Abstract
Freshwater mussels are sensitive to habitat and water quality, revealing the fastest rates of human-mediated global extinction among aquatic animals. These animals are especially diverse in tropical Asia, the faunas of which are characterized by high levels of endemism. Here we describe four new species and four new subspecies of freshwater mussels from Myanmar. Leoparreysia whitteni sp. nov., the smallest representative of this genus, was discovered from the Ayeyarwady and Chindwin rivers. Radiatula myitthanensis sp. nov. and R. chindwinensis sp. nov. were recorded from the Chindwin Basin, and R. mouhoti haungthayawensis ssp. nov. has been discovered from the Haungthayaw River. Indochinella pugio has been revised with a description of three subspecies: I. pugio viridissima ssp. nov. from the Sittaung, Bilin and Bago rivers, I. pugio daweiensis ssp. nov. from the Dawei River, and I. pugio paradoxa ssp. nov. from the Haungthayaw River. Yaukthwa elongatula sp. nov., a peculiar species, conchologically resembling representatives of the genus Solenaia (Gonideinae) with ultra-elongated shell was found in the Chindwin Basin. Our records highlight that tropical Asia harbors numerous, but still overlooked local endemic lineages of freshwater bivalves, which may be on the brink of extinction due to the high anthropogenic and climate change impacts.
Subject terms: Zoology, Biogeography, Taxonomy
Introduction
Freshwater bivalves contribute to or provide a plethora of ecosystem functions and services1. Currently, freshwater bivalves are among the most threatened groups in the world with 40% of the species being near threatened, threatened or extinct2. The interior basin of the USA, Central America, Yangtze Basin, India and Southeast Asia are the most species-rich hotspots of freshwater bivalves at the global scale2–4.
Myanmar differs from other tropical Asian countries by a spectacular freshwater fauna with numerous local endemic taxa at the genus and species levels5–10. Biogeographically, most of country’s river basins, e.g. the Ayeyarwady, Bago, Sittaung, and Salween, belongs to the Western Indochina Subregion7. However, northwestern drainages of the Rakhine Coast, e.g. the Kaladan and Lemro rivers, seem to belong to the Indian Subregion, although the mussel fauna of those basins is still to be explored in detail11. Finally, the most eastern edge of the Shan State belongs to the Mekong River basin, and, hence, to the Sundaland Subregion7.
Bolotov et al.6 published the first integrative revision of the freshwater mussel fauna of Myanmar, with a description of two new genera, seven new species and one new subspecies. Two additional new endemic genera, Indochinella and Yaukthwa, have recently been introduced7,9. However, our current knowledge of freshwater mussels of Myanmar is far from being complete, with many lineages still waiting to be described. Further taxonomic research is necessary to develop a national conservation action plan for freshwater bivalves for Myanmar. This plan is urgently needed due to the current high rates of economic development and cropland expansion, leading to the rapid degradation of freshwater habitats12.
This study aims to describe four new species and four new subspecies of freshwater mussels from Myanmar. We introduce the new taxa on the basis of an integrative approach combining morphological, molecular and biogeographic evidences. This approach seems to be the most appropriate tool to uncover the diversity of freshwater mollusks in species-rich areas such as the Oriental Region6–9,13,14.
Results
Phylogeny
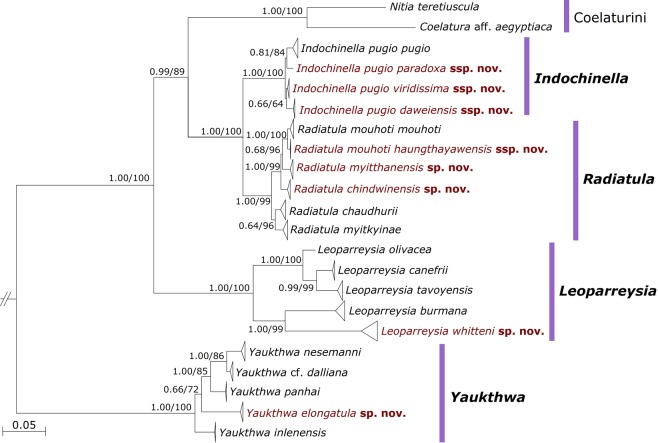
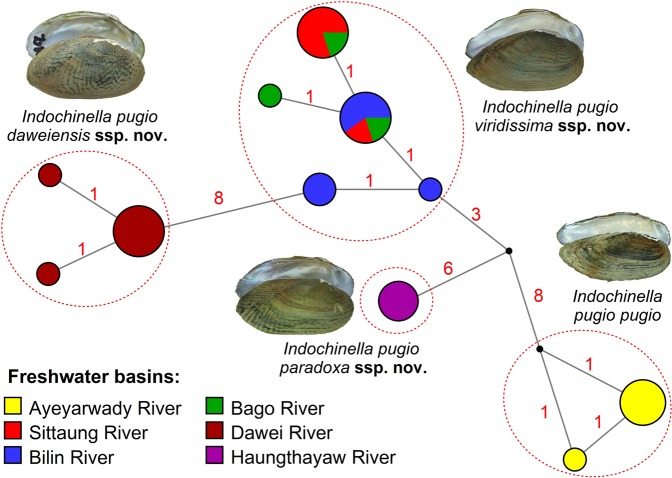
Searching with IQ-TREE and MrBayes based on a multi-locus molecular data set with the mitochondrial cytochrome c oxidase subunit I (COI), small ribosomal RNA (16S rRNA), and the nuclear large ribosomal RNA (28S rRNA) gene fragments returned well-resolved phylogenetic models of similar topology (Fig. 1). The four genera under discussion, i.e. Leoparreysia, Indochinella, Radiatula, and Yaukthwa, were recorded as robust, fully supported clades (BS ≥ 99%; BPP = 1.00). To check the relationship between closely related taxa within the genus Indochinella, we calculated a median-joining network that shows four distant haplogroups divided by corresponding river basins (Fig. 2).
Figure 1.
Bayesian phylogeny of the complete data set of mitochondrial and nuclear sequences (five partitions: three codons of COI + 16S rRNA + 28S rRNA) of the Unionidae genera under discussion: Leoparreysia, Radiatula, Indochinella, and Yaukthwa. Scale bar indicates the branch lengths. Black numbers near nodes are Bayesian posterior probabilities (BPP) of MrBayes v. 3.2.6/Ultrafast bootstrap support (BS) values of IQ-TREE. The names of novel taxa are in red. Outgroup is not shown.
Figure 2.
Median-joining haplotype network of the Indochinella subspecies based on the COI sequences (N = 29). The circle size is proportional to the number of available sequences belonging to a certain haplotype (smallest circle = one sequence). The small black dots represent hypothetical ancestral haplotypes. Red numbers near branches indicate the number of nucleotide substitutions between haplotypes. (Photos: Ekaterina S. Konopleva).
Species delimitation
The species delimitation analysis through the BEAST2 package STACEY with an initial grouping of taxa supported our hypothesis on 20 putative species-level lineages (probability = 0.80). Eight of these taxa are new to science and are described here (Tables 1–2; Figs 1–6). Each new taxon can be clearly distinguished from sister species or subspecies by molecular diagnosis (Table 2). Four lineages are assigned to valid species, i.e. Leoparreysia whitteni sp. nov., Radiatula myitthanensis sp. nov., R. chindwinensis sp. nov., and Yaukthwa elongatula sp. nov., because they share high levels of molecular divergence from sister clades (mean uncorrected COI p-distance = 3.2–9.3%). In contrast, four novel lineages sharing rather low molecular divergence from nearest neighbors (mean uncorrected COI p-distance = 1.4–1.6%) are introduced as the following subspecies: Indochinella pugio viridissima ssp. nov., I. pugio daweiensis ssp. nov., I. pugio paradoxa ssp. nov., and Radiatula mouhoti haungthayawensis ssp. nov. However, Indochinella pugio daweiensis ssp. nov. and I. pugio paradoxa ssp. nov. have one or two diagnostic nucleotide substitutions in the nuclear 28S rRNA gene fragment (Table 2) that usually indicates species-level differences between the Unionidae taxa.
Table 1.
Shell measurements and reference DNA sequences for the type series of new freshwater mussel taxa (Unionidae) from Myanmar.
| Taxon | Status of Specimen | Specimen Voucher* | Shell Length, mm | Shell Height, mm | Shell Width, mm | NCBI’s GenBank acc. nos. | ||
|---|---|---|---|---|---|---|---|---|
| COI | 16S rRNA | 28S rRNA | ||||||
| Leoparreysia whitteni sp. nov. | Holotype | biv449 | 28.7 | 19.5 | 12.2 | MK372449 | MK372476 | MK372508 |
| Paratype | biv349 | 27.9 | 17.6 | 10.3 | MK372411 | MK372459 | MK372489 | |
| Paratype | biv435 | 26.7 | 18.0 | 12.4 | MK372446 | MK372475 | MK372507 | |
| Radiatula myitthanensis sp. nov. | Holotype | biv337_3 | 41.0 | 21.7 | 15.5 | MK372396 | MK372450 | MK372477 |
| Paratype | biv348_1 | 42.4 | 22.1 | 15.5 | MK372409 | MK372457 | MK372487 | |
| Paratype | biv337_4 | 42.0 | 22.2 | 17.2 | n/a | n/a | n/a | |
| Paratype | biv337_8 | 35.5 | 19.4 | 13.8 | n/a | n/a | n/a | |
| Paratype | biv337_11 | 39.6 | 21.7 | 15.0 | n/a | n/a | n/a | |
| Paratype | biv337_17 | 42.9 | 22.4 | 16.0 | n/a | n/a | n/a | |
| Paratype | biv337_13 | 45.1 | 23.7 | 16.6 | n/a | n/a | n/a | |
| Paratype | biv337_20 | 38.6 | 21.3 | 14.5 | n/a | n/a | n/a | |
| Paratype | biv337_10 | 39.1 | 21.4 | 14.2 | n/a | n/a | n/a | |
| Paratype | biv337_12 | 37.6 | 21.7 | 14.8 | n/a | n/a | n/a | |
| Paratype | biv337_15 | 41.7 | 23.1 | 17.0 | n/a | n/a | n/a | |
| Radiatula mouhoti haungthayawensis ssp. nov. | Holotype | biv360_1 | 39.4 | 22.5 | 17.8 | MK372417 | MK372463 | MK372493 |
| Paratype | biv360_2 | 33.7 | 18.8 | 14.0 | MK372418 | MK372464 | MK372494 | |
| Paratype | biv360_3 | 31.3 | 18.0 | 14.2 | MK372419 | n/a | MK372495 | |
| Paratype | biv360_4 | 31.5 | 17.6 | 14.4 | n/a | n/a | n/a | |
| Paratype | biv360_5 | 30.2 | 17.4 | 12.5 | n/a | n/a | n/a | |
| Paratype | biv360_7 | 25.3 | 15.2 | 11.1 | n/a | n/a | n/a | |
| Paratype | biv360_8 | 26.4 | 15.3 | 10.7 | n/a | n/a | n/a | |
| Radiatula chindwinensis sp. nov. | Holotype | biv357_3 | 34.3 | 18.5 | 13.5 | MK372416 | n/a | n/a |
| Paratype | biv345_2 | 29.5 | 16.5 | 10.2 | MK372405 | MK372453 | MK372483 | |
| Paratype | biv348_2 | 34.5 | 19.5 | 13.8 | MK372410 | MK372458 | MK372488 | |
| Paratype | biv345_1 | 32.1 | 17.8 | 11.8 | MK372404 | MK372452 | MK372482 | |
| Paratype | biv357_2 | 33.8 | 18.6 | 15.0 | MK372415 | n/a | n/a | |
| Paratype | biv357_1 | 34.4 | 19.3 | 13.3 | MK372414 | MK372462 | MK372492 | |
| Paratype | biv357_4 | 28.7 | 16.7 | 13.1 | n/a | n/a | n/a | |
| Indochinella pugio viridissima ssp. nov. | Holotype | biv_251_3 | 34.8 | 18.6 | 14.1 | MF352244 | MF352314 | MF352372 |
| Paratype | biv_251_1 | 32.3 | 16.3 | 11.6 | MF352242 | MF352312 | MF352370 | |
| Paratype | biv_251_2 | 32.7 | 17.0 | 11.9 | MF352243 | MF352313 | MF352371 | |
| Paratype | biv371_1 | 44.3 | 21.3 | 16.1 | MK372426 | MK372468 | MK372500 | |
| Paratype | biv371_2 | 42.4 | 20.6 | 15.8 | MK372427 | MK372469 | MK372501 | |
| Paratype | biv371_3 | 44.8 | 21.2 | 14.9 | MK372428 | n/a | n/a | |
| Paratype | biv375_1 | 34.8 | 18.0 | 12.5 | MK372429 | n/a | n/a | |
| Paratype | biv375_2 | 32.2 | 16.3 | 11.5 | MK372430 | n/a | n/a | |
| Paratype | biv375_3 | 33.0 | 16.8 | 12.2 | MK372431 | n/a | n/a | |
| Paratype | biv377_2 | 35.4 | 17.9 | 13.0 | MK372432 | n/a | n/a | |
| Paratype | biv377_3 | 37.4 | 19.3 | 13.5 | MK372433 | n/a | n/a | |
| Paratype | biv381_3 | 45.8 | 23.8 | 16.9 | MK372434 | n/a | n/a | |
| Paratype | biv381_4 | 36.4 | 18.7 | 12.8 | MK372435 | n/a | n/a | |
| Paratype | biv386_2 | 37.0 | 20.1 | 14.1 | MK372436 | n/a | n/a | |
| Indochinella pugio daweiensis ssp. nov. | Holotype | biv_147_3 | 31.0 | 16.6 | 12.6 | KX865852 | KX865623 | KX865724 |
| Paratype | biv_147_10 | 27.9 | 14.8 | 12.2 | KX865853 | KX865624 | KX865725 | |
| Paratype | biv_147_18 | 26.1 | 13.6 | 9.9 | KX865854 | KX865625 | KX865726 | |
| Paratype | biv_148_4 | 32.3 | 17.5 | 14.2 | KX865855 | KX865626 | KX865727 | |
| Paratype | biv_148_7 | 32.6 | 17.5 | 14.8 | KX865856 | KX865627 | KX865728 | |
| Paratype | biv_148_15 | 37.4 | 19.6 | 15.7 | KX865857 | KX865628 | KX865729 | |
| Paratype | biv_147_30 | 30.4 | 16.4 | 12.2 | MK372395 | n/a | n/a | |
| Paratype | biv_147_1 | 32.0 | 16.6 | 13.3 | n/a | n/a | n/a | |
| Paratype | biv_147_2 | 30.3 | 16.3 | 11.9 | n/a | n/a | n/a | |
| Paratype | biv_147_4 | 31.8 | 17.4 | 12.0 | n/a | n/a | n/a | |
| Paratype | biv_148_2 | 36.9 | 20.4 | 14.9 | n/a | n/a | n/a | |
| Paratype | biv_148_3 | 38.0 | 20.0 | 15.5 | n/a | n/a | n/a | |
| Indochinella pugio paradoxa ssp. nov. | Holotype | biv361_1 | 36.1 | 17.3 | 12.5 | MK372420 | MK372465 | MK372496 |
| Paratype | biv361_2 | 35.6 | 18.0 | 13.1 | MK372421 | MK372466 | MK372497 | |
| Paratype | biv361_3 | 28.6 | 17.7 | 10.6 | MK372422 | MK372467 | MK372498 | |
| Paratype | biv361_4 | 29.0 | 16.4 | 10.8 | n/a | n/a | n/a | |
| Paratype | biv361_5 | 27.1 | 15.2 | 10.4 | n/a | n/a | n/a | |
| Paratype | biv361_6 | 26.6 | 14.8 | 9.9 | n/a | n/a | n/a | |
| Paratype | biv361_7 | 28.2 | 15.1 | 10.0 | n/a | n/a | n/a | |
| Paratype | biv361_8 | 23.3 | 13.1 | 8.2 | n/a | n/a | n/a | |
| Paratype | biv361_9 | 24.2 | 12.3 | 8.3 | n/a | n/a | n/a | |
| Yaukthwa elongatula sp. nov. | Paratype | biv341_2 | 64.3 | 28.1 | 13.2 | MK372400 | MK372451 | MK372480 |
| Paratype | biv346_3 | 51.3 | 24.2 | 12.4 | MK372408 | MK372456 | MK372486 | |
| Paratype | biv346_1 | 55.6 | 25.3 | 12.6 | MK372406 | MK372454 | MK372484 | |
| Paratype | biv344_1 | 58.6 | 26.8 | 14.9 | MK372402 | n/a | n/a | |
| Paratype | biv344_2 | 52.2 | 23.5 | 12.0 | n/a | n/a | n/a | |
| Paratype | biv344_3 | 35.1 | 16.4 | 8.2 | MK372401 | n/a | MK372481 | |
| Holotype | biv346_2 | 48.2 | 23.7 | 12.1 | MK372406 | MK372454 | MK372484 | |
| Paratype | biv341_3 | 49.2 | 21.3 | 12.5 | MK372401 | n/a | MK372481 | |
| Paratype | biv339_1 | 59.4 | 26.3 | 14.8 | MK372397 | n/a | MK372478 | |
| Paratype | biv339_2 | 57.0 | 23.2 | 14.3 | MK372398 | n/a | MK372479 | |
| Paratype | biv339_3 | 58.7 | 25.1 | 14.3 | MK372399 | n/a | n/a | |
| Paratype | biv339_7 | 51.2 | 24.7 | 13.1 | n/a | n/a | n/a | |
*Type series are deposited in the RMBH – Russian Museum of Biodiversity Hotspots, Federal Center for Integrated Arctic Research, Russian Academy of Sciences, Arkhangelsk, Russia. n/a – not available.
Table 2.
Molecular diagnoses of the new freshwater mussel taxa (Unionidae) from Myanmar.
| Taxon | Mean COI p-distance from the nearest neighbor, % | The nearest neighbor of new taxon | Fixed nucleotide differences based on the sequence alignment of congeners | ||
|---|---|---|---|---|---|
| COI | 16S rRNA | 28S rRNA | |||
| Leoparreysia whitteni sp. nov. | 9.29 | L. olivacea | 15 C, 56 C, 62 G, 74 A, 77 G, 92 A, 101 G, 128 C, 182 G, 227 C, 296 A, 308 G, 323 G, 353 G, 356 T, 407 C, 458 G, 524 A, 539 G, 554 G, 656 A | 12 C, 18 T, 20 C, 49 A, 51 T, 110 T, 144 C, 200 C, 233 C, 234 T, 238 G, 241 C, 248 A, 250 A, 264 C, 300 C, 324 C, 329 C | 127 С, 413 T, 432 T/Y, 454 T, 469 C, 470 G, 471 A, 484 G, 494 T, 523 G, 524 A, 531 T, 542 C, 550 C, 559 G, 567 G, 568 A, 584 T, 601 G, 621 G, 622 T, 623 T, 631 T, 647 T, 654 G, 655 T, 682 T, 686 A, 702 T, 709 A, 710 A, 747 C, 752 T, 760 C, 758 A |
| Radiatula myitthanensis sp. nov. | 3.21 | R. mouhoti | 389 A, 422 G, 425 G, 434 A, 443 C, 527 C, 578 A | 240 G, 317 C, 331 A | None |
| R. mouhoti haungthayawensis ssp. nov. | 1.36 | R. mouhoti | 35 C, 254 G | None | None |
| R. chindwinensis sp. nov. | 3.33 | R. mouhoti | 122 A, 212 A, 293 G, 308 A, 364 G, 449 C, 479 A | 53 G | None |
| Indochinella pugio viridissima ssp. nov. | 1.61 | I. pugio paradoxa ssp. nov. | 57 C | None | None |
| I. pugio daweiensis ssp. nov. | 1.59 | I. pugio viridissima ssp. nov. | 86 G, 110 G, 347 A, 402 C, 557 C, 635 C, 644 A, 659 C | 17 A, 314 C, 316 T, 324 T | 759 A |
| I. pugio paradoxa ssp. nov. | 1.61 | I. pugio viridissima ssp. nov. | 8 G, 291 C, 341 T, 404 G, 443 C, 608 G | 328 G | 507 C, 771 G |
| Yaukthwa elongatula sp. nov. | 8.28 | Y. inlenensis | 6 C, 15 C, 17 A, 35 T, 50 C, 56 C, 71 G, 95 A, 167 A, 182 C, 194 C, 206 T, 242 G, 248 C, 275 A, 296 A, 302 T, 319 C, 338 C, 365 C, 389 A, 425 A, 443 C, 506 C, 539 G, 542 A, 587 G, 629 A, 638 T | 154 A, 155 C, 238 G, 247 G, 249 T, 253 A, 270 G, 336 C, 342 T, 469 C | 210 G, 212 T, 496 A, 589 Del, 608 G |
Del = deletion mutation.
Figure 6.
Type localities and habitats of new freshwater mussel taxa from Myanmar. (A) Leoparreysia whitteni sp. nov.: Ayeyarwady River, Su Taung Seik area, Sagaing, Mandalay [type locality]. (B) Radiatula myitthanensis sp. nov.: Myit Tha (Manipur) River [type locality]. (C) R. mouhoti haungthayawensis ssp. nov.: Haungthayaw River upstream of Kawkareik town [type locality]. (D) R. chindwinensis sp. nov.: tributary of Nay Chin Sayar River, Chindwin Basin [type locality]. (E) Indochinella pugio viridissima ssp. nov.: Myit Kyi Pauk Stream, Sittaung Basin [type locality]. (F) I. pugio daweiensis ssp. nov.: Dawei River [type locality]. (G) I. pugio paradoxa ssp. nov.: Haungthayaw River upstream of Kawkareik town [type locality]. (H) Yaukthwa elongatula sp. nov.: Myit Tha (Manipur) River [habitat]. (Photos: Nyein Chan [A,C,G] and Ilya V. Vikhrev [B,D–F,H]).
Taxonomic account
Family Unionidae Rafinesque, 1820
Subfamily Parreysiinae Henderson, 1935
Type genus: Parreysia Conrad, 1853 (by original designation)
Tribe Leoparreysiini Vikhrev, Bolotov & Kondakov, 2017
Type genus: Leoparreysia Vikhrev, Bolotov & Aksenova, 2017 (by original designation)
Genus Leoparreysia Vikhrev, Bolotov & Aksenova, 2017
Type species: Leoparreysia canefrii Vikhrev, Bolotov & Kondakov, 2017 (by original designation)
Leoparreysia whitteni sp. nov.
Figures 3, 4A, 6A, Tables 1–2.
Figure 3.
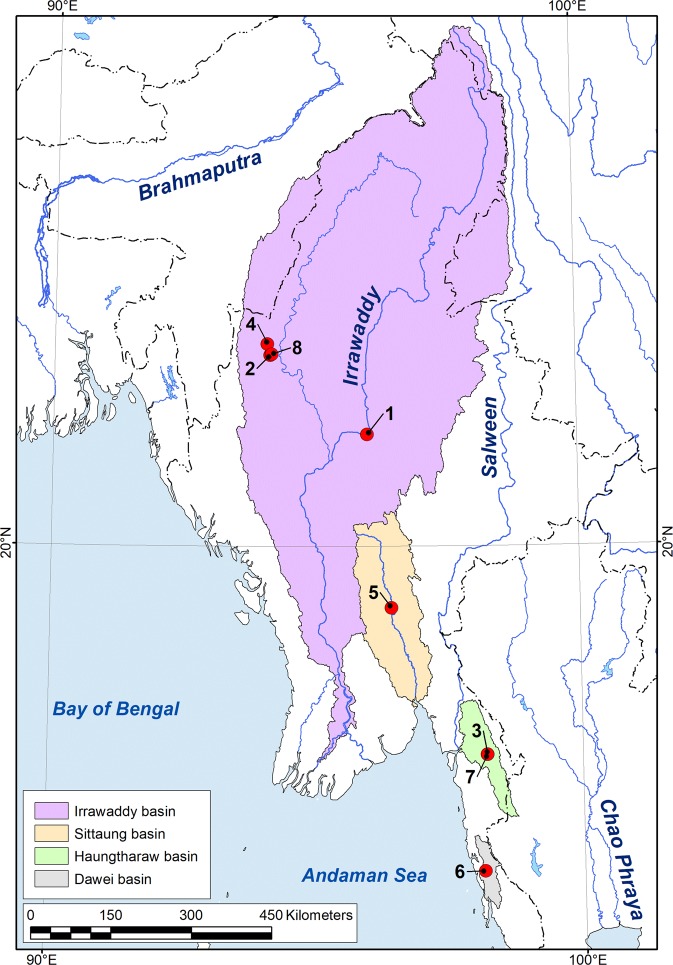
Map of the type localities of new freshwater mussel taxa from Myanmar. (1) Leoparreysia whitteni sp. nov.: Ayeyarwady River, Su Taung Seik area, Sagaing, Mandalay. (2) Radiatula myitthanensis sp. nov.: Myit Tha (Manipur) River. (3) R. mouhoti haungthayawensis ssp. nov.: Haungthayaw River upstream of Kawkareik town. (4) R. chindwinensis sp. nov.: tributary of Nay Chin Sayar River, Chindwin Basin. (5) Indochinella pugio viridissima ssp. nov.: Myit Kyi Pauk Stream, Sittaung Basin. (6) I. pugio daweiensis ssp. nov.: Dawei River. (7) I. pugio paradoxa ssp. nov.: Haungthayaw River upstream of Kawkareik town. (8) Yaukthwa elongatula sp. nov.: Chindwin River. The map was developed using ESRI ArcGIS 10 software (www.esri.com/arcgis). The topographic base of the map was compiled with Natural Earth Free Vector and Raster Map Data (www.naturalearthdata.com), GSHHG version 2.3.7 (www.soest.hawaii.edu/pwessel/gshhg)35, and the HydroSHEDS database (www.hydrosheds.org)36,37. (Map: Mikhail Yu. Gofarov).
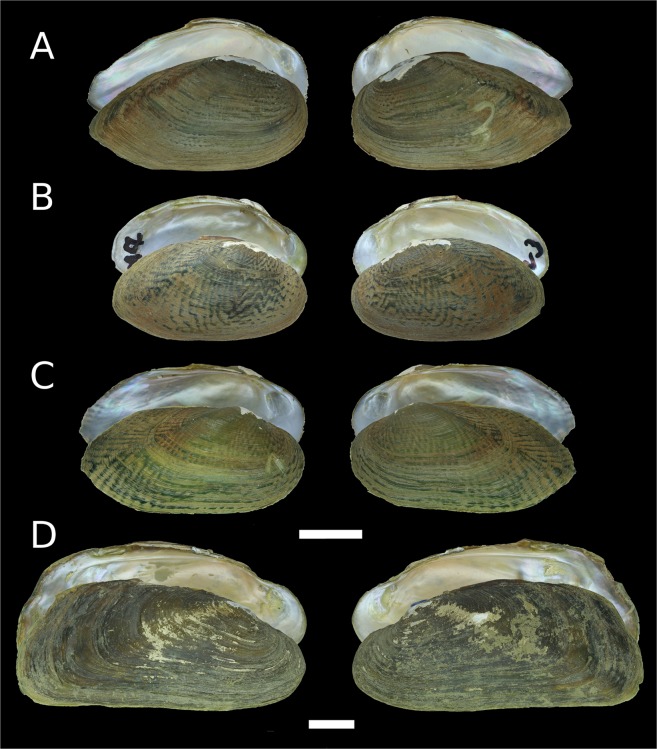
Figure 4.
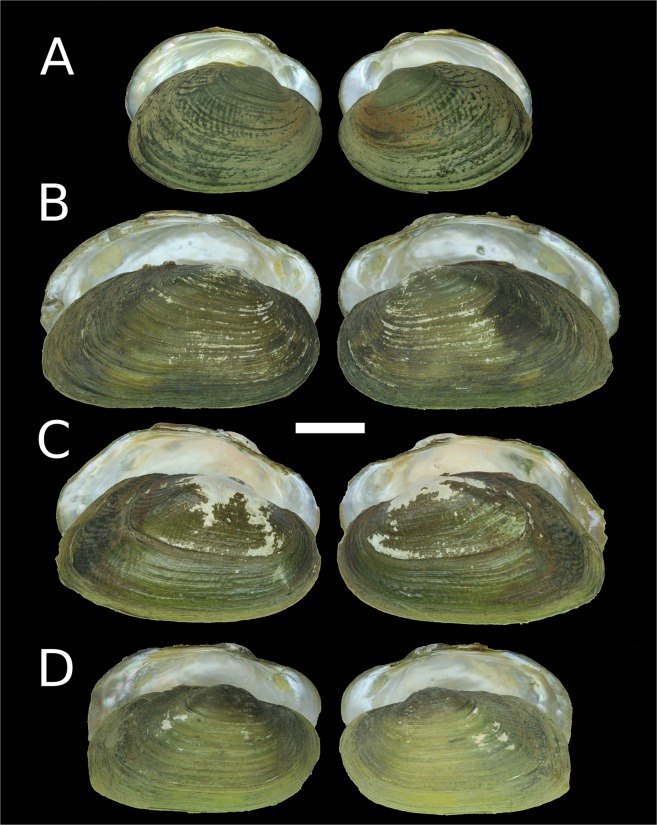
Shells of new freshwater mussel taxa from Myanmar. (A) Leoparreysia whitteni sp. nov. [holotype RMBH biv 449]. (B) Radiatula myitthanensis sp. nov. [holotype RMBH biv 337_3]. (C) R. mouhoti haungthayawensis ssp. nov. [holotype RMBH biv 360_1]. (D) R. chindwinensis sp. nov. [holotype RMBH biv357_3]. (Photos: Ekaterina S. Konopleva).
Type locality: Myanmar: Ayeyarwady River, Su Taung Seik area, Sagaing, Mandalay [21.8893°N, 95.9978°E].
Holotype: RMBH biv_449: Myanmar: Ayeyarwady River, Su Taung Seik area, Sagaing, Mandalay, 21.8893°N, 95.9978°E, 4.iii.2018, Bolotov, Vikhrev, Nyein Chan and local villagers leg.
Paratypes: Myanmar: Chindwin River, 23.1918°N, 94.3217°E, 2.ii.2018, 1 specimen (RMBH biv_349); Myanmar: Ayeyarwady River, Mandalay, 21.9574°N, 96.0510°E, 3.iii.2018, 1 specimen (RMBH biv_435), Bolotov, Vikhrev, Nyein Chan and local villagers leg.
Etymology: This species is named in memory of the late Dr. Tony Whitten (1953–2017), a famous conservation biologist, Fauna & Flora International – Asia Pacific, United Kingdom.
Diagnosis: The new species is morphologically most similar to Leoparreysia burmana (Blanford, 1869) but can be distinguished from it by a more elongated shell, and different location and size of umbo (small subinequilateral umbo in the new species vs. large strongly inequilateral umbo in L. burmana). Leoparreysia whitteni sp. nov. has a high genetic divergence from all other taxa in this genus by the COI, 16S rRNA and 28S rRNA gene fragments (Table 2).
Description: Very small mussel. Shell length 26.7–28.7 mm, height 17.6–19.5 mm, width 10.3–12.4 mm. Shell ovate, subinequilateral, slightly inflated and thick. Posterior margin broader than anterior margin, with arched bars along the slope. Strong v-shaped sculpture all over the shell surface (except for one specimen), but it is slightly visible or absent on umbo area. Periostracum yellow-green or brown-green, nacre whitish, somewhat shining. Pseudocardinal teeth strong and very indented, by two teeth in each valve, of typical Leoparreysia shape. Lateral teeth moderately short, curved, two teeth in left valve, one tooth in right valve. Anterior muscle attachment scar rounded, more or less deep. Posterior muscle attachment scar shallow.
Habitat and ecology: Silty-clay sites in large rivers (Fig. 6A).
Distribution: Ayeyarwady and Chindwin rivers, Ayeyarwady Basin, Myanmar.
Comments: There are only three available specimens of this species, each from a different locality. The specimen from the Chindwin River has smooth periostracum and no shell sculpture, only bars along the posterior margin.
Tribe Indochinellini Bolotov, Pfeiffer, Vikhrev & Konopleva, 2018
Type genus: Indochinella Bolotov, Pfeiffer, Vikhrev & Konopleva, 2018 (by original designation)
Genus Radiatula Simpson, 1900
Type species: Unio crispisulcatus Benson, 1862 (by original designation)
Radiatula myitthanensis sp. nov.
Figures 3, 4B, 6B, Tables 1–2.
Type locality: Myanmar: Myit Tha (Manipur) River [23.2285°N, 94.1434°E].
Holotype: RMBH biv337_3: Myanmar: Myit Tha (Manipur) River, 23.2285°N, 94.1434°E, 2.ii.2018, Bolotov, Vikhrev, Lopes-Lima, Nyein Chan and local villagers leg.
Paratypes: type locality, 2.ii.2018, 9 specimens (RMBH no. biv_337); Chindwin River, 23.1918°N, 94.3217°E, 2.ii.2018, 1 specimen (RMBH biv_348_1), Bolotov, Vikhrev, Lopes-Lima, Nyein Chan and local villagers leg.
Etymology: The name of this species is derived from its type locality, the Myit Tha River.
Diagnosis: This species is morphologically and genetically close to Radiatula mouhoti Vikhrev, Bolotov & Konopleva 2017, but differs from it by more elongated and rounded posterior margin, less developed posterior muscle scar and smaller umbo. Radiatula myitthanensis sp. nov. is also externally similar to R. chindwinensis sp. nov., but it differs from the latter species by a rounded posterior slope (vs. truncated posterior slope in R. chindwinensis sp. nov.) and curved ventral margin (vs. straighter ventral margin in R. chindwinensis sp. nov.). The new species also differs from all the congeners by fixed nucleotide substitutions in the COI and 16S rRNA gene fragments (Table 2).
Description: Shell length 35.5–45.1 mm, height 19.4–23.7 mm, width 13.8–17.2 mm. Shell ovate-elongated, subinequilateral, somewhat inflated and thick. Dorsal margin slightly curved, ventral margin straight. Anterior margin rounded; posterior slope smooth, margin covered by small wrinkles from the beak. The umbo area has poorly visible v-shaped sculpture, corrugated. Periostracum jade-green, concentrically striated; nacre yellow-whitish. Right valve with a single slightly curved lateral tooth and two pseudocardinal teeth, anterior tooth small and lamellar; posterior tooth rectangular and more or less indented. Left valve with two slightly curved lateral teeth and two pseudocardinal teeth, anterior tooth rectangular and ribbed, posterior tooth smaller and pyramidal. Anterior muscle attachment scar ovate, well pronounced. Posterior muscle attachment scar less well marked.
Habitat and ecology: Sites with clay substrate and large stones in large rivers (Fig. 6B).
Distribution: Myit Tha (Manipur) and Chindwin rivers, Ayeyarwady Basin, northwestern Myanmar.
Radiatula mouhoti haungthayawensis ssp. nov.
Figures 3, 4C, 6C, Tables 1–2.
Type locality: Myanmar: Haungthayaw River upstream of Kawkareik town [16.4714°N, 98.2182°E].
Holotype: RMBH biv360_1: Myanmar: Haungthayaw River upstream of Kawkareik town, 16.4714°N, 98.2183°E, 9.ii.2018, Nyein Chan leg.
Paratypes: type locality, 9.ii.2018, 6 specimens (RMBH nos. biv360_2, biv360_3, biv360_4, biv360_5, biv360_7, and biv360_8), Nyein Chan leg.
Etymology: The name of this subspecies refers to the Haungthayaw River, its type locality.
Diagnosis: The new subspecies is genetically close to Radiatula mouhoti Vikhrev, Bolotov & Konopleva, 2017, but is morphologically more similar to R. chaudhurii (Preston, 1912). Radiatula mouhoti haungthayawensis differs from the latter species by possessing a smoother periostracum, a stronger inflation, and a curved ventral margin. The new taxon also differs from the nominative subspecies by two diagnostic nucleotide substitutions in the COI gene fragment (Table 2).
Description: Small mussel. Shell length 26.4–39.4 mm, height 15.2–22.5 mm, width 10.7–17.8 mm. Shell elongated, thin, subinequilateral. Anterior end rounded, ventral and dorsal margin curved; posterior side truncated, covered by small wrinkles. The umbo corrugated, with w-shaped sculpture, continued along all over the shell surface. Periostracum olive-green with pinkish patches, concentrically striated; nacre blue-whitish with yellow umbo cavity. Right valve with a single slightly curved lateral tooth and two pseudocardinal teeth, anterior tooth reduced; posterior tooth pyramidal or more rectangular and can be slightly indented. Left valve with two slightly curved lateral teeth and two pseudocardinal teeth, anterior tooth rectangular, posterior tooth smaller and pyramidal. Anterior muscle attachment scar ovate and well-marked. Posterior muscle attachment scar rounded and less pronounced.
Habitat and ecology: This subspecies was collected from a single location, an upstream pool site of the river with sandy-clay substrate (Fig. 6C).
Distribution: Haungthayaw River, southeastern Myanmar.
Radiatula chindwinensis sp. nov.
Figures 3, 4D, 6D, Tables 1–2.
Type locality: Myanmar: tributary of Nay Chin Sayar River, Chindwin Basin [23.4160°N, 94.0875°E].
Holotype: RMBH biv 357_3: Myanmar: tributary of Nay Chin Sayar River, Chindwin Basin, 23.4160°N, 94.0875°E, 4.ii.2018, Bolotov, Vikhrev, Lopes-Lima, Nyein Chan and local villagers leg.
Paratypes: type locality, 4.ii.2018, 2 specimens (RMBH nos. biv357_1 and biv357_2); Myanmar: Myit Tha (Manipur) River, 23.2006°N, 94.2214°E, 2.ii.2018, 2 specimens (RMBH nos. biv345_1 and biv345_2); Myanmar: Chindwin River, 23.1918°N, 94.3217°E, 2.ii.2018, 1 specimen (RMBH biv348_2), Bolotov, Vikhrev, Lopes-Lima, Nyein Chan and local villagers leg.
Etymology: The new species is named after the Chindwin River, the largest tributary of the Ayeyarwady Basin, from which the species was collected.
Diagnosis: The new species is conchologically similar to Radiatula mouhoti and R. myitthanensis sp. nov., but can be distinguished from these taxa by more straight dorsal margin and smooth pseudocardinal teeth without marked sculpture (vs. more curved dorsal margin and strongly sculptured pseudocardinal teeth in R. mouhoti and R. myitthanensis sp. nov.). The new species also differs from all the congeners by fixed nucleotide substitutions in the COI and 16S rRNA gene fragments (Table 2).
Description: Small mussel. Shell length 28.7–34.5 mm, height 16.7–19.5 mm, width 10.2–15.0 mm. Shell elongate-ovate, rather thin, nearly equilateral in some specimens, not inflated. Anterior end rounded, slightly shifted upward at some specimens, ventral and dorsal margin straight or slightly curved; posterior side truncated, covered by small arched bars from umbo along the slope. Umbo corrugated, with unpronounced v-shaped sculpture, continued until the middle or ventral margin of the shell. Periostracum olive-green with yellow and dark regions, concentrically striated; nacre blue-whitish. Two lateral teeth on the left valve and a single tooth on the right valve, straight or slightly curved. Right valve with two pseudocardinal teeth, anterior tooth reduced; posterior tooth pyramidal or trapezoidal. Left valve with two pseudocardinal teeth, anterior tooth pyramidal or rectangular, posterior tooth smaller and pyramidal. Anterior muscle attachment scar ovate, rather deep. Posterior muscle attachment scar rounded and shallow.
Habitat and ecology: Clay sites in large rivers (Fig. 6D).
Distribution: Nay Chin Sayar, Myit Tha (Manipur) and Chindwin rivers, Ayeyarwady Basin, northwestern Myanmar.
Genus Indochinella Bolotov, Pfeiffer, Vikhrev & Konopleva, 2018
Type species: Unio pugio Benson, 1862 (by original designation)
Indochinella pugio viridissima ssp. nov.
= Oxynaia sp. ‘Taungoo’ sensu Bolotov et al. (2017): 106.
Figures 3, 5A, 6E, Tables 1–2.
Figure 5.
Shells of new freshwater mussel taxa from Myanmar. (A) Indochinella pugio viridissima ssp. nov. [holotype RMBH biv 251_3]. (B) I. pugio daweiensis ssp. nov. [holotype RMBH biv 147_3]. (C) I. pugio paradoxa ssp. nov. [holotype RMBH biv 361_1]. (D) Yaukthwa elongatula sp. nov. [paratype RMBH biv 341_2]. (Photos: Ekaterina S. Konopleva).
Type locality: Myanmar: Myit Kyi Pauk Stream, Sittaung Basin [18.9613°N, 96.4455°E].
Holotype RMBH biv_251_3: Myanmar: Myit Kyi Pauk Stream, Sittaung Basin, 18.9613°N, 96.4455°E, 26 November 2016, Vikhrev leg.
Paratypes: type locality, 26.xi.2016, 2 specimens (RMBH nos. biv_251_1 and biv_251_2), Vikhrev leg.; Myanmar: Bilin River, near Yhin Ohn village, 17.3306°N, 97.2418°E, 13.ii.2018, 3 specimens (RMBH nos. biv371_1, biv371_2, and biv_371_3); Myanmar: Thae Phyu Stream, Bilin River, 17.2757°N, 97.1274°E, 14.ii.2018, 3 specimens (RMBH nos. biv375_1, biv375_2, and biv375_3); Myanmar: Moeyungyi Lake, 17.5968°N, 96.5950°E, 2 specimens (RMBH biv biv377_2 and biv377_3), 17.ii.2018; Myanmar: Bago River, 17.5334°N, 96.3315°E, 18.ii.2018, 2 specimens (RMBH nos. biv381_3 and biv381_4); Myanmar: Moe Lut Stream, Bago Basin, 17.6011°N, 96.2861°E, 18.ii.2018, 1 specimen (RMBH biv386_2), Bolotov, Vikhrev, Nyein Chan and local villagers leg.
Etymology: The name of this subspecies refers to its olive-green periostracum.
Diagnosis: This subspecies is conchologically similar to Indochinella pugio pugio (Benson, 1862), but differs from it by shorter and higher shell, more pronounced and curved lateral teeth, moderately strong sculpture on shell disc, and a diagnostic nucleotide substitution in the COI gene fragment (Table 2).
Description: Small mussel. Shell length 32.2–45.8 mm, height 16.3–23.8, width 11.5–16.9 mm. Shell cuneiform, inequilateral, rather inflated. Posterior ridge oblique. Shell sculpture moderately strong. Periostracum smooth, olive-green, nacre whitish. Umbo not very prominent, corrugated, beak sculpture little pronounced. Left valve with two short lateral teeth and two ribbed pseudocardinal teeth. Right valve with single short curved lateral tooth and two pseudocardinal teeth, anterior tooth not pronounced, posterior tooth strong, ribbed and triangular. Umbo cavity rather deep. Anterior adductor scar well pronounced, funneled; posterior adductor scar shallow.
Habitat and ecology: Sites with clay bottom substrate in various rivers, streams and lakes (Fig. 6E).
Distribution: Sittaung, Bilin, and Bago River basins, Myanmar.
Indochinella pugio daweiensis ssp. nov.
= Scabies crispata sensu Bolotov et al. (2017): 6, Fig. 415.
= Oxynaia sp. ‘Tavoy’ sensu Bolotov et al. (2017): 106.
Figures 3, 5B, 6F, Tables 1–2.
Type locality: Myanmar: Dawei (Tavoy) River [14.5012°N, 98.1557°E].
Holotype RMBH biv_147_3: Myanmar: Dawei River, 14.5012°N, 98.1557°E, 26.iv.2015, Bolotov leg.
Paratypes: type locality, 26.iv.2015, 11 specimens (RMBH nos. biv147_10, biv147_18, biv148_4, biv148_7, biv148_15, biv147_30, biv147_1, biv147_2, biv147_4, biv148_2, and biv148_3), Bolotov leg.
Etymology: This local subspecies is named after Dawei River, in which the type series was collected.
Diagnosis: This taxon differs from the nominative subspecies in having oval-shaped shell, a more rounded posterior ridge and a more gradual posterior slope, distinct zigzag ridges across the shell disc (Fig. 5B), and fixed nucleotide substitutions in the COI, 16S rRNA and 28S rRNA gene fragments (Table 2). Perhaps, it is the most conchologically peculiar taxon within the genus.
Description: Very small mussel. Shell length 26.1–28.0 mm, height 11.5–20.4 mm, width 8.2–16.8 mm. Shell shape obovate, inequilateral, rather thick. Posterior ridge broader than the anterior ridge, oblique. Shell sculpture very strong, with distinct zigzag ridges across the shell disc. Periostracum sandy-brown with numerous dark-green zigzag ridges, nacre yellow-whitish. Umbo prominent, slightly corrugated, beak sculpture not very strong. Left valve with two short lateral teeth and two pseudocardinal teeth. Right valve with a single lateral tooth and blunt pseudocardinal tooth, anterior tooth not developed. Umbo cavity shallow and open. Anterior adductor scar marked, rounded or oval-shaped. Posterior adductor scar not pronounced, obovate.
Habitat and ecology: The subspecies is known only from its type series, which was collected from river sites with clay and gravel bottom substrate (Fig. 6F).
Distribution: Dawei River, southeastern Myanmar.
Indochinella pugio paradoxa ssp. nov.
Figures 3, 5C, 6G, Tables 1–2.
Type locality: Myanmar: Haungthayaw River upstream of Kawkareik town [16.4714°N, 98.2182°E].
Holotype: RMBH biv361_1: Myanmar: Haungthayaw River upstream of Kawkareik town, 16.47144°N, 98.21825°E, 9.ii.2018, Nyein Chan leg.
Paratypes: type locality, 9.ii.2018, 8 specimens (RMBH nos. biv361_2, biv361_3, biv361_4, biv361_5, biv361_6, biv361_7, biv361_8, and biv361_9), Nyein Chan leg.
Etymology: The name of this subspecies refers to the paradox that it has morphological features similar to the Scabies group but is closely related genetically with Indochinella pugio group.
Diagnosis: The subspecies is very similar morphologically to representatives of the genus Scabies Haas, 1911, e.g. S. crispata (Gould, 1843), but it genetically sisters to the Indochinella clade. The new taxon can be distinguished from other Indochinella pugio subspecies by strong radial and w-shaped sculpture of the shell, biangular shape of the posterior end, and by fixed nucleotide substitutions in the COI, 16S rRNA and 28S rRNA gene fragments (Table 2).
Description: Small mussel. Shell length 22.3–36.1 mm, height 12.3–18.0 mm, width 8.2–13.1 mm. Shell somewhat cuneiform, inequilateral, rather thin, slightly inflated. Anterior end rounded, dorsal and ventral margins almost straight or slightly curved, parallel to each other at some specimens. Posterior end biangular, from umbo to upper angle covered by ridges. Umbo slightly elevated with w-shaped sculpture; umbo area well-separated. Periostracum green, concentrically striated, laminiferous, with dark-green zigzag ridges in lower half of the shell; nacre white-yellow. Lateral teeth lamellar, thin, straight or slightly curved, a single tooth on right valve and two teeth on left valve. Two pseudocardinal teeth on right valve, anterior tooth lamellar, posterior tooth trapezoidal; two pseudocardinal teeth on left valve, anterior tooth rectangular and ribbed, anterior tooth small and pyramidal. Anterior muscle attachment scar rounded, well-visible. Posterior muscle attachment scar shallow.
Habitat and ecology: This species is known from the same locality as R. mouhoti haungthayawensis ssp. nov. (Fig. 6G).
Distribution: Haungthayaw River, southeastern Myanmar.
Subfamily Rectidentinae Modell, 1942
Type genus: Rectidens Simpson, 1900 (by original designation)
Tribe Contradentini Modell, 1942
Type genus: Contradens Haas, 1911 (by original designation)
Genus Yaukthwa Konopleva et al . , 2019
Type species: Yaukthwa nesemanni (Konopleva, Vikhrev & Bolotov, 2017) (by original designation)
Yaukthwa elongatula sp. nov.
Figures 3, 5D, 6H, Tables 1–2.
Type locality: Myanmar: Chindwin River [23.1918°N, 94.3217°E].
Holotype: RMBH biv346_2: Myanmar: Chindwin River, 23.1918°N, 94.3217°E, 2.ii.2018, Bolotov, Vikhrev, Lopes-Lima, Nyein Chan and local villagers leg.
Paratypes: type locality, 2.ii.2018, 2 specimens (RMBH nos. biv346_3 and biv346_1); Myanmar: Myit Tha (Manipur) River, 23.2006°N, 94.2214°E, 2.ii.2018, 3 specimens (RMBH nos. biv344_1, biv344_2, and biv344_3); Myanmar: Myit Tha (Manipur) River, 23.2448°N, 94.1661°E, 2.ii.2018, 2 specimens (biv341_2 and biv341_3); Myanmar: Myit Tha (Manipur) River, 23.2284°N, 94.1434°E, 2.ii.2018, 4 specimens (biv339_1, biv339_2, biv339_3, and biv339_7), Bolotov, Vikhrev, Lopes-Lima, Nyein Chan and local villagers leg.
Etymology: The name of this species refers to its elongated shell shape.
Diagnosis: The new species remotely resembles Yaukthwa nesemanni, but differs from it by much more elongated shell, broader posterior margin, narrower anterior margin, reduced pseudocardinal teeth, and fixed nucleotide substitutions in the COI, 16S rRNA and 28S rRNA gene fragments (Table 2).
Description: Medium-sized mussel. Shell length 35.1–64.3 mm, height 16.4–28.1 mm, width 8.2–14.9 mm. Shell trapezoidal, elongated, inequilateral, thin, not inflated. Anterior end rounded, very narrow at some specimens, dorsal side curved, ventral margin slightly concaved. Posterior end broader than anterior end, truncated. Umbo not projected, slightly elevated, strongly corrugated at some specimens; corrugation may cover almost entire shell. Periostracum from light- to dark-brown, nacre white-bluish with yellow regions. Lateral teeth very thin, slightly curved, a single tooth on right valve and two teeth on left valve. Pseudocardinal teeth reduced. Anterior muscle attachment scar oval or drop-like, visible. Posterior muscle attachment scar oval, shallow.
Habitat and ecology: Sites with hard clay and rocky bottom substrate in large, fast-flowing rivers (Fig. 6H).
Distribution: Myit Tha (Manipur) and Chindwin rivers, Ayeyarwady Basin, northwestern Myanmar.
Discussion
Taxonomic implications
Here, we introduce eight new mussel taxa belonging to the genera Leoparreysia, Radiatula, Indochinella, and Yaukthwa. Leoparreysia was established by us for a group of species from the Western Indochina Subregion that was previously placed within the Indian genus Parreysia because of their external similarity6. However, we found that the Leoparreysia members represent a separate phylogenetic clade of the Parreysiinae, which is distantly related to the true Parreysiini6. Leoparreysia whitteni sp. nov. appears to be the smallest representative of this group. This species was recorded from the Ayeyarwady Basin, which seems to be the primary evolutionary hotspot of Leoparreysiini diversity, with at least five valid species6.
Radiatula appears to be another endemic genus of Western Indochina6,7, while its sister groups, e.g. Scabies Haas, 1911, Unionetta Haas, 1955, and Harmandia Rochebrune, 1881, are distributed in the Sundaland Subregion14. In this study, we describe three additional members of Radiatula, i.e. R. myitthanensis sp. nov., R. chindwinensis sp. nov., and R. mouhoti haungthayawensis ssp. nov. Various Radiatula species are abundant throughout Myanmar6, while R. crispisulcata, the type species of this genus, is known only from old museum lots.
Indochinella was introduced by us for the Oxynaia pugio group as a monotypic genus, endemic to Western Indochina7. The pugio-group is characterized by a high genetic and morphological variability, indicating the presence of additional taxa6,7. In this study, we introduce three more subspecies-level lineages: Indochinella pugio viridissima ssp. nov. from the Sittaung, Bilin and Bago rivers, I. pugio daweiensis ssp. nov. from the Dawei River, and I. pugio paradoxa ssp. nov. from the Haungthayaw River (Fig. 2). The two latter subspecies differ from other taxa by one or two diagnostic substitutions in the nuclear 28S rRNA gene fragment (Table 2). This feature is remarkable, because such differences in the slowly evolving nuclear genes for freshwater mussels often correspond to species-level divergence16. Furthermore, these two subspecies are conchologically more similar to the Scabies taxa than to the nominative subspecies of I. pugio.
Yaukthwa is a species-rich genus in the subfamily Rectidentinae9. Historically, all Yaukthwa taxa were placed within the genus Trapezoideus Simpson, 1900 that was thought to comprise several widespread species, e.g. T. exolescens (Gould, 1843)17–19. However, based on a multi-locus phylogeny, Trapezoideus exolescens was found to be a member of the Parreysiinae13, and it has subsequently been transferred to its own genus, Trapezidens Bolotov, Vikhrev & Konopleva, 20176. A taxonomic revision of the tribe Contradentini revealed that the genus Trapezoideus is a monotypic taxon with a single species, T. foliaceus9. Several species from Western Indochina previously assigned to Trapezoideus6 were transferred to Yaukthwa9. This genus seems to be an endemic clade of Western Indochina, which includes a variety of endemic species with restricted distribution ranges, mostly in headwaters of rivers and streams6,9. Yaukthwa elongatula sp. nov. has a remarkable shell shape, which resembles the ultra-elongate representatives of the genus Solenaia (Gonideinae). This new species inhabits the Chindwin Basin, and it is associated with specific rocky and hard clay bottom sites.
While several recent integrative works6,7,9,14,15,20–22 have greatly improved our knowledge on the taxonomy and biogeography of freshwater mussels in Southeast Asia, many gaps in these fields are still to be filled. This novel study contributes to the further expansion and redefinition of the Oriental Unionidae. However, many nominal species of high importance for taxonomic and phylogenetic research, e.g. Pseudodon crebristriatus (Anthony, 1865) and Modellnaia siamensis Brandt, 1974, were not rediscovered by extensive field surveys during the last seven years6,7,9,14,15, and such taxa are in need of future research efforts.
Patterns of endemism
Previously, we have considered every large or medium-sized freshwater basin in the Oriental Region to be a separate evolutionary hotspot of the Unionidae fauna6,15. Pfeiffer et al.14 noted that this point of view does not consider the intra-basin heterogeneity, when several evolutionary hotspots can be recorded within a single drainage basin. Additionally, it was suggested that the proportion of single-drainage endemics proposed by Bolotov et al.15 may have been overestimated that underestimated the role of inter-basin faunal exchanges in shaping distribution patterns of freshwater mussels in Southeast Asia14. Based on the newly obtained results, we partly agree with those comments and propose an updated classification scheme for endemic freshwater mussel taxa from Southeast Asia as follows:
(1) Subregion endemics: widespread taxa, the range of which encompasses several freshwater drainages within a single biogeographic subregion. This group seems to contain a rather small number of taxa from Western Indochina, e.g. Lamellidens savadiensis (Nevill, 1877), Indochinella pugio viridissima ssp. nov., and Leoparreysia tavoyensis (Gould, 1843). A few examples of species with rather broad ranges are known from the Sundaland Subregion, e.g. Scabies crispata (Gould, 1843), S. phaselus (Lea, 1856), and S. mandarinus (Morelet, 1864)14. Several freshwater mussel species are widespread in rivers of the Malay Peninsula and northern Borneo20,21. Such multi-drainage distribution patterns were likely originated by dispersal events via direct connections between freshwater basins during the Late Pleistocene6,14,15.
(2) Single-drainage endemics. At first glance, the majority of freshwater mussels from Western Indochina may belong to this group, e.g. Leoparreysia whitteni sp. nov., L. burmana (Blanford, 1869), Radiatula mouhoti haungthayawensis ssp. nov., Indochinella pugio daweiensis ssp. nov., and I. pugio paradoxa ssp. nov.
(3) Intra-drainage endemics: taxa with local ranges corresponding to a certain section within a freshwater basin. Currently, members of this group from Western Indochina are poorly known, because our previous field surveys were primarily focused on sampling taxa from separate freshwater drainages6,15. Currently, a few species can be assigned to this group, e.g. Radiatula myitthanensis sp. nov., R. chindwinensis sp. nov., Yaukthwa elongatula sp. nov., Y. inlenensis Konopleva et al., 2019, and Y. paiensis Konopleva et al.9. However, a number of local endemics from other sites may be overlooked.
Methods
Data sampling
Samples of freshwater mussels were collected by hand from different water bodies throughout Myanmar using a rapid bioassessment approach of Cummings et al.23. A series of each mussel morphospecies from every study site has been collected. Soft tissue snips for DNA analyses were preserved in 96% ethanol immediately after collection.
Molecular data and phylogenetic analyses
New COI, 16S rRNA and 28S rRNA gene sequences were generated from 52 freshwater mussel specimens using a standard approach following published works6,9,15. The sequence data set and outgroup taxa are presented in Supplementary Table 1. The sequence alignment of each gene fragments was performed separately using the Muscle algorithm of MEGA724. The aligned data sets were concatenated into a multi-locus alignment. Absent sites were coded as missing data. Five partitions, i.e. three codons of COI + 16S rRNA + 28S rRNA, were used for phylogenetic analysis. We carried out maximum likelihood phylogenetic searches using web interface and server for IQ-TREE (W-IQ-TREE)25–27 with an automatic identification of the best-fit substitution model for each partition (Supplementary Table 2). To estimate the probability of internal branches, an ultrafast bootstrap (UFBoot) algorithm28 with 10,000 replicates was applied. Bayesian models were implemented in MrBayes v. 3.2.629 with two runs, each with three heated (temperature = 0.1) and one cold Markov chain (30,000,000 generations with sampling every 1000th generation) at the San Diego Supercomputer Center through the CIPRES Science Gateway30. The first 15% of trees were discarded as burn-in. Tracer v. 1.631 was used to check a convergence of the MCMC chains to a stationary distribution.
Species delimitation and diagnostics of new taxa
For preliminary delimitation of the putative species-level clades, we used BEAST2 v.2.5.132 with STACEY v.1.2.433 package. Each sequence in the multi-locus alignment (see above) was initially assigned to a prospective species unit. Nitia teretiuscula and Coelatura aff. aegyptiaca were used as outgroup (Supplementary Table 1). Five partitions (three codons of COI + 16S rRNA + 28S rRNA) were analyzed under a HKY substitution model and lognormal relaxed clock algorithm. The priors for the Birth Death Model were applied as follows: CollapseHeight = 0.001, Relative Death rate = 0.5, and Relative Collapse Weight = 0.5 using a beta prior. All parameters were estimated. Each run was conducted for 100,000,000 generations with sampling every 5000th tree and 10% burn-in. The calculations were performed at the San Diego Supercomputer Center through the CIPRES Science Gateway30. A matrix of putative species-level clusters based on SMC-trees generated by STACEY was constructed using SpeciesDelimitationAnalyser with initial 10% burn-in (www.indriid.com/software.html). An uncorrected COI mean p-distance to the nearest neighbor of each lineage was calculated in MEGA724. To check the putative subspecies-level units having a rather low molecular distance from nearest neighbors, we additionally used a network-based approach using Network v. 4.6.1.3 software with default settings34 that can reveal less prominent genetic differences between prospective taxa.
After a phylogenetic species delimitation, we estimated morphological differences between a new taxon and closely related (congeneric) taxa. The comparative analysis of the shell morphology was carried out with a special focus to the shell shape, structure of pseudocardinal and lateral teeth, shape of muscle attachment scars, and umbo position6,7,9,13. Three shell dimensions of each specimen, included in the type series of new taxa, i.e., the length, height, and width of the shell (all at the maximum diameter), were measured using calipers (±0.1 mm).
Finally, the molecular diagnosis of every new taxon was designed using fixed nucleotide substitutions, which were estimated for each gene separately using a Toggle Conserved Sites tool of MEGA724 at 50% level. For each new taxon, an alignment of congeneric haplotype sequences was performed using the Muscle algorithm implemented in MEGA724. All deleterious mutations were retained for the analyses.
Nomenclatural acts
The electronic edition of this article conforms to the requirements of the amended International Code of Zoological Nomenclature (ICZN), and hence the new names contained herein are available under that Code from the electronic edition of this article. This published work and the nomenclatural acts it contains have been registered in ZooBank (http://zoobank.org), the online registration system for the ICZN. The LSID for this publication is: urn:lsid:zoobank.org:pub:B017A518-0FBE-40F5-A2A4-8C13AC7E80BC. The electronic edition of this paper was published in a journal with an ISSN, and has been archived and is available from PubMed Central.
Supplementary information
Acknowledgements
The Russian Ministry of Science and Higher Education partly funded the fieldworks (E.S.K. and M.Y.G.) under project No. AAAA-A18-118012390161-9. The National Geographic Society partly funded the fieldworks (A.E.B., M.L.L., I.N.B., I.V.V., Z.L., and N.C.) under project No. NGS-274R-18. The Russian Ministry of Science and Higher Education supported the molecular analyses of freshwater mussel samples under project No. 6.2343.2017/4.6. Morphological study of the freshwater mussel samples by I.N.B., E.S.K. and I.V.V. was supported by the Ministry of Science and Higher Education of Russia, the Ministry of Europe and Foreign Affairs of France (MEAE), and the Ministry of Higher Education, Research and Innovation of France (MESRI) under project No. 05.616.21.0114 of the Hubert Curien Partnership (PHC) for the Franco-Russian Cooperation for Science and Technology (PHC Kolmogorov 2019). I.N.B. was supported by the Russian Foundation for Basic Research under grant No. 18-44-292001_r_mk. A.V.K. and M.Y.G. were funded by the Russian Foundation for Basic Research under grant 17-45-290066_r_a. I.V.V. and E.S.K. were funded by the Russian Foundation for Basic Research under grant No. 18-34-20033_mol_a_ved. M.L.-L. was funded by FCT – Foundation for Science and Technology under grant No. SFRH/BD/115728/2016. The Northern Arctic Federal University covered the article processing charge. We are grateful to the late Dr. Tony Whitten (Fauna & Flora International – Asia-Pacific), Mr. Frank Momberg (Fauna & Flora International – Myanmar Program, Myanmar), and the staff of the Department of Fisheries of the Ministry of Agriculture, Livestock and Irrigation of Myanmar for their great help during this study. Our research has been performed under the survey permission No. 5/6000/LFR(210/2018) dated on 23 January 2018 issued by the Ministry of Agriculture, Livestock and Irrigation of Myanmar and the export permission No. NWCD/CITES/9/5666/2018 dated on 28 June 2018 issued by the Forest Department of the Ministry of Environmental Conservation and Forestry of Myanmar. Two anonymous reviewers greatly helped us to improve an earlier version of the paper.
Author Contributions
I.N.B. developed the concept of the study. I.N.B., I.V.V., M.L.-L., Z.L., N.C. and T.W. collected samples. A.V.K., E.S.K. and A.A.T. designed and processed molecular analyses. E.S.K. performed morphological research and phylogenetic modeling. I.N.B. and E.S.K. wrote the paper, with input from I.V.V., M.L.-L., A.E.B., Z.L., N.C., A.V.K., M.Y.G., O.V.A. and T.W. All authors discussed the manuscript.
Data Availability
The type series of the new species are available in the RMBH – Russian Museum of Biodiversity Hotspots, Federal Center for Integrated Arctic Research, Russian Academy of Sciences, Arkhangelsk, Russia. The sequences generated in this study are available from GenBank. GenBank accession number and collecting locality for each specimen are presented in Supplementary Table 1.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-48528-z.
References
- 1.Vaughn CC. Ecosystem services provided by freshwater mussels. Hydrobiologia. 2018;810:15–27. doi: 10.1007/s10750-017-3139-x. [DOI] [Google Scholar]
- 2.Lopes-Lima M, et al. Conservation of freshwater bivalves at the global scale: diversity, threats and research needs. Hydrobiologia. 2018;810:1–14. doi: 10.1007/s10750-017-3486-7. [DOI] [Google Scholar]
- 3.Graf DL, Cummings KS. Review of the systematics and global diversity of freshwater mussel species (Bivalvia: Unionoida) Journal of Molluscan Studies. 2007;73:291–314. doi: 10.1093/mollus/eym029. [DOI] [Google Scholar]
- 4.Bogan AE. Global diversity of freshwater mussels (Mollusca, Bivalvia) in freshwater. Hydrobiologia. 2008;595:139–147. doi: 10.1007/s10750-007-9011-7. [DOI] [Google Scholar]
- 5.Kottelat M. A new genus and three new species of nemacheilid loaches from northern Irrawaddy drainage, Myanmar (Teleostei: Cypriniformes) Raffles Bulletin of Zoology. 2017;65:80–99. [Google Scholar]
- 6.Bolotov IN, et al. New taxa of freshwater mussels (Unionidae) from a species-rich but overlooked evolutionary hotspot in Southeast Asia. Scientific Reports. 2017;7:1–18. doi: 10.1038/s41598-017-11957-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolotov IN, et al. A new genus and tribe of freshwater mussel (Unionidae) from Southeast Asia. Scientific Reports. 2018;8:1–12. doi: 10.1038/s41598-018-28385-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolotov IN, et al. Discovery of Novaculina myanmarensis sp. nov. (Bivalvia: Pharidae: Pharellinae) closes the freshwater razor clams range disjunction in Southeast Asia. Scientific Reports. 2018;8:1–12. doi: 10.1038/s41598-018-34491-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konopleva ES, et al. A new genus and two new species of freshwater mussels (Unionidae) from western Indochina. Scientific Reports. 2019;9:1–14. doi: 10.1038/s41598-019-39365-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shu SS, Rogers DC, Chen XY, Sanoamuang LO. Streptocephalus diversity in Myanmar, with description of a new species (Branchiopoda, Anostraca) ZooKeys. 2018;734:1–12. doi: 10.3897/zookeys.734.21153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolotov IN, et al. Discovery of a silicate rock-boring organism and macrobioerosion in fresh water. Nature Communications. 2018;9:1–11. doi: 10.1038/s41467-018-05133-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, et al. Dramatic cropland expansion in Myanmar following political reforms threatens biodiversity. Scientific Reports. 2018;8:1–10. doi: 10.1038/s41598-018-34974-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konopleva ES, Bolotov IN, Vikhrev IV, Gofarov MY, Kondakov AV. An integrative approach underscores the taxonomic status of Lamellidens exolescens, a freshwater mussel from the Oriental tropics (Bivalvia: Unionidae) Systematics and Biodiversity. 2017;15:204–217. doi: 10.1080/14772000.2016.1249530. [DOI] [Google Scholar]
- 14.Pfeiffer JM, Graf DL, Cummings KS, Page LM. Molecular phylogeny and taxonomic revision of two enigmatic freshwater mussel genera (Bivalvia: Unionidae incertae sedis: Harmandia and Unionetta) reveals a diverse clade of Southeast Asian Parreysiinae. Journal of Molluscan Studies. 2018;84:404–416. doi: 10.1093/mollus/eyy028. [DOI] [Google Scholar]
- 15.Bolotov IN, et al. Ancient river inference explains exceptional Oriental freshwater mussel radiations. Scientific Reports. 2017;7:1–14. doi: 10.1038/s41598-017-02312-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Froufe E, et al. Phylogeny, phylogeography, and evolution in the Mediterranean region: News from a freshwater mussel (Potomida, Unionida) Molecular Phylogenetics and Evolution. 2016;100:322–332. doi: 10.1016/j.ympev.2016.04.030. [DOI] [PubMed] [Google Scholar]
- 17.Haas F. Superfamilia Unionacea. Das Tierreich. 1969;88:1–663. [Google Scholar]
- 18.Brandt RAM. The non-marine aquatic mollusca of Thailand. Archiv für Mollusckenkunde. 1974;105:1–423. [Google Scholar]
- 19.Subba Rao, N. V. Handbook of freshwater molluscs of India (Calcutta, 1989).
- 20.Zieritz A, et al. Factors driving changes in freshwater mussel (Bivalvia, Unionida) diversity and distribution in Peninsular Malaysia. Science of the Total Environment. 2016;571:1069–1078. doi: 10.1016/j.scitotenv.2016.07.098. [DOI] [PubMed] [Google Scholar]
- 21.Zieritz A, et al. Changes and drivers of freshwater mussel diversity and distribution in northern Borneo. Biological Conservation. 2018;219:126–137. doi: 10.1016/j.biocon.2018.01.012. [DOI] [Google Scholar]
- 22.Lopes-Lima M, et al. Phylogeny of the most species-rich freshwater bivalve family (Bivalvia: Unionida: Unionidae): Defining modern subfamilies and tribes. Molecular Phylogenetics and Evolution. 2017;106:174–191. doi: 10.1016/j.ympev.2016.08.021. [DOI] [PubMed] [Google Scholar]
- 23.Cummings, K. S., Jones, H. A. & Lopes-Lima, M. Rapid bioassessment methods for freshwater molluscs. In Larsen, T. H. (ed.) Core standardized methods for rapid biological field assessment. 185–207 (Conservation International, Arlington, 2016).
- 24.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen L-T, Schmidt HA, Haeseler, von A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research. 2016;44:W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chernomor O, von Haeseler A, Minh BQ. Terrace aware data structure for phylogenomic inference from supermatrices. Systematic Biology. 2016;65:997–1008. doi: 10.1093/sysbio/syw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: Improving the ultrafast bootstrap approximation. Molecular Biology and Evolution. 2017;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, M., Pfeiffer, W. & Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Gateway Computing Environments Workshop (GCE). 1–8 (IEEE, 2010).
- 31.Rambaut, A., Suchard, M. & Drummond, A. J. Tracer v.1.6 http://beast.bio.ed.ac.uk/sofware/tracer/ (2013).
- 32.Bouckaert R, et al. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Computational Biology. 2014;10:1–6. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones G. Algorithmic improvements to species delimitation and phylogeny estimation under the multispecies coalescent. Journal of Mathematical Biology. 2016;74:447–467. doi: 10.1007/s00285-016-1034-0. [DOI] [PubMed] [Google Scholar]
- 34.Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 35.Wessel P, Smith WHF. A global self-consistent, hierarchical, high-resolution shoreline database. Journal of Geophysical Research. 1996;101:8741–8743. doi: 10.1029/96JB00104. [DOI] [Google Scholar]
- 36.Lehner B, Grill G. Global river hydrography and network routing: baseline data and new approaches to study the world’s large river systems. Hydrological Processes. 2013;27:2171–2186. doi: 10.1002/hyp.9740. [DOI] [Google Scholar]
- 37.Lehner B, Verdin K, Jarvis A. New global hydrography derived from spaceborne elevation data. Eos. 2008;89:93–94. doi: 10.1029/2008EO100001. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The type series of the new species are available in the RMBH – Russian Museum of Biodiversity Hotspots, Federal Center for Integrated Arctic Research, Russian Academy of Sciences, Arkhangelsk, Russia. The sequences generated in this study are available from GenBank. GenBank accession number and collecting locality for each specimen are presented in Supplementary Table 1.