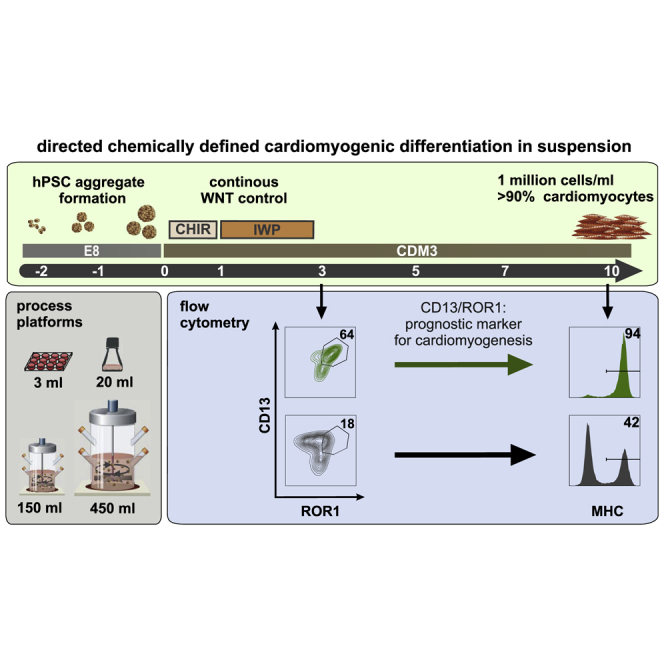
Summary
Aiming at clinical translation, robust directed differentiation of human pluripotent stem cells (hPSCs), preferentially in chemically defined conditions, is a key requirement. Here, feasibility of suspension culture based hPSC-cardiomyocyte (hPSC-CM) production in low-cost, xeno-free media compatible with good manufacturing practice standards is shown. Applying stirred tank bioreactor systems at increasing dimensions, our advanced protocol enables routine production of about 1 million hPSC-CMs/mL, yielding ∼1.3 × 108 CM in 150 mL and ∼4.0 × 108 CMs in 350–500 mL process scale at >90% lineage purity. Process robustness and efficiency is ensured by uninterrupted chemical WNT pathway control at early stages of differentiation and results in the formation of almost exclusively ventricular-like CMs. Modulated WNT pathway regulation also revealed the previously unappreciated role of ROR1/CD13 as superior surrogate markers for predicting cardiac differentiation efficiency as soon as 72 h of differentiation. This monitoring strategy facilitates process upscaling and controlled mass production of hPSC derivatives.
Keywords: human pluripotent stem cells, chemically defined process, cardiac differentiation, cardiomyocyte, suspension culture, stirred tank bioreactor, ROR1, WNT, process development, upscaling
Graphical Abstract
Highlights
-
•
Chemically defined hPSC cardiac differentiation applicable to stirred tank reactors
-
•
Protocol generates >90% purity of ventricular-like cardiomyocytes
-
•
Uninterrupted WNT pathway control enables superior cardiac mesoderm formation
-
•
Novel ROR1/CD13 combination as superior, predictive marker of cardiomyogenesis
Directed differentiation of human pluripotent stem cell cardiomyocytes (CMs) is required for clinical translation. Here, xeno-free suspension culture production in stirred bioreactors is shown to enable routine generation of ∼1.3 × 108 CMs/150 mL and ∼4.0 × 108 CMs/500 mL scale at >90% purity. Uninterrupted chemical WNT pathway control and the previously unappreciated role of ROR1/CD13 as predictive differentiation markers facilitate large-scale production of ventricular-like CMs.
Introduction
Progress in human pluripotent stem cell (hPSC) research has the potential to revolutionize regenerative medicine. Functional hPSC progenies may either fuel advanced in vitro assays for better drug development or replenish the loss of functional cells in diseased organs.
Given the high incidence of cardiac disorders, there have been substantial efforts in investigating cardiomyogenic differentiation of hPSCs. Stages of differentiation include early mesendoderm priming (Kempf et al., 2016), specification of cardiac progenitors (Soh et al., 2016), and directed differentiation into cardiomyocyte (CM) subtypes such as ventricular-, atrial- and nodal-like phenotypes (Devalla et al., 2015, Protze et al., 2017). Process specification was also accompanied by revealing more lineage-specific surface markers facilitating monitoring of differentiation stages and process optimization (reviewed in Skelton et al., 2017).
The field has also progressed from using recombinant factors toward chemical compounds for directing CM induction. These protocols typically aim at mimicking the biphasic pattern of WNT pathway upregulation and subsequent attenuation known from early heart development (Gonzalez et al., 2011, Lian et al., 2012, Tran et al., 2009, Ueno et al., 2007). Notably, chemical WNT pathway stimulators (particularly the GSK3 inhibitor CHIR99021 [CHIR]) or suppressors (including IWP2, IWR1, and Wnt-C59) have also been applied to specify other mesendodermal lineages including hepatocytes (Siller et al., 2015) and skeletal muscle cells (Shelton et al., 2014). This highlights process complexity due to the multiple spatiotemporally dependent roles of the WNT pathway in development. Moreover, we have recently demonstrated that, in response to CHIR stimulation, a complex pattern of paracrine factors, whose feedback-controlled concentration depends on the applied cell density, substantially modulates early primitive streak (PS)-like priming (Gaspari et al., 2018, Kempf et al., 2016). Thus, in addition to the well-studied impact of the CHIR dose, the cell density and the exact process timing have a dominant impact on hPSC differentiation.
Cell production in suspension culture by the differentiation of matrix-free hPSC aggregates is more compatible with process upscaling. It facilitates transition to stirred tank bioreactors favored for process control and optimization for conventional mammalian cell lines in the biotech industry. We and others demonstrated feasibility of suspension culture for both hPSC expansion (Abecasis et al., 2017, Kropp et al., 2016) and lineage differentiation, including successful CM, endothelial cell, and macrophage production (Ackermann et al., 2018, Chen et al., 2015, Fonoudi et al., 2015, Kempf et al., 2014, Olmer et al., 2018). However, whereas two-dimensional (2D) culture is restricted in their complexity, the number of process variables increases in 3D suspension culture. Besides the overall cell density, spherical aggregates (3D) increase in size over time (4D), thus constantly changing the physical and physiological parameters of the culture.
Multidimensional process parameters in combination with the known hPSC line-dependent properties often result into interexperimental variability. We have reported, for example, the average induction of ∼80% CMs in stirred suspension, but noted process variability ranging from <60% to >90% CM content (Kempf et al., 2014).
We thus performed systematic modifications of process parameters in this study by applying several culture platforms and numerous hPSC lines. By focusing on the critical early steps particularly on hPSC aggregation and the timing of chemical WNT modulation, a more robust and efficient protocol was developed. This includes the systematic usage of chemically defined media compatible with large-scale cell production and transition to good manufacturing practice standards. Applying molecular cell analysis in response to process modifications a novel surface marker, ROR1, is revealed which, in combination with CD13, is superior for predictive monitoring of cardiac mesoderm formation.
Results
WNT Pathway Inhibition Improves Priming toward Cardiac Mesoderm
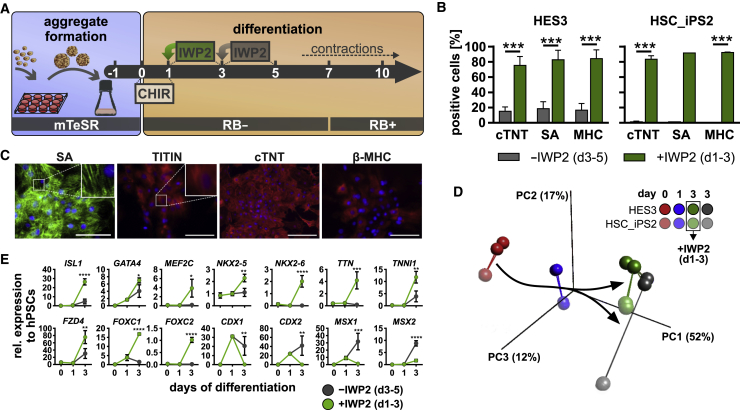
After kick-starting cardiac differentiation by 24 h of CHIR supplementation, WNT pathway attenuation is typically initiated after a 48- to 72-h gap (Kempf et al., 2014, Lian et al., 2012). In contrast to CHIR treatment, temporal patterning of WNT inhibition was less systematically investigated, particularly in suspension culture. We thus compared the effect of early (days 1–3) with that of the commonly applied late (days 3–5) supplementation of WNT inhibitors (schematic in Figure 1A). Flow cytometry (FC)-based endpoint analysis for CM-specific markers including cardiac troponin T (cTNT), sarcomeric actinin (SA), and myosin heavy chain (MHC) on day 10 revealed only low CM induction of ∼15%–18% (HES3) and <1.5% (HSC-iPS2), respectively (Figures 1B and S1A), when the WNT antagonist IWP2 was added on day 3. In contrast, starting inhibition on day 1 (i.e., immediately after CHIR removal) resulted in >75% CM content for both cell lines and all markers tested (Figure 1B). This was confirmed by qRT-PCR (Figure S1D) and immune cytology-based analysis of suspension-derived, plated cells (Figure 1C).
Figure 1.
Timed WNT Pathway Inhibition Primes Mesendoderm toward Cardiac Mesoderm
(A) Differentiation schematic. Aggregates generated in 12-well plates were transferred to agitated Erlenmeyer flasks 1 day prior (−1) to differentiation induction by CHIR (0); WNT inhibition was performed on day 1 or day 3 by IWP2.
(B) CM content (%) measured by FC on day 10 following WNT inhibition on day 1 ( ) and day 3 (
) and day 3 ( ) for cardiac troponin T (cTNT), sarcomeric actinin (SA), and myosin heavy chain (MHC). 5 and 4 independent experiments were performed for hESCs and HSC_iPS2 cells, respectively. p < 0.001.
) for cardiac troponin T (cTNT), sarcomeric actinin (SA), and myosin heavy chain (MHC). 5 and 4 independent experiments were performed for hESCs and HSC_iPS2 cells, respectively. p < 0.001.
(C) IF staining of plated cells (day 14 + 4) for structural cardiac markers SA, TITIN, cTNT, and β-MHC. Scale bars, 100 μm.
(D) PCA (σ/σmax = 0.184) of microarray analysis shows clustering of samples over time and a distinct cell line-dependent pattern for IWP2 treatment. Two independent experiments were performed for each cell line.
(E) Time course of selected cardiac/presomitic mesoderm markers. n = 4 independent experiments based on microarray signal intensities.
Error bars represent SEM. See also Figure S1.
Global gene expression analysis was conducted on day 0, day 1, and day 3 using HES3 (bright color code in Figure 1D) and HSC_iPS2 cells treated with (green dots) or without (gray dots) IWP2 on days 1–3 (Figure 1D). Principal component analysis showed expected clustering of day-0, -1, and -3 samples (Figures 1D and S1C). Notably, day-3 samples receiving no IWP2 (gray) appeared more distant to day 1 along the PC1 axis compared with IWP2-treated cells (green). This indicates that the (differentiation-induced) changes were less pronounced when IWP2 was supplemented, suggesting containment of differentiation progression by the inhibitor.
Interestingly, early WNT pathway inhibition appears to be essential to direct certain hPSC lines such as HSC_iPS2 toward the cardiomyogenic lineage, whereas the HES3 line seems to be less sensitive to external WNT inhibitors, in line with previous work (Kempf et al., 2014).
Gene ontology (GO) analysis of >10-fold regulated gene sets revealed that IWP2 treatment induced significant upregulation of genes related to the GO terms heart development and morphogenesis, while downregulated genes were associated with development and morphogenesis of the skeletal system, cartilage, bone, and cartilage, all resembling paraxial mesoderm derivatives (Figure S1B).
Patterning into cardiac versus presumptive presomitic mesoderm was confirmed by targeted heatmap analysis for associated GO terms, including osteoblast, skeletal muscle, mesenchyme, somite, and limb development. Remarkably, day-3 analysis of IWP-treated cells showed robust upregulation of key cardiac progenitor markers including ISL1, GATA4, MEF2C, NXK2-6 (Biben et al., 1998), FZD4 (Yoon et al., 2018), and FOXC1/2, as well as TTN and TNNI1 (Figure 1E). Conversely, markers described to inhibit cardiac mesoderm but to promote presomitic specification including CDX1/2 and MSX1/2 (Greber et al., 2010, Mendjan et al., 2014) were significantly upregulated in the absence of early IWP2 treatment (confirmed for CDX2 by FC; Figure S1E).
ROR1 Distinguishes Cardiac from Paraxial Mesoderm
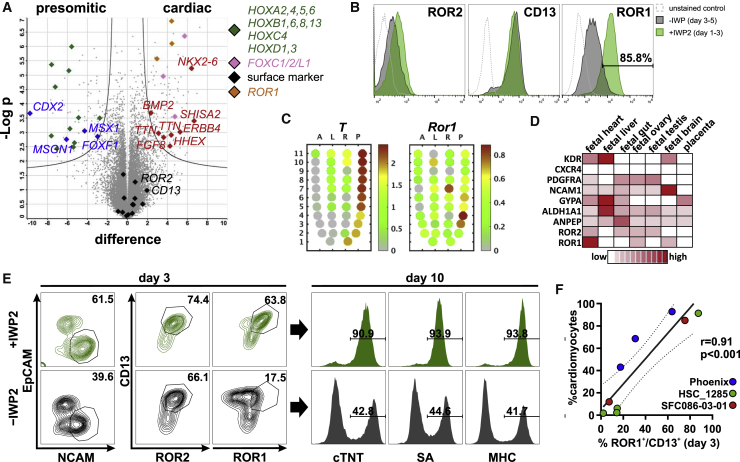
Multiple surface markers were reported to identify cardiac mesoderm during hPSC differentiation. We evaluated expression of these markers in our 3D system regarding their ability to discriminate cardiac versus presomitic mesoderm on day 3 of differentiation on gene and protein levels. Most established markers showed only marginal or low induction (KDR, CXCR4, and GYPA), while others were upregulated (PDGFRA, NCAM, CD13, and ROR2) but failed to discriminate between the two lineages (Figures 2A, 2B, S2A, and S2B). Interestingly, ROR1, in contrast to the structurally related protein ROR2 (60% homology in protein sequence), depicted significantly higher levels in cardiac compared with presomitic mesoderm (Figures 2A, 2B, S2A, and S2C). Studies in mice reported Ror2 expression throughout the entire PS (Matsuda et al., 2001) while recent data indicate distinct Ror1 expression in the anterior-to-mid PS only (Peng et al., 2016) (Figure 2C) suggesting a distinct role of Ror1 during mesendodermal patterning (note that T delineates the PS area of the epiblast of the gastrulating embryo; Figure 2C). Likewise, proteome data from human fetal tissues revealed prominent expression of ROR1 but not other established surface markers in the developing heart (Kim et al., 2014) (Figure 2D). Indeed, in contrast to the combination of CD13+/ROR2+ (r = 0.73), combining analysis of CD13+/ROR1+ by FC on day 3 and corresponding cardiomyocyte content on day 10 showed a more significant correlation (r = 0.9) (Figures 2E, 2F, and S2B), with CD13+ serving as panmesoderm marker to exclude the potential presence of pluripotent cells that also express ROR1 but lack CD13 (Figure S2A).
Figure 2.
ROR1 Serves as Predictive Surrogate Marker for Cardiomyocyte Formation
(A) Volcano plot comparing cardiac and presomitic mesoderm on day 3 of differentiation. Key genes for each condition are highlighted in respective colors, either grouped in categories ( ,
,  ,
,  ,
,  ) or individually (
) or individually ( ,
,  ). Note the detection of ROR1 (
). Note the detection of ROR1 ( ) in the cardiac mesoderm by three independent probes and the minor difference for established surface markers (
) in the cardiac mesoderm by three independent probes and the minor difference for established surface markers ( ), n = 4 independent experiments.
), n = 4 independent experiments.
(B) Representative FC histograms for ROR2, CD13, and ROR1 on day 3 with ( ) and without (
) and without ( ) IWP2 treatment on day 1.
) IWP2 treatment on day 1.
(C) Gene expression in the gastrulating mouse embryo (Peng et al., 2016) indicates Ror1 expression (right panel) in the anterior-to-mid PS region whereby T expression (left panel) increases toward the posterior PS.
(D) Pattern of mesodermal surface markers in human fetal tissues (Kim et al., 2014) indicates high ROR1 levels in the heart.
(E) Representative FC on day 3 for NCAM/EpCAM, CD13/ROR2 and CD13/ROR1 with/without addition of IWP2 on day 1 and corresponding CM content (cTNT, SA, MHC) on day 10.
(F) Correlation of ROR1+/CD13+ on day 3 and respective CM content on day 10 based on three different cell lines; each dot represents an independent biological repeat. r denotes Pearson's correlation coefficient. Dotted lines denote 95% confidence interval.
See also Figure S2.
Taken together, these data revealed ROR1 as a marker for cardiac mesoderm which, in combination with CD13, enables early prediction of cardiac differentiation efficiency.
Cell-Line-Independent Cardiomyogenesis in Chemically Defined Media
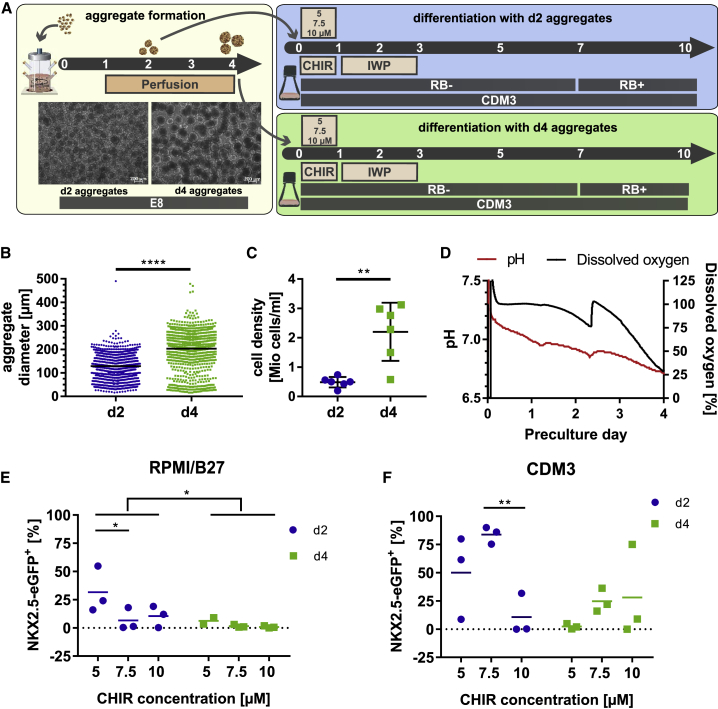
Considering upscaling of CM production for clinical translation, the development of cost-efficient xeno-free processes is essential. Based on successful hPSC expansion and maintenance of pluripotency in stirred suspension culture in chemically defined E8 (Chen et al., 2012, Kropp et al., 2016), it was applied to replace mTeSR used previously (Ludwig et al., 2006, Olmer et al., 2012, Silva et al., 2015) and in this work (Figures 1 and 2). For CM differentiation the chemically defined medium CDM3, recently established in 2D, was utilized (Burridge et al., 2014). Based on reports demonstrating the importance of the preculture strategy (Chen et al., 2015, Fonoudi et al., 2015, Kempf et al., 2014), we combined modified timing of aggregate generation in E8 with the improved WNT modulation described above. Aggregates were formed in E8 in stirred tank bioreactors for either 2 or 4 days before transition to either CDM3 or RPMI/B27 (routinely used previously by Kempf et al., 2014, Lian et al., 2012), and for differentiation in rotated Erlenmeyer flasks (schematic in Figure 3A).
Figure 3.
Day-2 Aggregates Derived in E8 Show Superior CM Formation in Chemically Defined Differentiation Medium
(A) Experimental scheme for optimizing expansion timing before differentiation. Microscopy of day 2/day 4 aggregates.
(B) Diameter assessment: evaluating >100 aggregates on day 2/day 4 from three independent runs. p < 0.0001.
(C) Cell density on day 2/day 4 of three independent runs in duplicates, respectively. p = 0.0019.
(D) Representative pH/DO curves of preculture.
(E and F) The impact of CHIR concentration and preculture duration. FC on day 10 revealed higher content of GFP+ CMs applying day-2 precultures in (E) RPMI/B27 (n = 3 independent experiments, preculture duration p =0.0262 and 5 versus 7.5 μM CHIR p = 0.0439) or (F) CDM3 (n = 3 independent experiments, 7.5 versus 70 μM CHIR p = 0.0095).
Error bars represent SEM. See also Figure S3.
An increase in average aggregate diameter from 128.5 ± 49.60 μm on day 2 to 203.2 ± 79.3 μm on day 4 was observed (Figure 3B and picture inset in Figure 3A) while the cell density increased by ∼4-fold (0.49 ± 0.07 × 106 and 2.2 ± 0.4 × 106 cells/mL, respectively; Figure 3C). Dissolved oxygen (DO) and pH (Figure 3D) values were substantially lower for day 4 than for day 2, demonstrating that, in addition to the mere change in aggregate diameters (suggested to impact on differentiation; Chen et al., 2015, Fonoudi et al., 2015), numerous other physical and physiological parameters that may impact on differentiation vary as well.
Cells' sensitivity to WNT pathway stimulation may also depend on the applied media. The CHIR concentration previously optimized to 7.5 μM in mTeSR/RPMI/B27 was thus reinvestigated (Kempf et al., 2014). Twenty-four hours of CHIR treatment at 5, 7.5, or 10 μM, respectively was immediately replaced by IWP2 for 48 h (days 1–3), and NKX2.5-eGFP expression was monitored as CM surrogate marker on day 10. For day-2 aggregates in RPMI/B27, 31.6% ± 11.8% of NKX2.5-eGFP+ cells were observed at 5 μM CHIR and even lower values were found for 7.5 μM and 10 μM CHIR, respectively (Figure 3E). For day-4 aggregates, the amount of GFP-expressing cells was also highest at 5 μM CHIR but the overall values were lower compared with day-2 aggregates (Figure 3E). Differentiation in CDM3 substantially improved NKX2.5-eGFP expression and was most robust combining E8-derived day-2 aggregates with 7.5 μM CHIR (83.8% ± 4.4%; Figure 3F). Day-2 aggregates outperformed day-4 cells over the entire CHIR range tested (Figure 3F).
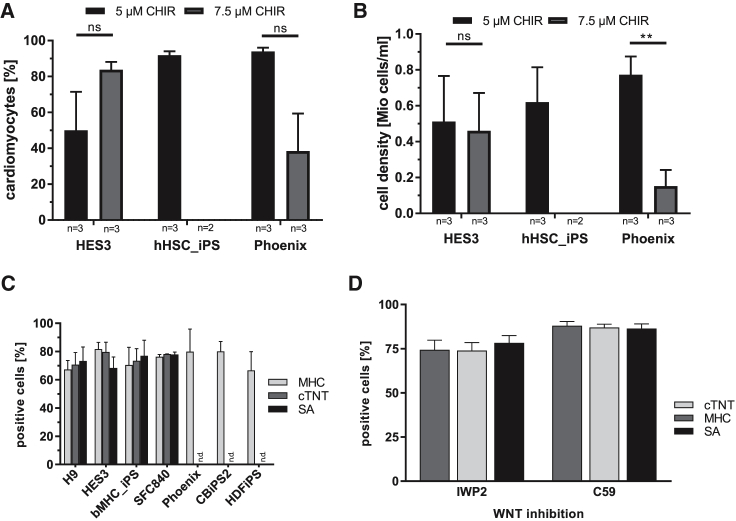
This chemically defined protocol (equivalent to schematic in Figure 3A) was tested with several lines (HES3-NKX2.5, HSC_iPS2, and Phoenix) with day-2 aggregates following differentiation in flasks at 5 μM and 7.5 μM CHIR.
In contrast to HES3-NKX2.5, 5 μM CHIR was optimal for HSC_iPS2 and Phoenix, inducing 91% and 93.7% ± 3.6% CMs, respectively (Figure 4A). CHIR at 7.5 μM also resulted in substantially lower cell yield during subsequent differentiation (Figures 4B, S3A, and S3B), eventually preventing endpoint analysis of HSC_iPS2 progenies. On the other hand, at 5 μM CHIR treatment, robust CM yields were observed for all cell lines tested (Figure 4B) whereby HES3-NKX2.5 were, again, less sensitive to the applied CHIR dose. Taken together, these data revealed that 5 μM CHIR is most appropriate for our chemically defined cardiac differentiation protocol.
Figure 4.
Robust Differentiation in Chemically Defined Conditions across Multiple hPSC Lines
(A and B) CM content (A) and respective cell numbers (B) on day 10 using bioreactor-derived aggregates from day-2 precultures of three cell lines differentiated in Erlenmeyer flasks in two or three independent experiments per cell line. (A) hESCs p = 0.1972, Phoenix cells p = 0.0575. (B) hESCs p = 0.8835, Pheonix cells p = 0.0067.
(C) Robustness of the chemically defined differentiation protocol was confirmed by FC for three markers with >70% CM found for seven cell lines tested in agitated 6-well plates in 2–5 independent experiments: hES3, bMHC_iPS, SFC840: n = 2; H9, CBiPS2, HDFiPS: n = 3; Phoenix: n = 5.
(D) Comparison of IWP2 versus Wnt-C59 added after 24 h of 5 μM CHIR treatment in three independent experiments revealed similar efficiencies in 6-well plates on day 10 for Phoenix.
Error bars represent SEM.
Additional human embryonic stem cell (hESC) and human iPSC (hiPSC) lines were assessed in a broadly available 6-well low-attachment platform revealing >70% CM induction across the seven cell lines tested (Figure 4C), supporting the general cell-line- and culture platform-independent utility of our protocol. Moreover, comparing the WNT pathway inhibitors IWP2 with Wnt-C59 using Phoenix showed comparable differentiation efficiencies (Figure 4D).
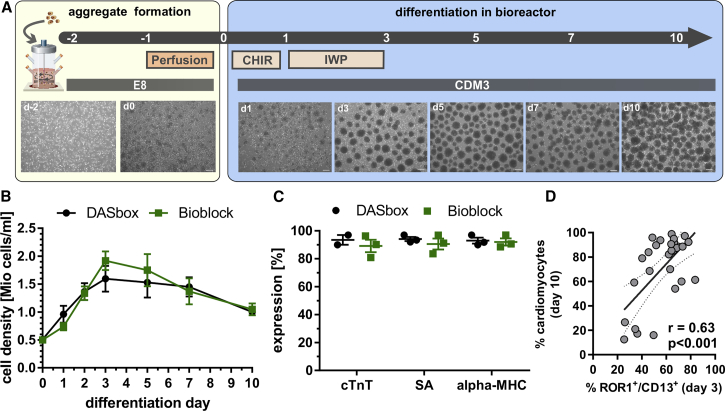
Process Upscaling in Stirred Tank Bioreactors Enables High Cell Yield at >90% CM Purity and Confirms Utility of CD13+/ROR1+ for Early Process Monitoring
Next, process upscaling in stirred tank bioreactors was performed, firstly in 150-mL (DASbox) and subsequently in up to 500-mL culture scale in a larger system (Bioblock). Applying the protocol in Figure 5A the cell density was adjusted to 0.5 × 106 cells/mL before induction of differentiation to consider the impact of the bulk cell density and interprocess reproducibility (Kempf et al., 2016).
Figure 5.
Process Upscaling in Stirred Tank Bioreactors Confirms Utility of CD13+/ROR1+ for Early Process Monitoring and Enables >90% CM Induction
(A) Scheme for differentiation in bioreactors and microscopic assessment of aggregates over time. Scale bars, 200 μm.
(B) Growth kinetics in DASbox (n = 3 independent experiments) and the Bioblock system (n = 3 independent experiments). Error bars represent SEM.
(C) Cardiac differentiation efficiency assessed by FC on day 10 for cTNT, SA, and MHC revealed >90% purity in DASbox and ∼90% in Bioblock (n = 3 independent experiments). Error bars represent SEM.
(D) Correlation of ROR1+/CD13+ on day 3 and respective CM content on day 10 of Phoenix in specific differentiation conditions resulting from upscale experiments. r denotes Pearson's correlation coefficient (n = 28 in seven independent experiments). Dotted line denotes 95% confidence interval.
See also Figure S4.
Growth kinetics revealed that the cell density peaked at >1.5 × 106 cells/mL on days 3–5 but declined thereafter, resulting in 1 ± 0.1 × 106 total cells/mL on day 10 corresponding to 0.93 ± 0.1 × 106 CMs/mL (Figures 5B and S4B), thus yielding ∼130 × 106 CMs per run in 150-mL scale.
On day 10, FC revealed a CM content of 93.5% ± 5.0%, 94,13% ± 2.8%, and 92.9% ± 3.64% for cTNT, MHC, and SA, respectively (Figures 5C and S4A), demonstrating high process efficiency and reproducibility.
This endpoint analysis for CM markers was correlated to CD13+/ROR1+ on day 3 for a large number of experiments across the entire optimization phase, including preliminary conditions leading to low CM contents (Figure 5D). The observed correlation coefficient of r = 0.63 may not seem ideal but the following must be considered: (1) the biological variance inherent to such complex biological systems, in particular when correlating two time points being 7 days apart; and (2) the transient nature of the CD13+/ROR1+ expression whereby slight variations in process timing and analysis may substantially impact on this correlation. With this in mind, the plot in Figure 5D highlights that the presence of low, i.e., ∼20%–40% of CD13+/ROR1+ cells, on day 3 is a strong hint for low CM content, typically <30% on day 10. In contrast, measurement of ∼60%–80% of CD13+/ROR1+ cells on day 3 typically resulted in 80%–98% CMs. As exemplarily depicted by inset pictures in the schematic (Figure 5A), the average cell aggregate diameter changed along bioreactor-based processes, rising from 112.8 ± 34.5 μm (day 0) to 214.5 ± 66.8 μm (day 10; Figure S4D). Moreover, online monitoring of pH and DO revealed typical “zigzag” curves between individual medium changes (Figure S4C). After day 3, the pH drop pattern was flatter in line with both decreasing cell numbers and the expected switch from a highly glycolytic metabolism established for the pluripotent state (Cliff et al., 2017, Moussaieff et al., 2015) toward more oxidative phosphorylation (and thus reduced lactate production) in differentiating cells. This was confirmed by glucose/lactate assessment (Figure S4E) for specific yield coefficient (Y) calculation, which was highest during the first days of differentiation and dropped strongly between days 2 and 5 (Figure S4F).
We next performed a proof of concept for up to 3-fold process upscaling, i.e., from 150- toward 500-mL scale in a larger stirred tank reactor system (Bioblock) revealing high similarity for key parameters including growth kinetics, cell yields, and CM content, whereas a somewhat smaller mean aggregate diameter was observed at a larger scale (Figure 5F). The green bars in Figure 5C show that 89.13% ± 7.8%, 90.53% ± 6.7%, and 92.03% ± 4.4% of cTnT-, SA-, and MHC-expressing CMs, respectively, were observed on day 10. The cell density was 1.05 ± 0.19 × 106 cells/mL (green line in Figure 5B). Together these data demonstrate platform- and process-scale-independent robustness.
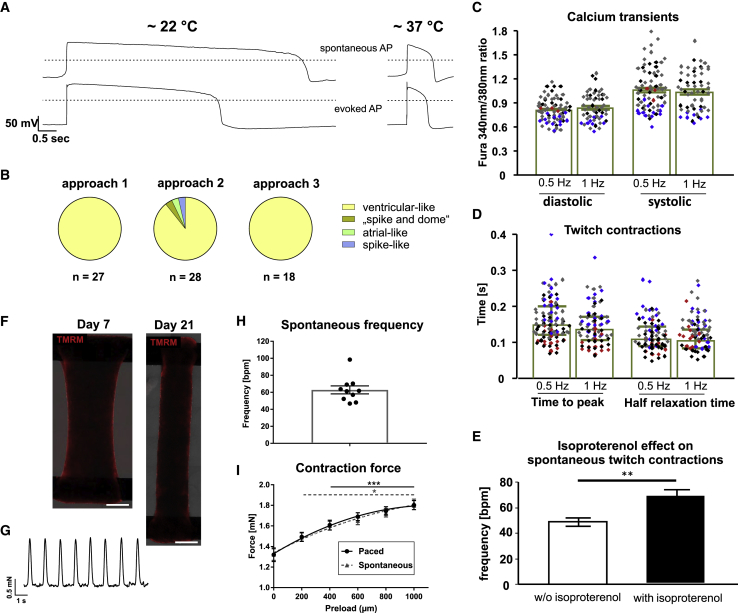
Qualitative Analysis Reveals High Purity of Functional Ventricular Cardiomyocytes
Assessing CMs’ electrophysiological properties revealed 70 out of 73 action potentials (APs) with long plateau phase and steep phase-3 repolarization (physiological APs found at 37°C; Figures 6A, 6B, S5A, and S5B), reflecting a ventricular-like phenotype with only three deviating APs noted in three independent differentiations (Figures 6A, 6B, S5A, and S5C–S5E).
Figure 6.
Characterization of CMs Generated by Chemically Defined Conditions
(A and B) Electrophysiology: (A) representative APs indicating ventricular-like CMs recorded at room temperature (∼22°C) or ∼37°C. Spontaneous APs (upper) compared with evoked APs (lower) of the same cells, elicited by short stimuli from resting potentials of −80 mV; dotted lines: 0 mV. CM phenotype distribution: (B) in three independent differentiations of Phoenix, 3 out of 73 messured cells deviated from the ventricular-like AP pattern.
(C–E) Single CM analysis: (C) systolic/diastolic calcium ratio at 0.5-/1-Hz stimulation; colors ( ,
, ,
, ,
, ) indicate four independent experiments (n = 57–71). (D) Time to peak of twitch contractions and half relaxation time at 0.5/1 Hz; four independent experiments, n = 74–93. Error bars denote median ± IQR. (E) Isoproterenol-induced upregulation of beating frequency (n = 5 in one experiment).
) indicate four independent experiments (n = 57–71). (D) Time to peak of twitch contractions and half relaxation time at 0.5/1 Hz; four independent experiments, n = 74–93. Error bars denote median ± IQR. (E) Isoproterenol-induced upregulation of beating frequency (n = 5 in one experiment).
(F–I) BCT analysis from two independent CM batches. (F) Tetramethylrhodamine methyl ester (TMRM) stain illustrates homogeneous CM distribution on day 7/day 21 of tissue formation. Scale bars, 1 mm. (G–I) Mechanical and electrophysiological properties of BCTs measured on day 21 (n = 10). (G) Example of spontaneous contraction pattern and (H) statistical analysis. (I) Force of spontaneous and paced contractions recorded over increasing preload.
Error bars represent SEM. See also Figures S5 and S6.
For intracellular calcium transients and unloaded twitch contraction recordings (Figures 6C and 6D), plated CMs were electrically stimulated (Mutig et al., 2013). The averaged diastolic calcium ratio at 0.5 Hz (Figure 6C) was 0.8 ± 0.15 and thus higher than 0.6 ± 0.1 previously found in hESC-CMs (Mutig et al., 2013, Weber et al., 2016), indicating a possibly lower activity of sarcoplasmic/endoplasmic calcium reticulum ATPase and Na+/Ca2+ exchanger in hiPSC-CMs in this study. This may be due to the shorter maturation time in culture (i.e., days 14–19 here versus day 35 in Weber et al., 2016). At both electrical stimulation frequencies, systolic calcium ratios (Figure 6C) were higher compared with (Weber et al., 2016) but the amplitude of calcium release (calculated as systolic peak/diastolic ratio) at 0.5 Hz was only slightly elevated (0.26 ± 0.18 versus 0.25 ± 0.1 in Weber et al., 2016). Time to peak and half relaxation times (Figure 6D) of twitch contractions were similar to historical data (Weber et al., 2016).
Notably, only 22% of all measured CMs were spontaneously contracting in the pacing-free interval of the acquisition protocol (Figure S6F). This is ∼16% less than previously noted in hESC-CMs using the same analysis (Weber et al., 2016), indicating that (without applying explicit maturation) we here derived CMs exhibiting a less contractile, more ventricular-like phenotype, a property of working myocardium.
Moreover, prolonged cultivation of CM aggregates in 3D resulted in progressively increasing expressions of ventricular myosin light chain 2 (MLC2, Figures S6I and S6J) as indicated for prolonged cultivation of seeded CMs in 2D (Burridge et al., 2014, Lian et al., 2012).
Upon electrically evoked and spontaneous twitch contractions (Figures 6E and S6C–S6E), isoproterenol slightly increased the contraction amplitude and marked shortening of time to peak and half relaxation times (expected positive inotropic and lusitropic effects, Figures S6D and S6E), and a significant increase of the beating frequency of spontaneously contracting CMs from 48.8 ± 3.26 to 68.4 ± 4.4 beats/min (expected positive chronotropic adrenergic effects; Figures 6E and S6C). Moreover, endothelin-1 treatment for 24 h induced a typical increase in cell surface area of plated CMs (Carlson et al., 2013; Figures S6A and S6B).
Finally, our resulting CMs were used for bioartificial cardiac tissue (BCT) generation (Kempf et al., 2014, Kensah et al., 2013). After 21 days of cultivation, tissues displayed homogeneous distribution of aligned CMs and synchronized spontaneous beating at 62.8 ± 4.7 beats/min and contraction-relaxation times (tC80%) of 146.1 ± 6.0 and 181.8 ± 8.1 ms, respectively (Figures 6F–6I, S6G, and S6H). In response to increased preload, both spontaneous and electrically paced forces increased significantly to 1.82 ± 0.04 and 1.80 ± 0.04 mN, respectively (Figure 6I). We also observed frequency-dependent acceleration of relaxation: compared with the initial relaxation time at 1 Hz, tR80% was reduced by 28% or 51% at 2 Hz or 3 Hz, respectively (Figures S6G and S6H).
Together, this highlights the typical physiological and mechanical properties of our hPSC-CMs indicating pronounced ventricular-like features.
Discussion
Directed cardiac differentiation has been continuously improved over the past decade, progressing toward simpler, more robust, and scalable conditions (Kempf et al., 2014, Lian et al., 2012, Yang et al., 2008). In this study we introduce an integrated, chemically defined differentiation platform enabling the production of human cardiomyocytes in 3D aggregate culture suitable for upscaling in controlled stirred tank bioreactors.
Previous studies demonstrated a substantial impact of preculture conditions (Kempf et al., 2014) at the pluripotent hPSC aggregate stage (Kempf et al., 2014) and the early differentiation phase (Faial et al., 2015, Kempf et al., 2016, Mendjan et al., 2014) on cardiomyogenesis. Culture conditions were revealed that do support an accelerated differentiation progression including efficient PS formation and mesoderm induction, but ultimately failed to direct efficient cardiomyogenesis (Kempf et al., 2014, Kempf et al., 2016, Laco et al., 2018, Mendjan et al., 2014).
We hypothesized here that, despite thoroughly directing hPSC priming toward precardiac mesoderm (i.e., by controlling cell aggregation and preculture timing, cell density adaptation ahead of differentiation, exact CHIR dosing, and incubation timing), the “48-h gap” of external WNT pathway control (which is inherent to the previous differentiation protocol, i.e., between CHIR deprivation after 24 h and before IWP addition at 72 h) is prone to perturbing lineage identity. Such perturbation is likely promoted by the complex paracrine environment and multiple signaling pathway activity in hPSCs downstream of CHIR addition (Gaspari et al., 2018, Kempf et al., 2016) and may lead to both a more broad induction of PS-like cell identities and less controlled progression to (posterior) mesoderm fates, i.e., paraxial mesoderm progenies.
We thus considered recent findings on mesoderm specification (Loh et al., 2016) to more stringently control hPSC priming to a defined transition point of lateral mesoderm specification (by meticulously defined preculture and CHIR treatment). This is followed by efficient transition of the entire progenitor population toward cardiac mesoderm (rather than undesired branching toward, e.g., limb bud mesoderm; Loh et al., 2016) by immediate chemical control of WNT pathway inhibition. The success of this immediate CHIR-to-IWP transition strategy is reflected in early gene expression data revealing suppression of CDX2 and MSX1, established inhibitors of cardiac mesoderm formation (Mendjan et al., 2014, Rao et al., 2016).
Opposing the gene expression patterns specific to cardiac mesoderm (resulting from immediate IWP addition on day 1) versus presomitic mesoderm (achieved by the lack of IWP before day 3) allowed us to evaluate several known cardiac/mesodermal progenitor markers including KDR, PDGFRα, SSEA1, NCAM, ROR2, and CD13 (Ardehali et al., 2013, Evseenko et al., 2010, Kattman et al., 2011, Lee et al., 2017, Nelson et al., 2008, Yang et al., 2008, Yoon et al., 2018) regarding their suitability for early process monitoring and eventually predicting functional CM formation at later stages. Interestingly, none of the established markers was well suited to discriminate early cardiac from presomitic mesoderm. Instead, we identified ROR1 in combination with CD13 (to exclude presence of ROR1-expressing undifferentiated cells; see Figure S2A) as being more informative in distinguishing between cardiac and presomitic mesoderm. This marker combination on day 3 was indeed more prognostic for indicating CM content on day 10, as demonstrated for several hPSC lines and culture platforms (Figures 2F and 5D).
ROR1 and ROR2 are part of a WNT5A-ROR-DVL pathway in limb development, where both receptors are partially redundant (Ho et al., 2012). However, in line with our data, distinct ROR1 expression was reported in the presumptive cardiac PS region in birds and mammals (Alev et al., 2010, Kinder et al., 1999, Peng et al., 2016) and in embryonic heart tissue as well (Kim et al., 2014). This distinct pattern may hint toward a broader functional role of ROR1 during early heart development, potentially involving non-canonical WNT signaling via FZD4 during lateral plate mesoderm formation (Grumolato et al., 2010, Yoon et al., 2018). In summary, we show that ROR1 may be exploited to identify primed hPSC states and predict lineage commitment, complementing recent findings on cKIT, FZD4, and GARP (Bargaje et al., 2017, Loh et al., 2016, Yoon et al., 2018).
A number of recently published studies progress toward the mass production of hPSC-CMs in stirred suspension culture (Chen et al., 2015, Fonoudi et al., 2015). In notable accordance, we confirm the role of preculture duration; 2 days of aggregation in E8 was optimally equivalent in our setup to Chen et al., whereas Fonoudi et al. favored prolonged cultivation for 5 days (Chen et al., 2015, Fonoudi et al., 2015). However, both studies suggested that the time-dependent increase in aggregate diameter, rather than preculture duration itself, is a predominant factor. Our previous studies also indicate that the hPSC aggregate diameter and, in particular, the process-dependent pattern of aggregate formation and growth, is important (Kempf et al., 2014, Kempf et al., 2015). However, whereas Chen et al. and Fonoudi et al. in unison support a median hPSC aggregate diameter of ∼175–220 μm as being optimal, we here found a median diameter of ∼125 μm (preculture day 2) being advantageous for cardiomyogenesis as compared with ∼200 μm (on day 4 or preculture; Figures 3B–3F). This comparison of different studies highlights that probably none of these process parameters (such as preculture duration, aggregate diameter, and inoculation cell density) represents an absolute requirement or a universal process-independent parameter for controlling directed lineage differentiation. These are rather correlative observations, which have to be interpreted within the context of the respective process. Interestingly, the group of Oh recently investigated the proliferative status of hPSCs regarding their impact on the cardiac differentiation efficiency (Laco et al., 2018). Results suggest that monitoring defined molecular markers of cell proliferation may indeed be a more universal, culture platform-independent approach to preassess hPSCs' compatibility with cardiac differentiation, although additional validation is required.
Regarding process yields, Chen et al. (2015) published ∼0.2 million CMs/mL whereas Fonoudi et al. (2015) reported ∼0.8 million CMs/mL in their approach (Chen et al., 2015, Fonoudi et al., 2015). We here show the induction of ∼1 million CMs/mL in process dimensions from 150 to 500 mL across two bioreactor platforms at increasing dimension, ultimately resulting in the production of up to ∼450 × 106 CMs in a single process run. Using a non-directed differentiation approach followed by antibiotic selection, Correia et al. (2014) reported the production of 2.3 × 106 CMs/mL in WAVE bioreactors compared with 0.64 × 106 in stirred tank bioreactors (Correia et al., 2014). In another concept applying microcarriers, Ting et al. (2014) generated up to 1.6 × 106 CMs/mL. A meaningful cross-comparison of these studies is problematic, given the great heterogeneity of bioreactor platforms and the overall experimental environment. Importantly, our protocol described here is one of the first demonstrating chemically defined conditions in scalable suspension culture inducing a high CM content of >90%.
Notably, functional characterization by numerous complementary methods revealed a predominantly ventricular-like CMs phenotype, which may support in vitro disease modeling and pharmacological assays as well as envisioned regenerative therapies of heart repair (Liu et al., 2018).
Experimental Procedures
For standard methods and prepublished techniques, please refer to Supplemental Experimental Procedures.
Cell Culture
The cell lines HES3 (HES3-MIXL1wt/eGFP; Davis et al., 2008, HES3-NKX2.5wt/eGFP; Elliott et al., 2011), H9, HSC_iPS2 (hHSC_F1285_T-iPS2; MHHi006-A; Hartung et al., 2013), Phoenix (HSC_ADCF_SeV-iPS2; MHHi001-A; Haase et al., 2017, Kempf et al., 2016), CBiPS2 (hCBiPS2; Haase et al., 2009), bMHC_iPS (hF_HCMR723G_4F_iPS_Klon2_MOI1), SFC840 (SFC840-03-01, STBCi026-B; Christoffersson et al., 2018; derived by the StemBANCC initiative http://stembancc.org/), HDFiPS (HDF-iPSC; Wattanapanitch et al., 2014), and CDX2_iPS (CDX2Venus knockin reporter, MHHi007-A-1; Malysheva et al., 2018) were maintained in mTeSR (STEMCELL Technologies) or E8 medium as described previously (Kempf et al., 2015). In brief, cells were passaged every 3.5 days using Accutase (Life Technologies) and reseeded at 0.4–0.5 × 104 cells/mL on Geltrex-coated (Life Technologies) flasks in mTeSR1 (STEMCELL Technologies) or E8 medium including 10 μM Y-27632 (LU Hannover; Palecek et al., 2011). Cells were maintained at 37°C and 5% CO2 and were regularly tested for mycoplasma contamination using the MycoAlert Mycoplasma Detection Kit (Lonza).
Differentiation was conducted as illustrated in Figure 1A and as previously described (Kempf et al., 2014). At chemically defined conditions, cells were cultivated in E8 and differentiated in CDM3 (Burridge et al., 2014; for details see Supplemental Experimental Procedures).
Bioreactor Differentiation in Chemically Defined Conditions
Chemically defined differentiation in Erlenmeyer flasks (20 mL working volume) and in stirred tank bioreactors (working volume: 150 mL/DASbox Mini Bioreactor System or 350–500 mL/Bioblock DASGIP Parallel Bioreactor System) was performed as depicted in Figures 3A and 5A, respectively (Eppendorf). After established inoculation/preculture (Kropp et al., 2016) cell density was determined after 48 h and adjusted to 5 × 105 cells/mL for differentiation in CDM3 (Burridge et al., 2014) supplemented with 5 μM CHIR and 5 μM Y-27632. Precisely 24 h later the medium was replaced by CDM3 plus 5 μM IWP2, and pure CDM3 was added 48 h later and every 2–3 days thereafter. Aggregate diameter, cell density, and metabolites were analyzed as previously published (Kropp et al., 2016).
Statistics
Data are presented as mean ± standard error of the mean. Unless otherwise noted, statistical significance was calculated using Student's t tests. To take account of multiplicity in group comparisons, one- or two-way ANOVA followed by Bonferroni's post test were conducted. Statistical significance is denoted by ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ns (not significant) in the figures.
Author Contributions
C.H., H.K., K.S., and R.Z. designed the experiments. C.H., A.F., W.L., M.S., N.W., F.O., J.d.l.R., F.P., S.B., S.W., S.M., K.S., and H.K. performed the experiments. C.H., K.S., M.S., N.W., J.d.l.R., I.G., M.F., H.K., and R.Z. analyzed and interpreted the data. T.K., I.G., M.F., K.W., U.M., and R.Z. provided conceptual advice and financial support. C.H., H.K., N.W., M.F., I.G., and R.Z. wrote the article.
Acknowledgments
We would like to thank A. Kirschning and G. Dräger for providing Y-27632 and CHIR99021, A. Haase for providing various iPSC lines, S. Malysheva for providing the cell line CDX2_iPS, E. Stanley and A. Elefanty for providing the HES3-NKX2.5 and HES3-MIXL1 cell lines, A. Francino and F. Navarro-Lopez (Hospital Clinic, University of Barcelona, Spain) for providing skin fibroblasts from a patient with hypertrophic cardiomyopathy to generate the cell line bMHC_iPS, and T. Holler and A. Lingk for supporting and performing immunofluorescence. We also thank the RCU Transcriptomics at Hannover Medical School for performing the microarray experiments. R.Z. received funding from the German Research Foundation (DFG; Cluster of Excellence REBIRTH EXC 62/2, ZW64/4-1 as well as KFO311 and ZW64/7-1), German Ministry for Education and Science (BMBF, grants: 13N14086, 01EK1601A, 01EK1602A), StemBANCC (support from the Innovative Medicines Initiative joint undertaking under grant agreement no. 115439-2), resources of which are composed of financial contributions from the European Union's Seventh Framework Programme (FP7/2007-2013) and EFPIA companies' kind contribution, and the European Union H2020 program to the project TECHNOBEAT (grant 66724). H.K. received funding from Joachim Herz Stiftung and MHH Hannover (HiLF grant). T.K. received funding from the German Research Foundation (DFG TK1187/21-1).
Published: July 25, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.06.004.
Contributor Information
Henning Kempf, Email: henningkempf@outlook.com.
Robert Zweigerdt, Email: zweigerdt.robert@mh-hannover.de.
Accession Numbers
The microarray data were deposited in the ArrayExpress database under accession number ArrayExpress: E-MTAB-6828.
Supplemental Information
References
- Abecasis B., Aguiar T., Arnault E., Costa R., Gomes-Alves P., Aspegren A., Serra M., Alves P.M. Expansion of 3D human induced pluripotent stem cell aggregates in bioreactors: bioprocess intensification and scaling-up approaches. J. Biotechnol. 2017;246:81–93. doi: 10.1016/j.jbiotec.2017.01.004. [DOI] [PubMed] [Google Scholar]
- Ackermann M., Kempf H., Hetzel M., Hesse C., Hashtchin A.R., Brinkert K., Schott J.W., Haake K., Kuhnel M.P., Glage S. Bioreactor-based mass production of human iPSC-derived macrophages enables immunotherapies against bacterial airway infections. Nat. Commun. 2018;9:5088. doi: 10.1038/s41467-018-07570-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alev C., Wu Y., Kasukawa T., Jakt L.M., Ueda H.R., Sheng G. Transcriptomic landscape of the primitive streak. Development. 2010;137:2863–2874. doi: 10.1242/dev.053462. [DOI] [PubMed] [Google Scholar]
- Ardehali R., Ali S.R., Inlay M.A., Abilez O.J., Chen M.Q., Blauwkamp T.A., Yazawa M., Gong Y., Nusse R., Drukker M. Prospective isolation of human embryonic stem cell-derived cardiovascular progenitors that integrate into human fetal heart tissue. Proc. Natl. Acad. Sci. U S A. 2013;110:3405–3410. doi: 10.1073/pnas.1220832110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargaje R., Trachana K., Shelton M.N., McGinnis C.S., Zhou J.X., Chadick C., Cook S., Cavanaugh C., Huang S., Hood L. Cell population structure prior to bifurcation predicts efficiency of directed differentiation in human induced pluripotent cells. Proc. Natl. Acad. Sci. U S A. 2017;114:2271–2276. doi: 10.1073/pnas.1621412114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biben C., Hatzistavrou T., Harvey R.P. Expression of NK-2 class homeobox gene Nkx2-6 in foregut endoderm and heart. Mech. Dev. 1998;73:125–127. doi: 10.1016/s0925-4773(98)00037-9. [DOI] [PubMed] [Google Scholar]
- Burridge P.W., Matsa E., Shukla P., Lin Z.C., Churko J.M., Ebert A.D., Lan F., Diecke S., Huber B., Mordwinkin N.M. Chemically defined generation of human cardiomyocytes. Nat. Methods. 2014;11:855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson C., Koonce C., Aoyama N., Einhorn S., Fiene S., Thompson A., Swanson B., Anson B., Kattman S. Phenotypic screening with human iPS cell-derived cardiomyocytes: HTS-compatible assays for interrogating cardiac hypertrophy. J. Biomol. Screen. 2013;18:1203–1211. doi: 10.1177/1087057113500812. [DOI] [PubMed] [Google Scholar]
- Chen V.C., Couture S.M., Ye J., Lin Z., Hua G., Huang H.I., Wu J., Hsu D., Carpenter M.K., Couture L.A. Scalable GMP compliant suspension culture system for human ES cells. Stem Cell Res. 2012;8:388–402. doi: 10.1016/j.scr.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Chen V.C., Ye J., Shukla P., Hua G., Chen D., Lin Z., Liu J.C., Chai J., Gold J., Wu J. Development of a scalable suspension culture for cardiac differentiation from human pluripotent stem cells. Stem Cell Res. 2015;15:365–375. doi: 10.1016/j.scr.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffersson J., Meier F., Kempf H., Schwanke K., Coffee M., Beilmann M., Zweigerdt R., Mandenius C.F. A cardiac cell outgrowth assay for evaluating drug compounds using a cardiac spheroid-on-a-chip device. Bioengineering (Basel) 2018;5 doi: 10.3390/bioengineering5020036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliff T.S., Wu T., Boward B.R., Yin A., Yin H., Glushka J.N., Prestegaard J.H., Dalton S. MYC controls human pluripotent stem cell fate decisions through regulation of metabolic flux. Cell Stem Cell. 2017;21:502–516.e9. doi: 10.1016/j.stem.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia C., Serra M., Espinha N., Sousa M., Brito C., Burkert K., Zheng Y., Hescheler J., Carrondo M.J., Saric T. Combining hypoxia and bioreactor hydrodynamics boosts induced pluripotent stem cell differentiation towards cardiomyocytes. Stem Cell Rev. 2014;10:786–801. doi: 10.1007/s12015-014-9533-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R.P., Ng E.S., Costa M., Mossman A.K., Sourris K., Elefanty A.G., Stanley E.G. Targeting a GFP reporter gene to the MIXL1 locus of human embryonic stem cells identifies human primitive streak-like cells and enables isolation of primitive hematopoietic precursors. Blood. 2008;111:1876–1884. doi: 10.1182/blood-2007-06-093609. [DOI] [PubMed] [Google Scholar]
- Devalla H.D., Schwach V., Ford J.W., Milnes J.T., El-Haou S., Jackson C., Gkatzis K., Elliott D.A., Chuva de Sousa Lopes S.M., Mummery C.L. Atrial-like cardiomyocytes from human pluripotent stem cells are a robust preclinical model for assessing atrial-selective pharmacology. EMBO Mol. Med. 2015;7:394–410. doi: 10.15252/emmm.201404757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott D.A., Braam S.R., Koutsis K., Ng E.S., Jenny R., Lagerqvist E.L., Biben C., Hatzistavrou T., Hirst C.E., Yu Q.C. NKX2-5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat. Methods. 2011;8:1037–1040. doi: 10.1038/nmeth.1740. [DOI] [PubMed] [Google Scholar]
- Evseenko D., Zhu Y., Schenke-Layland K., Kuo J., Latour B., Ge S., Scholes J., Dravid G., Li X., MacLellan W.R. Mapping the first stages of mesoderm commitment during differentiation of human embryonic stem cells. Proc. Natl. Acad. Sci. U S A. 2010;107:13742–13747. doi: 10.1073/pnas.1002077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faial T., Bernardo A.S., Mendjan S., Diamanti E., Ortmann D., Gentsch G.E., Mascetti V.L., Trotter M.W., Smith J.C., Pedersen R.A. Brachyury and SMAD signalling collaboratively orchestrate distinct mesoderm and endoderm gene regulatory networks in differentiating human embryonic stem cells. Development. 2015;142:2121–2135. doi: 10.1242/dev.117838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonoudi H., Ansari H., Abbasalizadeh S., Larijani M.R., Kiani S., Hashemizadeh S., Zarchi A.S., Bosman A., Blue G.M., Pahlavan S. A universal and robust integrated platform for the scalable production of human cardiomyocytes from pluripotent stem cells. Stem Cells Transl. Med. 2015;4:1482–1494. doi: 10.5966/sctm.2014-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspari E., Franke A., Robles-Diaz D., Zweigerdt R., Roeder I., Zerjatke T., Kempf H. Paracrine mechanisms in early differentiation of human pluripotent stem cells: insights from a mathematical model. Stem Cell Res. 2018;32:1–7. doi: 10.1016/j.scr.2018.07.025. [DOI] [PubMed] [Google Scholar]
- Gonzalez R., Lee J.W., Schultz P.G. Stepwise chemically induced cardiomyocyte specification of human embryonic stem cells. Angew. Chem. 2011;50:11181–11185. doi: 10.1002/anie.201103909. [DOI] [PubMed] [Google Scholar]
- Greber B., Wu G., Bernemann C., Joo J.Y., Han D.W., Ko K., Tapia N., Sabour D., Sterneckert J., Tesar P. Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell. 2010;6:215–226. doi: 10.1016/j.stem.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Grumolato L., Liu G., Mong P., Mudbhary R., Biswas R., Arroyave R., Vijayakumar S., Economides A.N., Aaronson S.A. Canonical and noncanonical Wnts use a common mechanism to activate completely unrelated coreceptors. Genes Dev. 2010;24:2517–2530. doi: 10.1101/gad.1957710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase A., Gohring G., Martin U. Generation of non-transgenic iPS cells from human cord blood CD34(+) cells under animal component-free conditions. Stem Cell Res. 2017;21:71–73. doi: 10.1016/j.scr.2017.03.022. [DOI] [PubMed] [Google Scholar]
- Haase A., Olmer R., Schwanke K., Wunderlich S., Merkert S., Hess C., Zweigerdt R., Gruh I., Meyer J., Wagner S. Generation of induced pluripotent stem cells from human cord blood. Cell Stem Cell. 2009;5:434–441. doi: 10.1016/j.stem.2009.08.021. [DOI] [PubMed] [Google Scholar]
- Hartung S., Schwanke K., Haase A., David R., Franz W.M., Martin U., Zweigerdt R. Directing cardiomyogenic differentiation of human pluripotent stem cells by plasmid-based transient overexpression of cardiac transcription factors. Stem Cell Dev. 2013;22:1112–1125. doi: 10.1089/scd.2012.0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho H.Y., Susman M.W., Bikoff J.B., Ryu Y.K., Jonas A.M., Hu L., Kuruvilla R., Greenberg M.E. Wnt5a-Ror-Dishevelled signaling constitutes a core developmental pathway that controls tissue morphogenesis. Proc. Natl. Acad. Sci. U S A. 2012;109:4044–4051. doi: 10.1073/pnas.1200421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman S.J., Witty A.D., Gagliardi M., Dubois N.C., Niapour M., Hotta A., Ellis J., Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Kempf H., Kropp C., Olmer R., Martin U., Zweigerdt R. Cardiac differentiation of human pluripotent stem cells in scalable suspension culture. Nat. Protoc. 2015;10:1345–1361. doi: 10.1038/nprot.2015.089. [DOI] [PubMed] [Google Scholar]
- Kempf H., Olmer R., Haase A., Franke A., Bolesani E., Schwanke K., Robles-Diaz D., Coffee M., Gohring G., Drager G. Bulk cell density and Wnt/TGFbeta signalling regulate mesendodermal patterning of human pluripotent stem cells. Nat. Commun. 2016;7:13602. doi: 10.1038/ncomms13602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf H., Olmer R., Kropp C., Ruckert M., Jara-Avaca M., Robles-Diaz D., Franke A., Elliott D.A., Wojciechowski D., Fischer M. Controlling expansion and cardiomyogenic differentiation of human pluripotent stem cells in scalable suspension culture. Stem Cell Rep. 2014;3:1132–1146. doi: 10.1016/j.stemcr.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensah G., Roa Lara A., Dahlmann J., Zweigerdt R., Schwanke K., Hegermann J., Skvorc D., Gawol A., Azizian A., Wagner S. Murine and human pluripotent stem cell-derived cardiac bodies form contractile myocardial tissue in vitro. Eur. Heart J. 2013;34:1134–1146. doi: 10.1093/eurheartj/ehs349. [DOI] [PubMed] [Google Scholar]
- Kim M.S., Pinto S.M., Getnet D., Nirujogi R.S., Manda S.S., Chaerkady R., Madugundu A.K., Kelkar D.S., Isserlin R., Jain S. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinder S.J., Tsang T.E., Quinlan G.A., Hadjantonakis A.K., Nagy A., Tam P.P. The orderly allocation of mesodermal cells to the extraembryonic structures and the anteroposterior axis during gastrulation of the mouse embryo. Development. 1999;126:4691–4701. doi: 10.1242/dev.126.21.4691. [DOI] [PubMed] [Google Scholar]
- Kropp C., Kempf H., Halloin C., Robles-Diaz D., Franke A., Scheper T., Kinast K., Knorpp T., Joos T.O., Haverich A. Impact of feeding strategies on the scalable expansion of human pluripotent stem cells in single-use stirred tank bioreactors. Stem Cells Transl. Med. 2016;5:1289–1301. doi: 10.5966/sctm.2015-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laco F., Woo T.L., Zhong Q., Szmyd R., Ting S., Khan F.J., Chai C.L.L., Reuveny S., Chen A., Oh S. Unraveling the inconsistencies of cardiac differentiation efficiency induced by the GSK3beta inhibitor CHIR99021 in human pluripotent stem cells. Stem Cell Rep. 2018;10:1851–1866. doi: 10.1016/j.stemcr.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Protze S.I., Laksman Z., Backx P.H., Keller G.M. Human pluripotent stem cell-derived atrial and ventricular cardiomyocytes develop from distinct mesoderm populations. Cell Stem Cell. 2017;21:179–194.e4. doi: 10.1016/j.stem.2017.07.003. [DOI] [PubMed] [Google Scholar]
- Lian X., Hsiao C., Wilson G., Zhu K., Hazeltine L.B., Azarin S.M., Raval K.K., Zhang J., Kamp T.J., Palecek S.P. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. U S A. 2012;109:E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.W., Chen B., Yang X., Fugate J.A., Kalucki F.A., Futakuchi-Tsuchida A., Couture L., Vogel K.W., Astley C.A., Baldessari A. Human embryonic stem cell-derived cardiomyocytes restore function in infarcted hearts of non-human primates. Nat. Biotechnol. 2018;36:597–605. doi: 10.1038/nbt.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh K.M., Chen A., Koh P.W., Deng T.Z., Sinha R., Tsai J.M., Barkal A.A., Shen K.Y., Jain R., Morganti R.M. Mapping the pairwise choices leading from pluripotency to human bone, heart, and other mesoderm cell types. Cell. 2016;166:451–467. doi: 10.1016/j.cell.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig T.E., Bergendahl V., Levenstein M.E., Yu J., Probasco M.D., Thomson J.A. Feeder-independent culture of human embryonic stem cells. Nat. Methods. 2006;3:637–646. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- Malysheva S.V., Wunderlich S., Haase A., Gohring G., Martin U., Merkert S. Generation of a human CDX2 knock-in reporter iPSC line (MHHi007-A-1) to model human trophoblast differentiation. Stem Cell Res. 2018;30:117–121. doi: 10.1016/j.scr.2018.05.015. [DOI] [PubMed] [Google Scholar]
- Matsuda T., Nomi M., Ikeya M., Kani S., Oishi I., Terashima T., Takada S., Minami Y. Expression of the receptor tyrosine kinase genes, Ror1 and Ror2, during mouse development. Mech. Dev. 2001;105:153–156. doi: 10.1016/s0925-4773(01)00383-5. [DOI] [PubMed] [Google Scholar]
- Mendjan S., Mascetti V.L., Ortmann D., Ortiz M., Karjosukarso D.W., Ng Y., Moreau T., Pedersen R.A. NANOG and CDX2 pattern distinct subtypes of human mesoderm during exit from pluripotency. Cell Stem Cell. 2014;15:310–325. doi: 10.1016/j.stem.2014.06.006. [DOI] [PubMed] [Google Scholar]
- Moussaieff A., Rouleau M., Kitsberg D., Cohen M., Levy G., Barasch D., Nemirovski A., Shen-Orr S., Laevsky I., Amit M. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab. 2015;21:392–402. doi: 10.1016/j.cmet.2015.02.002. [DOI] [PubMed] [Google Scholar]
- Mutig N., Geers-Knoerr C., Piep B., Pahuja A., Vogt P.M., Brenner B., Niederbichler A.D., Kraft T. Lipoteichoic acid from Staphylococcus aureus directly affects cardiomyocyte contractility and calcium transients. Mol. Immunol. 2013;56:720–728. doi: 10.1016/j.molimm.2013.07.007. [DOI] [PubMed] [Google Scholar]
- Nelson T.J., Faustino R.S., Chiriac A., Crespo-Diaz R., Behfar A., Terzic A. CXCR4+/FLK-1+ biomarkers select a cardiopoietic lineage from embryonic stem cells. Stem Cells. 2008;26:1464–1473. doi: 10.1634/stemcells.2007-0808. [DOI] [PubMed] [Google Scholar]
- Olmer R., Engels L., Usman A., Menke S., Malik M.N.H., Pessler F., Gohring G., Bornhorst D., Bolten S., Abdelilah-Seyfried S. Differentiation of human pluripotent stem cells into functional endothelial cells in scalable suspension culture. Stem Cell Rep. 2018;10:1657–1672. doi: 10.1016/j.stemcr.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmer R., Lange A., Selzer S., Kasper C., Haverich A., Martin U., Zweigerdt R. Suspension culture of human pluripotent stem cells in controlled, stirred bioreactors. Tissue engineering part C. Methods. 2012;18:772–784. doi: 10.1089/ten.tec.2011.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palecek J., Zweigerdt R., Olmer R., Martin U., Kirschning A., Drager G. A practical synthesis of Rho-Kinase inhibitor Y-27632 and fluoro derivatives and their evaluation in human pluripotent stem cells. Org. Biomol. Chem. 2011;9:5503–5510. doi: 10.1039/c1ob05332a. [DOI] [PubMed] [Google Scholar]
- Peng G., Suo S., Chen J., Chen W., Liu C., Yu F., Wang R., Chen S., Sun N., Cui G. Spatial transcriptome for the molecular annotation of lineage fates and cell identity in mid-gastrula mouse embryo. Dev. Cell. 2016;36:681–697. doi: 10.1016/j.devcel.2016.02.020. [DOI] [PubMed] [Google Scholar]
- Protze S.I., Liu J., Nussinovitch U., Ohana L., Backx P.H., Gepstein L., Keller G.M. Sinoatrial node cardiomyocytes derived from human pluripotent cells function as a biological pacemaker. Nat. Biotechnol. 2017;35:56–68. doi: 10.1038/nbt.3745. [DOI] [PubMed] [Google Scholar]
- Rao J., Pfeiffer M.J., Frank S., Adachi K., Piccini I., Quaranta R., Arauzo-Bravo M., Schwarz J., Schade D., Leidel S. Stepwise clearance of repressive roadblocks drives cardiac induction in human ESCs. Cell Stem Cell. 2016;18:341–353. doi: 10.1016/j.stem.2015.11.019. [DOI] [PubMed] [Google Scholar]
- Shelton M., Metz J., Liu J., Carpenedo R.L., Demers S.P., Stanford W.L., Skerjanc I.S. Derivation and expansion of PAX7-positive muscle progenitors from human and mouse embryonic stem cells. Stem Cell Rep. 2014;3:516–529. doi: 10.1016/j.stemcr.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller R., Greenhough S., Naumovska E., Sullivan G.J. Small-molecule-driven hepatocyte differentiation of human pluripotent stem cells. Stem Cell Rep. 2015;4:939–952. doi: 10.1016/j.stemcr.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M.M., Rodrigues A.F., Correia C., Sousa M.F., Brito C., Coroadinha A.S., Serra M., Alves P.M. Robust expansion of human pluripotent stem cells: integration of bioprocess design with transcriptomic and metabolomic characterization. Stem Cells Transl. Med. 2015;4:731–742. doi: 10.5966/sctm.2014-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelton R.J.P., Kamp T.J., Elliott D.A., Ardehali R. Biomarkers of human pluripotent stem cell-derived cardiac lineages. Trends Mol. Med. 2017;23:651–668. doi: 10.1016/j.molmed.2017.05.001. [DOI] [PubMed] [Google Scholar]
- Soh B.S., Ng S.Y., Wu H., Buac K., Park J.H., Lian X., Xu J., Foo K.S., Felldin U., He X. Endothelin-1 supports clonal derivation and expansion of cardiovascular progenitors derived from human embryonic stem cells. Nat. Commun. 2016;7:10774. doi: 10.1038/ncomms10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting S., Chen A., Reuveny S., Oh S. An intermittent rocking platform for integrated expansion and differentiation of human pluripotent stem cells to cardiomyocytes in suspended microcarrier cultures. Stem Cell Res. 2014;13:202–213. doi: 10.1016/j.scr.2014.06.002. [DOI] [PubMed] [Google Scholar]
- Tran T.H., Wang X., Browne C., Zhang Y., Schinke M., Izumo S., Burcin M. Wnt3a-induced mesoderm formation and cardiomyogenesis in human embryonic stem cells. Stem Cells. 2009;27:1869–1878. doi: 10.1002/stem.95. [DOI] [PubMed] [Google Scholar]
- Ueno S., Weidinger G., Osugi T., Kohn A.D., Golob J.L., Pabon L., Reinecke H., Moon R.T., Murry C.E. Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc. Natl. Acad. Sci. U S A. 2007;104:9685–9690. doi: 10.1073/pnas.0702859104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattanapanitch M., Klincumhom N., Potirat P., Amornpisutt R., Lorthongpanich C., U-pratya Y., Laowtammathron C., Kheolamai P., Poungvarin N., Issaragrisil S. Dual small-molecule targeting of SMAD signaling stimulates human induced pluripotent stem cells toward neural lineages. PLoS One. 2014;9:e106952. doi: 10.1371/journal.pone.0106952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber N., Schwanke K., Greten S., Wendland M., Iorga B., Fischer M., Geers-Knorr C., Hegermann J., Wrede C., Fiedler J. Stiff matrix induces switch to pure beta-cardiac myosin heavy chain expression in human ESC-derived cardiomyocytes. Basic Res. Cardiol. 2016;111:68. doi: 10.1007/s00395-016-0587-9. [DOI] [PubMed] [Google Scholar]
- Yang L., Soonpaa M.H., Adler E.D., Roepke T.K., Kattman S.J., Kennedy M., Henckaerts E., Bonham K., Abbott G.W., Linden R.M. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- Yoon C., Song H., Yin T., Bausch-Fluck D., Frei A.P., Kattman S., Dubois N., Witty A.D., Hewel J.A., Guo H. FZD4 marks lateral plate mesoderm and signals with NORRIN to increase cardiomyocyte induction from pluripotent stem cell-derived cardiac progenitors. Stem Cell Rep. 2018;10:87–100. doi: 10.1016/j.stemcr.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.