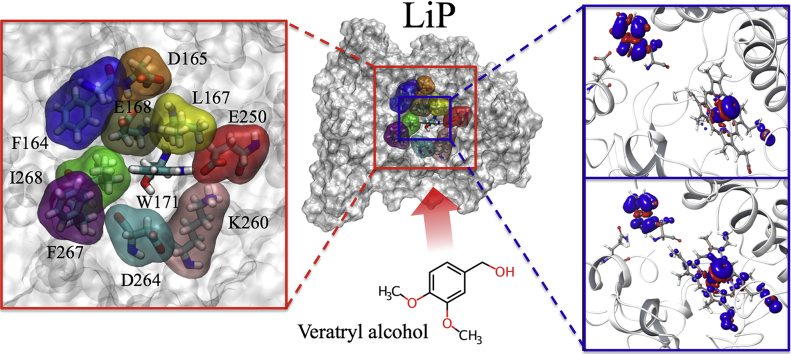
Abstract
Lignin peroxidase (LiP) and its natural substrate veratryl alcohol (VA) play a crucial role in lignin degradation by white-rot fungi. Understanding the molecular determinants for the interaction of this enzyme with its substrates is essential in the rational design of engineered peroxidases for biotechnological application. Here, we combine computational and experimental approaches to analyze the interaction of Phanerochaete chrysosporium LiP (isoenzyme H8) with VA and its radical cation (VA•+, resulting from substrate oxidation by the enzyme). Interaction energy calculations at semiempirical quantum mechanical level (SQM) between LiP and VA/VA•+ enabled to identify those residues at the acidic environment of catalytic Trp171 involved in the main interactions. Then, a battery of variants, with single and multiple mutations at these residues (Glu168, Asp165, Glu250, Asp264, and Phe267), was generated by directed mutagenesis, and their kinetics parameters were estimated on VA and two additional substrates. The experimental results show that Glu168 and Glu250 are crucial for the binding of VA, with Glu250 also contributing to the turnover of the enzyme. The experimental results were further rationalized through new calculations of interaction energies between VA/VA•+ and LiP with each of the single mutations. Finally, the delocalization of spin density was determined with quantum mechanics/molecular mechanics calculations (QM/MM), further supporting the contribution of Glu250 to VA oxidation at Trp171.
Keywords: Phanerochaete chrysosporium, Lignin peroxidase, Veratryl alcohol, Sited-directed mutagenesis, Interaction energy, SQM, QM/MM
Graphical Abstract
1. Introduction
Lignin peroxidase (LiP; EC 1.11.1.14) is a heme peroxidase classified in class-II of the peroxidase-catalase superfamily [1]. LiP plays a key role in the fungal delignification of lignocellulosic biomass for carbon recycling in land ecosystems [2]. LiP and other ligninolytic enzymes have gained interest because of their potential applications in lignocellulose valorization for the production of biofuels and chemicals [3]. For the latter, the elucidation of the mechanism whereby LiP modulates its catalytic properties has become a focus of substantial importance.
The catalytic cycle of LiP is similar to classical peroxidases, such as horseradish peroxidase, with one molecule of H2O2 oxidizing the resting state enzyme (with ferric heme) by withdrawing two electrons and forming compound I, the most reactive enzyme intermediate [4]. Compound I contains an oxo-ferryl (Fe4+ = O) species and a porphyrin cation radical, and it is reduced by one electron from a substrate (typically veratryl alcohol, VA) generating compound II (Fe4+ = O heme intermediate), which in turn can be further reduced with another substrate molecule. In this way, the enzyme returns to its resting state. The best characterized LiP corresponds to isoenzyme H8 (LiPH8) of the model ligninolytic fungus Phanerochaete chrysosporium. Doyle et al. [5] described two oxidation sites in this enzyme: the typical heme-cavity of heme peroxidases and the Trp171 on the enzyme surface. Oxidation in the heme cavity is achieved by direct contact with the activated cofactor through an access channel. Meanwhile, oxidation on the surface involves Trp171, which is activated as a radical species by Compound I subtraction of one of its electrons through a long-range electron transfer (LRET) mechanism [6].
Direct evidence about which radical species is formed at Trp171 has not been reported to date. Electron paramagnetic resonance (EPR) of peroxide-activated LiPH8 variants E250Q and E168Q/E250Q identified a neutral tryptophanyl radical (Trp•) similar to that reported in versatile peroxidase (VP), [7,8] a second member of class-II peroxidases [1]. Bernini et al. [9] performed quantum mechanics/molecular mechanics (QM/MM) calculations by which they argued that the cationic tryptophanyl radical (TrpH•+) is the most favorable species existing in LiP, where the negative electrostatic potential in the environment of Trp171 would stabilize this cationic species. It has been proposed that Glu250, which forms a hydrogen bond with the indolic N atom of Trp171 in LiPH8 crystal structure [10], is responsible for Trp171H•+ deprotonation. On the other hand, previous QM/MM and molecular dynamics (MD) simulations in related VP also evidenced this stepwise mechanism [11]. After the electron transfer from VP Trp164 (homologous to LiPH8 Trp171) to heme, Glu243 (homologous to LiPH8 Glu250) would deprotonate the cation radical intermediate (Trp•+) to form the neutral radical (Trp•) detected by EPR of this enzyme [7].
In this work, we report a theoretical and experimental analysis of residues participating in the binding/stabilization of VA and VA•+ at the LiP surface, where the importance of the residues predicted in energy calculations is analyzed by site-directed mutagenesis, steady-state kinetic measurements, and interaction energy profiles at semiempirical quantum mechanics level.
2. Material and Methods
2.1. General System Setup and MD Parameters
The tridimensional structure of P. chrysosporium LiPH8, corresponding to LiPA gene in the sequenced fungal genome (https://genome.jgi.doe.gov/Phchr2/Phchr2.home.html), was taken from Protein Data Bank (PDB code: 1LLP) [10] and prepared using the Protein Preparation Wizard tool from the Schrödinger suite [12]. Protonation states were adjusted at pH 3.0 with PROPKA [13]. Glu168 and Asp264 (that were protonated by PROPKA) were manually deprotonated in agreement with Recabarren et al. [14]. The heme group was modeled as compound I according to the OPLS-2005 force field parameters. For this, the formal charge of iron was set to +3 (Fe+3) and it was connected via zero-order bonds to 6 atoms: the indolic nitrogen of proximal His176, the four nitrogen atoms of the porphyrin ring (two of these nitrogens were set with formal charge of −1) and the distal oxygen atom with zero formal charge coordinating Fe+3 [14].
Then, six MD simulations of 50 ns were carried out for LiPH8 and its D165N, E168Q, E250Q, D264N and F267 in silico variants, which were generated with the “mutate residue” option in Maestro [15]. A minimization protocol for relaxing the structure of each mutant was performed using the MACROMODEL software package [16] applying the steepest descent minimization method with a convergence criterion of energy gradient to <0.05 kJ/Å·mol. The MD simulations were performed as previously described [14], with details of the protocol used given in the supporting information section. For each MD simulation one frame was saved every 50 ps to carry out calculations of the solvent accessible surface area (SASA) [17] around residue Trp171 using a script (Script S1) in tool command language (TCL) included in the visual molecular dynamics (VMD) software version 1.9.2 for Linux [18]. The enzyme structures obtained from these MD simulations were used for the quantum semiempirical interaction energy calculations described below.
2.2. LiP-VA Interaction Energy at Semiempirical Quantum Mechanical (SQM) Level
Firstly, interaction energies (ΔE) of VA and its cation radical with each amino acid of the solvent-exposed reactive site of LiPH8 (residues located at the Trp171 environment) were calculated using a computational strategy implemented by Avila-Salas et al. [19] which couples a Monte Carlo conformational sampling [20] and ΔE calculations at the SQM level [[19], [20], [21], [22]] (Fig. S1). With this strategy it is possible to quickly evaluate the energy contribution of each residue to the enzyme-substrate binding affinity. Thereby, similar to previous reports [19], ten frames from equilibrium conformations of the LiPH8 wild type, and mutants, MD simulation systems were used. Representative frames were selected every 3 ns, starting from 20 ns, based on the analysis of the root-mean-square deviation (RMSD) values of the stable backbone of proteins as shown in Fig. S2a. For each frame, the following amino acids located within 5.5 Å from Trp171 were selected: Phe164, Asp165, Leu167, Glu168, Glu250, Lys260, Asp264, Phe267, and Ile268. This selected region presented a stable backbone RMSD along the MD of LiPH8 and its mutants (Fig. S2b), nevertheless the sampled structures represented different spatial conformations of the VA binding site.
The ΔE for the different enzyme-substrate complexes (represented here as molecule1-molecule2, respectively) was calculated at SQM level. Briefly: i) molecule1 (enzyme) is fixed to remain static in the origin of its cartesian coordinates; ii) then, molecule2 (substrate) is moved and reoriented about molecule1 using a translation vector and rotation matrix until the van der Waals (vdW) surfaces of each molecule touch each other [20]; iii) for each new molecule1-molecule2 complex, its single-point energy (one self-consistent field (SCF) cycle) is calculated using the parameterized method number 7 (PM7) [23] implemented in MOPAC2016 version 16.111 L for Linux [24], which provides a good approximation in the calculation of both geometry and heat of formation, reducing the average unsigned errors (AUEs) [25]; and iv) finally, the heat of formation energy (E) for the complex is extracted from the previous SCF calculation and the SCF calculations of each non-interacting fragment in order to obtain the ∆E according to the following Eq. (1)
| (1) |
The process (steps i-iv) is repeated to generate thousands of different complexes, which generate a probability distribution (interaction energies vs their relative frequencies) similar to Boltzmann distribution, from which the ΔE value with the highest relative frequency is selected. This interaction energy is associated with a given spatial location of molecule1 with respect to molecule2. Finally, the averaged interaction energy values (Fig. S1, step 6) were used for comparison against experimental residual activity.
In order to perform the conformational sampling, molecule1 represented a full molecular region including each one of the amino acids selected (including Trp171) in the active site of wild-type system (Fig. S1, step 3) and molecule2 represented one hundred thousand different orientations of VA located until 11 Å around molecule1 (Fig. S1, step 4). This cutoff distance was chosen to generate a considerable number of VA poses around the selected region (which considers residues at the reactive space around Trp171). The conformational sampling considers the steric hindrance of the residues located around molecule1. Then, only the VA postures generated in the conformational sampling and located <5.5 Å away from each amino acid (AA) were selected to calculate the ΔE for the AA-VA and AA-VA•+ complexes. This calculation was carried out considering the steps (iii) and (iv) of the previously detailed methodology. Finally, the averaged ΔE value was obtained taking into consideration the results generated from the calculations in the 10 MD frames of each molecular system (Fig. S1, step 7).
In a second study, the five amino acids with the best ΔE against VA were selected to gain more evidence about the changes in their ΔE values when mutated, and to estimate their energy and structural relationships with the kinetic parameters of the enzymes. Then, molecule1 represented the molecular region (REG) with the selected residues around Trp171 (including the latter). These mutated regions (MREG) were subtracted from MD simulations of the D165N, E168Q, E250Q, D264N, and F267I mutants, applying a similar procedure as described for the wild type system. As in the first protocol, molecule2 represented one hundred thousand different orientations of VA, or VA•+, around the full molecular region (molecule1). The molecular regions considered Trp171 in its different states: non-radical, radical cation (TrpH•+) and neutral radical (Trp•). The averaged ΔE values for these mutant-VA complexes were obtained according to steps (iii) and (iv) of the previously detailed methodology. These calculations would allow us to evaluate how the residues (original and mutated) would impact VA affinity against wild-type LiPH8 and its variants at the active site and, thereby, comparing these computational results with the effect of those mutations on LiP experimental enzymatic activity.
Finally, 100 complexes of wild-type LiPH8, and its variants, interacting with VA with lower ΔE values were selected in order to carry out the characterization of their intermolecular interactions (π-π interactions, hydrogen bonds, among others) using BIOVIA Discovery Studio Visualizer software version 2017 R2 (for Windows) [26].
2.3. QM/MM Spin Density Calculations
For describing the electronic structure of compound I in LiPH8 and E250Q variant, representative frames from MD simulations of the LiPH8 and E250Q systems were extracted. Water molecules, VA substrate, and Na+ ions were deleted and a minimization protocol using Polak-Ribiere Conjugate Gradient (PRCG) method [27] was applied until the convergence criterion was met (0.05 kJ/Å·mol on energy gradient). The resulting structures were used as input for QM/MM optimization calculations using Qsite 5.7 (Schrödinger) [28]. The quartet electronic state of compound I was calculated applying unrestricted density functional theory with the M06-L functional [29] in combination with the lacvp* basis set for the QM region and the OPLS-2005 force field for the classical region, following similar protocols used in other studies [30]. The iron coordination sphere was included in the QM region (porphyrin ring with propionate groups, side chain of His176, and oxygen atom bounded to Fe+3 at ≈1.6 Å from). In subsequent calculations, Trp171 was added to the QM region and its oxidation by compound I was evaluated. Thus, the QM region included 122 atoms in QM/MM calculations. The QM/MM cuts in the respective amino acids were applied between α and β carbon atoms and the valences were saturated with hydrogen atoms (link-atom approach). Spin density maps were calculated and atomic spin densities were estimated with the Mulliken partition method [31] for evaluating electron delocalization.
2.4. Site-Directed Mutagenesis
Mutations in P. chrysosporium LiPH8 (yielding the D165N, E168Q, E250Q, D264N, F267I, E168Q/E250Q, D165N/E168Q, D264N/F267I, and D165N/E168Q/D264N/F267I variants) and Pleurotus eryngii allelic variant VPL2 (yielding the E243Q variant) were introduced by polymerase chain reaction (PCR) using the QuikChange™ site-directed mutagenesis kit (La Jolla, CA, USA). Each PCR reaction contained 20 ng of a DNA template, 500 μM of each dNTP, 125 ng of direct and reverse primers, 2.5 units of PfuTurbo polymerase and manufacturer's buffer reaction. The plasmids pET23b-LiPH8 and pFLAG1-VPL2 containing the coding DNA sequences of isoenzymes LiPH8 (GenBank Y00262) and VPL2 (GenBank AF007222) [32] were used as templates for single and some double mutations (LiPH8 D165N/E168Q and D264N/F267I). These mutants were obtained using primers that contained de corresponding mutations (all primers are included in Table S1). The LiPH8 E168Q/E250Q double mutant and the only quadruple mutant were obtained using, respectively, pET23b-LiPH8/E168Q and pET23b-LiPH8/D264N/F267I plasmids as templates, and the corresponding primers. Reaction conditions were as follows: (i) a start cycle at 95 °C for 1 min; (ii) 18 cycles at 95 °C for 50 s, 55 °C for 50 s, and 68 °C for 10 min; (iii) a final cycle of 10 min at 68 °C. Digestion with endonuclease DpnI of the mutagenic PCR reactions were performed during ≈1 h at 37 °C for digesting methylated parental no mutated DNA. The samples were quickly purified with QIAquick® PCR Purification Kit (#28104, Qiagen) before transforming the plasmids obtained from mutagenic PCR into chemically competent cells of Escherichia coli DH5α.
2.5. Enzyme Production, In Vitro Activation and Purification
Wild-type recombinant and mutated variants of LiPH8 and VPL2 were produced in E. coli BL21 (DE2)pLysS and W3110 strains, respectively. Cells were grown in Terrific broth medium [33] at 37 °C until an OD600 0.5 to 0.6 (≈3 h). Then, protein expression was induced with 1 mM isopropyl-β-D-thiogalactopyranoside and cells were grown for another 6 h at 37 °C. Cells were harvested by centrifugation at 4000 ×g for 8 min. The apoenzymes accumulated in inclusion bodies were recovered by solubilization in 50 mM Tris/HCl (pH 8) containing 8 M urea, 1 mM dithiothreitol and 1 mM ethylene-diamine tetraacetic acid. In vitro refolding was performed as reported by Ayuso-Fernández et al. [34] and Pérez-Boada et al. [35] for LiP and VP, respectively.
The active enzymes were purified by ion-exchange chromatography using a Resource-Q column (GE-Healthcare, USA) coupled to an ÄKTA fast protein liquid chromatography (FPLC) system. Concentrated samples of the refolding process were loaded onto the column in 10 mM sodium tartrate (pH 5.5) containing 1 mM of CaCl2 at 1 mL/min flow. The correctly folded proteins retained at pH 5.5 were eluted with 0–300 mM NaCl gradient at 2 mL/min. The fractions containing the properly folded proteins were pooled and dialyzed against 10 mM sodium tartrate (pH 5.0) and stored at −80 °C. The purity grade of proteins was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). UV–visible spectra in the 200–700 nm range were used to confirm the correct folding of the enzymes (Soret band at ≈407 nm and small maxima at ≈504 nm and ≈637 nm corresponding to charge transfer transition bands CT2 and CT1, respectively [36]). Enzymes were quantified using the extinction coefficient ε408 = 168 mM−1 cm−1 for LiPH8 and ε407 = 150 mM−1 cm−1 for VPL2 [37,38].
2.6. Enzyme Kinetics
The steady-state kinetic constants of LiPH8, VPL2 and their mutated variants were estimated from absorbance changes during the oxidation of four substrates at the optimal oxidation pH in 0.1 M tartrate at 25 °C using a Shimadzu UV-1800 spectrophotometer. The measurements were initiated by the addition of H2O2 to a final concentration of 0.25 mM. Oxidation of VA to veratraldehyde (ε310 9.3 mM−1·cm−1), 2,6-dimethoxyphenol (DMP) to dimeric coerulignone (ε469 55 mM−1·cm−1), and 2,2′-azino-bis[3-ethylbenzothiazoline-6-sulfonate] (ABTS) to ABTS cation radical (ε436 29.3 mM−1·cm−1) were assayed at pH 3; while the oxidation of Mn2+ to Mn3+ (tartrate complex ε238 6.5 mM−1·cm−1) was assayed at pH 5. The enzymatic activities were measured as initial velocities taken from linear increments of absorbance due to the appearance of the reaction product. Values for Michaelis-Menten constant (Km) and maximal enzyme turnover (kcat) with its standard errors were calculated by fitting the experimental measurements to the Michaelis-Menten model incorporated in SigmaPlot 12.0 software (Systat Software Inc., California). Catalytic efficiency (kcat/Km) values with their standard errors were obtained fitting the experimental data to the normalized Michaelis-Menten equation:
| (2) |
3. Results and Discussion
3.1. Study of LiP-VA Interaction Energies
To quickly evaluate the LiP-VA binding, the ΔE values at a SQM level were calculated. LiPH8 (wild type) was modeled using a molecular region that considered the active site residues located <5.5 Å around Trp171, and VA was modeled both in its neutral and radical cationic form. The results shown in Table 1 demonstrate that more favorable (more negative) values of ΔE were obtained for the substrate considered in its radical cationic state. Moreover, in all cases, the better-averaged ΔE values were with Trp171 and the neighbor residues Asp165, Asp264, Glu250, Glu168 and Phe267.
Table 1.
Averaged ΔE values calculated using SQM approach between VA (non-radical and cation radical) and each amino acid (AA) located around LiPH8 Trp171 (<5.5 Å far).
| AA | Average interaction energy (ΔE in kcal·mol−1) |
|
|---|---|---|
| AA/VA | AA/VA•+ | |
| Trp171 | −11.9 ± 0.2 | −13.8 ± 0.4 |
| Phe164 | −7.4 ± 0.4 | −21.5 ± 0.6 |
| Asp165 | −10.5 ± 0.2 | −87.1 ± 0.7 |
| Leu167 | −7.4 ± 0.3 | −14.3 ± 0.5 |
| Glu168 | −10.2 ± 0.2 | −75.7 ± 0.6 |
| Glu250 | −10.3 ± 0.2 | −76.5 ± 0.5 |
| Lys 260 | −10.3 ± 0.3 | 30.1 ± 0.7 |
| Asp264 | −10.4 ± 0.3 | −83.1 ± 0.4 |
| Phe267 | −7.9 ± 0.3 | −22.7 ± 0.6 |
| Ile268 | −5.5 ± 0.2 | −13.6 ± 0.3 |
3.2. LiP (and VP) Directed Variants
To clarify the role of the above amino acids in the catalytic cycle of LiP, site-directed mutagenesis and kinetic experiments were performed, as described below. The residues with the better ΔE values with VA and VA•+ identified above - i.e. Asp165, Glu168, Glu250 and Asp264, followed by Phe267 - were substituted by site-directed mutagenesis in single variants, where the new residues introduced resulted in worse (less negative) interaction energies (Table S2) without disrupting the protein structure. The kinetic constants of the selected single variants are shown in Table 2.
Table 2.
Kinetic constants (Km, μM; kcat, s−1; and kcat/Km, s−1 mM−1) of wild-type LiPH8 and its single and multiple variants oxidizing VA, DMP and ABTS (means and 95% confidence limits).
| Wild-type LiPH8LiPH8 | Mutated variants |
|||||
|---|---|---|---|---|---|---|
| D165N | E168Q | E250Q | D264N | |||
| VA | Km | 73.0 ± 3.9 | 80.9 ± 4.1 | 348.0 ± 37.0 | 221.0 ± 22.0 | 90.1 ± 5.5 |
| kcat | 20.9 ± 0.2 | 12.5 ± 0.2 | 17.1 ± 0.7 | 5.2 ± 0.1 | 20.0 ± 0.3 | |
| kcat/Km | 282.0 ± 15.0 | 155.0 ± 8.0 | 49.2 ± 5.6 | 23.5 ± 2.4 | 222.0 ± 14.0 | |
| DMP | Km | 38.1 ± 5.8 | 42.7 ± 2.4 | 50.2 ± 3.9 | 39.3 ± 3.2 | 33.4 ± 1.0 |
| kcat | 9.9 ± 0.4 | 5.3 ± 0.1 | 7.0 ± 0.2 | 1.5 ± 0.0 | 10.1 ± 0.1 | |
| kcat/Km | 260.0 ± 41.0 | 214.0 ± 7.0 | 139.0 ± 12.0 | 38.2 ± 3.1 | 302.0 ± 10.0 | |
| ABTS | Km | 26.7 ± 1.2 | 30.3 ± 0.6 | 26.4 ± 1.8 | 37.2 ± 4.1 | 14.5 ± 0.7 |
| kcat | 19.9 ± 0.3 | 15.7 ± 0.1 | 16.2 ± 0.3 | 9.2 ± 0.3 | 22.5 ± 0.4 | |
| kcat/Km | 745.0 ± 35.0 | 520.0 ± 3.0 | 614.0 ± 43.0 | 247.0 ± 28.0 | 1550.0 ± 80.0 | |
| Mutated variants |
||||||
|---|---|---|---|---|---|---|
| F267I | D165N/E168Q | E168Q/E250Q | D264N/F267I | D165N/E168Q/D264N/F267I | ||
| VA | Km | 233.0 ± 20.0 | 368.0 ± 34.0 | 2160.0 ± 470.0 | 273.0 ± 24.0 | 2550.0 ± 628.0 |
| kcat | 15.7 ± 0.3 | 11.6 ± 0.3 | 7.6 ± 0.6 | 13.5 ± 0.3 | 20.7 ± 1.8 | |
| kcat/Km | 67.3 ± 6.0 | 31.5 ± 3.0 | 3.5 ± 0.8 | 49.4 ± 4.4 | 8.1 ± 2.1 | |
| DMP | Km | 64.2 ± 10.3 | 78.3 ± 10.5 | 75.4 ± 11.4 | 78.3 ± 6.8 | 81.8 ± 12.3 |
| kcat | 7.3 ± 0.4 | 9.6 ± 0.4 | 5.3 ± 0.3 | 9.2 ± 0.5 | 10.9 ± 0.5 | |
| kcat/Km | 114.0 ± 19.0 | 123.0 ± 17.0 | 70.3 ± 11.3 | 245.0 ± 22.0 | 133.0 ± 21.0 | |
| ABTS | Km | 28.8 ± 1.2 | 38.3 ± 3.4 | 5.4 ± 0.5 | 22.6 ± 1.9 | 5.9 ± 0.8 |
| kcat | 19.4 ± 0.2 | 26.2 ± 0.7 | 13.2 ± 0.3 | 29.3 ± 0.7 | 15.2 ± 0.8 | |
| kcat/Km | 674.0 ± 29.0 | 684.0 ± 63.0 | 2460.0 ± 220.0 | 1290.0 ± 120.0 | 2610.0 ± 390.0 | |
The results show that the (Km) for VA increases with the neutralization of the partial negative charge of acidic residues like Glu168 and Glu250 (3 and ~5 folds, respectively) indicating lower substrate affinity. Moreover, LiPH8 turnover with VA is affected by mutation of Glu250 and Asp165 (4-fold kcat decrease for E250Q and 1.5-fold for D165N). Smith et al. [39] described the Fe(IV) = O Trp• reactive intermediate for VA oxidation in the LiPH8 E250Q variant. The reactivity of this variant is evaluated here through steady-state kinetics using VA and two additional substrates: DMP, representing phenolic units of lignin; and ABTS, a low redox-potential dye. Similarly to the results for VA oxidation, DMP and ABTS oxidation was affected in terms of turnover if Glu250 was mutated. On the contrary, the single mutation of the other acidic residues surrounding Trp171 did not change the kinetics for DMP or ABTS oxidation.
On the other hand, the aromaticity of Phe267 seems to be important in the binding of VA and DMP [40]. If the electron-rich π system of this residue is removed (for instance, mutated by an isoleucine), the Km values for substrates VA and DMP increase 3 and 1.5 folds, respectively. The π-stacking interaction established between aromatic rings of substrates and enzyme aromatic residues is not feasible in the F267I variant.
To gain insight about the potential cooperative effects in molecular interactions between different LiP residues with substrates, double and quadruple mutants were also developed (Table 2). According to our results, some double mutants exhibited synergistic effects for the oxidation of substrates. For instance, the E168Q/E250Q variant showed strong differences in the VA kinetic parameters compared with wild-type LiPH8: 30-fold increase in the Km, and 3-fold decrease in the kcat. The increased Km value in the E250Q and E168Q single variants is reflected in the double mutant with a higher Km value, together with kcat value similar to that found for single E250Q variant.
The D165N/E168Q/D264N/F267I quadruple and E168Q/E250Q double variants exhibits better efficiencies in ABTS oxidation, mainly due to a 5-fold higher affinity. The reduction of the negative charge around Trp171, and the removal of the bulky Phe267, minimizes the repulsive electrostatic and steric interactions with the anionic and bulky ABTS substrate. Fernández-Fueyo et al. [41] described similar results when charges in the propionate channel of Ceriporiopsis subvermispora MnP6 were neutralized.
To validate our kinetic results, the mutant homologous to E250Q in LiPH8 was produced in Versatile Peroxidase (VPL2) from P. eryngii (E243Q), which is another ligninolytic enzyme belonging to class II family of the peroxidase-catalase superfamily [1]. This enzyme presents 57% sequence homology and some similar catalytic properties with respect to LiP, due to the conserved catalytic tryptophan residue on the surface of the enzyme. The kinetic parameters for the oxidation of VA, DMP, ABTS, and Mn2+ in VPL2 are shown in Table 3. In agreement with the results obtained for LiPH8 mutant E250Q, VPL2 enzyme turnover (kcat) decreased for the oxidation of VA, DMP and ABTS when Glu243 was mutated (the kcat value decreased 4, 3, and 2 folds, respectively). However, the oxidation of Mn2+ was not affected since it takes place in another region of the enzyme [42]. These results indicate the involvement of homologous residues Glu250 and Glu243 in the catalytic mechanism of LiP and VP, respectively.
Table 3.
Kinetic constants (Km, μM; kcat, s−1; and kcat/Km, s−1 mM−1) of wild-type VP (allelic variant VPL2) and its E243Q variant oxidizing VA, DMP, ABTS and Mn2+ (means and 95% confidence limits).
| Wild-type VPL2 | E243Q variant | ||
|---|---|---|---|
| VA | Km | 2750.0 ± 50.0 | 2270.0 ± 150.0 |
| kcat | 6.6 ± 0.0 | 1.6 ± 0.0 | |
| kcat/Km | 2.4 ± 0.0 | 0.7 ± 0.0 | |
| DMP | Km | 18.0 ± 1.0 | 18.3 ± 1.3 |
| kcat | 7.1 ± 0.1 | 2.6 ± 0.0 | |
| kcat/Km | 403.0 ± 24.0 | 142.0 ± 10.0 | |
| ABTS | Km | 7.9 ± 0.9 | 7.3 ± 0.6 |
| kcat | 15.4 ± 0.6 | 7.8 ± 2.5 | |
| kcat/Km | 1950.0 ± 240.0 | 1070.0 ± 390.0 | |
| Mn2+ | Km | 268.0 ± 20.0 | 224.0 ± 10.0 |
| kcat | 282.1 ± 6.1 | 248.0 ± 3.0 | |
| kcat/Km | 1050.0 ± 80.0 | 1100.0 ± 50.0 |
3.3. SQM Study of VA Interaction Energy With the LiP Variants
In order to provide a rational (theoretical) explanation for the above kinetic experimental results, the interaction energies of VA (in its non-radical and radical cationic states) against LiPH8, including each of the five single variants in Table 2, were investigated at SQM level. The averaged ΔE values obtained between VA and LiP regions (REG) formed by wild-type residues (including Trp171) selected around 5.5 Å of Trp171, and LiP regions that include each one of the five point mutations (MREG), are shown in Table 4. For the estimation of ΔE values between the reaction substrate (VA) and the REG and MREG systems, we considered the two possible radical species formed by Trp171 (TrpH•+ and Trp•) according to Bernini et al. [9]. More favorable averaged ΔE values were obtained for TrpH•+, when compared to Trp• (Table 4). However, the most favorable (negative) ΔE values were observed when the intermolecular interaction between the product of the reaction (VA•+) and the already reduced tryptophan residue (Trp) was considered. Fuenzalida-Valdivia et al. [43] reported a comparative analysis for enzyme-substrate interaction in the systems LiP-VA and LiP-VA•+ during long MDS with simulation times of 1 μs. In these simulations, the authors observed that the LiP-VA•+ system was mainly stabilized by electrostatic interactions, whereas the main stabilization in the other system came from vdW interactions. Moreover, the most negative energy values were observed for VA•+ instead of VA. Together, all these pieces of evidence may explain why LiPH8 is very efficient stabilizing VA•+ at its surface, which is in agreement with the experimental detection of a LiP-VA•+ complex by EPR [44].
Table 4.
Averaged ΔE values calculated using SQM methods between VA (neutral and cation radical) and the wild-type and mutated regions (REG and MREG, respectively) constituted by amino acids located at <5.5 Å from LiPH8 Trp171, which was included in its non-radical (Trp171), radical cation (Trp171•+) and neutral radical (Trp171•) forms. Averaged SASA values for Trp171 are also shown.
| LiP regionsa | Average interaction energy (ΔE in kcal·mol−1) |
Residual Activityb (%) | Trp171 SASAc (Å2) | ||
|---|---|---|---|---|---|
| Trp•/VA | TrpH•+/VA | Trp/VA•+ | |||
| REG(wild type) | −10.4 ± 0.4 | −29.6 ± 0.4 | −161.9 ± 0.2 | 100 ± 0.9 | 399 ± 0.3 |
| MREG(D165N) | −5.5 ± 0.3 | −15.3 ± 0.3 | −124.3 ± 0.3 | 59.0 ± 0.4 | 385 ± 0.4 |
| MREG(E168Q) | −8.1 ± 0.2 | −24.4 ± 0.2 | −138.4 ± 0.3 | 68.2 ± 1.3 | 394 ± 0.3 |
| MREG(E250Q) | −4.1 ± 0.2 | −5.1 ± 0.3 | −109.1 ± 0.2 | 24.0 ± 0.4 | 379 ± 0.2 |
| MREG(D264N) | −9.8 ± 0.4 | −22.7 ± 0.2 | −145.8 ± 0.4 | 92.3 ± 1.8 | 404 ± 0.4 |
| MREG(F267I) | −6.5 ± 0.2 | −18.1 ± 0.4 | −135.9 ± 0.5 | 71.0 ± 1.7 | 390 ± 0.3 |
Regions located at <5.5 Å from Trp171.
The residual activity was evaluated in 0.1 M sodium tartrate buffer pH 3.0 using 0.31 mM VA (saturated concentration), 0.25 mM H2O2 and 0.01 μM of enzyme. Residual activity (%) was calculated as the quotient between enzyme activity of each mutant and wild type LiP, and then multiplied by 100.
Averaged SASA of Trp171 considering the steric hindrance at the different (wild-type and mutated) LiP regions.
For all analyzed MREG systems, the point mutations caused a partial reduction in the averaged ΔE values (Table 4) when compared to wild-type LiPH8. In the results for the system with the lowest averaged values, MREG(Trp/VA•+), it is observed that the E250Q, D165N, F267I, E168Q, and D264N mutations caused a reduction in the averaged ΔE values of 32%, 23%, 16%, 14%, and 9%, respectively. Interestingly, a good correlation between these computational ΔE values and the residual activity percentages for the six enzyme-substrate complexes could be established (Fig. S3a). The loss of VA affinity, in the case of some mutants, was correctly predicted by the ΔE calculations. Similar studies supplied evidence that substrate binding differences affect the enzyme catalysis [45]. Therefore, the SQM methodology enabled us to explain the experimental results obtained with the different LiP variants. Moreover, it provides averaged ΔE values that allow us to predict specific mutations that would cause changes in the enzymatic activity (as the E250Q mutant in the present study).
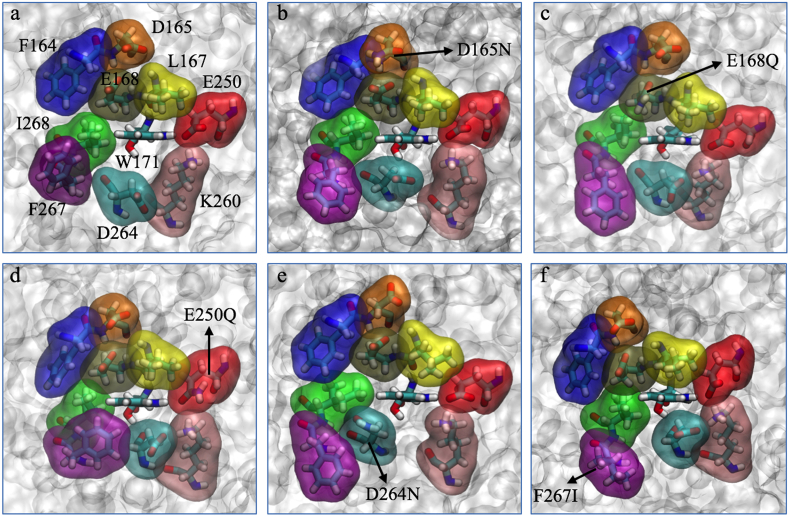
On the other hand, the SASA values for Trp171 in the mutated systems E250Q, D165N, F267I and E168Q in comparison with the wild-type system (Fig. 1a) showed a partial reduction of 8%, 4%, 2% and 1%, respectively (Table 4). This is because (as shown in Fig. 1b–d and f) these mutations caused compaction of the space around Trp171. On the contrary, the D264N mutation (Fig. 1e) generated an expansion of the residues around Trp171. This produced an increase in the SASA values (Table 4) and a consequent improvement in the Trp171/VA•+ interaction energy value compared to the other analyzed mutants. A correlation between the experimental kcat and the SASA values was observed with r2 0.89 (Fig. S3 b). These results suggest that the accessibility of Trp171 towards the substrate is crucial for catalysis, since the electron transfer reaction has to take place at an adequate orientation and distance.
Fig. 1.
Residues located at <5.5 Å from Trp171 in wild-type LiPH8 (a), and its in silico mutated systems (b-f).
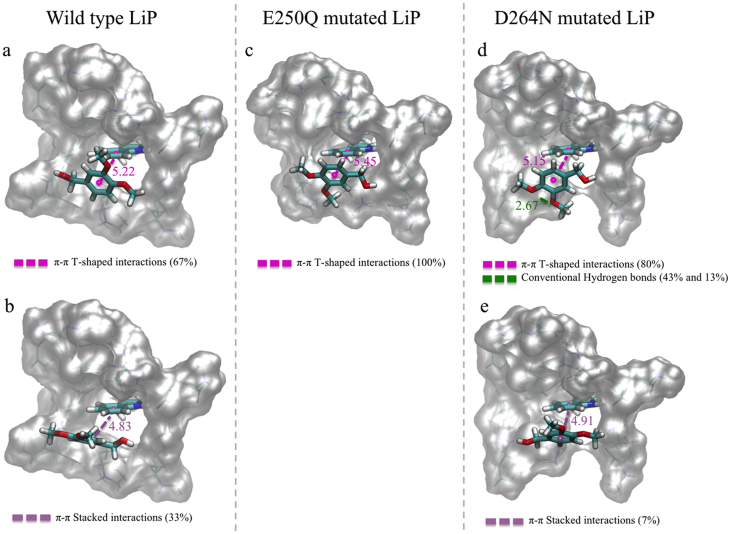
Finally, 100 complexes with the lower ΔE values were selected to analyze the main intermolecular interactions occurring in the Trp171/VA•+ complexes of the wild-type (Fig. 2a and b), E250Q mutated (the mutant that worsens the LiPH8 enzymatic activity) (Fig. 2c) and D264N mutated (the mutant where LiPH8 enzymatic activity is not affected) (Fig. 2d and e) LiP systems. The analysis showed the presence of π-π T-shaped interactions between the aromatic ring in VA and the indole ring in Trp171 in the three systems (Fig. 2a–d) but in different percentages (67%, 100%, 80%, respectively). This difference may be due to the presence of other types of interactions in a lower percentage (Fig. 2). The slightly less compacted cavity around Trp171 in wild-type LiPH8 and its D264N variant allowed 33% and 7% additional π-π stacked interactions, respectively (Fig. 2b and e). Additionally, 43% of conventional hydrogen bonds between the asparagine residue and VA were identified (Fig. 2d). From these H-bonds, 13% corresponded to complexes only with this type of interaction and 30% corresponded to complexes that stabilize at the same time with H-bonds and π-π T-shaped interactions. These intermolecular interactions contributed to ΔE values similar to those observed in the wild-type LiP. The compacted cavity, and greatest steric hindrance around Trp171, in the E250Q variant only allowed π-π T-shaped interactions at greater distances (differences about 0.25 Å on average). Therefore, these π-π interactions were less stable than those generated with the other mutants and the wild type enzyme. This should contribute to unfavorable interaction energy with VA•+ in those molecular systems.
Fig. 2.
Main π-π stacking intermolecular interactions in the LiPH8 Trp171-VA•+ complexes with the best interaction energies, corresponding to: (a,b) wild-type LiP region, (c) E250Q variant region, and (d, e) D264N variant region.
3.4. Spin Density QM/MM Calculations
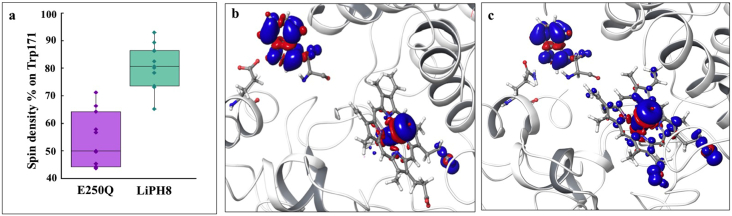
The enzyme kinetic data presented in Tables 2 and 3 suggest the implication of a hydrogen bond interaction between the indolic nitrogen of catalytic tryptophan and a close glutamic acid (Glu250 in LiPH8 and Glu243 in VPL2) on the reactivity of LiP and VP enzymes. Therefore, additional spin density QM/MM calculations were performed in order to investigate the electronic density distribution on Trp171 upon oxidation by compound I in wild-type LiPH8 and its E250Q variant.
The spin density percentage with respect to one unpaired electron on Trp171 is shown in Fig. 3a. It is possible to observe a more pronounced electron delocalization from the heme group towards Trp171 in wild-type LiPH8 (~80%) than in its E250Q variant (~55%) when averaged values of ten structures for each system were analyzed. Fig. 3b and c exhibit the spin density maps for ferromagnetic quartet state. As expected, the complex Fe O located two electrons in the mixed metal dxz/dyz and oxygen px/py orbitals [46]. The main difference is the delocalization of the third unpaired electron. In wild-type LiPH8, the unpaired electron is mostly localized over Trp171, meanwhile in the E250Q mutant, a lower electron localization is observed on this residue, which is compensated by a larger localization on the porphyrin ring, especially on the propionate groups (Fig. 3c).
Fig. 3.
QM/MM electron spin distribution on LiPH8 Trp171 and heme cofactor. (a) Boxplot representation of Trp171 spin density in wild-type LiPH8 and its E250Q variant. (b) Spin density for the three unpaired electrons in LiPH8 (c) Spin density for the three unpaired electron in E250Q.
Electron delocalization estimated by spin density maps has demonstrated to correlate well with the electron transfer driving force to determine the stability of the unpaired electron on the donor or acceptor's molecular orbitals [[47], [48], [49]]. Also, the spin delocalization is comparable to electron affinity and one of the criteria for any peroxidase to be a good catalyst is its capability to accept electrons efficiently [50]. Thus, spin densities calculated through the QM/MM approach provide quantitative information that demonstrates that the mutation on Glu250 implies an increase of electron affinity on Trp171. Therefore, the enzymatic reactivity of LiP decreased because compound I in the E250Q variant would extract one electron from Trp171 with less efficiency than the wild-type LiPH8.
4. Conclusions
VA, a secreted secondary metabolite of P. chrysosporium and other white-rot fungi [51], has proven to play a central role in LiP catalysis because, among other possible roles, prevents inactivation by hydrogen peroxide [52], and helps oxidation of other substrates contributing to catalytic cycle closure [53,54]. Therefore, many investigations have focused on the study of the different mechanisms in which LiP and VA interact. Moreover, VA has been suggested to act as a redox mediator through its cation radical (VA•+). The diffusion of this radical has been recently challenged [55] and, instead, the hypothesis that catalytic processes mediated by VA are guided through the formation of the LiP-VA•+ complex has taken importance and is more accepted in present days [56].
Interaction energies calculated at the quantum semiempirical level of theory allowed us to analyze enzyme-substrate interactions in the environment of catalytic Trp171. SQM was applied for study the interactions of LiP with its natural substrate VA and its primary product of reaction, VA in its radical cation state. ΔE values demonstrated that interaction between the residues located within 5.5 Å from Trp171 was more favorable with VA•+ than with VA. Moreover, the final complex between the reduced tryptophan and the reaction product (Trp-VA•+) presented better energy values than the initial complex between VA and the Trp radical in its neutral or cationic forms (TrpH•+/Trp•). These results justify the successful detection of the LiP-VA•+ complex by EPR [44]. VA•+ stabilization at the LiP surface would: i) increase enzyme turnover by stabilizing the product formed after one-electron abstraction by Trp171 (the final product, veratraldehyde, would result from rapid reaction of VA•+ with O2); and ii) make possible its participation in lignin degradation as a redox mediator, as mentioned above [55].
Mutation of the five residues with better averaged ΔE revealed that Glu168 and Glu250 are important residues participating in the binding of substrate and, additionally, Glu250 is a key residue in the electron transfer to the enzyme, as advanced by Smith et al. [57]. When the latter residue is mutated, the turnover of the enzyme decreases 4-fold, and the enzyme affinity is also modified. Rationalization of these results established that Trp171 in this variant is less accessible from the solvent (low SASA), and the analysis of Trp171 oxidation in LiP compound I by QM/MM calculations demonstrated that Glu250 participates in stabilizing the reactive tryptophanyl radical, with the Trp171 spin density decreasing in the E250Q variant.
Declaration of Competing Interests
There are no conflicts to declare.
Acknowledgments
J.A.M. and F.A.S. acknowledge the financial support from FONDECYT projects 1140618 and 3170909, respectively. R.R. and J.O.R acknowledge support from the doctoral fellowships CONICYT-PFCHA/Folios 21130949, and 21160905, respectively. A.T.M. thanks the financial support of the EnzOx2 (H2020-BBI-PPP-2015-2-720297, https://www.enzox2.eu) EU project. The authors thank the “Academic Writing Center” initiative at Programa de Idiomas from Universidad de Talca.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2019.07.002.
Contributor Information
Jans Alzate-Morales, Email: jalzate@utalca.cl.
Angel T. Martínez, Email: atmartinez@cib.csic.es.
Appendix A. Supplementary data
Supplementary material
References
- 1.Zámocký M., Hofbauer S., Schaffner I., Gasselhuber B., Nicolussi A., Soudi M., et al. Independent evolution of four heme peroxidase superfamilies. Arch Biochem Biophys. 2015;574:108–119. doi: 10.1016/j.abb.2014.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martínez A.T., Ruiz-Dueñas F.J., Martínez M.J., del Río J.C., Gutiérrez A. Enzymatic delignification of plant cell wall: from nature to mill. Curr Opin Biotechnol. 2009;20:348–357. doi: 10.1016/j.copbio.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Martinez A.T., Camarero S., Ruiz-Dueñas F.J., Martínez M.J. The royal society of chemistry. 2018. Biological lignin degradation; pp. 199–225. [DOI] [Google Scholar]
- 4.Harvey P.J., Palmer J.M., Schoemaker H.E., Dekker H.L., Wever R. Pre-steady-state kinetic study on the formation of compound I and II of ligninase. Biochim Biophys Acta (BBA)/Protein Struct Mol. 1989;994:59–63. doi: 10.1016/0167-4838(89)90062-9. [DOI] [PubMed] [Google Scholar]
- 5.Doyle W.A., Blodig W., Veitch N.C., Piontek K., Smith A.T. Two substrate interaction sites in lignin peroxidase revealed by site-directed mutagenesis. Biochemistry. 1998;37:15097–15105. doi: 10.1021/bi981633h. [DOI] [PubMed] [Google Scholar]
- 6.Acebes S., Ruiz-Dueñas F.J., Toubes M., Sáez-Jiménez V., Pérez-Boada M., Lucas M.F., et al. Mapping the long-range electron transfer route in ligninolytic Peroxidases. J Phys Chem B. 2017;121:3946–3954. doi: 10.1021/acs.jpcb.7b00835. [DOI] [PubMed] [Google Scholar]
- 7.Pogni R., Baratto M.C., Teutloff C., Giansanti S., Ruiz-Dueñas F.J., Choinowski T., et al. A tryptophan neutral radical in the oxidized state of versatile Peroxidase from Pleurotus eryngii: a combined multifrequency EPR and density functional theory study. J Biol Chem. 2006;281:9517–9526. doi: 10.1074/jbc.M510424200. [DOI] [PubMed] [Google Scholar]
- 8.Smith A.T., Doyle W.A., Dorlet P., Ivancich A. Spectroscopic evidence for an engineered, catalytically active trp radical that creates the unique reactivity of lignin peroxidase. Proc Natl Acad Sci. 2009;106:16084–16089. doi: 10.1073/pnas.0904535106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernini C., Pogni R., Basosi R., Sinicropi A. The nature of tryptophan radicals involved in the long-range electron transfer of lignin peroxidase and lignin peroxidase-like systems: insights from quantum mechanical/molecular mechanics simulations. Proteins Struct Funct Bioinforma. 2012;80:1476–1483. doi: 10.1002/prot.24046. [DOI] [PubMed] [Google Scholar]
- 10.Choinowski T., Blodig W., Winterhalter K.H., Piontek K. The crystal structure of lignin peroxidase at 1.70 å resolution reveals a hydroxy group on the C(β) of tryptophan 171: a novel radical site formed during the redox cycle. J Mol Biol. 1999;286:809–827. doi: 10.1006/jmbi.1998.2507. [DOI] [PubMed] [Google Scholar]
- 11.Bernini C., Pogni R., Ruiz-Dueñas F.J., Martínez A.T., Basosi R., Sinicropi A. EPR parameters of amino acid radicals in P. eryngii versatile peroxidase and its w164y variant computed at the qm/mm level. Phys Chem Chem Phys. 2011;13:5078–5098. doi: 10.1039/c0cp02151b. [DOI] [PubMed] [Google Scholar]
- 12.Schrödinger Release 2017 Protein Preparation Wizard; Epik, Schrödinger, LLC, New York, NY, 2016; Impact, Schrödinger, LLC, New York, NY, 2016; Prime, Schrödinger, LLC, New York, NY, 2017.
- 13.Olsson M.H.M., Søndergaard C.R., Rostkowski M., Jensen J.H. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J Chem Theory Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 14.Recabarren R., Alzate-Morales J., Fuenzalida-Valdivia I. Studying the binding mechanisms of veratryl alcohol to P. chrysosporium lignin peroxidase: insights from theoretical approaches. Theor Chem Acc. 2016;135:1–12. doi: 10.1007/s00214-016-1828-6. [DOI] [Google Scholar]
- 15.Maestro, Schrödinger, LLC, New York, NY, 2017.
- 16.MacroModel, Schrödinger, LLC, New York, NY, 2017.
- 17.Durham E., Dorr B., Woetzel N., Staritzbichler R., Meiler J. Solvent accessible surface area approximations for rapid and accurate protein structure prediction. J Mol Model. 2009;15:1093–1108. doi: 10.1007/s00894-009-0454-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Humphrey Dalke A., Schulten K. VMD - visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 19.Avila-Salas F., Sandoval C., Caballero J., Guinez-Molinos S., Santos L.S., Cachau R.E., et al. Study of interaction energies between the pamam dendrimer and nonsteroidal anti-inflammatory drug using a distributed computational strategy and experimental analysis by ESI-MS/MS. J Phys Chem B. 2012;116:2031–2039. doi: 10.1021/jp2069122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan C.F., Olafson B.D., Blanco M., Hsu S.L. Application of molecular simulation to derive phase diagrams of binary mixtures. Macromolecules. 1992;25:3667–3676. doi: 10.1021/ma00040a010. [DOI] [Google Scholar]
- 21.Durán-Lara E.F., López-Cortés X.A., Castro R.I., Avila-Salas F., González-Nilo F.D., Laurie V.F., et al. Experimental and theoretical binding affinity between polyvinylpolypyrrolidone and selected phenolic compounds from food matrices. Food Chem. 2015;168:464–470. doi: 10.1016/j.foodchem.2014.07.048. [DOI] [PubMed] [Google Scholar]
- 22.Durán-Lara E.F., Ávila-Salas F., Galaz S., John A., Maricán A., Gutiérrez M., et al. Nano-detoxification of organophosphate agents by PAMAM derivatives. J Braz Chem Soc. 2015;26:580–591. doi: 10.5935/0103-5053.20150013. [DOI] [Google Scholar]
- 23.Stewart J.J.P. Optimization of parameters for semiempirical methods. VI: more modifications to the nddo approximations and re-optimization of parameters. J Mol Model. 2013;19:1–32. doi: 10.1007/s00894-012-1667-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart J.J.P. Colorado Springs; CO, USA: 2016. MOPAC2016, Stewart computational chemistry.http://OpenMOPAC.net [Google Scholar]
- 25.Yilmazer N.D., Korth M. Comparison of molecular mechanics, semi-empirical quantum mechanical, and density functional theory methods for scoring protein-ligand interactions. J Phys Chem B. 2013;117:8075–8084. doi: 10.1021/jp402719k. [DOI] [PubMed] [Google Scholar]
- 26.Accelrys Sofware Inc . 2018. BIOVIA Discovery studio visualizer sofware. [Google Scholar]
- 27.Polak E., Ribiere G. Note sur la cenvergence de méthodes de directions conjuguées. ESAIM Math Model Numer Anal - Modélisation Math Anal Numérique. 1969;3:35–43. [Google Scholar]
- 28.QSite, Schrödinger, LLC, New York, NY, 2017.
- 29.Zhao Y., Truhlar D.G. A new local density functional for main-group thermochemistry, transition metal bonding, thermochemical kinetics, and noncovalent interactions. J Chem Phys. 2006;125 doi: 10.1063/1.2370993. [DOI] [PubMed] [Google Scholar]
- 30.Linde D., Pogni R., Marina C., Lucas F., Guallar V., Baratto M.C., et al. Catalytic surface radical in dye-decolorizing peroxidase: a computational, spectroscopic and site-directed mutagenesis study. Biochem J. 2015;466:253–262. doi: 10.1042/BJ20141211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulliken R.S. Electronic population analysis on LCAO-MO molecular wave functions. I. J Chem Phys. 1955;23:1833–1840. doi: 10.1063/1.1740588. [DOI] [Google Scholar]
- 32.Sáez-Jiménez V., Rencoret J., Rodríguez-Carvajal M.A., Gutiérrez A., Ruiz-Dueñas F.J., Martínez A.T. Role of surface tryptophan for peroxidase oxidation of nonphenolic lignin. Biotechnol Biofuels. 2016;9:1–13. doi: 10.1186/s13068-016-0615-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J., Fritsch E., Maniatis T. 2nd ed. Cold Spring Harbor Laboratory Press; New Year, NY: 1989. Molecular cloning: a laboratory manual. [Google Scholar]
- 34.Ayuso-Fernández I., Martínez A.T., Ruiz-Dueñas F.J. Experimental recreation of the evolution of lignin-degrading enzymes from the Jurassic to date. Biotechnol Biofuels. 2017;10:1–13. doi: 10.1186/s13068-017-0744-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pérez-Boada M., Doyle W.A., Ruiz-Dueñas F.J., Martínez M.J., Martínez A.T., Smith A.T. Expression of Pleurotus eryngii versatile peroxidase in Escherichia coli and optimisation of in vitro folding. Enzyme Microb Technol. 2002;30:518–524. doi: 10.1016/S0141-0229(02)00008-X. [DOI] [Google Scholar]
- 36.Dunford H.B., Peroxidases Heme. vol. 2. 2016. Royal society of chemistry. Cambridge, UK. [DOI] [Google Scholar]
- 37.Tien M., Kirk T.K. Lignin peroxidase of Phanerochaete chrysosporium. Methods Enzymol. 1988;161:238–249. doi: 10.1016/0076-6879(88)61025-1. [DOI] [Google Scholar]
- 38.Ruiz-Dueñas F.J., Martínez M.J., Martínez A.T. Molecular characterization of a novel peroxidase isolated from the ligninolytic fungus Pleurotus eryngii. Mol Microbiol. 1999;31:223–235. doi: 10.1046/j.1365-2958.1999.01164.x. [DOI] [PubMed] [Google Scholar]
- 39.Smith A.T., Doyle W.A., Dorlet P., Ivancich A. Spectroscopic evidence for an engineered, catalytically active trp radical that creates the unique reactivity of lignin peroxidase. Proc Natl Acad Sci U S A. 2009;106:16084–16089. doi: 10.1073/pnas.0904535106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sollewijn Gelpke M.D., Lee J., Gold M.H. Lignin Peroxidase oxidation of veratryl alcohol: effects of the mutants H82A, Q222A, W171A, and F267L. Biochemistry. 2002;41:3498–3506. doi: 10.1021/bi011930d. [DOI] [PubMed] [Google Scholar]
- 41.Fernández-Fueyo E., Acebes S., Ruiz-Dueñas F.J., Martínez M.J., Romero A., Medrano F.J., et al. Structural implications of the c-terminal tail in the catalytic and stability properties of manganese peroxidases from ligninolytic fungi. Acta Crystallogr Sect D Biol Crystallogr. 2014;70:3253–3265. doi: 10.1107/S1399004714022755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pérez-Boada M., Ruiz-Dueñas F.J., Pogni R., Basosi R., Choinowski T., Martínez M.J., et al. Versatile peroxidase oxidation of high redox potential aromatic compounds: site-directed mutagenesis, spectroscopic and crystallographic investigation of three long-range electron transfer pathways. J Mol Biol. 2005;354:385–402. doi: 10.1016/j.jmb.2005.09.047. [DOI] [PubMed] [Google Scholar]
- 43.Fuenzalida-Valdivia I., Recabarren R., Alzate-Morales J. Universidad de Talca; Talca, Chile: 2016. Molecular dynamics analysis of the binding mechanism of veratryl alcohol at the protein surface of Lignin peroxidase (P. chrysosporium) and its Mutants E168Q and D264N. 2nd Bioinforma; pp. 1–2. Meet., Oct. 13-14. [DOI] [Google Scholar]
- 44.Khindaria a Nie G., Aust S.D. Detection and characterization of the lignin peroxidase compound II-veratryl alcohol cation radical complex. Biochemistry. 1997;36:14181–14185. doi: 10.1021/bi9715730. [DOI] [PubMed] [Google Scholar]
- 45.Tiwari M.K., Kalia V.C., Kang Y.C., Lee J.K. Role of a remote leucine residue in the catalytic function of polyol dehydrogenase. Mol Biosyst. 2014;10:3255–3263. doi: 10.1039/c4mb00459k. [DOI] [PubMed] [Google Scholar]
- 46.Bathelt C.M., Mulholland A.J., Harvey J.N. QM/MM studies of the electronic structure of the compound i intermediate in cytochrome c peroxidase and ascorbate peroxidase. Dalton Trans. 2005:3470–3476. doi: 10.1039/b505407a. [DOI] [PubMed] [Google Scholar]
- 47.Artz K., Williams J.C., Allen J.P., Lendzian F., Rautter J., Lubitz W. Relationship between the oxidation potential and electron spin density of the primary electron donor in reaction centers from Rhodobacter sphaeroides. Biophys Approv. 1997;94:13582–13587. doi: 10.1073/pnas.94.25.13582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lucas M.F., Monza E., Jørgensen L.J., Ernst H.A., Piontek K., Bjerrum M.J., et al. Simulating substrate recognition and oxidation in laccases: from description to design. J Chem Theory Comput. 2017;13:1462–1467. doi: 10.1021/acs.jctc.6b01158. [DOI] [PubMed] [Google Scholar]
- 49.Monza E., Lucas M.F., Camarero S., Alejaldre L.C., Martínez A.T., Guallar V. Insights into laccase engineering from molecular simulations: toward a binding-focused strategy. J Phys Chem Lett. 2015;6:1447–1453. doi: 10.1021/acs.jpclett.5b00225. [DOI] [PubMed] [Google Scholar]
- 50.De Visser S.P. What affects the quartet-doublet energy splitting in peroxidase enzymes? J Phys Chem A. 2005;109:11050–11057. doi: 10.1021/jp053873u. [DOI] [PubMed] [Google Scholar]
- 51.Shimada M., Nakatsubo F., Kirk T.K., Higuchi T. Biosynthesis of the secondary metabolite veratryl alcohol in relation to lignin degradation in Phanerochaete chrysosporium. Arch Microbiol. 1981;129:321–324. [Google Scholar]
- 52.Tonon F., Odier E. Influence of veratryl alcohol and hydrogen peroxide on ligninase activity and ligninase production by Phanerochaete chrysosporium. Appl Environ Microbiol. 1988;54:466–472. doi: 10.1128/aem.54.2.466-472.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koduri R., Tien M. Oxidation of guaiacol by lignin peroxidase. J Biol Chem. 1995;270:22254–22258. doi: 10.1074/jbc.270.38.22254. [DOI] [PubMed] [Google Scholar]
- 54.Tien M., Ma D. Oxidation of 4-methoxymandelic acid by lignin peroxidase. J Biol Chem. 1997;272:8912–8917. doi: 10.1074/jbc.272.14.8912. [DOI] [PubMed] [Google Scholar]
- 56.Khindaria A., Yamazaki I., Aust S.D. Stabilization of the veratryl alcohol cation radical by lignin peroxidase. Bichemistry. 1996;35:6418–6424. doi: 10.1021/bi9601666. [DOI] [PubMed] [Google Scholar]
- 55.Houtman C.J., Maligaspe E., Hunt C.G., Fernández-Fueyo E., Martínez A.T., Hammel K.E. Fungal lignin Peroxidase does not produce the veratryl alcohol cation radical as a diffusible ligninolytic oxidant. J Biol Chem. 2018;293:4702–4712. doi: 10.1074/jbc.RA117.001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith A., Doyle W. 2006. Engineered peroxidases with veratryl Alcohol Oxidase Activity. [WO 2006/114616 A1] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material