Abstract
The mechanistic target of rapamycin complex 1 (mTORC1) controls cell growth and metabolism in response to various environmental inputs, especially amino acids. In fact, the activity of mTORC1 is highly sensitive to changes in amino acid levels. Over past decades, a variety of proteins have been identified as participating in the mTORC1 pathway regulated by amino acids. Classically, the Rag guanosine triphosphatases (GTPases), which reside on the lysosome, transmit amino acid availability to the mTORC1 pathway and recruit mTORC1 to the lysosome upon amino acid sufficiency. Recently, several sensors of leucine, arginine, and S-adenosylmethionine for the amino acid-stimulated mTORC1 pathway have been coming to light. Characterization of these sensors is requisite for understanding how cells adjust amino acid sensing pathways to their different needs. In this review, we summarize recent advances in amino acid sensing mechanisms that regulate mTORC1 activity and highlight these identified sensors that accurately transmit specific amino acid signals to the mTORC1 pathway.
Keywords: Mechanistic target of rapamycin complex 1 (mTORC1), Amino acid, Sensor, Lysosome, Rag GTPases
1. Introduction
The mechanistic target of rapamycin complex 1 (mTORC1), evolutionally conserved from yeast to mammalian, acts as a central signaling hub that integrates nutrients, growth factors, and energy to regulate many anabolic and catabolic processes, ultimately controlling cell growth and metabolism (Chantranupong et al., 2015; Saxton and Sabatini, 2017). Deregulation of mTORC1 frequently happens in pathophysiological conditions, especially in cancer.
The target of rapamycin (TOR) gene was originally identified in yeast mutants resistant to rapamycin which binds to the peptidyl-prolyl-isomerase FKBP12 (Heitman et al., 1991; Koltin et al., 1991). Subsequently, four groups independently discovered a protein which is a homolog of the yeast TOR directly interacting with FKBP12-rapamycin in the mammal, and named the protein FKBP12-rapamycin-associated protein, rapamycin and FKBP12 target 1, rapamycin target 1, and mammalian TOR (mTOR), respectively (Brown et al., 1994; Chiu et al., 1994; Sabatini et al., 1994; Sabers et al., 1995). Early functional studies showed that mTOR carries out kinase activity to directly phosphorylate ribosomal protein S6 kinase 1 (S6K1) and eukaryotic translation initiation factor 4E-binding protein 1 (4EBP1), which can be regulated by amino acids (Burnett et al., 1998; Hara et al., 1998). However, alterations in amino acid sufficiency or rapamycin failed to change the kinase activity of mTOR, which implies that mTOR might function as a complex.
In addition to mTOR, mTORC1 is composed of regulatory associated protein of mTOR (Raptor) (Hara et al., 2002; Kim et al., 2002), mammalian lethal with SEC13 protein 8 (mLST8, also named as GβL) (Kim et al., 2003), proline-rich Akt substrate of 40 kDa (PRAS40) (Oshiro et al., 2007; Sancak et al., 2007; Vander Haar et al., 2007; Wang et al., 2007), and dishevelled, Egl-10 and pleckstrin (DEP) domain containing mTOR-interacting protein (DEPTOR) (Peterson et al., 2009). Recently, study of the architecture of human mTORC1 revealed that mTORC1 is a dimer of heterotrimer mTOR-Raptor-mLST8 (Aylett et al., 2016; Yang et al., 2016). Co-crystal structure of PRAS40 and mTOR-mLST8 demonstrated that PRAS40 also binds with FKBP12-rapamycin complex binding domain and mLST8 WD40 domain to achieve its inhibition of mTORC1 (Yang et al., 2017).
Amino acids serve as building blocks for protein and polypeptide synthesis, and molecular signal transducers to regulate diverse metabolic processes in animals (Layman et al., 2015). Defects in amino acid sensing pathways such as the mTORC1 pathway are linked to human diseases including immunodeficiency syndrome, cancers, and Birt-Hogg-Dube syndrome (Shimobayashi and Hall, 2016). In addition, amino acids have been proved to efficiently activate mTORC1. This increases the phosphorylation of substrates S6K1 and 4EBP1 to stimulate anabolic processes such as protein synthesis, and unc-51 like autophagy activating kinase 1 (ULK1) complex to suppress autophagy (Jewell et al., 2013). However, different amino acids regulate mTORC1 in distinct ways. Here, we discuss the precise mechanism of specific amino acid sensing in the regulation of the mTORC1 pathway.
2. Regulation of the mTORC1 pathway
2.1. Key sites of amino acid signaling to mTORC1
2.1.1 Plasma membrane
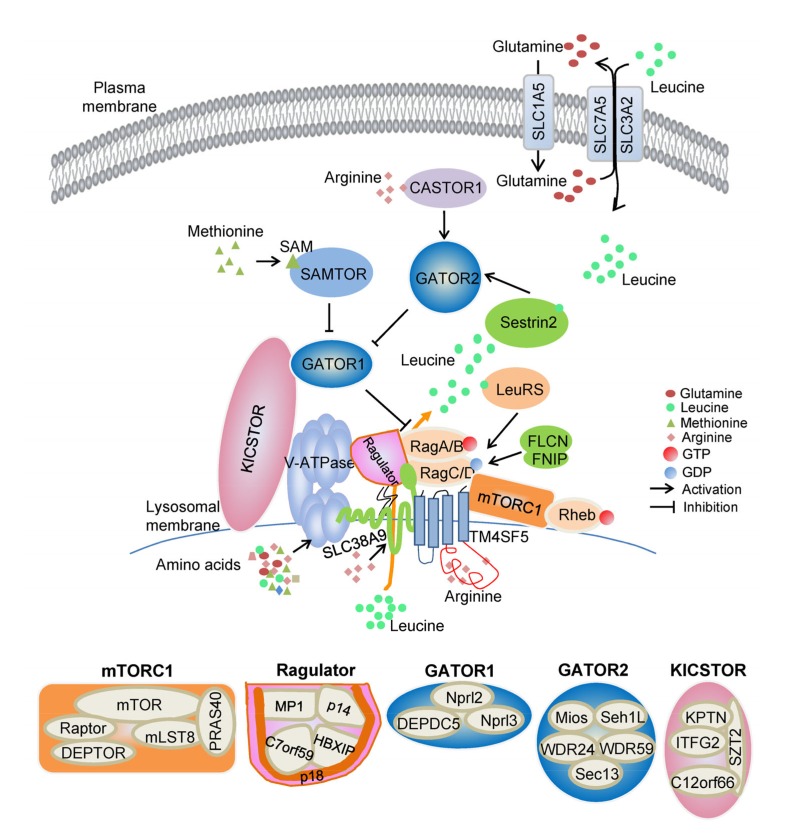
Amino acid transporters at the plasma membrane, which are responsible for amino acid exchange between the extracellular and intracellular microenvironments, play a role in the regulation of mTORC1 activity (Taylor, 2014). The solute carrier family (SLC) 1 member 5 (SLC1A5), a high-affinity glutamine transporter, is required for mTORC1 activation. SLC7A5 forms a heterodimer with SLC3A2 and the heterodimer functions as the bidirectional antiporter for the exchange of intracellular L-glutamine which is accumulated by SLC1A5 for extracellular branched chain amino acids such as leucine, contributing to activation of the mTORC1 pathway eventually (Nicklin et al., 2009) (Fig. 1).
Fig. 1.
Sensors for the mTORC1 pathway regulated by amino acids
Schematic shows amino acid-sensing pathway upstream of mTORC1, including several complexes that control Rag GTPases, and amino acid sensors Sestrin2, LARS, CASTOR1, SLC38A9, and TM4SF5, as well as SAM sensor SAMTOR. mTORC1: mechanistic target of rapamycin complex 1; SLC1A5: solute carrier family (SLC) 1 member 5; CASTOR1: cellular arginine sensor for mTORC1 protein 1; SAM: S-adenosylmethionine; SAMTOR: SAM sensor upstream of mTORC1; V-ATPase: vacuolar H+-adenosine triphosphatase; FLCN: folliculin; FNIP: FLCN-interacting protein; LARS: leucyl-tRNA synthetase; TM4SF5: transmembrane 4 L six family member 5; Raptor: regulatory-associated protein of mTOR; DEPTOR: dishevelled, Egl-10 and pleckstrin (DEP) domain containing mTOR-interacting protein; mLST8: mammalian lethal with SEC13 protein 8; PRAS40: proline-rich Akt substrate of 40 kDa; MP1: mitogen-activated protein kinase (MAPK) kinase 1 binding partner 1; HBXIP: hepatitis B virus X-interacting protein; GATOR1/2: GAP activity toward the Rag GTPases 1/2; Nprl2: nitrogen permease regulator 2-like protein; DEPDC5: DEP domain containing 5; Mios: meiosis regulator for oocyte development; Seh1L: Sec13-like protein; WDR24: WD repeat-containing protein 24; KICSTOR: named for kaptin (KPTN), integrin-α FG-GAP repeat-containing protein 2 (ITFG2), C12orf66, and seizure threshold 2 (SZT2)-containing regulator of mTORC1; GTP: guanosine triphosphate; GDP: guanosine diphosphate
2.1.2 Golgi apparatus
Golgi apparatus was reported as a potential platform for the activation of mTORC1 by amino acids (Thomas et al., 2014; Fan et al., 2016). Mechanistically, small guanosine triphosphatases (GTPases) Ras-related protein Rab-1A (Rab1A) and Ras homolog enriched in brain (Rheb) in coordination activate mTORC1 independently of Rag. Rab1A mediates the amino acid signal to mTORC1 on the Golgi apparatus, where Rheb localizes and activates mTORC1 (Thomas et al., 2014). However, how amino acid signal is transmitted to Rab1A is unclear, and amino acid sensors for Rab1A-and Golgi-dependent mTORC1 pathway need further investigation.
2.1.3 Lysosome
The roles of lysosome in nutrient degradation, recycling, and signaling cooperate to regulate fundamental cellular processes such as nutrient sensing (Perera and Zoncu, 2016). Classically, amino acid signals transmitted to mTORC1 originate from the lysosomes (Sancak et al., 2010; Zoncu et al., 2011). Cell-free reconstitution of intact lysosomes and mTORC1 induced colocalization of lysosomes and mTORC1 upon amino acid sufficiency. However, lysosomes which have defects in amino acid accumulation inhibit translocation of mTORC1. These results suggested that the lysosome is the major site of amino acid signaling to mTORC1. RNA interference (RNAi) of lysosomal proteins in Drosophila cells discovered that vacuolar H+-adenosine triphosphatase (V-ATPase) is required for amino acid-dependent lysosomal recruitment and activation of mTORC1. V-ATPase interacts with Ragulator, Rag GTPases, and SLC38A9, involving both amino acid sensing and efflux from lysosome (Zoncu et al., 2011; Abu-Remaileh et al., 2017; Wyant et al., 2017). Together, lysosomal amino acid signaling to mTORC1 requires a V-ATPase-associated inside-out mechanism.
2.2. Tuberous sclerosis complex-Rheb GTPase
The Rheb and Ras-related guanosine triphosphate (GTP)-binding (Rag) proteins in coordination regulate mTORC1 activity on the lysosomal surface in response to a range of environmental cues including amino acids, growth factors, and energy levels (Saito et al., 2005; Buerger et al., 2006; Kim et al., 2008; Sancak et al., 2008). Tuberous sclerosis complex (TSC), composed of TSC1, TSC2, and TBC1D7, functions as a GTPase-activating protein (GAP) for GTP-loading Rheb which binds directly with and allosterically stimulates mTORC1 (Manning et al., 2002; Tee et al., 2003; Long et al., 2005; Dibble et al., 2012; Yang et al., 2017). Growth factors promote protein kinase B (PKB)/Akt-mediated phosphorylation of TSC2, contributing to dissociation of TSC2 from the lysosome to activate Rheb and mTORC1 (Inoki et al., 2002; Potter et al., 2002; Menon et al., 2014). In contrast, energy starvation stimulates adenosine monophosphate (AMP)-activated protein kinase (AMPK)-induced phosphorylation of TSC2, enhancing the interaction of TSC2-Rheb to inhibit Rheb and mTORC1 (Inoki et al., 2003). In addition to these inputs, amino acids are thought to be presented to mTORC1 in a completely different way. Under amino acid sufficiency, the Rag GTPases recruit mTORC1 to the lysosomal surface (Kim et al., 2008; Sancak et al., 2008). Surprisingly, arginine and growth factors collectively promote TSC dissociation from the lysosome thereby activating Rheb and mTORC1 (Carroll et al., 2016). Under amino acid starvation, TSC relocates to the lysosome through the recruitment of Rag GTPases and stimulates GTP hydrolysis from Rheb, resulting in complete inactivation of mTORC1 (Demetriades et al., 2014). In conclusion, TSC-Rheb is essential for mTORC1 mediated by various microenvironmental factors, including amino acids.
2.3. Rag GTPases
The landmark discovery in understanding the regulation of mTORC1 pathway by amino acids is the identification of Rag GTPases that mediate amino acid signaling to mTORC1. The Rag GTPases have four Rag proteins in the mammal: functionally equivalent RagA or RagB (RagA/B) forms a heterodimer with functionally equivalent RagC or RagD (RagC/D) (Schürmann et al., 1995; Sekiguchi et al., 2001). In yeast, Gtr1, a homolog of RagA/B, interacts with Gtr2, a homolog of RagC/D (Hirose et al., 1998; Nakashima et al., 1999). Using chemical purification of the Raptor in HEK293 cells (Sancak et al., 2008) and RNAi screen of 132 GTPases in Drosophila cells (Kim et al., 2008), Rag GTPases were identified as participating in mTORC1 mediated by amino acids. Loss of the Rag GTPases in the mammal and Drosophila cells caused mTORC1 inactivity in response to amino acid sufficiency (Kim et al., 2008; Sancak et al., 2008). Neonatal mice with a constitutively active allele of RagA which stimulates the activity of mTORC1 in nutrient starvation failed to induce autophagy and survive the fasting periods (Efeyan et al., 2013). Overall, the Rag GTPases play a central role in the amino acid-mediated mTORC1 pathway (Fig. 1).
Importantly, the guanine nucleotide-loading status of the Rag heterodimer controls Rag activity and regulates mTORC1 lysosomal localization and activity. When cells are rich in amino acids, activated Rag heterodimer in which RagA/B is loaded with GTP while RagC/D is loaded with guanosine diphosphate (GDP) (RagA/BGTP-RagC/DGDP) enhances the Rag GTPases-Raptor interaction, and recruits mTORC1 to the lysosome (Kim et al., 2008; Sancak et al., 2008, 2010). Instead, GDP-loaded RagA/B in complex with GTP-loaded RagC/D (RagA/BGDP-RagC/DGTP) inhibits mTORC1 activity. A recent study has shed light on a “locking mechanism” of the nucleotide-binding status of the Rag heterodimer (Shen et al., 2017). In brief, loading GTP with one subunit of the Rag heterodimer inhibits the GTP-loading and triggers GDP-loading with another subunit. If unexpected GTP loading with both subunits occurs, the subunit with the later-loaded GTP induces faster hydrolysis. In addition, nutrient-induced alteration of the nucleotide-loading state of Rag heterodimer as an affinity switch governs the dynamic interactions between Rag GTPases, mTORC1, and Ragulator (Lawrence et al., 2018). RagA/BGTP-RagC/DGDP exhibits higher affinity for mTORC1, but reduces lysosomal association with Ragulator, leading to a decrease of the lifetime of mTORC1 on the lysosome. However, although RagA/BGDP-RagC/DGTP shows higher affinity for Ragulator, it is unable to capture mTORC1 dispersed in cytoplasm.
2.4. Regulation of the Rag GTPases
The guanine nucleotide-loading status of GTPases is regulated by multiple factors including GAPs that stimulate GTP hydrolysis, guanine nucleotide exchange factors (GEFs) that facilitate GDP dissociation, and the guanine nucleotide dissociation inhibitors that prevent GDP dissociation.
2.4.1 GEFs for the Rag GTPases
To identify the interacting proteins of the Rag GTPases, Ragulator was discovered as a pentameric complex of p18, p14, mitogen-activated protein kinase (MAPK) kinase 1 binding partner 1 (MP1), C7orf59, and hepatitis B virus X-interacting protein (HBXIP), also known as late endosomal and lysosomal adaptor and MAPK and mTOR activator (LAMTOR) complex of LAMTOR1 to LAMTOR5, respectively (Sancak et al., 2010; Bar-Peled et al., 2012). With the absence of Ragulator components in cells, the Rag GTPases fail to reside on the lysosome, and recruit mTORC1 to the lysosomal surface, leading to the mTORC1 pathway inactivation (Sancak et al., 2010; Bar-Peled et al., 2012). Myristoyl and palmitoyl modifications of LAMTOR1 are required for Ragulator specific localization to the lysosome (Nada et al., 2009). Structure of the complex Rag GTPases and Ragulator (Rag-Ragulator) demonstrated that C-terminal domains of Rags directly bind with LAMTOR2-LAMTOR3, and LAMTOR1 bridges LAMTOR2-LAMTOR3 with LAMTOR4-LAMTOR5 and enhances Rag-Ragulator interaction (de Araujo et al., 2017; Su et al., 2017; Yonehara et al., 2017; Zhang et al., 2017). Thus, Ragulator tethers the Rag GTPases to the lysosomal membrane.
The nucleotide loading state of the Rag GTPases is also important for its interaction with Ragulator. Nucleotide-free Rag GTPases bind to Ragulator more strongly than nucleotide-loading. Ragulator accelerates both GTP and GDP dissociation from RagA/B (Bar-Peled et al., 2012). Moreover, silencing LAMTOR2, RagB is unable to load GTP under a leucine stimulation condition (Lee et al., 2018). These results indicate that Ragulator is a GEF for RagA/B. However, a recent study has shed light on the mechanism activating the Rag GTPases by SLC38A9 and Ragulator which function as GEFs for RagA and RagC, respectively (Shen and Sabatini, 2018). First, Ragulator accelerates GTP release from RagC, not RagB, thus opening up the nucleotide-binding pocket. Second, SLC38A9 promotes GDP release from RagA.
2.4.2 GATOR functions as a GAP for RagA/B
GAP activity toward the Rag GTPases (GATOR) complex is an octamer of two subunits including GATOR1 (DEP domain containing 5 (DEPDC5), nitrogen permease regulator 2-like protein (Nprl2), Nprl3) and GATOR2 (meiosis regulator for oocyte development (Mios), Sec13, Sec13-like protein (Seh1L), WD repeat-containing protein 24 (WDR24), WDR59). Lack of components of GATOR1 and GATOR2 caused mTORC1 to be insensitive to amino acid deprivation and sufficiency, respectively, indicating that GATOR1 and GATOR2 are functionally contrary regulators for mTORC1 (Bar-Peled et al., 2013). GATOR1 binds to, and stimulates GTP hydrolysis of RagA/B, suggesting that GATOR1 is a GAP for RagA/B (Bar-Peled et al., 2013). The architecture of GATOR1 and GATOR1-Rag visualized by cyro-electron microscopy delineated that Nprl2 connects DEPDC5 and Nprl3, and DEPDC5 directly binds to RagA (Shen et al., 2018). However, further biochemical assays showed that Nprl2-Nprl3, not DEPDC5, receives amino acid signals from GATOR2 and executes the GAP activity to RagA. Interestingly, an inhibitory mode of GATOR1-Rag interaction has been identified such that a high-affinity binding of DEPDC5 and RagA prevents Nprl2-Nprl3-mediated GAP activity toward RagA.
Several factors are involved in GATOR1-Rag interaction. KICSTOR (named for kaptin (KPTN), integrin-α FG-GAP repeat-containing protein 2 (ITFG2), C12orf66 and seizure threshold 2 (SZT2)-containing regulator of mTORC1) was discovered to bind and recruit GATOR1 complex on the lysosomal membrane regardless of amino acid levels. Loss of SZT2 released the inhibitory function of GATOR1 by dissociation from Rags, which contributes to mTORC1 persistent activation upon amino acid deprivation, showing that KICSTOR negatively regulates mTORC1 through promoting GATOR1-Rag interaction (Peng et al., 2017; Wolfson et al., 2017). In contrast to KICSTOR, tyrosine kinase Src positively regulates mTORC1 through disrupting GATOR1-Rag interaction (Pal et al., 2018).
Ubiquitination plays a role in GATOR1-Rag interaction. E3 ubiquitin ligases S-phase kinase-associated protein 2 (Skp2) and RING finger protein 152 (RNF152) mediate K63-linked ubiquitination of RagA and recruit GATOR1 to the lysosomal surface in an amino acid-dependent manner (Deng et al., 2015; Jin et al., 2015). Deng et al. (2015) suggested that amino acid starvation increases RNF152-mediated RagA ubiquitination and its interaction with GATOR1 which inhibits RagA GTPase activity, eventually leading to mTORC1 inactivity. Surprisingly, Jin et al. (2015) revealed that, conversely, replenishing amino acid after starvation promotes Skp2-mediated RagA ubiquitination and its interaction with GATOR1. This is likely a negative feedback to control mTORC1 hyperactivity. In addition, E3 ubiquitin ligase Kelch-like protein 22 (KLHL22)-induced K48-linked polyubiquitination and degradation of DEPDC5 release the inhibitory function of GATOR1, contributing to activating mTORC1 under a condition of amino acid repletion (Chen et al., 2018).
2.4.3 FLCN-FNIP functions as a GAP for RagC/D
Folliculin (FLCN), an evolutionally conserved protein, interacts with FLCN-interacting protein 1/2 (FNIP1/2) and is involved in the mTORC1 pathway (Baba et al., 2006; Hasumi et al., 2008; Takagi et al., 2008). Subsequent studies showed that FLCN-FNIP shifts to the lysosome and binds with the Rag GTPases in the absence of amino acids. Loss of FLCN-FNIP in cells suppressed lysosomal recruitment of mTORC1 and desensitized the mTORC1 pathway to amino acids, indicating that FLCN is necessary for mTORC1 regulation by amino acids (Petit et al., 2013; Tsun et al., 2013). Tsun et al. (2013) further suggested that FLCN-FNIP functions as a GAP for RagC/D and promotes GTP hydrolysis of RagC/D.
3. Sensors for the mTORC1 pathway regulated by amino acids
What properties should a sensor have in an amino acid-stimulated mTORC1 pathway? First, sensors should bind directly with substrates at a physiologically relevant concentration (Table 1). Second, sensor-substrate interaction should be necessary for its function in the mTORC1 pathway. Third, sensors should directly or indirectly signal amino acid availability to the mTORC1 pathway. Several sensors have been identified for the mTORC1 pathway stimulated by leucine, arginine, and methionine (Fig. 1).
Table 1.
Affinities of sensors for their substrates including leucine, arginine, SAM, and methionine and corresponding intracellular and lysosomal concentrations of those substrates
| Amino acid/metabolite | Binding protein | K d/K i/IC50 (μmol/L) | Intracellular concentration in HEK293T (μmol/L) |
Lysosomal concentration in HEK293T (μmol/L) |
||
| Amino acid repletion | Amino acid starvation | Amino acid repletion | Amino acid starvation | |||
| Leucine | Sestrin1 | 10–15 | 192.55 | 20.25 | 53.99 | 48.09 |
| Sestrin2 | 20 | |||||
| LARS | 45 | |||||
| Arginine | CASTOR1 | 30 | 344.29 | 38.68 | 119.42 | 73.66 |
| SLC38A9 | 100–200 | |||||
| TM4SF5 | 10.5–37.9 | |||||
| SAM | SAMTOR | 7 | 33.24 | Unknown | 3.63 | Unknown |
| Methionine | Unknown | Unknown | 157.48 | 19.49 | 118.18 | 53.77 |
SAM: S-adenosylmethionine; LARS: leucyl-tRNA synthetase; CASTOR1: cellular arginine sensor for mTORC1 protein 1; SLC38A9: solute carrier family 38 member 9; TM4SF5: transmembrane 4 L six family member 5; SAMTOR: SAM sensor upstream of mTORC1; K d/K i/IC50: dissociation constant or inhibition constant or half maximal inhibitory concentration. Data from Chen et al. (2011), Han et al. (2012), Chantranupong et al. (2016), Wolfson et al. (2016), Abu-Remaileh et al. (2017), Gu et al. (2017), Wyant et al. (2017), and Jung et al. (2019)
3.1. Sensors for arginine-mediated mTORC1
Arginine can significantly regulate mTORC1 signaling, and deprivation of arginine strongly suppresses mTORC1 in various cell types. Recently, arginine sensors have been found for the mTORC1 pathway, including CASTOR1, SLC38A9, and TM4SF5.
3.1.1 CASTOR1
Cellular arginine sensor for mTORC1 protein 1/2 (CASTOR1/2), initially named GATS protein-like 3/2, was found to be a GATOR2-interacting protein and cytosolic arginine sensor (Huttlin et al., 2015; Chantranupong et al., 2016). Sequence analysis revealed that CASTOR1 has two conversed aspartate kinase, chorismate mutase, tyrA (ACT) domains which have important roles in small molecule binding, such as amino acids (Grant, 2006; Chantranupong et al., 2016), while structural analysis suggested that CASTOR1 has four ACT domains (Gai et al., 2016; Saxton et al., 2016b; Xia et al., 2016). Co-immunoprecipitation assay suggested that CASTOR1 and CASTOR2 could form homodimers and heterodimers, but only CASTOR1 dimers, including CASTOR1-CASTOR1 and CASTOR1-CASTOR2, interact with GATOR2 in an arginine-sensitive manner. Under arginine depletion condition, GATOR2 binds to the CASTOR1 dimers. In contrast, re-addition of arginine triggers dissociation of GATOR2 from CASTOR1 dimers (Chantranupong et al., 2016). In addition, dimerization of CASTOR1 is important for its interaction with GATOR2 and the mTORC1 pathway in response to arginine (Saxton et al., 2016b). Further binding assay revealed that arginine binds directly to CASTOR1 dimers to disrupt CASTOR1-GATOR2 interaction (Chantranupong et al., 2016). Loss of CASTOR1 resulted in the mTORC1 being insensitive to arginine deprivation. Also, a CASTOR1 mutant that could not bind to arginine has a defect in transmitting arginine sufficiency to the mTORC1. Altogether, CASTOR1 is an arginine sensor for the mTORC1 pathway.
3.1.2 SLC38A9
According to the transporter classification database, SLC38A9 belongs to the amino acid/auxin permease family which is characterized by 10 or 11 transmembrane domains (Saier et al., 2016). In addition to the 11 transmembrane domains, SLC38A9 has a distinct about 110-residue cytosolic N-terminal domain (SLC38A9N) responsible for its interaction with Rag-Ragulator (Jung et al., 2015; Rebsamen et al., 2015; Wang et al., 2015). Overexpression of SLC38A9 or SLC38A9N renders mTORC1 activity resistant to amino acid starvation, especially arginine or leucine starvation. Lacking SLC38A9 confers arginine, but not leucine, insensitivity on mTORC1 signaling. Biochemical analysis suggested that SLC38A9 interacts with Rag-Ragulator, which is dependent on SLC38A9N and sensitive to arginine. Interestingly, SLC38A9N also binds to Rag-Ragulator, and the interaction is insensitive to amino acid. Altogether, these results demonstrated that SLC38A9 is a candidate for an arginine sensor.
The following study validated that SLC38A9 is a bona fide arginine sensor for mTORC1 (Wyant et al., 2017). Mutants of SLC38A9 that could not bind to arginine or Rag-Ragulator fail to convey arginine sufficiency to mTORC1 in the SLC38A9-null cells. In vitro binding assay showed that arginine strengthens the interaction of SLC38A9 with Rag-Ragulator at a lysosomal arginine concentration of 100–200 μmol/L. However, in cells, arginine blunts the interaction of SLC38A9 with Rag-Ragulator (Wang et al., 2015). Recent work has shed light on the relationship between SLC38A9, Ragulator, and Rag GTPases (Shen and Sabatini, 2018). SLC38A9 preferentially binds to RagA/BGDP-RagC/DGTP heterodimer. Consequently, arginine deprivation which promotes the formation of RagA/BGDP-RagC/DGTP enhances interaction of SLC38A9 with Rag-Ragulator in cells. In vitro binding assay indicated that arginine promotes the interaction of Rag GTPases with SLC38A9. GDP-loaded RagA associates more strongly with SLC38A9 than GTP-loaded RagA. These results elucidated that the nucleotide state of Rag is the dominant effect on the mTORC1 pathway.
Apart from being an arginine sensor for the mTORC1 pathway, SLC38A9 is also an effluxer for several essential amino acids, especially for leucine. Interestingly, lysine promotes the interaction of SLC38A9 and Rag-Ragulator, indicating that lysine could regulate mTORC1 through SLC38A9 (Wyant et al., 2017). In addition, SLC38A9 plays a crucial role in the lysosomal cholesterol-activated mTORC1 pathway (Castellano et al., 2017). Altogether, SLC38A9 is a complicated and multifunctional protein, and future work may focus on the exploration of high-resolution structures and other crucial functions of SLC38A9.
3.1.3 TM4SF5
Interestingly, a very recent study identified transmembrane 4 L six family member 5 (TM4SF5) as a potential lysosomal arginine sensor for the mTORC1 pathway (Jung et al., 2019). TM4SF5 is an N-glycosylated protein with four transmembrane domains, two extracellular loops, an intracellular loop, and two cytosolic terminal tails (Wright et al., 2000). TM4SF5, especially the extracellular loops which are in the lysosomal lumen, binds to arginine with an affinity that is compatible with the lysosome arginine concentration. Furthermore, the TM4SF5 mutant that could not bind to arginine has a defect in activating the mTORC1 pathway upon arginine sufficiency. Loss of TM4SF5 could partially inhibit the mTORC1 pathway upon arginine sufficiency. Thus, TM4SF5 is a lysosomal arginine sensor. Surprisingly, TM4SF5 interacts with another arginine sensor SLC38A9, and Jung et al. (2019) established a new model that TM4SF5 senses and signals lysosomal arginine to SLC38A9 which mediates arginine efflux, contributing to activation of the mTORC1 pathway. However, the relationship between TM4SF5 and other components of mTORC1 such as Rag GTPases, GATOR, Ragulator, and FLCN-FNIP remains elusive.
3.2. Sensors for leucine-mediated mTORC1
Leucine is an essential amino acid that serves as not only a building block for protein synthesis but also a signal molecule that regulates protein metabolism, including autophagy (Grinde and Seglen, 1981; Wu et al., 2012; Yan et al., 2012) and the mTORC1 pathway (Hara et al., 1998; Gao et al., 2015). Leucine regulates the mTORC1 pathway through leucine sensors, such as leucyl-tRNA synthetase (LARS) and Sestrin2 (Han et al., 2012; Wolfson et al., 2016), and leucine metabolite acetyl-coenzyme A which promotes EP300-mediated acetylation of the Raptor (Son et al., 2019).
3.2.1 LARS
LARS catalyzes the ligation of leucine and tRNALeu to form leucyl-tRNALeu (Park et al., 2005). Interestingly, LARS was identified as a regulator for leucine-stimulated TORC1 from yeast to the mammal (Bonfils et al., 2012; Han et al., 2012). In the mammal, LARS specifically and directly binds to RagD GTPase, and functions as a GAP for RagD in a leucine-dependent fashion (Han et al., 2012). In addition, LARS likely functions as a leucine sensor for the mTORC1. The Michaelis constant (K m) of LARS for leucine activation is about 45 μmol/L (Chen et al., 2011). Silence of LARS renders mTORC1 resistant to leucine and unable to translocate to the lysosome. LARS mutant without leucylation activity leads to mTORC1 being less sensitive to leucine (Han et al., 2012). Interestingly, He et al. (2018) found that LARS signals intracellular leucine to mTORC1 through direct leucylation on lysine 142 (K142) of RagA. K142 leucylation regulates the interaction of RagA with Ragulator and GATOR1, and promotes the formation of GTP-loaded RagA which has higher GTPase activity. This result indicated that aminoacyl-tRNA synthetases act as potential amino acid sensors by direct lysine aminoacylation on the specific target proteins in response to amino acid sufficiency.
3.2.2 Sestrins
Sestrins, highly conserved proteins, have been proved to play crucial roles in oxidative damage and mTOR signaling (Ho et al., 2016). Sestrin1 and Sestrin2 were initially identified as p53 target genes (Buckbinder et al., 1994; Velasco-Miguel et al., 1999; Budanov et al., 2002), involved in cellular stress and mTOR signaling (Budanov and Karin, 2008; Lee et al., 2010). Sestrin1 and Sestrin2 were previously considered to inhibit the mTORC1 pathway through the activation of AMPK and TSC (Budanov and Karin, 2008). However, subsequent studies found that Sestrin2 still suppresses the mTORC1 pathway in the absence of AMPK and TSC (Parmigiani et al., 2014; Peng et al., 2014), indicating that AMPK and TSC are not necessary for Sestrin2-mediated inhibition of mTORC1. Indeed, Sestrin2 negatively regulates mTORC1 in a GATOR-and Rag GTPase-dependent manner (Chantranupong et al., 2014; Parmigiani et al., 2014; Peng et al., 2014). Sestrin2 binds to GATOR2, and functions upstream of the Rag GTPases and GATOR1 in the amino acid-sensing mTORC1 pathway (Chantranupong et al., 2014).
Amino acid deprivation obviously promotes Sestrin2-GATOR2 interaction, but leucine restimulation disrupts the interaction in cells and in vitro (Wolfson et al., 2016). As well as leucine, methionine and isoleucine stimulation both disrupt Sestrin2-GATOR2 interaction at concentrations about 50 and 100 μmol/L, respectively, much higher than the 5 μmol/L of leucine (Wolfson et al., 2016). The following evidence demonstrates that Sestrin2 is a leucine sensor for the mTORC1 pathway. First, Sestrin2 binds directly to leucine at an affinity that is compatible with the concentration at which mTORC1 is activated. Second, cells lacking Sestrins confer leucine insensitivity on the mTORC1. Third, Sestrin2 mutants that cannot bind with leucine are unable to transmit leucine sufficiency to the mTORC1 (Wolfson et al., 2016). The crystal structure of Sestrin2-leucine further supports the concept that Sestrin2 is a leucine sensor (Saxton et al., 2016c). However, it was argued that the crystal structure of Sestrin2 in the presence of leucine is highly similar to the absence of leucine (Kim et al., 2015; Lee et al., 2016; Saxton et al., 2016c). The authors analyzed the apo-Sestrin2 structure (Kim et al., 2015) and the 3.0 Å structure of Sestrin2 which was purified without adding exogenous leucine (Saxton et al., 2016a). However, both structures have leucine present, indicating that the apo-Sestrin2 structure is elusive.
3.3. Sensors for methionine-mediated mTORC1
SAMTOR was originally named C7orf60, but was renamed S-adenosylmethionine (SAM) sensor upstream of mTORC1 according to its functions in the mTORC1 pathway (Gu et al., 2017). SAMTOR was identified as an interacting protein of GATOR1 and KICSTOR through the BioPlex database (https://bioplex.hms.harvard.edu). Overexpression of SAMTOR inhibits mTORC1, indicating that SAMTOR is a negative regulator for the mTORC1. Sequence analysis predicted that SAMTOR has a SAM-binding domain. Indeed, SAM binds SAMTOR and disrupts the interaction of SAMTOR with GATOR1 and KICSTOR (Gu et al., 2017). SAM is derived from methionine, and methionine starvation reduces SAM concentration (Gu et al., 2017; Quinlan et al., 2017). Accordingly, methionine starvation strengthens the interaction of SAMTOR with GATOR1 and KICSTOR, and SAM stimulation reverses the effect. In addition, Sutter et al. (2013) reported that methionine and SAM inhibit autophagy through TORC1, but SAM, not methionine, is sensed directly by TORC1. Thus, these results indicated that SAMTOR likely serves as a SAM sensor for the methionine-mediated mTORC1. Loss of SAMTOR renders mTORC1 insensitive to methionine deprivation. Importantly, SAMTOR mutants that cannot bind to SAM are incapable of transmitting methionine sufficiency to the mTORC1 (Gu et al., 2017).
4. Perspectives
mTORC1 is not equally responsive to individual amino acids; arginine and leucine are particularly efficient for mTORC1 activation (Hara et al., 1998). How arginine and leucine are sensed by the mTORC1 pathway has been partially elucidated through the discovery of their own specific sensors CASTOR1, SLC38A9, TM4SF5, Sestrin2, and LARS (Han et al., 2012; Rebsamen et al., 2015; Wang et al., 2015; Chantranupong et al., 2016; Wolfson et al., 2016; Wyant et al., 2017; Jung et al., 2019). However, knowing other amino acid sensors for the mTORC1 pathway and whether these sensors are limited to specific intracellular organelle, cell type, tissue, or even species remain elusive.
Glutamine activates mTORC1 both independently and dependently on the Rag GTPases (Durán et al., 2012; Stracka et al., 2014; Jewell et al., 2015). In the Rag-independent manner, ADP ribosylation factor (Arf1) GTPase promotes mTORC1 translocation to lysosomes and activates mTORC1 in response to glutamine (Stracka et al., 2014; Jewell et al., 2015). Thus, regulators of Arf1 GTPase have the potential to be glutamine sensors. Lysine seems to have similar functions to those of arginine (Liu et al., 2012; Wyant et al., 2017). Lysine deprivation is less inhibitory to mTORC1 than that of leucine and arginine (Hara et al., 1998; Wyant et al., 2017). SLC38A9 contributes to conveying arginine and lysine sufficiency to the mTORC1 pathway, but it is more sensitive to arginine (Wyant et al., 2017). Instead, lysosomal amino acid transporter 1 (LAAT-1) transports lysosomal arginine and lysine, while it is more susceptible to lysine (Liu et al., 2012). It is possible that LAAT-1 is not only a transporter, but also a sensor for lysine.
In addition, increasing numbers of small GTPases containing Rags, Rheb, Arf1, Rab, Ras-related protein Ral-A, and Ras homolog gene family Rho are involved in mTORC1 activity (Nguyen et al., 2017). Classically, the Rag GTPases reside on the lysosomal surface and recruit mTORC1 to the lysosome in response to amino acid sufficiency (Sancak et al., 2008). It will be interesting to discovery how other GTPases regulate mTORC1 subcellular localization, and how these different proteins are responsive to and integrate amino acid signals. The intracellular organelle-specific amino acid sensors may be linked to small GTPases which localize to various intracellular organelles including lysosome, mitochondria, endoplasmic reticulum, and Golgi apparatus.
Acknowledgments
We thank all YAN laboratory members for suggestions and critical reading of the manuscript.
Footnotes
Project supported by the National Natural Science Foundation of China (Nos. 31520103915, 31730090, and 31322053) and the Hubei Provincial Natural Science Foundation of China (No. 2018CFA020)
Contributors: Xiu-zhi LI and Xiang-hua YAN analyzed the literatures and wrote the manuscript. Both authors have read and approved the final manuscript.
Compliance with ethics guidelines: Xiu-zhi LI and Xiang-hua YAN declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by either of the authors.
References
- 1.Abu-Remaileh M, Wyant GA, Kim C, et al. Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science. 2017;358(6364):807–813. doi: 10.1126/science.aan6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aylett CHS, Sauer E, Imseng S, et al. Architecture of human mTOR complex 1. Science. 2016;351(6268):48–52. doi: 10.1126/science.aaa3870. [DOI] [PubMed] [Google Scholar]
- 3.Baba M, Hong SB, Sharma N, et al. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc Natl Acad Sci USA. 2006;103(42):15552–15557. doi: 10.1073/pnas.0603781103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bar-Peled L, Schweitzer LD, Zoncu R, et al. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150(6):1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bar-Peled L, Chantranupong L, Cherniack AD, et al. A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340(6136):1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonfils G, Jaquenoud M, Bontron S, et al. Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol Cell. 2012;46(1):105–110. doi: 10.1016/j.molcel.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 7.Brown EJ, Albers MW, Shin TB, et al. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369(6483):756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 8.Buckbinder L, Talbott R, Seizinger BR, et al. Gene regulation by temperature-sensitive p53 mutants: identification of p53 response genes. Proc Natl Acad Sci USA. 1994;91(22):10640–10644. doi: 10.1073/pnas.91.22.10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Budanov AV, Karin M. p53 target genes Sestrin1 and Sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134(3):451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Budanov AV, Shoshani T, Faerman A, et al. Identification of a novel stress-responsive gene Hi95 involved in regulation of cell viability. Oncogene. 2002;21(39):6017–6031. doi: 10.1038/sj.onc.1205877. [DOI] [PubMed] [Google Scholar]
- 11.Buerger C, DeVries B, Stambolic V. Localization of Rheb to the endomembrane is critical for its signaling function. Biochem Biophys Res Commun. 2006;344(3):869–880. doi: 10.1016/j.bbrc.2006.03.220. [DOI] [PubMed] [Google Scholar]
- 12.Burnett PE, Barrow RK, Cohen NA, et al. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci USA. 1998;95(4):1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carroll B, Maetzel D, Maddocks OD, et al. Control of TSC2-Rheb signaling axis by arginine regulates mTORC1 activity. eLife, 5:e11058. 2016 doi: 10.7554/eLife.11058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castellano BM, Thelen AM, Moldavski O, et al. Lysosomal cholesterol activates mTORC1 via an SLC38A9-Niemann-Pick C1 signaling complex. Science. 2017;355(6331):1306–1311. doi: 10.1126/science.aag1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chantranupong L, Wolfson RL, Orozco JM, et al. The Sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell Rep. 2014;9(1):1–8. doi: 10.1016/j.celrep.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chantranupong L, Wolfson RL, Sabatini DM. Nutrient-sensing mechanisms across evolution. Cell. 2015;161(1):67–83. doi: 10.1016/j.cell.2015.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chantranupong L, Scaria SM, Saxton RA, et al. The CASTOR proteins are arginine sensors for the mTORC1 pathway. Cell. 2016;165(1):153–164. doi: 10.1016/j.cell.2016.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, Ou YH, Yang YY, et al. KLHL22 activates amino-acid-dependent mTORC1 signalling to promote tumorigenesis and ageing. Nature. 2018;557(7706):585–589. doi: 10.1038/s41586-018-0128-9. [DOI] [PubMed] [Google Scholar]
- 19.Chen X, Ma JJ, Tan M, et al. Modular pathways for editing non-cognate amino acids by human cytoplasmic leucyl-tRNA synthetase. Nucleic Acids Res. 2011;39(1):235–247. doi: 10.1093/nar/gkq763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiu MI, Katz H, Berlin V. RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc Natl Acad Sci USA. 1994;91(26):12574–12578. doi: 10.1073/pnas.91.26.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Araujo MEG, Naschberger A, Fürnrohr BG, et al. Crystal structure of the human lysosomal mTORC1 scaffold complex and its impact on signaling. Science. 2017;358(6361):377–381. doi: 10.1126/science.aao1583. [DOI] [PubMed] [Google Scholar]
- 22.Demetriades C, Doumpas N, Teleman AA. Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell. 2014;156(4):786–799. doi: 10.1016/j.cell.2014.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng L, Jiang C, Chen L, et al. The ubiquitination of RagA GTPase by RNF152 negatively regulates mTORC1 activation. Mol Cell. 2015;58(5):804–818. doi: 10.1016/j.molcel.2015.03.033. [DOI] [PubMed] [Google Scholar]
- 24.Dibble CC, Elis W, Menon S, et al. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol Cell. 2012;47(4):535–546. doi: 10.1016/j.molcel.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durán RV, Oppliger W, Robitaille AM, et al. Glutaminolysis activates Rag-mTORC1 signaling. Mol Cell. 2012;47(3):349–358. doi: 10.1016/j.molcel.2012.05.043. [DOI] [PubMed] [Google Scholar]
- 26.Efeyan A, Zoncu R, Chang S, et al. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature. 2013;493(7434):679–683. doi: 10.1038/nature11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan SJ, Snell C, Turley H, et al. PAT4 levels control amino-acid sensitivity of rapamycin-resistant mTORC1 from the Golgi and affect clinical outcome in colorectal cancer. Oncogene. 2016;35(23):3004–3015. doi: 10.1038/onc.2015.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gai ZC, Wang Q, Yang C, et al. Structural mechanism for the arginine sensing and regulation of CASTOR1 in the mTORC1 signaling pathway. Cell Discov, 2:16051. 2016 doi: 10.1038/celldisc.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao HN, Hu H, Zheng N, et al. Leucine and histidine independently regulate milk protein synthesis in bovine mammary epithelial cells via mTOR signaling pathway. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2015;16(6):560–572. doi: 10.1631/jzus.B1400337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grant GA. The ACT domain: a small molecule binding domain and its role as a common regulatory element. J Biol Chem. 2006;281(45):33825–33829. doi: 10.1074/jbc.R600024200. [DOI] [PubMed] [Google Scholar]
- 31.Grinde B, Seglen PO. Leucine inhibition of autophagic vacuole formation in isolated rat hepatocytes. Exp Cell Res. 1981;134(1):33–39. doi: 10.1016/0014-4827(81)90460-2. [DOI] [PubMed] [Google Scholar]
- 32.Gu X, Orozco JM, Saxton RA, et al. SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science. 2017;358(6364):813–818. doi: 10.1126/science.aao3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han JM, Jeong SJ, Park MC, et al. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012;149(2):410–424. doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- 34.Hara K, Yonezawa K, Weng QP, et al. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273(23):14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 35.Hara K, Maruki Y, Long XM, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110(2):177–189. doi: 10.1016/S0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 36.Hasumi H, Baba M, Hong SB, et al. Identification and characterization of a novel folliculin-interacting protein FNIP2. Gene. 2008;415(1-2):60–67. doi: 10.1016/j.gene.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He XD, Gong W, Zhang JN, et al. Sensing and transmitting intracellular amino acid signals through reversible lysine aminoacylations. Cell Metab. 2018;27(1):151–166e6. doi: 10.1016/j.cmet.2017.10.015. [DOI] [PubMed] [Google Scholar]
- 38.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253(5022):905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 39.Hirose E, Nakashima N, Sekiguchi T, et al. RagA is a functional homologue of S. cerevisiae Gtr1p involved in the Ran/Gsp1-GTPase pathway. J Cell Sci. 1998;111(Pt 1):11–21. doi: 10.1242/jcs.111.1.11. [DOI] [PubMed] [Google Scholar]
- 40.Ho A, Cho CS, Namkoong S, et al. Biochemical basis of Sestrin physiological activities. Trends Biochem Sci. 2016;41(7):621–632. doi: 10.1016/j.tibs.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huttlin EL, Ting L, Bruckner RJ, et al. The BioPlex network: a systematic exploration of the human interactome. Cell. 2015;162(2):425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Inoki K, Li Y, Zhu TQ, et al. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4(9):648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 43.Inoki K, Zhu TQ, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115(5):577–590. doi: 10.1016/S0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 44.Jewell JL, Russell RC, Guan KL. Amino acid signalling upstream of mTOR. Nat Rev Mol Cell Biol. 2013;14(3):133–139. doi: 10.1038/nrm3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jewell JL, Kim YC, Russell RC, et al. Differential regulation of mTORC1 by leucine and glutamine. Science. 2015;347(6218):194–198. doi: 10.1126/science.1259472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin GX, Lee SW, Zhang X, et al. Skp2-mediated RagA ubiquitination elicits a negative feedback to prevent amino-acid-dependent mTORC1 hyperactivation by recruiting GATOR1. Mol Cell. 2015;58(6):989–1000. doi: 10.1016/j.molcel.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung J, Genau HM, Behrends C. Amino acid-dependent mTORC1 regulation by the lysosomal membrane protein SLC38A9. Mol Cell Biol. 2015;35(14):2479–2494. doi: 10.1128/MCB.00125-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jung JW, Macalino SJY, Cui MH, et al. Transmembrane 4 L six family member 5 senses arginine for mTORC1 signaling. Cell Metab. 2019;29(6):1306–1319e7. doi: 10.1016/j.cmet.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 49.Kim DH, Sarbassov DD, Ali SM, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110(2):163–175. doi: 10.1016/S0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 50.Kim DH, Sarbassov DD, Ali SM, et al. GβL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell. 2003;11(4):895–904. doi: 10.1016/S1097-2765(03)00114-X. [DOI] [PubMed] [Google Scholar]
- 51.Kim E, Goraksha-Hicks P, Li L, et al. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10(8):935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim H, An S, Ro SH, et al. Janus-faced Sestrin2 controls ROS and mTOR signalling through two separate functional domains. Nat Commun, 6:10025. 2015 doi: 10.1038/ncomms10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koltin Y, Faucette L, Bergsma DJ, et al. Rapamycin sensitivity in Saccharomyces cerevisiae is mediated by a peptidyl-prolyl cis-trans isomerase related to human FK506-binding protein. Mol Cell Biol. 1991;11(3):1718–1723. doi: 10.1128/MCB.11.3.1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lawrence RE, Cho KF, Rappold R, et al. A nutrient-induced affinity switch controls mTORC1 activation by its Rag GTPase-Ragulator lysosomal scaffold. Nat Cell Biol. 2018;20(9):1052–1063. doi: 10.1038/s41556-018-0148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Layman DK, Anthony TG, Rasmussen BB, et al. Defining meal requirements for protein to optimize metabolic roles of amino acids. Am J Clin Nutr. 2015;101(6):1330S–1338S. doi: 10.3945/ajcn.114.084053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee JH, Budanov AV, Park EJ, et al. Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science. 2010;327(5970):1223–1228. doi: 10.1126/science.1182228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee JH, Cho US, Karin M, et al. Sestrin regulation of TORC1: is Sestrin a leucine sensor? Sci Signal. 2016;9(431):re5. doi: 10.1126/scisignal.aaf2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee M, Kim JH, Yoon I, et al. Coordination of the leucine-sensing Rag GTPase cycle by leucyl-tRNA synthetase in the mTORC1 signaling pathway. Proc Natl Acad Sci USA. 2018;115(23):E5279–E5288. doi: 10.1073/pnas.1801287115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu B, Du HW, Rutkowski R, et al. LAAT-1 is the lysosomal lysine/arginine transporter that maintains amino acid homeostasis. Science. 2012;337(6092):351–354. doi: 10.1126/science.1220281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Long XM, Lin Y, Ortiz-Vega S, et al. Rheb binds and regulates the mTOR kinase. Curr Biol. 2005;15(8):702–713. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- 61.Manning BD, Tee AR, Logsdon MN, et al. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/Akt pathway. Mol Cell. 2002;10(1):151–162. doi: 10.1016/S1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 62.Menon S, Dibble CC, Talbott G, et al. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell. 2014;156(4):771–785. doi: 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nada S, Hondo A, Kasai A, et al. The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK-ERK pathway to late endosomes. EMBO J. 2009;28(5):477–489. doi: 10.1038/emboj.2008.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakashima N, Noguchi E, Nishimoto T. Saccharomyces cerevisiae putative G protein, Gtr1p, which forms complexes with itself and a novel protein designated as Gtr2p, negatively regulates the Ran/Gsp1p G protein cycle through Gtr2p. Genetics. 1999;152(3):853–867. doi: 10.1093/genetics/152.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nguyen TP, Frank AR, Jewell JL. Amino acid and small GTPase regulation of mTORC1. Cell Logist. 2017;7(4):e1378794. doi: 10.1080/21592799.2017.1378794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nicklin P, Bergman P, Zhang BL, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136(3):521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oshiro N, Takahashi R, Yoshino KI, et al. The proline-rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mammalian target of rapamycin complex 1. J Biol Chem. 2007;282(28):20329–20339. doi: 10.1074/jbc.M702636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pal R, Palmieri M, Chaudhury A, et al. Src regulates amino acid-mediated mTORC1 activation by disrupting GATOR1-Rag GTPase interaction. Nat Commun. 2018;9(1):4351. doi: 10.1038/s41467-018-06844-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Park SG, Ewalt KL, Kim S. Functional expansion of aminoacyl-tRNA synthetases and their interacting factors: new perspectives on housekeepers. Trends Biochem Sci. 2005;30(10):569–574. doi: 10.1016/j.tibs.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 70.Parmigiani A, Nourbakhsh A, Ding BX, et al. Sestrins inhibit mTORC1 kinase activation through the GATOR complex. Cell Rep. 2014;9(4):1281–1291. doi: 10.1016/j.celrep.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peng M, Yin N, Li MO. Sestrins function as guanine nucleotide dissociation inhibitors for Rag GTPases to control mTORC1 signaling. Cell. 2014;159(1):122–133. doi: 10.1016/j.cell.2014.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peng M, Yin N, Li MO. SZT2 dictates GATOR control of mTORC1 signalling. Nature. 2017;543(7645):433–437. doi: 10.1038/nature21378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Perera RM, Zoncu R. The lysosome as a regulatory hub. Annu Rev Cell Dev Biol. 2016;32:223–253. doi: 10.1146/annurev-cellbio-111315-125125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peterson TR, Laplante M, Thoreen CC, et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137(5):873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Petit CS, Roczniak-Ferguson A, Ferguson SM. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol. 2013;202(7):1107–1122. doi: 10.1083/jcb.201307084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Potter CJ, Pedraza LG, Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nat Cell Biol. 2002;4(9):658–665. doi: 10.1038/ncb840. [DOI] [PubMed] [Google Scholar]
- 77.Quinlan CL, Kaiser SE, Bolaños B, et al. Targeting S-adenosylmethionine biosynthesis with a novel allosteric inhibitor of Mat2A. Nat Chem Biol. 2017;13(7):785–792. doi: 10.1038/nchembio.2384. [DOI] [PubMed] [Google Scholar]
- 78.Rebsamen M, Pochini L, Stasyk T, et al. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature. 2015;519(7544):477–481. doi: 10.1038/nature14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sabatini DM, Erdjument-Bromage H, Lui M, et al. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78(1):35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 80.Sabers CJ, Martin MM, Brunn GJ, et al. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J Biol Chem. 1995;270(2):815–822. doi: 10.1074/jbc.270.2.815. [DOI] [PubMed] [Google Scholar]
- 81.Saier MH, Jr, Reddy VS, Tsu BV, et al. The Transporter Classification Database (TCDB): recent advances. Nucleic Acids Res. 2016;44(D1):D372–D379. doi: 10.1093/nar/gkv1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saito K, Araki Y, Kontani K, et al. Novel role of the small GTPase Rheb: its implication in endocytic pathway independent of the activation of mammalian target of rapamycin. J Biochem. 2005;137(3):423–430. doi: 10.1093/jb/mvi046. [DOI] [PubMed] [Google Scholar]
- 83.Sancak Y, Thoreen CC, Peterson TR, et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25(6):903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 84.Sancak Y, Peterson TR, Shaul YD, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320(5882):1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sancak Y, Bar-Peled L, Zoncu R, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141(2):290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168(6):960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Saxton RA, Knockenhauer KE, Schwartz TU, et al. The apo-structure of the leucine sensor Sestrin2 is still elusive. Sci Signal. 2016;9(446):ra92. doi: 10.1126/scisignal.aah4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Saxton RA, Chantranupong L, Knockenhauer KE, et al. Mechanism of arginine sensing by CASTOR1 upstream of mTORC1. Nature. 2016;536(7615):229–233. doi: 10.1038/nature19079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Saxton RA, Knockenhauer KE, Wolfson RL, et al. Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science. 2016;351(6268):53–58. doi: 10.1126/science.aad2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schürmann A, Brauers A, Maßmann S, et al. Cloning of a novel family of mammalian GTP-binding proteins (RagA, RagBs, RagB1) with remote similarity to the Ras-related GTPases. J Biol Chem. 1995;270(48):28982–28988. doi: 10.1074/jbc.270.48.28982. [DOI] [PubMed] [Google Scholar]
- 91.Sekiguchi T, Hirose E, Nakashima N, et al. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J Biol Chem. 2001;276(10):7246–7257. doi: 10.1074/jbc.M004389200. [DOI] [PubMed] [Google Scholar]
- 92.Shen K, Sabatini DM. Ragulator and SLC38A9 activate the Rag GTPases through noncanonical GEF mechanisms. Proc Natl Acad Sci USA. 2018;115(38):9545–9550. doi: 10.1073/pnas.1811727115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shen K, Choe A, Sabatini DM. Intersubunit crosstalk in the Rag GTPase heterodimer enables mTORC1 to respond rapidly to amino acid availability. Mol Cell. 2017;68(3):552–565e8. doi: 10.1016/j.molcel.2017.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shen K, Huang RK, Brignole EJ, et al. Architecture of the human GATOR1 and GATOR1-Rag GTPases complexes. Nature. 2018;556(7699):64–69. doi: 10.1038/nature26158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shimobayashi M, Hall MN. Multiple amino acid sensing inputs to mTORC1. Cell Res. 2016;26(1):7–20. doi: 10.1038/cr.2015.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Son SM, Park SJ, Lee H, et al. Leucine signals to mTORC1 via its metabolite acetyl-coenzyme A. Cell Metab. 2019;29(1):192–201e7. doi: 10.1016/j.cmet.2018.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stracka D, Jozefczuk S, Rudroff F, et al. Nitrogen source activates TOR (target of rapamycin) complex 1 via glutamine and independently of Gtr/Rag proteins. J Biol Chem. 2014;289(36):25010–25020. doi: 10.1074/jbc.M114.574335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Su MY, Morris KL, Kim DJ, et al. Hybrid structure of the RagA/C-ragulator mTORC1 activation complex. Mol Cell. 2017;68(5):835–846e3. doi: 10.1016/j.molcel.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sutter BM, Wu X, Laxman S, et al. Methionine inhibits autophagy and promotes growth by inducing the SAM-responsive methylation of PP2A. Cell. 2013;154(2):403–415. doi: 10.1016/j.cell.2013.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Takagi Y, Kobayashi T, Shiono M, et al. Interaction of folliculin (Birt-Hogg-Dubé gene product) with a novel Fnip1-like (FnipL/Fnip2) protein. Oncogene. 2008;27(40):5339–5347. doi: 10.1038/onc.2008.261. [DOI] [PubMed] [Google Scholar]
- 101.Taylor PM. Role of amino acid transporters in amino acid sensing. Am J Clin Nutr. 2014;99(1):223S–230S. doi: 10.3945/ajcn.113.070086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tee AR, Manning BD, Roux PP, et al. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 2003;13(15):1259–1268. doi: 10.1016/S0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 103.Thomas JD, Zhang YJ, Wei YH, et al. Rab1A is an mTORC1 activator and a colorectal oncogene. Cancer Cell. 2014;26(5):754–769. doi: 10.1016/j.ccell.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tsun ZY, Bar-Peled L, Chantranupong L, et al. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell. 2013;52(4):495–505. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vander Haar E, Lee SI, Bandhakavi S, et al. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9(3):316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 106.Velasco-Miguel S, Buckbinder L, Jean P, et al. PA26, a novel target of the p53 tumor suppressor and member of the GADD family of DNA damage and growth arrest inducible genes. Oncogene. 1999;18(1):127–137. doi: 10.1038/sj.onc.1202274. [DOI] [PubMed] [Google Scholar]
- 107.Wang LF, Harris TE, Roth RA, et al. PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J Biol Chem. 2007;282(27):20036–20044. doi: 10.1074/jbc.M702376200. [DOI] [PubMed] [Google Scholar]
- 108.Wang SY, Tsun ZY, Wolfson RL, et al. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 2015;347(6218):188–194. doi: 10.1126/science.1257132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wolfson RL, Chantranupong L, Saxton RA, et al. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science. 2016;351(6268):43–48. doi: 10.1126/science.aab2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wolfson RL, Chantranupong L, Wyant GA, et al. KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature. 2017;543(7645):438–442. doi: 10.1038/nature21423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wright MD, Rudy GB, Ni J. The L6 membrane proteins–a new four-transmembrane superfamily. Protein Sci. 2000;9(8):1594–1600. doi: 10.1110/ps.9.8.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wu H, Wang FL, Hu SL, et al. MiR-20a and miR-106b negatively regulate autophagy induced by leucine deprivation via suppression of ULK1 expression in C2C12 myoblasts. Cell Signal. 2012;24(11):2179–2186. doi: 10.1016/j.cellsig.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 113.Wyant GA, Abu-Remaileh M, Wolfson RL, et al. mTORC1 activator SLC38A9 is required to efflux essential amino acids from lysosomes and use protein as a nutrient. Cell. 2017;171(3):642–654e12. doi: 10.1016/j.cell.2017.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xia J, Wang R, Zhang TL, et al. Structural insight into the arginine-binding specificity of CASTOR1 in amino acid-dependent mTORC1 signaling. Cell Discov, 2:16035. 2016 doi: 10.1038/celldisc.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yan XH, Sun QM, Ji J, et al. Reconstitution of leucine-mediated autophagy via the mTORC1-Barkor pathway in vitro. Autophagy. 2012;8(2):213–221. doi: 10.4161/auto.8.2.18563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yang HJ, Jiang XL, Li BR, et al. Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature. 2017;552(7685):368–373. doi: 10.1038/nature25023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yang HR, Wang J, Liu MJ, et al. 4.4 Å resolution Cryo-EM structure of human mTOR complex 1. Protein Cell. 2016;7(12):878–887. doi: 10.1007/s13238-016-0346-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yonehara R, Nada S, Nakai T, et al. Structural basis for the assembly of the Ragulator-Rag GTPase complex. Nat Commun. 2017;8(1):1625. doi: 10.1038/s41467-017-01762-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang TL, Wang R, Wang ZJ, et al. Structural basis for Ragulator functioning as a scaffold in membrane-anchoring of Rag GTPases and mTORC1. Nat Commun. 2017;8(1):1394. doi: 10.1038/s41467-017-01567-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zoncu R, Bar-Peled L, Efeyan A, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H+-ATPase. Science. 2011;334(6056):678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]