Abstract
Diabetic cardiomyopathy (DCM) is a vital cause of fatalities in diabetic patients. The programmed death of cardiomyocytes and inflammation critically contribute to cardiac hypertrophy and fibrosis in DCM. Furthermore, circular RNA (circRNA) is a key regulator of various diseases. However, the role of circRNAs in DCM remains to be elucidated. Our previous study found that pyroptosis was markedly activated in the cardiomyocytes subjected to high-glucose conditions, and miR-214-3p regulated the expression of caspase-1. The aim of this study was to elucidate whether circRNA is involved in DCM pyroptosis via the miR-214-3p/caspase-1 pathway. Herein, we identified that hsa_circ_0076631, named caspase-1-associated circRNA (CACR), was increased both in high-glucose-treated cardiomyocytes and in the serum of diabetic patients. CACR also sponged an endogenous miR-214-3p to sequester and inhibit its expression. CACR knockdown in cardiomyocytes counteracted high-glucose-induced caspase-1 activation. Conversely, miR-214-3p knockdown partially abolished the beneficial effects of CACR silencing on pyroptosis in cardiomyocytes. Therefore, this study elucidated that CACR might be a novel therapeutic target via the CACR/miR-214-3p/caspase-1 pathway in DCM.
Keywords: circular RNA, miR-214-3p, caspase-1, diabetic cardiomyopathy, pyroptosis
Introduction
Diabetic cardiomyopathy (DCM), characterized by myocardial structural and functional dysfunction exclusive of hypertension, coronary artery disease, and other cardiac pathologies, is a critical cause of fatalities in chronic diabetes mellitus patients.1 Hyperglycemia, insulin resistance, increased fatty acid metabolism, microcirculatory changes, myocardial fibrosis, cardiomyocyte death, inflammation, and calcium overload collectively contribute to the pathophysiology of DCM.2 Among these causes, cardiomyocyte death, which initiates the cardiac dysfunction, is considered the most basic pathological change of DCM.3, 4 However, the mechanisms involved in DCM remain to be elucidated.
Pyroptosis, which is pro-inflammatory programmed cell death, depends mainly on the activity of caspase-1.5, 6, 7, 8 Previous studies have demonstrated that pyroptosis is activated in DCM.4, 9, 10 Hyperglycemia can lead to a chronic low-grade inflammation state in the body by inducing high levels of reactive oxygen species (ROS), which stimulate nucleotide-binding oligomerization domain-like receptor pyrin domain containing 3 (NLRP3) inflammasome activation.11 This process cleaves pro-caspase-1 into the active caspase-1 form. Then, the activated caspase-1 facilitates the cleavage of downstream cytokines, such as pro-interleukin-1β (pro-IL-1β) and pro-IL-18, into their mature forms.12, 13 Particularly, mature IL-1β and IL-18 are the common initiating points of many pathophysiological processes in DCM, including cardiomyocyte apoptosis and fibroblast activation.14, 15, 16, 17 Therefore, myocardial pyroptosis plays a vital role in DCM. However, the mechanisms underlying the regulation of pyroptosis in DCM are still unclear.
Modern molecular biology techniques indicate that 98% of the genome cannot code proteins, and these molecules are called non-coding RNAs (ncRNAs). MicroRNAs (miRNAs) have been shown to regulate many physiological processes via sponging mRNAs. Our previous study revealed that miR-214-3p has binding sites with caspase-1 and could regulate pyroptosis in DCM.18 However, how to regulate miR-214-3p in DCM requires further clarification.
Circular RNA (circRNA) is a widely studied type of ncRNA that is highly conserved in mammals.19 Different from linear RNAs, circRNAs have closed loop structures without 3′ and 5′ ends. Previous studies have revealed that circRNAs can regulate gene expression, especially when they act as competing endogenous RNAs (ceRNAs) targeting miRNAs.20 CircRNAs have been demonstrated to participate in the occurrence and development of various diseases, including tumors, cardiovascular diseases, diabetes, and diabetic complications.21, 22, 23, 24 However, there is no research on the relationship between circRNAs and pyroptosis in DCM.
In this study, a bioinformatics prediction indicated that hsa_circ_0076631 regulated caspase-1 by targeting miR-214-3p. Here, we characterize the expression of hsa_circ_0076631 in DCM and determine its functions and mechanisms in the regulation of DCM.
Results
miR-214-3p Regulates caspase-1 Expression in High-Glucose-Treated Cardiomyocytes
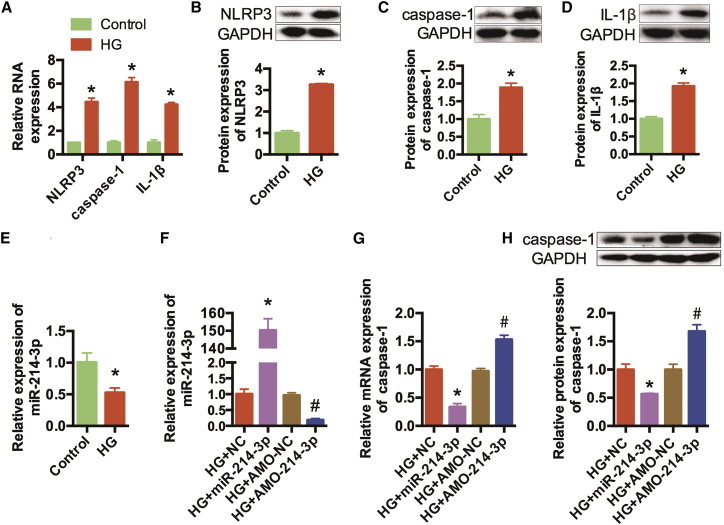
Our previous study found that high-glucose conditions activated pyroptosis in cardiomyocytes. We cultured AC16 cells, a human cardiac cell line, in high-glucose medium to simulate diabetes conditions. qRT-PCR and western blotting indicated that the expression levels of NLRP3, caspase-1, and IL-1β were increased in the high-glucose (HG) group (Figures 1A–1D). Therefore, pyroptosis was activated in high-glucose-treated cardiomyocytes.
Figure 1.
miR-214-3p Inhibited Pyroptosis Activation in High-Glucose-Treated AC16 Cells
(A) qRT-PCR was conducted to detect the expression levels of NLRP3, caspase-1, and IL-1β in AC16 cells. (B–D) Western blots were conducted to detect the protein expression levels of NLRP3 (B), caspase-1 (C), and IL-1β(Δ). GAPDH was detected as the internal control. (E) qRT-PCR was conducted to detect the expression levels of miR-214-3p in AC16 cells. *p < 0.05 versus the control group. (F) High-glucose-treated AC16 cells were transfected with NC, miR-214-3p mimics, AMO-NC, and AMO-214-3p, and miR-214-3p expression levels were detected by qRT-PCR. (G) The relative mRNA expression levels of caspase-1 were detected. (H) Western blots were performed to detect the protein expression of caspase-1. n = 3. *p < 0.05 versus the HG+NC group, #p < 0.05 versus the HG+AMO-NC group.
A previous study indicated that miR-214-3p had potential binding sites on caspase-1 and regulated pyroptosis in cataracts.25 We also confirmed that miR-214-3p can regulate caspase-1 in high-glucose-treated cardiomyocytes.18 In this experiment, we detect the expression of miR-214-3p in DCM models. The results indicated that miR-214-3p was evidently reduced in the HG group (Figure 1E). Then, function acquisition and deletion experiments were performed. High-glucose-treated AC16 cells were transfected separately with negative control (NC), miR-214 mimics, antisense oligonucleotide (AMO)-NC, or AMO for miR-214-3p (AMO-214-3p), and the transfection efficiencies were proven by qRT-PCR (Figure 1F). qRT-PCR and western blotting indicated that the expression level of caspase-1 was decreased significantly after transfection with miR-214-3p mimics, and this effect was reversed by AMO-214-3p (Figures 1G and 1H). Collectively, these findings indicated that caspase-1 is a target gene of miR-214-3p in high-glucose-treated cardiomyocytes.
CACR Silencing Regulates caspase-1 and miR-214-3p in Cardiomyocytes
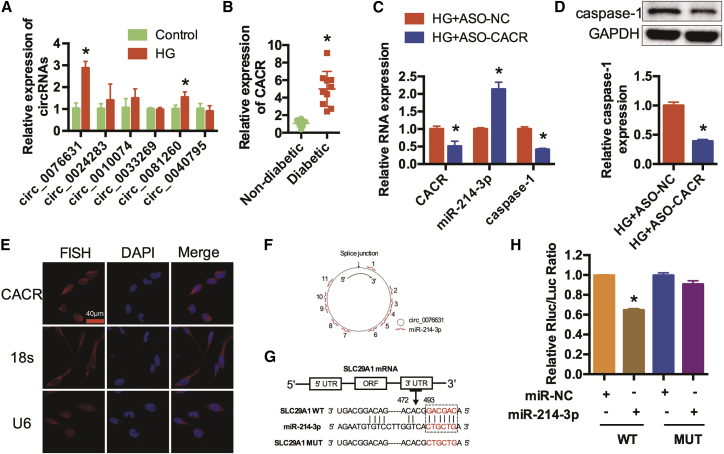
Emerging evidence has demonstrated that circRNAs regulate miRNA expression and function as competing endogenous sponges. Consequently, we predicted potential circRNAs that possibly bind to miR-214-3p by bioinformatics analysis. Six candidates were predicted to have potential binding sites with miR-214-3p via the MiRanda, Pita, and RNAhybrid databases. qRT-PCR was performed to detect their expression in high-glucose-treated cardiomyocytes (Figure 2A). Among them, hsa_circ_0076631, we named caspase-1-associated circRNA (CACR), was increased in the HG group. In addition, qRT-PCR indicated that CACR was increased in the serum of diabetic patients (Figure 2B). CACR was silenced to further determine its function in high-glucose-treated AC16 cells. qRT-PCR showed that silencing CACR obviously increased miR-214-3p expression levels and decreased caspase-1 expression levels (Figure 2C). Similarly, the protein expression level of caspase-1 was remarkably reduced in high-glucose-treated AC16 cells transfected with antisense oligonucleotides for CACR (ASO-CACR) (Figure 2D). In addition, we detected the protein expression levels of caspase-3 to assess the effects of CACR on apoptosis. The results showed that silencing CACR slightly inhibited apoptosis in high-glucose-treated AC16 cells (Figure S1).
Figure 2.
CACR Silencing Regulates Caspase-1 and miR-214-3p in Cardiomyocytes
(A) qRT-PCR was used to detect six candidate circRNAs that have potential binding sites with miR-214-3p in AC16 cells. n = 3. (B) CACR expression levels in the serum of non-diabetic patients and diabetic patients. n = 10. (C) High-glucose-treated AC16 cells were transfected with ASO-NC and ASO-CACR. The relative expression levels of CACR, miR-214-3p, and caspase-1 were detected by qRT-PCR. (D) Relative caspase-1 protein expression was detected in AC16 cells. (E) FISH assays were performed to detect the location of CACR in AC16 cells. CACR, 18 s, U6, red; nuclei, blue. Scale bar, 40 μm. (F) CACR contains 11 potential binding sites complementary to miR-214-3p as analyzed by bioinformatics programs. (G) SLC29A1 is the corresponding linear sequence of CACR. The binding sequence of SLC29A1 and miR-214-3p and the mutant sequence of SLC29A1 are shown. (H) Wide-type (WT) and mutant type (MUT) SLC29A1 were cloned downstream of the luciferase vector and co-transfected with miR-NC and miR-214-3p mimics into HEK293 cells. Luciferase activity was detected using a luciferase assay. The relative Rluc/Luc ratio is shown. n = 3. *p < 0.05.
Fluorescence in situ hybridization (FISH) assays indicated that hsa_circ_0076631 is located in both the nucleus and the cytoplasm (Figure 2E). The CACR isoform located at chr6: 44194999-44200165 in the human genome is SLC29A1. We found that CACR contained 11 potential binding sites on miR-214-3p (Figure 2F). These binding sites were cloned into a luciferase vector, and miR-214-3p mimics were co-transfected into HEK293T cells. Luciferase activity was significantly suppressed when co-transfected with miR-214-3p mimics for position of 472-493, whereas the miR-214-3p target site mutation reversed this repression (Figures 2G and 2H). In summary, these results revealed that CACR could regulate caspase-1 expression and function as a ceRNA of miR-214-3p in cardiomyocytes.
CACR Silencing Alleviates Caspase-1 via miR-214-3p in Cardiomyocytes
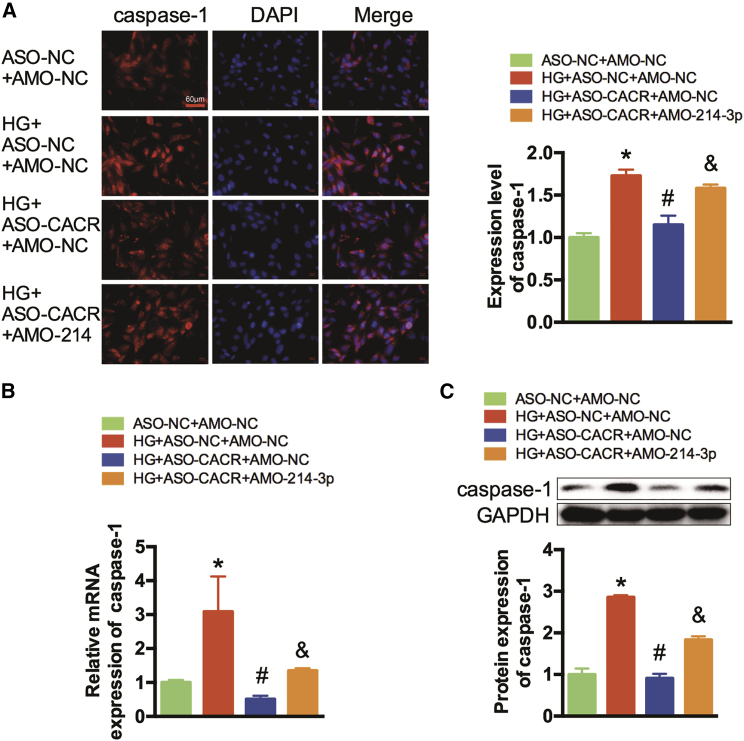
We further investigated the role of miR-214-3p in the effect of CACR expression on caspase-1 in cardiomyocytes. Immunofluorescence staining showed that after transfection with ASO-CACR, caspase-1 expression levels were significantly decreased in high-glucose-treated AC16 cells, while this inhibition was attenuated by co-transfection with AMO-214-3p (Figure 3A). Simultaneously, qRT-PCR and western blotting also reflected similar trends (Figures 3B and 3C). In addition, we also found that the protein expression level of caspase-1 was higher in the HG+ASO-NC+AMO-214-3p group than in the HG+ASO-NC+AMO-NC and HG+ASO-CACR+AMO-214-3p groups (Figure S2). Therefore, we ruled out the possibility that AMO-214-3p affects pyroptosis and demonstrated that CACR regulates pyroptosis by sponging miR-214-3p.
Figure 3.
CACR Regulates Caspase-1 Expression via Sponging miR-214-3p in Cardiomyocytes
(A) Immunofluorescence analysis was performed to detect the expression level of caspase-1. A representative image is shown in the left panel. Caspase-1, red; nuclei, blue. Scale bar, 60 μm. The calculated fluorescence intensity is shown in the right panel. (B) The relative mRNA expression levels of caspase-1 were detected by qRT-PCR. (C) Western blots were performed to detect the protein expression of caspase-1. n = 3. *p < 0.05 compared with the ASO-NC+AMO-NC group, #p < 0.05 compared with the HG+ASO-NC+AMO-NC group, &p < 0.05 compared with the HG+ASO-CACR+AMO-NC group.
CACR Regulates Pyroptosis via the miR-214-3p/caspase-1 Pathway in AC16 Cells
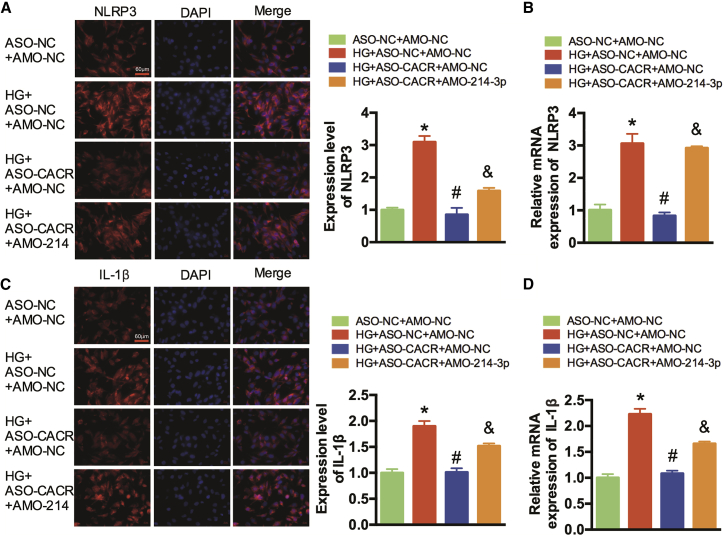
We also detected other major factors in the pyroptosis signaling pathway. Immunofluorescence staining and qRT-PCR showed that the expression levels of NLRP3 and IL-1β were significantly decreased after silencing ASO-CACR, but this improvement was abolished after transfection with AMO-214-3p (Figures 4A–4D). In addition, the NLRP3 mRNA expression level in HG+ASO-NC+AMO-214-3p group was detected to rule out the effect of AMO-214-3p on NLRP3 (Figure S3).
Figure 4.
CACR Regulates Pyroptosis via the miR-214-3p/Caspase-1 Pathway in AC16 Cells
(A) Immunofluorescence analysis was performed to detect the expression levels of NLRP3. A representative image is shown in the left panel. NLRP3, red; DAPI, blue. Scale bar, 60 μm. The calculated fluorescence intensity is shown in the right panel. (B) The relative expression levels of NLRP3 were detected by qRT-PCR. (C) Immunofluorescence analysis was performed to detect the expression levels of IL-1β. (D) The relative expression levels of IL-1β were detected by qRT-PCR. n = 3. *p < 0.05 compared with the ASO-NC+AMO-NC group, #p < 0.05 compared with the HG+ASO-NC+AMO-NC group, &p < 0.05 compared with the HG+ASO-CACR+AMO-NC group.
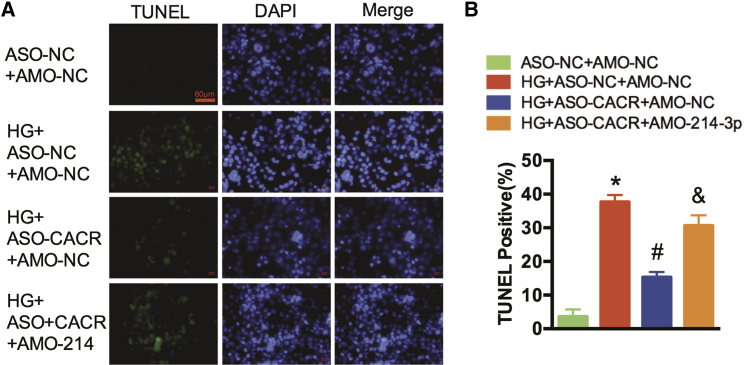
Moreover, TUNEL staining was conducted to detect myocardial cell death. The number of TUNEL-positive cells were reduced after silencing CACR, whereas these reductions were reversed after transfection with AMO-214-3p (Figures 5A and 5B).
Figure 5.
The CACR/miR-214-3p Pathway Regulates DNA Fracture in AC16 Cells
(A) A representative image of the TUNEL assay is shown. TUNEL-positive cells, green; nuclei, blue. Scale bar, 60 μm. (B) TUNEL-positive cells were analyzed. n = 3. *p < 0.05 compared with the ASO-NC+AMO-NC group, #p < 0.05 compared with the HG+ASO-NC+AMO-NC group, &p < 0.05 compared with the HG+ASO-CACR+AMO-NC group.
Discussion
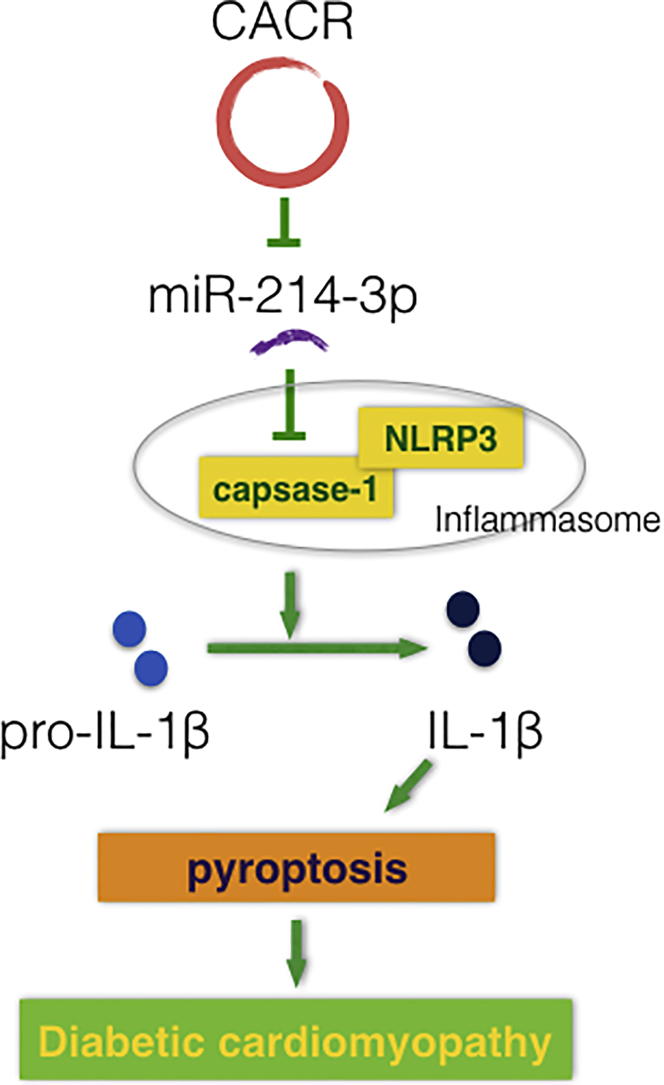
Although circRNAs appear to act as ncRNAs, they have recently attracted great attention due to their critical roles in the pathogenesis of many disorders. However, little is known about the role of circRNAs in the process of pyroptosis in DCM. In this study, we illustrated that CACR is highly expressed in high-glucose-treated cardiomyocytes and in the serum of diabetic patients. CACR silencing inhibited caspase-1 expression via sponging miR-214-3p, consequently suppressing cardiomyocyte inflammation and death. We speculate that CACR might be a new therapeutic target for DCM (Figure 6).
Figure 6.
Proposed Model of the Function of CACR in Regulating Pyroptosis via miR-214-3p in High-Glucose-Treated Cardiomyocytes
Previous studies have fully confirmed the important role of miRNAs in various diseases.26 In addition, circRNAs can target miRNAs to regulate diseases. Therefore, circRNAs are considered potential diagnostic markers and therapeutic targets for many diseases. Unlike other ncRNAs, circRNAs are evolutionarily conserved and resistant to nuclease digestion. Their half-life is over 48 h, which makes circRNAs more stable than other ncRNAs.21, 27, 28 Because of the above characteristics, it is very important to study the role of circRNA in diseases. circRNAs have been shown to be associated with a variety of cardiovascular diseases.29 Wang et al.30 indicated that circRNA HRCR could sponge miR-223 and reverse cardiac hypertrophy and heart failure. Recent studies revealed that in DCM, circ_000203 exacerbated myocardial fibrosis by increasing Col1a2 and connective tissue growth factor (CTGF) via miR-26b-5p.31 circRNA_010567 also promotes myocardial fibrosis via the miR-141/TGF-β1 pathway in DCM.32 However, there are no reports about circRNAs in the regulation of inflammation and pyroptosis in DCM. Here, we demonstrated that a new circRNA, CACR, was increased in DCM. Thus, CACR is considered a vital regulator of DCM and should be further investigated.
Hyperglycemia contributes to myocardial injury directly or indirectly and causes left ventricular dysfunction, cardiac fibrosis, hypertrophy, and arrhythmia.33 Cell death and inflammation are recognized as vital mechanisms of DCM.34, 35 Pyroptosis is a type of programmed cell death associated with inflammation36 and is characterized by pore formation in the membrane, cell swelling, osmotic lysis, and intracellular pro-inflammatory cytokine leakage.37, 38 In the present study, pyroptosis was evidently activated in high-glucose-induced cardiomyocytes, and CACR levels were increased. In addition, silencing CACR attenuated pyroptosis significantly. Therefore, we speculate that silencing CACR has a cardiac protective effect via weakening pyroptosis. On the other hand, apoptosis is another type of programmed cell death. We also found that apoptosis was slightly inhibited after silencing CACR. This effect may be due to the inhibition of inflammation after pyroptosis is suppressed.
A ceRNA hypothesis that circRNAs regulate the expression of mRNAs at the post-transcriptional level by competing for binding to shared miRNAs has been proposed.39, 40, 41, 42, 43 Comprehending RNA cross-talk will provide important novel information about gene regulatory networks in the pathogenesis of DCM. Herein, bioinformatics analysis was performed to predict potential miRNAs and circRNAs that might participate in caspase-1 regulatory networks. After the luciferase assay and functional acquisition and deletion, we clarified that CACR could bind to miR-214-3p and function as a ceRNA to regulate the expression of caspase-1 during DCM. In conclusion, the CACR/miR-214-3p/caspase-1 regulatory network acts as a balance regulator of the inflammation and cell death of cardiomyocytes and provides novel insight into DCM.
There are some limitations to this research. First, gene sequencing revealed that there was no homologous gene of human CACR in mouse. For that reason, animal experiments were not performed, and only human AC16 cells were used for the in vitro experiments. Second, the cardiac tissues from patients with DCM could not be collected clinically, so more accurate histological validation could not be performed. However, we detected differences in the expression of CACR in the serum of diabetic and non-diabetic patients. This finding suggests that CACR may serve as a clinical biomarker of DCM. We believe that circRNAs derived from humans are more representative for studying the mechanisms of DCM in humans.
In summary, we found that CACR was increased in DCM and was shown to be a regulator of DCM. Silencing CACR significantly alleviated pyroptosis in high-glucose-treated cardiomyocytes. Notably, CACR functions as a ceRNA, which may sequester miR-214-3p and thus relieve its suppressive effect on caspase-1 expression. Here, we have provided a new mechanism for the treatment of DCM. These findings will promote a better understanding of the mechanisms of DCM and facilitate precise gene diagnosis and accurate treatment of DCM.
Materials and Methods
Ethics Statement and Clinical Sample Collection
Serum samples were collected from non-diabetic subjects and patients with type 2 diabetes mellitus at the Department of Endocrinology of the Second Affiliated Hospital of Harbin Medical University (n = 10 in each group). Then, total serum RNA was extracted. The clinical characteristics of the patients are shown in Table S1. All patients signed informed consent in advance. The research was approved by the Ethics Committee of Harbin Medical University and was carried out in accordance with the Declaration of Helsinki.
Cell Culture
Human cardiomyocyte AC16 cells were obtained from the Biological Sciences Institutes of Shanghai and were maintained in DMEM containing 10% fetal bovine serum and 1% penicillin/streptomycin (HyClone, Logan, UT, USA) at 37°C in 5% CO2 air. AC16 cells were treated with 5.5 mM glucose (control) and 30 mM glucose (HG) for 24 h. The control group was also treated with a corresponding amount of mannitol for osmolarity.
Transfection
AC16 cells were transiently transfected at 60%–75% confluence with an antisense oligonucleotides for CACR (ASO-CACR), miR-214-3p mimics (miR-214-3p), or AMO-214-3p (RIBOBIO, Guangzhou, China) using X-treme GENE transfection reagent (Roche, Mannheim, Germany). Negative controls for ASO-CACR (ASO-NC), miR-214-3p mimics (NC), and AMO-214-3p (AMO-NC) were utilized as controls. AC16 cells were cultured in 12-well plates and transfected with a 50-μL transfection system according to the manufacturer’s protocol. The interfering RNA sequences are shown in Table S2.
RNA Isolation and Real-Time qPCR
RNA samples from human serum and AC16 cells were isolated using TRIzol LS or TRIzol (Invitrogen). A NanoDrop was used to detect the purity and concentration of the RNA. cDNAs were synthesized by using a Toyobo reverse transcription reagent kit. Then, real-time qPCR was performed using an ABI 7500 fast machine (Applied Biosystems, CA, USA) with Toyobo SYBR qPCR mix reagent (Toyobo, Osaka, Japan). The sequences of the primer pairs are listed in Table S2. The expression of miR-214-3p was normalized to U6, and the others were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The relative quantification of gene expression levels was determined by the 2−ΔΔCT method.
Western Blot Analysis
Total protein samples from AC16 cells were harvested and extracted using RIPA lysis buffer. The protein samples were prepared using loading buffer and separated by 10% SDS-PAGE. Then, the proteins were transferred onto nitrocellulose membranes and subsequently blocked with 5% non-fat milk dissolved in PBS for 2 h at room temperature. Next, the membranes were incubated with primary antibodies against NLRP3 (Boster Biological Technology, Wuhan, China; catalog #BA3677), caspase-1 (Cell Signaling Technology, MA, USA; catalog #2225S), IL-1β (R&D Systems; catalog #MAB201), and GAPDH (ZSGB-BIO, Beijing, China; catalog #TA-08) (1:1,000) at 4°C overnight, followed by goat anti-mouse immunoglobulin G (IgG) (ZSGB-BIO, Beijing, China; catalog #ZB-2305) or anti-rabbit IgG (ZSGB-BIO, Beijing, China; catalog #ZB-2301) for 1 h at room temperature. GAPDH was used as an internal control. Western blotting bands were measured by Quantity One software.
Immunofluorescence Staining
AC16 cells were fixed with 4% buffered paraformaldehyde in PBS for 20 min at room temperature. The cells were then washed with PBS buffer. Blocking buffer containing 1% BSA and 0.1% Triton X was used to block the cells for 1 h at room temperature. Then the cells were treated with 500 μL goat serum for 2 h and incubated with primary antibodies against NLRP3, caspase-1, and IL-1β (1:200) at 4°C overnight. Next, the cells were incubated with secondary antibody for 1 h at room temperature. The nuclei were stained with DAPI (Beyotime, Shanghai, China) for 20 min. Images were captured using a fluorescence microscope (Nikon 80i, Otawara, Tochigi, Japan) and analyzed using ImageJ software.
TUNEL Staining
DNA fragmentation in cardiomyocytes was detected using a cell death detection kit (Roche, Indianapolis, IN, USA) according to the corresponding instructions. DAPI was used to stain the nuclei. Finally, the stained sections were examined under a fluorescence microscope (Eclipse 80i; Nikon, Japan).
Luciferase Assay
The corresponding linear sequence of hsa_circ_0076631 is SLC29A1, as shown in the circBase software. A dual-luciferase vector, SLC29A1, 3′ UTRs with miR-214-3p binding sites, and mutated 3′ UTRs were constructed by RIBOBIO (Guangzhou, China). The target fragments were cloned into the vector using the restriction enzymes XhoI and NotI. HEK293 cells were transfected with the wild-type vector, mutation vector, and miR-214-3p mimics or NC using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After 48 h of transfection, the dual-luciferase reporter assay system (Promega Biotech) was used to detect the luciferase activity.
FISH
To detect the location of CACR, AC16 cells were plated into a 24-well plate. When the cells reached 60%–70% confluency, they were fixed in 4% paraformaldehyde for 10 min at room temperature and then permeabilized with 0.5% Triton X-100 on ice for 5 min. The cells were added to a pre-hybridization solution and incubated at 37°C for 30 min. Then, the solution was changed to a hybridization buffer containing a Cy3-labeled FISH probe for CACR and the internal controls 18 s and U6, and the samples were incubated at 37°C overnight in the dark. After that, the cells were washed with sodium citrate (SCC) and PBS. DAPI was used to stain the nuclei for 10 min. Mounting medium was added, and the cells were detected using a microscope (Nikon, Japan).
Data Analysis
All data in this study are expressed as the mean ± SD as calculated with GraphPad Prism 6. Statistical significances between two measurements were determined by unpaired Student’s t tests, and one-way ANOVA was used to determine the significance among different groups. A value of p < 0.05 was considered statistically significant.
Author Contributions
F.Y., Y.B., and L.W. designed the experiments; F.Y. and A.L. analyzed the data and wrote the manuscript; Y.Q. and A.L. performed western blotting; H.C. and J.L. performed cell culture and transfection; Y.W. and E.Y. performed FISH; Y.Y. and X.D. performed qRT-PCR; H.L. collected the serums of patients; Y.L. performed TUNEL staining and immunofluorescence staining.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was generously supported by grants from the National Natural Science Foundation of China (grant no. 81770809 to L.W. and grant no. 81673426 to Y.B.), Bethune-Merck Diabetes Research Foundation (grant no. G2017044 to L.W.), and Graduate Innovation Fund of Harbin Medical University (YJSCX2017-59HYD to F.Y.).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.06.026.
Contributor Information
Yunlong Bai, Email: baiyunlong@ems.hrbmu.edu.cn.
Lihong Wang, Email: nd6688@163.com.
Supplemental Information
References
- 1.Aneja A., Tang W.H., Bansilal S., Garcia M.J., Farkouh M.E. Diabetic cardiomyopathy: insights into pathogenesis, diagnostic challenges, and therapeutic options. Am. J. Med. 2008;121:748–757. doi: 10.1016/j.amjmed.2008.03.046. [DOI] [PubMed] [Google Scholar]
- 2.Bugger H., Abel E.D. Molecular mechanisms of diabetic cardiomyopathy. Diabetologia. 2014;57:660–671. doi: 10.1007/s00125-014-3171-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu L., Ding W.Y., Zhao J., Wang Z.H., Zhong M., Zhang W., Chen Y.G., Zhang Y., Li L., Tang M.X. Activin receptor-like kinase 7 mediates high glucose-induced H9c2 cardiomyoblast apoptosis through activation of Smad2/3. Int. J. Biochem. Cell Biol. 2013;45:2027–2035. doi: 10.1016/j.biocel.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 4.Luo B., Huang F., Liu Y., Liang Y., Wei Z., Ke H., Zeng Z., Huang W., He Y. NLRP3 Inflammasome as a Molecular Marker in Diabetic Cardiomyopathy. Front. Physiol. 2017;8:519. doi: 10.3389/fphys.2017.00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fink S.L., Cookson B.T. Apoptosis, pyroptosis, and necrosis: mechanistic description of dead and dying eukaryotic cells. Infect. Immun. 2005;73:1907–1916. doi: 10.1128/IAI.73.4.1907-1916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kroemer G., Galluzzi L., Vandenabeele P., Abrams J., Alnemri E.S., Baehrecke E.H., Blagosklonny M.V., El-Deiry W.S., Golstein P., Green D.R., Nomenclature Committee on Cell Death 2009 Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guarda G., So A. Regulation of inflammasome activity. Immunology. 2010;130:329–336. doi: 10.1111/j.1365-2567.2010.03283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu Q., Jiang Y., Zhang W., Xu C., Du W., Tuguzbaeva G., Qin Y., Li A., Zhang L., Sun G. Pyroptosis is involved in the pathogenesis of human hepatocellular carcinoma. Oncotarget. 2016;7:84658–84665. doi: 10.18632/oncotarget.12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X., Du N., Zhang Q., Li J., Chen X., Liu X., Hu Y., Qin W., Shen N., Xu C. MicroRNA-30d regulates cardiomyocyte pyroptosis by directly targeting foxo3a in diabetic cardiomyopathy. Cell Death Dis. 2014;5:e1479. doi: 10.1038/cddis.2014.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo B., Li B., Wang W., Liu X., Xia Y., Zhang C., Zhang M., Zhang Y., An F. NLRP3 gene silencing ameliorates diabetic cardiomyopathy in a type 2 diabetes rat model. PLoS ONE. 2014;9:e104771. doi: 10.1371/journal.pone.0104771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo B., Li B., Wang W., Liu X., Liu X., Xia Y., Zhang C., Zhang Y., Zhang M., An F. Rosuvastatin alleviates diabetic cardiomyopathy by inhibiting NLRP3 inflammasome and MAPK pathways in a type 2 diabetes rat model. Cardiovasc. Drugs Ther. 2014;28:33–43. doi: 10.1007/s10557-013-6498-1. [DOI] [PubMed] [Google Scholar]
- 12.Lamkanfi M., Dixit V.M. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 13.Schroder K., Zhou R., Tschopp J. The NLRP3 inflammasome: a sensor for metabolic danger? Science. 2010;327:296–300. doi: 10.1126/science.1184003. [DOI] [PubMed] [Google Scholar]
- 14.Santiago J.J., McNaughton L.J., Koleini N., Ma X., Bestvater B., Nickel B.E., Fandrich R.R., Wigle J.T., Freed D.H., Arora R.C., Kardami E. High molecular weight fibroblast growth factor-2 in the human heart is a potential target for prevention of cardiac remodeling. PLoS ONE. 2014;9:e97281. doi: 10.1371/journal.pone.0097281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Somanna N.K., Yariswamy M., Garagliano J.M., Siebenlist U., Mummidi S., Valente A.J., Chandrasekar B. Aldosterone-induced cardiomyocyte growth, and fibroblast migration and proliferation are mediated by TRAF3IP2. Cell. Signal. 2015;27:1928–1938. doi: 10.1016/j.cellsig.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Guo X., Xue M., Li C.J., Yang W., Wang S.S., Ma Z.J., Zhang X.N., Wang X.Y., Zhao R., Chang B.C., Chen L.M. Protective effects of triptolide on TLR4 mediated autoimmune and inflammatory response induced myocardial fibrosis in diabetic cardiomyopathy. J. Ethnopharmacol. 2016;193:333–344. doi: 10.1016/j.jep.2016.08.029. [DOI] [PubMed] [Google Scholar]
- 17.Palomer X., Salvadó L., Barroso E., Vázquez-Carrera M. An overview of the crosstalk between inflammatory processes and metabolic dysregulation during diabetic cardiomyopathy. Int. J. Cardiol. 2013;168:3160–3172. doi: 10.1016/j.ijcard.2013.07.150. [DOI] [PubMed] [Google Scholar]
- 18.Yang F., Qin Y., Wang Y., Li A., Lv J., Sun X., Che H., Han T., Meng S., Bai Y., Wang L. LncRNA KCNQ1OT1 Mediates Pyroptosis in Diabetic Cardiomyopathy. Cell. Physiol. Biochem. 2018;50:1230–1244. doi: 10.1159/000494576. [DOI] [PubMed] [Google Scholar]
- 19.Pamudurti N.R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., Hanan M., Wyler E., Perez-Hernandez D., Ramberger E. Translation of CircRNAs. Mol. Cell. 2017;66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beermann J., Piccoli M.T., Viereck J., Thum T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 21.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H.D., Jiang L.H., Sun D.W., Hou J.C., Ji Z.L. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25:1–7. doi: 10.1007/s12282-017-0793-9. [DOI] [PubMed] [Google Scholar]
- 23.Jiang G., Ma Y., An T., Pan Y., Mo F., Zhao D., Liu Y., Miao J.N., Gu Y.J., Wang Y., Gao S.H. Relationships of circular RNA with diabetes and depression. Sci. Rep. 2017;7:7285. doi: 10.1038/s41598-017-07931-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan X., Weng X., Zhao Y., Chen W., Gan T., Xu D. Circular RNAs in Cardiovascular Disease: An Overview. Biomed Res. Int. 2017;2017 doi: 10.1155/2017/5135781. 5135781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin X., Jin H., Shi Y., Guo Y., Zhang H. Long Non-Coding RNA KCNQ1OT1 Promotes Cataractogenesis via miR-214 and Activation of the Caspase-1 Pathway. Cell. Physiol. Biochem. 2017;42:295–305. doi: 10.1159/000477330. [DOI] [PubMed] [Google Scholar]
- 26.He L., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., Hammond S.M. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang T., Pan W., Hu J., Zhang Z., Li G., Liang Y. Circular RNAs in Metabolic Diseases. Adv. Exp. Med. Biol. 2018;1087:275–285. doi: 10.1007/978-981-13-1426-1_22. [DOI] [PubMed] [Google Scholar]
- 28.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan X., Weng X., Zhao Y., Chen W., Gan T., Xu D. Circular RNAs in Cardiovascular Disease: An Overview. BioMed Res. Int. 2017;2017:5135781. doi: 10.1155/2017/5135781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K., Long B., Liu F., Wang J.X., Liu C.Y., Zhao B., Zhou L.Y., Sun T., Wang M., Yu T. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 31.Tang C.M., Zhang M., Huang L., Hu Z.Q., Zhu J.N., Xiao Z., Zhang Z., Lin Q.X., Zheng X.L., -Yang M. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci. Rep. 2017;7:40342. doi: 10.1038/srep40342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou B., Yu J.W. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-β1. Biochem. Biophys. Res. Commun. 2017;487:769–775. doi: 10.1016/j.bbrc.2017.04.044. [DOI] [PubMed] [Google Scholar]
- 33.Goyal B.R., Mehta A.A. Diabetic cardiomyopathy: pathophysiological mechanisms and cardiac dysfuntion. Hum. Exp. Toxicol. 2013;32:571–590. doi: 10.1177/0960327112450885. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y.H., Cai L. Diabetes/obesity-related inflammation, cardiac cell death and cardiomyopathy. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2006;31:814–818. [PubMed] [Google Scholar]
- 35.Diamant M., Lamb H.J., Smit J.W., de Roos A., Heine R.J. Diabetic cardiomyopathy in uncomplicated type 2 diabetes is associated with the metabolic syndrome and systemic inflammation. Diabetologia. 2005;48:1669–1670. doi: 10.1007/s00125-005-1821-4. [DOI] [PubMed] [Google Scholar]
- 36.Broz P. Immunology: Caspase target drives pyroptosis. Nature. 2015;526:642–643. doi: 10.1038/nature15632. [DOI] [PubMed] [Google Scholar]
- 37.de Zoete M.R., Palm N.W., Zhu S., Flavell R.A. Inflammasomes. Cold Spring Harb. Perspect. Biol. 2014;6:a016287. doi: 10.1101/cshperspect.a016287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shalini S., Dorstyn L., Dawar S., Kumar S. Old, new and emerging functions of caspases. Cell Death Differ. 2015;22:526–539. doi: 10.1038/cdd.2014.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Denzler R., Agarwal V., Stefano J., Bartel D.P., Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell. 2014;54:766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bai Y., Zhang L., Jiang Y., Ju J., Li G., Xu J., Jiang X., Zhang P., Lang L., Sadkovaya O. Identification and Functional Verification of MicroRNAs in the Obese Rat With Erectile Dysfunction. Sex. Med. 2017;5:e261–e271. doi: 10.1016/j.esxm.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang B., Lin H., Xiao J., Lu Y., Luo X., Li B., Zhang Y., Xu C., Bai Y., Wang H. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat. Med. 2007;13:486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 43.Li X., Edwards M., Swaney K.F., Singh N., Bhattacharya S., Borleis J., Long Y., Iglesias P.A., Chen J., Devreotes P.N. Mutually inhibitory Ras-PI(3,4)P2 feedback loops mediate cell migration. Proc. Natl. Acad. Sci. USA. 2018;115:E9125–E9134. doi: 10.1073/pnas.1809039115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.