Abstract
Honey bees are agriculturally important, both as pollinators and by providing products such as honey. The sustainability of beekeeping is at risk through factors of global change such as habitat loss, as well as through the spread of infectious diseases. In China and other parts of Asia, beekeepers rely both on native Apis cerana and non-native Apis mellifera, putting bee populations at particular risk of disease emergence from multi-host pathogens. Indeed, two important honey bee parasites have emerged from East Asian honey bees, the mite Varroa destructor and the microsporidian Nosema ceranae. As V. destructor vectors viral bee diseases, we investigated whether another key bee pathogen, Deformed Wing Virus (DWV), may also have originated in East Asian honey bee populations. We use a large-scale survey of apiaries across China to investigate the prevalence and seasonality of DWV in managed A. mellifera and A. cerana colonies, showing that DWV-A prevalence was higher in A. mellifera, with a seasonal spike in prevalence in autumn and winter. Using phylogenetic and population genetic approaches, we show that while China and East Asian DWV isolates show comparatively high levels of genetic diversity, these bee populations are not a source for the current global DWV epidemic.
Subject terms: Ecological epidemiology, Evolutionary ecology, Evolutionary genetics
Introduction
Honey bees play a vital role in agriculture and food security through the pollination of crops as well as the production of honey and other related goods such as royal jelly and honey bee-collected pollen. While the proportion of global agricultural production relying upon animal pollination has nearly doubled in the last 50 years1, apiculture increasingly fails to provide sufficient pollination services in Europe2 and farmers in parts of Southwestern China have reportedly had to rely on hand-pollination due to pollinator declines3, a practice which is not economically viable. China alone currently hosts more than 9 million bee colonies and 180,000 beekeepers4. Honey bee populations in China are undergoing rapid changes, with a strong drive to intensify apiculture and increase the number of managed honey bees5, and a concurrent reduction in managed and wild A. cerana in line with the increasing use of A. mellifera6. Beekeepers in America and Europe have experienced high colony mortality in recent years7,8, which threaten the sustainability of beekeeping operations.
While habitat loss and pesticide exposure certainly play a major role, declines in honey bees have been exacerbated by emerging diseases9,10. Several field studies have linked overwinter mortality of honey bee hives to increased titers of Deformed Wing Virus (DWV)11–14, which in turn are driven by the presence of the ectoparasitic mite Varroa destructor (hereafter referred to as ‘Varroa’)15–17. DWV is a globally distributed single-stranded RNA Picornavirus that can infect honey bees and other insects such as bumble bees18,19. DWV and other pathogens can be transmitted between species through direct and indirect contact, such as by foraging on contaminated flowers or through behavior such as robbing (reviewed in19,20).
Varroa, itself an emerging parasite that originated in the East Asian honey bee A. cerana, feeds on the honey bee’s fat body21. It can thus transmit virus particles directly into the bee’s body cavity22. Field and laboratory studies have shown that the presence of Varroa leads to an increase in prevalence and titre of DWV in honey bees15–17. Prior to the emergence of this vector, DWV was considered largely avirulent in honey bees23. The high DWV titers associated with Varroa infestations15,17 result in an increase in virulence: high titer DWV infections may result in the wing deformities giving this virus its name (reviewed in18). The virus also reduces survival in individual infected workers as well as increasing mortality in the colony’s overwinter workforce24,25, thereby reducing honey bee fitness. DWV is currently undergoing a global epidemic, driven by Varroa-infested European honey bee populations26. The combined spread of DWV and its emerging vector Varroa has been facilitated by the unregulated global movement of honey bees between countries and the resulting DWV pandemic is spreading into wild pollinators24,26, threatening insect pollination.
Globally, the European honey bee Apis mellifera is currently predominantly used in apiculture and pollination service. In Asia, and in particular in China, beekeepers use the indigenous honey bee A. cerana in addition to non-native A. mellifera. The Asian honey bee has coevolved with Varroa mites (Varroa jacobsoni and V. destructor) and possesses behavioural27–30 and physiological resistance mechanisms against these mites31, which may protect them from the current global DWV epidemic26. At the same time, East Asia is the epicentre of disease emergence for bees: both the Varroa mite and the gut parasite Nosema ceranae have jumped from the Asian honey bee A. cerana to the European honey bee A. mellifera in East Asia in the last century and have spread globally. This raises the question of whether East Asian honey bee populations may serve as a source or reservoir for other emerging bee pathogens such as DWV.
Here, we test the hypothesis that Chinese managed A. cerana populations are part of the phylogenetic source population for the current global Varroa-associated DWV epidemic, as suggested by this Asian bee population’s role in the emergence of N. ceranae and in particular DWV’s novel vector, the Varroa mite V. destructor. Given A. cerana’s relative resistance to V. destructor, we expect lower DWV prevalence than in A. mellifera. If Chinese managed honey bees serve as a source for the current global DWV epidemic, we expect to find higher ancestral genetic variation in this DWV population. To test these predictions, we use a large-scale multi-year survey of both European and Asian honey bees across China. Using phylogenetic and population genetic approaches, we show that while China and East Asian DWV isolates indeed show comparatively high levels of genetic diversity, these bee populations are not the ancestral source for the current global DWV epidemic.
Results
Prevalence
50 A. cerana hives from 24 apiaries and 117 A. mellifera hives from 61 apiaries were sampled across 22 Chinese provinces over a 2 year period between April 2015 and March 2017 (see Tables S1, S2 for colony- and apiary-level data). The estimated true colony-level prevalence (i.e. the percentage of DWV-positive colonies) of DWV-A in Chinese A. mellifera populations reaches 45.7% (95% Confidence Interval (CI) 35.6–56.2%), whereas the true colony-level prevalence of DWV-A for A. cerana is estimated only at 5.6% (0–18.3%); this estimation takes into account the uncertainty of PCR-based pathogen detection, conservatively assuming a PCR assay sensitivity and specificity of 95%32. DWV is a viral complex, consisting of at least 3 viral strains, the globally distributed DWV-A, as well as DWV-B (originally described as Varroa Destructor Virus-1 (VDV-1) in V. destructor33) and the rare DWV-C34. Sequence analysis confirmed that these samples belong to DWV-A. We additionally screened for DWV-B using specific primers for the rdrp-gene. We found DWV-B in two A. mellifera colonies out of a total of 117, resulting in an estimated true colony-level prevalence of 0–0.83%; we found no evidence for DWV-B presence in the 50 A. cerana colonies tested. A phylogenetic tree (Fig. S1) shows that the non-heterozygous Chinese DWV-B isolate SC-3 was highly similar to the known European DWV-B isolates.
We tested whether apiary-level DWV-A prevalence (i.e. the number of positive vs. negative colonies per apiary) was affected by host species or season (modeled as a continuous trait) using Generalised linear mixed models (GLMMs) with binomial error distribution and logit link function, including the location of origin and the collection year as random effects. The minimum adequate model (MAM) was identified through model simplification using ANOVA and removal of non-significant terms. Measuring apiary-level prevalence per location and time-point, we found that both the host species (ANOVA, χ2 = 9.2649, d.f. = 1, p < 0.01) and the season in which samples were collected (ANOVA, χ2 = 9.7502, d.f. = 1, p < 0.01; modeled as a continuous variable) had a significant effect on prevalence, see Fig. 1. These factors did not significantly interact (ANOVA, χ2 = 0.2086, d.f. = 1, p = 0.65). We found that the region samples were collected in (North, Northeast, Northwest, East, Central and South China) did not significantly affect prevalence (ANOVA, χ2 = 7.2353, d.f. = 5, p = 0.2037). These results – significant effects of host species (ANOVA, χ2 = 11.469, d.f. = 1, p < 0.001) and season (ANOVA, χ2 = 19.399, d.f. = 3, p < 0.001) on prevalence, but no significant interaction (ANOVA, χ2 = 2.4559, d.f. = 3, p = 0.48) - were also replicated when modeling season as a discrete trait rather than a continuous trait in order to perform post-hoc tests, however for convergence reasons, year was not included as a random factor in these models. Prevalence was not only higher in A. mellifera than in A. cerana colonies (z = 2.827, p < 0.01), but was also found to be higher in autumn than in spring and summer (zspring = −2.956, pspring < 0.05; zsummer = −3.419, pspring < 0.01) as well as higher in winter than summer (z = 2.913, p < 0.05) using multiple comparisons of means with Tukey contrasts.
Figure 1.
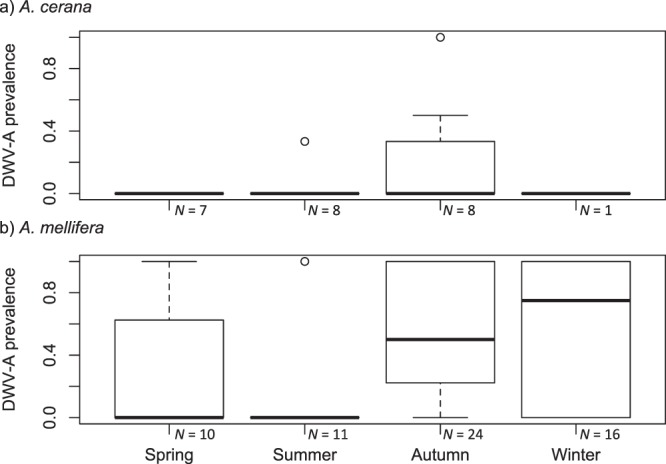
Apiary-level DWV-A prevalence in A. cerana and A. mellifera across seasons; Sample sizes indicate the number of apiaries per site per species (Napiaries = 85, Nhives = 167). Both host species and season affect prevalence, with prevalence significantly higher in autumn than in spring and summer, and in winter than in summer.
Several previous reports include measures of DWV prevalence in one or both species in East Asia35–42 (see Table S3). Across all studies, the mean prevalence in A. cerana is 22% and 74% in A. mellifera. Testing the studies that measured prevalence in both species simultaneously, we find that prevalences are significantly different between species across all studies (glm with binomial error function, z = 7.22, p < 0.001).
Phylogenetic and population genetic analysis
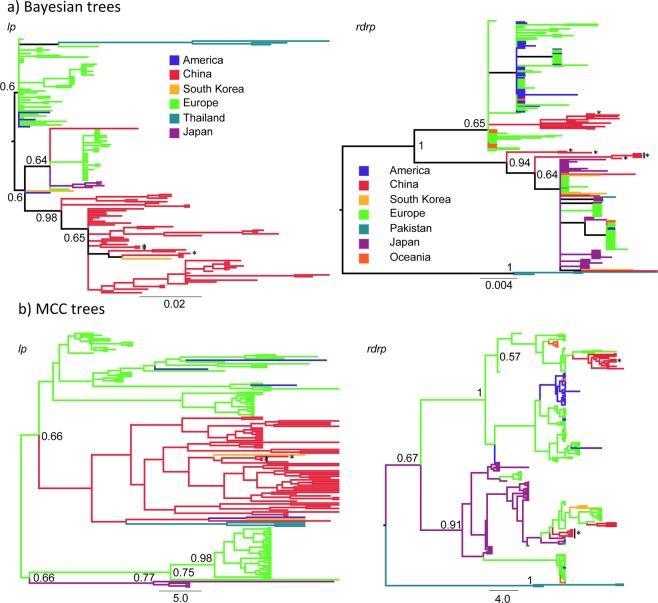
We reconstructed the DWV-A phylogeny independently via three gene fragments (lp-, rdrp- and vp3), using all available comparative DWV-A sequences on Genbank (Fig. 2; see supplementary information for trees including Genbank accession numbers) from both A. mellifera and A. cerana. A Bayesian approach (Fig. 2a) showed that the lp- and rdrp-fragments produced relatively well resolved phylogenies, whereas the vp3-fragment contained too little genetic variation to resolve DWV-A population structure beyond the split between the globally distributed DWV-A isolates and divergent isolates from Pakistan26 (see Fig. S2). We therefore restricted further population genetic analysis to the phylogenetically informative lp- and rdrp-fragments. For the rdrp-fragment, Chinese samples fall in two distinct clades. For the lp-fragment on the other hand, the maximum clade credibility (MCC) tree shows Chinese and South Korean samples in a single well-supported clade; this result is also largely supported by the Bayesian phylogeny, which does not take geographic origin into account. While it is important to note that the alignments on which these phylogenetic trees are based contain different individual samples both globally and as collected for this study within China, several samples from the same Apiary (ZJ-1&2; HuB-5&7; XJ-4; JS-2; HeB-1&2; LN-5) are included for both gene fragments; in the lp-tree, these DWV sequences form a monophyletic clade, whereas the rdrp-tree shows that two viral sequences of different phylogentic origins are present in the same apiary.
Figure 2.
Phylogenetic trees for lp-, rdrp- and vp3-fragments; (a) Bayesian trees (b). MCC trees constructed with TreeAnnotator based on BEAST runs. Star symbols indicate samples isolated from A. cerana in China. All trees were midpoint-rooted. Posterior support (MCC trees) and Bayesian probabilities (Bayesian trees) above 0.5 are indicated up to the 3rd node from the midpoint root; horizontal bars indicate the time scale in years. In the MCC trees, the branches are colored according to the lineages’ inferred geographic origin. See supplementary information (Figs S5–S8) for enlarged tree figures including sample names/Genbank accession numbers.
We found no evidence for genetic differentiation between DWV sequences isolated from A. cerana or A. mellifera (lp-fragment: NA.cerana = 3, NA.mellifera = 127, rdrp-fragment: NA.cerana = 8, NA.mellifera = 184). As shown in Fig. 2, samples from Chinese A. cerana apiaries are nested within other samples isolated from China. A group of 7 isolates from A. cerana in the rdrp-fragment (JX679473-JX679480) cluster together (see black vertical bar on rdrp-trees in Fig. 2); however, these samples from Yunnan province come from a single Genbank submission that did not include corresponding samples from A. mellifera and lend no support for genetic differentiation between DWV-A populations in A. cerana and A. mellifera without further investigation. All subsequent analysis combine DWV sequences from both host species.
The different phylogenies for the Chinese isolates seen in the two fragments (Fig. 2) are also reflected in their demographic parameters as reconstructed with the coalescent sampler BEAST43. Exponential population growth is supported for the Chinese samples within the lp-fragment (mean doubling time 7.66 years, 95% Highest Posterior Density (HPD) 4.53–20.75), while the 95% HPD for the exponential growth rate overlaps with 0 for the rdrp-fragment (95% HPD −0.03–0.23). An excess of low frequency polymorphism was detected in both fragments using a coalescent simulation of Tajima’s D in DNAsp v.544 (Dlp = −1.69, plp < 0.05, 95% Confidence Interval −1.598–1.946; Drdrp = −1.75, prdrp > 0.01, 95% Confidence Interval −1.565–1.855), also indicating potential population size expansion in the Chinese DWV population. For both fragments, the most recent common ancestor of the Chinese DWV-A population is reconstructed about 3 decades ago (lp-fragment: 33 years, HPD 21–48 years, rdrp-fragment: 31 years, HPD 20–45 years; root height: lp-fragment: 47 years, HPD 37–60 years, rdrp-fragment: 38 years, HPD 24–57 years).
Measuring population differentiation based on the proportion of between-population nucleotide differences45, we found moderate levels of population differentiation between China, East Asia (Japan and South Korea as well as, for the lp-fragment, Thailand), Europe and America (Kstlp = 0.12, Kstrdrp = 0.27, p < 0.001) in accordance with the phylogenetic trees. Across all three fragments, samples from China and East Asia (Japan, South Korea and, for the lp-fragment, Thailand) consistently showed the highest nucleotide diversities (see Table 1). Nucleotide diversity π was typically ~2–3 times higher for these populations than for the samples from Europe and America. Discrete trait analysis in BEAST, modelling the geographic origin as a discrete trait, however indicated that China is not a source population, but a sink population with Europe serving as a source for the Chinese DWV-A population (Bayes Factor = 17.14 and 499.84 for the lp- and rdrp-fragments respectively; see Tables S4, S5 for Bayes Factors between all populations). The phylogenetic trees (Fig. 2) indicate no geographic clustering within China based on the available genetic information. There is also no evidence for isolation by distance between samples from China based on geographic distance and Euclidian genetic distance (Mantel-test, 999 permutations, lp-fragment p = 0.51, rdrp-fragment p = 0.154).
Table 1.
Nucleotide diversity π per fragment, number in bracket indicates sample size.
| lp | rdrp | vp3 | |
|---|---|---|---|
| China | 0.057 [61] | 0.027 [25] | 0.021 [34] |
| East Asia | 0.064 [69] | 0.021 [40] | 0.016 [10] |
| America | 0.021 [4] | 0.006 [36] | 0.006 [27] |
| Europe | 0.034 [67] | 0.018 [109] | 0.009 [41] |
Discussion
This multi-year survey of A. mellifera and A. cerana apiaries across China shows that, while DWV-A is genetically more diverse in China and East Asia than in the rest of the global populations sampled, China represents a sink population, rather than the source of this global epidemic. DWV is predominantly a disease of non-native European honey bees in China; DWV-A prevalence was significantly higher in A. mellifera (45.7% DWV-positive colonies) than in A. cerana (5.6%), with prevalences increasing at the end of the season. The emerging DWV-B was rare in A. mellifera and absent in A. cerana.
In combination with previous studies of DWV in A. mellifera and A. cerana in East Asia35–42, we can conclude that DWV is more prevalent in the European honey bee than in the Asian honey bee in East Asia. This lower prevalence may be due to a reduced exposure of Asian honey bees to DWV vectored by Varroa mites. A. cerana has co-evolved with Varroa ectoparasites; in contrast to A. mellifera, Asian honey bees can control mite populations by grooming of adult bees, removing infested brood and by effectively restricting Varroa reproduction to drone brood27,28.
The present study shows that there is no evidence for genetic differentiation between DWV isolates from A. mellifera and A. cerana (Fig. 2). This corroborates findings from other smaller studies from China40,42,46 and Japan39. There is thus no evidence that A. cerana harbours a distinct DWV-A-clade or acts as a reservoir of infection. To uncover the full viral flora of Asian honey bees, including testing whether these populations harbor divergent DWV strains and whether they may be the ancestral host of DWV pre-dating the global emergence of DWV-A in concert with the global spread of Varroa26, next-generation sequencing studies are needed47. Next-generation sequencing studies allow an unbiased de-novo approach to discover the true viral diversity in host species.
This study was carried out across two years, sampling throughout the year. This revealed strong seasonality (Fig. 1), with prevalence highest at the end of the season in autumn and winter. This matches patterns reported from studies in France48, Denmark49 and the United States50. Both Tentcheva et al.48 and Francis et al.49 report high and increasing prevalence in worker honey bees with the highest prevalence reported in autumn (the studies did not include workers collected in winter). Using a microarray, Runckel et al.50 found low prevalence of DWV with sporadic incidences only in autumn and winter. This seasonal change in prevalence, with reduced prevalence in spring and summer, may be caused by increased mortality of DWV-infected workers in the cold season. Natsopoulou et al.25 showed an increase in over-winter workforce mortality in colonies infected by DWV-B (note no DWV-A was detected in this field study). An increase in mortality of infected workers would lead to a reduction in the detected prevalence in spring. Additionally, seasonality in DWV prevalence could also be linked to seasonal population dynamics or behavioural changes in V. destructor, with an increase in phoretic mite density in adult bees typically reported in autumn51. Other factors that may contribute to this strong seasonality may be environmental stressors, such as forage availability and diversity (e.g.52,53), and a build-up of exposure to chemicals throughout the season54, which may lead to a trade-off in resistance to DWV via the bees’ immune system55.
Comparing Chinese and East Asian DWV populations to the global DWV data, it is evident that these populations harbor higher genetic diversity than other populations, especially the very well sampled European population (Table 1). Comparatively high genetic diversity is a potential indicator of an ancestral population, which could support two non-exclusive hypotheses that: a) these populations represent the ancestral source population of the current DWV-A population, spread in conjunction with its emergent vector V. destructor; b) the high genetic diversity is maintained or generated because of the diversity in Chinese honey bee species and their co-evolved ectoparasitic mite species6,56. In contrast, the Hawaiian Varroa invasion has demonstrated that the introduction of Varroa as a viral vector can lead to a drastic reduction in DWV’s genetic variation within A. mellifera15. However, a phylogenetic reconstruction using geographic origin as a discrete trait clearly shows China and neighboring South Korea to be sink rather than source populations for global DWV-A. The most recent common ancestors date within the last 30–50 years, several decades after the acquisition and spread of the Varroa mite26. Thus we find no support for the hypothesis that the current DWV-A epidemic has its origin in East Asian honey bee and Varroa populations. Instead, these results, in line with the global geographic reconstruction of the DWV-A epidemic26, point towards a non-Asian origin of the current epidemic, with Varroa serving as an amplifier rather than the ancestral source of the epidemic. The comparatively high genetic variation in China is compatible with a recent population expansion of the Chinese DWV-population; there is little evidence that the Chinese DWV populations show inherently higher evolutionary rates (sup Fig. S3, S4). The comparatively high genetic variation has a recent origin within the last 4 decades, not pre-dating the spread of non-native A. mellifera or its acquisition of Varroa as a viral vector in China6. Whether the higher genetic variation within Chinese DWV populations may relate to the higher species-level diversity of Chinese honey bees and their co-evolved mites will need to be addressed by surveys including not only A. mellifera, A. cerana and V. destructor, but also the wild species (A. florea, A. dorsata, A. andreniformis, A. laboriosa) and their respective mites6,56.
The DWV-A phylogeny within China also shows a potential for recombination and divergent evolution within its genome: whereas the Chinese isolates are part of a well-supported single clade in the lp-fragment, isolates are distributed over two well-supported clades for the rdrp-fragment. This pattern is indicative of recombination within the DWV genome. While the present study cannot directly show recombination within individuals, 6 isolates that are placed on different clades for the two fragments come from the same apiaries, which are likely to be genetically highly similar. This pattern is reminiscent of the DWV-A/B recombinants found in field studies in Israel57 and the UK58,59 and may indicate that recombination may play a role in DWV evolution not only between DWV-A and DWV-B22 but also within DWV-A itself.
DWV is a species complex consisting of at least 3 distinct subtypes34, with DWV-A being globally distributed26. While we find sequence evidence for the presence of DWV-B in China in this study, this subtype showed a prevalence of less than 1% and was not found in A. cerana. Previous studies that conducted phylogenetic analysis have only detected DWV-A in China40,42,46 and Japan39. However, it has to be noted that earlier studies used primers designed to amplify DWV-A and may thus have underestimated the prevalence of DWV-B60. Given the rapid population expansion of DWV-B in Europe25,61 and its recent emergence and spread in North America22, Chinese honey bees may be at the cusp of a DWV-B epidemic. This is of particular concern given the increased virulence of DWV-B in European honey bees25,62. In addition, this viral type, like DWV-A24, also shows potential spillover into bumblebees in the presence of Varroa61. This potential spread is of particular concern in China, which is a threatened hotspot for honey bee and bumble bee biodiversity63. The potential spread of DWV-B in China may allow testing whether competition between viral strains leads to a reduction in within-strain genetic diversity, an alternative hypothesis for the high genetic diversity found in Chinese DWV populations.
In conclusion, our results show Chinese honey bee populations are experiencing a DWV-A epidemic with exponential viral population expansion, but that this viral population is not a source of the current global epidemic26. We thus find no evidence that the globally spreading DWV-A strain has switched host from A. cerana to A. mellifera with its vector, the Varroa mite, nor that this currently globally dominant strain or the emerging DWV-B have originated in East Asia. This raises the questions of how the introduction of the non-native A. mellifera and the acquisition of Varroa as a viral vector in Asia have affected viral epidemiology in Asian wild and managed honey bee populations; addressing these questions calls for surveys of wild and managed populations using de novo next-generation sequencing approaches to uncover the full viral diversity in these populations. Our results show that the emerging virulent DWV-B25,60,62,64 is also present in Chinese A. mellifera. These findings call for urgent surveillance work on DWV-B in East Asia, to prevent and mitigate its further spread into apiculture and wild populations. The potential spread of DWV-B to China is a further demonstration that the unregulated movement of honey bee queens and colonies across borders and geographic regions needs to be curbed to prevent the spread of emerging pathogens, a risk which needs to the considered in the global movement of all live animal and plant material.
Methods
Samples
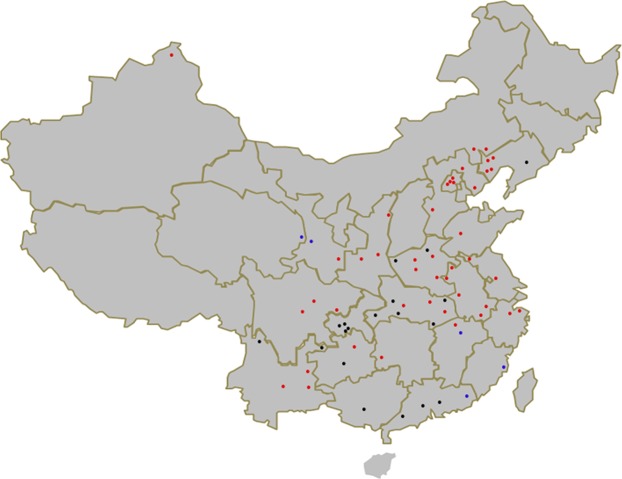
50 A. cerana and 117 A. mellifera hives across 22 Chinese provinces were sampled from March 2015 to February 2017, including Anhui, Jiangsu, Shandong, Zhejiang, Hebei, Beijing, Shannxi, Qinghai, Gansu, Xinjiang, Sichuan, Guizhou, Inner Mogolia, Chongqing, Hubei, Jiangxi, Henan, Liaoning, Guangdong, Fujian, Yunnan and Guangxi (Fig. 3; Table S1), with ~ fifty adult worker bees per hive being taken for further analysis.
Figure 3.
Sampling locations; A. mellifera sites are indicated in red, A. cerana in black and sites where both A. mellifera and A. cerana were collected are indicated in blue.
Clinical signs of virus infection were not found in these colonies; thus, these colonies were considered to be capable of honey production, although they were reported either to have shown the presence of crawling bees or to have undergone important worker losses previously65,66. All of the above colonies were maintained consistent with guidelines for beekeeping practice and, in the case of A. mellifera, regularly monitored and treated with acaricides for mites. There was no effect on colony development reported due to mite infestation during the previous year. Living bee samples were placed in a small iron wire cage for direct transport or in 50 mL sterile tubes with dry ice for transport to the laboratory, where the samples were stored at −80 °C prior to use.
RNA extraction
All individual samples were submerged in liquid nitrogen and ground into a fine powder using a pestle and mortar; this material was used in Trizol-based RNA extractions as described in Diao et al.67. The obtained RNA was dissolved in 20 µL of sterile water and stored at −80 °C prior to analysis. The quantity and purity of the RNA were measured using a Nanodrop spectrophotometer (Thermo Scientific, Beijing, China), with samples included for further analysis if OD 260/280 values were in the range of 1.8–2.0. The cDNA was synthesized using M-MLV reverse transcriptase with oligo dT primers according to the manufacturer’s instructions (Thermo, USA).
RT-PCR amplification
Pooled samples of ~50 individuals were initially screened for DWV using RT-PCR according to standard procedures at the Institute of Apicultural Research. To specifically test for the presence of the two most common DWV strains, DWV-A and DWV-B, we used strain specific primers for DWV-A (DWV-F1a GGA AAC ATC TGG AAT TAG CGA CAA) and DWV-B (VDV-F1a GAA AAC ATT TGG AAT TAG CAA CGA C) respectively, with the conserved reverse primer DWV-VDV 7aR (AAT CCG TGA ATA TAG TGT GAG G)68. Only 2 out of 167 colonies were confirmed to be infected with DWV-B using specific primers and Sanger sequencing and we therefore focused further analysis on DWV-A.
Following the detection of positive hives via the pooled samples, DWV was amplified from 3 or 5 individuals per hive (see Table S1) using primers for three fragments (lp, vp3 and rdrp)26 (see Table S6); these fragments were chosen based on the availability of comparative and informative sequence data on Genbank. The PCR products were sequenced by BGI (BGI Company, Shenzhen). The sequences of the lp, vp3 and rdrp-fragments have been deposited in GenBank under accession numbers MF431915-431945 (lp-fragment), MF144195-144203, MF351971-351972 and MF431886-431914 (vp3-fragment) and MF667713-667747 (rdrp-fragment).
Prevalence measures and statistical analysis
To take account of PCR assay efficiency and sensitivity (conservatively set at 95%)32, we used the R library epiR v.0.9–8269 and the function epi.prev to calculate the true prevalence with 95% confidence based on methods in Blaker70. We used R v. 3.5.3. in all analyses71. To test whether DWV-A prevalence at the apiary-level (the number of DWV-A-positive and negative hives in a location at a given sampling time-point) was affected by host species, and season, we used the lme4 package (v1.1-12)72 to run generalised linear mixed models (GLMMs) with binomial error distribution and a logit link function. The full model included the two-way interaction between the fixed effects host species (A. cerana and A. mellifera) and season (spring (March–May), summer (June–August), autumn (September–November) and winter (December–February) modeled either as a continuous or discrete trait). Location and year were included as random factors when modeling season as a continuous factor; year was excluded as a random factor when modeling season as a discrete trait to allow the model to converge. The minimum adequate model (MAM) was identified through removal of non-significant terms and comparison of models using ANOVA. We additionally tested whether the geographic origin by region (Center: Henan, Hubei, Jianxi; East: Anhui, Jiangsu, Shandong, Zhejiang; North: Beijing, Hebei; Northeast: Liaoning; Northwest: Chongqing, Gansu, Guizhou, Inner Mongolia, Qinghai, Sichuan, Xinjang; South: Guangdong, Guangxi, Yunnan) affected DWV-A prevalence, but due to a lack of convergence could not test for interactions with species and season. We used the glht function in the multicomp package v. 1.4–1073 for multiple comparisons of means using Tukey conrasts.
Sequence analysis
Using BLAST74 and a cutoff of 90% sequence similarity, we identified all available comparative DWV-A sequences on Genbank which included country of origin, host organism and year of collection for phylogenetic analysis and created alignments in Geneious 6.1.8, optimizing the alignment length to increase the number of isolates from China. We constructed 3 alignments, lp-fragment (n = 143, 287 bp; 56 samples from China, including 14 new samples as well as isolates MF036686 and samples from Zhejiang (HG779848-9; 50; 52; 56-8; 60-61), Hainan (JF346640-54) and Shaanxi provinces (MF134371-83)); vp3-fragment (n = 135, 273 bp; 34 samples from China, including 20 new samples as well as MF036686 and samples from Shaanxi province (MF134371-83)); and rdrp-fragment (n = 224, 252 bp; 25 samples from China including MF036686 and samples from Yunnan province (JX679473-80)). We confirmed that there was no recombination within fragments at a p-value of 0.05, using the GENECONV75, MaxChi76, BootScan77 and SiScan78 algorithms in the Rdp4 package (v4.56)77.
Phylogenetic reconstruction and population genetics
Beast 1.843 was used for phylogenetic reconstruction with tip dates, with discrete trait models with asymmetric substitution models for geographic location; the sample size and genetic variation contained in the alignments was not sufficient to include host species as a trait. Jmodeltest (v.2.1)79,80 was used to determine suitable substitution models for each fragment based on the Bayesian Information Criterion (lp-fragment: TrN + G; vp3- and rdrp-fragments HKY + G) and substitution rates were partioned between the 1st & 2nd and 3rd codon positions. The evolutionary rate prior was set for each fragment according to Wilfert et al.26 (lp-fragment: normal prior with a mean of 0.91 × 10−3 (stdev = 0.24 × 10−3); vp3-fragment: normal prior with a mean of 1.85 × 10−3 (stdev = 0.35 × 10−3); rdrp-fragment: lognormal prior with a mean in real space of 1.28 × 10−3 (log(stdev) = 0.325)). Demographic and molecular clock rates were set according to model comparison via the path sampling maximum likelihood estimator; exponential growth was preferred for all fragments, with an exponential relaxed clock for the lp- and vp3-fragment and a lognormal relaxed clock for the rdrp-fragment. All models were checked for convergence in Tracer (v1.6) and run long enough to obtain effective sample sizes > 200 for all parameters, with a 10% burn-in, sampling the chain at equal distances to obtain a total of 10,000 trees per analysis. We produced Maximum Clade Credibility (MCC) trees (TreeAnnotator (v1.8.4)) to infer host ancestral state probabilities. Well supported rates for migration routes between geographic regions were identified using a Bayes factors analysis (SPREAD v1.0.681), with a Bayes Factor of 3 as a cut-off. Phylogenetic trees were also produced for each alignment using MrBayes 3.2.6.
Using DNASPv5.10.144 we calculated Tajima’s D to test for an excess of rare polymorphisms. Additionally, we calculated Kst, a measure of population differentiation based on the proportion of between-population nucleotide differences45 between China, East Asia (South Korea and Japan), Europe and America as well as the nucleotide diversity π. To test for isolation by distance between samples collected for this study in China, we calculated individual Euclidean genetic distances using the package adegenet82 in R; we used individual-based distances as many locations were represented by single sequences. We tested for isolation by distance via a Mantel test implemented following Legendre and Legendre83 in the R package vegan84 of the correlation between geographic distance and Euclidean genetic distance using 999 permutations.
Supplementary information
Acknowledgements
We acknowledge the support of The National Natural Science Foundation of China (31572471) to CH and the Agricultural Science and Technology Innovation Program of CAAS (CAAS-ASTIP-2017-IAR) to Q.Y.D., as well as The Royal Society (IE161462) and BBSRC (BB/P025854/1) to L.W.
Author Contributions
C.S.H. and L.W. conceived the project. Q.Y.D., D.H.Y., H.X.Z., S.D. and X.L.W. conducted the field sampling. D.H.Y., H.X.Z., S.D. and X.L.W. performed the lab work. L.W. conducted the data analysis. C.S.H. and L.W. wrote the manuscript.
Data Availability
Sequence data have been deposited in GenBank under accession numbers MF431915-431945 (lp-fragment), MF144195-144203, MF351971-351972 and MF431886-431914 (vp3-fragment) and MF667713-667747 (rdrp-fragment).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Qingyun Diao and Dahe Yang contributed equally.
Contributor Information
Chunsheng Hou, Email: houchunsheng@caas.cn.
Lena Wilfert, Email: lena.wilfert@uni-ulm.de.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-48618-y.
References
- 1.Aizen MA, Harder LD. The global stock of domesticated honey bees is growing slower than agricultural demand for pollination. Curr. Biol. 2009;19:915–918. doi: 10.1016/j.cub.2009.03.071. [DOI] [PubMed] [Google Scholar]
- 2.Breeze TD, et al. Agricultural policies exacerbate honeybee pollination service supply-demand mismatches across Europe. PLoS ONE. 2014;9:e82996. doi: 10.1371/journal.pone.0082996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Partap U, Ya T. The human pollinators of fruit crops in Maoxian County, Sichuan, China. Mountain Res. Dev. 2012;32:176–186. doi: 10.1659/MRD-JOURNAL-D-11-00108.1. [DOI] [Google Scholar]
- 4.Wu, Q. & Ou, H. Analysis of the restriction factor of Guangxi province beekeeping. Apicult. China2, www.cnki.com.cn/Article/CJFDTOTAL-ZGYF201706039 (2017).
- 5.Zheng H-Q, Wei W-T, Hu F-L. Beekeeping industry in China. Bee World. 2011;88:41–44. doi: 10.1080/0005772X.2011.11417406. [DOI] [Google Scholar]
- 6.Hepburn, H. R. & Radloff, S. E. Biogeography in Honeybees of Asia (eds Hepburn, H. R. & Radloff, S. E.) 51–67 (Springer Verlag, 2011).
- 7.De Graaf D, et al. Risk indicators affecting honeybee colony survival in Europe: one year of surveillance. Apidologie. 2016;47:348–378. doi: 10.1007/s13592-016-0440-z. [DOI] [Google Scholar]
- 8.van Engelsdorp D, Hayes J, Underwood RM, Pettis J. A survey of honey bee colony losses in the U.S., fall 2007 to spring 2008. PLoS One. 2008;3:e4071. doi: 10.1371/journal.pone.0004071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goulson D, Nicholls E, Botias C, Rotheray EL. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science. 2015;347:1435. doi: 10.1126/science.1255957. [DOI] [PubMed] [Google Scholar]
- 10.Vanbergen AJ, et al. Threats to an ecosystem service: pressures on pollinators. Frontiers Ecol. Envir. 2013;11:251–259. doi: 10.1890/120126. [DOI] [Google Scholar]
- 11.Highfield AC, et al. Deformed Wing Virus implicated in overwintering honeybee colony losses. Appl. Environ. Microbiol. 2009;75:7212–7220. doi: 10.1128/AEM.02227-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berthoud H, Imdorf A, Haueter M, Radloff S, Neumann P. J. Api. Res. 2010. Virus infections and winter losses of honey bee colonies (Apis mellifera) pp. 60–65. [Google Scholar]
- 13.Dainat B, Evans JD, Chen YP, Gauthier L, Neumann P. Dead or Alive: Deformed Wing Virus and Varroa destructor reduce the life span of winter honeybees. Appl. Env. Microbiol. 2012;78:981–987. doi: 10.1128/AEM.06537-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genersch E, et al. The German bee monitoring project: a long term study to understand periodically high winter losses of honey bee colonies. Apidologie. 2010;41:332–352. doi: 10.1051/apido/2010014. [DOI] [Google Scholar]
- 15.Martin SJ, et al. Global honey bee viral landscape altered by a parasitic mite. Science. 2012;336:1304–1306. doi: 10.1126/science.1220941. [DOI] [PubMed] [Google Scholar]
- 16.Mondet F, de Miranda JR, Kretzschmar A, Le Conte Y, Mercer AR. On the front line: Quantitative virus dynamics in honeybee (Apis mellifera L.) colonies along a new expansion front of the parasite Varroa destructor. PLoS Path. 2014;10:e1004323. doi: 10.1371/journal.ppat.1004323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nazzi F, et al. Synergistic parasite-pathogen interactions mediated by host immunity can drive the collapse of honeybee colonies. PLoS Path. 2012;8:e1002735. doi: 10.1371/journal.ppat.1002735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Miranda JR, Genersch E. Deformed wing virus. J. Invert. Pathol. 2010;103:S48–S61. doi: 10.1016/j.jip.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 19.Manley R, Boots M, Wilfert L. Emerging viral disease risk to pollinating insects: ecological, evolutionary and anthropogenic factors. J. Appl. Ecol. 2015;52:331–340. doi: 10.1111/1365-2664.12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McMahon DP, Wilfert L, Paxton RJ, Brown MJF. Emerging viruses in bees: From molecules to ecology. Adv. Vir. Res. 2018;101:251–291. doi: 10.1016/bs.aivir.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 21.Ramsey SD, et al. Varroa destructor feeds primarily on honey bee fat body tissue and not hemolymph. PNAS. 2019;116:1792–1801. doi: 10.1073/pnas.1818371116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryabov EV, et al. A virulent strain of Deformed Wing Virus (DWV) of honeybees (Apis mellifera) prevails after Varroa destructor-mediated, or in vitro, transmission. PLoS Path. 2014;10:e1004230. doi: 10.1371/journal.ppat.1004230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bailey, L. & Ball, B. Honeybee Pathology. (Academic Press 1991).
- 24.Fürst MA, McMahon DP, Osborne JL, Paxton RJ, Brown MJF. Disease associations between honeybees and bumblebees as a threat to wild pollinators. Nature. 2014;506:364–366. doi: 10.1038/nature12977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Natsopoulou ME, et al. The virulent, emerging genotype B of Deformed wing virus is closely linked to overwinter honeybee worker loss. Sci. Rep. 2017;7:5242. doi: 10.1038/s41598-017-05596-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilfert L, et al. Deformed wing virus is a recent global epidemic in honeybees driven by Varroa mites. Science. 2016;351:594–597. doi: 10.1126/science.aac9976. [DOI] [PubMed] [Google Scholar]
- 27.Büchler R, Drescher W, Tornier I. Grooming behaviour of Apis cerana, Apis mellifera and Apis dorsata and its effect on the parasitic mites Varroa jacobsoni and Tropilaelaps clareae. Exp. Appl. Acar. 1992;16:313–319. doi: 10.1007/BF01218573. [DOI] [Google Scholar]
- 28.Rath W. Co-adaptation of Apis cerana Fabr. and Varroa jacobsoni Oud. Apidologie. 1999;30:97–110. doi: 10.1051/apido:19990202. [DOI] [Google Scholar]
- 29.Pritchard DJ. J. Api. Res. 2016. Grooming by honey bees as a component of Varroa resistant behavior; pp. 38–48. [Google Scholar]
- 30.Ji T, Yin L, Liu Z, Shen F, Shen J. High-throughput sequencing identification of genes involved with Varroa destructor resistance in the eastern honeybee, Apis cerana. Gen. Mol. Res. 2014;13:9086–9096. doi: 10.4238/2014.October.31.24. [DOI] [PubMed] [Google Scholar]
- 31.Wang, X. et al. The difference of the nutritional components in hemolymph of honeybee larva may affected the host preference of Varroa destructor. Chinese J. Vet. Med. 4, http://lib.cqvip.com/Qikan/Article/Detail?id=669522978 (2016).
- 32.Reiczigel J, Foldi J, Ozsvari L. Exact confidence limits for prevalence of a disease with imperfect diagnostic test. Epid. Inf. 2010;138:1674–1678. doi: 10.1017/S0950268810000385. [DOI] [PubMed] [Google Scholar]
- 33.Ongus JR, et al. Complete sequence of a picorna-like virus of the genus Iflavirus replicating in the mite Varroa destructor. J. Gen. Virol. 2004;85:3747–3755. doi: 10.1099/vir.0.80470-0. [DOI] [PubMed] [Google Scholar]
- 34.Mordecai GJ, Wilfert L, Martin SJ, Jones IM, Schroeder DC. Diversity in a honey bee pathogen: first report of a third master variant of the Deformed Wing Virus quasispecies. ISME J. 2015;10:1264–1273. doi: 10.1038/ismej.2015.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ai HX, Yan X, Han RC. Occurrence and prevalence of seven bee viruses in Apis mellifera and Apis cerana apiaries in China. J. Invertebr. Pathol. 2012;109:160–164. doi: 10.1016/j.jip.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 36.Choe SE, et al. Prevalence and distribution of six bee viruses in Korean Apis cerana populations. J. Invertebr. Pathol. 2012;109:330–333. doi: 10.1016/j.jip.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 37.Ding G, et al. Prevalence of honeybee viruses in different regions of China and Argentina. Rev. Sci. Tech. Off. Int. Epi. 2016;35:825–833. doi: 10.20506/rst.35.3.2572. [DOI] [PubMed] [Google Scholar]
- 38.Forsgren E, et al. Preliminary observations on possible pathogen spill-over from Apis mellifera to Apis cerana. Apidologie. 2015;46:265–275. doi: 10.1007/s13592-014-0320-3. [DOI] [Google Scholar]
- 39.Kojima Y, et al. Infestation of Japanese native honey bees by tracheal mite and virus from non-native European honey bees in Japan. Microb. Ecol. 2011;62:895–906. doi: 10.1007/s00248-011-9947-z. [DOI] [PubMed] [Google Scholar]
- 40.Li JL, et al. The prevalence of parasites and pathogens in Asian honeybees Apis cerana in China. PLoS ONE. 2012;7:e47955. doi: 10.1371/journal.pone.0047955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thu HT, et al. Prevalence of bee viruses among Apis cerana populations in Vietnam. J. Api. Res. 2016;55:379–385. doi: 10.1080/00218839.2016.1251193. [DOI] [Google Scholar]
- 42.Yañez O, et al. Potential for virus transfer between the honey bees Apis mellifera and A. cerana. J. Api. Res. 2015;54:179–191. doi: 10.1080/00218839.2015.1128145. [DOI] [Google Scholar]
- 43.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 45.Hudson RR, Boos DD, Kaplan NL. A statistical test for detecting geographic subdivision. Mol. Biol. Evol. 1992;9:138–151. doi: 10.1093/oxfordjournals.molbev.a040703. [DOI] [PubMed] [Google Scholar]
- 46.Yang B, Peng G, Li T, Kadowaki T. Molecular and phylogenetic characterization of honey bee viruses, Nosema microsporidia, protozoan parasites, and parasitic mites in China. Ecol. Evol. 2013;3:298–311. doi: 10.1002/ece3.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Webster CL, et al. The discovery, distribution, and evolution of viruses associated with Drosophila melanogaster. PLoS. Biol. 2015;13:e1002210. doi: 10.1371/journal.pbio.1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tentcheva D, et al. Prevalence and seasonal variations of six bee viruses in Apis mellifera L. and Varroa destructor mite populations in France. Appl. Env. Mirobiol. 2004;70:7185–7191. doi: 10.1128/AEM.70.12.7185-7191.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Francis RM, Nielsen SL, Kryger P. Varroa-Virus interaction in collapsing honey bee colonies. PLoS One. 2013;8:e57540. doi: 10.1371/journal.pone.0057540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Runckel C, et al. Temporal analysis of the honey bee microbiome reveals four novel viruses and seasonal prevalence of known viruses, Nosema, and Crithidia. PLoS ONE. 2011;6:e20656. doi: 10.1371/journal.pone.0020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeGrandi-Hoffman G, et al. Population growth of Varroa destructor (Acari: Varroidae) in honey bee colonies is affected by the number of foragers with mites. Exp. Appl. Acar. 2016;69:21–34. doi: 10.1007/s10493-016-0022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Di Pasquale G, et al. Variations in the availability of pollen resources affect honey bee health. PLoS ONE. 2016;11:e0162818. doi: 10.1371/journal.pone.0072016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smart M, Pettis J, Rice N, Browning Z, Spivak M. Linking measures of colony and individual honey bee health to survival among apiaries exposed to varying agricultural land use. PLoS ONE. 2016;11:e0152685. doi: 10.1371/journal.pone.0152685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boyle NK, Sheppard WS. A scientific note on seasonal levels of pesticide residues in honey bee worker tissues. Apidologie. 2017;48:128–130. doi: 10.1007/s13592-016-0455-5. [DOI] [Google Scholar]
- 55.Di Prisco G, et al. Neonicotinoid clothianidin adversely affects insect immunity and promotes replication of a viral pathogen in honey bees. PNAS. 2013;110:18466–18471. doi: 10.1073/pnas.1314923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Warrit, N. & Lekprayoon, C. Asian Honeybee Mites in Honeybees of Asia (eds Hepburn, H. R. & Radloff, S. E.) 347–368 (Springer Verlag, 2011).
- 57.Zioni N, Soroker V, Chejanovsky N. Replication of Varroa Destructor Virus 1 (VDV-1) and a Varroa Destructor Virus 1-Deformed Wing Virus recombinant (VDV-1-DWV) in the head of the honey bee. Virology. 2011;417:106–112. doi: 10.1016/j.virol.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 58.Moore J, et al. Recombinants between Deformed Wing Virus and Varroa Destructor Virus-1 may prevail in Varroa destructor-infested honeybee colonies. J. Gen. Virol. 2011;92:156–161. doi: 10.1099/vir.0.025965-0. [DOI] [PubMed] [Google Scholar]
- 59.Wang H, et al. Sequence recombination and conservation of Varroa Destructor Virus-1 and Deformed Wing Virus in field collected honey bees (Apis mellifera) PLoS ONE. 2013;8:e74508. doi: 10.1371/journal.pone.0074508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Manley, R. Emerging viral diseases of pollinating insects, University of Exeter (2017).
- 61.Manley R, et al. Knock-on community impacts of a novel vector: spillover of emerging DWV-B from Varroa-infested honeybees to wild bumblebees. Ecol. Lett. 2019;22:1306–1315. doi: 10.1111/ele.13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McMahon DP, et al. Elevated virulence of an emerging viral genotype as a driver of honeybee loss. Proc. Roy. Soc. B. 2016;283:20160811. doi: 10.1098/rspb.2016.0811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Teichroew JL, et al. Is China’s unparalleled and understudied bee diversity at risk? Biol. Cons. 2017;210:19–28. doi: 10.1016/j.biocon.2016.05.023. [DOI] [Google Scholar]
- 64.Ryabov EV, et al. Recent spread of Varroa destructor virus-1, a honey bee pathogen, in the United States. Sci. Rep. 2017;7:17447. doi: 10.1038/s41598-017-17802-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hou C, Li B, Deng S, Chu Y, Diao Q. Diagnosis and distribution of the Apis mellifera filamentous virus (AmFV) in honey bees (Apis mellifera) in China. Ins. Soc. 2017;64:597–603. doi: 10.1007/s00040-017-0569-4. [DOI] [Google Scholar]
- 66.Hou C, Li B, Luo Y, Deng S, Diao Q. First detection of Apis mellifera filamentous virus in Apis cerana cerana in China. J. Invertebr. Pathol. 2016;138:112–115. doi: 10.1016/j.jip.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 67.Diao QY, et al. Enhancement of chronic bee paralysis virus levels in honeybees acute exposed to imidacloprid: A Chinese case study. Sci. Tot. Env. 2018;630:487–494. doi: 10.1016/j.scitotenv.2018.02.258. [DOI] [PubMed] [Google Scholar]
- 68.McMahon DP, et al. A sting in the spit: widespread cross-infection of multiple RNA viruses across wild and managed bees. J. Anim. Ecol. 2015;84:615–624. doi: 10.1111/1365-2656.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stevenson, M. et al. epiR: Tools for the Analysis of Epidemiological Data, https://CRAN.R-project.org/package=epiR (2019).
- 70.Blaker H. Confidence curves and improved exact confidence intervals for discrete distributions. Can. J. Statistics. 2000;28:783–798. doi: 10.2307/3315916. [DOI] [Google Scholar]
- 71.R Core Team. R: A Language and Environment for Statistical Computing. http://www.R-project.org (2016).
- 72.Bates, D., Mächler, M., Bolker, B. & Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Software67; 10.18637/jss.v067.i01 (2015).
- 73.Hothorn T, Bretz F, Westfall P. Simultaneous Inference in General Parametric Models. Biometrical J. 2008;50:346–363. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- 74.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 75.Padidam M, Sawyer S, Fauquet CM. Possible emergence of new Geminiviruses by frequent recombination. Virology. 1999;265:218–225. doi: 10.1006/viro.1999.0056. [DOI] [PubMed] [Google Scholar]
- 76.Maynard Smith J. Analyzing the mosaic structure of genes. J. Mol. Evol. 1992;34:126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- 77.Martin DP, Posada D, Crandall KA, Williamson C. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res. Hum. Retrovir. 2005;21:98–102. doi: 10.1089/aid.2005.21.98. [DOI] [PubMed] [Google Scholar]
- 78.Gibbs MJ, Armstrong JS, Gibbs AJ. Sister-Scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics. 2000;16:573–582. doi: 10.1093/bioinformatics/16.7.573. [DOI] [PubMed] [Google Scholar]
- 79.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 81.Bielejec F, Rambaut A, Suchard MA, Lemey P. SPREAD: spatial phylogenetic reconstruction of evolutionary dynamics. Bioinformatics. 2011;27:2910–2912. doi: 10.1093/bioinformatics/btr481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jombart T, Ahmed I. adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics. 2011;27:3070–3071. doi: 10.1093/bioinformatics/btr521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Legendre, P. & Legendre, L. Numerical Ecology. Vol. 24 (Elsevier Science, 1998).
- 84.Oksanen, J. et al. vegan: Community Ecology Package. R package version 2.4-5, https://CRAN.R-project.org/package=vegan (2017).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data have been deposited in GenBank under accession numbers MF431915-431945 (lp-fragment), MF144195-144203, MF351971-351972 and MF431886-431914 (vp3-fragment) and MF667713-667747 (rdrp-fragment).