Abstract
Recent studies show p85α up-regulates epidermal growth factor (EGF) receptor, thereby promoting malignant cell transformation and migration in normal mouse embryonic fibroblasts (MEFs). However, the potential role of p85α in human bladder cancer (BC) remains unknown. Here, we show that p85α is down-regulated in BC tumor tissues. Ectopic expression of p85α inhibited cell invasion, but not migration, whereas p85α knockdown promoted invasion in BC cells, revealing that p85α inhibits BC invasion. Overexpression of kinase-deficient p110 in T24 T(p85α) cells inhibited BC cell migration, but not invasion, suggesting that the inhibition of p85α on invasion is independent of PI3K activity. The effect of p85α on inhibiting BC invasion was mediated by the inactivation of MMP-2 concomitant with the up-regulation of TIMP-2 and down-regulation of MMP-14. Mechanistic studies revealed c-Jun inactivation was associated with p85α knockdown-induced MMP-14 expression, and down-regulated miR-190, leading to ATG7 mRNA degradation. This suppressed the autophagy-dependent removal of TIMP-2 in human BC cells. The present results identify a novel function of p85α and clarify the mechanisms underlying its inhibition of BC invasion, providing insight into the role of p85α in normal and cancer cells.
Abbreviations: EGF, Epidermal growth factor receptor; BC, Bladder cancer; MMPs, Matrix metalloproteinases; TIMP-2, Tissue inhibitor of metalloproteinase-2; PI3K, Phosphatidylinositol 3-kinase; EGFR, Epidermal growth factor receptor; MEFs, Mouse embryonic fibroblasts; ATG, Autophagy-related
Introduction
Bladder cancer (BC) is the most common cause of death in patients with urinary system malignancies. The incidence and mortality of BC have increased in recent decades [1]. BC is a heterogeneous disease, and 70% of patients present with superficial tumors, which tend to recur but are generally not life-threatening, whereas 30% present with muscle-invasive disease associated with a high risk of death from distant metastases [2]. Tumor metastasis is the primary cause of death in BC, and is associated with a 5-year survival rate of approximately 8.1% [1]. Although different markers are associated with disease progression, including depth of invasion, stage, and grade, definitive clinical prognostic markers are lacking, and the cellular mechanism underlying disease progression remains unclear [3], [4].
Matrix metalloproteinases (MMPs) are zinc-dependent endopeptidases with a conserved domain structure that are secreted in a latent (pro) form and activated by proteolytic removal of the NH2-terminal propeptide [5]. MMPs promote cancer progression by catalyzing the degradation of the extracellular matrix, which allows cancer cells to migrate out of the primary tumor to form metastases [6], [7]. In BC, MMP-2 activation is higher in invasive than in superficial tumors [8]. MMP-2 is secreted in its latent form (pro-MMP-2) and activated on the cell surface, which is mediated by two molecules: MMP-14, a membrane type 1 MMP that can induce MMP-2 activation [9], and tissue inhibitor of metalloproteinase-2 (TIMP-2), which has a dual role in the regulation of MMP-14–induced MMP-2 activation [10], [11]. However, little is known about their upstream regulators. Therefore, exploring the mechanisms underlying MMP-2 activation is important.
p85α is a class IA phosphatidylinositol 3-kinase (PI3K) regulatory subunit that binds, stabilizes, and inhibits the p110 catalytic subunit to allow receptor tyrosine kinase activation [12]. p85α up-regulates epidermal growth factor receptor (EGFR) expression, thereby promoting malignant cell transformation and migration in normal mouse embryonic fibroblasts (MEFs) in response to EGF [13]. However, p85α mRNA expression is down-regulated in human prostate, lung, ovarian, bladder, and liver cancers, consistent with the proposed tumor suppressor role of p85α. p85α acts as a tumor suppressor by negatively regulating growth factor signaling in the liver [14]. In response to the loss of PTEN expression, monomeric p85α negatively regulates PI3K signaling to suppress tumor growth [15]. The p85α monomer also has important functions independent of its regulation of PI3K, including the detachment of insulin receptor substrate proteins and positive regulation of PTEN function [16]. High p85α protein expression is significantly correlated with invasive breast cancer [17]. Therefore, the downstream regulators involved in human BC progression need to be identified.
Recent work from our group demonstrated that p85α is down-regulated in BC, and inhibits human BC invasion via a PI3K-independent pathway in vitro. In the present study, we investigated the effect of p85α down-regulation on the invasive ability of human BC cells. The results indicated that p85α regulates cancer cell invasion by inhibiting MMP-2 activation through the suppression of MMP-14 mRNA transcription and TIMP-2 protein degradation via the c-Jun/Talin2/ATG7/autophagy axis.
Materials and Methods
Cell Culture and Reagents
T24 T cells were described in our previous publication [18] and cultured in DMEM:F-12 = 1:1 with 5% FBS. UMUC3 cells were described in our previous studies [19], [20] and cultured in DMEM with 10% FBS (ATLANTA, Flowery Branch, GA, USA). Cells were maintained in a humidified incubator at 37 °C and a 5% CO2 atmosphere. Antibodies against p85α (Abcam, Cambridge, MA, USA); BECLIN1, ATG3, ATG5/12, ATG7, LC3, HUR, and RhoA (Cell Signaling Technology Inc., Beverly, MA, USA); Sp1, E2F1, β-actin, and α-tubulin (Santa Cruz Biotechnology, Santa Cruz, CA, USA); and MMP-14, TIMP-2, SESN2, and p62 (GeneTex, Inc., CA, USA) were used in the study.
Plasmids and Stable Cell Transfection
The shRNA targeting human p85α was purchased from Open Biosystems (Lafayette, CO, USA). The p85α overexpression plasmid was bought from Addgene (Cambridge, MA, USA). Constructs for p110* and p110*∆kin overexpression were gifts from Professor Williams LT (Cardiovascular Research Institute and Daiichi Research Center, University of California, San Francisco, CA, USA.), and the TAM67 plasmid, a well-characterized dominant-negative c-Jun mutant, was described in our previous study [21]. The talin2 promoter luciferase reporter was kindly provided by Dr. Fei Chen, (Department of Pharmaceutical Sciences, Wayne State University, Detroit, MI, USA). shRNA constructs targeting ATG7 (shATG7), and the antisense plasmid for miR-190 were purchased from Open Biosystems (Pittsburg, PA, USA). All plasmids were prepared using the Plasmid Preparation/Extraction Maxi kit from QIAGEN (Valencia, CA, USA). Cell transfections were performed with PolyJet™ DNA in vitro Transfection Reagent (SignaGen Laboratories, Rockville, MD, USA) according to the manufacturer's instructions. For stable transfections, cell cultures were subjected to hygromycin B, G418, or puromycin selection according to the resistance of plasmids, and surviving cells were pooled as stable mass transfectants.
Western Blot Analysis
Whole cell extracts were prepared using cell lysis buffer (10 mM Tris–HCl, pH 7.4, 1% SDS, and 1 mM Na3VO4) as described in our previous study [22]. Aliquots containing 50 μg of protein were resolved by SDS-PAGE, transferred to a PVDF membrane, and probed with the indicated primary antibodies followed by AP-conjugated secondary antibody. Signals were detected using the enhanced chemifluorescence western blotting system as described in a previous report [18]. The images were acquired by scanning with a phosphoimager (Typhoon FLA 7000 imager; Pittsburgh, PA, USA).
Luciferase Promoter Reporter Assay
ATG7/talin2 promoter luciferase reporters were transiently transfected into cultured cells. Twenty-four hours after the transfection, luciferase activity was determined using the Luciferase Assay System kit (Promega, Madison, WI, USA). The results were normalized to the internal TK signal. All experiments were performed in triplicate, and the results were expressed as the mean ± standard error (SE).
Cell Invasion Assay
The invasion kit was purchased from BD Falcon 353,047 (Franklin Lakes, NJ, USA). The invasion assay was performed according to the manufacturer's instructions in normal cell culture serum. Cells were seeded in Transwell chambers and fixed with 3.7% formalin for 2 min, washed twice with PBS, transferred to 100% methanol for 20 min, washed twice again, and finally stained by Giemsa (1:20 diluted with PBS) at room temperature for 15 min in the dark. After staining, the cells were washed twice with PBS, and non-invaded cells were scraped off with a cotton swab (PBS wetted) four times. Images were captured under an Olympus DP71, and the number of cells was calculated using Image J software as described previously [18], [23].
RT-PCR
Total RNA was extracted using the TRIzol reagent as described in the manufacturer's instructions (Invitrogen, Grand Island, NY, USA). Total RNA (5 μg) was used for first-strand cDNA synthesis with oligodT primers using the SuperScript IV First-Strand Synthesis system (Invitrogen). The primers used for PCR amplification were as follows: human ATG7, forward: 5′-GCC AAG ATC TCC TAC TCC A-3′, reverse: 5′-CAG AAG TAG CAG CCA AGC TTG T-3′; human MMP-14, forward: 5′-TTG GAC TGT CAG GAA TGA GG-3′, reverse: 5′-GCA GCA CAA AAT TCT CCG TG-3′; human TIMP2, forward: 5′-TAC GGC AGC AAG TCC AAT-3′, reverse: 5′-CCG CTC AAA TAC CTT CAC A-3′; human MMP-2, forward: 5′-CAA GTG GGA CAA GAA CCA GA-3′, reverse: 5′-CCA AAG TTG ATC ATG ATG T-3′; human GAPDH, forward: 5′-AGA AGG CTG GGG CTC ATT TG-3′, reverse: 5′-AGG GGC CAT CCA CAG TCT TC-3′; and human Talin2, forward: 5′-TGG TCA AAT CGG CCT CAG CA-3′, reverse: 5′-CCC TGA ACG GAG GCA TTG GC-3′.
Quantitative RT-PCR
RNA extraction from cells was performed with the miRNeasy Mini Kit (QIAGEN). Total RNA (1 μg) was used for reverse transcription. Analysis of human miR-190 (5′-TAT GTT TGA TAT ATT AGG T-3′) was performed using the miScript PCR system (QIAGEN) by the QuantStudio Real-time PCR system (Applied Biosystems). The primer for miRNA assays was purchased from Invitrogen, and U6 was used as the control. The initial activation was performed at 95 °C for 15 min, followed by 40 cycles of denaturation at 95 °C for 15 s, annealing at 55 °C for 30 s, and extension at 70 °C for 30 s. The data were analyzed as described previously.
Statistical Analysis
Student's t-test was used to determine the significance between groups. P < .05 was considered as a significant difference between groups.
Results
p85α is Down-Regulated in BC and Inhibits Human BC Cell Invasion in a PI3K-Independent Manner
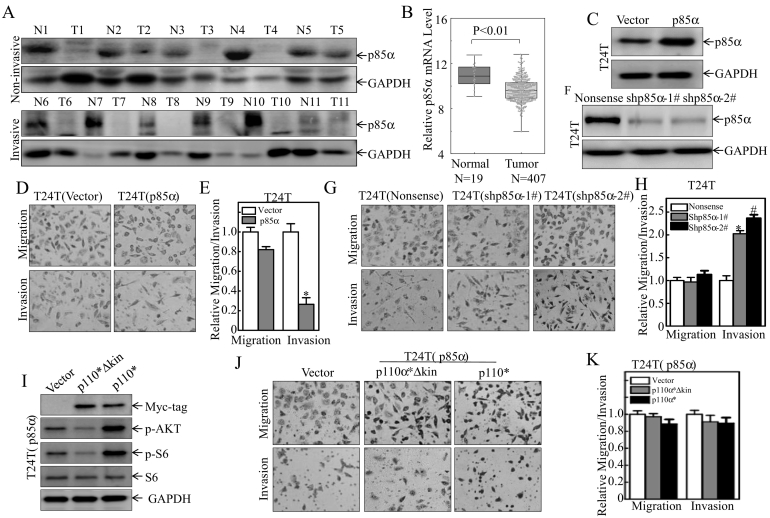
The Cancer Genome Atlas data and analysis of normal and tumor tissue samples from 11 patients with non-invasive and invasive BC showed that p85α expression was lower in tumor than in normal tissues (Figure 1, A and B), revealing the potential association of p85α with BC invasion. To test this notion, stable T24 T transfectants overexpressing p85α were established (Figure 1,C), and ectopic expression of p85α significantly impaired the invasive ability of T24 T cells without affecting migration, indicating that p85α is involved in regulation of the invasiveness of BC (Figure 1, D and E). This notion was further supported by the results obtained from using short-hairpin RNA (shRNA)-mediated knockdown of p85α expression in T24 T cells, which enhanced invasive abilities of T24 T cells (Figure 1, F–H). Transfection of T24 T (p85α) cells with p110*, an active form of p110, and p110*∆kin, a kinase-deficient form of p110*, which is a constitutively active chimera that contains the iSH2 domain of p85α fused to the NH2 terminus of p110 by means of a flexible glycine linker, had no effect on the invasive capacity of cells (Figure 1, I–K), suggesting that p85α inhibits human BC cell invasion in a PI3K-independent manner.
Figure 1.
p85α is down-regulated in bladder cancers in vivo and inhibits human bladder cancer cell invasion independently of PI3K in vitro.
(A) Eleven pairs of normal bladder and tumor tissue samples were analyzed. (B) The expression of p85α was analyzed in human bladder cancer and normal samples using The Cancer Genome Atlas (TCGA) cohort. (C, F, & I) Cell extracts from the stable transfectants T24 T(p85α), T24 T(shp85α-1#), T24 T(shp85α-2#), T24 T(p85α/p110*∆kin), T24 T(p85α/p110*), and the respective vector controls were subjected to Western blot analysis to determine the expression of p85α and the Myc-tag using the indicated antibodies. GAPDH was used as the protein loading control. (D, E, G, H, J & K) The invasive abilities of T24 T(p85α), T24 T(vector), T24 T(shp85α-1#), T24 T(shp85α-2#), T24 T(Nonsense), T24 T(p85α/p110*∆kin), T24 T(p85α/p110*), and T24 T(p85α), were determined using BD BioCoat™ Matrigel™ Invasion Chambers. Migration ability was determined using the empty insert membrane without the Matrigel. Invasion ability was assessed using the same system except for the inclusion of Matrigel. The invasion rate was normalized to the insert control according to the manufacturer's instructions. “*” indicates a significant difference in invasion abilities in T24 T(p85α) vs. T24 T(vector), and T24 T(shp85α) vs. T24 T(Nonsense) cells (P < .05). The results are presented as the mean ± SD from three independent experiments.
ATG7-Mediated Autophagy is Essential for p85α Inhibition of BC Cell Invasion
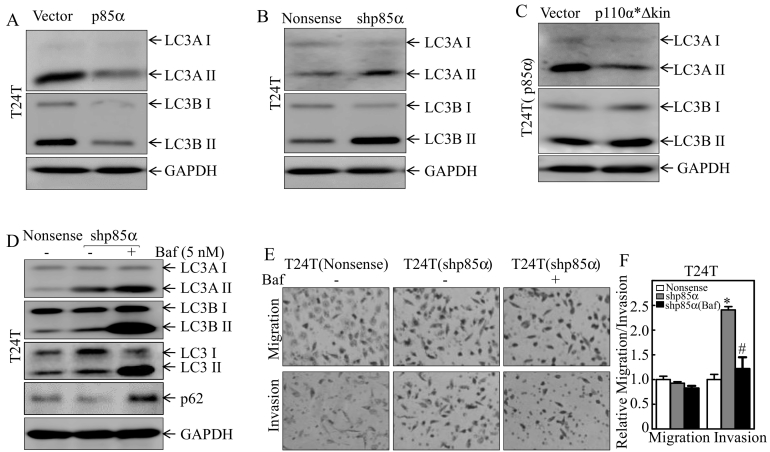
Ectopic expression of p85α in T24 T cells suppressed the conversion of both autophagy markers, LC3A and LC3B, from LC3-I to LC3-II (Figure 2A), whereas shRNA-mediated silencing of p85α promoted the conversion of LC3 from LC3-I to LC3-II (Figure 2B). These results reveal that p85α has an inhibitory effect on human BC cell autophagy. Consistent with results observed on cell invasive abilities, transfection of p110*∆kin into T24 T(p85α) cells did not restore autophagic level in T24 T(p85α) cells (Figure 2C). Moreover, the inhibition of autophagy by 5 nM of bafilomycin A1 (Baf) in T24 T(shp85α) cells specific suppressed invasion without affecting migration of T24 T(shp85α) cells (Figure 2, D–F), indicating that p85α specific inhibits human BC invasion by suppressing autophagy.
Figure 2.
p85α inhibition of human bladder cancer invasion is associated with its suppression of autophagy in human BC cells.
(A–C) The indicated cells were seeded into 6-well plates and cultured till 80–90% confluent. The cell extracts were subjected to Western blot analysis with the indicated antibodies. GAPDH was used as the protein loading control. (D) The indicated stable transfectants were treated with or without Baf (5 nM) for 24 h, and cell extracts were subjected to Western blot analysis with the indicated antibodies. (E and F) Invasion was evaluated using BD BioCoat™ Matrigel™ Invasion Chambers in the presence of either vehicle or the indicated concentration of Baf. “*” indicates a significant difference between control and shp85α transfectants, and “#” indicates a significant difference compared with the vehicle control group (P < .05). The results are presented as the mean ± SD from three independent experiments.
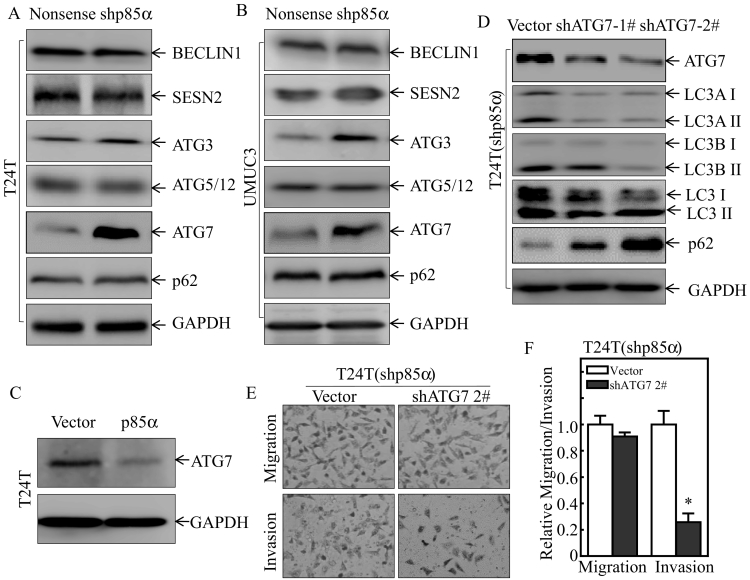
The autophagy-related (ATG) protein family, including ATG3, ATG7, and ATG12/ATG5, regulates cell autophagy in many experimental systems [24]. BECLIN1 is a critical upstream regulator of autophagy [25]. In addition, SESN2 and p62 are also associated with autophagy pathway [26]. Therefore, the effect of p85α on the expression of various ATG family members and other related molecules was determined in two pairs of cells, T24 T(Nonsense) vs. T24 T(shp85α) cells, and UMUC3(Nonsense) vs. UMUC3(shp85α) cells. The results indicated that p85α inhibited ATG7 expression, whereas it had no consistent effect on other related proteins (Figure 3, A and B). ATG7 expression was lower in T24 T(p85α) cells than in T24 T(Vector) cells (Figure 3C). shRNA-mediated knockdown of ATG7 in T24 T(shp85α) cells impaired the autophagy in T24 T(shp85α) cells (Figure 3D). Consistently, ATG7 knockdown also abolished the increase in invasive ability in T24 T(shp85α) cells (Figure 3, E and F). These results reveal that down-regulation of ATG7 mediates p85α inhibition of autophagy and in turn inhibiting BC invasion.
Figure 3.
p85α inhibition of ATG7 mediated its suppression of autophagy in human BC cells.
(A–B) Whole cells lysates from the indicated transfectants were subjected to Western blot analysis of the autophagy pathway, and GAPDH was used as the protein loading control. (C) Whole cell lysates were subjected to Western blot analysis. GAPDH was used as the protein loading control. (D) ATG7 knockdown plasmids were stably transfected into T24 T(shp85α) cells, and whole cells lysates were subjected to Western blot analysis. GAPDH was used as the protein loading control. (E and F) The invasive abilities of the indicated stable transfectants and their scramble vector transfectants were evaluated using BD BioCoat™ Matrigel™ Invasion Chambers. “*” indicates a significant difference in invasion ability between T24 T(shp85α/Vector) and T24 T(shp85α/shATG72#) (P < .05). The results are presented as the mean ± SD from three independent experiments.
p85α-Mediated miR-190 Down-Regulation via Inhibition of Talin2 Transcription Regulates ATG7 mRNA Stability
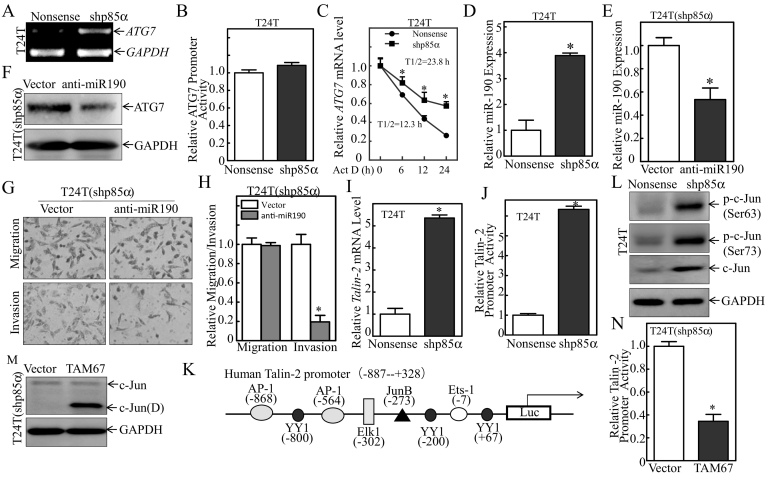
To evaluate the molecular mechanisms underlying the effect of p85α on down-regulating ATG7 expression, ATG7 mRNA levels were examined in T24 T(Nonsense) and T24 T(shp85α) cells (Figure 4A). Knockdown of p85α up-regulated ATG7 mRNA in T24 T(shp85α) cells compared with T24 T(Nonsense) cells. p85α knockdown had no effect on the promoter transcriptional activity (Figure 4B). The results indicate that p85α does not inhibit ATG7 expression at the transcriptional level, which further implies the possible regulation of ATG7 mRNA stability by p85α. To test this hypothesis, T24 T cells expressing Nonsense/shATG7 were treated with actinomycin D (Act D) for different times and ATG7 mRNA degradation rates were evaluated. The results showed that p85α knockdown did increase ATG7 mRNA stability (Figure 4C), indicating that ATG7 expression is regulated at mRNA degradation level by p85α in human BC cells.
Figure 4.
p85α-mediated miR-190 down-regulation associated with inhibition of Talin2 transcription regulated ATG7 mRNA stability.
(A) Total RNA was extracted from the indicated stable transfectants using TRIzol. ATG7 mRNA was determined by RT-PCR using specific primers. GAPDH was used as an internal control. (B) T24 T(Nonsense) and T24 T(shp85α) cells were transfected with an ATG7 promoter-driven luciferase reporter together with pRL-TK. The transfectants were seeded into 96-well plates to determine ATG7 promoter transcriptional activity. pRL-TK was used as an internal control to normalize the transfection efficiency. The results are presented as the mean ± SD from three replicate assays. (C) T24 T(Nonsense) and T24 T(shp85α) cells were seeded into 6-well plates. After synchronization, the cells were treated with Actinomycin D (Act D) for the indicated time points, and total RNA was isolated and subjected to qRT-PCR for analysis of mRNA level. (D) miR-190 levels were evaluated by qRT-PCR. The results are presented as the mean ± SD from triplicate experiments. “*” indicates a significant increase compared with the nonsense transfectant (P < .05). (E) A miR-190 inhibitor plasmid was stably transfected into T24 T(shp85α) cells, and the expression efficiency was determined by qRT-PCR. The results are presented as the mean ± SD of triplicate experiments. “*” indicates a significant decrease compared with T24 T(shp85α/vector) cells (P < .05). (F) T24 T(shp85α/vector) and T24 T(shp85α/anti-miR-190) cells were subjected to Western blotting for detection of the indicated proteins. GAPDH was used as the protein loading control. (G and H) The invasive abilities of T24 T(shp85α/vector) and T24 T(shp85α/anti-miR-190) cells were determined using a Transwell invasion assay. Migration ability was determined using the empty insert membrane without the Matrigel, and invasion was evaluated using the same system except for the addition of Matrigel. The invasion rate was normalized to the insert control according to the manufacturer's instructions, and the results were presented as the number of invasive T24 T(shp85α/anti-miR-190) cells relative to invasive T24 T(shp85α/vector) transfectants. “*” indicates a significant difference in invasion ability between T24 T(shp85α/vector) and T24 T(shp85α/anti-miR-190) cells (P < .05). (I) the Talin2 mRNA expression level was evaluated by qRT-PCR in T24 T(Nonsense) and T24 T(shp85α) cells. (J) Talin2 promoter luciferase activity was evaluated by the Dual-Luciferase Reporter Assay System. The results are presented as the mean ± SD of triplicate experiments. “*” indicates a significant increase compared with nonsense cells (P < .05). (K) Schematic representation of the transcription factor binding sites of the human Talin2 gene promoter. (L) The expression of transcription factors was analyzed by Western blotting with GAPDH as the protein loading control. (M) TAM67 was stably transfected into T24 T(shp85α) cells, and cell extracts from T24 T(shp85α/vector) and T24 T(shp85α/TAM67) were subjected to Western blot analysis for detection of the indicated proteins. GAPDH was used as the protein loading control. (N) The Dual-Luciferase Reporter Assay System was used to evaluate talin2 promoter luciferase activity. The results are expressed as the mean ± SD of triplicate experiments. “*” indicates a significant decrease compared with nonsense cells (P < .05).
MicroRNAs (miRNAs), a class of noncoding small RNAs, bind to the 3′ untranslated region (3′-UTR) of target genes and inhibit protein translation [27] or induce target mRNA degradation in human cells. We most recently identify miR-190 as a highly up-regulated miRNA at the transcriptional level in BC tissues and cell lines, and show that it binds to the ATG7 3′-UTR to regulate its stability. To determine whether miR-190 regulated p85α-dependent ATG7 expression, miR-190 levels were analyzed in T24 T(Nonsense) and T24 T(shp85α) cells (Figure 4D). The results of quantitative reverse-transcription PCR (qRT-PCR) indicated that miR-190 was significantly up-regulated upon knockdown of p85α, indicating that miR-190 may be negatively regulated by p85α. To evaluate the effect of miR-190 on ATG7 expression, we generated stable T24 T(shp85α) transfectants expressing anti-miR-190, which showed over 2-fold lower expression of miR-190 than the corresponding empty vector transfectants (Figure 4E). Inhibition of miR-190 in T24 T(shp85α) cells dramatically reduced ATG7 expression (Figure 4F), suggesting that miR-190 targeted ATG7 mRNA. Assessment of the effect of miR-190 on the invasive capacity of BC cells showed that inhibition of miR-190 suppressed T24 T(shp85α) cell invasion (Figure 4, G and H).
miR-190 is conserved and located in the intronic region of its host gene, Talin2, in mice, rats, and humans (14), and talin2 regulates the expression of miR-190. We therefore measured the mRNA levels of Talin2 (Figure 4I), and showed that Talin2 mRNA expression was significantly higher in T24 T(shp85α) cells than in T24 T(Nonsense) cells. Then, the promoter activity of Talin2 was evaluated and compared between T24 T(Nonsense) and T24 T(shp85α) cells. As shown in Figure 4J, knockdown of p85α significantly increased the promoter activity of Talin2, indicating that p85α inhibits the transcription of Talin2/miR-190. We next performed a bioinformatics scan on the promoter region of Talin2, which identified several potential binding sites for transcription factors in the Talin2 promoter region, including binding sites for Ets1, YY1, AP-1, and Elk-1 (Figure 4K). To define the specific transcription factors involved in the regulation of Talin2, the expression of these transcription factors was examined in T24 T(Nonsense) and T24 T(shp85α) cells as indicated in Figure 4L. The c-Jun protein and c-Jun phosphorylation (ser63/73) levels were increased in p85α knockdown cells, consistent with the alteration of Talin2 mRNA and promoter activity in those transfectants. Therefore, the c-Jun dominant-negative mutant TAM67 was transfected into T24 T(shp85α) cells to determine the potential contribution of c-Jun to the activity of the Talin2 promoter (Figure 4M). Ectopic expression of TAM67 successfully blocked Talin2 promoter activity (Figure 4N), suggesting that c-Jun plays an important role in the p85α-mediated down-regulation of Talin2 mRNA. Taken together, these results demonstrate that p85α inhibits c-Jun and in turn suppresses Talin2/miR-190 transcription, thereby down-regulating miR-190 expression in human BC cells.
p85α Inhibits Human BC Invasion by Regulating MMP-2 Activation
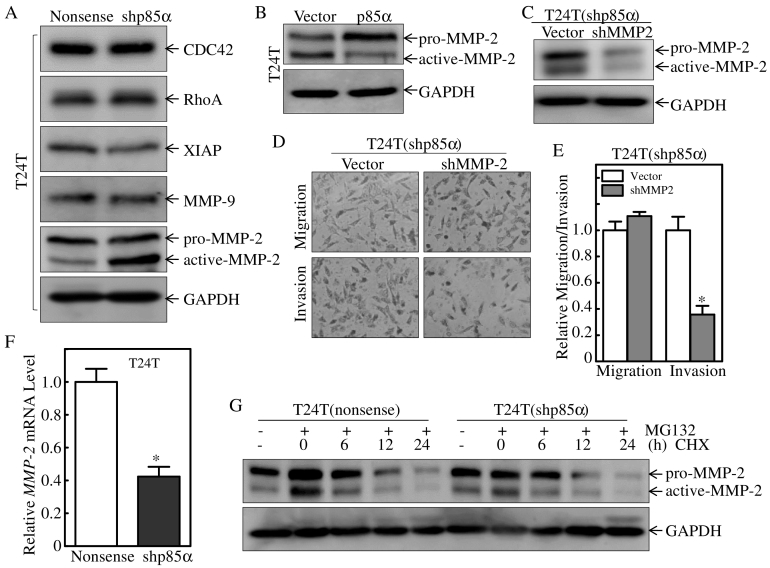
Many molecules have been implicated in BC cell invasion. Cdc42 plays a crucial role in regulating migration, and Rho GTPases are key players in this process [28]. Previous work from our group showed that XIAP regulates cell motility through its effect on actin polymerization and cytoskeleton formation [29]. MMPs play important roles in tumor spread [8], and MMP-9 expression and MMP-2 activity affect the progression, invasion, and metastasis of various cancers [30]. Detection of molecules involved in BC invasion in T24 T(Nonsense) and T24 T(shp85α) cells showed that activated MMP-2, but not pro-MMP2, was specifically up-regulated in T24 T(shp85α) cells compared with T24 T(Nonsense) cells, whereas other molecules did not correlate with the effect of p85α on down-regulating human BC invasion (Figure 5A). Taken together with the down-regulation of activated-MMP-2 and up-regulation of pro-MMP2 in T24 T(p85α) cells compared with T24 T(Vector) cells (Figure 5B), these results suggested that p85α plays an important role in modulating MMP-2 activation. To determine whether p85α inhibits human BC invasion by down-regulating MMP-2 activation, T24 T cells expressing shp85α/Vector and shp85α/shMMP2 were subjected to cell invasion assays. The results showed that MMP-2 knockdown decreased the invasive ability of T24 T(shp85α) cells (Figure 5, C–E), indicating that MMP-2 is crucial for invasion of T24 T(shp85α) cells, and further suggesting that p85α inhibits human BC invasion by specific decreasing MMP-2 activation. Our results also showed that MMP-2 mRNA level was down-regulated in T24 T(shP85α) cells (Figure 5F), which is inconsistent with MMP-2 protein level, while the MMP-2 protein degradation rates were comparable between T24 T(Nonsense) and T24 T (shp85α) cells (Figure 5G). Collectively, our results reveal that p85α inhibits human BC invasion by inhibiting MMP-2 activation, rather than increasing total MMP-2 protein expression.
Figure 5.
p85α inhibits human bladder cancer invasion by suppression of MMP-2 activation.
(A and B) The indicated cells were seeded into 6-well plates and cultured until 80–90% confluent. The cell extracts were then subjected to Western blot analysis with the indicated antibodies. GAPDH was used as the protein loading control. (C) Extracts from T24 T(shp85α/Vector) and T24 T(shp85α/shMMP2) cells were subjected to Western blot analysis for detection of the indicated proteins. GAPDH was used as the protein loading control. (D and E) The invasive abilities of the indicated stable transfectants and the corresponding scramble vector control transfectants were evaluated using BD BioCoat™ Matrigel™ Invasion Chambers. “*” indicates a significant difference in invasive ability between T24 T(shp85α/Vector) and T24 T(shp85α/shMMP2) cells (P < .05). The results are presented as the mean ± SD from three independent experiments. (F) MMP-2 mRNA expression levels were evaluated by qRT-PCR in T24 T(Nonsense) and T24 T(shp85α) cells. (G) The indicated stable transfectants were pretreated with or without MG132 (10 μM) for 6 h, and then treated with cycloheximide (CHX, 100 μg/ml) for the indicated times. The cell extracts were subjected to Western blotting for determination of MMP-2 degradation, and GAPDH was used as the protein loading control.
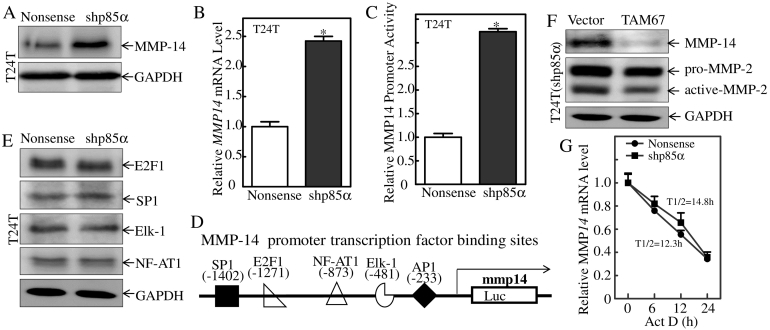
MMP-14 mRNA Transcription is a p85α Downstream Effector Promoting MMP-2 Activation
MMP-2 is secreted in its latent form (pro-MMP-2) and activated on the cell surface. MMP-14, also known as membrane type 1 MMP, can induce MMP-2 activation. We therefore analyzed the expression of the MMP-14 protein in T24 T(Nonsense) and T24 T(shp85α) cells, which showed that knockdown of p85α markedly up-regulated MMP-14 (Figure 6A). Consistently with protein levels, MMP-14 mRNA levels were also higher in T24 T(shp85α) cells than in T24 T(Nonsense) cells (Figure 6B). Next, we compared the promoter activity of MMP-14 between T24 T(Nonsense) and T24 T(shp85α) cells. As shown in Figure 6C, knockdown of p85α significantly increased MMP-14 promoter activity in human T24 T cells. A bioinformatics scan of the promoter region of MMP-14 identified several potential binding sites for transcription factors, including binding sites for E2F1, SP1, NFAT-1, AP-1, and Elk-1 (Figure 6D). To define the specific transcription factors involved in the regulation of MMP-14, the expression of these transcription factors was assessed in T24 T(Nonsense) and T24 T(shp85α) cells and there was no remarkable alteration of these transcription factors shown in Figure 6E. Taken together with our results obtained from observation of p85α on c-Jun as indicated in Figure 4L, the up-regulated levels of c-Jun protein and c-Jun phosphorylation (ser63/73) in p85α knockdown cells were consistent with the alteration of MMP-14 in the transfectants. Therefore, the effect of c-Jun on the expression of MMP-14 was assessed in T24 T(shp85α) cells transfected with the c-Jun dominant-negative mutant TAM67 (Figure 6F). Ectopic expression of TAM67 decreased MMP-14 protein levels and activated MMP-2 level (Figure 6F), suggesting that the inhibition of c-Jun plays a role in the p85α-mediated suppression of MMP-14 mRNA. Given our results showing that MMP-14 mRNA degradation rates did not show the observable difference between the T24 T(Nonsense) and T24 T(shp85α) transfectants (Figure 6G), our results demonstrated that p85α inhibits c-Jun and down-regulates MMP-14 mRNA transcription, thereby reducing MMP-2 activation in human BC cells.
Figure 6.
MMP-14 mRNA transcription is a p85α downstream effector that mediates MMP-2 activation.
(A) The indicated cell extracts were subjected to Western blot analysis with the indicated antibodies. GAPDH was used as the protein loading control. (B) MMP-14 mRNA expression levels were evaluated by qRT-PCR in T24 T(Nonsense) and T24 T(shp85α) cells. (C) MMP-14 promoter luciferase activity was evaluated using the Dual-Luciferase Reporter Assay System. The results are expressed as the mean ± SD of triplicate experiments. “*” indicates a significant increase compared with nonsense cells (P < .05). (D) Schematic representation of transcription factor binding sites in the human MMP-14 gene promoter. (E) Transcription factor expression was evaluated by Western blotting with GAPDH as the protein loading control. (E) Extracts from T24 T(shp85α/vector) and T24 T(shp85α/TAM67) cells were subjected to Western blot analysis for detection of the indicated proteins with GAPDH as the protein loading control. (F) T24 T(Nonsense) and T24 T(shp85α) cells were seeded into 6-well plates. After synchronization, the cells were treated with Act D for the indicated times. Total RNA was isolated and subjected to qRT-PCR for analysis of degradation.
Autophagy-Mediated TIMP-2 Degradation Down-Regulates Activated MMP-2, Thereby Inhibiting BC Cell Invasion
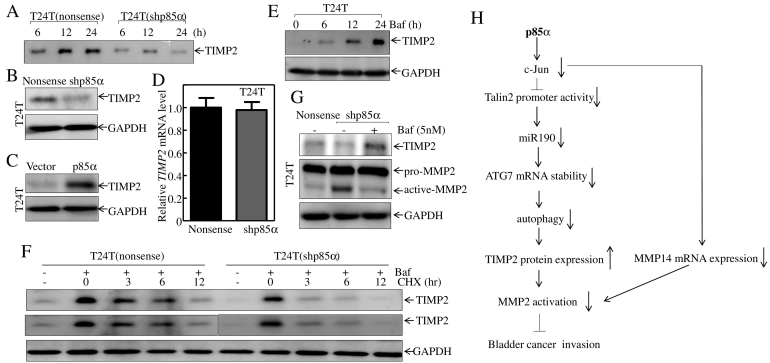
The inhibitory effect of TIMP-2 is mediated by its COOH-terminal domain, which interacts with the COOH-terminal region of MMP-2, thereby preventing it from interacting with MMP-14 and suppressing MMP-2 activation [31]. Assessment of TIMP-2 protein expression in the supernatant showed a decrease in secreted TIMP-2 in T24 T(shp85α) cells compared with the levels in T24 T(Nonsense) cells, indicating that p85α regulated activated MMP-2 by modulating TIMP-2 secretion (Figure 7A). The decrease in TIMP-2 secretion in the supernatant of T24 T(shp85α) cells could be explained by two mechanisms: an effect on TIMP-2 expression or alterations in the secretory pathway. Assessment protein expression of TIMP-2 in T24 T(Nonsense)/T24 T(shp85α) cells and T24 T(Vector)/T24 T(p85α) cells both showed the corresponding changes in the two pairs of cells (Figures 7, B and C), indicating that the increase of TIMP-2 in the extracellular environment was caused by the p85-mediated regulation of intracellular TIMP-2 expression.
Figure 7.
Timp-2 protein degradation is a p85α downstream effector that mediates MMP-2 activation.
(A) Extracellular proteins were concentrated from culture supernatants of T24 T(Nonsense) and T24 T(shp85α) cells at different times as indicated, and Western blotting was performed to detect TIMP-2 protein expression. (B and C) Extracts from the indicated cells were subjected to Western blot analysis with the indicated antibodies. GAPDH was used as a protein loading control. (D) TIMP-2 mRNA expression levels were evaluated by qRT-PCR in T24 T(Nonsense) and T24 T(shp85α) cells. (E) T24 T cells were seeded into 6-well plates and treated with Baf (5 nM) for the indicated times. The cell extracts were then subjected to Western blotting with the indicated antibodies. GAPDH was used as the protein loading control. (F) Stable transfectants were pretreated with or without Baf (5 nM) for 12 h, and then treated with cycloheximide (CHX, 100 μg/ml) for the indicated times. The cell extracts were subjected to Western blotting for determination of TIMP-2 degradation. GAPDH was used as the protein loading control. The middle panel was derived from top panel, in which we tried to adjust the intensity of TIMP2 protein band accumulated by Baf in T24 T(shp85α) cells was comparable with that observed in T24 T(Nonsense) cells. (G) The indicated stable transfectants were treated with or without Baf (5 nM) for 24 h, and cell extracts were subjected to Western blot analysis with the indicated antibodies. (H) Schematic diagram showing the proposed mechanisms underlying p85α inhibition of human bladder cancer invasion in a PI3K-independent manner.
Protein expression is regulated at the mRNA and protein levels. However, RT-PCR results showed no difference in TIMP-2 mRNA levels between T24 T(Nonsense) and T24 T(shp85α) cells (Figure 7D), suggesting that p85α-mediated TIMP-2 modulation did not occur at the mRNA level. The expression of TIMP-2 was increased at different times in response to treatment of cells with autophagy inhibitor Baf (Figure 7E). Further, T24 T(Nonsense) and T24 T(shp85α) cells were pre-treated with or without an autophagy inhibitor Baf for 12 h to accumulate the TIMP-2 protein. Cells were then treated with CHX at different times to assess TIMP-2 degradation rates in the two cells. As shown in top panel of Fig. 7F, TIMP2 protein band accumulated by Baf in T24 T(shp85α) cells was lower than that observed in T24 T(Nonsense) cells, and this resulted in a little concern on assessing the difference of TIMP2 protein degradation rates between two transfectants. To address this concern, the second panel of Figure 7F was derived from top panel by separating TIMP2 protein bands in two transfectants, in which we adjusted the intensity of TIMP2 protein band accumulated by Baf in T24 T(shp85α) cells was comparable with that observed in T24 T(Nonsense) cells. Thus, the difference of TIMP2 protein degradation rates could be easily to observe in two transfectants. The results showed that p85α knockdown increase in TIMP-2 protein degradation in comparison to T24 T(Nonsense) cells (Figure 7F), indicating that p85α expression inhibits TIMP-2 protein degradation through an autophagy-dependent mechanism in human BC cells, and increased TIMP-2 interacts with MMP-14, and in turn reducing MMP-2 activation and resulting in suppression of invasion of human BC cells.
Discussion
PI-3 K pathways control many physiological functions and cellular processes, including cell growth, survival, motility, and metabolism [32], [33], [34]. However, the role of p85α in BC invasion remains unexplored. The present study is the first to show that p85α is down-regulated in human BCs and plays an important role in MMP-2 activation in a PI3K-independent manner, therefore is a strong negative regulator of BC invasion. We showed that p85α inhibits MMP-2 activation by suppressing TIMP-2 protein degradation and MMP-14 transcription. C-Jun inhibition by p85α results in C-Jun downstream regulated gene MMP-14 transcription attenuation and Talin2/miR-190 transcription inhibition. miR-190 inhibition leads to its less binding to the 3′-UTR of ATG7 and therefore increasing ATG7 mRNA degradation and further inhibiting the autophagy-dependent degradation of TIMP-2. The inhibition of MMP-14 transcription and increased TIMP-2 protein degradation together inhibits MMP-2 activation and BC invasion. This study is the first demonstration that p85α inhibits cancer cell invasion by negatively regulating MMP-2 activation in human BC cells.
p85α is a class IA PI3K regulatory subunit that interacts with the p110 catalytic subunit and inhibits PI3K pathways [35], [36], [37]. The PI3K activity-independent function of p85α was reported previously by different groups including ours [38], [39], [40]. For instance, p85α modulates cell responses by regulating the activation of signaling molecules such as Cdc42 in a PI3K-independent manner [39]. We previously reported the occurrence of p85α-associated and PI3K-independent cell death in response to UV radiation (UVR), and identified a novel p85α/NFAT3/TNFα signaling pathway that mediates cellular apoptotic responses under certain stress conditions [40]. The present study showed, for the first time, that p85α down-regulation is associated with BC cell invasion. Ectopic expression of p110* and p110*∆kin, the kinase-deficient form of p110 [41], did not rescue the invasive ability, suggesting that the function of p85α in BC invasion is PI3K-independent.
The present study demonstrated the involvement of autophagy in the regulation of cancer cell invasion and migration. In glioblastoma stem cells, autophagy inhibition or knockdown of the autophagy regulator p62 decreases invasion and migration in vitro and leads to metabolic defects [42]. Induction of autophagy by toll-like receptors promotes the secretion of pro-invasive factors, including IL-6, in lung cancer cells, supporting the role of autophagy as a determinant of pro-invasive secretion [43]. Here, we showed that p85α inhibits human BC invasion by suppression of ATG7-mediated autophagy. Previous work from our group showed that miR-190 binds to the 3′-UTR of ATG7, promoting ATG7 mRNA degradation [44]. In the present study, we showed that p85α down-regulated miR-190 expression, thereby regulating ATG7 mRNA stability. Talin2 is the host gene of miR-190, and its promoter controls the expression of miR-190 [45], [46]. Assessment of the promoter activity of Talin2 led to the conclusion that p85α regulates Talin2 at the transcriptional level. Further experiments showed that c-Jun regulated Talin2 transcription and mediated the effect of p85α on miR-190 down-regulation. Taken together, the present data suggest a new model in which p85α inhibits human BC invasion via the c-Jun/miR-190/ATG7/autophagy axis, which differs from its PI3K/AKT/mTOR pathway-dependent inhibition of autophagy by ULK-Atg13-FIP200 complexes [47], [48] and the regulation of cancer cell proliferation [49], [50].
MMP-2 was identified as a downstream target mediating the inhibitory effect of p85α. MMP-2 activation status is implicated in the progression, invasion, and metastasis of various cancers. In the present study, activated MMP-2 was the only protein induced by knockdown of p85α among a series of invasion-related proteins. Low levels of activated MMP-2 were associated with decreased BC invasion. MMP-14 (also known as membrane type 1 MMP) and TIMP-2 are important regulators for MMP-2 activation. MMP-14 can induce MMP-2 activation by cleaving the propeptide of pro-MMP-2 [51], and TIMP-2 has a dual role in regulating MMP-14–induced MMP-2 activation [31]. TIMP-2 binds to MT1-MMP to form a binary complex that acts as a receptor for pro-MMP-2. A free MT1-MMP molecule in close proximity cleaves the propeptide of pro-MMP-2, generating an intermediate species. Further proteolysis of the propeptide through an autocatalytic mechanism generates the fully active enzyme. However, when expressed at high levels, TIMP-2 binds to and inhibits MT1-MMP activity, preventing pro-MMP-2 activation [52]. In the present study, MMP-14 was up-regulated in T24 T(shp85α) cells concomitant with decreased TIMP-2 secretion, which can generate the tri-molecular complex required for proteolysis of the propeptide. Further research demonstrated that TIMP-2 protein degradation was mediated by autophagy, and treatment with an autophagy inhibitor increased TIMP-2 and decreased MMP-2 activation. The present study is the first to show that autophagy can mediate TIMP-2 protein degradation, thereby decreasing activated MMP-2 and inhibiting BC cell invasion.
In summary, we present a novel PI3K-independent function of p85α in regulating cancer cell invasion through the inhibition of MMP-2 activation mediated by the coordinated inhibition of TIMP-2 protein degradation and MMP-14 transcription through the c-Jun/Talin2/ATG7/autophagy axis. This pathway provides new insight into the mechanisms underlying the effect of p85α on regulating cancer cell invasion in a PI3K-independent manner.
Acknowledgments
Acknowledgements
We thank Dr. Williams LT from Cardiovascular Research Institute and Daiichi Research Center, University of California, San Francisco, for the gift of constructs of p110* and p110*∆kin; We also thank that Dr. Fei Chen from Department of Pharmaceutical Sciences, Wayne State University, for providing the talin2 promoter luciferase reporter. This work was partially supported by grants from the Natural Science Foundation of China (NSFC81872587 and NSFC81702530); and the Key Discipline of Zhejiang Province in Medical Technology (First Class, Category A).
Conflicts of Interest Statement
Neither this paper nor any similar paper has been or will be submitted to or published in any other scientific journal. All authors are aware and agree to the content of the paper and to their being listed as an author on the manuscript. There is no conflict of interest or competing financial interests for all authors.
Contributor Information
Haishan Huang, Email: haishan_333@163.com.
Chuanshu Huang, Email: Chuanshu.huang@nyulangone.org.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Kaufman DS, Shipley WU, Feldman AS. Bladder cancer. Lancet. 2009;374:239–249. doi: 10.1016/S0140-6736(09)60491-8. [DOI] [PubMed] [Google Scholar]
- 3.Gorin MA, Verdone JE, van der Toom E, Bivalacqua TJ, Allaf ME, Pienta KJ. Circulating tumour cells as biomarkers of prostate, bladder, and kidney cancer. Nat Rev Urol. 2017;14:90–97. doi: 10.1038/nrurol.2016.224. [DOI] [PubMed] [Google Scholar]
- 4.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verma RP, Hansch C. Matrix metalloproteinases (MMPs): chemical-biological functions and (Q)SARs. Bioorg Med Chem. 2007;15:2223–2268. doi: 10.1016/j.bmc.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 6.Mook OR, Frederiks WM, Van Noorden CJ. The role of gelatinases in colorectal cancer progression and metastasis. Biochim Biophys Acta. 2004;1705:69–89. doi: 10.1016/j.bbcan.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 7.Kargozaran H, Yuan SY, Breslin JW, Watson KD, Gaudreault N, Breen A, Wu MH. A role for endothelial-derived matrix metalloproteinase-2 in breast cancer cell transmigration across the endothelial-basement membrane barrier. Clin Exp Metastasis. 2007;24:495–502. doi: 10.1007/s10585-007-9086-6. [DOI] [PubMed] [Google Scholar]
- 8.Davies B, Waxman J, Wasan H, Abel P, Williams G, Krausz T, Neal D, Thomas D, Hanby A, Balkwill F. Levels of matrix metalloproteases in bladder cancer correlate with tumor grade and invasion. Cancer Res. 1993;53:5365–5369. [PubMed] [Google Scholar]
- 9.Hernandez-Barrantes S, Bernardo M, Toth M, Fridman R. Regulation of membrane type-matrix metalloproteinases. Semin Cancer Biol. 2002;12:131–138. doi: 10.1006/scbi.2001.0421. [DOI] [PubMed] [Google Scholar]
- 10.Jezierska A, Motyl T. Matrix metalloproteinase-2 involvement in breast cancer progression: a mini-review. Med Sci Monit. 2009;15:RA32–40. [PubMed] [Google Scholar]
- 11.Butler GS, Butler MJ, Atkinson SJ, Will H, Tamura T, Schade van Westrum S, Crabbe T, Clements J, d'Ortho MP, Murphy G. The TIMP2 membrane type 1 metalloproteinase "receptor" regulates the concentration and efficient activation of progelatinase A. A kinetic studyJ Biol Chem. 1998;273:871–880. doi: 10.1074/jbc.273.2.871. [DOI] [PubMed] [Google Scholar]
- 12.Yu J, Zhang Y, McIlroy J, Rordorf-Nikolic T, Orr GA, Backer JM. Regulation of the p85/p110 phosphatidylinositol 3′-kinase: stabilization and inhibition of the p110alpha catalytic subunit by the p85 regulatory subunit. Mol Cell Biol. 1998;18:1379–1387. doi: 10.1128/mcb.18.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie Q, Guo X, Gu J, Zhang L, Jin H, Huang H, Li J, Huang C (2016). p85alpha promotes nucleolin transcription and subsequently enhances EGFR mRNA stability and EGF-induced malignant cellular transformation.Oncotarget 7, 16636–16649. [DOI] [PMC free article] [PubMed]
- 14.Taniguchi CM, Winnay J, Kondo T, Bronson RT, Guimaraes AR, Aleman JO, Luo J, Stephanopoulos G, Weissleder R, Cantley LC. The phosphoinositide 3-kinase regulatory subunit p85alpha can exert tumor suppressor properties through negative regulation of growth factor signaling. Cancer Res. 2010;70:5305–5315. doi: 10.1158/0008-5472.CAN-09-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo J, Cantley LC. The negative regulation of phosphoinositide 3-kinase signaling by p85 and it's implication in cancer. Cell Cycle. 2005;4:1309–1312. doi: 10.4161/cc.4.10.2062. [DOI] [PubMed] [Google Scholar]
- 16.Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol. 2006;7:85–96. doi: 10.1038/nrm1837. [DOI] [PubMed] [Google Scholar]
- 17.Zhou W, An G, Wei P, Chen W. Significance of p85 expression as a prognostic factor for patients with breast cancer. Oncol Lett. 2014;8:1657–1661. doi: 10.3892/ol.2014.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin H, Yu Y, Hu Y, Lu C, Li J, Gu J, Zhang L, Huang H, Zhang D, Wu XR. Divergent behaviors and underlying mechanisms of cell migration and invasion in non-metastatic T24 and its metastatic derivative T24T bladder cancer cell lines. Oncotarget. 2015;6:522–536. doi: 10.18632/oncotarget.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang HY, Shariat SF, Sun TT, Lepor H, Shapiro E, Hsieh JT, Ashfaq R, Lotan Y, Wu XR. Persistent uroplakin expression in advanced urothelial carcinomas: implications in urothelial tumor progression and clinical outcome. Hum Pathol. 2007;38:1703–1713. doi: 10.1016/j.humpath.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang Y, Cao Z, Hou Q, Ma C, Yao C, Li J, Wu XR, Huang C. Cyclin d1 downregulation contributes to anticancer effect of isorhapontigenin on human bladder cancer cells. Mol Cancer Ther. 2013;12:1492–1503. doi: 10.1158/1535-7163.MCT-12-0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang D, Song L, Li J, Wu K, Huang C. Coordination of JNK1 and JNK2 is critical for GADD45alpha induction and its mediated cell apoptosis in arsenite responses. J Biol Chem. 2006;281:34113–34123. doi: 10.1074/jbc.M602821200. [DOI] [PubMed] [Google Scholar]
- 22.Zhu J, Zhang J, Huang H, Li J, Yu Y, Jin H, Li Y, Deng X, Gao J, Zhao Q. Crucial role of c-Jun phosphorylation at Ser63/73 mediated by PHLPP protein degradation in the cheliensisin a inhibition of cell transformation. Cancer Prev Res (Phila) 2014;7:1270–1281. doi: 10.1158/1940-6207.CAPR-14-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang G, Wu AD, Huang C, Gu J, Zhang L, Huang H, Liao X, Li J, Zhang D, Zeng X. Isorhapontigenin (ISO) Inhibits Invasive Bladder Cancer Formation In Vivo and Human Bladder Cancer Invasion In Vitro by Targeting STAT1/FOXO1 Axis. Cancer Prev Res (Phila) 2016;9:567–580. doi: 10.1158/1940-6207.CAPR-15-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papinski D, Kraft C. Atg1 kinase organizes autophagosome formation by phosphorylating Atg9. Autophagy. 2014;10:1338–1340. doi: 10.4161/auto.28971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Codogno P, Mehrpour M, Proikas-Cezanne T. Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat Rev Mol Cell Biol. 2011;13:7–12. doi: 10.1038/nrm3249. [DOI] [PubMed] [Google Scholar]
- 26.Ding WX, Yin XM. Mitophagy: mechanisms, pathophysiological roles, and analysis. Biol Chem. 2012;393:547–564. doi: 10.1515/hsz-2012-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Che X, Huang C. microRNA, Cancer and Cancer Chemoprevention. Curr Mol Pharmacol. 2012;5:362–371. [PubMed] [Google Scholar]
- 28.Raftopoulou M, Hall A. Cell migration: Rho GTPases lead the way. Dev Biol. 2004;265:23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Liu J, Zhang D, Luo W, Yu Y, Yu J, Li J, Zhang X, Zhang B, Chen J, Wu XR. X-linked inhibitor of apoptosis protein (XIAP) mediates cancer cell motility via Rho GDP dissociation inhibitor (RhoGDI)-dependent regulation of the cytoskeleton. J Biol Chem. 2011;286:15630–15640. doi: 10.1074/jbc.M110.176982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar B, Koul S, Petersen J, Khandrika L, Hwa JS, Meacham RB, Wilson S, Koul HK. p38 mitogen-activated protein kinase-driven MAPKAPK2 regulates invasion of bladder cancer by modulation of MMP-2 and MMP-9 activity. Cancer Res. 2010;70:832–841. doi: 10.1158/0008-5472.CAN-09-2918. [DOI] [PubMed] [Google Scholar]
- 31.Shen Q, Lee ES, Pitts RL, Wu MH, Yuan SY. Tissue inhibitor of metalloproteinase-2 regulates matrix metalloproteinase-2-mediated endothelial barrier dysfunction and breast cancer cell transmigration through lung microvascular endothelial cells. Mol Cancer Res. 2010;8:939–951. doi: 10.1158/1541-7786.MCR-09-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong JT, Yu J, Wang HJ, Shi Y, Zhao TS, He BX, Qiao B, Feng ZW (2017). Effects of endoplasmic reticulum stress on the autophagy, apoptosis, and chemotherapy resistance of human breast cancer cells by regulating the PI3K/AKT/mTOR signaling pathway.Tumour Biol 39, 1010428317697562. [DOI] [PubMed]
- 33.Crumbaker M, Khoja L, Joshua AM. AR Signaling and the PI3K Pathway in Prostate Cancer. Cancers (Basel) 2017;9 doi: 10.3390/cancers9040034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun C, Zhang Z, He P, Zhou Y, Xie X. Involvement of PI3K/Akt pathway in the inhibition of hepatocarcinoma cell invasion and metastasis induced by SASH1 through downregulating Shh-Gli1 signaling. Int J Biochem Cell Biol. 2017;89:95–100. doi: 10.1016/j.biocel.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 35.de la Cruz-Herrera CF, Baz-Martinez M, Lang V, El Motiam A, Barbazan J, Couceiro R, Abal M, Vidal A, Esteban M, Munoz-Fontela C Conjugation of SUMO to p85 leads to a novel mechanism of PI3K regulation. Oncogene. 2016;35:2873–2880. doi: 10.1038/onc.2015.356. [DOI] [PubMed] [Google Scholar]
- 36.De Gregorio G, Coppa A, Cosentino C, Ucci S, Messina S, Nicolussi A, D'Inzeo S, Di Pardo A, Avvedimento EV, Porcellini A. The p85 regulatory subunit of PI3K mediates TSH-cAMP-PKA growth and survival signals. Oncogene. 2007;26:2039–2047. doi: 10.1038/sj.onc.1210011. [DOI] [PubMed] [Google Scholar]
- 37.Bousquet C, Guillermet-Guibert J, Saint-Laurent N, Archer-Lahlou E, Lopez F, Fanjul M, Ferrand A, Fourmy D, Pichereaux C, Monsarrat B. Direct binding of p85 to sst2 somatostatin receptor reveals a novel mechanism for inhibiting PI3K pathway. EMBO J. 2006;25:3943–3954. doi: 10.1038/sj.emboj.7601279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia Z, Silio V, Marques M, Cortes I, Kumar A, Hernandez C, Checa AI, Serrano A, Carrera AC. A PI3K activity-independent function of p85 regulatory subunit in control of mammalian cytokinesis. EMBO J. 2006;25:4740–4751. doi: 10.1038/sj.emboj.7601324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jimenez C, Portela RA, Mellado M, Rodriguez-Frade JM, Collard J, Serrano A, Martinez AC, Avila J, Carrera AC. Role of the PI3K regulatory subunit in the control of actin organization and cell migration. J Cell Biol. 2000;151:249–262. doi: 10.1083/jcb.151.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song L, Li J, Ye J, Yu G, Ding J, Zhang D, Ouyang W, Dong Z, Kim SO, Huang C. p85alpha acts as a novel signal transducer for mediation of cellular apoptotic response to UV radiation. Mol Cell Biol. 2007;27:2713–2731. doi: 10.1128/MCB.00657-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu Q, Klippel A, Muslin AJ, Fantl WJ, Williams LT. Ras-dependent induction of cellular responses by constitutively active phosphatidylinositol-3 kinase. Science. 1995;268:100–102. doi: 10.1126/science.7701328. [DOI] [PubMed] [Google Scholar]
- 42.Galavotti S, Bartesaghi S, Faccenda D, Shaked-Rabi M, Sanzone S, McEvoy A, Dinsdale D, Condorelli F, Brandner S, Campanella M. The autophagy-associated factors DRAM1 and p62 regulate cell migration and invasion in glioblastoma stem cells. Oncogene. 2013;32:699–712. doi: 10.1038/onc.2012.111. [DOI] [PubMed] [Google Scholar]
- 43.Zhan Z, Xie X, Cao H, Zhou X, Zhang XD, Fan H, Liu Z. Autophagy facilitates TLR4- and TLR3-triggered migration and invasion of lung cancer cells through the promotion of TRAF6 ubiquitination. Autophagy. 2014;10:257–268. doi: 10.4161/auto.27162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu J, Tian Z, Li Y, Hua X, Zhang D, Li J, Jin H, Xu J, Chen W, Niu B. ATG7 promotes bladder cancer invasion via autophagy-mediated increased ARHGDIB mRNA stability. Advanced Science. 2019;6 doi: 10.1002/advs.201801927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng H, Chu J, Zeng Y, Loh HH, Law PY. Yin Yang 1 phosphorylation contributes to the differential effects of mu-opioid receptor agonists on microRNA-190 expression. J Biol Chem. 2010;285:21994–22002. doi: 10.1074/jbc.M110.112607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heras-Sandoval D, Perez-Rojas JM, Hernandez-Damian J, Pedraza-Chaverri J. The role of PI3K/AKT/mTOR pathway in the modulation of autophagy and the clearance of protein aggregates in neurodegeneration. Cell Signal. 2014;26:2694–2701. doi: 10.1016/j.cellsig.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 48.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 50.Liu B, Cheng Y, Liu Q, Bao JK, Yang JM. Autophagic pathways as new targets for cancer drug development. Acta Pharmacol Sin. 2010;31:1154–1164. doi: 10.1038/aps.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Onogi A, Naruse K, Sado T, Tsunemi T, Shigetomi H, Noguchi T, Yamada Y, Akasaki M, Oi H, Kobayashi H. Hypoxia inhibits invasion of extravillous trophoblast cells through reduction of matrix metalloproteinase (MMP)-2 activation in the early first trimester of human pregnancy. Placenta. 2011;32:665–670. doi: 10.1016/j.placenta.2011.06.023. [DOI] [PubMed] [Google Scholar]