reliability of the CCSD/daDZ α data in the QM7b database.
Validation of the CCSD/daDZ α Data
Since α describes the response of a molecule to an applied electric field, an accurate and reliable treatment of this quantity is particularly sensitive to the description of the underlying electronic structure as well as the quality of the basis set. The highest level α values provided in this work were computed with LR-CCSD, a sophisticated wavefunction-based method that consistently yields highly accurate α values for equilibrium and non-equilibrium molecular geometries when used with sufficiently large (and sufficiently diffuse) basis sets16–18,24. To account for the basis set incompleteness error, which is almost always larger than the contributions from higher-order (e.g., beyond doubles) excitations in coupled-cluster theory16–19, we employed the daDZ basis set. Although daDZ is a double-ζ basis set containing a moderate number of polarization functions, the incorporation of two sets of augmented functions (i.e., double augmentation) significantly reduces the basis set incompleteness error in the prediction of α. To validate the accuracy of our CCSD/daDZ calculations, we performed a series of calculations using the larger daTZ basis set19, which is arguably the largest Dunning-style basis that can be used to compute α for the molecules in the QM7b database without significant supercomputer resources. To proceed with this technical validation, we used the FPS algorithm29,30 to choose the 100 most diverse molecules in the QM7b database (which we denote as the FPS-100 database). Due to the prohibitively large computational cost associated with LR-CCSD calculations with the daTZ basis set, we were only able to compute α for the 24 smallest molecules (by number of basis functions) in the FPS-100 database. A statistical error analysis of the αiso and αaniso values for these 24 molecules is provided in Table 5, and a more extensive discussion regarding the basis set convergence of our CCSD/daDZ calculations can be found in the main text and Supplementary Information of Ref.23. From Table 5, one can immediately see that the CCSD/daDZ αiso values have similar MSE, MAE, and RMSE values of ≈0.20 Bohr3, which corresponds to a MAPE of ≈0.4%. For αaniso, a measure of the anisotropy in the α tensor, we report slightly larger errors corresponding to a MAPE of . When compared to the errors made by the hybrid DFT functionals employed in this work (with CCSD/daDZ as the reference), namely 2.5% (B3LYP/daDZ) and 1.3% (SCAN0/daDZ) for αiso and 9.3% (B3LYP/daDZ) and 7.9% (SCAN0/daDZ) for αaniso, we conclude that the basis set incompleteness errors in our reference α values are significantly smaller (see Table 4). As such, the CCSD/daDZ α tensors presented in this work should be accurate and reliable enough for use in the development (and assessment) of next-generation force fields, density functionals, and quantum chemical methodologies, as well as machine-learning based approaches for predicting this fundamental response property.
Table 5.
Statistical error analysis of the CCSD/daDZ isotropic (αiso) and anisotropic (αaniso) polarizabilities in the FPS-100 database (i.e., the first 100 molecules in the QM7b database chosen by the FPS algorithm) computed with respect to the CCSD/daTZ level.
| α | MSE (MSPE) | MAE (MAPE) | RMSE (RMSPE) |
|---|---|---|---|
| α iso | 0.22 (0.42) | 0.22 (0.43) | 0.26 (0.50) |
| α aniso | −0.25 (−0.79) | 0.27 (0.87) | 0.33 (1.15) |
Due to the increased computational cost associated with computing α at the CCSD/daTZ level, a subset of the FPS-100 database (which includes the 24 molecules with the smallest number of basis functions) was considered during this analysis.
ISA-Tab metadata file
Acknowledgements
Y.Y., K.U.L. and R.A.D. acknowledge support from Cornell University through start-up funding. D.M.W. and M.C. acknowledge support from the European Research Council (Horizon 2020 Grant Agreement No. 677013-HBMAP). M.C. and A.G. acknowledge funding by the MPG-EPFL Center for Molecular Nanoscience and the NCCR MARVEL, funded by the Swiss National Science Foundation. This research used resources of the Argonne Leadership Computing Facility at Argonne National Laboratory, which is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-06CH11357. This research also used resources of the National Energy Research Scientific Computing Center (NERSC), a U.S. Department of Energy Office of Science User Facility operated under Contract No. DE-AC02-05CH11231. This work was also supported by a grant from the Swiss National Supercomputing Centre (CSCS) under Project ID s843, and by computer time from the EPFL scientific computing centre.
Author Contributions
Y.Y., K.U.L. and R.A.D. designed and performed all polarizability calculations. D.M.W., A.G. and M.C. implemented and performed the farthest point sampling of the QM7b database. All authors contributed to the design of the AlphaML showcase database, analyzed the data, and contributed to the writing of the manuscript.
Code Availability
As mentioned above, three different software packages were utilized in this work. Psi4 v1.134 is freely available from its official website38 Q-Chem v5.035 and FHI-AIMS32 must be downloaded from their official sites39,40 with a signed license.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Michele Ceriotti, Email: michele.ceriotti@epfl.ch.
Robert A. DiStasio, Jr., Email: distasio@cornell.edu
ISA-Tab metadata
is available for this paper at 10.1038/s41597-019-0157-8.
References
- 1.Stone, A. The Theory of Intermolecular Forces 2nd edn (Oxford University Press, 2016).
- 2.Hermann J, DiStasio RA, Jr., Tkatchenko A. First-principles models for van der Waals interactions in molecules and materials: concepts, theory, and applications. Chem. Rev. 2017;117:4714–4758. doi: 10.1021/acs.chemrev.6b00446. [DOI] [PubMed] [Google Scholar]
- 3.Grimme, S. In The Chemical Bond: Chemical Bonding Across the Periodic Table. (eds Frenking, G. & Shaik, S.) Ch. 16 (Wiley-VCH, 2014).
- 4.Shen YR. Surface properties probed by second harmonic and sum-frequency generation. Nature. 1989;337:519–525. doi: 10.1038/337519a0. [DOI] [Google Scholar]
- 5.Morita A, Hynes JT. A theoretical analysis of the sum frequency generation spectrum of the water surface. J. Chem. Phys. 2000;258:371–390. [Google Scholar]
- 6.Luber S, Iannuzzi M, Hutter J. Raman spectra from ab initio molecular dynamics and its application to liquid S-methyloxirane. J. Chem. Phys. 2014;141:094503. doi: 10.1063/1.4894425. [DOI] [PubMed] [Google Scholar]
- 7.Medders GR, Paesani F. Dissecting the molecular structure of the air/water interface from quantum simulations of the sum-frequency generation spectrum. J. Am. Chem. Soc. 2016;138:3912–3919. doi: 10.1021/jacs.6b00893. [DOI] [PubMed] [Google Scholar]
- 8.Sprik M, Klein ML. A polarizable model for water using distributed charge sites. J. Chem. Phys. 1988;89:7556–7560. doi: 10.1063/1.455722. [DOI] [Google Scholar]
- 9.Fanourgakis GS, Xantheas SS. Development of transferable interaction potentials for water. V. Extension of the flexible, polarizable, Thole-type model potential (TTM3-F, v. 3.0) to describe the vibrational spectra of water clusters and liquid water. J. Chem. Phys. 2008;128:074506. doi: 10.1063/1.2837299. [DOI] [PubMed] [Google Scholar]
- 10.Ponder JW, et al. Current status of the AMOEBA polarizable force field. J. Phys. Chem. B. 2010;114:2549–2564. doi: 10.1021/jp910674d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Medders GR, Babin V, Paesani F. Development of a “first-principles” water potential with flexible monomers. III. Liquid phase properties. J. Chem. Theory Comput. 2014;10:2906–2910. doi: 10.1021/ct5004115. [DOI] [PubMed] [Google Scholar]
- 12.Bereau T, DiStasio RA, Jr., Tkatchenko A, von Lilienfeld OA. Non-covalent interactions across organic and biological subsets of chemical space: physics-based potentials parametrized from machine learning. J. Chem. Phys. 2018;148:241706. doi: 10.1063/1.5009502. [DOI] [PubMed] [Google Scholar]
- 13.Monkhorst HJ. Calculation of properties with the coupled-cluster method. Int. J. Quantum Chem. 1977;12:421–432. doi: 10.1002/qua.560120850. [DOI] [Google Scholar]
- 14.Koch H, Jørgensen P. Coupled cluster response functions. J. Chem. Phys. 1990;93:3333–3344. doi: 10.1063/1.458814. [DOI] [Google Scholar]
- 15.Christiansen O, Jørgensen P, Hättig C. Response functions from fourier component variational perturbation theory applied to a time-averaged quasienergy. Int. J. Quantum Chem. 1998;68:1–52. doi: 10.1002/(SICI)1097-461X(1998)68:1<1::AID-QUA1>3.0.CO;2-Z. [DOI] [Google Scholar]
- 16.Hammond JR, Govind N, Kowalski K, Autschbach J, Xantheas SS. Accurate dipole polarizabilities for water clusters n = 2–12 at the coupled-cluster level of theory and benchmarking of various density functionals. J. Chem. Phys. 2009;131:214103. doi: 10.1063/1.3263604. [DOI] [PubMed] [Google Scholar]
- 17.Hammond JR, de Jong WA, Kowalski K. Coupled-cluster dynamic polarizabilities including triple excitations. J. Chem. Phys. 2008;128:224102. doi: 10.1063/1.2929840. [DOI] [PubMed] [Google Scholar]
- 18.Lao KU, Jia J, Maitra R, DiStasio RA., Jr. On the geometric dependence of the molecular dipole polarizability in water: a benchmark study of higher-order electron correlation, basis set incompleteness error, core electron effects, and zero-point vibrational contributions. J. Chem. Phys. 2018;149:204303. doi: 10.1063/1.5051458. [DOI] [PubMed] [Google Scholar]
- 19.Woon DE, Dunning TH., Jr. Gaussian basis sets for use in correlated molecular calculations. IV. Calculation of static electrical response properties. J. Chem. Phys. 1994;100:2975–2988. doi: 10.1063/1.466439. [DOI] [Google Scholar]
- 20.Blum LC, Reymond J-L. 970 million druglike small molecules for virtual screening in the chemical universe database GDB-13. J. Am. Chem. Soc. 2009;131:8732–8733. doi: 10.1021/ja902302h. [DOI] [PubMed] [Google Scholar]
- 21.Rupp M, Tkatchenko A, Müller K-R, von Lilienfeld OA. Fast and accurate modeling of molecular atomization energies with machine learning. Phys. Rev. Lett. 2012;108:058301. doi: 10.1103/PhysRevLett.108.058301. [DOI] [PubMed] [Google Scholar]
- 22.Montavon G, et al. Machine learning of molecular electronic properties in chemical compound space. New J. Phys. 2013;15:095003. doi: 10.1088/1367-2630/15/9/095003. [DOI] [Google Scholar]
- 23.Wilkins DM, et al. Accurate molecular polarizabilities with coupled cluster theory and machine learning. Proc. Natl. Acad. Sci. USA. 2019;116:3401–3406. doi: 10.1073/pnas.1816132116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christiansen O, Gauss J, Stanton JF. Frequency-dependent polarizabilities and first hyperpolarizabilities of CO and H2O from coupled cluster calculations. Chem. Phys. Lett. 1999;305:147–155. doi: 10.1016/S0009-2614(99)00358-9. [DOI] [Google Scholar]
- 25.Becke AD. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993;98:5648–5652. doi: 10.1063/1.464913. [DOI] [Google Scholar]
- 26.Stephens PJ, Devlin FJ, Chabalowski CF, Frisch MJ. Ab Initio calculation of vibrational absorption and circular dichroism spectra using density functional force fields. J. Chem. Phys. 1994;98:11623–11627. doi: 10.1021/j100096a001. [DOI] [Google Scholar]
- 27.Hui K, Chai J-D. SCAN-based hybrid and double-hybrid density functionals from models without fitted parameters. J. Chem. Phys. 2016;144:044114. doi: 10.1063/1.4940734. [DOI] [PubMed] [Google Scholar]
- 28.The QM7b Dataset, http://quantum-machine.org/datasets (2013).
- 29.Imbalzano G, et al. Automatic selection of atomic fingerprints and reference configurations for machine-learning potentials. J. Chem. Phys. 2018;148:241730. doi: 10.1063/1.5024611. [DOI] [PubMed] [Google Scholar]
- 30.Bartók AP, Kondor R, Csányi G. On representing chemical environments. Phys. Rev. B. 2013;87:184115. doi: 10.1103/PhysRevB.87.184115. [DOI] [Google Scholar]
- 31.Perdew JP, Burke K, Ernzerhof M. Generalized gradient approximation made simple. Phys. Rev. Lett. 1996;77:3865–3868. doi: 10.1103/PhysRevLett.77.3865. [DOI] [PubMed] [Google Scholar]
- 32.Blum V, et al. Ab initio molecular simulations with numeric atom-centered orbitals. Comput. Phys. Commun. 2009;180:2175–2196. doi: 10.1016/j.cpc.2009.06.022. [DOI] [Google Scholar]
- 33.Yang Y, 2019. Quantum mechanical static dipole polarizabilities in the QM7b and AlphaML showcase databases. Materials Cloud. [DOI] [PMC free article] [PubMed]
- 34.Parrish RM, et al. Psi4 1.1: an open-source electronic structure program emphasizing automation, advanced libraries, and interoperability. J. Chem. Theory Comput. 2017;13:3185–3197. doi: 10.1021/acs.jctc.7b00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shao Y, et al. Advances in molecular quantum chemistry contained in the Q-Chem 4 program package. Mol. Phys. 2015;113:184–215. doi: 10.1080/00268976.2014.952696. [DOI] [Google Scholar]
- 36.Feller D. The role of databases in support of computational chemistry calculations. J. Comput. Chem. 1996;17:1571–1586. doi: 10.1002/(SICI)1096-987X(199610)17:13<1571::AID-JCC9>3.0.CO;2-P. [DOI] [Google Scholar]
- 37.Schuchardt KL, et al. Basis set exchange: a community database for computational sciences. J. Chem. Inf. Model. 2007;47:1045–1052. doi: 10.1021/ci600510j. [DOI] [PubMed] [Google Scholar]
- 38.The PSI4 Project. Psi4: Open-Source Quantum Chemistry, http://www.psicode.org (2017).
- 39.Q-Chem Inc. Quantum Computational Software; Molecular Modeling; Visualization, http://www.q-chem.com (2015).
- 40.Theory Department of the Fritz-Haber-Institut der Max-Planck-Gesellschaft. FHI-aims, https://aimsclub.fhi-berlin.mpg.de (2009).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Yang Y, 2019. Quantum mechanical static dipole polarizabilities in the QM7b and AlphaML showcase databases. Materials Cloud. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
As mentioned above, three different software packages were utilized in this work. Psi4 v1.134 is freely available from its official website38 Q-Chem v5.035 and FHI-AIMS32 must be downloaded from their official sites39,40 with a signed license.