Abstract
Production of reactive oxygen species (ROS) is a conserved immune response primarily mediated by NADPH oxidases (NOXs), also known in plants as respiratory burst oxidase homologs (RBOHs). Most microbe-associated molecular patterns (MAMPs) trigger a very fast and transient ROS burst in plants. However, recently, we found that lipopolysaccharides (LPS), a typical bacterial MAMP, triggered a biphasic ROS burst. In this study, we isolated mutants defective in LPS-triggered biphasic ROS burst (delt) in Arabidopsis, and cloned the DELT1 gene that was shown to encode RBOHD. In the delt1-2 allele, the antepenultimate residue, glutamic acid (E919), at the C-terminus of RBOHD was mutated to lysine (K). E919 is a highly conserved residue in NADPH oxidases, and a mutation of the corresponding residue E568 in human NOX2 has been reported to be one of the causes of chronic granulomatous disease. Consistently, we found that residue E919 was indispensable for RBOHD function in the MAMP-induced ROS burst and stomatal closure. It has been suggested that the mutation of this residue in other NADPH oxidases impairs the protein’s stability and complex assembly. However, we found that the E919K mutation did not affect RBOHD protein abundance or the ability of protein association, suggesting that the residue E919 in RBOHD might have a regulatory mechanism different from that of other NOXs. Taken together, our results confirm that the antepenultimate residue E is critical for NADPH oxidases and provide a new insight into the regulatory mechanisms of RBOHD.
Keywords: Reactive oxygen species (ROS), NADPH oxidase (NOX), Microbe associated molecular pattern (MAMP), Lipopolysaccharides (LPS), Respiratory burst oxidase homolog D (RBOHD)
1. Introduction
In plants, the first layer of innate immunity is initiated by the recognition of conserved microbe-associated molecular patterns (MAMPs), such as bacterial flagellin, lipopolysaccharides (LPS), elongation factor Tu (EF-Tu), and peptidoglycan or fungal chitooligosaccharides (COs) (Boller and Felix, 2009; Dodds and Rathjen, 2010). MAMPs are recognized by surface-localized pattern recognition receptors, leading to MAMP-triggered immunity which prevents microbial entry, growth, and colonization (Antolín-Llovera et al., 2012; Macho and Zipfel, 2014).
Production of reactive oxygen species (ROS) is one of the rapid immune responses after perception of MAMPs (Mehdy, 1994; Lamb and Dixon, 1997). ROS not only directly reduce microbe viability, but also serve as local and systemic signaling molecules to activate additional immune responses, such as stomatal closure (Melotto et al., 2006; Mittler et al., 2011; Qi et al., 2017). The MAMP-induced ROS burst is produced in the apoplast and is often very fast and transient (Baker and Orlandi, 1995; Lamb and Dixon, 1997). Recently, we found LPS, the major component of the outer membrane of Gram-negative bacteria, could trigger a biphasic ROS burst, in which the first ROS burst is similar to that induced by other MAMPs, and the second shows slow and prolonged kinetics (Shang-Guan et al., 2018). However, the molecular mechanism underlying the LPS-triggered biphasic ROS burst is poorly understood.
Nicotinamide adenine dinucleotide phosphate (NADPH) oxidases (NOXs) are mainly responsible for ROS production during immune responses, and are highly conserved in mammals, plants, and fungi (Lambeth, 2004; Bedard and Krause, 2007). In mammals, NADPH oxidases consist of seven members, NOX1–NOX5 and dual oxidases DUOX1 and DUOX2, whereas Arabidopsis has ten members belonging to the respiratory burst oxidase homolog (RBOH) family (Bedard and Krause, 2007; Kawahara et al., 2007; Sumimoto, 2008). All NADPH oxidases are plasma-and/or endo-membrane enzymes, and contain a C-terminal dehydrogenase domain that binds flavin adenine dinucleotide (FAD) and NADPH, and a functional oxidase domain responsible for superoxide production by transferring electrons from NADPH to oxygen (Lambeth, 2004; Sumimoto, 2008). The resulting superoxide then can be converted to hydrogen peroxide by superoxide dismutase (Vignais, 2002). In terms of the molecular mechanisms underlying the activation of NADPH oxidase, the best characterized member is the phagocytic NOX2 (Segal et al., 2012; Singel and Segal, 2016). In phagocytes, NOX2 is complexed with p22phox, and its activation requires translocation of cytosolic regulatory proteins to the membrane and their assembly with the NOX2 complex to facilitate electron transfer (Sumimoto, 2008; Leto et al., 2009). Mutations of the NOX2 complex and its regulatory proteins result in chronic granulomatous disease, a rare congenital immunodeficiency disorder (Heyworth et al., 2003; Kuhns et al., 2010). In natural variants of X+-linked chronic granulomatous disease, characterized by defective enzyme activity but normal gene expression, mutation sites in the NOX2 gene are highly correlated with the FAD/NADPH binding region, suggesting that the C-terminal region of NADPH oxidases plays a critical role in the immune responses (Stasia and Li, 2008; Debeurme et al., 2010).
Unlike NOX2, plant RBOH proteins have an additional N-terminal region with calcium binding EF-hand motifs, which are absent in NOX1–4, but present in NOX5 and DUOXs (Bedard et al., 2007; Suzuki et al., 2011). In plants, NADPH oxidases regulate a wide variety of biological processes, including plant development and responses to biotic or abiotic stresses (Marino et al., 2012; Suzuki et al., 2012). The MAMP-induced ROS burst is primarily regulated by RBOHD, as rbohD mutants do not produce ROS in response to flagellin, EF-Tu, or chitin treatment (Torres et al., 2002; Chinchilla et al., 2007). Activation of RBOHD upon MAMP treatment has been extensively studied, including the roles of phosphorylation, the binding of calcium ions, and phosphatidic acid (Ogasawara et al., 2008; Zhang et al., 2009; Kadota et al., 2014; Li et al., 2014). All these regulatory sites locate at the N-terminal region of RBOHD. However, the regulation of the C-terminal FAD/NADPH-binding region is unknown.
In this study, delt (defective in LPS-triggered ROS burst) was isolated by a genetic screen for Arabidopsis mutants that showed reduced ROS burst after LPS treatment. We cloned DELT1 gene by map-based cloning and whole genome sequencing approaches, and found that DELT1 encoded RBOHD protein. In the delt1-2 allele, a G-to-A mutation converts the Glu-919 residue to Lys (E919K) in RBOHD. E919 is located two amino acids before the C-terminal stop codon of RBOHD, and is highly conserved in NOX family members. We found that the RBOHD transcript level and protein abundance were not altered in delt1-2 mutants compared to wild type. In addition, the E919K mutation did not affect the subcellular localization and oligomerization of RBOHD. However, the E919 residue was indispensable for RBOHD function in LPS-induced stomatal closure.
2. Materials and methods
2.1. Plant materials and growth conditions
Aequorin-expressing transgenic Arabidopsis was kindly provided by Marc KNIGHT (Knight and Knight, 1995), and rbohD (CS9555) mutant seeds were obtained from the Arabidopsis Biological Resource Center (Ohio State University, USA). Arabidopsis seeds were sterilized with 10% (0.1 g/mL) sodium hypochlorite, and then grown on 1/2 Murashige-Skoog (MS) agar plates in a growth chamber (model A1000AR, Conviron) under a 16-h photoperiod, 75% humidity, at 22 °C. Ten-day-old seedlings were transferred to pots of peat-based compost (Klasmann-Deilmann, Germany) and grown under a 14-h photoperiod for the ROS assay or 10-h photoperiod for the stomatal assay.
2.2. Chemiluminescence assay of ROS production
ROS production was measured using a chemiluminescence assay as previously described (Liang et al., 2013). Briefly, leaf disks (0.2 cm2) from mature leaves were punched and the chemiluminescent signal was recorded for 24 h or 1 h using a Photek camera HRPCS5 (Photek Ltd., UK). LPS (Sigma, USA), elf26 (a synthetic polypeptide that corresponds to EF-Tu, GenScript, China), chitooctaose (IsoSep, Sweden), and flg22 (a 22-amino acid synthetic polypeptide that corresponds to Pseudomonas syringae flagellin, GenScript, China) were dissolved in sterilized doble distilled H2O (ddH2O). The lipid A (the active moiety of LPS) was extracted as previously described (Shang-Guan et al., 2018).
2.3. Map-based cloning of DELT1-1 gene
The delt1-1 mutants were crossed to the Landsberg erecta (Ler) ecotype, and the generated F1 was self-pollinated to produce F2 seeds. A mapping population which showed reduced ROS production after LPS treatment was isolated in the F2 generation. Genomic DNA was extracted from each individual for linkage analysis. The polymerase chain reaction (PCR) markers were developed using the Arabidopsis Mapping Platform (https://www.arabidopsis.org/browse/Cereon/ help.jsp), and the primers used are listed in Table S1.
2.4. Whole genome sequencing
The delt1-1 mutants were backcrossed to ColQ, and the generated F1 was self-pollinated to produce F2 seeds. Individuals with or without ROS production after LPS treatment were pooled together, respectively, and subjected to whole genome sequencing using Illumina HiSeq (1gene Company, China). Illumina short reads generated from the bulked DNA samples were filtered by their Phred quality score and aligned to the wild-type reference sequence using Burrows-Wheeler Aligner (BWA) software (http://bio-bwa.sourceforge.net/bwa.shtml). The alignment data were converted to sequence alignment map (SAM)/binary alignment map (BAM) files using SAMtools (https://sourceforge.net/projects/samtools), and low-quality single nucleotide polymorphisms (SNPs) were excluded with a Coval filter.
2.5. RNA isolation and qRT-PCR
Total RNA was extracted from 10-d-old seedlings using a RNeasy kit (Tiangen, China) according to the manufacturer’s instructions. First-strand complementary DNA (cDNA) was synthesized from 2 μg RNA using reverse transcriptase (Promega, USA). SYBR Green Master Mix (Vazyme, China) was used for quantitative real-time PCR (qRT-PCR) reactions. The relative levels of gene expression were calculated using the 2−∆∆ C T method with ELONGATION FACTOR1α (EF1α) as an internal control. All primers used for qRT-PCR are listed in Table S1.
2.6. Gene cloning and plasmid construction
All primers used for gene cloning are listed in Table S1. The coding sequences of RBOHD, RBOHDE919K, and Botrytis-induced kinase 1 (BIK1) were amplified and cloned into the pDONR-Zeo plasmid by BP cloning (Invitrogen, USA). After the inserts were verified by sequencing, they were cloned into the destination plasmids: pGWB5 for subcellular localization, pCAMBIA-GW-nLUC and pCAMBIA-GW-cLUC for the split luciferase complementation (SLC) assay, and pEarly gate 201-nYFP and pEarly gate 201-cYFP for the bimolecular fluorescence complementation (BiFC) analysis.
2.7. Agrobacterium tumefaciens-mediated transient protein expression in Nicotiana benthamiana
Transient protein expression in N. benthamiana was performed as previously described (Liao et al., 2017).
2.8. SLC assay
The SLC assay was performed as previously described (Liao et al., 2017). To detect the activity of luciferase, 0.5 mmol/L luciferin (Promega, USA) was sprayed on the leaves and the signal was captured with a Photek camera HRPCS5 (Photek Ltd., UK).
2.9. BiFC analysis
The BiFC assay was performed as previously described (Liao et al., 2017). Fluorescence was imaged using a confocal laser scanning microscope (Zeiss, Germany).
2.10. Protein extraction and immunoblot analysis
Total protein extraction and immunoblot analysis were performed as previously described (Liao et al., 2017). Buffer (50 mmol/L Tris-HCl pH 5.7, 2% sodium dodecyl sulfate (SDS), and 1 mmol/L phenylmethylsulfonyl fluoride (PMSF)) was used for total protein extraction. Plasma membrane proteins were extracted using the Brij-58 method (Zhang and Peck, 2011). Briefly, 5 g fresh weight of seedlings were ground and dissolved in 5 mL buffer B (330 mmol/L sucrose, 50 mmol/L 4-(2-hydroxyethyl)-1-pipera zineethanesulfonic acid (HEPES)/KOH pH 7.5, 50 mmol/L Na4P2O7, 25 mmol/L NaF, 5% (50 g/L) glycerol, 0.5% (5 g/L) polyvinyl pyrrolidone, 10 mmol/L ethylenediaminetetraacetic acid (EDTA), 1 mmol/L Na2MoO4, 1 mmol/L PMSF, 10 μ mol/L leupeptin, 1 nmol/L calyculin A, and 3 mmol/L dithiothreitol (DTT)). After clearing the homogenate of cell debris by centrifugation for 10 min at 10 000g at 4 °C, crude microsomes (CMs) were isolated from the supernatant by ultracentrifugation for 30 min at 100 000g at 4 °C, then dissolved in buffer C (buffer B without DTT but with 0.02% (0.2 g/L) Brij-58) and kept on ice for 45 min. These CMs within buffer C were centrifuged again for 30 min at 100 000g at 4 °C. The resulting pellets were dissolved in Tris-HCl pH 7.5 to yield the plasma membrane-enriched fraction. RBOHD was detected with anti-RBOHD antibody (Agrisera, Sweden), and plasma membrane protein chitin elicitor receptor kinase 1 (CERK1) was used as a loading control (Cao et al., 2014; Liao et al., 2017).
2.11. Stomatal closure assay
Abaxial epidermal strips of fully expanded rosette leaves from 5-week-old plants were peeled and floated on 10 mmol/L 2-(N-morpholino)ethanesulfonic acid hydrate (MES)/KOH buffer (pH 6.2) for 30 min to close stomata. The peels were then transferred to opening buffer (10 mmol/L MES/KOH, 50 mmol/L KCl, pH 6.15). After incubation under white light (125 μmol/(m2∙s)) for 3 h, 100 nmol/L flg22 and 10 μg/mL lipid A were added, and 1 h after treatment photos were taken under a light microscope (Nikon, Japan). Stomatal apertures were measured using ImageJ software 1.52 (National Institutes of Health, USA).
3. Results
3.1. Isolation of delt mutants
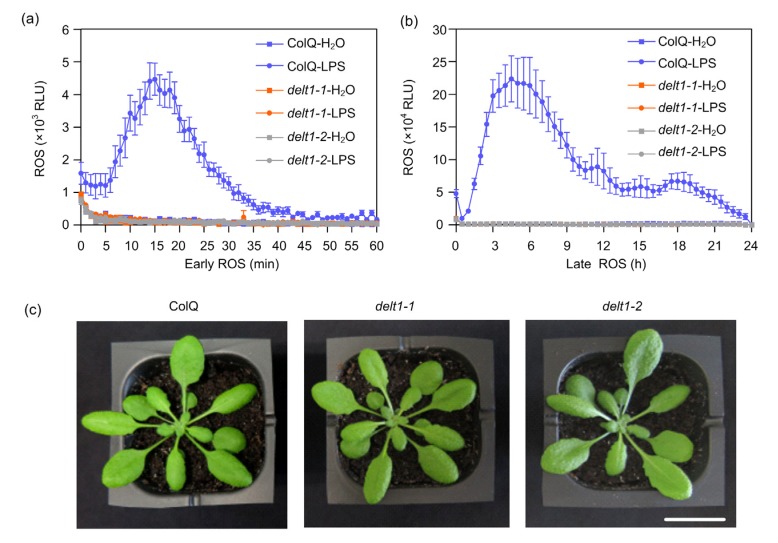
To identify the signaling components required for LPS-triggered biphasic ROS burst, we performed a genetic screen to identify mutants that exhibited reduced ROS production after LPS treatment. A previously ethylmethylsulfonate mutagenized Arabidopsis ColQ (stable transgenic Col-0 with aequorin insertion) seedling population was used for mutant isolation (Choi et al., 2014). Aequorin does not affect luminol-based ROS measurement. More than ten mutants were isolated from about 10 000 M2 individuals, and two of these mutants showed no biphasic ROS burst after LPS treatment and were selected to be characterized in detail (Figs. 1a and 1b). These two mutants were indistinguishable from wild-type plants when grown in soil (Fig. 1c). Complementation tests between these two mutants indicated that they were allelic to each other (Fig. S1a and S1b), and so are named delt1-1 and delt1-2 hereafter. The F1 genotype of delt1-2×delt1-1 was confirmed by sequencing the DELT1-1 mutation site after DELT1 was cloned (Fig. S1c). The delt1-1 mutant was backcrossed to ColQ, and the LPS-triggered ROS burst was evaluated in the resulting F1 and F2 population. All eight tested F1 plants showed a level of ROS similar to that of ColQ plants after LPS treatment. Among the 145 tested F2 plants, 33 plants showed no biphasic ROS after LPS treatment. The remainder were similar to wild type (with ROS: without ROS=112:33, χ 2=0.39, P>0.01), indicating that the DELT1-1 mutation is recessive in a single nuclear gene. Similarly, DELT1-2 is also a single recessive mutation. The original delt1-1 and delt1-2 mutants were backcrossed to ColQ twice, and homozygous progenies of F3 or subsequent generations were used in all experiments described below.
Fig. 1.
Isolation of the delt1-1 and delt1-2 mutants
(a, b) The delt1-1 and delt1-2 mutants showed deficiency in biphasic reactive oxygen species (ROS) production after LPS (50 μg/mL) treatment. ROS were monitored using a chemiluminescence assay and signal was recorded for 24 h after LPS treatment (b). To show the early ROS burst, a graph was plotted from 0 to 1 h (a). The data are shown as mean±standard error (SE, n=8). (c) Mature plants of delt1-1 and delt1-2 are indistinguishable from those of ColQ. Photos were taken four weeks after germination. Bar=1 cm. RLU: relative light unit
3.2. Mapping of delt1 mutant
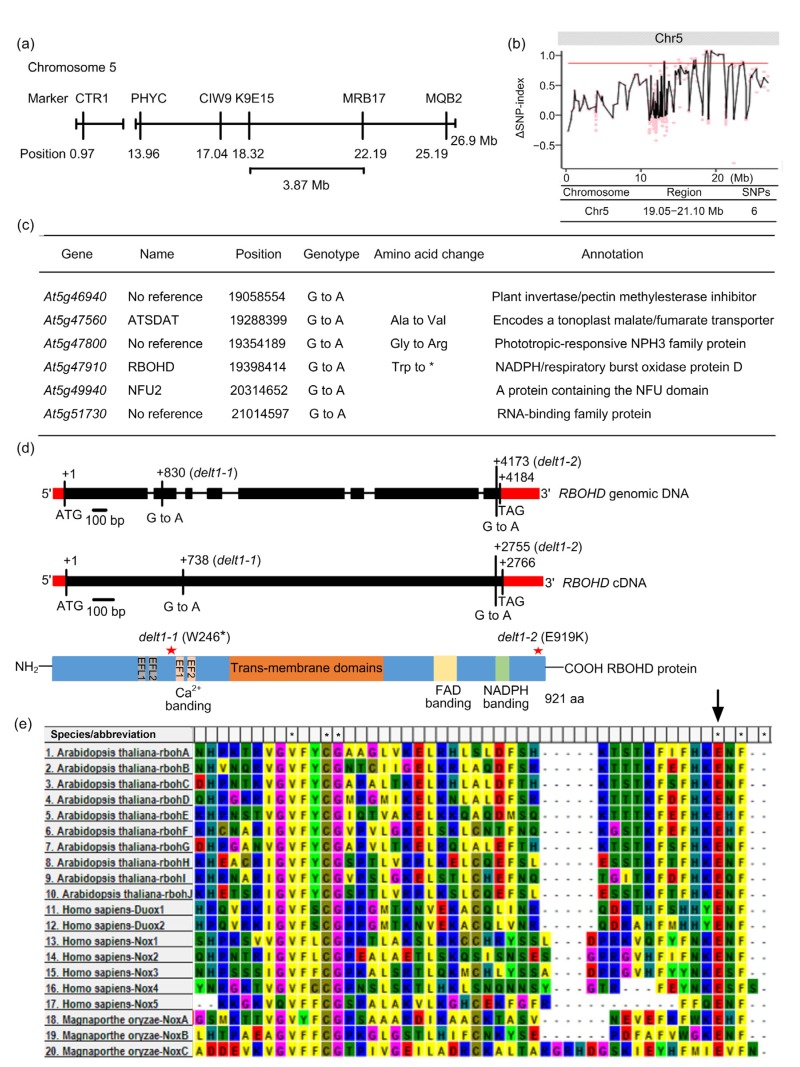
To genetically map DELT1-1, we generated an F2 population derived from the cross between delt1-1 and Ler. We roughly mapped the position of DELT1-1 to the short arm of chromosome 5 between two molecular markers, K9E15 and MRB17, which are 3.87 Mb apart (Fig. 2a). We identified an SNP in delt1-1 compared to wild type using a whole genome sequencing approach. The F2 progenies generated from the backcross between delt1-1 and ColQ were divided into two pools according to their mutant or wild-type phenotype. The genomic DNA from 22 individuals was pooled together and subjected to whole genome sequencing. After filtering for detected SNPs, we found a 2.05-Mb region on chromosome 5 with a relatively high SNP index, which located within our mapping region, indicating linkage with the delt1-1 phenotype (Fig. 2b). Three nonsynonymous SNPs were found in the linkage region and one mutation was in the At5g47910 gene, encoding an RBOHD protein (Fig. 2c). The delt1-1 mutant harbors a G-to-A mutation at nucleotide 830 of RBOHD (Fig. 2d), which results in a substitution of residue Trp-246 by a stop codon (W246*) (Fig. 2e). We then amplified RBOHD using delt1-2 genomic DNA. The sequencing results revealed a G-to-A mutation at nucleotide 4137 in the DELT1-2 genomic DNA and 2755 in the cDNA (Fig. 2d), which converts residue Glu-919 of RBOHD to Lys (E919K) in delt1-2 mutants (Fig. 2d).
Fig. 2.
Mapping of delt1 mutant
DELT1 encodes the RBOHD. (a) The DELT1-1 mutation was mapped to chromosome 5 (Chr5). (b) ΔSNP-index plot of Chr5 generated by whole genome sequencing. Pink dots correspond to each ΔSNP-index, and the black line represents the average values of ΔSNP-index of 1 Mb intervals with a 100-kb increment. The candidate region (19.05–21.10 Mb) is above the cutoff red line. (c) Six SNPs were identified in the candidate region. (d) Schematic representation of RBOHD gene and RBOHD protein. The mutation sites in delt1-1 and delt1-2 are indicated. Red and black boxes represent untranslated regions and exons, respectively. Thin lines indicate introns. (e) Sequence alignment of the RBOHD C-terminal region. Sequences were downloaded from the National Center for Biotechnology Information (NCBI) and aligned by MEGA software 5.0. The conserved residue Glu (e) is marked by an arrow
RBOHD is an evolutionarily conserved protein widely distributed in most eukaryotic organisms. E919 is located two amino acids before the C-terminal stop codon of RBOHD. The sequence alignment of RBOH family proteins demonstrated that E919 was not only highly conserved in all ten RBOH proteins in Arabidopsis, but also conserved in human and fungal NOXs (Fig. 2e). This implies that residue E919 plays a conserved and critical role in maintaining the function of NADPH oxidases.
3.3. delt1-1 and delt1-2 mutations are allelic to rbohD null mutations
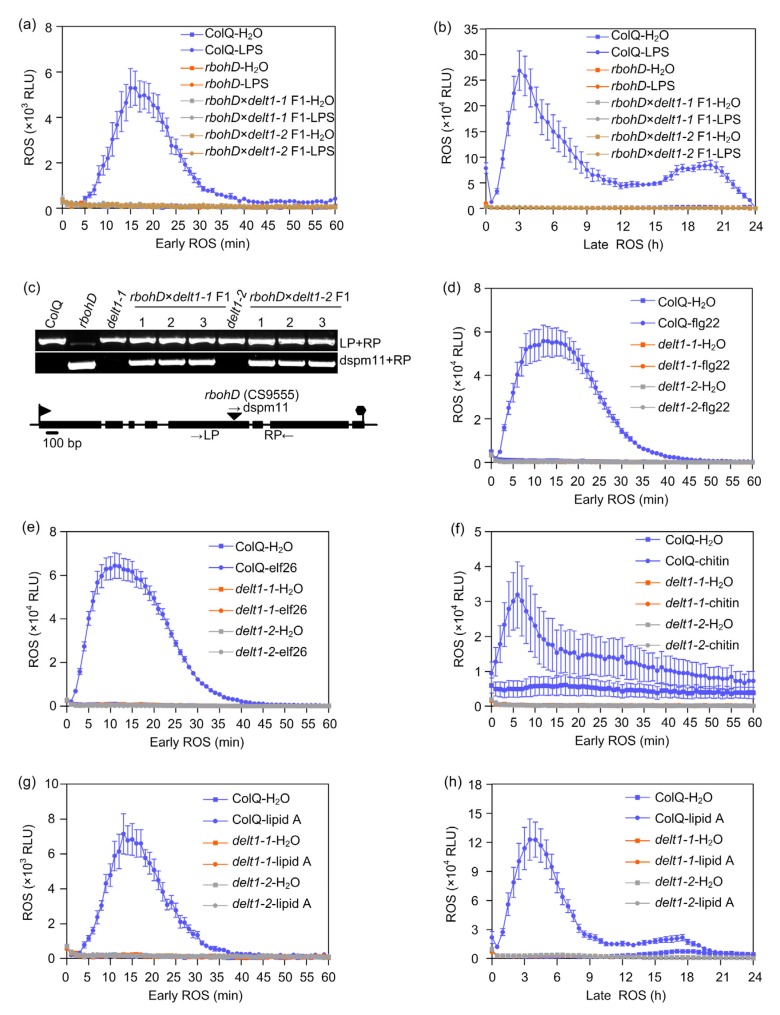
A null mutation in RBOHD, rbohD T-DNA insertion (CS9555), results in loss of ROS production upon MAMP treatment (Torres et al., 2002; Kadota et al., 2014; Li et al., 2014). To test the possible allelism between delt1 and rbohD, we crossed rbohD T-DNA mutants with delt1-1 and delt1-2 mutants, respectively. Results showed that all 16 F1 plants from rbohD×delt1-1 and rbohD×delt1-2 had no ROS burst after LPS treatment (Figs. 3a and 3b). Genotyping with T-DNA insertion indicated that F1 individuals contained the T-DNA fragments, suggesting that the cross was successful (Fig. 3c). Similar to the rbohD T-DNA mutants, delt1-1 and delt1-2 also showed no ROS burst after flg22, chitin, elf26, and lipid A treatments (Figs. 3d–3h), suggesting that delt1-1 and delt1-2 are loss-of-function mutants of RBOHD.
Fig. 3.
delt1-1 and delt1-2 mutations are allelic to rbohD null mutations
(a, b) delt1-1 and delt1-2 are allelic to rbohD T-DNA mutants. F1 individuals were generated from homozygous delt1-1 and delt1-2 pollinated with rbohD pollen. ROS were monitored using a chemiluminescence assay after 50 μg/mL LPS treatment. Signal was recorded for 24 h after LPS treatment (b). To show the early ROS burst, a graph was plotted from 0 to 1 h (a). The data are shown as means±standard error (SE, n=8). (c) Genotyping of the T-DNA insertion of rbohD confirmed that the cross was successful. The triangle indicates the rbohD T-DNA insertion site. (d–h) The delt1-1 and delt1-2 mutants showed no ROS production after flg22, elf26, chitin, or lipid A treatment. ROS were monitored using a chemiluminescence assay after 100 nmol/L flg22 (d), 500 nmol/L elf26 (e), 500 nmol/L chitin (f), and 10 μg/mL lipid A (g and h) treatment. The data are shown as mean±standard error (SE, n=8). LP: left primer; RP: right primer; RLU: relative light unit
3.4. E919K mutation does not affect RBOHD transcript and protein levels
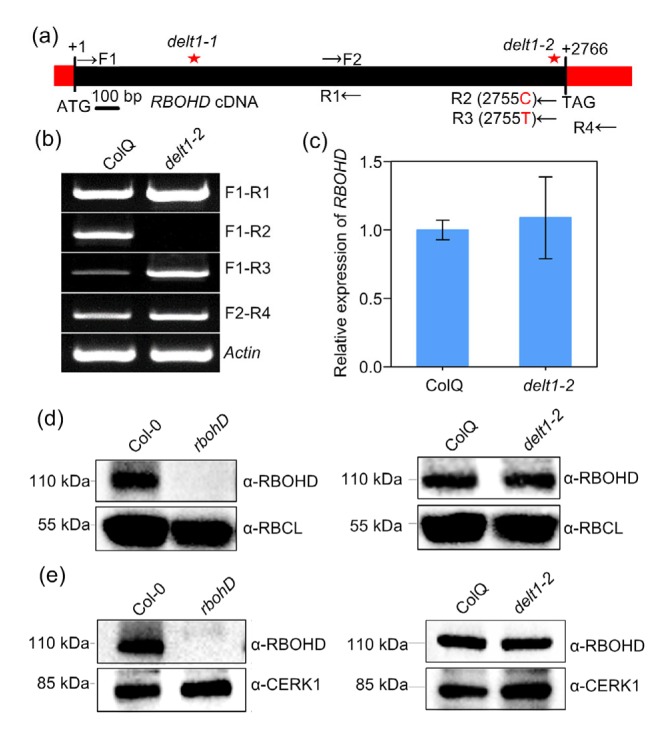
It is easy to understand why delt1-1 is a loss of function mutant of RBOHD since it contains a premature stop codon, which might result in the elimination of RBOHD transcripts because of the nonsense-mediated mRNA decay mechanism in eukaryotes (Peccarelli and Kebaara, 2014). We then focused on the delt1-2 mutants, in which E919K substitution abolishes RBOHD function in MAMP-induced ROS production. We examined the transcript levels of RBOHD by RT-PCR and qRT-PCR. RBOHD transcripts were not altered in delt1-2 mutants when a short fragment was amplified (Figs. 4a and 4b; F1-R1 primer pair). Surprisingly, we could not amplify the full-length RBOHD transcripts from delt1-2 mutants by RT-PCR when wild-type reverse primer was used (Fig. 4b; F1-R2 primer pair). The DELT1-2 mutation site (G2755A for cDNA) is within the R2 primer region, and therefore we suspected that it may reduce PCR efficiency. Therefore, we designed a reverse primer complementary to the G2755A mutation which enabled us to amplify full-length DELT1-2 (Fig. 4b; F1-R3 primer pair). Consistently, the reverse primer at the 3' untranslated region (UTR) was able to amply DELT1-2 transcripts (Fig. 4b; F2-R4 primer pair). qRT-PCR also confirmed that the transcript level of RBOHD was not affected by the DELT1-2 mutation (Fig. 4c).
Fig. 4.
RBOHD transcript and protein abundance in delt1-2 mutants
(a) Diagram of primer position used in (b). (b, c) RBOHD transcript levels. Total RNA was prepared from leaves of 7-d-old plants. RBOHD transcript levels were determined by semi-quantitative (b) and quantitative (c) RT-PCR. EF-1α was used as the reference gene. Values are mean±standard error (SE) of three biological repeats in (c). (d, e) RBOHD protein abundance. Total (d) and plasma membrane (e) proteins were extracted and detected by immunoblot analysis with anti-RBOHD antibody. The rbohD T-DNA null allele was used to verify the specificity of anti-RBOHD antibody. Rubisco (RBCL) and chitin elicitor receptor kinase 1 (CERK1) were used as loading controls. The experiment was carried out twice with similar results
Next, we examined whether RBOHD protein stability was affected by the E919K substitution in delt1-2 mutants. The protein abundance of RBOHD was detected by immunoblot analysis in a total protein extraction with an anti-RBOHD antibody. Antibody specificity was verified using rbohD T-DNA mutants (Fig. 4d). The predicted molecular mass of the full-length RBOHD is around 103 kDa. A specific band around 110 kDa was detected in Col-0 but not rhohD mutants, indicating that the anti-RBOHD antibody could specifically detect RBOHD (Fig. 4d). Compared to ColQ, RBOHD protein abundance in delt1-2 mutants was not significantly reduced (Fig. 4d). RBOHD is a membrane protein, and therefore we performed an immunoblot analysis using the enriched plasma membrane fraction. Similarly, RBOHD protein in delt1-2 mutants showed a level equal to that in ColQ (Fig. 4e). Taken together, these results show the E919K mutation of RBOHD does not affect its transcript or protein level.
3.5. E919K mutation does not affect RBOHD subcellular localization
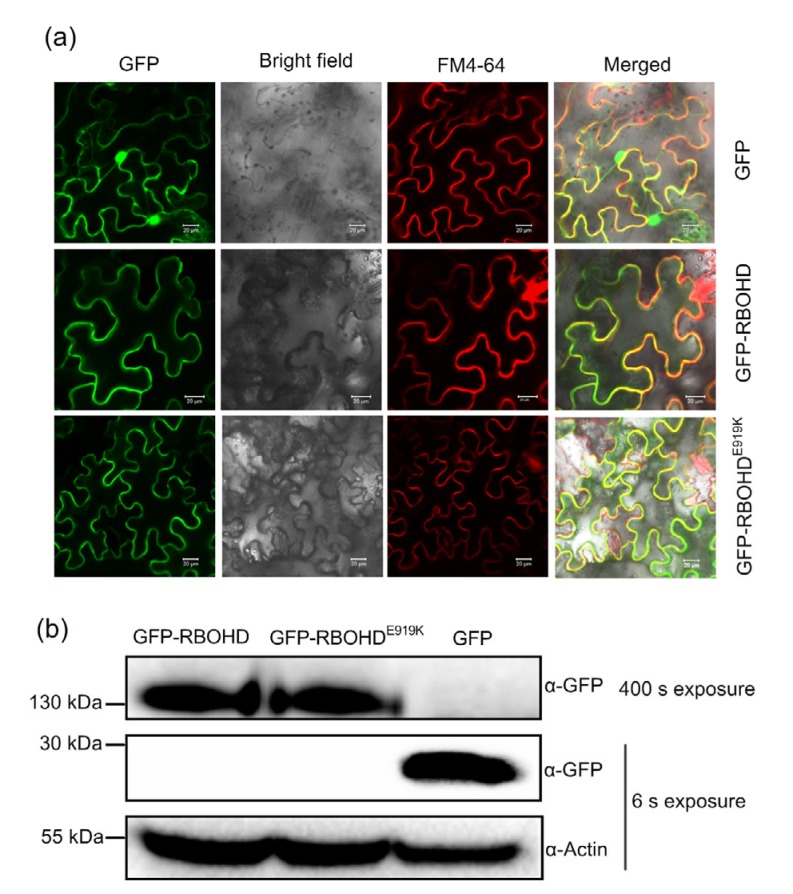
To examine whether E19K affects RBOHD subcellular localization, green fluorescence protein (GFP) was fused to the N-termini of RBOHD and RBOHDE919K and then transiently expressed in N. benthamiana. Both GFP-RBOHD and GFP-RBOHDE919K showed strong green fluorescence signals in the plasma membrane, as they were colocalized with the membrane marker FM4-64 (Fig. 5a). The protein size was verified by immunoblot analysis with an anti-GFP antibody. The GFP-RBOHD and GFP-RBOHDE919K proteins were significantly less abundant than the free GFP proteins, but clear bands with a predicted size around 130 kDa were shown after a long exposure (Fig. 5b). All these results suggest that the E919K mutation does not affect RBOHD subcellular localization.
Fig. 5.
RBOHD subcellular localization
E919K does not affect RBOHD subcellular localization. (a) RBOHD and RBOHDE919K were expressed on the plasma membrane. Green fluorescence protein (GFP)-RBOHD and GFP-RBOHDE919K fusion proteins were transiently expressed in Nicotiana benthamiana leaves via agroinfiltration. The plasma membrane was stained with the membrane marker FM4-64. Images were taken 3 d after infiltration by a confocal laser scanning microscope. Scale bar represents 20 μm. (b) GFP-RBOHD and GFP-RBOHDE919K were correctly expressed. Total proteins were extracted from leaves expressing GFP-RBOHD and GFP-RBOHDE919K in (a). Immunoblot analysis was performed using anti-GFP antibody. Actin was used as a loading control. The experiment was carried out three times with similar results
3.6. E919K mutation does not affect RBOHD association with proteins
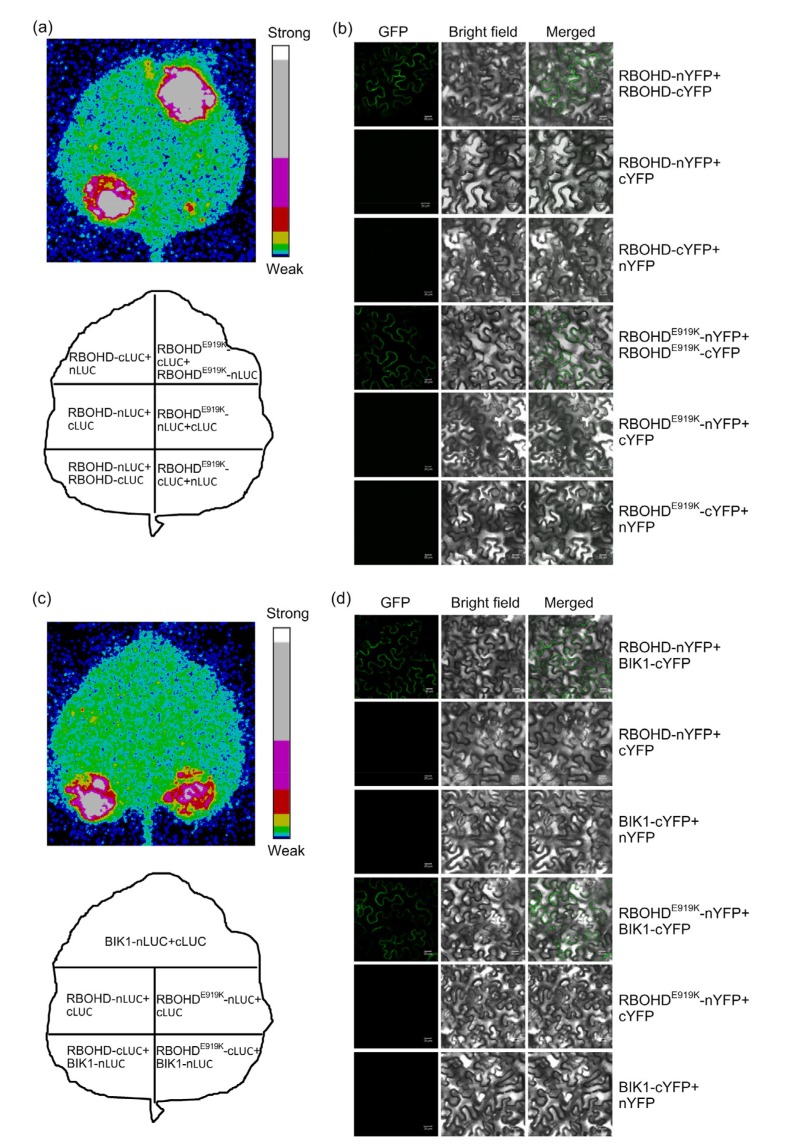
It has been suggested that the C-terminal dehydrogenase domain is required for oligomerization of NADPH oxidase (Kawahara et al., 2011). Therefore, using an SLC assay, we examined whether RBOHD was associated with itself in N. benthamiana. Strong luminescent signal could be detected when RBOHD-nLUC and RBOHD-cLUC were co-expressed, as well as when RBOHDE919K-nLUC and RBOHDE919K-cLUC were co-infiltrated, suggesting that E919K substitution does not affect the self-association of RBOHD (Fig. 6a). RBOHD self-association was confirmed using the BiFC assay (Fig. 6b). During the MAMP-induced ROS burst, RBOHD interacts directly with BIK1, a member of the receptor like cytosolic kinase, acting as a central signaling component which activates RBOHD through phosphorylation (Kadota et al., 2014; Li et al., 2014). We found that the E919K mutation did not impair RBOHD-BIK1 association (Figs. 6c and 6d). Taken together, these results suggest that E919 is not required for RBOHD oligomerization and RBOHD-BIK1 interaction.
Fig. 6.
RBOHD oligomerization and RBOHD‒BIK1 interaction
E919K does not affect RBOHD oligomerization and RBOHD‒BIK1 interaction. (a, c) Detection of protein–protein interactions by the split luciferase complementation (SLC) assay. RBOHD, RBOHDE919K, and Botrytis-induced kinase 1 (BIK1) were fused to the N-or C-terminal portion of luciferase (nLUC or cLUC), and the indicated fusion proteins were co-expressed in Nicotiana benthamiana. Images were taken using a Photek camera 3 d after infiltration. (b, d) Detection of protein–protein interactions by the bimolecular fluorescence complementation (BiFC) assay. RBOHD, RBOHDE919K, and BIK1 were fused to the N-or C-terminal portion of yellow fluorescence protein (nYFP or cYFP), and the indicated fusion proteins were co-expressed in N. benthamiana. Images were taken using a confocal laser scanning microscope 3 d after infiltration. Scale bar represents 20 μm. These experiments were carried out twice with similar results. GFP: green fluorescence protein
3.7. E919 of RBOHD is critical for its function in stomatal closure
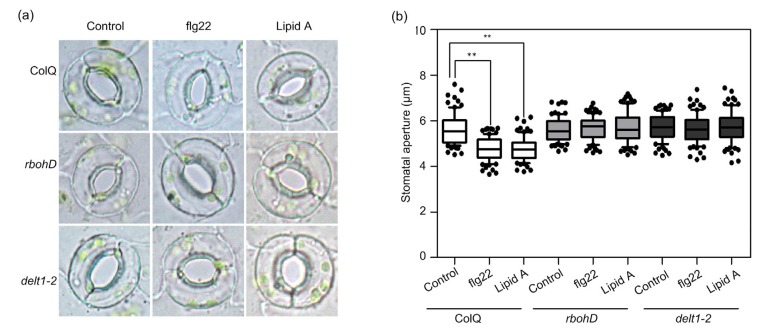
RBOHD has been shown to play an important role in the MAMP-induced stomatal closure. Therefore, we wondered whether E919 of RBOHD was required for stomatal closure in response to flg22 and lipid A. Lipid A (10 μg/mL) could induce stomatal closure at a level equivalent to 100 nmol/L flg22, and the rbohD T-DNA mutants failed to close stomata after flg22 and lipid A treatments (Figs. 7a and 7b). Similar to the rbohD T-DNA line, delt1-2 mutants also lost sensitivity to flg22 and lipid A. All these results suggest that E919 of RBOHD is critical for its function in stomatal closure.
Fig. 7.
Effects of flg22 and lipid A treatments on stomatal closure
delt1-2 mutants fail to close stomata in response to flg22 and lipid A. (a) Representative photographs of stomata; (b) The median of the stomatal aperture. The stomatal assay was performed using epidermal strips and stomatal aperture was measured 1 h after flg22 or lipid A treatment. The values of the stomatal aperture are expressed in microns and represented in the box plot where the box is bound by the 25th to 75th percentiles, whiskers span the 10th to 90th percentiles, and the line in the middle is the median. The individual points represent outliers. Data are represented as mean±standard error (SE, n=80). Asterisks indicate a significant difference from the control treatment (Student’s t-test, ** P<0.01)
4. Discussion
The antepenultimate residue at the C terminus of NADPH oxidase RBOHD, E919, is highly conserved in NOX family members (Kawahara et al., 2007; Magnani et al., 2017). Mutation of the corresponding residue E568 in human NOX2 leads to chronic granulomatous disease (Verhoeven, 1997; Heyworth et al., 2003; Kuhns et al., 2010). Consistently, our results suggest that residue E919 is critical for RBOHD function in the MAMP-induced ROS burst and stomatal closure.
Although residue E568 is indispensable for NOX2, how this residue affects NOX2 function is still unclear. Mutation of E568 in NOX2 correlates with the lack of oxidase activity, FAD incorporation, and translocation of cytosolic factors (Debeurme et al., 2010). E568 of NOX2 is not the binding site of NADPH and FAD. However, it has been predicted that mutation of E568 in NOX2 alters the structural orientation of the C-terminus, and indirectly impairs the protein’s ability to bind to substrates and other complex subunits (Debeurme et al., 2010). Unlike NOX2, the comparable cytosolic subunits of RBOHD have not yet been identified in Arabidopsis (Liu and He, 2016). It has been shown that the C-terminus of NOX5 is required for the binding of other proteins which facilitate NOX5 oligomerization (Kawahara et al., 2011; Chen et al., 2015), and the C-terminus of rice OsRBOHB could interact with its N-terminus to mediate a direct intramolecular interaction (Oda et al., 2010). These studies suggest that the C-terminus of NADPH oxidase might play a role in oligomerization or protein‒protein interaction. However, in this study, we found that mutation of E919 did not impair RBOHD oligomerization or RBOHD–BIK1 interaction. In addition, the sequence of the C-terminal dehydrogenase domain was very similar to that of members of the ferredoxin NADP+ reductase (FNR) family (Kawahara et al., 2007). The mutation of the corresponding residue in the FNR of peas impairs protein stability (Calcaterra et al., 1995), but we found that mutation of E919 in RBOHD had no effect on protein accumulation. Taken together, the most likely explanation for the impairment caused by E919K substitution of RBOHD is that it might disrupt the ability of the C-terminal dehydrogenase domain to bind to NADPH. However, this hypothesis awaits experimental verification in vivo by some sophisticated techniques, such as NADPH-based fluorescence lifetime imaging (Niesner et al., 2008; Leben et al., 2018).
RBOHD plays a predominant role in MAMP-induced ROS production, and phosphorylation is one of the most important regulatory mechanisms for RBOHD upon MAMP perception. MAMP receptors activate BIK1 which subsequently phosphorylates residues S39, S339, S343, and S347 of RBOHD in a calcium independent manner (Kadota et al., 2014; Li et al., 2014). The elevation of Ca2+ in cytoplasm activates Ca2+-dependent protein kinases which can phosphorylate residues S133, S148, S163, and S347 of RBOHD (Dubiella et al., 2013; Kadota et al., 2014). Recently, a mitogen-activated protein 4 (MAP4) kinase has been reported that mainly phosphorylates residue S347 of RBOHD to promote a flg22-induced ROS burst (Zhang et al., 2018). Similar to the MAMP-induced ROS burst, phosphorylation of RBOHD is also required for damage-associated molecular pattern (DAMP)-induced ROS bursts, for example, caused by extracellular ATP (eATP) (Choi et al., 2014). Upon eATP treatment, its receptor LecRK-I.9 (DORN1) kinase could directly phosphorylate the residues S22 and T24 of RBOHD to trigger an ROS burst (Chen et al., 2017). Other than phosphorylation, RBOHD is also activated by intracellular Ca2+ and phosphatidic acid (Ogasawara et al., 2008; Zhang et al., 2009). However, all these regulatory sites are located in the N-terminal region of RBOHD, and how the activation of the N-terminal region coordinates the binding of the C-terminal domain to NADPH and directs the electron flow across plasma membrane from NADPH to oxygen is still unknown. It will be interesting to examine whether the N-terminal modification of RBOHD alters the structural orientation of the C-terminal region, and whether residue E919 plays a role in this process.
ROS is also produced during effector triggered immunity (ETI), but the kinetics of ROS burst during ETI is very different from that in MAMP-triggered immunity (MTI) (Doke, 1983; Lamb and Dixon, 1997). MTI-ROS shows a fast and transient pattern, whereas ETI-ROS shows a slow and prolonged pattern. In terms of ROS kinetics, LPS-triggered biphasic ROS burst resembles that produced upon incompatible pathogen treatment (Lamb and Dixon, 1997; Shang-Guan et al., 2018). It has long been known that RBOHD is also required for ETI-ROS (Torres et al., 2002). Similarly, in this study we found that RBOHD was indispensable for LPS-triggered late ROS production. Interestingly, a quantitative phosphoproteomics study comparing pathogen associated molecular pattern (PAMP)-triggered immunity (PTI) and ETI found that residues S343 and S347 of RBOHD were necessary for both MTI-ROS and ETI-ROS (Kadota et al., 2019). This prompts the question of how RBOHD is regulated to trigger different kinetics of ROS burst. Do transcriptional regulation, posttranscriptional modification, or other interacting factors mediate the specificity? Therefore, further study on the regulation of RBOHD upon LPS treatment could be useful for deciphering the molecular mechanisms of different ROS kinetics.
5. Conclusions
RBOHD plays the predominant role in the production of ROS in plant immunity. Although the molecular mechanism underlying the activation of RBOHD has been extensively studied, all known regulatory sites locate at the N-terminal region of RBOHD. In this study, we found that residue E919 in the C-terminal region of RBOHD was critical for MAMP-induced ROS burst and stomatal closure. Unlike the function of the corresponding residue in other NADPH oxidases, we found that the E919K mutation did not affect RBOHD protein abundance or the ability of protein association, suggesting that the C-terminal region of RBOHD might have an uncommon regulatory mechanism.
Acknowledgments
We thank Dr. Kun JIANG (College of Life Science, Zhejiang University, Hangzhou, China) for the instruction of the stomatal closure assay.
List of electronic supplementary materials
Primer sequences used in this study
Allelic mutants delt1-1 and delt1-2
Footnotes
Project supported by the National Natural Science Foundation of China (No. 31622006) and the Postdoctoral Science Foundation of China (Nos. 2018M630683 and 2018T110601)
Contributors: Yan LIANG planned and designed the research; Qiu-ying LI and Ping LI performed most of the experiments; Nang MYINT PHYU SIN HTWE and Ke-ke SHANGGUAN contributed to the cloning of DELT1. Qiu-ying LI and Ping LI wrote the draft manuscript and Yan LIANG revised the manuscript. All authors read and approved the final manuscript. Therefore, all authors have full access to all the data in the study and take responsibility for the integrity and security of the data.
Electronic supplementary materials: The online version of this article (https://doi.org/10.1631/jzus.B1900105) contains supplementary materials, which are available to authorized users
Compliance with ethics guidelines: Qiu-ying LI, Ping LI, Nang MYINT PHYU SIN HTWE, Ke-ke SHANGGUAN, and Yan LIANG declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Antolín-Llovera M, Ried MK, Binder A, et al. Receptor kinase signaling pathways in plant‒microbe interactions. Annu Rev Phytopathol. 2012;50:451–473. doi: 10.1146/annurev-phyto-081211-173002. [DOI] [PubMed] [Google Scholar]
- 2.Baker CJ, Orlandi EW. Active oxygen in plant pathogenesis. Annu Rev Phytopathol. 1995;33:299–321. doi: 10.1146/annurev.py.33.090195.001503. [DOI] [PubMed] [Google Scholar]
- 3.Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. 2007;87(1):245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 4.Bedard K, Lardy B, Krause KH. NOX family NADPH oxidases: not just in mammals. Biochimie. 2007;89(9):1107–1112. doi: 10.1016/j.biochi.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 5.Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 6.Calcaterra NB, Pico GA, Orellano EG, et al. Contribution of the FAD binding site residue tyrosine 308 to the stability of pea ferredoxin-NADP+ oxidoreductase. Biochemistry. 1995;34(39):12842–12848. doi: 10.1021/bi00039a045. [DOI] [PubMed] [Google Scholar]
- 7.Cao YR, Liang Y, Tanaka K, et al. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife, 3: e03766. 2014 doi: 10.7554/elife.03766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen DQ, Cao YR, Li H, et al. Extracellular ATP elicits DORN1-mediated RBOHD phosphorylation to regulate stomatal aperture. Nat Commun. 2017;8(1):2265. doi: 10.1038/s41467-017-02340-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen F, Haigh S, Yu YF, et al. Nox5 stability and superoxide production is regulated by C-terminal binding of Hsp90 and CO-chaperones. Free Radical Biol Med. 2015;89:793–805. doi: 10.1016/j.freeradbiomed.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chinchilla D, Zipfel C, Robatzek S, et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448(7152):497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- 11.Choi J, Tanaka K, Liang Y, et al. Extracellular ATP, a danger signal, is recognized by DORN1 in Arabidopsis . Biochem J. 2014;463(3):429–437. doi: 10.1042/bj20140666. [DOI] [PubMed] [Google Scholar]
- 12.Debeurme F, Picciocchi A, Dagher MC, et al. Regulation of NADPH oxidase activity in phagocytes: relationship between FAD/NADPH binding and oxidase complex assembly. J Biol Chem. 2010;285(43):33197–33208. doi: 10.1074/jbc.m110.151555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant–pathogen interactions. Nat Rev Genet. 2010;11(8):539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 14.Doke N. Involvement of superoxide anion generation in the hypersensitive response of potato tuber tissues to infection with an incompatible race of Phytophthora infestans and to the hyphal wall components. Physiol Plant Pathol. 1983;23(3):345–357. doi: 10.1016/0048-4059(83)90019-x. [DOI] [Google Scholar]
- 15.Dubiella U, Seybold H, Durian G, et al. Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc Natl Acad Sci USA. 2013;110(21):8744–8749. doi: 10.1073/pnas.1221294110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heyworth PG, Cross AR, Curnutte JT. Chronic granulomatous disease. Curr Opin Immunol. 2003;15(5):578–584. doi: 10.1016/S0952-7915(03)00109-2. [DOI] [PubMed] [Google Scholar]
- 17.Kadota Y, Sklenar J, Derbyshire P, et al. Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol Cell. 2014;54(1):43–55. doi: 10.1016/j.molcel.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 18.Kadota Y, Liebrand TWH, Goto Y, et al. Quantitative phosphoproteomic analysis reveals common regulatory mechanisms between effector- and PAMP-triggered immunity in plants. New Phytol. 2019;221(4):2160–2175. doi: 10.1111/nph.15523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawahara T, Quinn MT, Lambeth JD. Molecular evolution of the reactive oxygen-generating NADPH oxidase (Nox/Duox) family of enzymes. BMC Evol Biol, 7:109. 2007 doi: 10.1186/1471-2148-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawahara T, Jackson HM, Smith SM, et al. Nox5 forms a functional oligomer mediated by self-association of its dehydrogenase domain. Biochemistry. 2011;50(12):2013–2025. doi: 10.1021/bi1020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knight H, Knight MR. Recombinant aequorin methods for intracellular calcium measurement in plants. Methods Cell Biol. 1995;49:201–216. doi: 10.1016/s0091-679x(08)61455-7. [DOI] [PubMed] [Google Scholar]
- 22.Kuhns DB, Alvord WG, Heller T, et al. Residual NADPH oxidase and survival in chronic granulomatous disease. N Engl J Med. 2010;363(27):2600–2610. doi: 10.1056/NEJMoa1007097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamb C, Dixon RA. The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:251–275. doi: 10.1146/annurev.arplant.48.1.251. [DOI] [PubMed] [Google Scholar]
- 24.Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol. 2004;4(3):181–189. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- 25.Leben R, Ostendorf L, van Koppen S, et al. Phasor-based endogenous NAD(P)H fluorescence lifetime imaging unravels specific enzymatic activity of neutrophil granulocytes preceding NETosis. Int J Mol Sci. 2018;19(4):1018. doi: 10.3390/ijms19041018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leto TL, Morand S, Hurt D, et al. Targeting and regulation of reactive oxygen species generation by Nox family NADPH oxidases. Antioxid Redox Signal. 2009;11(10):2607–2619. doi: 10.1089/ars.2009.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li L, Li M, Yu LP, et al. The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe. 2014;15(3):329–338. doi: 10.1016/j.chom.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 28.Liang Y, Cao YR, Tanaka K, et al. Nonlegumes respond to rhizobial Nod factors by suppressing the innate immune response. Science. 2013;341(6152):1384–1387. doi: 10.1126/science.1242736. [DOI] [PubMed] [Google Scholar]
- 29.Liao DH, Cao YR, Sun X, et al. Arabidopsis E3 ubiquitin ligase PLANT U-BOX13 (PUB13) regulates chitin receptor LYSIN MOTIF RECEPTOR KINASE5 (LYK5) protein abundance. New Phytol. 2017;214(4):1646–1656. doi: 10.1111/nph.14472. [DOI] [PubMed] [Google Scholar]
- 30.Liu YK, He CZ. Regulation of plant reactive oxygen species (ROS) in stress responses: learning from AtRBOHD. Plant Cell Rep. 2016;35(5):995–1007. doi: 10.1007/s00299-016-1950-x. [DOI] [PubMed] [Google Scholar]
- 31.Macho AP, Zipfel C. Plant PRRs and the activation of innate immune signaling. Mol Cell. 2014;54(2):263–272. doi: 10.1016/j.molcel.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 32.Magnani F, Nenci S, Fananas EM, et al. Crystal structures and atomic model of NADPH oxidase. Proc Natl Acad Sci USA. 2017;114(26):6764–6769. doi: 10.1073/pnas.1702293114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marino D, Dunand C, Puppo A, et al. A burst of plant NADPH oxidases. Trends Plant Sci. 2012;17(1):9–15. doi: 10.1016/j.tplants.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 34.Mehdy MC. Active oxygen species in plant defense against pathogens. Plant Physiol. 1994;105(2):467–472. doi: 10.1104/pp.105.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melotto M, Underwood W, Koczan J, et al. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006;126(5):969–980. doi: 10.1016/j.cell.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 36.Mittler R, Vanderauwera S, Suzuki N, et al. ROS signaling: the new wave? Trends Plant Sci. 2011;16(6):300–309. doi: 10.1016/j.tplants.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 37.Niesner R, Narang P, Spiecker H, et al. Selective detection of NADPH oxidase in polymorphonuclear cells by means of NAD(P)H-based fluorescence lifetime imaging. J Biophys, 2008, 2008:602639. 2008 doi: 10.1155/2008/602639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oda T, Hashimoto H, Kuwabara N, et al. Structure of the N-terminal regulatory domain of a plant NADPH oxidase and its functional implications. J Biol Chem. 2010;285(2):1435–1445. doi: 10.1074/jbc.m109.058909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogasawara Y, Kaya H, Hiraoka G, et al. Synergistic activation of the Arabidopsis NADPH oxidase AtrbohD by Ca2+ and phosphorylation. J Biol Chem. 2008;283(14):8885–8892. doi: 10.1074/jbc.m708106200. [DOI] [PubMed] [Google Scholar]
- 40.Peccarelli M, Kebaara BW. Regulation of natural mRNAs by the nonsense-mediated mRNA decay pathway. Eukaryot Cell. 2014;13(9):1126–1135. doi: 10.1128/EC.00090-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qi JS, Wang JL, Gong ZZ, et al. Apoplastic ROS signaling in plant immunity. Curr Opin Plant Biol. 2017;38:92–100. doi: 10.1016/j.pbi.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 42.Segal BH, Grimm MJ, Khan ANH, et al. Regulation of innate immunity by NADPH oxidase. Free Radical Biol Med. 2012;53(1):72–80. doi: 10.1016/j.freeradbiomed.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shang-Guan K, Wang M, Htwe NMPS, et al. Lipopolysaccharides trigger two successive bursts of reactive oxygen species at distinct cellular locations. Plant Physiol. 2018;176(3):2543–2556. doi: 10.1104/pp.17.01637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singel KL, Segal BH. NOX2-dependent regulation of inflammation. Clin Sci. 2016;130(7):479–490. doi: 10.1042/CS20150660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stasia MJ, Li XJ. Genetics and immunopathology of chronic granulomatous disease. Semin Immunopathol. 2008;30(3):209–235. doi: 10.1007/s00281-008-0121-8. [DOI] [PubMed] [Google Scholar]
- 46.Sumimoto H. Structure, regulation and evolution of Nox-family NADPH oxidases that produce reactive oxygen species. FEBS J. 2008;275(13):3249–3277. doi: 10.1111/j.1742-4658.2008.06488.x. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki N, Miller G, Morales J, et al. Respiratory burst oxidases: the engines of ROS signaling. Curr Opin Plant Biol. 2011;14(6):691–699. doi: 10.1016/j.pbi.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 48.Suzuki N, Koussevitzky S, Mittler R, et al. ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 2012;35(2):259–270. doi: 10.1111/j.1365-3040.2011.02336.x. [DOI] [PubMed] [Google Scholar]
- 49.Torres MA, Dangl JL, Jones JDG. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci USA. 2002;99(1):517–522. doi: 10.1073/pnas.012452499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verhoeven AJ. The NADPH oxidase: lessons from chronic granulomatous disease neutrophils. Ann N Y Acad Sci. 1997;832:85–92. doi: 10.1111/j.1749-6632.1997.tb46239.x. [DOI] [PubMed] [Google Scholar]
- 51.Vignais PV. The superoxide-generating NADPH oxidase: structural aspects and activation mechanism. Cell Mol Life Sci. 2002;59(9):1428–1459. doi: 10.1007/s00018-002-8520-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang MX, Chiang YH, Toruño TY, et al. The MAP4 kinase SIK1 ensures robust extracellular ROS burst and antibacterial immunity in plants. Cell Host Microbe. 2018;24(3):379–391e5. doi: 10.1016/j.chom.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang YY, Zhu HY, Zhang Q, et al. Phospholipase Dα1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis . Plant Cell. 2009;21(8):2357–2377. doi: 10.1105/tpc.108.062992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang ZJ, Peck SC. Simplified enrichment of plasma membrane proteins for proteomic analyses in Arabidopsis thaliana . Proteomics. 2011;11(9):1780–1788. doi: 10.1002/pmic.201000648. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer sequences used in this study
Allelic mutants delt1-1 and delt1-2