Abstract
Circular RNAs (circRNAs) are a novel category of non-coding RNAs, and they have been identified to participate in glioma tumorigenesis. Here we investigated the functions of circRNA circSCAF11 in glioma genesis, and we unveiled its molecular mechanism in the pathophysiological process. Expression levels of circSCAF11, miR-421, and SP1 mRNA were measured using RT-PCR. Proteins were measured using western blotting. The tumor phenotypes of glioma cells were detected using flow cytometry and Cell Counting Kit-8 (CCK-8), transwell, and xenograft mouse assays. The combination within circSCAF11, miR-421, and SP1 was validated using luciferase reporter assay or RNA pull-down assay. The binding of transcription factor SP1 with vascular endothelial cell growth factor A (VEGFA) promoter was inspected using chromatin immunoprecipitation (ChIP). circSCAF11 expression was found to be significantly upregulated in the glioma tissue specimens and cell lines. The ectopic overexpression of circSCAF11 was closely correlated with the poor clinical outcome of glioma patients. Functionally, knockdown of circSCAF11 inhibited the proliferation, invasion, and tumor growth and induced the G0/G1 phase arrest. Mechanically, circSCAF11 positively regulated the SP1 expression through sponging miR-421. Moreover, transcription factor SP1 activated the transcription of VEGFA, constructing the circSCAF11/miR-421/SP1/VEGFA axis in the glioma genesis. The findings in this research illustrate that circSCAF11 accelerates glioma tumorigenesis through the miR-421/SP1/VEGFA axis, providing a potential target for circRNA and glioma treatment.
Keywords: glioma, circular RNA, circSCAF11, miR-421, SP1, VEGFA
Introduction
Glioma is the most common intracranial primary cancer with extraordinarily high morbidity and mortality worldwide.1, 2, 3, 4 In spite of the common methods for clinical treatment, such as surgery, radiotherapy, and chemotherapy, the long-term effects and postoperative outcomes for the patients with glioma are still dissatisfactory.5, 6 Emerging theories support this view that the abnormity of genetic molecular could cause the glioma genesis.7, 8, 9 With the exception of those traditional methods, the disorders of genetic and epigenetic regulation could take part in the pathophysiological process and act as the vital effectors.
Circular RNA (circRNA) is a novel type of noncoding RNA with a covalently closed loop, which is generated by the back-splicing of pre-mRNA.10, 11, 12, 13 More newly identified circRNAs have been found using high-throughput sequencing and via further functional validation.14, 15, 16 For example, circRNA circMMP9 is upregulated in glioma cells and acts as an oncogene to promote the proliferation, migration, and invasion of glioma cells, via targeting miR-124/cyclin-dependent kinase 4 (CDK4).17 Another example, circ-FBXW7 is abundantly expressed in the normal human brain, and FBXW7-185aa upregulation inhibits the proliferation and cell cycle in vitro and in vivo.18
In this report, we identify a novel circRNA, circSCAF11 (hsa_circ_0098619), in the glioma tissue and cells. The assays reveal that circSCAF11 is markedly upregulated in the glioma tissue and cells, and the functional assay reveals the critical function of circSCAF11 on glioma cells’ phenotype.
Results
circSCAF11 Is Upregulated in the Glioma Tissue and Cells
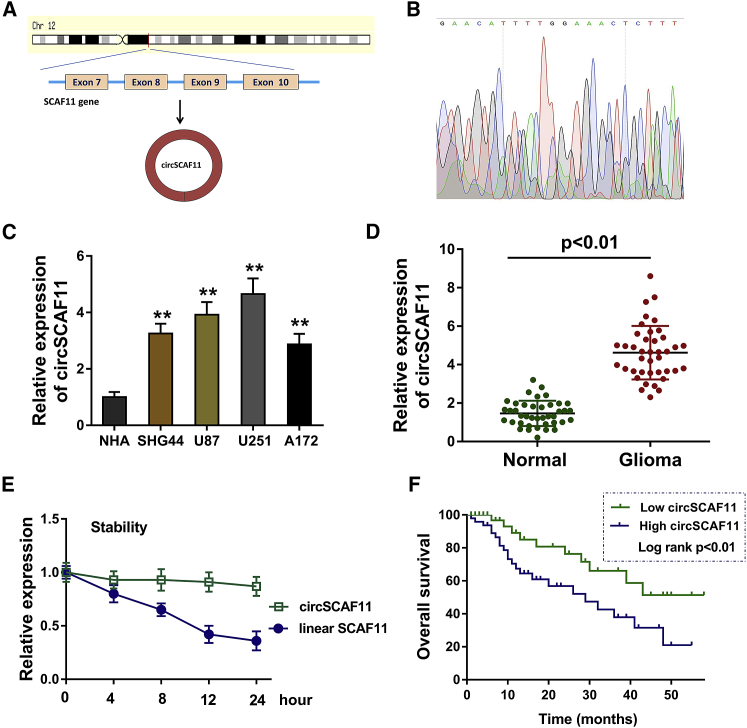
circSCAF11 is transcribed from the exons (7–10) of the SCAF11 gene. The ID for the circSCAF11 is hsa_circ_0098619 (circBase, chr12: 46325288–46328288) (Figure 1A). The Sanger sequencing experiments were performed to validate the sequence on the junction sites of circSCAF11 (Figure 1B). The junction sites of circSCAF11 are GAACATTTTG and GAAACTCTTT. RT-PCR showed that circSCAF11 was upregulated in the glioma cells (SHG44, U87, U251, and A172) as compared to the normal cell lines (Figure 1C). In the clinical glioma specimens (Table 1), RT-PCR showed that circSCAF11 was upregulated in the glioma tissue as compared to the normal tissue (Figure 1D). When the RNA extraction was treated with actinomycin D, the linear SCAF11 RNA was markedly decreased, while the circular SCAF11 RNA was much more stable (Figure 1E). The prognosis analysis by the Kaplan-Meier test showed that the higher circSCAF11 expression indicated the poorer prognosis and low survival rate when comparing with others (Figure 1F). Overall, this finding suggests that circSCAF11 is upregulated in the glioma tissue and cells.
Figure 1.
circSCAF11 Is Upregulated in the Glioma Tissue and Cells
(A) circSCAF11 is transcribed from the exons (7–10) of the SCAF11 gene (circBase, hsa_circ_0098619, chr12: 46325288–46328288). (B) Sanger sequencing validated the sequence on the junction sites of circSCAF11. (C) RT-PCR showed the circSCAF11 level in the glioma cells (SHG44, U87, U251, and A172). (D) circSCAF11 was upregulated in the glioma tissue as compared to the normal tissue. (E) When the RNA extraction was treated with actinomycin D, the linear SCAF11 RNA and circular SCAF11 RNA were measured using RT-PCR. (F) The prognosis analysis by the Kaplan-Meier test showed the prognosis and survival rate of patients with glioma with different circSCAF11 expressions. **p < 0.01.
Table 1.
Relationship between circSCAF11 and Clinicopathological Characteristics of Patients with Glioma
| Variable | n = 40 | circSCAF11 Expression |
p Value | |
|---|---|---|---|---|
| Low = 20 | High = 20 | |||
| Gender | ||||
| Male | 22 | 12 | 9 | 0.612 |
| Female | 18 | 8 | 11 | |
| Age | ||||
| <50 years | 17 | 9 | 8 | 0.153 |
| ≥50 years | 23 | 11 | 12 | |
| WHO Grading | ||||
| I-II | 19 | 9 | 10 | 0.178 |
| III-IV | 21 | 11 | 10 | |
| Tumor Size | ||||
| <3cm | 13 | 7 | 6 | 0.015∗ |
| ≥3cm | 27 | 13 | 14 | |
| KPS | ||||
| ≥80 | 23 | 11 | 12 | 0.317 |
| <80 | 17 | 9 | 8 | |
p < 0.05 represents statistical differences. KPS, Karnofsky performance score; WHO, World Health Organization.
Knockdown of circSCAF11 Represses the Glioma Proliferation, Invasion, and Tumor Growth In Vitro and In Vivo
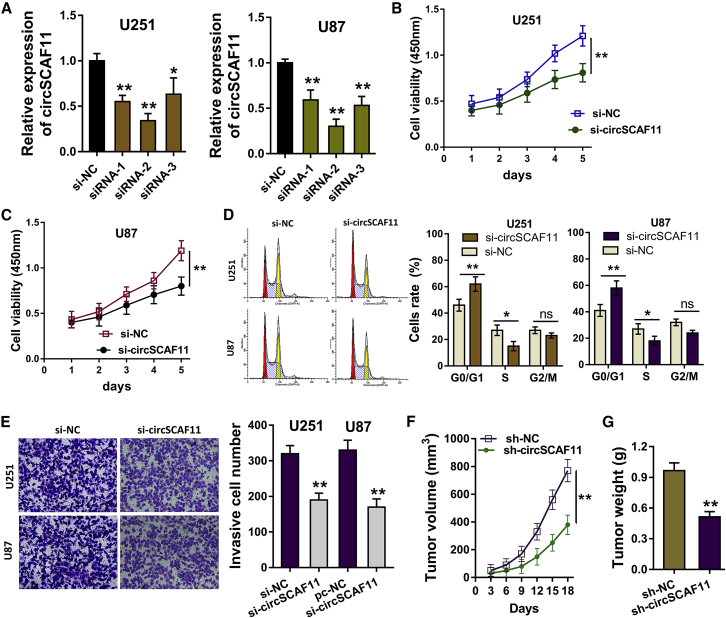
The expression and abundance of circSCAF11 were found to be upregulated in the glioma tissue and cells; thus, the silencing of circSCAF11 was constructed using oligonucleotide transfection (Figure 2A). The proliferative ability of glioma cells (U251 and U87) by Cell Counting Kit-8 (CCK-8) assay indicated that the circSCAF11 silencing inhibited the proliferation of glioma cells, presenting the obstruction of circSCAF11 knockdown (Figures 2B and 2C). Flow cytometry for the cycle analysis indicated the cycle arrest of glioma cells (U251 and U87) at the G0/G1 phase triggered by the circSCAF11 knockdown (Figure 2D). Transwell chamber for the invasion assay indicated the inhibition of invasive ability of glioma cells caused by the circSCAF11 knockdown (Figure 2E). To identify the role of circSCAF11 on tumor growth, xenograft in vivo assay was carried out, showing decreasing tumor volume caused by circSCAF11 knockdown (Figure 2F), as well as tumor weight (Figure 2G). Taken together, these results conclude that the knockdown of circSCAF11 represses the glioma proliferation, invasion, and tumor growth in vitro and in vivo.
Figure 2.
Knockdown of circSCAF11 Represses the Glioma Proliferation, Invasion, and Tumor Growth In Vitro and In Vivo
(A) The silencing of circSCAF11 was constructed using the oligonucleotide transfection in glioma cells (U251 and U87). (B and C) U251 (B) and U87 (C) cells were detected by CCK-8 assay, indicating the inhibition on the proliferation of glioma cells caused by circSCAF11 silencing. (D) Flow cytometry for the cycle analysis indicated the cycle arrest of glioma cells (U251 and U87) at the G0/G1 phase. (E) Transwell chamber for the invasion assay indicated the inhibition of invasive ability of glioma cells. (F and G) Tumor (F) volume and (G) weight of xenograft in vivo assay showed the decreasing tumor volume and weight. **p < 0.01, *p < 0.05.
circSCAF11 Is Targeted by miR-421 as the miRNA Sponge
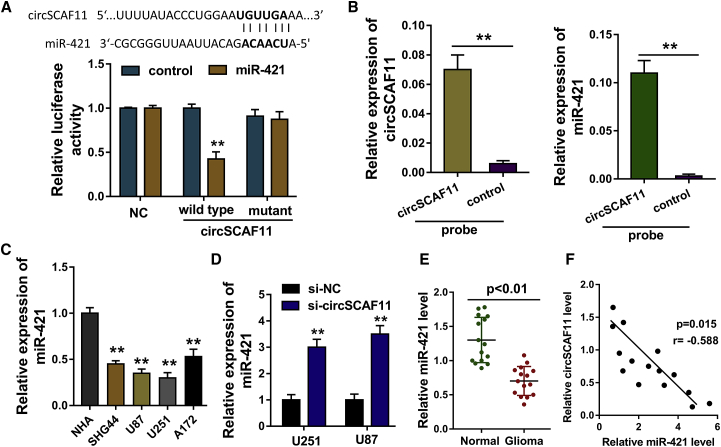
On account of the biogenesis of circSCAF11 generated from exons of the SCAF11 gene, we assumed that circSCAF11 could act as the microRNA (miRNA) sponge to regulate the glioma tumor phenotype. Bioinformatics tool Circular RNA Interactome (https://circinteractome.nia.nih.gov/) indicated that, among the candidate miRNAs, miR-421 acted as the downstream target of circSCAF11, which was validated by luciferase assay (Figure 3A). RNA pull-down assay was performed using the biotinylated circRNA probe, indicating that this probe could significantly pull down the circSCAF11 in the U251 cells (Figure 3B, left). Besides, miR-421 was also pulled down by the circSCAF11 probe (Figure 3B, right). In the glioma cells, RT-PCR showed that miR-421 was markedly decreased in these cells (Figure 3C). Moreover, when the circSCAF11 was knocked down, the miR-421 was increased (Figure 3D). In the glioma biopsies, miR-421 expression was found to be decreased compared to normal controls (Figure 3E). Pearson’s correlation analysis presented that miR-421 was negatively correlated to circSCAF11 in the patients with glioma (Figure 3F). Overall, these data prove that circSCAF11 is targeted by miR-421 as the miRNA sponge.
Figure 3.
circSCAF11 Was Targeted by the miR-421 as the miRNA Sponge
(A) Bioinformatics tool CircInteractome (https://circinteractome.nia.nih.gov/) indicated that miR-421 acted as the downstream target of circSCAF11, which was validated by luciferase assay. (B) RNA pull-down assay with biotinylated circRNA probe indicated the levels of circSCAF11 and miR-421 in the U251 cells. (C) RT-PCR showed the miR-421 level in glioma cells. (D) RT-PCR showed the miR-421 level after the circSCAF11 silencing. (E) miR-421 expression was measured using RT-PCR in the glioma biopsies. (F) Pearson’s correlation analysis presented that miR-421 was negatively correlated with circSCAF11 in the patients with glioma. **p < 0.01.
circSCAF11 Positively Regulates SP1 via Sponging miR-421
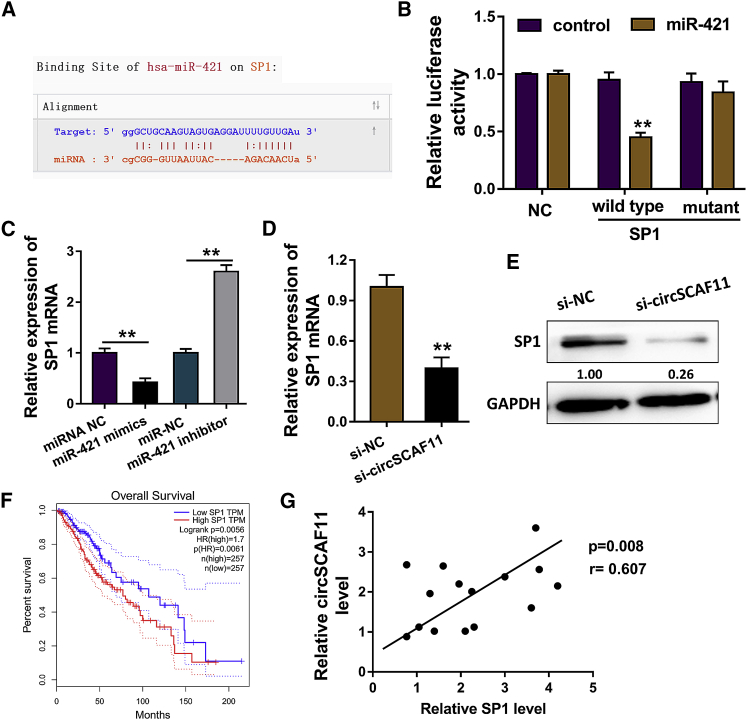
Bioinformatics analysis revealed that SP1 might act as the target of miR-421 (Figure 4A). Luciferase assay showed that SP1 could availably combine with miR-421 (Figure 4B). RT-PCR indicated that miR-421 mimics receded the SP1 mRNA, while miR-421 inhibitor enforced the SP1 mRNA level (Figure 4C). Then, when circSCAF11 was silenced, SP1 mRNA level was decreased (Figure 4D). Western blot showed that SP1 protein was decreased when circSCAF11 was silenced (Figure 4E). The prognosis analysis by the Kaplan-Meier test showed that the higher SP1 expression indicated the poorer prognosis and low survival rate comparing with others (Figure 4F). Pearson’s correlation analysis presented that SP1 was positively correlated with circSCAF11 in the patients with glioma (Figure 4G). Overall, these results support that circSCAF11 positively regulates SP1 via sponging miR-421.
Figure 4.
circSCAF11 Positively Regulates SP1 via Sponging miR-421
(A) Bioinformatics analysis revealed that SP1 might act as the target of miR-421. (B) Luciferase assay showed that SP1 could availably combine with miR-421. (C) RT-PCR indicated the SP1 mRNA expression after the miR-421 mimic transfection or miR-421 inhibitor transfection. (D) SP1 mRNA level was decreased when circSCAF11 was silenced. (E) Western blot showed the SP1 protein. (F) Kaplan-Meier test showed the prognosis analysis of patients with glioma. (G) Pearson’s correlation analysis presented the positive correlation within SP1 and circSCAF11. **p < 0.01.
SP1 Promotes the Transcription of VEGFA in Glioma Cells
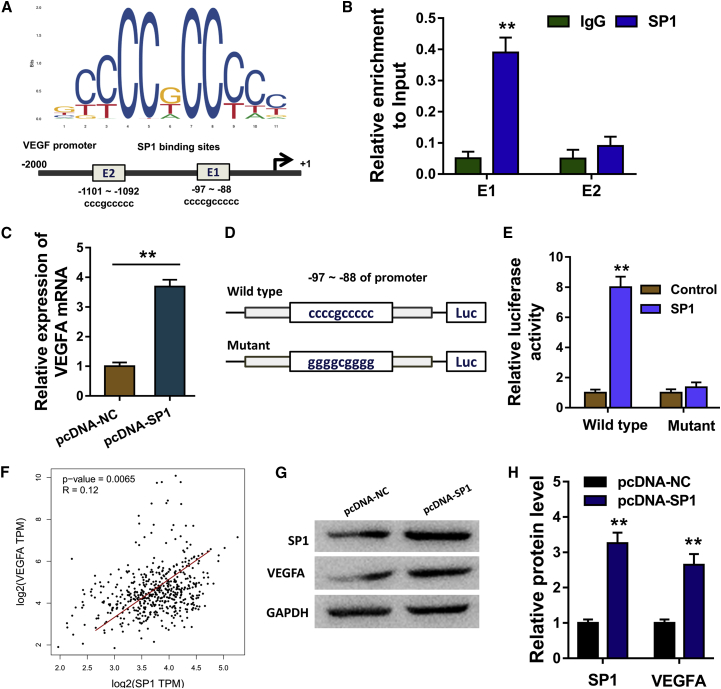
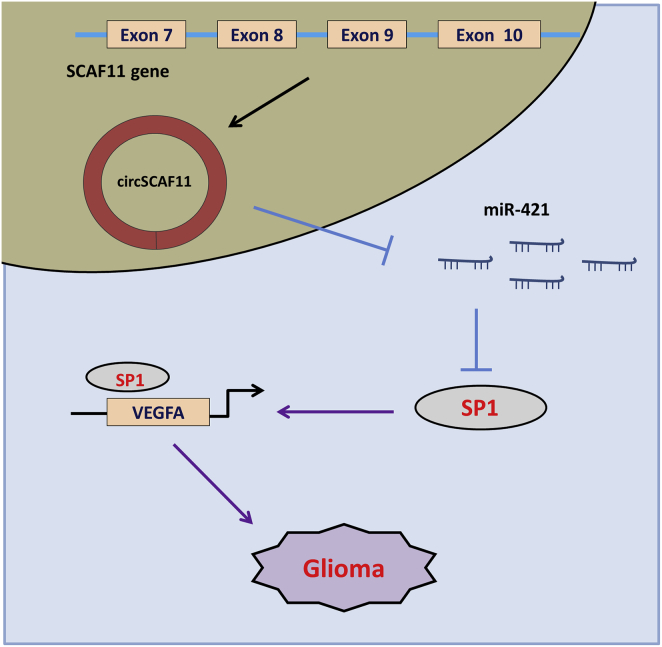
SP1 is a critical transcription factor in the human tumorigenesis. Thus, to identify the regulation of transcription factor SP1 in the glioma oncogenesis, we tried to identify its targeted gene. With the help of the JASPAR database (http://jaspar.genereg.net/), we found that vascular endothelial cell growth factor A (VEGFA) acted as the targeted gene for SP1. There are several binding sites within the SP1 and VEGFA promoter region (Figure 5A). Chromatin immunoprecipitation (ChIP) demonstrated that the SP1 antibody could be effectively precipitated with the region (−97 to −88) of the VEGFA promoter, instead of another region (−1,101 to −1,092) (Figure 5B). RT-PCR showed that SP1 overexpression plasmid could increase the VEGFA mRNA in the glioma cells (Figure 5C). The wild-type and mutant sequences of the region (−97 to −88) of the VEGFA promoter were constructed, and luciferase gene reporter assay showed that the wild-type sequence of the VEGFA promoter region could bind with the SP1 (Figures 5D and 5E). Based on The Cancer Genome Atlas (TCGA) database, the expression of VEGFA was positively correlated with that of SP1 in the patients with glioma (Figure 5F). Western blot analysis indicated that SP1 plasmid transfection could enforce the VEGFA protein expression (Figures 5G and 5H). Overall, the results indicated that circSCAF11 promotes the glioma tumorigenesis through the miR-421/SP1/VEGFA axis (Figure 6).
Figure 5.
SP1 Promotes the Transcription of VEGFA in Glioma Cells
(A) The JASPAR database (http://jaspar.genereg.net/) found that there are several binding sites within the SP1 and VEGFA promoter region. (B) Chromatin immunoprecipitation (ChIP) demonstrated that the SP1 antibody could be effectively precipitated with the region (−97 to −88) of the VEGFA promoter. (C) RT-PCR showed that SP1 overexpression plasmid could increase the VEGFA mRNA in the glioma cells. (D and E) The (D) wild-type and mutant sequences of the region (−97 to −88) of the VEGFA promoter were constructed, and (E) luciferase gene reporter assay showed that the wild-type sequence of the VEGFA promoter region could bind with the SP1. (F) Based on TCGA database, the expression of VEGFA was positively correlated with that of SP1 in the patients with glioma. (G and H) Western blot (G) and quantitative (H) analysis indicated that SP1 plasmid transfection could enforce the VEGFA protein expression.
Figure 6.
circSCAF11 Promotes Glioma Tumorigenesis through the miR-421/SP1/VEGFA Axis
Discussion
Recently, the extensive distribution and functional regulation of circRNAs have been wildly validated in human cancers.19, 20, 21, 22, 23 For the glioma, there are several circRNAs with explicit functions, such as circ-FBXW7, hsa_circ_0046701, and hsa_circ_0007534 among others.18, 24, 25 In this research, we discovered and identified a novel circRNA, circSCAF11, in glioma cells. However, the underlying mechanism by which circSCAF11 regulates glioma genesis is still ambiguous.
The circSCAF11 is generated from the exons 7–10 of the SCAF11 gene, which has been confirmed by the Sanger sequence. The level of circSCAF11 was measured using RT-PCR, which showed that circSCAF11 was significantly upregulated in the glioma cells. Overall, this overexpression indicated the poor prognosis of patients with glioma. Functional investigation unveiled the inhibition of glioma cells’ tumor phenotype induced by the circSCAF11 silencing. This finding suggested that circSCAF11 might act as an oncogene in the glioma carcinogenesis.
Up to now, the major researchers have adopted the view that circRNAs function as the miRNA sponge to absorb the miRNA to regulate the tumorigenesis.26, 27, 28, 29 circRNAs are characterized by the covalently closed loop, which could resist the actinomycin D digestion and weaken the miRNAs. This cascade reaction is described as competing endogenous RNA (ceRNA). For example, circRNA circ-ABCB10 acts as the sponge of miR-1271 to regulate breast cancer proliferation, and miR-1271 rescued the function of circ-ABCB10 on breast cancer cells.30 Another example, eIF4A3-induced circMMP9 promotes the proliferation and invasion through miR-124 and its targets, including cyclin-dependent kinase 4 and aurora kinase.17 In this study, circSCAF11 functions as a sponge for miR-421, and circSCAF11 could absorb miR-421 abundance in glioma cells.
SP1 acts as the target of miR-421; besides, it could activate the transcription of VEGFA in the glioma cells. SP1 is a critical transcription factor in human cancers, regulating multiple pathological processes. For instance, transcription factor SP1 could increase activity and expression of matrix metalloproteinase-2 (MMP-2), and overexpression of SP1 increased the invasiveness of glioma cells, representing a valuable prognostic marker for glioma and tumor invasion.31 In the glioblastoma, SP1 acts as a critical stemness-related transcriptional factor to enhance the temozolomide-resistance for the cells.32 This research discovered the transcription promotion of SP1 for the VEGFA, which could accelerate the progression of glioma.
In this study, the transcription factor SP1 could bind with the promoter region of VEGFA to increase its transcription level. VEGFA is a critical member of vascular endothelial growth factor in humans. It has been clearly certified that VEGFA contributes to the angiogenesis in glioma. The ability to form new blood vessels in the glioma could supply nutrient-carrying blood to the tumor tissue to promote the growth. Since the acceleration of VEGFA is vital for glioma angiogenesis, the positive regulation of circSCAF11/miR-421/SP1/VEGFA could support the angiogenesis and tumorigenesis of glioma.
Conclusions
Taken together, this research presents the critical roles of circSCAF11 in glioma. circSCAF11 could positively regulate the SP1 expression via sponging miR-421, and then SP1 could activate the VEGFA transcription. These findings provide not only a new mechanism for the research of glioma angiogenesis but also effective targets for the molecular therapy of glioma.
Materials and Methods
Ethical Approval and Consent to Participate
The study was conducted in accordance with the Declaration of Helsinki principles. It was approved by the Medical Research Ethics Committee of Tangdu Hospital.
Clinical Tissue Samples
Clinical glioma tissue samples were collected, including cancer tissue and adjacent normal tissue, from patients with glioma who underwent surgery in the Tangdu Hospital. Based on the inclusion criteria, the individuals who received chemotherapy or radiotherapy were excluded. The brain tissues were resected and stored at −80°C. This clinical research was approved by the Ethics Committees of Tangdu Hospital.
Cell Line Culture and Transfection
Human glioma cell lines (A172, U251, U87, and SHG44) and normal human astrocytes (NHAs) were purchased from the Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). Glioma cell lines and normal astrocytes were maintained in RPMI Medium 1640 (Gibco, Invitrogen, Carlsbad, CA) supplemented with heat-inactivated 10% FBS (fetal bovine serum) (Gibco) and 100 U/mL penicillin and 100 mg/mL streptomycin (Invitrogen) in a humidified incubator at 37°C with 5% CO2. Specific oligonucleotides for the small interfering RNA (siRNA) or mimics were synthesized by RiboBio (Guangzhou, China) and transfected using Lipofectamine 2000, according to the manufacturer’s protocol (Invitrogen).
RNA Isolation and qRT-PCR
RNA was totally extracted from the cells and tissue using the with TRIzol reagent (1 mL) (Invitrogen) based on the manufacturer’s protocol. The testing for miRNA extraction was mirVana miRNA isolation kit (Ambion, Austin, TX, USA). After isolation, the RNA concentration in the RNA solution was determined using NanoDrop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and stored at −80°C for further use. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous control. Primers used for the qRT-PCR analysis are presented in Table S1.
Western Blotting
Cells were cultured in 6-well plates (5 × 105 cells/well). Total proteins were obtained using the radio immunoprecipitation assay (RIPA) (Sigma-Aldrich, St. Louis, MO, USA) and then moved to the 12% separation SDS-PAGE. Proteins were transferred to nitrocellulose filter membranes (Hybond, Escondido, CA, USA) with Tris-buffered saline plus Tween (TBST). The membrane was blocked with 5% skim milk powder and incubated with primary antibody (anti-SP1, 1:1,000, Abcam). The blots were determined using an EZ-ECL chemiluminescence and visualized using Immuno Star LD (Wako Pure Chemical, Osaka, Japan).
CCK-8 Assay
The proliferation was tested by the CCK-8 assay kit (Dojindo Japan). Briefly, cells were seeded in 96-well plates (1 × 103 cells/well). Cells were cultured in FBS-free medium, and then CCK-8 reagent (10 μL) was added and the absorbance was measured at 450 nm at the indicated time points (1, 2, 3, 4, and 5 days).
Flow Cytometry Analysis
Glioma cells (4 × 105/well) were harvested and precipitated using centrifugation. The precipitated cells were washed with PBS. Cells were stained with fluorescein isothiocyanate propidium iodide using CycleTEST PLUS DNA Reagent Kit (BD Biosciences). The distribution of cells was analyzed using a flow cytometer (FACScan, BD Biosciences) equipped with CellQuest software (BD Biosciences).
Transwell Invasion Assay
Cell invasion was evaluated by performing a transwell invasion assay. Briefly, the 12-well transwell chamber (8-mm pore, Corning) was pre-coated with Matrigel (100 μL, Becton Dickinson). The glioma cells (1 × 105 cells) were seeded onto the chamber. Medium with serum was added into the lower floor, and the medium without serum was added to the upper floor. Finally, the invaded cells through the membrane were fixed and then stained with crystal violet. The quantification of invasive cells was determined and counted from five random areas under high-power microscope (Olympus, Tokyo, Japan).
Luciferase Reporter Assay
circSCAF11 wild-type with potential miR-421-binding sites or mutants of each site, as well as the VEGFA promoter region sequence sites, was amplified and cloned into the psi-CHECK-2 vector (Promega, Madison, WI, USA). Then, cells were co-transfected with luciferase plasmids and miR-421 or control miRNA. After 48-h transfection, the luciferase activities of firefly and Renilla were measured with Dual-Luciferase Reporter Assay System (Promega).
RNA Pull-down
RNA pull-down was performed using the Biotin RNA Labeling Mix (Roche, Shanghai, China). In brief, the biotin-labeled circRNA and blank bound to streptavidin magnetic beads and bound with protein. The lysate was administered with RNA-bound beads for immunoprecipitation. The RNA-protein beads mixture was incubated for 1 h at 4°C with rotation. The RNA complexes combining on the beads were washed three times and boiled in SDS buffer. Finally, extracted and eluted with elution buffer (50 mM HEPES, 5 mM EDTA, 100 mM NaCl, 1% SDS, and 10 mM DTT), the streptavidin beads were collected and analyzed by qRT-PCR.
ChIP Assay
ChIP assay was performed using Magna ChIP Chromatin Immunoprecipitation Kit according to the manual (Millipore, Billerica, MA, USA). Cross-linked cells were sonicated to fragments (200–1,000 bp). Specific antibodies for SP1 (ab59257, Abcam) or FLAG were administered to precipitate DNA-protein complexes. The precipitated RNAs with proteins were quantified and detected by real-time qRT-PCR with SYBR-Green incorporation (Applied Biosystems, Foster City, CA, USA). Immunoglobulin G (IgG) acted as the negative control. The primer sequences for the promoter are presented in Table S1.
Xenograft Animal Assay
BALB/c nude mice (6 weeks old) were purchased and randomly divided into two groups and nourished under pathogen-free conditions. Glioma cells (U251) were stably transfected with lentiviral vector for the silencing of circSCAF11, as well as the control. U251 cells were subcutaneously injected into the right back of mice with 1 × 106 cells/mousce. Then 3 weeks later, the tumor size was measured and the weight was measured. The animal assay was manipulated according to protocols, and the scheme was approved by the Tangdu Hospital Medical Experimental Animal Care Commission.
Statistical Analysis
Statistical analysis was performed using the SPSS software package (version 17.0) and GraphPad Prism (version 6.0). The Kaplan-Meier method was performed for the survival analysis, and the differences between patients were calculated by the log rank test. Student’s t test or a chi-square test was performed for the statistical significance with a p value <0.05.
Author Contributions
Q.M., S.L., and Y.L. performed the assays. S.Z., J.J., Y.Z., and C.G. assisted with the assays. Y.S. and B.L. wrote and revised the paper.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
The authors have no acknowledgments.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.06.022.
Contributor Information
Bei Liu, Email: liubei_xian@yeah.net.
Yang Sun, Email: sunyang.edu@aliyun.com.
Supplemental Information
References
- 1.Barbagallo D., Caponnetto A., Brex D., Mirabella F., Barbagallo C., Lauretta G., Morrone A., Certo F., Broggi G., Caltabiano R. CircSMARCA5 Regulates VEGFA mRNA Splicing and Angiogenesis in Glioblastoma Multiforme Through the Binding of SRSF1. Cancers (Basel) 2019;11:E194. doi: 10.3390/cancers11020194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wank M., Schilling D., Schmid T.E., Meyer B., Gempt J., Barz M., Schlegel J., Liesche F., Kessel K.A., Wiestler B. Human Glioma Migration and Infiltration Properties as a Target for Personalized Radiation Medicine. Cancers (Basel) 2018;10:E456. doi: 10.3390/cancers10110456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen T.C., da Fonseca C.O., Schönthal A.H. Intranasal Perillyl Alcohol for Glioma Therapy: Molecular Mechanisms and Clinical Development. Int. J. Mol. Sci. 2018;19:E3905. doi: 10.3390/ijms19123905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Ierschot F., Bastiaanse R., Miceli G. Evaluating Spelling in Glioma Patients Undergoing Awake Surgery: a Systematic Review. Neuropsychol. Rev. 2018;28:470–495. doi: 10.1007/s11065-018-9391-7. [DOI] [PubMed] [Google Scholar]
- 5.Dworkin M., Mehan W., Niemierko A., Kamran S.C., Lamba N., Dietrich J., Martinez-Lage M., Oh K.S., Batchelor T.T., Wen P.Y. Increase of pseudoprogression and other treatment related effects in low-grade glioma patients treated with proton radiation and temozolomide. J. Neurooncol. 2019;142:69–77. doi: 10.1007/s11060-018-03063-1. [DOI] [PubMed] [Google Scholar]
- 6.Noorlag L., De Vos F.Y., Kok A., Broekman M.L.D., Seute T., Robe P.A., Snijders T.J. Treatment of malignant gliomas with ketogenic or caloric restricted diets: A systematic review of preclinical and early clinical studies. Clin. Nutr. 2018 doi: 10.1016/j.clnu.2018.10.024. S0261-5614(18)32519-6. [DOI] [PubMed] [Google Scholar]
- 7.Liu H., Li C., Yang J., Sun Y., Zhang S., Yang J., Yang L., Wang Y., Jiao B. Long noncoding RNA CASC9/miR-519d/STAT3 positive feedback loop facilitate the glioma tumourigenesis. J. Cell. Mol. Med. 2018;22:6338–6344. doi: 10.1111/jcmm.13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu H., Zhou M., Sheng Z., Chen Y., Yeh C.K., Chen W., Liu J., Liu X., Yan F., Zheng H. Theranostic nanosensitizers for highly efficient MR/fluorescence imaging-guided sonodynamic therapy of gliomas. J. Cell. Mol. Med. 2018;22:5394–5405. doi: 10.1111/jcmm.13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun X., Wang J., Huang M., Chen T., Chen J., Zhang F., Zeng H., Xu Z., Ke Y. STAT3 promotes tumour progression in glioma by inducing FOXP1 transcription. J. Cell. Mol. Med. 2018;22:5629–5638. doi: 10.1111/jcmm.13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbagallo D., Caponnetto A., Cirnigliaro M., Brex D., Barbagallo C., D’Angeli F., Morrone A., Caltabiano R., Barbagallo G.M., Ragusa M. CircSMARCA5 Inhibits Migration of Glioblastoma Multiforme Cells by Regulating a Molecular Axis Involving Splicing Factors SRSF1/SRSF3/PTB. Int. J. Mol. Sci. 2018;19:E480. doi: 10.3390/ijms19020480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu J., Zhao W., Wang Z., Xiang X., Zhang S., Liu L. Long non-coding RNA SNHG20 promotes the tumorigenesis of oral squamous cell carcinoma via targeting miR-197/LIN28 axis. J. Cell. Mol. Med. 2019;23:680–688. doi: 10.1111/jcmm.13987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L., Fu J., Zhou Y. Circular RNAs and Their Emerging Roles in Immune Regulation. Front. Immunol. 2018;9:2977. doi: 10.3389/fimmu.2018.02977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang N., Li G., Li X., Xu L., Chen M. Circ5379-6, a circular form of tumor suppressor PPARα, participates in the inhibition of hepatocellular carcinoma tumorigenesis and metastasis. Am. J. Transl. Res. 2018;10:3493–3503. [PMC free article] [PubMed] [Google Scholar]
- 14.Nan A., Chen L., Zhang N., Jia Y., Li X., Zhou H., Ling Y., Wang Z., Yang C., Liu S., Jiang Y. Circular RNA circNOL10 Inhibits Lung Cancer Development by Promoting SCLM1-Mediated Transcriptional Regulation of the Humanin Polypeptide Family. Adv. Sci. (Weinh) 2018;6 doi: 10.1002/advs.201800654. 1800654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang X., Li Y., Liu Y., Xu X., Wang Y., Yan Y., Zhou W., Yang J., Wei W. Novel circular RNA expression profile of uveal melanoma revealed by microarray. Chin. J. Cancer Res. 2018;30:656–668. doi: 10.21147/j.issn.1000-9604.2018.06.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 17.Wang R., Zhang S., Chen X., Li N., Li J., Jia R., Pan Y., Liang H. EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis. Mol. Cancer. 2018;17:166. doi: 10.1186/s12943-018-0911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F., Huang N., Yang X., Zhao K., Zhou H. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J. Natl. Cancer Inst. 2018;110:304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao W., Ma X., Liu L., Chen Q., Liu Z., Zhang Z., Ma S., Wang Z., Li H., Wang Z., Wu J. SNHG20: A vital lncRNA in multiple human cancers. J. Cell. Physiol. 2019;234:14519–14525. doi: 10.1002/jcp.28143. [DOI] [PubMed] [Google Scholar]
- 20.Qu S., Liu Z., Yang X., Zhou J., Yu H., Zhang R., Li H. The emerging functions and roles of circular RNAs in cancer. Cancer Lett. 2018;414:301–309. doi: 10.1016/j.canlet.2017.11.022. [DOI] [PubMed] [Google Scholar]
- 21.Kristensen L.S., Hansen T.B., Venø M.T., Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng S., Zhou H., Feng Z., Xu Z., Tang Y., Li P., Wu M. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol. Cancer. 2017;16:94. doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu J., Qi X., Liu L., Hu X., Liu J., Yang J., Yang J., Lu L., Zhang Z., Ma S. Emerging Epigenetic Regulation of Circular RNAs in Human Cancer. Mol. Ther. Nucleic Acids. 2019;16:589–596. doi: 10.1016/j.omtn.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li G.F., Li L., Yao Z.Q., Zhuang S.J. Hsa_circ_0007534/miR-761/ZIC5 regulatory loop modulates the proliferation and migration of glioma cells. Biochem. Biophys. Res. Commun. 2018;499:765–771. doi: 10.1016/j.bbrc.2018.03.219. [DOI] [PubMed] [Google Scholar]
- 25.Li G., Yang H., Han K., Zhu D., Lun P., Zhao Y. A novel circular RNA, hsa_circ_0046701, promotes carcinogenesis by increasing the expression of miR-142-3p target ITGB8 in glioma. Biochem. Biophys. Res. Commun. 2018;498:254–261. doi: 10.1016/j.bbrc.2018.01.076. [DOI] [PubMed] [Google Scholar]
- 26.Rong D., Lu C., Zhang B., Fu K., Zhao S., Tang W., Cao H. CircPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR-296-5p. Mol. Cancer. 2019;18:25. doi: 10.1186/s12943-019-0958-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Shi F., Shi Z., Zhao Y., Tian J. CircRNA hsa-circ-0014359 promotes glioma progression by regulating miR-153/PI3K signaling. Biochem. Biophys. Res. Commun. 2019;510:614–620. doi: 10.1016/j.bbrc.2019.02.019. [DOI] [PubMed] [Google Scholar]
- 28.Xu J.Z., Shao C.C., Wang X.J., Zhao X., Chen J.Q., Ouyang Y.X., Feng J., Zhang F., Huang W.H., Ying Q. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis. 2019;10:175. doi: 10.1038/s41419-019-1382-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X., Wang S., Wang H., Cao J., Huang X., Chen Z., Xu P., Sun G., Xu J., Lv J., Xu Z. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol. Cancer. 2019;18:20. doi: 10.1186/s12943-018-0935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang H.F., Zhang X.Z., Liu B.G., Jia G.T., Li W.L. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am. J. Cancer Res. 2017;7:1566–1576. [PMC free article] [PubMed] [Google Scholar]
- 31.Guan H., Cai J., Zhang N., Wu J., Yuan J., Li J., Li M. Sp1 is upregulated in human glioma, promotes MMP-2-mediated cell invasion and predicts poor clinical outcome. Int. J. Cancer. 2012;130:593–601. doi: 10.1002/ijc.26049. [DOI] [PubMed] [Google Scholar]
- 32.Chang K.Y., Huang C.T., Hsu T.I., Hsu C.C., Liu J.J., Chuang C.K., Hung J.J., Chang W.C., Tsai K.K., Chuang J.Y. Stress stimuli induce cancer-stemness gene expression via Sp1 activation leading to therapeutic resistance in glioblastoma. Biochem. Biophys. Res. Commun. 2017;493:14–19. doi: 10.1016/j.bbrc.2017.09.095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.