Abstract
Hepatocyte nuclear factor 4-alpha (HNF4α) is a highly conserved member of nuclear receptor superfamily of ligand-dependent transcription factors that is expressed in liver and gastrointestinal organs (pancreas, stomach, and intestine). In liver, HNF4α is best known for its role as a master regulator of liver-specific gene expression and essential for adult and fetal liver function. Dysregulation of HNF4α expression has been associated with many human diseases such as ulcerative colitis, colon cancer, maturity-onset diabetes of the young, liver cirrhosis, and hepatocellular carcinoma. However, the precise role of HNF4α in the etiology of these human pathogenesis is not well understood. Limited information is known about the role of HNF4α isoforms in liver and gastrointestinal disease progression. There is, therefore, a critical need to know how disruption of the expression of these isoforms may impact on disease progression and phenotypes. In this review, we will update our current understanding on the role of HNF4α in human liver and gastrointestinal diseases. We further provide additional information on possible use of HNF4α as a target for potential therapeutic approaches.
Keywords: Hepatocyte nuclear factor 4-alpha, Liver cirrhosis, Hepatocellular carcinoma, Viral hepatitis, Gastrointestinal tract, Colorectal carcinoma, Transcription factor
Core tip: Our current understanding of the molecular etiology of human liver and gastrointestinal diseases is limited and there is a critical need to explore novel hypotheses and risk factors that may contribute to these diseases. Hepatocyte nuclear factor 4-alpha (HNF4α) has been well recognized as an important transcription factor that regulates gene expression involved in the differentiation of liver and gastrointestinal cells. Dysregulation of HNF4α function is associated with many diseases related to these cells. Here we attempt to update our understanding on the role of HNF4α in the pathogenesis of these diseases for use as target for better therapeutic modality.
INTRODUCTION
Hepatocyte nuclear factor 4-alpha (HNF4α) is a transcription factor with important roles in liver and gastrointestinal tract development, hepatocyte differentiation, and lipid and glucose metabolism[1]. The HNF4α gene is located on chromosome 20, with transcription regulated by two promoters (P1 and P2) and alternative splicing variants, resulting in 9 distinct isoforms (α1-α9)[1,2]. Adult hepatocytes exclusively express P1 isoforms, while both promoters are active in intestinal epithelia within distinct compartments[1,2]. The importance of HNF4α in development is highlighted by embryonic lethality of gene knockout in mice[3]. Targeted knockout in colonic epithelium disrupts architecture, decreases enterocyte numbers and goblet cell maturation, and perturbs transcriptional profiles[4], while liver-targeted knockout results in hepatomegaly with altered liver architecture, and decreased glycogen storage[5]. Furthermore, expression of HNF4α in mesenchymal stem cells is sufficient to induce epithelioid changes and some hepatocyte functionality including urea production and albumin secretion[6].
The regulation of HNF4α expression, activity, and localization is highly complex (reviewed in[1,7]), reflective of downstream transcri-ptional networks with diverse functional roles including drug metabolism, bile acid synthesis and conjugation, lipid homeostasis, gluconeogenesis, ureagenesis, cell adhesion, proliferation, and apoptosis[7]. Regulation of HNF4α occurs at multiple levels: Epigenetic[8,9]; tran-scriptional, including promoter regulation, transcript secondary structure, and microRNA-mediated inhibition[7,10]; and post-translational, including protein phosphorylation, degradation, and nuclear localization[1,11,12]. HNF4α transcription factor binding sites are also widely dispersed in the human genome, as evidenced by changes in mRNA levels of ~2500 genes upon over-expression of HNF4α in cell culture[13]. HNF4α expression and activity are altered in numerous disease states involving multiple organ systems, and immunohistochemical detection in the clinical setting has potential diagnostic and prognostic value. This review article updates the understanding of HNF4α role in liver and gastrointestinal pathogenesis and presents potential therapeutic approaches and strategies for possible treatment based on HNF4α involvement.
HNF4ALPHA ACTIVITY IN LIVER PATHOGENESIS
HNF4α perturbations in disease states have been most extensively investigated in the context of liver disease. Most of the major liver diseases have been associated with altered HNF4α expression, isoform ratios, and localization, including inflammation, alcoholic liver disease, non-alcoholic fatty liver disease, fibrosis and cirrhosis, viral hepatitis, and the hepatocellular carcinoma (Table 1). Most commonly, HNF4α expression at the protein and transcript levels is decreased across liver diseases, an observation substantiated by human, animal model, and cell culture-based studies in many cases (Table 2). Altered expression of HNF4α in response to relatively non-specific stimuli, such as inflammation[14] and injury-induced acute phase response[15], as well as in a remarkable spectrum of disease suggests centrality of HNF4α in response to most modes of hepatocyte injury and stress resulting in de-differetion state of liver function (Table 1, Figure 1)[16]. There is, therefore, a critical need to understand the role of HNF4α in the molecular epidemiology of the de-differentiation state of liver function across these liver diseases for the potential restoration of normal liver function.
Table 1.
Expression of hepatocyte nuclear factor 4alpha and variants in various disease states
| Disease state | HNF4α expression | Organism | Model | Ref. |
| Development | ||||
| Hypertriglyceridemia (preterm infants, children) | Decreased, P1 promoter methylation | Human | - | [8,9] |
| Intrauterine growth restriction | Decreased, hypermethylated promoter | Human | - | [73] |
| High fat diet (in utero) | Increased | Mouse | HFD | [87] |
| Metformin exposure (in utero) | Increased, hypomethylated promoter | Mouse | Metformin | [74] |
| Liver | ||||
| Acute phase response | Decreased activity | Mouse | Injury | [15] |
| Inflammation (IL-1β) | Decreased | Human, mouse | Hepatoma cells | [14] |
| TNFα-induced hepatotoxicity | Decreased | Human, Mouse | Hepatoma cells | [88,89] |
| Alcoholic liver disease | Decreased | Mouse | Ethanol, MCD diets | [17,89] |
| α1-antitrypsin | Decreased | Mouse | Human ATZ | [90] |
| Fibrosis | Decreased | Human | Hepatocytes | [24] |
| Cirrhosis | Decreased | Human, rat | DEN | [41,91] |
| Hepatocellular carcinoma | P2 increased, P1 decreased | Human, rat | DEN | [32,41,59] |
| Decreased, expressed in metastases, increased P1:P2 | Human, mouse, rat | HCC cells, DEN | [13,31,33,40,41,59,92] | |
| Hepatitis B virus | Decreased | Human | Hepatoma cells | [46,47] |
| Hepatitis C virus | Increased | Human | Hepatoma cells | [93,94] |
| Decreased | Human, mouse | Hepatocytes, hepatoma cells, HCV+ HCC | [35,95] | |
| Hepatitis E virus | Increased phosphorylation, cytoplasmic retention | Human | Hepatoma cells | [48] |
| Non-alcoholic steatohepatitis | Modestly increased | Human | - | [21] |
| Decreased, cytoplasmic retention | Mouse | db/db or HFD | [18,22] | |
| Iron overload | Decreased | Human, mouse | Hepatoma cells, iron rich diet | [49] |
| Endocrine | ||||
| Type 2 diabetes | Decreased, P1 promoter hypermethylation | Human | - | [72] |
| Increased in liver | mouse | HFD, steptozotocin | [76] | |
| Mature onset diabetes of the young (MODY) | HNF4α variants | Human | - | [66-69] |
| Obesity | HNF4α variant | Human | - | [79] |
| Islet cell hypoxia | Decreased | mouse | ob/ob | [96] |
| Intestine | ||||
| Micrbiome colonization | Decreased | Zebrafish | - | [52] |
| IBD | Decreased | Human, mouse | - | [56,57] |
| Crohn disease | HNF4α variant, decreased | Human | - | [55,56,98] |
| Ulcerative colitis | HNF4α variant | Human | - | [53,54] |
| Colorectal carcinoma | Decreased or cytoplasmic retention, altered P1:P2, expressed in metastases | Human, mouse | Mutagen | [59,99,100] |
| Intestinal type ampullary carcinoma | Increased | Human | - | [10] |
| Upper GI | ||||
| Gastric carcinoma | Increased, altered P1:P2 | Human | Carcinoma cells | [59,62,78,102] |
| Barrett esophagus (intestinal metaplasia) | Increased | Human, mouse | Explant | [63,64] |
| Stomach intestinal metaplasia | Increased | Human | - | [59] |
| Kidney | ||||
| Renal cell carcinoma | Decreased, expressed in metastases, increased P1:P2 | Human | - | [59,103,104] |
| Atypical fanconi syndrome | HNF4α variant | Human | - | [66] |
| Lung | ||||
| Mucinous adenocarcinomas | Increased in some | Human | - | [82,100] |
| Ovary | ||||
| Ovarian mucionous neoplasm | Expressed | Human | - | [81] |
| Heart | ||||
| Cardiac fibrosis | Increased | Mouse | Angiotensin II | [83] |
IL-1β: Interleukin 1beta; MCD: Methionine-choline deficient; HFD: High fat diet; TNFα: Tumor necrosis factor alpha; ATZ: Mutant Z form of alpha1-antitrypsin deficiency (ATD); HCC: Hepatocellular carcinoma; HCV: Hepatitis C virus; IBD: Inflammatory bowel disease.
Table 2.
Effects of experimental perturbations of hepatocyte nuclear factor 4alpha
| HNF4α manipulation | Organism | Model | Effect | Ref. |
| Development | ||||
| Overexpression of α1D | Human | iPSCs | Promoted endoderm differentiation | [105] |
| Liver | ||||
| Overexpression | Human | Marrow-derived mesenchymal stem cells | Epithelioid changes, glycogen storage, albumin secretion | [6] |
| Rat | DEN | Suppressed carcinogenesis, suppressed EMT, decreased fibrosis, restored hepatic function in cirrhosis | [5,26,41] | |
| Rat | mst1/2 conditional mutant | Reduced liver size and HCC proliferative indices | [31] | |
| Mouse | Hepatoma cell xenograft | Decreased tumorigenesis and proliferation | [39,40] | |
| Mouse | Acute liver failure | Increased survival, urea production | [86] | |
| Knockdown | Human | Hepatocyte culture, HBV infection | Increased hepcidin expression, impaired transcription and replication of HBV, transformation and tumorigenicity in mice | [13,36,42,43,106] |
| Liver targeted knockdown | Mouse | - | Increased hepatocyte proliferation and promitogenic transcription | [38] |
| Expression of α7 only | Mouse | - | Steatosis, downregulation of CAR | [75] |
| Liver targeted knockout | Mouse | DEN | Upregulation of miR-194 and -192, reduced transcriptional response to extracellular matrix rigidity, increased hepatocyte proliferation, HCC risk, steatosis | [20,24,107,108] |
| Intestine | ||||
| Overexpression | Mouse, rat | Embryonal carcinoma cells, co-culture | Differentiation to polarized epithelium, tight junction proteins | [109,110] |
| Exon swapping, P1 or P2 only | Mouse | DSS | Altered enterocyte migration, ion transport, barrier function, susceptibility to DSS colitis and associated cancer | [2] |
| Intestine targeted knockout | Mouse | DSS | Transcription profiles similar to IBD, altered embyonic development, Paneth cell alterations, susceptibility to colitis | [50,56,111,112] |
| Intestine targeted knockout of P1 and P2 | Mouse | - | Spontaneous intestinal inflammation | [57] |
| Dominant negative expression | Mouse, Human | Enterocytes | Decreased expression of apolipoprotein A | [113] |
| Upper GI | ||||
| Esophagus overexpression | Mouse | Esophageal explants | Induced partial columnar cell phenotype | [63] |
| Stomach overexpression | Human | Gastric carcinoma cells | Resistance to multiple chemotherapeutics | [102] |
| Stomach knockdown | Human | Gastric carcinoma cells | Increased susceptibility to chemotherapeutics | [102] |
| Stomach targeted knockout | Mouse | - | Reduced chief cell size, epithelial proliferation and migration | [61] |
| Endocrine | ||||
| HNF4α7 only expression | Mouse | - | Dyslipidemia, mild steatosis | [75] |
| HNF4α1 only expression | Mouse | - | Impaired glucose tolerance, hyperinsulinemia | [75] |
| Knockout in pancreatic beta cells | Mouse | - | Reduced glucose stimulated insulin secretin, similar to MODY | [65,70,71] |
| Overexpression in pancreatic beta cells | Human | islet cells | Induced cell cycle entry without expansion | [114] |
iPSCs: Induced pluripotent stem cells; DEN: Diethylnitrosoamine; HBV: Hepatitis B virus; DSS: Dextran sodium sulfate; HNF4alpha7: Hepatocyte nuclear factor 4alpha7; HNF4alpha1: Hepatocyte nuclear factor 4alpha1.
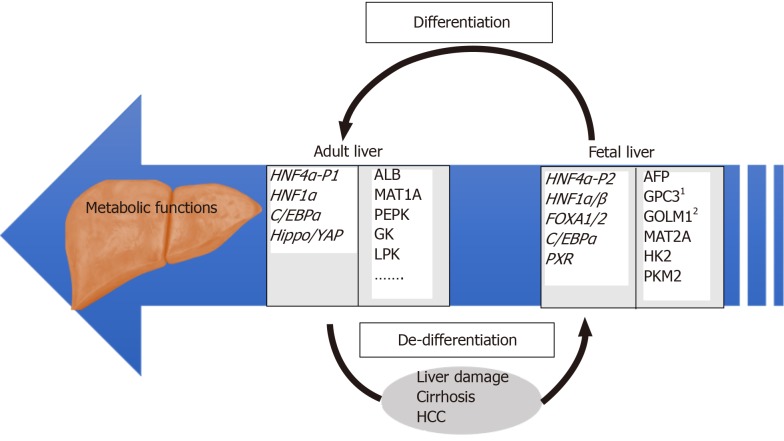
Figure 1.
Overview of regulatory and target genes involved in differentiated and de-differentiated stages of liver development. Examples of relevant HNF4α target genes identified by our group[27] are individually numbered.
Alcoholic liver disease
Expression of HNF4α and carboxylesterase 1 (CES1), an enzyme involved in trigly-ceride metabolism, was reduced in patients with alcoholic steatohepatitis and mice with methionine and choline-deficient diet-induced inflammation[17]. Alcohol repressed both HNF4α and CES1 expression in primary hepatocyte cell culture. Knockout of CES1 in mice exacerbated alcohol-induced steatosis and steatohepatitis, as well as diet-induced liver inflammation[17].
Non-alcoholic fatty liver disease
HNF4α has important roles in liver lipid and lipoprotein metabolism[18]. Hepatocyte-targeted knockout of HNF4α in mice resulted in lipid accumulation, changes in VLDL secretion and bile acid uptake, and alterations in peripheral blood cholesterol and triglycerides[19]. Acute knockout of HNF4α in adult mice resulted in hepatocyte proliferation and vacuolization, and hepatomegaly[20].
HNF4α mRNA and protein levels were decreased in patients with non-alcoholic steatohepatitis (NASH), as well as in cultured hepatocytes and in livers of mice with genetic obesity (ob/ob) or on high fat diet[18]. Network analysis of transcriptomic data in patients with non-alcoholic steatohepatitis identified HNF4α as a central regulator, although transcription of HNF4α itself was not significantly altered[21]. In contrast to a prior study[18], limited immunohistochemistry on 12 liver specimens of patients with NASH, non-alcoholic fatty liver disease (NAFLD) activity (NAS) score 5-7, showed minimal increased immunoreactivity (24%-40% positive cells) compared to a single control (17% positive cells)[21]. Cytoplasmic retention of HNF4α in high fat diet mice with steatosis has also been observed, corresponding to reduced transcription of target genes and HNF4α phosphorylation by protein kinase C isotypes[22]. Taken together, these studies consistently indicate a significant decrease in HNF4α activity in NAFLD, although observations of HNF4α expression levels and localization are less uniform. This could be related to disruption of the transcription factor network responsible for the de-defferentiated state, which is partially controlled by HNF4α activity (Figure 1).
Hepatic fibrosis and cirrhosis
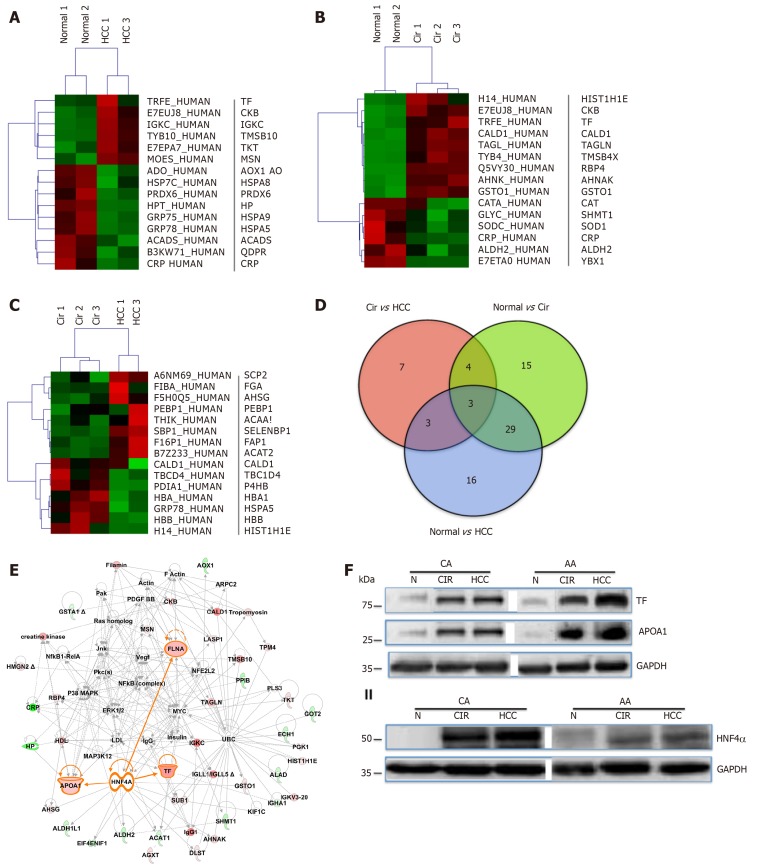
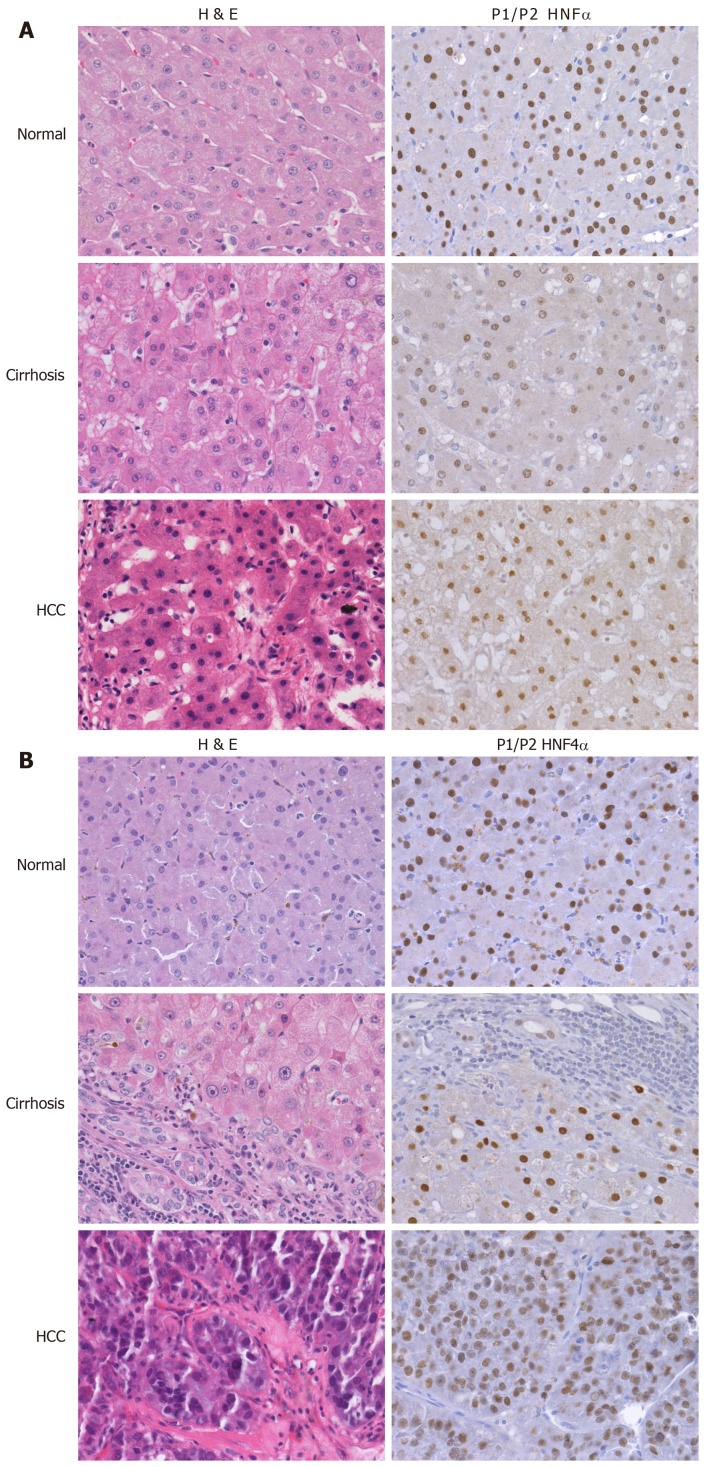
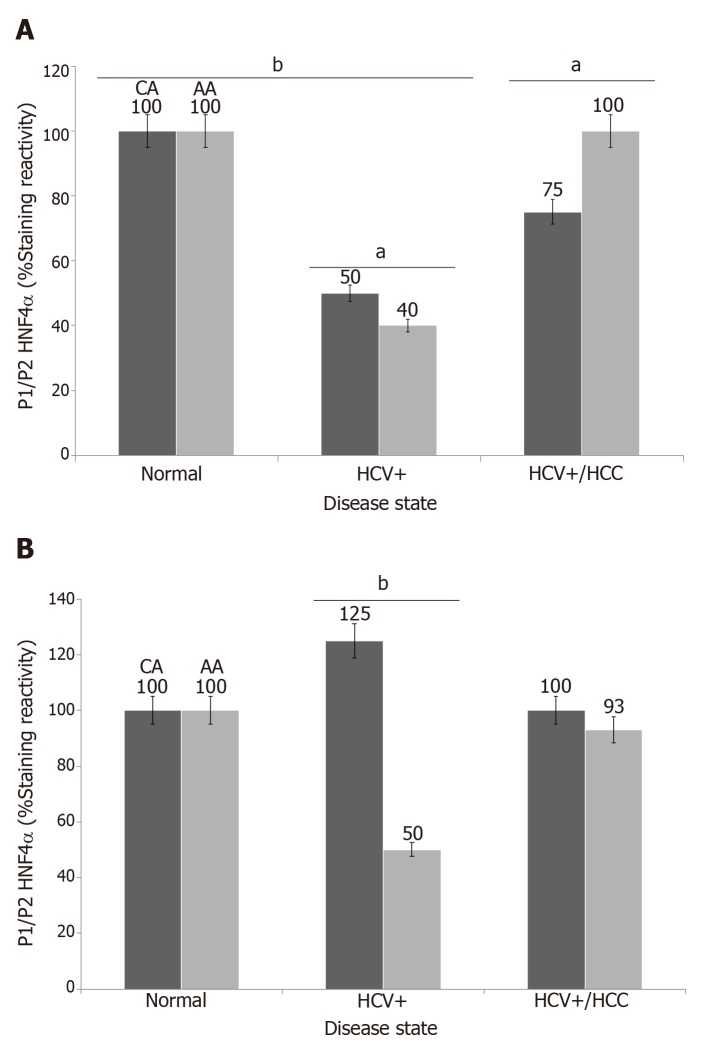
To explore and establish an understanding of the molecular basis for decreased HNF4α activity across US racial population, we performed a mass spectrometry-based proteomics study using clinical tissue samples from Caucasian Americans (CA) and African Americans (AA) and demonstrated, for the first time, that differentially expressed proteins (DEPs) in cirrhotic livers are actually distinct from hepatocellular carcinoma (HCC) and the expression of these proteins are also racially dependent [23]. For example, Figure 2C shows the heat map (truncated) of DEPs between cirrhotic and HCC groups, and that there is a high degree of interaction between HNF4α (a focus hub) and some of these DEPs like serotransferrin (TF) and apolipo-protein lipase A1(APOA1) (Figure 2E). Note also that the level of TF and APOA1 proteins in AA cirrhotic and HCC samples (Figure 2F, upper panel) is equal as in CA protein samples. Furthermore, the levels of HNF4α protein in AA samples are lower compared to CA samples (Figure 2F, lower panel). It is known that AA patients with cirrhotic liver and HCC usually have elevated levels of serum markers of iron stores and altered cholesterol and triglyceride levels[24,25], hence the levels of both are elevated in AA samples. The expression of both TF and APOA1 genes is known to be regulated by the transcription factor HNF4α[26]. The differential expression of HNF4α has been shown in colitis and colitis-associated colon cancer[2], and more recently by our group[27] in cirrhotic livers and HCC. In our study, we used immunohistoche-mistry to validate the expression of HNF4α isoforms in HCV cirrhotic livers and HCC tissues among CA and AA tissue samples (Figure 3). As shown in Figures 4A and 4B, the staining reactivity of P1- HNF4α isoforms are lower in cirrhotic HCV livers of AAs (grey bars) as compared to CAs (black bars), whereas the observed increase in P1/P2- HNF4α staining (Figure 4A) but not P1 staining (Figure 4B) in HCC for AAs relative to CAs suggests a potential involvement of P2- HNF4α.
Figure 2.
Differentially expressed proteins in normal and liver disease states. Heat maps of differentially expressed proteins (DEPs) (truncated) that were selected following supervised analysis (A) Normal vs. Cirrhosis, (B) Normal vs. Hepatocellular carcinoma, (C) Cirrhosis vs. Hepatocellular carcinoma, and (D) Venn Diagram comparing the significantly DEPs identified. (E) Interactive Network Analysis of DEPs in cirrhosis and hepatocellular carcinoma as compared to normal shows HNF4α as a focus hub to many DEPs. (F) A representative of immunoblot analysis of TF and APOA1 (upper panel), HNF4α (lower panel) in tissue samples of AA & CA. GAPDH was used as a loading control, as published in[23].
Figure 3.
Differential expression of hepatocyte nuclear factor 4alpha in cirrhotic and hepatocellular carcinoma livers. Representative H&E and P1/P2 HNF4α stained samples of HCV cirrhotic and hepatocellular carcinoma of Caucasian (A) and African American (B) tissue samples, as published in[27].
Figure 4.
Immunoreactivity of hepatocyte nuclear factor 4alpha isoforms in cirrhotic and hepatocellular carcinoma livers. Data are presented as the mean ± standard error (n=4 tissue sections from 24 paraffin embedded tissue blocks). Data were evaluated for stastistical significance by one-way analysis of variance and are represented as follows: aP<0.05, bP<0.001as compared to normal for P1/P2 HNF4α (A) and P1- HNF4α (B). Black bar (CA) = Caucasian Americans; Gray bar (AA) = African Americans, as published in[27]
Increased extracellular matrix rigidity, as shown in CCl4 or 5-diethoxycarbonyl-1,4-dihydrocollidine (DDC) induced fibrosis in rat livers, modulates hepatocyte function and cytoskeletal arrangement in part through inhibition of the HNF4α transcriptional network[28]. HNF4α transcriptional repression in the context of cell culture was prevented by treatment with Rho-dependent kinase (ROCK) inhibitor. In rats with hepatic fibrosis induced by dimethylnitrosamine (DEN) or bile duct ligation, forced hepatic expression of HNF4α decreased fibrosis in improved liver function[29]. Similarly, forced re-expression of HNF4α improved functionality in isolated hepatocytes and reversed liver failure in a CCl4-induced rat model[30]. These studies indicate that HNF4α expression is decreased in hepatic fibrosis, and forced expression in this setting appears to promote hepatocyte and liver function.
Hepatocellular carcinoma
A number of studies have examined HNF4α expression patterns in hepatocellular carcinoma (HCC), including differences in P1 and P2 promoter-derived isoforms. Total HNF4α transcripts were lower in 224 cases of HCC than 220 controls[31]. Rats with DEN-induced hepatocarcinogenesis exhibit decreased hepatic HNF4α expre-ssion[29]. However, tissue microarray immunohistochemistry on 615 human HCCs showed inverse correlation of P2 and P1 HNF4α[32]. High P2 HNF4α expression correlated with poor differentiation, vascular invasion, and shorter overall patient survival. Conversely, relatively high P1 HNF4α immunoreactivity in HCC correlated with better differentiation and longer overall survival among a small cohort of 16 patients[33]. However, decreased expression of HNF4α is not uniform in HCC. A series of 196 human HCCs in a heterogeneous background of liver disease, showed 52 (26%) were positive for intense-to-moderate immunoreactivity to HNF4α[34].
HNF4α expression in HCC has been linked to Hippo pathway signaling [31]. Tissue microarray IHC on 75 HCCs revealed increased immunoreactivity for yes-associated protein 1 (YAP1), and lower HNF4α than adjacent tissues[31]. The YAP1/ HNF4α ratio increased with high Edmondson grade. HNF4α appears to be degraded in a proteasome-dependent pathway in the presence of YAP1, and expression of HNF4α in cultured cells or mice with YAP-mediated HCC (mst1/2 conditional mutant) resulted in decreased liver size, transcription of YAP-TEAD target genes, and Ki67 proliferative indices.
Transient knockdown of HNF4α initiates transformation of immortalized hepato-cytes through a feedback loop involving miR-24, IL6R, STAT3, miR-124, and miR-629[35,36]. Hepatocytes with knockdown of HNF4α or overexpression of either miR-24 or miR-629 (both HNF4α suppressors) were capable of tumor formation in nude mice[36]. Delivery of miR-124, a transcriptional target of HNF4α, suppressed tumor growth in HCC xenografts and DEN-treated mice. As a corollary in cell culture, knockdown of HNF4α in hepatoma cells also promoted transcription of genes related to the epithelial-mesenchymal transition (EMT) and neoplasia[37,38].
Conversely, overexpression of HNF4α in hepatoma cell lines induced differen-tiation into hepatocytes and suppressed HCC growth and metastases[39,40]. Forced HNF4α expression in a rat model of DEN-induced liver carcinoma reduced carcinogenesis and decreased EMT[41]. Expression in fibroblasts actually induced a mesenchymal-to-epithelial transition[5].
HNF4α directly interacts with the promoter and induces expression of apoptosis signal-regulating kinase 1 (ASK1)[13]. RT-PCR of human HCC and surrounding non-neoplastic tissue revealed downregulation of HNF4α in 45 of 60 cases (75%) and corresponding ASK1 downregulation in 44 of 50 cases (73%)[13]. Low ASK1 or HNF4α mRNA levels correlated with larger tumor size and advanced stage. Low ASK1 mRNA also correlated with shorter patient survival, in part due to correlation with tumor size, and ASK1 injection directly into xenograft tumors or systemically in mice reduced growth of HCC[13].
Viral hepatitis
The viral hepatitis are associated with decreased HNF4α and/or activity. The hepatitis B (HBV) viral genome contains an HNF4α binding motif in the promoter core, and viral transcription and regulation are dependent on hepatocyte HNF4α[42-45]. Interleukin 35 enhanced HBV replication through enhanced HNF4α binding to the core promoter, an effect impaired by promoter mutation or knockdown of HNF4α expression. However, HBV infection or overexpression of vial protein HBx in hepa-toma cells reduced HNF4α expression and downstream transcriptional targets[46,47].
Electroporation of hepatoma cells with HCV DNA lead to viral replication and decrease in HNF4α mRNA and protein, as well as decreased downstream transcriptional targets[35]. Overexpression of viral proteins including core protein or non-structural proteins (NS5A) was sufficient to significantly decrease HNF4α expression. A transcriptome comparison of hepatocellular carcinomas associated with HCV infection in either African American or Caucasian groups identified differential expression of HNF4α target genes such as SAA1[27]. Immunohistochemistry demon-strated decreased HNF4α expression in HCV positive cirrhosis and hepatocellular carcinoma (n = 72) relative to normal livers, although levels of suppression varied by ethnicity[27].
Hepatitis E virus open reading frame 3 (ORF3) in cultured hepatoma cells resulted in increased HNF4α a phosphorylation, impaired nuclear translocation, and down-regulation of target genes[48]. There was no detected effect on HNF4α expression.
Iron overload
Iron overload in an iron-rich diet mouse model reduced HNF4α and miR-122 in liver[49]. Liver-targeted adenovirus delivery and overexpression of miR-122 resulted in reduced hepatic inflammation but did not significantly affect iron overload.
HNF4ALPHA ACTIVITY IN COLON PATHOGENESIS
HNF4α plays an important role in colon development[50], and has been implicated in intestinal epithelial differentiation, lipid metabolism, and epithelial junctions[1,51]. Expression levels appear to be negatively regulated by gut microbiota, evidenced by a zebrafish model[52]. Altered HNF4α expression and activity, as well as germline variants, have been associated with inflammatory bowel disease (IBD) and colorectal carcinoma[2].
Inflammatory bowel disease
Genome-wide associations studies have linked HNF4α variants with susceptibility to ulcerative colitis in two independent studies[53,54]. An HNF4α P2 promoter single nucleotide polymorphism has also been associated with childhood-onset Crohn disease[55]. In addition to germline variants, HNF4α transcripts were significantly decreased in intestinal biopsies from patients with IBD[56].
Intestine targeted knockout of HNF4α in mice increased susceptibility to dextran sulfate sodium (DSS) induced colitis[1,56]. In another study, knockout of P1 and P2 isoforms of HNF4α in mice resulted in spontaneous intestinal inflammation that worsened with time, leading to epithelial injury, crypt hyperplasia, and prolife-ration[57]. HNF4α derived from P1 or P2 promoters have distinct effects on colonic epithelium, as demonstrated with an exon swapping mouse model[2]. Mice expressing only P1 promoter-derived α1 isoform HNF4α developed fewer and smaller tumors than wild type mice after treatment with DSS and azoxymethane (AOM), and less susceptibility to DSS-induced colitis. In contrast, expression of only P2 promoter-derived α7 isoform HNF4α resulted in greater tumor load and number than wild type mice and were highly sensitive to DSS-induced colitis[2]. HNF4α directly modulated expression of Na+/H+ exchanger isoform 3 (NHE3), which has been implicated in IBD pathogenesis[58].
Colorectal carcinoma
Isoform-specific HNF4α antibody immunohistochemistry on 18 colorectal carcinomas demonstrated uniform immunoreactivity for P2 and 5/18 (28%) positive for P1[59]; a similar pattern was observed in metastases to lung. Another immunohistochemical study of 450 human colorectal carcinomas revealed either loss or cytoplasmic locali-zation of P1 HNF4α in ~80% of tumors[60]. This pattern appears to be attributable, at least in part, to interaction of HNF4α and Src kinase. Src-mediated phosphorylation of an N-terminal HNF4α tyrosine, present in P1 but not P2 isoforms, influences HNF4α protein stability, transactivation function, and nuclear localization[60]. Consis-tent with HNF4α P1 downregulation being and important feature of colorectal carcinomas, mice expressing only α7 isoform (P2 promoter) HNF4α developed greater tumor load and tumor size than wild type mice in a DSS and azoxymethane (AOM) model[2]. Conversely, expression of only the α1 isoform (P1 promoter) resulted in fewer and small tumors than wild type mice.
HNF4ALPHA ACTIVITY IN UPPER GASTROINTESTINAL TRACT PATHOGENESIS
Gastric epithelial development and maintenance are dependent on intact HNF4α. Stomach targeted knockout of HNF4α alters gastric epithelial architecture, with changes including reduced chief cell size and endoplasmic reticulum content, increased proliferation of the stem cell zone, and altered mucous neck cell migra-tion[61].
Transcriptomic analysis of 22 human gastric carcinoma specimens and non-neoplastic controls identified upregulation of HNF4α in carcinoma[62]. P1 promoter HNF4α isoforms were detected in 8 of 14 gastric carcinomas by immunohisto-chemistry, while normal gastric mucosa had positive immunoreactivity for P2 isoforms only[59]. Knockdown of HNF4α in gastric carcinoma cell lines and xenograft mouse models reduced tumor growth and angiogenesis[62]. Metformin reduced HNF4α expression in gastric carcinoma cell lines and mouse xenografts, and significantly impaired xenograft tumor growth when systemically administered[62].
HNF4α expression appears to be involved in intestinal metaplasia of the upper gastrointestinal tract. Aberrant P1 promoter-driven HNF4α immunoreactivity was observed in gastric intestinal metaplasia, although the number of cases tested is unknown[59]. While HNF4α is not expressed in normal squamous epithelia of the esophagus, HNF4α was expressed along with CDX-2 in esophageal goblet cell metaplasia (Barrett esophagus)[63]. A gene expression profiling study also identified enrichment of HNF4α expression among Barrett esophagus specimens[64]. Overexpression of HNF4α in adult mouse esophageal explants resulted in decreased squamous marker such as p63 and induced an expression profile (CK8, E-cadherin, and villin positive) suggestive of a columnar phenotype[13].
HNF4ALPHA ACTIVITY IN PANCREAS AND ENDOCRINE PATHOGENESIS
HNF4α variants have been well described as causing maturity onset diabetes of the young 1 (MODY1), characterized by diminished glucose-stimulated insulin secretion and susceptibility to type 2 diabetes[65-69]. In mouse models of pancreatic β cell HNF4α knockout, there was a similar reduction of glucose stimulation of insulin secre-tion[65,70,71]. The underlying mechanism is related to endoplasmic reticulum homeostasis in pancreatic islet cells[65].
Variation in HNF4α expression has also been observed in patients with type 2 diabetes and metabolic syndrome. For example, a monozygotic twin study identified HNF4α P1 promoter hypermethylation as a significant correlate to type 2 diabetes [72]. P1 promoter methylation has also been linked to hypertriglyceridemia in preterm infants and children, as well as intrauterine growth restriction, indicating that epigenetic modulation of HNF4α is linked to the metabolic syndrome[8,9,73]. In contrast, fetuses of metformin-treated mice exhibited hypomethylation of promoter and increased expression of HNF4a[74]. Exon swap mice expressing only HNF4α7 were dyslipidemic with mild hepatic steatosis, and HNF4α1 only expression led to impaired glucose tolerance and hyperinsulinemia[75]. A large genome-wide meta-analysis also identified variants at the HNF4α locus in association with obesity[76]. HNF4α and transcriptional target genes were increased in livers of mice with type 2 diabetes in a model of high fat diet followed by streptozotocin injection[76]. In pan-creatic cancer cells, HNF4α has been shown to promote resistance to gemcitabine by downregulating hENT1[77]. Therefore, targeting HNF4α might reverse gemcitabine resistance and provide novel treatment strategies for pancreatic adenocarcinoma.
DIAGNOSTIC AND PROGNOSTIC UTILITY OF HNF4ALPHA
HNF4α has been proposed as an immunohistochemical marker useful for pathologic differential diagnosis and prognosis in specific circumstances. HNF4α immunohisto-chemistry can be helpful in distinguishing gastric primary adenocarcinoma (essentially uniformly positive) from metastatic breast carcinoma (rarely positive)[78-80]. A tissue microarray study of 348 lung adenocarcinomas identified 54 cases with positive immunoreactivity for HNF4α, with enrichment among mucinous adenocarcinomas[81]. HNF4α positivity was associated with shorter overall survival among patients with lung adenocarcinoma[81]. However, an independent study of 1021 non-small cell lung carcinomas identified only 20 cases (2% overall, 4% of adenocarcinomas, 11% of mucinous adenocarcinomas) with positive immunorea-ctivity for HNF4α, and these correlated with absence of lymph node metastases and lower clinical stage[81]. A high percentage of metastatic colorectal carcinomas in lung specimens were positive for HNF4α, but performance of this marker was not a significant improvement over the more commonly used CDX2 and CK20 in this context[81]. HNF4α is also not specific to adenocarcinomas from the GI tract, as HCCs, renal cell carcinomas, and ovarian mucinous neoplasms may be expected to exhibit positive immunoreactivity in some cases[59,80].
Promoter-specific HNF4α antibodies for immunohistochemistry have also revealed substantial variation in neoplastic immunoreactivity for isoforms[59]. For example, 10 of 10 renal cell carcinomas examined were positive for P1 and negative for P2 HNF4α, whereas gastric and colorectal carcinomas were uniformly positive for P2 and variably positive for P1 HNF4α[59].
Hepatocellular carcinomas with β-catenin activation are associated with relatively favorable prognosis, and often exhibit significantly higher uptake of the magnetic resonance contrast agent gadoxetic acid disodium[34]. HNF4α expression correlated with nuclear β-catenin immunoreactivity and expression of the contrast agent transporter OATP1B3, as well as tumor differentiation, indicating potential utility in HCC prognostication.
HNF4ALPHA ACTIVITY AS A THERAPEUTIC POTENTIAL
Given the widespread expression of HNF4α, and demonstrable roles in development, homeostasis, and disease in multiple tissue types, systemically administered direct inhibitors or activators might be expected to exhibit significant undesirable effects. An illustrative example is the opposing effects of HNF4α on fibrosis in cardiac and liver tissue. In cardiac tissue, HNF4α is downstream of AMPK, and upregulation of expression was seen in an angiotensin II-induced cardiac fibrosis mouse model[82]. Metformin inhibition or knockout of AMPK reduced cardiac fibrosis, in part by preventing increased HNF4α expression. In contrast, liver fibrosis-associated with decreased HNF4α transcription regulation was restored by forced re-expression of HNF4α which led to reduced fibrosis and reversal fatal liver failure in a rat model[26]. These data suggest that de-differentiation state of liver function likely the cause of terminal liver failure and that resetthing the transcription factor network has therapeutic potential for correcting liver failure.
Exposure to flavonoids appears to affect HNF4α expression and activity, although mechanisms underlying these phenomena are unclear. The flavonoid luteolin impaired HBV replication and particle release from cultured HepG2 cells while suppressing HNF4α transcription and reduced viral antigen detection in peripheral blood in a mouse model of acute HBV infection[43]. Treatment of HepG2 cells with the flavonoid derivative 4’-nitro-6-hydroxyflavone reduced expression of the HNF4α target gene microsomal triglyceride transfer protein (MTP) in a transcriptional reporter system[83].
HNF4α antagonists have been described and demonstrated to impair transcription factor activity and exhibit cytotoxicity in human HCC cell lines and xenograft mouse models[84]. The HNF4α antagonist BI6015 also decreased survival of multiple gastric carcinoma cells lines in culture[62]. EC-50 values were estimated in the low micromolar range, but dose-response was non-sigmoidal[62]. The specificity of these compounds, as well as toxicity to non-neoplastic tissues, remains to be fully examined.
Pharmacological manipulation of HNF4α regulatory pathways and transcriptional targets holds promise for therapeutic development. For example, systemic treatment with the AMPK inhibitor metformin impaired gastric carcinoma tumor growth in a xenograft mouse model[62]. Similarly, metformin reduced cardiac fibrosis in an angiotensin II mouse model, an effect correlating to decreased HNF4α expression[82]. Systemic administration of the HNF4α transcriptional target miR-124 suppressed hepatocellular carcinoma growth in xenograft and DEN-treated mouse models[36].
Engineered cellular therapies with manipulation of HNF4α have been explored in few studies. Conditioned media of mesenchymal stem cells (MSCs) stably expressing HNF4α inhibited proliferation of SK-Hep-1 and HepG2 cells in culture, and intravenous injection of HNF4α-expressing MSCs into nude mice xenograft models reduced tumor size[85]. Peritoneal injection of immortalized hepatocytes overex-pressing HNF4α improved survival and serologic liver enzyme markers in a D-galactosamine rat model of acute liver failure, as compared to immortalized hepatocyte controls[86].
CONCLUSION
HNF4α is a highly conserved member of nuclear receptors superfamily of ligand-dependent transcription factors that is expressed in liver, stomach, intestine, pancreas and kidney. HNF4α is known for its role in the liver where it is a master regulator of liver-specific gene expression and essential for adult and fetal liver function (Figure 1). Dysregulation of HNF4α transcriptional activity is linked to several pathological disorders, such as liver cirrhosis, hepatocellular carcinoma, Maturity Onset Diabetes of the Young 1 (MODY1), colitis and colon cancer. Although there are growing evidences for the role of different HNF4α isoforms in the pathogenesis of these diseases, the exact molecular epidemiology and the molecular mechanisms involved are yet to be established. It is anticipated that the identification of specific interacting partners associate with these isoforms in each disease state is essential for differential expression of target genes, and hence signaling pathways. In turns, these targets could be used as diagnostic tools and for the treatment of diseases liked to transcriptional dysregulation of HNF4α.
Footnotes
Conflict-of-interest statement: No potential conflicts of interest. No financial support.
Manuscript source: Invited manuscript
Peer-review started: March 26, 2019
First decision: April 30, 2019
Article in press: July 19, 2019
Specialty type: Gastroenterology and hepatology
Country of origin: United States
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ding MX, Tuna N S-Editor: Gong ZM L-Editor: A E-Editor: Wu YXJ
Contributor Information
Matthew M Yeh, Department of Pathology, University of Washington School of Medicine, Seattle, WA 98195, United States.
Dustin E Bosch, Department of Medicine, University of Washington School of Medicine, Seattle, WA 98195, United States.
Sayed S Daoud, Department of Pharmaceutical Sciences, Washington State University Health Sciences, Spokane, WA 99210, United States. daoud@wsu.edu.
References
- 1.Babeu JP, Boudreau F. Hepatocyte nuclear factor 4-alpha involvement in liver and intestinal inflammatory networks. World J Gastroenterol. 2014;20:22–30. doi: 10.3748/wjg.v20.i1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chellappa K, Deol P, Evans JR, Vuong LM, Chen G, Briançon N, Bolotin E, Lytle C, Nair MG, Sladek FM. Opposing roles of nuclear receptor HNF4α isoforms in colitis and colitis-associated colon cancer. Elife. 2016;5 doi: 10.7554/eLife.10903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen WS, Manova K, Weinstein DC, Duncan SA, Plump AS, Prezioso VR, Bachvarova RF, Darnell JE., Jr Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev. 1994;8:2466–2477. doi: 10.1101/gad.8.20.2466. [DOI] [PubMed] [Google Scholar]
- 4.Pentney RJ, Gratton A. Effects of local delta and mu opioid receptor activation on basal and stimulated dopamine release in striatum and nucleus accumbens of rat: an in vivo electrochemical study. Neuroscience. 1991;45:95–102. doi: 10.1016/0306-4522(91)90106-x. [DOI] [PubMed] [Google Scholar]
- 5.Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G, Kaestner KH, Rossi JM, Zaret KS, Duncan SA. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet. 2003;34:292–296. doi: 10.1038/ng1175. [DOI] [PubMed] [Google Scholar]
- 6.Hu X, Xie P, Li W, Li Z, Shan H. Direct induction of hepatocyte-like cells from immortalized human bone marrow mesenchymal stem cells by overexpression of HNF4α. Biochem Biophys Res Commun. 2016;478:791–797. doi: 10.1016/j.bbrc.2016.08.026. [DOI] [PubMed] [Google Scholar]
- 7.Lu H. Crosstalk of HNF4α with extracellular and intracellular signaling pathways in the regulation of hepatic metabolism of drugs and lipids. Acta Pharm Sin B. 2016;6:393–408. doi: 10.1016/j.apsb.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwon EJ, Lee HA, You YA, Park H, Cho SJ, Ha EH, Kim YJ. DNA methylations of MC4R and HNF4α are associated with increased triglyceride levels in cord blood of preterm infants. Medicine (Baltimore) 2016;95:e4590. doi: 10.1097/MD.0000000000004590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon EJ, You YA, Park B, Ha EH, Kim HS, Park H, Kim YJ. Association between the DNA methylations of POMC, MC4R, and HNF4A and metabolic profiles in the blood of children aged 7-9 years. BMC Pediatr. 2018;18:121. doi: 10.1186/s12887-018-1104-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo S, Lu H. Conjunction of potential G-quadruplex and adjacent cis-elements in the 5' UTR of hepatocyte nuclear factor 4-alpha strongly inhibit protein expression. Sci Rep. 2017;7:17444. doi: 10.1038/s41598-017-17629-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vető B, Bojcsuk D, Bacquet C, Kiss J, Sipeki S, Martin L, Buday L, Bálint BL, Arányi T. The transcriptional activity of hepatocyte nuclear factor 4 alpha is inhibited via phosphorylation by ERK1/2. PLoS One. 2017;12:e0172020. doi: 10.1371/journal.pone.0172020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Viollet B, Kahn A, Raymondjean M. Protein kinase A-dependent phosphorylation modulates DNA-binding activity of hepatocyte nuclear factor 4. Mol Cell Biol. 1997;17:4208–4219. doi: 10.1128/mcb.17.8.4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang CF, Wen LZ, Yin C, Xu WP, Shi B, Zhang X, Xie WF. Apoptosis signal-regulating kinase 1 mediates the inhibitory effect of hepatocyte nuclear factor-4α on hepatocellular carcinoma. Oncotarget. 2016;7:27408–27421. doi: 10.18632/oncotarget.8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simó R, Barbosa-Desongles A, Hernandez C, Selva DM. IL1β down-regulation of sex hormone-binding globulin production by decreasing HNF-4α via MEK-1/2 and JNK MAPK pathways. Mol Endocrinol. 2012;26:1917–1927. doi: 10.1210/me.2012-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bauzá G, Miller G, Kaseje N, Wang Z, Sherburne A, Agarwal S, Burke PA. Injury-induced changes in liver specific transcription factors HNF-1α and HNF-4α. J Surg Res. 2012;175:298–304. doi: 10.1016/j.jss.2011.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berasain C, Avila MA. Regulation of hepatocyte identity and quiescence. Cell Mol Life Sci. 2015;72:3831–3851. doi: 10.1007/s00018-015-1970-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J, Xu Y, Li Y, Jadhav K, You M, Yin L, Zhang Y. Carboxylesterase 1 Is Regulated by Hepatocyte Nuclear Factor 4α and Protects Against Alcohol- and MCD diet-induced Liver Injury. Sci Rep. 2016;6:24277. doi: 10.1038/srep24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Zalzala M, Xu J, Li Y, Yin L, Zhang Y. A metabolic stress-inducible miR-34a-HNF4α pathway regulates lipid and lipoprotein metabolism. Nat Commun. 2015;6:7466. doi: 10.1038/ncomms8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonzo JA, Ferry CH, Matsubara T, Kim JH, Gonzalez FJ. Suppression of hepatocyte proliferation by hepatocyte nuclear factor 4α in adult mice. J Biol Chem. 2012;287:7345–7356. doi: 10.1074/jbc.M111.334599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baciu C, Pasini E, Angeli M, Schwenger K, Afrin J, Humar A, Fischer S, Patel K, Allard J, Bhat M. Systematic integrative analysis of gene expression identifies HNF4A as the central gene in pathogenesis of non-alcoholic steatohepatitis. PLoS One. 2017;12:e0189223. doi: 10.1371/journal.pone.0189223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu D, Chen G, Pan M, Zhang J, He W, Liu Y, Nian X, Sheng L, Xu B. High fat diet-induced oxidative stress blocks hepatocyte nuclear factor 4α and leads to hepatic steatosis in mice. J Cell Physiol. 2018;233:4770–4782. doi: 10.1002/jcp.26270. [DOI] [PubMed] [Google Scholar]
- 23.Dillon ST, Bhasin MK, Feng X, Koh DW, Daoud SS. Quantitative proteomic analysis in HCV-induced HCC reveals sets of proteins with potential significance for racial disparity. J Transl Med. 2013;11:239. doi: 10.1186/1479-5876-11-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ioannou GN, Dominitz JA, Weiss NS, Heagerty PJ, Kowdley KV. Racial differences in the relationship between hepatitis C infection and iron stores. Hepatology. 2003;37:795–801. doi: 10.1053/jhep.2003.50147. [DOI] [PubMed] [Google Scholar]
- 25.Samantray J, Zambare S, Seyoum B, Abou-Samra AB. Glucose control and lipid metabolism in African American patients with type 2 diabetes mellitus and chronic hepatitis C viral infection. Endocr Pract. 2011;17:363–368. doi: 10.4158/EP10175.OR. [DOI] [PubMed] [Google Scholar]
- 26.Mogilenko DA, Dizhe EB, Shavva VS, Lapikov IA, Orlov SV, Perevozchikov AP. Role of the nuclear receptors HNF4 alpha, PPAR alpha, and LXRs in the TNF alpha-mediated inhibition of human apolipoprotein A-I gene expression in HepG2 cells. Biochemistry. 2009;48:11950–11960. doi: 10.1021/bi9015742. [DOI] [PubMed] [Google Scholar]
- 27.Yeh MM, Boukhar S, Roberts B, Dasgupta N, Daoud SS. Genomic variants link to hepatitis C racial disparities. Oncotarget. 2017;8:59455–59475. doi: 10.18632/oncotarget.19755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Desai SS, Tung JC, Zhou VX, Grenert JP, Malato Y, Rezvani M, Español-Suñer R, Willenbring H, Weaver VM, Chang TT. Physiological ranges of matrix rigidity modulate primary mouse hepatocyte function in part through hepatocyte nuclear factor 4 alpha. Hepatology. 2016;64:261–275. doi: 10.1002/hep.28450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yue HY, Yin C, Hou JL, Zeng X, Chen YX, Zhong W, Hu PF, Deng X, Tan YX, Zhang JP, Ning BF, Shi J, Zhang X, Wang HY, Lin Y, Xie WF. Hepatocyte nuclear factor 4alpha attenuates hepatic fibrosis in rats. Gut. 2010;59:236–246. doi: 10.1136/gut.2008.174904. [DOI] [PubMed] [Google Scholar]
- 30.Nishikawa T, Bell A, Brooks JM, Setoyama K, Melis M, Han B, Fukumitsu K, Handa K, Tian J, Kaestner KH, Vodovotz Y, Locker J, Soto-Gutierrez A, Fox IJ. Resetting the transcription factor network reverses terminal chronic hepatic failure. J Clin Invest. 2015;125:1533–1544. doi: 10.1172/JCI73137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai WY, Lin LY, Hao H, Zhang SM, Ma F, Hong XX, Zhang H, Liu QF, Ye GD, Sun GB, Liu YJ, Li SN, Xie YY, Cai JC, Li BA. Yes-associated protein/TEA domain family member and hepatocyte nuclear factor 4-alpha (HNF4α) repress reciprocally to regulate hepatocarcinogenesis in rats and mice. Hepatology. 2017;65:1206–1221. doi: 10.1002/hep.28911. [DOI] [PubMed] [Google Scholar]
- 32.Cai SH, Lu SX, Liu LL, Zhang CZ, Yun JP. Increased expression of hepatocyte nuclear factor 4 alpha transcribed by promoter 2 indicates a poor prognosis in hepatocellular carcinoma. Therap Adv Gastroenterol. 2017;10:761–771. doi: 10.1177/1756283X17725998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lazarevich NL, Shavochkina DA, Fleishman DI, Kustova IF, Morozova OV, Chuchuev ES, Patyutko YI. Deregulation of hepatocyte nuclear factor 4 (HNF4)as a marker of epithelial tumors progression. Exp Oncol. 2010;32:167–171. [PubMed] [Google Scholar]
- 34.Kitao A, Matsui O, Yoneda N, Kozaka K, Kobayashi S, Koda W, Minami T, Inoue D, Yoshida K, Yamashita T, Yamashita T, Kaneko S, Takamura H, Ohta T, Ikeda H, Sato Y, Nakanuma Y, Harada K, Kita R, Gabata T. Gadoxetic acid-enhanced magnetic resonance imaging reflects co-activation of β-catenin and hepatocyte nuclear factor 4α in hepatocellular carcinoma. Hepatol Res. 2018;48:205–216. doi: 10.1111/hepr.12911. [DOI] [PubMed] [Google Scholar]
- 35.Vallianou I, Dafou D, Vassilaki N, Mavromara P, Hadzopoulou-Cladaras M. Hepatitis C virus suppresses Hepatocyte Nuclear Factor 4 alpha, a key regulator of hepatocellular carcinoma. Int J Biochem Cell Biol. 2016;78:315–326. doi: 10.1016/j.biocel.2016.07.027. [DOI] [PubMed] [Google Scholar]
- 36.Hatziapostolou M, Polytarchou C, Aggelidou E, Drakaki A, Poultsides GA, Jaeger SA, Ogata H, Karin M, Struhl K, Hadzopoulou-Cladaras M, Iliopoulos D. An HNF4α-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. 2011;147:1233–1247. doi: 10.1016/j.cell.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Q, Pu M, Zhao G, Dai B, Bian Z, Tang H, Chen C, Liu W, Qu X, Shen L, Tao K. Tg737 regulates epithelial-mesenchymal transition and cancer stem cell properties via a negative feedback circuit between Snail and HNF4α during liver stem cell malignant transformation. Cancer Lett. 2017;402:52–60. doi: 10.1016/j.canlet.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 38.Walesky C, Gunewardena S, Terwilliger EF, Edwards G, Borude P, Apte U. Hepatocyte-specific deletion of hepatocyte nuclear factor-4α in adult mice results in increased hepatocyte proliferation. Am J Physiol Gastrointest Liver Physiol. 2013;304:G26–G37. doi: 10.1152/ajpgi.00064.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin C, Lin Y, Zhang X, Chen YX, Zeng X, Yue HY, Hou JL, Deng X, Zhang JP, Han ZG, Xie WF. Differentiation therapy of hepatocellular carcinoma in mice with recombinant adenovirus carrying hepatocyte nuclear factor-4alpha gene. Hepatology. 2008;48:1528–1539. doi: 10.1002/hep.22510. [DOI] [PubMed] [Google Scholar]
- 40.Lazarevich NL, Cheremnova OA, Varga EV, Ovchinnikov DA, Kudrjavtseva EI, Morozova OV, Fleishman DI, Engelhardt NV, Duncan SA. Progression of HCC in mice is associated with a downregulation in the expression of hepatocyte nuclear factors. Hepatology. 2004;39:1038–1047. doi: 10.1002/hep.20155. [DOI] [PubMed] [Google Scholar]
- 41.Ning BF, Ding J, Yin C, Zhong W, Wu K, Zeng X, Yang W, Chen YX, Zhang JP, Zhang X, Wang HY, Xie WF. Hepatocyte nuclear factor 4 alpha suppresses the development of hepatocellular carcinoma. Cancer Res. 2010;70:7640–7651. doi: 10.1158/0008-5472.CAN-10-0824. [DOI] [PubMed] [Google Scholar]
- 42.Tao NN, Gong R, Chen X, He L, Ren F, Yu HB, Chen J, Ren JH. Interleukin-35 stimulates hepatitis B virus transcription and replication by targeting transcription factor HNF4α. J Gen Virol. 2018;99:645–654. doi: 10.1099/jgv.0.001050. [DOI] [PubMed] [Google Scholar]
- 43.Bai L, Nong Y, Shi Y, Liu M, Yan L, Shang J, Huang F, Lin Y, Tang H. Luteolin Inhibits Hepatitis B Virus Replication through Extracellular Signal-Regulated Kinase-Mediated Down-Regulation of Hepatocyte Nuclear Factor 4α Expression. Mol Pharm. 2016;13:568–577. doi: 10.1021/acs.molpharmaceut.5b00789. [DOI] [PubMed] [Google Scholar]
- 44.Tang H, McLachlan A. Transcriptional regulation of hepatitis B virus by nuclear hormone receptors is a critical determinant of viral tropism. Proc Natl Acad Sci USA. 2001;98:1841–1846. doi: 10.1073/pnas.041479698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He F, Chen EQ, Liu L, Zhou TY, Liu C, Cheng X, Liu FJ, Tang H. Inhibition of hepatitis B Virus replication by hepatocyte nuclear factor 4-alpha specific short hairpin RNA. Liver Int. 2012;32:742–751. doi: 10.1111/j.1478-3231.2011.02748.x. [DOI] [PubMed] [Google Scholar]
- 46.Liu H, Lou G, Li C, Wang X, Cederbaum AI, Gan L, Xie B. HBx inhibits CYP2E1 gene expression via downregulating HNF4α in human hepatoma cells. PLoS One. 2014;9:e107913. doi: 10.1371/journal.pone.0107913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu Q, Liu HO, Liu YD, Liu WS, Pan D, Zhang WJ, Yang L, Fu Q, Xu JJ, Gu JX. Decreased expression of hepatocyte nuclear factor 4α (Hnf4α)/microRNA-122 (miR-122) axis in hepatitis B virus-associated hepatocellular carcinoma enhances potential oncogenic GALNT10 protein activity. J Biol Chem. 2015;290:1170–1185. doi: 10.1074/jbc.M114.601203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chandra V, Holla P, Ghosh D, Chakrabarti D, Padigaru M, Jameel S. The hepatitis E virus ORF3 protein regulates the expression of liver-specific genes by modulating localization of hepatocyte nuclear factor 4. PLoS One. 2011;6:e22412. doi: 10.1371/journal.pone.0022412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li M, Tang Y, Wu L, Mo F, Wang X, Li H, Qi R, Zhang H, Srivastava A, Ling C. The hepatocyte-specific HNF4α/miR-122 pathway contributes to iron overload-mediated hepatic inflammation. Blood. 2017;130:1041–1051. doi: 10.1182/blood-2016-12-755967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garrison WD, Battle MA, Yang C, Kaestner KH, Sladek FM, Duncan SA. Hepatocyte nuclear factor 4alpha is essential for embryonic development of the mouse colon. Gastroenterology. 2006;130:1207–1220. doi: 10.1053/j.gastro.2006.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stegmann A, Hansen M, Wang Y, Larsen JB, Lund LR, Ritié L, Nicholson JK, Quistorff B, Simon-Assmann P, Troelsen JT, Olsen J. Metabolome, transcriptome, and bioinformatic cis-element analyses point to HNF-4 as a central regulator of gene expression during enterocyte differentiation. Physiol Genomics. 2006;27:141–155. doi: 10.1152/physiolgenomics.00314.2005. [DOI] [PubMed] [Google Scholar]
- 52.Davison JM, Lickwar CR, Song L, Breton G, Crawford GE, Rawls JF. Microbiota regulate intestinal epithelial gene expression by suppressing the transcription factor Hepatocyte nuclear factor 4 alpha. Genome Res. 2017;27:1195–1206. doi: 10.1101/gr.220111.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.UK IBD Genetics Consortium. Barrett JC, Lee JC, Lees CW, Prescott NJ, Anderson CA, Phillips A, Wesley E, Parnell K, Zhang H, Drummond H, Nimmo ER, Massey D, Blaszczyk K, Elliott T, Cotterill L, Dallal H, Lobo AJ, Mowat C, Sanderson JD, Jewell DP, Newman WG, Edwards C, Ahmad T, Mansfield JC, Satsangi J, Parkes M, Mathew CG; Wellcome Trust Case Control Consortium 2, Donnelly P, Peltonen L, Blackwell JM, Bramon E, Brown MA, Casas JP, Corvin A, Craddock N, Deloukas P, Duncanson A, Jankowski J, Markus HS, Mathew CG, McCarthy MI, Palmer CN, Plomin R, Rautanen A, Sawcer SJ, Samani N, Trembath RC, Viswanathan AC, Wood N, Spencer CC, Barrett JC, Bellenguez C, Davison D, Freeman C, Strange A, Donnelly P, Langford C, Hunt SE, Edkins S, Gwilliam R, Blackburn H, Bumpstead SJ, Dronov S, Gillman M, Gray E, Hammond N, Jayakumar A, McCann OT, Liddle J, Perez ML, Potter SC, Ravindrarajah R, Ricketts M, Waller M, Weston P, Widaa S, Whittaker P, Deloukas P, Peltonen L, Mathew CG, Blackwell JM, Brown MA, Corvin A, McCarthy MI, Spencer CC, Attwood AP, Stephens J, Sambrook J, Ouwehand WH, McArdle WL, Ring SM, Strachan DP. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet. 2009;41:1330–1334. doi: 10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Sommeren S, Visschedijk MC, Festen EA, de Jong DJ, Ponsioen CY, Wijmenga C, Weersma RK. HNF4α and CDH1 are associated with ulcerative colitis in a Dutch cohort. Inflamm Bowel Dis. 2011;17:1714–1718. doi: 10.1002/ibd.21541. [DOI] [PubMed] [Google Scholar]
- 55.Marcil V, Sinnett D, Seidman E, Boudreau F, Gendron FP, Beaulieu JF, Menard D, Lambert M, Bitton A, Sanchez R, Amre D, Levy E. Association between genetic variants in the HNF4A gene and childhood-onset Crohn's disease. Genes Immun. 2012;13:556–565. doi: 10.1038/gene.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahn SH, Shah YM, Inoue J, Morimura K, Kim I, Yim S, Lambert G, Kurotani R, Nagashima K, Gonzalez FJ, Inoue Y. Hepatocyte nuclear factor 4alpha in the intestinal epithelial cells protects against inflammatory bowel disease. Inflamm Bowel Dis. 2008;14:908–920. doi: 10.1002/ibd.20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Darsigny M, Babeu JP, Dupuis AA, Furth EE, Seidman EG, Lévy E, Verdu EF, Gendron FP, Boudreau F. Loss of hepatocyte-nuclear-factor-4alpha affects colonic ion transport and causes chronic inflammation resembling inflammatory bowel disease in mice. PLoS One. 2009;4:e7609. doi: 10.1371/journal.pone.0007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muthusamy S, Jeong JJ, Cheng M, Bonzo JA, Kumar A, Gonzalez FJ, Borthakur A, Dudeja PK, Saksena S, Malakooti J. Hepatocyte nuclear factor 4α regulates the expression of intestinal epithelial Na+/H+ exchanger isoform 3. Am J Physiol Gastrointest Liver Physiol. 2018;314:G14–G21. doi: 10.1152/ajpgi.00225.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tanaka T, Jiang S, Hotta H, Takano K, Iwanari H, Sumi K, Daigo K, Ohashi R, Sugai M, Ikegame C, Umezu H, Hirayama Y, Midorikawa Y, Hippo Y, Watanabe A, Uchiyama Y, Hasegawa G, Reid P, Aburatani H, Hamakubo T, Sakai J, Naito M, Kodama T. Dysregulated expression of P1 and P2 promoter-driven hepatocyte nuclear factor-4alpha in the pathogenesis of human cancer. J Pathol. 2006;208:662–672. doi: 10.1002/path.1928. [DOI] [PubMed] [Google Scholar]
- 60.Chellappa K, Jankova L, Schnabl JM, Pan S, Brelivet Y, Fung CL, Chan C, Dent OF, Clarke SJ, Robertson GR, Sladek FM. Src tyrosine kinase phosphorylation of nuclear receptor HNF4α correlates with isoform-specific loss of HNF4α in human colon cancer. Proc Natl Acad Sci USA. 2012;109:2302–2307. doi: 10.1073/pnas.1106799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moore BD, Khurana SS, Huh WJ, Mills JC. Hepatocyte nuclear factor 4α is required for cell differentiation and homeostasis in the adult mouse gastric epithelium. Am J Physiol Gastrointest Liver Physiol. 2016;311:G267–G275. doi: 10.1152/ajpgi.00195.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang HR, Nam S, Kook MC, Kim KT, Liu X, Yao H, Jung HR, Lemos R, Jr, Seo HH, Park HS, Gim Y, Hong D, Huh I, Kim YW, Tan D, Liu CG, Powis G, Park T, Liang H, Kim YH. HNF4α is a therapeutic target that links AMPK to WNT signalling in early-stage gastric cancer. Gut. 2016;65:19–32. doi: 10.1136/gutjnl-2014-307918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Colleypriest BJ, Burke ZD, Griffiths LP, Chen Y, Yu WY, Jover R, Bock M, Biddlestone L, Quinlan JM, Ward SG, Mark Farrant J, Slack JM, Tosh D. Hnf4α is a key gene that can generate columnar metaplasia in oesophageal epithelium. Differentiation. 2017;93:39–49. doi: 10.1016/j.diff.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J, Qin R, Ma Y, Wu H, Peters H, Tyska M, Shaheen NJ, Chen X. Differential gene expression in normal esophagus and Barrett's esophagus. J Gastroenterol. 2009;44:897–911. doi: 10.1007/s00535-009-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moore BD, Jin RU, Lo H, Jung M, Wang H, Battle MA, Wollheim CB, Urano F, Mills JC. Transcriptional Regulation of X-Box-binding Protein One (XBP1) by Hepatocyte Nuclear Factor 4α (HNF4Α) Is Vital to Beta-cell Function. J Biol Chem. 2016;291:6146–6157. doi: 10.1074/jbc.M115.685750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Improda N, Shah P, Güemes M, Gilbert C, Morgan K, Sebire N, Bockenhauer D, Hussain K. Hepatocyte Nuclear Factor-4 Alfa Mutation Associated with Hyperinsulinaemic Hypoglycaemia and Atypical Renal Fanconi Syndrome: Expanding the Clinical Phenotype. Horm Res Paediatr. 2016;86:337–341. doi: 10.1159/000446396. [DOI] [PubMed] [Google Scholar]
- 67.Bacon S, Kyithar MP, Condron EM, Vizzard N, Burke M, Byrne MM. Prolonged episodes of hypoglycaemia in HNF4A-MODY mutation carriers with IGT. Evidence of persistent hyperinsulinism into early adulthood. Acta Diabetol. 2016;53:965–972. doi: 10.1007/s00592-016-0890-9. [DOI] [PubMed] [Google Scholar]
- 68.Stanik J, Skopkova M, Brennerova K, Danis D, Rosolankova M, Salingova A, Bzduch V, Klimes I, Gasperikova D. Congenital hyperinsulinism and glycogenosis-like phenotype due to a novel HNF4A mutation. Diabetes Res Clin Pract. 2017;126:144–150. doi: 10.1016/j.diabres.2017.02.014. [DOI] [PubMed] [Google Scholar]
- 69.Thomas H, Jaschkowitz K, Bulman M, Frayling TM, Mitchell SM, Roosen S, Lingott-Frieg A, Tack CJ, Ellard S, Ryffel GU, Hattersley AT. A distant upstream promoter of the HNF-4alpha gene connects the transcription factors involved in maturity-onset diabetes of the young. Hum Mol Genet. 2001;10:2089–2097. doi: 10.1093/hmg/10.19.2089. [DOI] [PubMed] [Google Scholar]
- 70.Gupta RK, Vatamaniuk MZ, Lee CS, Flaschen RC, Fulmer JT, Matschinsky FM, Duncan SA, Kaestner KH. The MODY1 gene HNF-4alpha regulates selected genes involved in insulin secretion. J Clin Invest. 2005;115:1006–1015. doi: 10.1172/JCI200522365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miura A, Yamagata K, Kakei M, Hatakeyama H, Takahashi N, Fukui K, Nammo T, Yoneda K, Inoue Y, Sladek FM, Magnuson MA, Kasai H, Miyagawa J, Gonzalez FJ, Shimomura I. Hepatocyte nuclear factor-4alpha is essential for glucose-stimulated insulin secretion by pancreatic beta-cells. J Biol Chem. 2006;281:5246–5257. doi: 10.1074/jbc.M507496200. [DOI] [PubMed] [Google Scholar]
- 72.Ribel-Madsen R, Fraga MF, Jacobsen S, Bork-Jensen J, Lara E, Calvanese V, Fernandez AF, Friedrichsen M, Vind BF, Højlund K, Beck-Nielsen H, Esteller M, Vaag A, Poulsen P. Genome-wide analysis of DNA methylation differences in muscle and fat from monozygotic twins discordant for type 2 diabetes. PLoS One. 2012;7:e51302. doi: 10.1371/journal.pone.0051302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Einstein F, Thompson RF, Bhagat TD, Fazzari MJ, Verma A, Barzilai N, Greally JM. Cytosine methylation dysregulation in neonates following intrauterine growth restriction. PLoS One. 2010;5:e8887. doi: 10.1371/journal.pone.0008887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Deng J, Mueller M, Geng T, Shen Y, Liu Y, Hou P, Ramillapalli R, Taylor HS, Paidas M, Huang Y. H19 lncRNA alters methylation and expression of Hnf4α in the liver of metformin-exposed fetuses. Cell Death Dis. 2017;8:e3175. doi: 10.1038/cddis.2017.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Briançon N, Weiss MC. In vivo role of the HNF4alpha AF-1 activation domain revealed by exon swapping. EMBO J. 2006;25:1253–1262. doi: 10.1038/sj.emboj.7601021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun SI, Deng SI, Gustafsson S, Mägi R, Ganna A, Wheeler E, Feitosa MF, Justice AE, Monda KL, Croteau-Chonka DC, Day FR, Esko T, Fall T, Ferreira T, Gentilini D, Jackson AU, Luan J, Randall JC, Vedantam S, Willer CJ, Winkler TW, Wood AR, Workalemahu T, Hu YJ, Lee SH, Liang L, Lin DY, Min JL, Neale BM, Thorleifsson G, Yang J, Albrecht E, Amin N, Bragg-Gresham JL, Cadby G, den Heijer M, Eklund N, Fischer K, Goel A, Hottenga JJ, Huffman JE. Huffman JE, Jarick I, Johansson Å, Johnson T, Kanoni S, Kleber ME, König IR, Kristiansson K, Kutalik Z, Lamina C, Lecoeur C, Li G, Mangino M, McArdle WL, Medina-Gomez C, Müller-Nurasyid M, Ngwa JS, Nolte IM, Paternoster L, Pechlivanis S, Perola M, Peters MJ, Preuss M, Rose LM, Shi J, Shungin D, Smith AV, Strawbridge RJ, Surakka I, Teumer A, Trip MD, Tyrer J, Van Vliet-Ostaptchouk JV, Vandenput L, Waite LL, Zhao JH, Absher D, Asselbergs FW, Atalay M, Attwood AP, Balmforth AJ, Basart H, Beilby J, Bonnycastle LL, Brambilla P, Bruinenberg M, Campbell H, Chasman DI, Chines PS, Collins FS, Connell JM, Cookson WO, de Faire U, de Vegt F, Dei M, Dimitriou M, Edkins S, Estrada K, Evans DM, Farrall M, Ferrario MM, Ferrières J, Franke L, Frau F, Gejman PV, Grallert H, Grönberg H, Gudnason V, Hall AS, Hall P, Hartikainen AL, Hayward C, Heard-Costa NL, Heath AC, Hebebrand J, Homuth G, Hu FB, Hunt SE, Hyppönen E, Iribarren C, Jacobs KB, Jansson JO, Jula A, Kähönen M, Kathiresan S, Kee F, Khaw KT, Kivimäki M, Koenig W, Kraja AT, Kumari M, Kuulasmaa K, Kuusisto J, Laitinen JH, Lakka TA, Langenberg C, Launer LJ, Lind L, Lindström J, Liu J, Liuzzi A, Lokki ML, Lorentzon M, Madden PA, Magnusson PK, Manunta P, Marek D, März W, Mateo Leach I, McKnight B, Medland SE, Mihailov E, Milani L, Montgomery GW, Mooser V, Mühleisen TW, Munroe PB, Musk AW, Narisu N, Navis G, Nicholson G, Nohr EA, Ong KK, Oostra BA, Palmer CN, Palotie A, Peden JF, Pedersen N, Peters A, Polasek O, Pouta A, Pramstaller PP, Prokopenko I, Pütter C, Radhakrishnan A, Raitakari O, Rendon A, Rivadeneira F, Rudan I, Saaristo TE, Sambrook JG, Sanders AR, Sanna S, Saramies J, Schipf S, Schreiber S, Schunkert H, Shin SY, Signorini S, Sinisalo J, Skrobek B, Soranzo N, Stančáková A, Stark K, Stephens JC, Stirrups K, Stolk RP, Stumvoll M, Swift AJ, Theodoraki EV, Thorand B, Tregouet DA, Tremoli E, Van der Klauw MM, van Meurs JB, Vermeulen SH, Viikari J, Virtamo J, Vitart V, Waeber G, Wang Z, Widén E, Wild SH, Willemsen G, Winkelmann BR, Witteman JC, Wolffenbuttel BH, Wong A, Wright AF, Zillikens MC, Amouyel P, Boehm BO, Boerwinkle E, Boomsma DI, Caulfield MJ, Chanock SJ, Cupples LA, Cusi D, Dedoussis GV, Erdmann J, Eriksson JG, Franks PW, Froguel P, Gieger C, Gyllensten U, Hamsten A, Harris TB, Hengstenberg C, Hicks AA, Hingorani A, Hinney A, Hofman A, Hovingh KG, Hveem K, Illig T, Jarvelin MR, Jöckel KH, Keinanen-Kiukaanniemi SM, Kiemeney LA, Kuh D, Laakso M, Lehtimäki T, Levinson DF, Martin NG, Metspalu A, Morris AD, Nieminen MS, Njølstad I, Ohlsson C, Oldehinkel AJ, Ouwehand WH, Palmer LJ, Penninx B, Power C, Province MA, Psaty BM, Qi L, Rauramaa R, Ridker PM, Ripatti S, Salomaa V, Samani NJ, Snieder H, Sørensen TI, Spector TD, Stefansson K, Tönjes A, Tuomilehto J, Uitterlinden AG, Uusitupa M, van der Harst P, Vollenweider P, Wallaschofski H, Wareham NJ, Watkins H, Wichmann HE, Wilson JF, Abecasis GR, Assimes TL, Barroso I, Boehnke M, Borecki IB, Deloukas P, Fox CS, Frayling T, Groop LC, Haritunian T, Heid IM, Hunter D, Kaplan RC, Karpe F, Moffatt MF, Mohlke KL, O'Connell JR, Pawitan Y, Schadt EE, Schlessinger D, Steinthorsdottir V, Strachan DP, Thorsteinsdottir U, van Duijn CM, Visscher PM, Di Blasio AM, Hirschhorn JN, Lindgren CM, Morris AP, Meyre D, Scherag A, McCarthy MI, Speliotes EK, North KE, Loos RJ, Ingelsson E. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat Genet. 2013;45:501–512. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun Q, Xu W, Ji S, Qin Y, Liu W, Hu Q, Zhang Z, Liu M, Yu X, Xu X. Role of hepatocyte nuclear factor 4 alpha in cell proliferation and gemcitabine resistance in pancreatic adenocarcinoma. Cancer Cell Int. 2019;19:49. doi: 10.1186/s12935-019-0767-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wei S, Zhang M, Yu Y, Xue H, Lan X, Liu S, Hatch G, Chen L. HNF-4α regulated miR-122 contributes to development of gluconeogenesis and lipid metabolism disorders in Type 2 diabetic mice and in palmitate-treated HepG2 cells. Eur J Pharmacol. 2016;791:254–263. doi: 10.1016/j.ejphar.2016.08.038. [DOI] [PubMed] [Google Scholar]
- 79.Jucá PCFC, Corrêa S, Vignal GM, Accioly MTS, Lustosa SAS, Abdelhay E, Matos D. HNF4A expression as a potential diagnostic tool to discriminate primary gastric cancer from breast cancer metastasis in a Brazilian cohort. Diagn Pathol. 2017;12:43. doi: 10.1186/s13000-017-0635-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koyama T, Sekine S, Taniguchi H, Tsuda H, Ikegami M, Hano H, Kushima R. Hepatocyte nuclear factor 4A expression discriminates gastric involvement by metastatic breast carcinomas from primary gastric adenocarcinomas. Hum Pathol. 2011;42:1777–1784. doi: 10.1016/j.humpath.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 81.van der Post RS, Bult P, Vogelaar IP, Ligtenberg MJ, Hoogerbrugge N, van Krieken JH. HNF4A immunohistochemistry facilitates distinction between primary and metastatic breast and gastric carcinoma. Virchows Arch. 2014;464:673–679. doi: 10.1007/s00428-014-1574-x. [DOI] [PubMed] [Google Scholar]
- 82.Nakajima N, Yoshizawa A, Nakajima T, Hirata M, Furuhata A, Sumiyoshi S, Rokutan-Kurata M, Sonobe M, Menju T, Miyamoto E, Chen-Yoshikawa TF, Date H, Haga H. GATA6-positive lung adenocarcinomas are associated with invasive mucinous adenocarcinoma morphology, hepatocyte nuclear factor 4α expression, and KRAS mutations. Histopathology. 2018;73:38–48. doi: 10.1111/his.13500. [DOI] [PubMed] [Google Scholar]
- 83.Chen R, Feng Y, Wu J, Song Y, Li H, Shen Q, Li D, Zhang J, Lu Z, Xiao H, Zhang Y. Metformin attenuates angiotensin II-induced TGFβ1 expression by targeting hepatocyte nuclear factor-4-α. Br J Pharmacol. 2018;175:1217–1229. doi: 10.1111/bph.13753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Inoue J, Ikeda S, Kanayama T, Sato R. The flavonoid derivative 4'-nitro-6-hydroxyflavone suppresses the activity of HNF4α and stimulates the degradation of HNF4α protein through the activation of AMPK. Biosci Biotechnol Biochem. 2017;81:1548–1552. doi: 10.1080/09168451.2017.1325316. [DOI] [PubMed] [Google Scholar]
- 85.Kiselyuk A, Lee SH, Farber-Katz S, Zhang M, Athavankar S, Cohen T, Pinkerton AB, Ye M, Bushway P, Richardson AD, Hostetler HA, Rodriguez-Lee M, Huang L, Spangler B, Smith L, Higginbotham J, Cashman J, Freeze H, Itkin-Ansari P, Dawson MI, Schroeder F, Cang Y, Mercola M, Levine F. HNF4α antagonists discovered by a high-throughput screen for modulators of the human insulin promoter. Chem Biol. 2012;19:806–818. doi: 10.1016/j.chembiol.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu N, Zhang YL, Wang HT, Li DW, Dai HJ, Zhang QQ, Zhang J, Ma Y, Xia Q, Bian JM, Hang HL. Overexpression of hepatocyte nuclear factor 4α in human mesenchymal stem cells suppresses hepatocellular carcinoma development through Wnt/β-catenin signaling pathway downregulation. Cancer Biol Ther. 2016;17:558–565. doi: 10.1080/15384047.2016.1177675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hang HL, Liu XY, Wang HT, Xu N, Bian JM, Zhang JJ, Xia L, Xia Q. Hepatocyte nuclear factor 4A improves hepatic differentiation of immortalized adult human hepatocytes and improves liver function and survival. Exp Cell Res. 2017;360:81–93. doi: 10.1016/j.yexcr.2017.08.020. [DOI] [PubMed] [Google Scholar]
- 88.McCurdy CE, Bishop JM, Williams SM, Grayson BE, Smith MS, Friedman JE, Grove KL. Maternal high-fat diet triggers lipotoxicity in the fetal livers of nonhuman primates. J Clin Invest. 2009;119:323–335. doi: 10.1172/JCI32661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou Z, Kang X, Jiang Y, Song Z, Feng W, McClain CJ, Kang YJ. Preservation of hepatocyte nuclear factor-4alpha is associated with zinc protection against TNF-alpha hepatotoxicity in mice. Exp Biol Med (Maywood) 2007;232:622–628. [PubMed] [Google Scholar]
- 90.Kang X, Zhong W, Liu J, Song Z, McClain CJ, Kang YJ, Zhou Z. Zinc supplementation reverses alcohol-induced steatosis in mice through reactivating hepatocyte nuclear factor-4alpha and peroxisome proliferator-activated receptor-alpha. Hepatology. 2009;50:1241–1250. doi: 10.1002/hep.23090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Piccolo P, Annunziata P, Soria LR, Attanasio S, Barbato A, Castello R, Carissimo A, Quagliata L, Terracciano LM, Brunetti-Pierri N. Down-regulation of hepatocyte nuclear factor-4α and defective zonation in livers expressing mutant Z α1-antitrypsin. Hepatology. 2017;66:124–135. doi: 10.1002/hep.29160. [DOI] [PubMed] [Google Scholar]
- 92.Berasain C, Herrero JI, García-Trevijano ER, Avila MA, Esteban JI, Mato JM, Prieto J. Expression of Wilms' tumor suppressor in the liver with cirrhosis: relation to hepatocyte nuclear factor 4 and hepatocellular function. Hepatology. 2003;38:148–157. doi: 10.1053/jhep.2003.50269. [DOI] [PubMed] [Google Scholar]
- 93.Kalkuhl A, Kaestner K, Buchmann A, Schwarz M. Expression of hepatocyte-enriched nuclear transcription factors in mouse liver tumours. Carcinogenesis. 1996;17:609–612. doi: 10.1093/carcin/17.3.609. [DOI] [PubMed] [Google Scholar]
- 94.Li X, Jiang H, Qu L, Yao W, Cai H, Chen L, Peng T. Hepatocyte nuclear factor 4α and downstream secreted phospholipase A2 GXIIB regulate production of infectious hepatitis C virus. J Virol. 2014;88:612–627. doi: 10.1128/JVI.02068-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Qadri I, Iwahashi M, Kullak-Ublick GA, Simon FR. Hepatocyte nuclear factor (HNF) 1 and HNF4 mediate hepatic multidrug resistance protein 2 up-regulation during hepatitis C virus gene expression. Mol Pharmacol. 2006;70:627–636. doi: 10.1124/mol.106.023499. [DOI] [PubMed] [Google Scholar]
- 96.Wang Z, Ceniccola K, Florea L, Wang BD, Lee NH, Kumar A. Viral non-coding RNA inhibits HNF4α expression in HCV associated hepatocellular carcinoma. Infect Agent Cancer. 2015;10:19. doi: 10.1186/s13027-015-0014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sato Y, Tsuyama T, Sato C, Karim MF, Yoshizawa T, Inoue M, Yamagata K. Hypoxia reduces HNF4α/MODY1 protein expression in pancreatic β-cells by activating AMP-activated protein kinase. J Biol Chem. 2017;292:8716–8728. doi: 10.1074/jbc.M116.767574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Franke A, Hampe J, Rosenstiel P, Becker C, Wagner F, Häsler R, Little RD, Huse K, Ruether A, Balschun T, Wittig M, Elsharawy A, Mayr G, Albrecht M, Prescott NJ, Onnie CM, Fournier H, Keith T, Radelof U, Platzer M, Mathew CG, Stoll M, Krawczak M, Nürnberg P, Schreiber S. Systematic association mapping identifies NELL1 as a novel IBD disease gene. PLoS One. 2007;2:e691. doi: 10.1371/journal.pone.0000691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bollrath J, Phesse TJ, von Burstin VA, Putoczki T, Bennecke M, Bateman T, Nebelsiek T, Lundgren-May T, Canli O, Schwitalla S, Matthews V, Schmid RM, Kirchner T, Arkan MC, Ernst M, Greten FR. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell. 2009;15:91–102. doi: 10.1016/j.ccr.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 100.Kriegsmann M, Harms A, Longuespée R, Muley T, Winter H, Kriegsmann K, Kazdal D, Goeppert B, Pathil A, Warth A. Role of conventional immunomarkers, HNF4-α and SATB2, in the differential diagnosis of pulmonary and colorectal adenocarcinomas. Histopathology. 2018;72:997–1006. doi: 10.1111/his.13455. [DOI] [PubMed] [Google Scholar]
- 101.Ehehalt F, Rümmele P, Kersting S, Lang-Schwarz C, Rückert F, Hartmann A, Dietmaier W, Terracciano L, Aust DE, Jahnke B, Saeger HD, Pilarsky C, Grützmann R. Hepatocyte nuclear factor (HNF) 4α expression distinguishes ampullary cancer subtypes and prognosis after resection. Ann Surg. 2011;254:302–310. doi: 10.1097/SLA.0b013e31821994a8. [DOI] [PubMed] [Google Scholar]
- 102.Ma Y, Wei X, Wu Z. HNF-4α promotes multidrug resistance of gastric cancer cells through the modulation of cell apoptosis. Oncol Lett. 2017;14:6477–6484. doi: 10.3892/ol.2017.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sel S, Ebert T, Ryffel GU, Drewes T. Human renal cell carcinogenesis is accompanied by a coordinate loss of the tissue specific transcription factors HNF4 alpha and HNF1 alpha. Cancer Lett. 1996;101:205–210. doi: 10.1016/0304-3835(96)04136-5. [DOI] [PubMed] [Google Scholar]
- 104.Lenburg ME, Liou LS, Gerry NP, Frampton GM, Cohen HT, Christman MF. Previously unidentified changes in renal cell carcinoma gene expression identified by parametric analysis of microarray data. BMC Cancer. 2003;3:31. doi: 10.1186/1471-2407-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hanawa M, Takayama K, Sakurai F, Tachibana M, Mizuguchi H. Hepatocyte Nuclear Factor 4 Alpha Promotes Definitive Endoderm Differentiation from Human Induced Pluripotent Stem Cells. Stem Cell Rev. 2017;13:542–551. doi: 10.1007/s12015-016-9709-x. [DOI] [PubMed] [Google Scholar]
- 106.Shi W, Wang H, Zheng X, Jiang X, Xu Z, Shen H, Li M. HNF-4alpha Negatively Regulates Hepcidin Expression Through BMPR1A in HepG2 Cells. Biol Trace Elem Res. 2017;176:294–304. doi: 10.1007/s12011-016-0846-5. [DOI] [PubMed] [Google Scholar]
- 107.Morimoto A, Kannari M, Tsuchida Y, Sasaki S, Saito C, Matsuta T, Maeda T, Akiyama M, Nakamura T, Sakaguchi M, Nameki N, Gonzalez FJ, Inoue Y. An HNF4α-microRNA-194/192 signaling axis maintains hepatic cell function. J Biol Chem. 2017;292:10574–10585. doi: 10.1074/jbc.M117.785592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Walesky C, Edwards G, Borude P, Gunewardena S, O'Neil M, Yoo B, Apte U. Hepatocyte nuclear factor 4 alpha deletion promotes diethylnitrosamine-induced hepatocellular carcinoma in rodents. Hepatology. 2013;57:2480–2490. doi: 10.1002/hep.26251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chiba H, Gotoh T, Kojima T, Satohisa S, Kikuchi K, Osanai M, Sawada N. Hepatocyte nuclear factor (HNF)-4alpha triggers formation of functional tight junctions and establishment of polarized epithelial morphology in F9 embryonal carcinoma cells. Exp Cell Res. 2003;286:288–297. doi: 10.1016/s0014-4827(03)00116-2. [DOI] [PubMed] [Google Scholar]
- 110.Lussier CR, Babeu JP, Auclair BA, Perreault N, Boudreau F. Hepatocyte nuclear factor-4alpha promotes differentiation of intestinal epithelial cells in a coculture system. Am J Physiol Gastrointest Liver Physiol. 2008;294:G418–G428. doi: 10.1152/ajpgi.00418.2007. [DOI] [PubMed] [Google Scholar]
- 111.Sartor RB, Wu GD. Roles for Intestinal Bacteria, Viruses, and Fungi in Pathogenesis of Inflammatory Bowel Diseases and Therapeutic Approaches. Gastroenterology. 2017;152:327–339.e4. doi: 10.1053/j.gastro.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Babeu JP, Darsigny M, Lussier CR, Boudreau F. Hepatocyte nuclear factor 4alpha contributes to an intestinal epithelial phenotype in vitro and plays a partial role in mouse intestinal epithelium differentiation. Am J Physiol Gastrointest Liver Physiol. 2009;297:G124–G134. doi: 10.1152/ajpgi.90690.2008. [DOI] [PubMed] [Google Scholar]
- 113.Archer A, Sauvaget D, Chauffeton V, Bouchet PE, Chambaz J, Pinçon-Raymond M, Cardot P, Ribeiro A, Lacasa M. Intestinal apolipoprotein A-IV gene transcription is controlled by two hormone-responsive elements: a role for hepatic nuclear factor-4 isoforms. Mol Endocrinol. 2005;19:2320–2334. doi: 10.1210/me.2004-0462. [DOI] [PubMed] [Google Scholar]
- 114.Rieck S, Zhang J, Li Z, Liu C, Naji A, Takane KK, Fiaschi-Taesch NM, Stewart AF, Kushner JA, Kaestner KH. Overexpression of hepatocyte nuclear factor-4α initiates cell cycle entry, but is not sufficient to promote β-cell expansion in human islets. Mol Endocrinol. 2012;26:1590–1602. doi: 10.1210/me.2012-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]