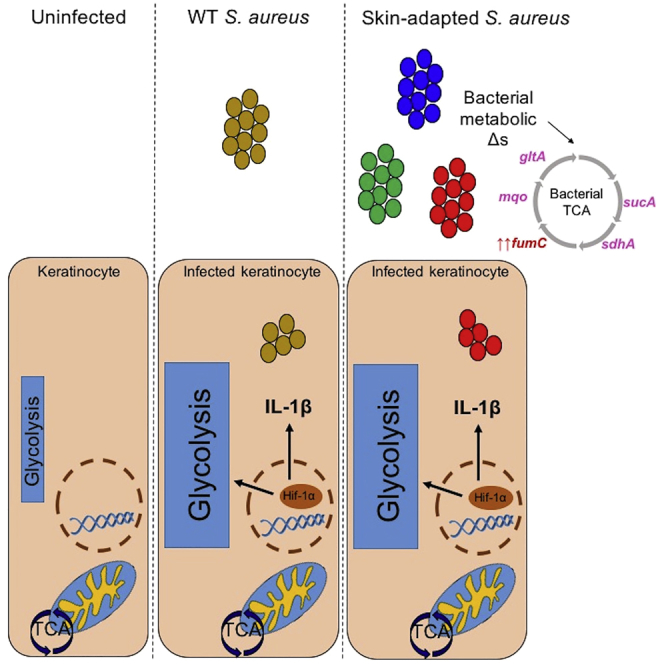
Summary
Staphylococcus aureus is the most common cause of skin and soft tissue infections, yet the bacterial genetic changes associated with adaptation to human skin are not well characterized. S. aureus strains isolated from patients with chronic skin colonization and intermittent infection were used to determine the staphylococcal genotypes or phenotypes associated with adaptation to human skin. We demonstrate that polymorphisms in metabolic genes, particularly those involved in the tricarboxylic acid cycle, the fumarate-succinate axis, and the generation of terminal electron transporters, are unexpectedly common. These skin-adapted strains activated glycolysis and hypoxia-inducible factor-1α, interleukin (IL)-1β, and IL-18 release from keratinocytes and promoted dermatopathology equivalent to a methicillin-resistant Staphylococcus aureus USA300 control in a murine model of infection. However, in contrast to USA300, a skin-adapted isolate failed to generate protection from a secondary infectious challenge. Within the context of human skin, there appears to be selection for S. aureus metabolic adaptive changes that promote glycolysis and maintain pathogenicity.
Subject Areas: Bacteriology, Microbial Genetics, Microbiome
Graphical Abstract
Highlights
-
•
Staphylococcus aureus is a metabolically adaptive organism
-
•
Skin-adapted isolates harbor mutations in fumC and other metabolic genes
-
•
Novel metabolic gene variants were identified in skin-adapted strains
Bacteriology; Microbial Genetics; Microbiome
Introduction
Staphylococcus aureus is a ubiquitous organism that both colonizes human skin as an innocuous component of the commensal flora and causes invasive infection. The many surface proteins and toxins expressed by S. aureus that contribute to its ability to cause skin infection have been well characterized (Geoghegan et al., 2018), as has the inflammatory response that these proteins elicit. Less well understood is how S. aureus adapts to the human skin and maintains a state of chronic colonization and infection. To determine how S. aureus adapts to human skin, we studied a collection of isolates from children with atopic dermatitis (AD) who are colonized and repeatedly infected with S. aureus (Guzik et al., 2005, Park et al., 2013, Totte et al., 2016). Although many host factors have been found to contribute to S. aureus infection in AD (Kaesler et al., 2014, Krishna and Miller, 2012, Sehra et al., 2010), we were interested in determining the changes in the organisms that are associated with the ability to adapt to skin. Specific S. aureus clonal complexes, agr genotypes, and virulence factors have not been consistently linked to the pathogenesis of infection or colonization in AD (Benito et al., 2016, Fleury et al., 2017, Geoghegan et al., 2018, Yeung et al., 2011). The activation of a robust inflammatory response mediated by interleukin (IL)-1β is thought to be important in the clearance of S. aureus skin infection (Miller et al., 2007), but the contribution of specific proinflammatory toxins or surface proteins has not been linked to these infections.
The metabolic pathways used by S. aureus in the context of human skin are closely linked to inflammation. Immune cells as well as keratinocytes rapidly alter their metabolic activities upon bacterial stimulation using glycolysis to generate ATP and induce hypoxia-inducible factor (HIF) 1α, an important metabolic and proinflammatory transcription factor (Tannahill et al., 2013). S. aureus must use glycolysis and stimulate keratinocyte glycolysis to establish skin infection as demonstrated by the inability of Δpyk mutants that are defective in glycolysis to infect skin (Vitko et al., 2015, Wickersham et al., 2017). Staphylococcal induction of keratinocyte glycolysis, even in the absence of toxin production, is sufficient to induce HIF1-α signaling and production of pro-IL-1β, a major proinflammatory cytokine (Wickersham et al., 2017). Activation of the inflammasome and IL-1β production in epithelial stem cells has been associated with epigenetic changes that confer protection from secondary challenge (Netea et al., 2016). This epigenetic reprogramming may be mediated by HIF-1α signaling (Cheng et al., 2014) and fumarate accumulation that modifies histone deacetylase activity and amplifies the cytokine response upon reinfection (Arts et al., 2016). Murine skin infection with virulent S. aureus confers local protection from reinfection at the same site, due in part to increased macrophage populations in the skin that become primed and confer protective immunity to subsequent infection (Chan et al., 2017, Chan et al., 2018). We postulated that the organisms associated with infection and colonization in patients with AD are selected for their ability to evade local immune clearance mechanisms and that this selective process involves metabolic adaptation to the skin.
In the studies detailed in this report, we characterized a group of 10 S. aureus isolates associated with chronic colonization or infection from patients with AD. Although no consistent changes in the genes associated with pathogen-associated molecular pattern or toxin expression were found, there was a striking accumulation of genetic polymorphisms or variants and changes in expression of genes that affect the tricarboxylic acid (TCA) cycle enzymes and terminal electron acceptors, metabolic changes that were found to have important immunological consequences.
Results
S. aureus Isolates Adapted to Human Skin Are Genotypically Diverse
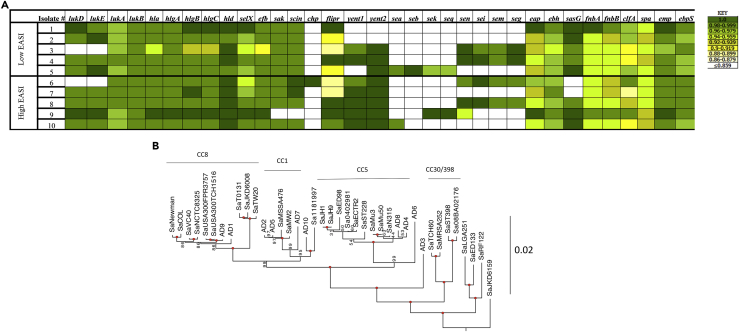
From a collection of 134 S. aureus strains isolated from 79 patients, we selected 10 isolates from patients with the five highest and five lowest Eczema Area and Severity Index (EASI) scores as representing strains that had adapted to human skin. These skin-adapted strains (AD 1–10) were used for whole-genome sequencing, transcriptomic analysis, and phenotypic analysis (Table 1). Screening assays for selected virulence factors indicated substantial heterogeneity and a lack of correlation with clinical score. All the skin-adapted strains contained genes encoding many major toxins and adhesins, including hla, fibronectin-binding protein, and clumping factor (Figure 1A). The hla gene was present in all isolates, whereas in AD3, the gene contained a premature stop codon, which correlated with decreased Hla expression (Soong et al., 2015). AD3 and AD6 also lacked genes for the leukotoxin LukED. Genes for arcA-ACME, speG-ACME, lukF-PVL, and lukS-PVL were not present in any of these isolates, suggesting that none of the clinical isolates had the classical suite of accessory genes found in many USA300 strains. Whole-genome phylogenies were constructed using both SNP-based and amino acid-based approaches along with 30 previously sequenced, publically available isolates to contextualize genetic diversity (Figure 1B, Data S1 and S2). The results of the phylogenetic analysis, which agreed with clonal complex groupings as determined from multilocus sequence typing (MLST), showed a sporadic distribution mostly among CC8, CC1, and CC5, without clustering by clinical EASI score.
Table 1.
Characteristics of S. aureus Strains Isolated from Patients with Atopic Dermatitis
| Isolate # | EASI Score |
Culture Site | Bacterial Burden | MRSA? | MLST(CC; spa Type) | Antibiotic Resistance | |
|---|---|---|---|---|---|---|---|
| Low EASI | AD1 | 4.5 | S | Few | MSSA | ST8(CC8; t1892) | EM |
| AD2 | 0.2 | S | Moderate | MSSA | ST1(CC1; t922) | ||
| AD3 | 2.4 | P | Moderate | MRSA | ST22(CC22; new) | OX | |
| AD4 | 3.1 | N | Few | MSSA | ST5 (CC5; t002) | EM | |
| AD5 | 2 | S | Moderate | MSSA | ST15(CC15; new) | EM | |
| High EASI | AD6 | 33.3 | S | Heavy | MSSA | ST9 (CC109; t209) | EM |
| AD7 | 33.6 | S | Few | MSSA | ST? (CC188; t189) | ||
| AD8 | 40 | S | Heavy | MSSA | ST5 (CC5; t045) | ||
| AD9 | 28.5 | S | Few | MRSA | ST8 (CC8; t008) | CM, EM, LVX, OX, TC | |
| AD10 | 40 | S | Heavy | MSSA | ST? (CC6; new) | EM |
CC, clonal complex; CM, clindamycin; EM, erythromycin; LVX, levofloxacin; MRSA, methicillin-resistant Staphylococcus aureus; N, nares; OX, oxacillin; P, perianal; S, skin; TC, tetracycline.
S. aureus isolates from low and high EASI (Eczema Area and Severity Index) score groups listed with site of culture, methicillin sensitivity, Ridom type, MLST, antibiotic resistance, and associated clinical EASI score.
Spa types labeled as “new” could not be assigned to a recognized type in the Ridom database, but had characteristics consistent with spa variable repeat regions.
Figure 1.
S. aureus Strains Isolated from Patients with Atopic Dermatitis Are Diverse
(A) Variable presence of toxin, resistance, and adhesion-associated genes. Heatmap display of Basic Local Alignment Search Tool (BLAST) results for specific gene sequence presence in each isolate compared with the reference strain USA300 FPR3757. Sequence identity (%) presented in KEY (right).
(B) Maximum likelihood phylogenetic tree from a concatenated open reading frame dataset, rooted with SaMSHR1132 (branch not shown). Bacterial isolates from patients with atopic dermatitis analyzed with 30 previously sequenced “sign-post” genomes that are publicly available. Values on or near the node are bootstrap support percentages; values of 100% are indicated with a red circle. The branch length reference is in substitutions/site.
Skin-Adapted S. aureus Strains Have Variants in Metabolic Genes
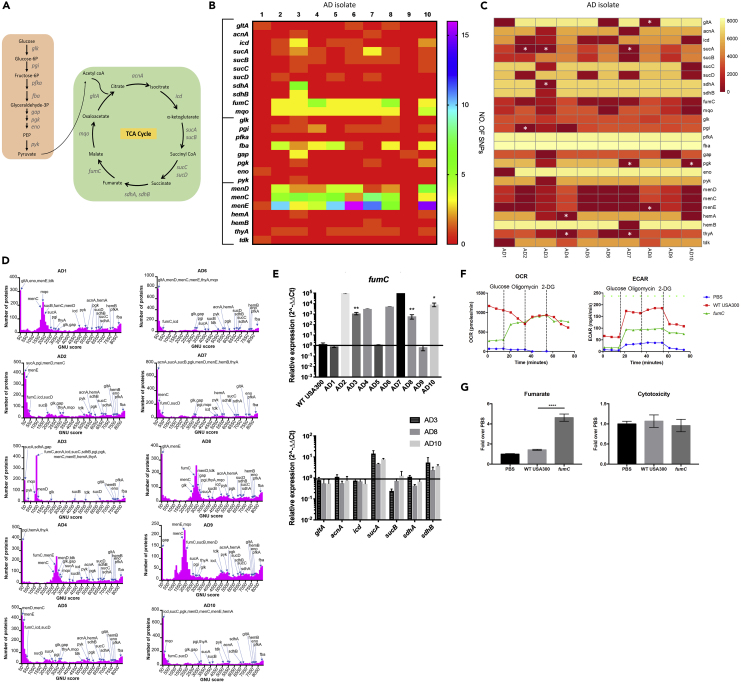
Whole-genome sequence data were analyzed to determine if adaptation to skin was reflected in the selection of specific mutations or variants (Figures 2A–2D). When compared with the sequence of the well-studied reference genome USA300 FPR3757, each of the clinical isolates had numerous nonsynonymous SNPs for key components of the TCA cycle, the glycolytic pathway, or terminal components of the electron transport chain, sites of mutation typically present in staphylococcal small-colony variants (SCVs) (Proctor et al., 2006) (Figures 2A and 2B). As a way to establish the novelty of the SNPs that we identified, we used a Gene Novelty Unit (GNU) score to determine how many times a specific protein variant had been previously identified in all available annotated S. aureus genomes in the database (8,524 genomes available at NCBI) (Figures 2C and 2D). The proteins from each isolate were matched with the S. aureus database, and each protein given a score (GNU score) that is the number of exact matches in the entire database. We asked how abundant each of the variants in metabolic genes identified in our clinical strains was in the entire database and generated a heatmap based on the GNU score for each metabolic protein (Figure 2C). We observed that some were entirely novel (GNU score = 0), whereas others were found repeatedly in the isolates in the database (GNU score > 7,000). This analysis highlights several sequence variants that are completely new as well as proteins that have higher levels of novelty (lower GNU scores) overall, especially those in the terminal components of the electron transport chain and those involved in the succinate-fumarate axis. There was a striking number of nonsynonymous sequence polymorphisms and novel protein variants involved in bacterial handling of oxidant stress, namely, the SCV-associated menadione genes, fumC, and fba encoding aldolase in the glycolytic pathway, which is involved in the production of glyceraldehyde 3-phosphate (glyceraldehyde-3P). Glyceraldehyde-3P is a product of the pentose phosphate shunt, a substrate for glycolysis, and an important source of the anti-oxidant NADH. Note that the presence of SNPs or novel protein variants does not necessarily reflect loss of function, but may in fact be associated with increased or decreased expression and function of that locus. Although we did note many SNPs and novel protein variants in menadione, hem, and mqo loci, these were not associated with obvious growth defects. The lack of growth defects is not unexpected as these strains were recovered from the clinical microbiology laboratory, which was less likely to identify SCVs under their routine culture conditions given the ability of SCVs to rapidly revert to a wild-type (WT) phenotype (Becker et al., 2006, Tuchscherr et al., 2011). The SNPs and protein variants identified in known SCV genes were more heterogeneous in the database (Figure 2C), suggesting that these are not likely novel to our clinical isolates.
Figure 2.
Skin-Adapted Isolates Harbor Metabolic Gene Variants
(A) Diagram of glycolysis and TCA cycle.
(B) Heatmap of number of nonsynonymous SNPs in skin-adapted isolates in TCA cycle, glycolytic, and SCV genes compared with the reference strain USA300 FPR3757.
(C) Heatmap of genes from 2B showing GNU score (number of exact protein matches [100% identity and 100% coverage]) in all known 8,524 S. aureus proteomes (as of 01/14/2019). Genes with 0 protein matches are indicated with a white asterisk.
(D) Ten histograms of GNU scores from 2C of all the proteins in each isolate. The x axis indicates the GNU score within a bin of 100 GNU scores in the S. aureus database, and the y axis indicates the number of proteins in the isolate's genome with a certain GNU score.
(E) qRT-PCR of TCA cycle genes in clinical S. aureus strains grown to exponential phase in lysogeny broth compared with USA300 WT LAC (representative graph of three individual experiments, n = 3 per condition). See also Table S3.
(F) Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) of uninfected HEKn cells and those stimulated with WT USA300 JE2 and fumC transposon mutant.
(G) Fumarate levels measured in cell media after 5-h infection in HEKn cells and associated cytotoxicity.
Each data point is the mean value ±SEM (n = 3) *p < 0.05, ∗∗p < 0.01, and ****p < 0.0001 by one-way (F) or two-way ANOVA (E).
To put the GNU scores of these genes into the context of the entire genome and to get a sense of how many proteins in the genome have a similar number of exact hits, we plotted GNU score histograms with the y axis displaying the number of proteins in the genome that have matches in the databases within a bin of 100 GNU scores. Thus, for the 10 skin-adapted isolates that we analyzed, all had variants in metabolic genes that were very novel with GNU scores of <50 (Figure 2D). We interpret this finding as an indication that such novel variants are more abundant in our collection of S. aureus from the skin, when compared with all the isolates from diverse sites that are represented in the database.
Eight of 10 skin-adapted strains had SNPs in fumC compared with USA300 and all 10 isolates had lower GNU scores at this locus; 5 of 10 were variants seen in less than 500 strains in the database, and the other 5 were present in less than 3,000 of 8,524 annotated genomes available at NCBI (Figures 2B and 2C). FumC was of interest as we noted its dramatic upregulation in our clinical isolates (Figure 2E) along with S. aureus isolates associated with chronic pulmonary infection (Gabryszewski et al., 2019). Moreover, SNPs in genes involved in succinate metabolism (sucA, B, C, D), immediately upstream of fumarate in the TCA cycle, were present in 8/10 of the clinical strains from skin. Of these, 3 were completely novel protein variants in the SucA enzyme (Figure 2A) that converts α ketoglutarate into succinyl CoA, the succinate, and hence fumarate, precursor. The clinical isolate AD3 also had a completely novel protein sequence for SdhA, the enzyme that catalyzes the conversion of succinate to fumarate.
The variants that we identified could be associated with either increased or decreased gene expression. The fumC locus was upregulated in 7/10 isolates by 1,000–100,000 fold compared with USA300 (Figure 2E). Fumarate can suppress glycolysis by binding glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Kornberg et al., 2018), a key component of the glycolytic pathway generating pyruvate from glucose. Increased fumC expression would limit fumarate production and facilitate glycolysis, which is required for S. aureus proliferation in skin (Wickersham et al., 2017). This was confirmed by demonstrating that a fumC mutant was associated with less induction of glycolysis in keratinocytes in vitro (Figure 2F) and significantly greater accumulation of fumarate, but no apparent effects on cytotoxicity (Figure 2G). Thus, it appears that a major selective pressure for S. aureus adaptation to skin is metabolic as a result of its need to use glycolysis to generate ATP and may account for increased fumC expression in some strains as well as the numerous polymorphisms in other genes involved in the TCA cycle.
Genetic Changes Associated with Metabolic Adaptation Variably Impact Cytokine Induction
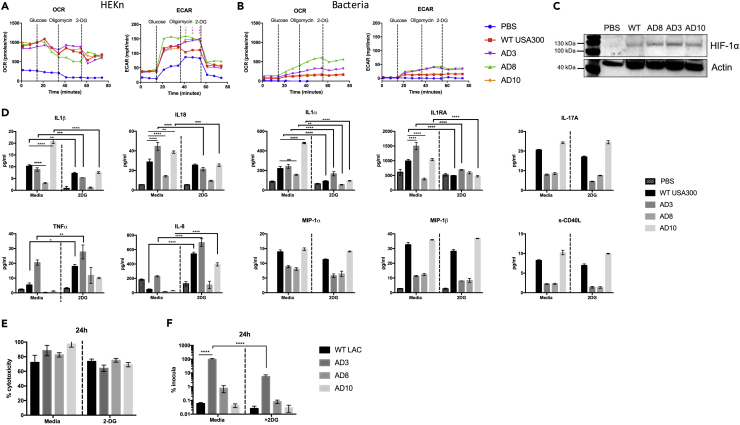
Given the metabolic genetic variants found in the skin isolates, we sought to determine the nature of the immune responses elicited in host keratinocytes and the association with glycolysis. We confirmed that three representative skin-adapted S. aureus strains and a USA300 control activate keratinocyte glycolysis (Figure 3A), a response attributed chiefly to keratinocyte metabolism (Figure 3B). The stabilization of HIF1α was stimulated by each of the S. aureus strains tested (Figure 3C). The IL-1 cytokines, IL-1β, IL-1α, IL-1RA, and IL-18, were each significantly suppressed in the absence of glycolysis (Figure 3D), an effect that was not due to cytotoxicity (Figure 3E) or differences in the uptake of the various strains by the keratinocytes (Figure 3F). The expression of the proinflammatory cytokines tumor necrosis factor (TNF)-α and IL-8, seemed to be compensatory, as their production was increased in keratinocytes treated with 2-deoxyglucose (2-DG). Induction of the monocyte or macrophage-specific cytokines and IL-17A was not dependent on glycolysis. Although we observed statistically significant differences in cytokine induction by the various strains, the biological relevance of these differences remains to be established.
Figure 3.
Skin-Adapted Strains Depend on Glycolysis to Stimulate Inflammatory Responses in Keratinocytes
(A and B) (A) OCR and ECAR of uninfected HEKns versus those stimulated with clinical AD strains and WT USA300 LAC S. aureus and (B) bacteria alone. Each data point is the mean value (n = 3) with *p < 0.05 by two-way ANOVA compared with WT.
(C) Immunoblots showing Hif1α in HEKn cells exposed to S. aureus for 4 h.
(D–F) (D) Cytokines measured by ELISA, and (E) cytoxicity and (F) intracellular persistence at 24 h in HEKn cells infected with S. aureus isolates and treated with and without 2-DG by lysostaphin protection assay. Each data point is the mean value ± SEM (n = 3 for cytotoxicity and cytokines, n = 9 for intracellular persistence), *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 by two-way ANOVA.
Metabolic Adaptation Impairs Protection against Secondary Infection
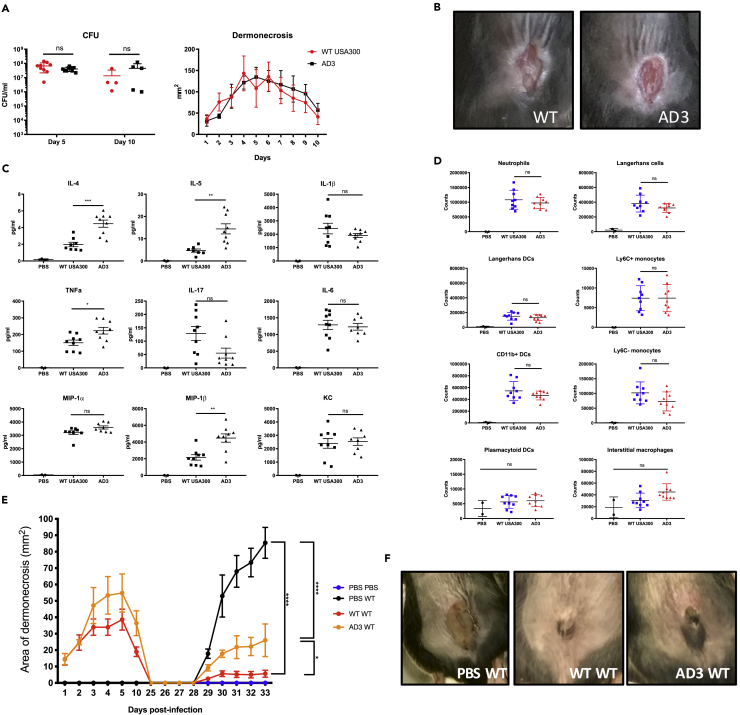
A murine model of cutaneous infection was used to compare the pathogenicity of an AD isolate and the USA300 LAC control strain. We studied the responses to infection caused by strain AD3, which had increased fumC and sucA expression as well as a completely novel amino acid sequence in SucA. Having first established that AD3 and USA300 had similar in vitro growth rates (Figure S1), we showed that both strains caused similar amounts of dermonecrosis and were cleared equally well over 10 days of infection (Figures 4A and 4B). AD3 elicited a significantly greater amount of TNFα, IL-4, and IL-5 (Figure 4C), the latter two cytokines being consistent with the Th2-skewed response characteristic of previous studies of strains from patients with AD (Mu et al., 2014). Immune cell recruitment into the skin lesions was also equivalent (Figure 4D).
Figure 4.
AD3 Generates Less Local Memory than USA300
(A) Bacterial load after 5 (n = 10) and 10 (n = 5) days, and dermonecrosis area over 10 days in mice intradermally infected with WT LAC or clinical isolate AD3.
(B) Images of mouse lesions after 5 days of infection.
(C) Cytokine measurements from skin biopsies of the infected sites after 5 days measured by ELISA.
(D) Immune cell recruitment 5 days after infection measured by fluorescence-activated cell sorting analysis. Results shown are pooled from two independent experiments.
(E) Area of dermonecrosis in mice infected with PBS, WT LAC, or AD3, followed by re-challenge at day 28 with WT LAC.
(F) Image of mouse lesions at day 33 from initial infection (5 days after challenge).
Each data point represents a mouse, and lines show mean values ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 determined by one- or two-way (E) ANOVA.
As the impact of the metabolic changes in the skin-adapted strains was not manifested in acute immunogenicity, we postulated that the accrued polymorphisms in metabolic genes might impact the generation of local immune memory affecting the induction of protection to a repeated infectious challenge. We compared the ability of WT USA300 and AD3 to induce local protective immunity against a secondary infection. Following complete resolution of the initial infection, caused by either USA300 LAC or AD3, the animals were re-challenged at the same site with WT USA300. Mice primed with WT USA300 infection developed memory and had minimal lesions upon re-challenge, whereas those initially infected with AD3 had significantly larger areas of dermonecrosis upon USA300 re-challenge (Figures 4E and 4F). Thus the genetic differences in AD3 likely impact the generation of local protection to secondary S. aureus challenge, even though the initial response to infection appears similar to a non-adapted USA300 strain.
Discussion
Based upon the genotypic and phenotypic characterization of S. aureus from chronically colonized patients, these studies illustrate that S. aureus dependence upon glycolysis for proliferation within the skin may promote the selection of strains with metabolic changes that promote glycolysis. We identified the accumulation of polymorphisms in genes likely to enhance glycolytic activity, limit the generation of fumarate, and in one case, prevent the development of local memory to secondary challenge. Our clinical strains were not phenotypically different from the isolates described in other reports characterizing strains from patients with AD (Byrd et al., 2017, Chiu et al., 2009, Kim et al., 2009), exhibiting substantial diversity and the presence of the genes encoding surface proteins and toxins typically associated with pathogenicity. However, genotypically, these isolates associated with chronic skin colonization or infection reflected a distinctive distribution of SNPs and protein variants in metabolic loci, many of which were not commonly found in the extensive collection of publically available S. aureus genomes. These multiple sequence variants in genes were associated with portions of the TCA cycle, as well as with loci involved in the production of terminal electron acceptors, including menadione and hemin loci, changes that are expected to promote glycolysis. Our analysis of protein variants from clinical isolates associated with skin colonization or infection compared with the S. aureus isolates in the NCBI suggests that the majority of the organisms in the database, which are unlikely to be skin adapted, rarely contain this same collection of variants in genes involved in succinate and fumarate metabolism and suggests that these variants are selected within the context of skin infection.
The fumC locus was targeted in the skin-associated S. aureus strains, and several of these isolates had protein variants that were infrequent in the S. aureus genomic database. Some of these protein variants were identified in strains with over 100-fold induction of fumC expression along with differences in gene expression in additional loci that contribute to succinate-fumarate homeostasis. The presence of these metabolic variants suggests that regulation of fumarate is especially important for S. aureus to persist in the skin, which is consistent with the S. aureus requirement for glycolysis to support its bioenergetic activity in the skin (Vitko et al., 2015, Wickersham et al., 2017). There are several possible roles for fumarate and fumC in S. aureus metabolism. The expression of fumC may be upregulated for fumarate utilization as a carbon source when glucose is limited (Michalik et al., 2017, Surmann et al., 2014), or fumarate may function to decrease oxidant stress, as it increases glutathione, a major oxidant trap within the cell. Another possibility is that the SNPs identified in our isolates may result in FumC inactivation leading to upregulation of fumC expression by the host cell to compensate for fumarate accumulation within the cell. Most consistent with our findings is fumarate suppression of aerobic glycolysis by binding directly to GAPDH, which provides substrates for glycolytic metabolism (Kornberg et al., 2018). Hence, increased fumC expression by S. aureus isolates may lead to increased fumarate hydrolysis and prevent suppression of glycolysis. Of note, fumarate is also associated with selective effects on CD4 versus CD8 T cell function and the induction of IL-4 production (Ghoreschi et al., 2011, Mills et al., 2018), an immunological phenotype characteristic of AD (Weidinger et al., 2018).
The reliance of S. aureus upon glycolysis for its bioenergetic needs in the setting of keratinocyte infection provides a unifying theme to help explain the diversity in the phenotypes and genotypes of strains associated with skin infection in humans. As previously established, staphylococcal induction of glycolysis is associated with the stabilization of HIF-1α and IL-1β production (Wickersham et al., 2017). Although not all the proinflammatory genes activated in keratinocytes by S. aureus were influenced by glycolysis, the major members of the IL-1 family including IL-1β, an important factor in an effective response to skin infection (Miller et al., 2007), were suppressed in the presence of 2-DG. Despite variability in the presence of toxin and other virulence genes, the inherent immunostimulatory response to S. aureus induction of glycolysis was sufficient to activate a robust immune response. This helps to explain how even agr mutants and strains lacking toxins could be associated with AD flares (Soong et al., 2015).
A major goal of this study was to establish the genetic changes in S. aureus that facilitate chronic skin infection. We demonstrate that there are metabolic changes in skin-adapted strains that function to promote staphylococcal glycolytic activity. We compared two unrelated strains with distinct phenotypes, a skin-adapted AD strain and a virulent USA300 LAC, which both activated similar immune responses and dermonecrosis, but had very different metabolic profiles. Although S. aureus is highly adapted to evade host immune responses, our studies indicate that adaptation to the local metabolic milieu is also important in the skin. Numerous epithelial and immune defects may contribute to the susceptibility of patients with AD to S. aureus infection. Our data indicate that the ability of S. aureus to adapt to the metabolic conditions imposed by skin is critical in establishing S. aureus colonization as well as susceptibility to reinfection.
Limitations of the Study
Comparing clinical strains genotypically is limited by the difficulty in establishing an appropriate reference strain for each clinical isolate, and thus the variability in the genotypic background cannot be entirely accounted for, which is why we developed a GNU score to better appreciate the significance of the SNPs identified in these strains. The phenotypic analysis of local immune memory was limited by using two strains with varying genetic backgrounds, and thus we can postulate but cannot deduce that the differing response is due to metabolic differences between a skin-adapted and WT strain.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This work was supported by NIH grant R01AI103854 to A.P., NIH grants 1K08AI101005 and 1R01AI137526-01 to P.J.P., NIH grant S10RR027050 to the Columbia Center for Translational Immunology Flow Cytometry Core, and NIH NIAID T32 Training Grant in Pediatric Infectious Diseases (AI007531).
Author Contributions
Conceptualization, K.P.A., T.W.F.L., A.P., P.J.P., C.L., and E.W.; Formal Analysis and Investigation, K.P.A., T.W.F.L, E.W., J.C., A.N., H.S., K.O., A.M.M., C.L., and P.J.P.; Writing – Original Draft, K.P.A. and A.P.; Writing – Review and Editing: K.P.A., T.W.F.L., A.P., P.J.P., and A.M.M.; Funding Acquisition, A.P. and P.J.P.; Supervision: P.J.P. and A.P.
Declaration of Interests
The authors declare no competing interests.
Published: September 27, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2019.07.037.
Supplemental Information
References
- Arts R.J., Novakovic B., Ter Horst R., Carvalho A., Bekkering S., Lachmandas E., Rodrigues F., Silvestre R., Cheng S.C., Wang S.Y. Glutaminolysis and fumarate accumulation integrate immunometabolic and epigenetic programs in trained immunity. Cell Metab. 2016;24:807–819. doi: 10.1016/j.cmet.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K., Laham N.A., Fegeler W., Proctor R.A., Peters G., von Eiff C. Fourier-transform infrared spectroscopic analysis is a powerful tool for studying the dynamic changes in Staphylococcus aureus small-colony variants. J. Clin. Microbiol. 2006;44:3274–3278. doi: 10.1128/JCM.00847-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito D., Aspiroz C., Gilaberte Y., Sanmartin R., Hernandez-Martin A., Alonso M., Gomez P., Lozano C., Torres C. Genetic lineages and antimicrobial resistance genotypes in Staphylococcus aureus from children with atopic dermatitis: detection of clonal complexes CC1, CC97 and CC398. J. Chemother. 2016;28:359–366. doi: 10.1179/1973947815Y.0000000044. [DOI] [PubMed] [Google Scholar]
- Byrd A.L., Deming C., Cassidy S.K.B., Harrison O.J., Ng W.I., Conlan S., Belkaid Y., Segre J.A., Kong H.H. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci. Transl. Med. 2017;9:eaal4651. doi: 10.1126/scitranslmed.aal4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan L.C., Chaili S., Filler S.G., Miller L.S., Solis N.V., Wang H., Johnson C.W., Lee H.K., Diaz L.F., Yeaman M.R. Innate immune memory contributes to host defense against recurrent skin and skin structure infections caused by methicillin-resistant Staphylococcus aureus. Infect. Immun. 2017;85 doi: 10.1128/IAI.00876-16. e00876–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan L.C., Rossetti M., Miller L.S., Filler S.G., Johnson C.W., Lee H.K., Wang H., Gjertson D., Fowler V.G., Jr., Reed E.F. Protective immunity in recurrent Staphylococcus aureus infection reflects localized immune signatures and macrophage-conferred memory. Proc. Natl. Acad. Sci. U S A. 2018;115:E11111–E11119. doi: 10.1073/pnas.1808353115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S.C., Quintin J., Cramer R.A., Shepardson K.M., Saeed S., Kumar V., Giamarellos-Bourboulis E.J., Martens J.H., Rao N.A., Aghajanirefah A. mTOR- and HIF-1alpha-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345:1250684. doi: 10.1126/science.1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu L.S., Ho M.S., Hsu L.Y., Tang M.B. Prevalence and molecular characteristics of Staphylococcus aureus isolates colonizing patients with atopic dermatitis and their close contacts in Singapore. Br. J. Dermatol. 2009;160:965–971. doi: 10.1111/j.1365-2133.2009.09038.x. [DOI] [PubMed] [Google Scholar]
- Fleury O.M., McAleer M.A., Feuillie C., Formosa-Dague C., Sansevere E., Bennett D.E., Towell A.M., McLean W.H.I., Kezic S., Robinson D.A. Clumping factor B promotes adherence of staphylococcus aureus to corneocytes in atopic dermatitis. Infect. Immun. 2017;85 doi: 10.1128/IAI.00994-16. e00994–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabryszewski S.J., Wong Fok Lung T., Annavajhala M.K., Tomlinson K.L., Riquelme S.A., Khan I.N., Noguera L.P., Wickersham M., Zhao A., Mulenos A.M. Metabolic adaptation in MRSA pneumonia. Am. J. Respir. Cell Mol. Biol. 2019;61:185–197. doi: 10.1165/rcmb.2018-0389OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan J.A., Irvine A.D., Foster T.J. Staphylococcus aureus and atopic dermatitis: a complex and evolving relationship. Trends Microbiol. 2018;26:484–497. doi: 10.1016/j.tim.2017.11.008. [DOI] [PubMed] [Google Scholar]
- Ghoreschi K., Brück J., Kellerer C., Deng C., Peng H., Rothfuss O., Hussain R.Z., Gocke A.R., Respa A., Glocova I. Fumarates improve psoriasis and multiple sclerosis by inducing type II dendritic cells. J. Exp. Med. 2011;208:2291–2303. doi: 10.1084/jem.20100977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzik T.J., Bzowska M., Kasprowicz A., Czerniawska-Mysik G., Wojcik K., Szmyd D., Adamek-Guzik T., Pryjma J. Persistent skin colonization with Staphylococcus aureus in atopic dermatitis: relationship to clinical and immunological parameters. Clinical and experimental allergy. J. Br. Soc. Allergy Clin. Immunol. 2005;35:448–455. doi: 10.1111/j.1365-2222.2005.02210.x. [DOI] [PubMed] [Google Scholar]
- Kaesler S., Volz T., Skabytska Y., Koberle M., Hein U., Chen K.M., Guenova E., Wolbing F., Rocken M., Biedermann T. Toll-like receptor 2 ligands promote chronic atopic dermatitis through IL-4-mediated suppression of IL-10. J. Allergy Clin. Immunol. 2014;134:92–99. doi: 10.1016/j.jaci.2014.02.017. [DOI] [PubMed] [Google Scholar]
- Kim D.W., Park J.Y., Park K.D., Kim T.H., Lee W.J., Lee S.J., Kim J. Are there predominant strains and toxins of Staphylococcus aureus in atopic dermatitis patients? Genotypic characterization and toxin determination of S. aureus isolated in adolescent and adult patients with atopic dermatitis. J. Dermatol. 2009;36:75–81. doi: 10.1111/j.1346-8138.2009.00592.x. [DOI] [PubMed] [Google Scholar]
- Kornberg M.D., Bhargava P., Kim P.M., Putluri V., Snowman A.M., Putluri N., Calabresi P.A., Snyder S.H. Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science. 2018;360:449–453. doi: 10.1126/science.aan4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna S., Miller L.S. Innate and adaptive immune responses against Staphylococcus aureus skin infections. Semin. Immunopathol. 2012;34:261–280. doi: 10.1007/s00281-011-0292-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalik S., Depke M., Murr A., Gesell Salazar M., Kusebauch U., Sun Z., Meyer T.C., Surmann K., Pförtner H., Hildebrandt P. A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions. Sci. Rep. 2017;7:9718. doi: 10.1038/s41598-017-10059-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L.S., Pietras E.M., Uricchio L.H., Hirano K., Rao S., Lin H., O'Connell R.M., Iwakura Y., Cheung A.L., Cheng G. Inflammasome-mediated production of IL-1beta is required for neutrophil recruitment against Staphylococcus aureus in vivo. J. Immunol. 2007;179:6933–6942. doi: 10.4049/jimmunol.179.10.6933. [DOI] [PubMed] [Google Scholar]
- Mills E.A., Ogrodnik M.A., Plave A., Mao-Draayer Y. Emerging understanding of the mechanism of action for dimethyl fumarate in the treatment of multiple sclerosis. Front. Neurol. 2018;9:5. doi: 10.3389/fneur.2018.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Z., Zhao Y., Liu X., Chang C., Zhang J. Molecular biology of atopic dermatitis. Clin. Rev. Allergy Immunol. 2014;47:193–218. doi: 10.1007/s12016-014-8415-1. [DOI] [PubMed] [Google Scholar]
- Netea M.G., Joosten L.A., Latz E., Mills K.H., Natoli G., Stunnenberg H.G., O'Neill L.A., Xavier R.J. Trained immunity: a program of innate immune memory in health and disease. Science. 2016;352:aaf1098. doi: 10.1126/science.aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.Y., Kim C.R., Huh I.S., Jung M.Y., Seo E.Y., Park J.H., Lee D.Y., Yang J.M. Staphylococcus aureus colonization in acute and chronic skin lesions of patients with atopic dermatitis. Ann. Dermatol. 2013;25:410–416. doi: 10.5021/ad.2013.25.4.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor R.A., von Eiff C., Kahl B.C., Becker K., McNamara P., Herrmann M., Peters G. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 2006;4:295–305. doi: 10.1038/nrmicro1384. [DOI] [PubMed] [Google Scholar]
- Sehra S., Yao Y., Howell M.D., Nguyen E.T., Kansas G.S., Leung D.Y., Travers J.B., Kaplan M.H. IL-4 regulates skin homeostasis and the predisposition toward allergic skin inflammation. J. Immunol. 2010;184:3186–3190. doi: 10.4049/jimmunol.0901860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soong G., Paulino F., Wachtel S., Parker D., Wickersham M., Zhang D., Brown A., Lauren C., Dowd M., West E. Methicillin-resistant Staphylococcus aureus adaptation to human keratinocytes. mBio. 2015;6 doi: 10.1128/mBio.00289-15. e00289–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surmann K., Michalik S., Hildebrandt P., Gierok P., Depke M., Brinkmann L., Bernhardt J., Salazar M.G., Sun Z., Shteynberg D. Comparative proteome analysis reveals conserved and specific adaptation patterns of Staphylococcus aureus after internalization by different types of human non-professional phagocytic host cells. Front. Microbiol. 2014;5:392. doi: 10.3389/fmicb.2014.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannahill G.M., Curtis A.M., Adamik J., Palsson-McDermott E.M., McGettrick A.F., Goel G., Frezza C., Bernard N.J., Kelly B., Foley N.H. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totte J.E., van der Feltz W.T., Hennekam M., van Belkum A., van Zuuren E.J., Pasmans S.G. Prevalence and odds of Staphylococcus aureus carriage in atopic dermatitis: a systematic review and meta-analysis. Br. J. Dermatol. 2016;175:687–695. doi: 10.1111/bjd.14566. [DOI] [PubMed] [Google Scholar]
- Tuchscherr L., Medina E., Hussain M., Volker W., Heitmann V., Niemann S., Holzinger D., Roth J., Proctor R.A., Becker K. Staphylococcus aureus phenotype switching: an effective bacterial strategy to escape host immune response and establish a chronic infection. EMBO Mol. Med. 2011;3:129–141. doi: 10.1002/emmm.201000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitko N.P., Spahich N.A., Richardson A.R. Glycolytic dependency of high-level nitric oxide resistance and virulence in Staphylococcus aureus. mBio. 2015;6 doi: 10.1128/mBio.00045-15. e00045–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidinger S., Beck L.A., Bieber T., Kabashima K., Irvine A.D. Atopic dermatitis. Nat. Rev. Dis. Primers. 2018;4:1. doi: 10.1038/s41572-018-0001-z. [DOI] [PubMed] [Google Scholar]
- Wickersham M., Wachtel S., Wong Fok Lung T., Soong G., Jacquet R., Richardson A., Parker D., Prince A. Metabolic stress drives keratinocyte defenses against Staphylococcus aureus infection. Cell Rep. 2017;18:2742–2751. doi: 10.1016/j.celrep.2017.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung M., Balma-Mena A., Shear N., Simor A., Pope E., Walsh S., McGavin M.J. Identification of major clonal complexes and toxin producing strains among Staphylococcus aureus associated with atopic dermatitis. Microbes Infect. 2011;13:189–197. doi: 10.1016/j.micinf.2010.10.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.