ABSTRACT
In addition to core Cas proteins, CRISPR-Cas loci often encode ancillary proteins that modulate the activity of the respective effectors in interference. Subtype VI-B1 CRISPR-Cas systems encode the Csx27 protein that down-regulates the activity of Cas13b when the type VI-B locus is expressed in Escherichia coli. We show that Csx27 belongs to an expansive family of proteins that contain four predicted transmembrane helices and are typically encoded in predicted operons with components of the bacterial natural transformation machinery, multidomain proteins that consist of components of the ubiquitin signaling system and proteins containing the ligand-binding WYL domain and a helix-turn-helix domain. The Csx27 family proteins are predicted to form membrane channels for ssDNA that might comprise the core of a putative novel, Ub-regulated system for DNA uptake and, possibly, degradation. In addition to these associations, a distinct subfamily of the Csx27 family appears to be a part of a novel, membrane-associated system for DNA modification. In Bacteroidetes, subtype VI-B1 systems might degrade nascent transcripts of foreign DNA in conjunction with its uptake by the bacterial cell. These predictions suggest several experimental directions for the study of type VI CRISPR-Cas systems and distinct mechanisms of foreign DNA uptake and degradation in bacteria.
Keywords: type VI CRISPR-Cas, Csx27, CRISPR ancillary proteins, membrane proteins, ubiquitin network, DNA modification, natural competence
Comparative genomic analysis predicts unexpected functional connections between CRISPR-Cas systems and DNA uptake and modification systems in bacteria.
INTRODUCTION
CRISPR-Cas are archaeal and bacterial adaptive immunity systems that show considerable diversity in their mechanisms of action, Cas protein repertoires and genomic loci architectures (Makarova et al. 2015; Mohanraju et al. 2016; Barrangou and Horvath 2017). At the top level, CRISPR-Cas systems split into Class 1, in which the effector module consists of multiple Cas proteins, and Class 2, in which the essential activities required for interference and, in many cases, also the CRISPR (cr) RNA maturation, are concentrated within a single, multidomain protein (Makarova et al. 2015; Koonin, Makarova and Zhang 2017). Class 2 CRISPR-Cas systems include the most common type II, with the Cas9 effector containing the RuvC and HNH nuclease domains, the rarer but highly diverse type V, with the Cas12 effectors containing only the RuvC nuclease domain and type VI in which the effector, Cas13, contains two higher eukaryotes and prokaryote nucleotide-binding (HEPN) RNase domains (Shmakov et al. 2017).
Type VI is the only known variety of CRISPR-Cas systems that exclusively target RNA (Abudayyeh et al. 2016; Smargon et al. 2017; Yan et al. 2018). Four type VI subtypes (VI-A,B,C and D) have been identified all of which are most common in the bacterial phylum Bacteroidetes. Their actual biological functions have not been characterized in any detail but the current hypothesis is that the cleavage of virus or plasmid transcripts by Cas13 induces its collateral RNase activity which induces cell dormancy or death (Abudayyeh et al. 2016; Koonin and Zhang 2017). The type VI loci all include the signature Cas13 effectors, large proteins that contain two RNase domains of the HEPN superfamily. The amino acid sequences of Cas13 proteins from different subtypes show only limited similarity to each other that is mostly confined to the HEPN domains. Nevertheless, the presence of two HEPN domains is a signature of type VI, so far not encountered in other proteins, and suggestive of a common origin of all type VI subtypes, conceivably, from a HEPN-containing toxin or a Csx1-like accessory protein of type III CRISPR-Cas (Shmakov et al. 2017). Many type VI systems also contain the adaptation module (cas1 and cas2 genes). Additionally, subtypes VI-B and VI-D loci encode accessory proteins that modulate the activity of the respective Cas13 effectors (Smargon et al. 2017; Yan et al. 2018).
In particular, Csx27 is an ancillary component of the subtype VI-B1 systems that has been shown to down-regulate the interference capacity of Cas13b1 when co-expressed in Escherichia coli (Smargon et al. 2017). Csx27 is confidently predicted to contain four transmembrane (TM) helices (Fig. 1) although, when expressed in E. coli, fluorescent protein-tagged Csx27 did not exhibit membrane localization (Smargon et al. 2017). Thus, the specific activity of Csx27 and the role of the predicted TM helices remain unknown. Recently, it has been shown that diverse (predicted) membrane proteins are stably associated with type III systems suggesting unexplored role of membranes in CRISPR activity as well as possible links between CRISPR-Cas and various signal transduction pathways (Shah et al. 2018; Shmakov et al. 2018). More generally, membrane proteins containing either two or four TM helices prominently figure in nucleotide-based signaling systems (Burroughs et al. 2015).
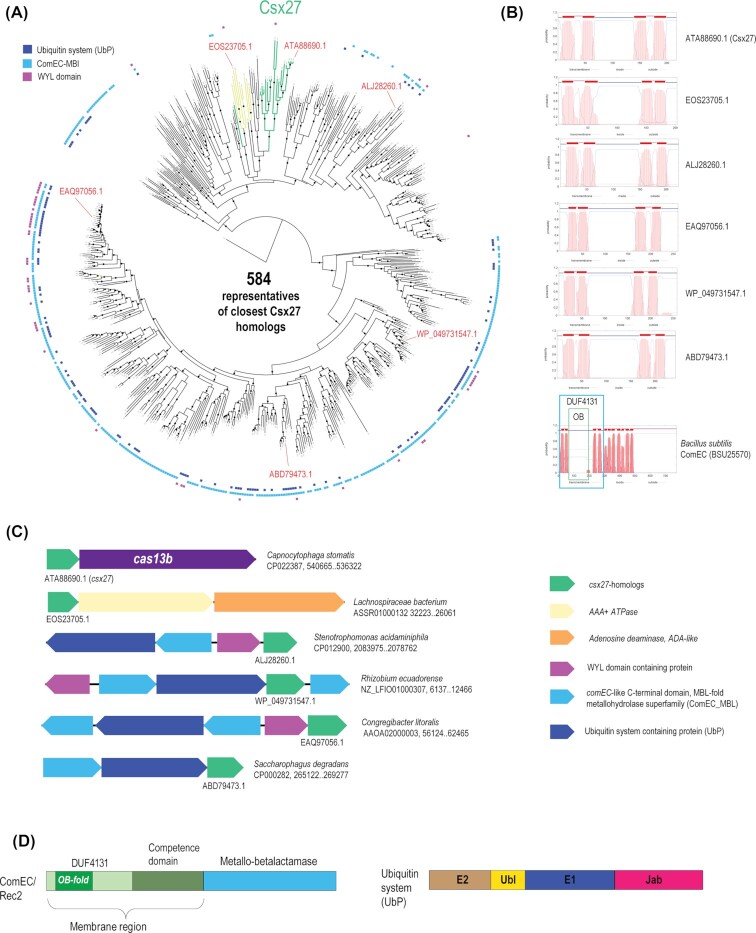
Figure 1.
The Csx27 protein family and associated genes. (A). The FastTree phylogenetic tree of Csx27 homologs overlaid with the pattern of occurrence of the three genes (ComEC-MBL domain, UbP and WYL-HTH) most frequently identified in the loci that encode Csx27 family proteins. The green branch includes the bona fide Csx27 from the subtype VI-b1 CRISPR-Cas systems, and the yellow branch corresponds to the proteins associated with AAA ATPase and adenosine deaminase. Red protein accessions specify proteins that were used to illustrate membrane topology prediction (B) and conserved neighborhood architectures (C). (B). TMHMM prediction for selected proteins. Plot show probability and location of TM helices along the protein. (C). Typical conserved neighborhoods. Genes are shown by directed arrows. Homologous genes are color coded according to the legend shown on the right. (D). Domain organization of ComEC and ubiquitin domain containing protein.
As part of our concerted efforts on comprehensive investigation of the links between CRISPR-Cas and various membrane-associated functions, in particular, signal transduction, we applied sensitive sequence analysis methods to identify membrane proteins homologous to Csx27 and explored their genome contexts, guided by the guilt-by-association principle. This analysis resulted in the identification of a network of predicted functional connections that, unexpectedly, link subtype VI-B CRISPR-Cas systems to bacterial natural competence and ubiquitin (Ub) signaling.
METHODS
Iterative profile searches using PSI-BLAST (Altschul et al. 1997), with a cut-off E-value of 0.0001, and composition-based statistics and low complexity filtering turned off, were employed to search for similar protein sequences in the non-redundant (NR) database. For loci annotation, PSI-BLAST against Conserved Domain Database (CDD) was used (Marchler-Bauer et al. 2015), with the cut-off E-value of 0.01 and low complexity filtering turned off. HHpred search with default parameters against Protein Data Bank (PDB) and CDD profile databases was used to search for remotely similar sequences (Soding 2005).
TM helices and topology of membrane proteins were predicted using TMHMM v. 2.0c with default parameters (Krogh et al. 2001). Protein secondary structure was predicted using Jpred 4 (Drozdetskiy et al. 2015). Multiple alignments of protein sequences were constructed using the MUSCLE program with default parameters (Edgar 2004). Approximate Maximum Likelihood phylogenetic trees were constructed using FastTree with default parameters (Price, Dehal and Arkin 2010).
RESULTS
To delineate the Csx27 family, we employed an iterative database search procedure. First, PSI-BLAST was run with the EKB54194.1 (Csx27) sequence from Bergeyella zoohelcum ATCC 43 767 as a query against the NR database, with the E-value threshold set at 10−4; this stringent cut-off was used to avoid inclusion of unrelated membrane proteins. All the proteins with similarity to Csx27 below this threshold were collected after second iteration, and several hits were used as queries for two iterations of PSI-BLAST search with the same threshold. All the hits from these searches were pooled, and the resulting protein set was filtered for size, retaining proteins of 200–400 aa, and for the presence of three to five predicted TM helices, in order to eliminate irrelevant membrane proteins and partial sequences. The remaining sequences were clustered using BLASTclust, with a 95% sequence identity threshold and a 90% sequence coverage threshold, to generate a non-redundant set of Csx27 homologs. The final set of Csx27 family representatives obtained through this procedure included 584 proteins. A multiple alignment of all these proteins (after discarding several poorly aligned sequences) was used to construct a phylogenetic tree (Fig. 1A).
Thus, Csx27 belongs to a large family of membrane proteins with a distinct architecture of which only a small subfamily is associated with subtype VI-B CRISPR-Cas systems (Fig. 1A). The Csx27 family shows a broad distribution among bacteria, with the majority identified in Proteobacteria, Firmicutes and Bacteroidetes. Most of the Csx27 homologs contain four predicted TM helices, with a large loop between TM2 and TM3 (Fig. 1B). The loop is predicted to adopt, mostly, an α-helical structure and shows no detectable similarity to any proteins outside the family. Membrane topology prediction indicates that the N-terminus of the Csx27 family proteins is located inside the cell and, accordingly, so is the long loop that can be expected to interact with the functional partners of these proteins (Fig. 1B).
We sought to infer the functions of the Csx27 family proteins using the ‘guilt by association’ approach (Aravind 2000; Galperin and Koonin 2000). To this end, gene neighborhoods were retrieved for all 584 family members (10 genes upstream and downstream of the gene encoding a Csx27 homolog) and annotated these loci using PSI-BLAST search against the CDD which includes position-specific scoring matrices for protein families from different databases (Table S1, Supporting Information). This search showed that, in the Csx27 neighborhoods, the four most frequent genes are: 1) ComA/ComEC (348 loci, some contain two genes of this family), 2) multidomain protein containing Ub pathway enzymes (Iyer, Burroughs and Aravind 2006), namely, E1, E2 and Jab family deubiquitinase (143 loci; hereafter UbP), 3) HTH_XRE family transcriptional regulator (104 loci) and 4) YafY/WYL (55 loci). Additionally, these loci include many genes involved in different forms of microbial defense and stress response, such as restriction-modification system components, Zn-dependent peptidase ImmA (M78 family) associated with toxin-antitoxin systems, a RloB-like protein, which is a component of abortive infection systems, and LexA, the SOS response activator, suggesting that many of the Csx27 homologs are encoded in defense islands (Table S1, Supporting Information). The genes for three proteins, namely, ComA/ComEC, UbP and YafY/WYL, are likely co-regulated, either comprising a single operon or using the same promoter in the case of divergent orientation (Fig. 1C).
ComEC is a key component of the bacterial natural transformation (competence) machinery, i.e. the dedicated pump that imports exogenous DNA into bacterial cells and facilitates its integration into the chromosome (Chen, Christie and Dubnau 2005; Matthey and Blokesch 2016). The ComA/ComEC protein contains a predicted nuclease domain of the metallo-beta-lactamase (MBL) superfamily that is fused to an uncharacterized domain denoted DUF4131 (DUF, Domain of Unknown Function) and the so called ‘competence’ membrane region (Baker et al. 2016; Pimentel and Zhang 2018) (Fig. 1D). The DUF4131 region contains four predicted TM helices and a large loop between TM2 and TM3 that is predicted to adopt a ssDNA-binding OB-fold. The ComEC protein forms the pore through which one strand of an exogenous DNA molecule is pumped into the bacterial whereas the second strand is degraded, likely, by the MBL. The uncharacterized ComEC homologs encoded in the same neighborhood with Csx27 family proteins contain only the MBL domain (hereafter ComEC-MBL) but lack counterparts to the other domains. Considering that the association of the ComEC-MBL gene with the gene for a Csx27 family protein is conserved in many bacterial genomes (Fig. 1A and C), it seems likely that the two proteins jointly perform functions analogous to those of ComEC, with Csx27 functionally substituting for DUF4131. Indeed, both Csx27 and DUF4131 are four TM proteins with the same predicted membrane topology although there is no detectable sequence similarity between these proteins and the cytosolic loops appear to be unrelated.
The bacterial genomic neighborhoods that encode ComEC-MBL and Csx27 family proteins also typically encode the UbP protein. In addition to the E1 and E2 domains of Ub ligase and the Jab deubiquitinase, we identified a highly diverged Ub domain, albeit with a marginal significance (with the query sequence WP_011798012.1 (amino acids 185–246) from Polaromonas naphthalenivorans, HHPred produced a hit to Ubiquitin-like domain of PDZ_GEF_RA (cd01785), with the probability 54%). Given the context, the size of the region in question and the secondary structure prediction, there is nevertheless little doubt that this is a bona fide Ub, and accordingly, the UbP protein contains a nearly full complement of the domain that comprise the Ub signaling network except for the E3 subunit of Ub ligase (Fig. 1D). Additionally, some of these neighborhoods encode proteins that consist of a ligand-binding WYL domain and an HTH domain. WYL domain proteins are poorly characterized but recently have been shown to regulate the activity of the effectors of subtype VI-D CRISPR-Cas systems as well as some type I systems (Makarova et al. 2014; Yan et al. 2018). The evolutionarily conserved association between ComEC-MBL, Csx27, UbP and WYL-HTH implies that all these proteins are functionally linked in an uncharacterized system of membrane transfer of DNA that is regulated via ubiquitylation and ligand binding by the WYL domain. Although, traditionally, the Ub network has been regarded as a quintessential eukaryotic signaling system, recently, a variety of Ub-like proteins and the cognate Ub ligases have been discovered in archaea and bacteria (Maupin-Furlow 2014). The functions of the prokaryotic Ub pathways have not been studied in detail compared to the eukaryotic counterparts, but it has been shown that they are involved in both protein modification and sulfur-transfer reactions, such as tRNA thiolation. A comprehensive analysis of the genomic neighborhoods of the prokaryotic Ub systems has revealed multiple associations including those with membrane proteins, restriction-modification modules and other defense systems, suggesting an untapped diversity of regulatory and signaling activities (Burroughs et al. 2015).
Apart from these connections to the putative novel variant of the natural transformation machinery, Csx27 genes in nine Firmicutes species are embedded in an unrelated neighborhood that includes genes for a predicted AAA + ATPase and an adenosine deaminase (Fig. 1C). These proteins can be predicted to jointly comprise a membrane-associated system for DNA modification and degradation. Interestingly, this configuration includes the closest homologs of the CRISPR-associated Csx27 within this family of membrane proteins (Fig. 1A).
In addition to these conserved genomic neighborhoods, many other Csx27-like genes are found in bacteriophage genomes (KFX26875.1), next to retrons, a distinct variety of prokaryotic retroelements (Lampson, Inouye and Inouye 2005) (e.g. AWR58724.1), or within genomic islands that include genes for integrases (Fig. 1A and Fig. S1, Supporting Information). Taken together, these observations show an association between Csx27 family genes and mobile genetic elements.
CONCLUSIONS
Comparative genomic analysis reported here shows that Csx27 protein, originally identified as an ancillary component of subtype VI-B CRISPR-Cas systems, belong to an expansive family of four TM proteins. The guilt by association approach suggests that these proteins form membrane channels for ssDNA and in that capacity comprise a key component of a previously uncharacterized DNA uptake system (Fig. 2A). The architecture of these systems appears to be similar to that of the canonical competence machinery except that Csx27 replaces DUF4131 as the membrane channel. Of particular interest is the finding that this putative version of bacterial competence machinery is consistently linked to Ub system components and WYL domain proteins, with the implication that, in this case, the process of DNA uptake and, possibly, subsequent degradation is subject to complex regulation. Independent of the association with the transformation machinery components, a distinct subfamily of the Csx27 family appears to be a component of a novel, membrane-associated system of DNA modification that could function as a form of defense against foreign DNA, perhaps, targeting it for degradation (Fig. 2B). The putative novel DNA uptake machinery is connected to type VI CRISPR-Cas through both Csx27 and the WYL domain proteins. Membrane association of Csx27 was not detected in E. coli, suggesting that additional factors present in Bacteroidetes, the native carriers of subtype VI-B systems, but not in E. coli, are required for the membrane integration of this protein. In Bacteroidetes, subtype VI-B systems might degrade nascent transcripts of foreign DNA in conjunction with its uptake by the bacterial cell (Fig. 2C). The experiments in E. coli, while not demonstrating membrane localization of Csx27, show that this protein down-regulates Cas13b activity(Smargon et al. 2017), suggesting that the two proteins directly interact, perhaps, via the long cytoplasmic loop of Csx27. This from of regulation can be expected to occur in the native host as well but, in that case, it would be modulated by additional components of the system, in particular, UbP. The hypotheses proposed here suggest several directions of experimental validation that could reveal novel membrane-associated mechanisms of DNA transport, modification and degradation in bacteria.
Figure 2.
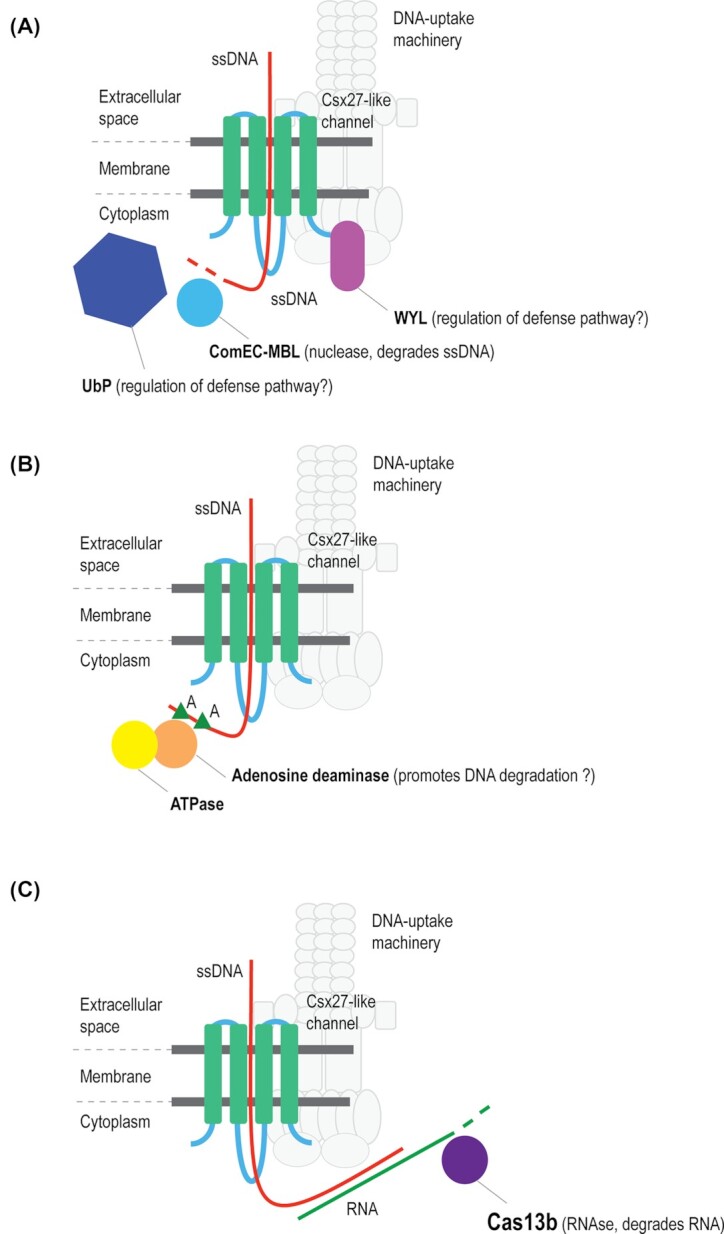
A hypothetical scheme of the functions of Csx27-like proteins as components of a novel DNA uptake machinery (A), a membrane-associated DNA modification system (B) and subtype VI-B1 CRISPR-Cas (C).
Author’s contributions
KSM performed research; KSM, LG, FZ and EVK analyzed the data; KSM and EVK wrote the manuscript that was read, edited and approved by all authors.
Supplementary Material
ACKNOWLEDGEMENTS
KSM and EVK are supported by intramural funds of the US Department of Health and Human Services (to National Library of Medicine). FZ is a New York Stem Cell Foundation–Robertson Investigator. FZ is supported by NIH grants (1R01- HG009761, 1R01-MH110049 and 1DP1-HL141201); the Howard Hughes Medical Institute; the New York Stem Cell, Simons, Paul G. Allen Family and Vallee Foundations; the Poitras Center for Affective Disorders Research at MIT; the Hock E. Tan and K. Lisa Yang Center for Autism Research at MIT; and J. and P. Poitras, and R. Metcalfe.
Conflicts of interest . None declared.
REFERENCES
- Abudayyeh OO, Gootenberg JS, Konermann Set al.. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AAet al.. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L. Guilt by association: contextual information in genome analysis. Genome Res. 2000;10:1074–7. [DOI] [PubMed] [Google Scholar]
- Baker JA, Simkovic F, Taylor HMet al.. Potential DNA binding and nuclease functions of ComEC domains characterized in silico. Proteins. 2016;84:1431–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Horvath P. A decade of discovery: CRISPR functions and applications. Nat Microbiol. 2017;2:17092. [DOI] [PubMed] [Google Scholar]
- Burroughs AM, Zhang D, Schaffer DEet al.. Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling. Nucleic Acids Res. 2015;43:10633–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I, Christie PJ, Dubnau D. The ins and outs of DNA transfer in bacteria. Science. 2005;310:1456–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdetskiy A, Cole C, Procter Jet al.. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015;43:W389–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Koonin EV. Who's your neighbor? New computational approaches for functional genomics. Nat Biotechnol. 2000;18:609–13. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Burroughs AM, Aravind L. The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like beta-grasp domains. Genome Biol. 2006;7:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 2017;37:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Zhang F. Coupling immunity and programmed cell suicide in prokaryotes: life-or-death choices. Bioessays. 2017;39:1–9. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne Get al.. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–80. [DOI] [PubMed] [Google Scholar]
- Lampson BC, Inouye M, Inouye S. Retrons, msDNA, and the bacterial genome. Cytogenet Genome Res. 2005;110:491–9. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Anantharaman V, Grishin NVet al.. CARF and WYL domains: ligand-binding regulators of prokaryotic defense systems. Front Genet. 2014;5:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OSet al.. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Derbyshire MK, Gonzales NRet al.. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2015;43:D222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthey N, Blokesch M. The DNA-uptake process of naturally competent vibrio cholerae. Trends Microbiol. 2016;24:98–110. [DOI] [PubMed] [Google Scholar]
- Maupin-Furlow JA. Prokaryotic ubiquitin-like protein modification. Annu Rev Microbiol. 2014;68:155–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanraju P, Makarova KS, Zetsche Bet al.. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016;353:aad5147. [DOI] [PubMed] [Google Scholar]
- Pimentel ZT, Zhang Y. Evolution of the natural transformation protein, ComEC, in bacteria. Front Microbiol. 2018;9:2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SA, Alkhnbashi OS, Behler Jet al.. Comprehensive search for accessory proteins encoded with archaeal and bacterial type III CRISPR-cas gene cassettes reveals 39 new cas gene families. RNA Biol. 2018,16:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Smargon A, Scott Det al.. Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol. 2017;15:169–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov SA, Makarova KS, Wolf YIet al.. Systematic prediction of genes functionally linked to CRISPR-Cas systems by gene neighborhood analysis. Proc Natl Acad Sci USA. 2018;115:E5307–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smargon AA, Cox DB, Pyzocha NKet al.. Cas13b is a type VI-B CRISPR-associated RNA-Guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol Cell. 2017;65: 618–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–60. [DOI] [PubMed] [Google Scholar]
- Yan WX, Chong S, Zhang Het al.. Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol Cell. 2018;70: 327–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.