Abstract
There is increasing recognition that the intestinal microbiota govern human well-being and prevent diseases. Intestinal colonization by antibiotic-resistant pathogens, however, can lead to the spread of resistance as well as serious infections. Extended-spectrum β-lactamase producing Enterobacteriaceae (ESBL-E) represent particularly dangerous pathogens, which are known to asymptomatically colonized the intestinal tract in the community. Here, we performed a 16s rRNA metagenomics sequence analysis to analyze differences in the microbiota composition between ESBL-E carriers and non-carriers in Thailand, where ESBL-E carriage rates are notoriously high. The most notable difference we detected was that the phylum Bacteroidetes, and in particular, the species Bacteroides uniformis, were significantly more abundant in ESBL-E non-carriers than carriers. The Shannon diversity index in non-carriers (5.10 ± 0.69) was also lower than that in ESBL-E carriers (5.39 ± 0.48) without statistical significance (p = 0.13). The overall beta diversity difference of the intestinal microbiota of ESBL-E carriers as compared to non-carriers was statistically significant (Adonis on weighted unifrac: R2=0.14, P =0.005). Furthermore, ESBL-E carriage was significantly lower in farmers than other occupations. Our findings suggest that a dynamic interaction exists between microbiota diversity and ESBL-E carriage, which is possibly driven by dietary composition and may be exploited using probiotic approaches to control the spread of ESBL-E.
Keywords: ESBL-E, metagenomics, intestinal microbiota, antibiotic resistance
1. Introduction
The human gastrointestinal tract is colonized by numerous bacteria. Some of them are opportunistic pathogens and may carry drug resistance genes [1]. This may lead to the rapid spread of drug-resistant pathogenic bacteria in the community. However, the prevalence of this worrisome intestinal carriage of drug-resistant bacteria is not well understood. Extended-spectrum β-lactamase-producing Enterobacteriaceae (ESBL-E) represent a particularly dangerous threat to public health, because these bacteria are resistant to most oxyimino-cephalosporins (e.g. ceftriaxone, cefotaxime and ceftazidime). Escherichia coli and Klebsiella pneumoniae are among the most frequent ESBL-producing isolates and resistance rates in these potentially highly dangerous pathogens are on the rise in particular in Asia [2, 3]. Many recent studies have reported that the healthy population is asymptomatically colonized with ESBL-E, with colonization frequencies ranging from low single-digit percentage rates, for example in the U.S. and Western Europe [4–6], to much higher frequencies, for example ~ 34% in Turkey [7], ~ 30% in Shanghai, China [8], and over 50% in some African and Asian countries including Thailand [9–11]. Due to the high ESBL-E colonization rates in those latter countries, travel to (sub)tropical regions has been recognized as a risk for acquiring ESBL-E colonization [12].
The intestinal microbiota provide an important host defense mechanism by inhibiting the colonization of potentially pathogenic microorganisms either via depletion of nutrients or production of inhibitory substances or conditions that can inhibit the growth of the invading pathogens. For example, Pultz et al. have shown that colonization with vancomycin-resistant enterococci (VRE) was inhibited by anaerobic microbiota in the colon, which by depleting nutrients within cecal contents limit the association of VRE with the mucus layer [13]. This phenomenon, termed “colonization resistance”, has the potential to be applied in a therapeutic probiotic manner to the prevention of overgrowth of indigenous pathogens and the inhibition of exogenously introduced foreign organisms [14].
Despite the increasing recognition of the role of the intestinal microbiota in controlling the colonization of specific pathogenic and antibiotic-resistant bacteria, there has been almost no effort yet to investigate this role regarding colonization by ESBL-E. To our knowledge, only one study has addressed this question. That study analyzed the population of a remote and isolated village in French Guyana with a very low ESBL-E colonization rate and found increased prevalence in non-ESBL-E carriers of four non-related genera, two of which belong to the Clostridiales [15].
Here, to address ESBL-E colonization as a global public health risk, we analyzed the microbiome of ESBL-E carriers versus non-carriers in Thailand, which is known to have high ESBL-E colonization rates. Furthermore, we performed the study on several rural community populations from the entire country to rule out an impact of potentially specific conditions endemic to geographically limited areas.
2. Materials and Methods
2.1. Fecal sample collection
Fecal samples of 200 healthy volunteers living in rural areas in the provinces of Chiang-Mai, Chaiyaphum, Ratchaburi and Surat-Thani, Thailand were collected. The four provinces were selected to represent geographically different regions of the country (Figure 1). All participants were over 20 years old, gave informed written consent to participate in the study (Approval No. Si773/2015, Siriraj Hospital), and had no history of gastrointestinal disease. They had neither received antibacterial treatment nor were hospitalized within the last three months prior to participating in this study.
Figure 1. Location of research sites.
2.2. ESBL–producing Enterobacteriaceae screening
Fecal samples were plated on both MacConkey agar supplemented with 1 mg/L cefotaxime (CTX-MacConkey) and MacConkey agar supplemented with 1 mg/L ceftazidime (CTZ-MacConkey), and then incubated at 37 °C for 16–18 hours. Bacterial colonies grown from both plates were confirmed for ESBL production by a disk diffusion method using cefotaxime and ceftazidime with and without clavulanic acid according to CLSI guidelines [16]. ESBL-producing isolates were then identified on the species level using MALDI-TOF MS analysis on a Bruker Microflex LT/SH instrument according to the manufacturer’s protocol.
2.3. DNA extraction and 16s rRNA gene sequencing
Genomic DNA was extracted using QIAamp DNA stool Minikits (Qiagen) according to the manufacturer’s instructions, amplifying the V4 region of the 16S rRNA genes. The DNA was quantified and processed for 16s rRNA paired-end sequencing, which was performed by Illumina (San Diego, California) using the Illumina MiSeq system as previously described [17].
2.4. Taxonomic, diversity and statistical analysis
All MiSeq sequences were processed using the 16S QIIME Paired-End pipeline implemented in the Nephele platform of the National Institute of Allergy and Infectious Diseases (NIAID) (release 1.6, which uses QIIME 1.9.1) [18]. Operational Taxonomic Units (OTUs) were picked with QIIME’s uclust-based [19] open-reference OTU picking protocol [20] and the taxonomic assignment was performed against the Greengenes 13_8 reference sequence set [21] at 99% similarity. Alpha diversity was calculated by using Chao1 and Shannon [22] and compared across groups with a nonparametric t test with 999 permutations. Beta diversity calculations were performed with QIIME’s implementations of weighted and unweighted UniFrac [23, 24] by using exactly 36146 randomly selected sequences per sample. In addition, principle coordinate analysis (PCoA) was performed to compare groups of samples based on unifrac distance. We evaluated statistical significance of sample groups by the adonis method. Comparisons of significantly different OTUs across sample groups were performed with negative binomial DESeq2 available via the Calypso web server [25].
Other statistical analysis was performed using GraphPadPrism version 7; all ranges show the standard deviation (S.D.).
2.6. LDA Effect Size (LEfSe) analysis
We performed LEfSe analysis [26] to identify the biomarker bacterial taxa at different taxonomic levels found in the metagenomes predicted by PICRUSt. The LEfSe analysis was performed using the online Galaxy interface.
3. Results
3.1. Characterization of healthy volunteers
Of 200 Thai healthy volunteers living in four rural regions of Thailand, 108 (54%) individual volunteers were found to be ESBL-E carriers. In order to analyze differences in the microbiota composition between ESBL-E carriers and non-carriers in Thailand, 40 fecal samples were randomly selected from fecal samples of 200 samples for 16s rRNA metagenomics sequence analysis. The median age of ESBL-E carriers was 45.22 years (range 26–60 years) and that of non-carriers was 59.72 years (range 20–85 years) (Tab. 1). There were no statistically significant differences in the analyzed sociodemographic variables, except that the majority (63.6%) of ESBL-E carriers were general employees (P=0.0002) whereas the majority (72.2%) of non-carriers were farmers (P=0.0003). Furthermore, participants from the non-carrier group reported a significantly higher use of antibiotics without prescription (P=0.0248) in the 9 months preceding the study. However, according to the exclusion criteria of our study, they did not take antibiotics in 3 months prior to the study.
Table 1.
Baseline characteristics between ESBL-E carriers and non-carriers
| Baseline characteristics | ESBL-E carriage status | ||
|---|---|---|---|
| Carrier (N=22) |
Non-carrier (N=18) |
P valuea | |
| Sociodemographic data | |||
| Age, mean ± SD (years) | 45.22 ± 11.12 | 59.72 ± 20.21 | b |
| Age, range (years) | 26–60 | 20–85 | b |
| Female gender | 15 | 15 | nsd |
| Farmer | 3 | 13 | 0.0003 |
| General employee | 14 | 1 | 0.0002 |
| Grocer | 2 | 3 | ns |
| Unemployed | 3 | 1 | ns |
| Medical historyc | |||
| Used antibiotics without prescription | 7 | 13 | 0.0248 |
| Admitted to a hospital | 4 | 1 | ns |
| Stopped taking antibiotic early | 16 | 17 | ns |
Results were analyzed by Fisher’s exact test.
Statistical testing was not performed on matching variables.
Medical history 12 to 3 months prior to the study.
ns, not significant
3.2. Taxonomic composition of intestinal microbiota in ESBL-E carriers and non-carriers
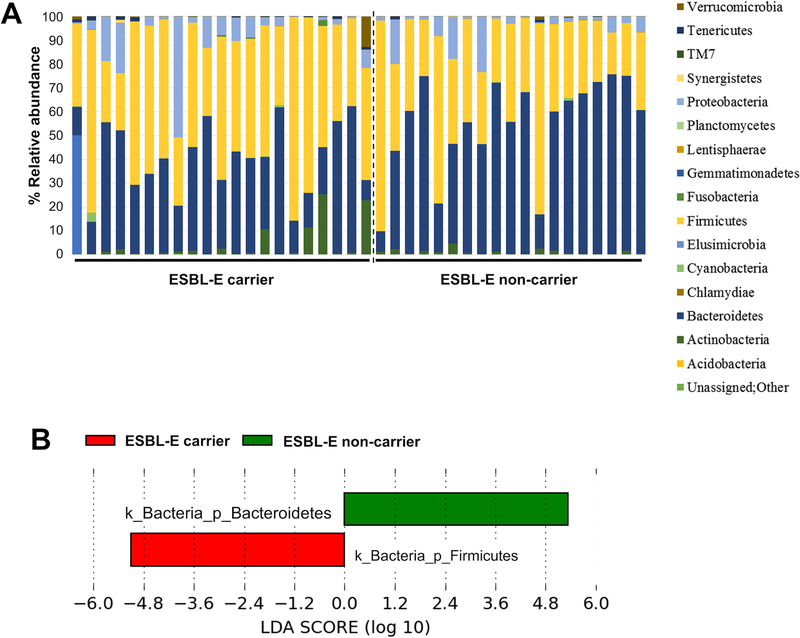
A total of 8,396,814 non-chimeric reads were obtained with an average of 209,920 ± 64,458 per sample. Taxonomic composition of the intestinal microbiota was analyzed by using QIIME. Figure 2A shows taxon abundance and sample composition. The phyla Bacteroidetes (46.94%) and Firmicutes (43.6%) were most abundant, followed by Proteobacteria (6.29%), Actinobacteria (2.25%), Tenericutes (0.28%) and Fusobacteria (0.1%). The taxa with a significant difference in abundance between ESBL-E carriers and non-carriers were then identified by using the LEfSe algorithm (Fig. 2B), revealing that ESBL-E carriers had a significantly higher abundance of Firmicutes (P <0.05), while Bacteroidetes were significantly more abundant in non-carriers (P <0.05). The Negative Binomial DESeq2 test was used to identify significant differences between groups on the species level (Tab. 2). Abundance of Bacteroides uniformis was significantly higher in the non-carrier group, while Coprococcus eutactus, Collinsella aerofaciens, Akkermansia muciniphila, and Pseudomonas fragi showed higher abundance in the carrier group.
Figure 2. Relative taxa abundance of the intestinal microbiota in ESBL-E carriers and non-carriers.
(A) Taxa abundance was determined by 16S rRNA paired-end sequencing using the Illumina MiSeq system. (B) Taxa with a log LDA (linear discriminant analysis) score above 3.00. as determined by using LEfSe.
Table 2.
Species with significantly differential abundance between ESBL-E carriers and non-carriers
| Taxa | ESBL-E carriage status (mean)a | ||||
|---|---|---|---|---|---|
| Carrier | Non-carrier | P Bonferronib | FDRb | P valuec | |
| Coprococcus eutactus | 623.82 | 277.78 | 0.044 | 0.012 | 0.0017 |
| Collinsella aerofaciens | 3454.73 | 957.33 | 0.048 | 0.012 | 0.0019 |
| Bacteroides uniformis | 3504.5 | 9139.5 | 0.18 | 0.035 | 0.007 |
| Akkermansia muciniphila | 975.77 | 285 | 1 | 0.17 | 0.043 |
| Pseudomonas fragi | 297.41 | 10 | 1 | 0.17 | 0.048 |
Negative Binomial DESeq2 analysis
Both the Bonferroni correction (P Bonferroni) and the False-Discovery-Rate (FDR) were applied to reduce the chances of obtaining false-positive results.
After the Negative Binomial DESeq2 analysis, calculated P-values were adjusted for multiple testing by P Bonferroni and FDR.
3.3. Comparison of the diversity of the intestinal microbiota between ESBL-E carriers and non-carriers
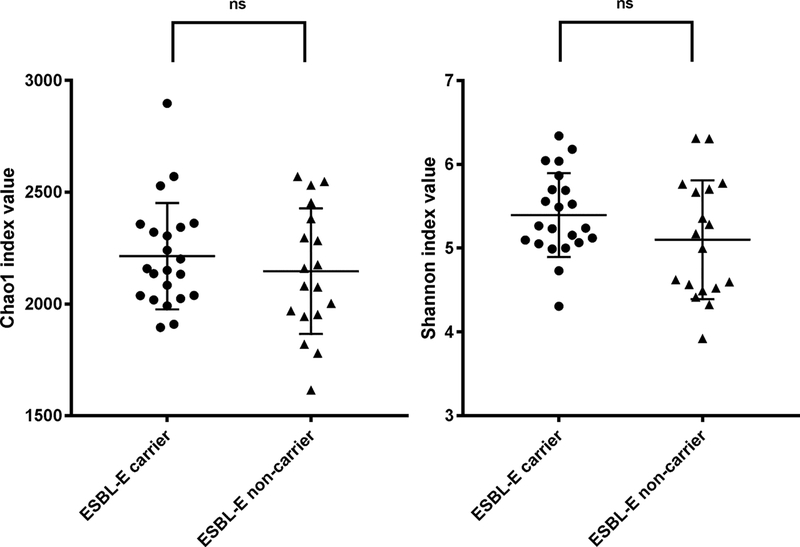
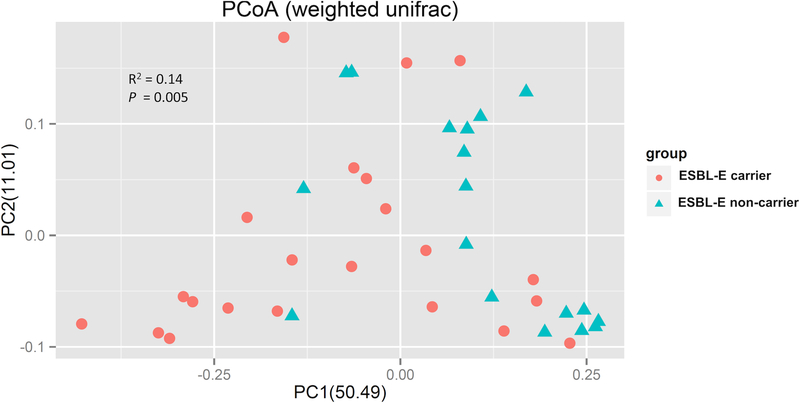
We performed a comparison of alpha diversity (i.e., the diversity within each group) between ESBL-E carriers and non-carriers. Chao1 and Shannon diversity index values from all operational taxonomic units (OTU) data were used to quantify the bacterial diversity of each group (Fig. 3). The Chao1 species richness estimator value in non-carriers (2146.47 ± 273.10) was slightly lower than that in ESBL-E carriers (2213.64 ± 232.58), but the difference was not statistically significant (p = 0.43). The Shannon diversity index in non-carriers (5.10 ± 0.69) was also lower than that in ESBL-E carriers (5.39 ± 0.48) without statistical significance (p = 0.13). In addition, beta diversity analysis (comparing the diversity between the two groups) was performed by using the weighted UniFrac analysis. This analysis indicated clear clustering of ESBL carriers along the first principle coordination (PC1), representing 50.49% of intersample variance (Fig. 4). The overall beta diversity difference of the intestinal microbiota of ESBL-E carriers as compared to non-carriers was statistically significant (Adonis on weighted unifrac: R2 =0.14, P =0.005). These results indicate that ESBL-E carriage is not associated with altered abundance of taxa, but with the underlying phylogenetic diversity.
Figure 3. Microbiome alpha diversity in ESBL-E carrier and non-carriers.
Alpha diversity was calculated for species richness by the Chao1 and for both richness and evenness by the Shannon diversity index. Differences in Chao1 richness estimator values and Shannon index values were analyzed using a nonparametric t test with 999 permutations. Data are presented as a scatter plot with mean and standard deviation (S.D.).
Fig 4. Principal Coordinate Analysis (PCoA) plot based on weighted UniFrac metrics for samples from ESBL-E carriers and non-carriers.
Each point represents an individual and is plotted in a distance matrix showing the relative diversity. Red circles, ESBL carrier; blue triangle, non-carrier. Statistical analysis was by the adonis method.
4. Discussion
ESBL-E colonization presents a source for the spread of dangerous antibiotic-resistant microorganisms. ESBL-E carriage is on the rise in the community, and especially high in Asia [9–11]. Except for one geographically very limited study in a remote village in South America with low incidence of ESBL-E colonization [15], this is the first study to analyze the role of the intestinal microbiota in determining carriage status of ESBL-E. Our study was performed in subjects from rural areas covering an entire country and ruling out recent antibiotic exposure. Due to the high incidence of ESBL-E carriage in our study (54 %), which is in accordance with other reports from Asia [10, 11], our results give previously unavailable information on the underpinnings of community ESBL-E antibiotic resistance carriage as a public health risk.
Notably, although participants who used antimicrobial agents and had a history of hospitalization in the previous 3 months were excluded, we still observed a very high incidence of ESBL-E carriers. This suggests that the high colonization rates previously observed in Thai communities [10] may not be due to the recent antibiotic exposure; however, they may be due to complex and multifactorial factors, including the dose, duration, and spectrum of administered antibiotics, the immune status of the host, and their microbiome.
The main purpose of our study was, however, to detect a possible association of intestinal carriage of ESBL-E with the composition of the intestinal microbiota. We found that the phyla Bacteroidetes and Firmicutes were generally most dominant in the investigated rural population. This is in accordance with several studies that have also detected Bacteroidetes and Firmicutes as main constituents of the intestinal microbiota, including in Thai populations [27–29]. Notably, we observed a significant difference between the ESBL-E carriers and non-carriers groups in beta but not alpha diversity, implying that the main difference is the relative abundance of some bacterial community members.
The Firmicutes, and on the species level, C. eutactus, C. aerofaciens, A. muciniphila, and P. fragi showed significantly higher abundance among ESBL-E carriers. While the subjects in our study did not report any signs of acute intestinal disease, C. eutactus, C. aerofaciens and A. muciniphila have been reported as associated with intestinal inflammation or infection [30–32]. It has been well established that intestinal inflammation may change the composition of the microbiota, disrupt colonization resistance, and enhance pathogen growth [33]. Furthermore, it has been reported that host-mediated inflammation altered the colonic microbial community and promoted the overgrowth of either resident or introduced bacteria, particularly of the Enterobacteriaceae family [34]. These findings are in accordance with the notion that ESBL-E carriers may suffer from sub-acute inflammation that leads to intestinal dysbiosis [33, 34].
Contrastingly, we detected significantly lower abundance of Bacteroidetes, and on the species level, B. uniformis, in ESBL-E carriers. This association may also be due to intestinal inflammation, as in a murine inflammatory model, an enhanced inflammatory host response was responsible for a dramatic decrease in the Bacteroides population [34]. Bacteroides, the dominant genus of the intestinal microbiota, can directly inhibit intestinal pathogens by competing for nutrients or producing inhibitory substances [35]. Bacteroides thetaiotaomicron can consume carbohydrates needed for Citrobacter rodentium, which contributes to the competitive exclusion of this pathogen from the intestinal lumen [36]. Moreover, Bacteroidetes are associated with protection against Salmonella Typhimurium-induced colitis [37] and Helicobacter hepaticus-induced colitis in murine model [38]. Thus, our finding suggesting that Bacteroidetes may play a significant role in inhibiting ESBL-E carriage is in accordance with the discussed reports on probiotic functions of Bacteroidetes.
The subjects in our study had not received any antibiotic treatment and did not visit any hospital or healthcare setting within at least the three months prior to the study. The participants from the non-carrier group reported significantly more use of antibiotics without prescription within a year but not 3 months prior to the study. However, this is not a likely reason for the observed difference in taxa abundance, because generally normalization of the intestinal microbiota is observed within three months upon short-term antibiotic exposure [39]. On the other hand, any possible long-term antibiotic exposure would not likely lead to the observed differences in the bacterial communities [39–41].
Another notable finding of our study is the observed significantly lower ESBL-E carriage rates in farmers. Dietary conditions impact the composition of the intestinal microbiota, and it is quite likely that these differ in farmers from other occupations. Interestingly, Bacteroidetes have been reported to be significantly more abundant in the nasal and oral microbiota of farmers than non-farmers, suggesting that this may also be true for the intestine [42]. Recently, non-toxigenic Bacteroidetes was shown to have powerful health benefits to the host, and was recommended as a probiotic candidate (43, 44), These findings may lead to the future identification of specific molecules controlling ESBL-E carriage to potentially be used in probiotic therapeutic approaches to limit ESBL-E colonization. Together, our findings indicate that ESBL-E carriage may be controlled by promoting the abundance of anaerobic bacteria, particularly probiotic intestinal bacteria, possibly by dietary conditions and may support the notion of probiotic intervention to control the spread of antibiotic resistance (45).
Funding:
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID), U.S. National Institutes of Health (NIH) (project number ZIA AI000904, M.O.) and the Thailand Research Fund through the Royal Golden Jubilee PhD Program (grant number PHD/0072/2557, to P.P. and P.K.). P.K. was also supported by the Faculty of Medicine Siriraj Hospital, Mahidol University, Grant Number (IO) R015833012 and P.P. by the Graduate Partnership Program of the NIH. The funders had no role in study design; in the collection, analysis and interpretation of data; in the writing of the report; or in the decision to submit the article for publication.
Footnotes
Competing Interests: None
Ethical Approval: Approval No. Si773/2015, Faculty of Medicine Siriraj Hospital, Mahidol University
References
- [1].Salyers AA, Gupta A, Wang Y. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 2004;12:412–6. [DOI] [PubMed] [Google Scholar]
- [2].Jean SS, Coombs G, Ling T, Balaji V, Rodrigues C, Mikamo H, et al. Epidemiology and antimicrobial susceptibility profiles of pathogens causing urinary tract infections in the Asia-Pacific region: Results from the Study for Monitoring Antimicrobial Resistance Trends (SMART), 2010–2013. Int J Antimicrob Agents. 2016;47:328–34. [DOI] [PubMed] [Google Scholar]
- [3].Jean SS, Hsueh PR, Group SA-P. Distribution of ESBLs, AmpC beta-lactamases and carbapenemases among Enterobacteriaceae isolates causing intra-abdominal and urinary tract infections in the Asia-Pacific region during 2008–14: results from the Study for Monitoring Antimicrobial Resistance Trends (SMART). J Antimicrob Chemother. 2017;72:166–71. [DOI] [PubMed] [Google Scholar]
- [4].Islam S, Selvarangan R, Kanwar N, McHenry R, Chappell JD, Halasa N, et al. Intestinal Carriage of Third-Generation Cephalosporin-Resistant and Extended-Spectrum beta-Lactamase-Producing Enterobacteriaceae in Healthy US Children. J Pediatric Infect Dis Soc. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].van Duijkeren E, Wielders CCH, Dierikx CM, van Hoek A, Hengeveld P, Veenman C, et al. Long-term carriage of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in the general population in the Netherlands. Clin Infect Dis. 2017. [DOI] [PubMed] [Google Scholar]
- [6].Ulstad CR, Solheim M, Berg S, Lindbaek M, Dahle UR, Wester AL. Carriage of ESBL/AmpC-producing or ciprofloxacin non-susceptible Escherichia coli and Klebsiella spp. in healthy people in Norway. Antimicrob Resist Infect Control. 2016;5:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hazirolan G, Mumcuoglu I, Altan G, Ozmen BB, Aksu N, Karahan ZC. Fecal carriage of extended-spectrum beta-lactamase and ampc beta-lactamase-producing enterobacteriaceae in a turkish community. Niger J Clin Pract. 2018;21:81–6. [DOI] [PubMed] [Google Scholar]
- [8].Ni Q, Tian Y, Zhang L, Jiang C, Dong D, Li Z, et al. Prevalence and quinolone resistance of fecal carriage of extended-spectrum beta-lactamase-producing Escherichia coli in 6 communities and 2 physical examination center populations in Shanghai, China. Diagn Microbiol Infect Dis. 2016;86:428–33. [DOI] [PubMed] [Google Scholar]
- [9].Kiratisin P, Chattammanat S, Sa-Nguansai S, Dansubutra B, Nangpatharapornthawee P, Patthamalai P, et al. A 2-year trend of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Thailand: an alert for infection control. Trans R Soc Trop Med Hyg. 2008;102:460–4. [DOI] [PubMed] [Google Scholar]
- [10].Niumsup PR, Tansawai U, Na-Udom A, Jantapalaboon D, Assawatheptawee K, Kiddee A, et al. Prevalence and risk factors for intestinal carriage of CTX-M-type ESBLs in Enterobacteriaceae from a Thai community. Eur J Clin Microbiol Infect Dis. 2018;37:69–75. [DOI] [PubMed] [Google Scholar]
- [11].Kurz MS, Bayingana C, Ndoli JM, Sendegeya A, Durst A, Pfuller R, et al. Intense pre-admission carriage and further acquisition of ESBL-producing Enterobacteriaceae among patients and their caregivers in a tertiary hospital in Rwanda. Trop Med Int Health. 2017;22:210–20. [DOI] [PubMed] [Google Scholar]
- [12].Woerther PL, Andremont A, Kantele A. Travel-acquired ESBL-producing Enterobacteriaceae: impact of colonization at individual and community level. J Travel Med. 2017;24:S29–S34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pultz NJ, Stiefel U, Subramanyan S, Helfand MS, Donskey CJ. Mechanisms by which anaerobic microbiota inhibit the establishment in mice of intestinal colonization by vancomycin-resistant Enterococcus. J Infect Dis. 2005;191:949–56. [DOI] [PubMed] [Google Scholar]
- [14].Lawley TD, Walker AW. Intestinal colonization resistance. Immunology. 2013;138:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gosalbes MJ, Vazquez-Castellanos JF, Angebault C, Woerther PL, Ruppe E, Ferrus ML, et al. Carriage of Enterobacteria Producing Extended-Spectrum beta-Lactamases and Composition of the Gut Microbiota in an Amerindian Community. Antimicrob Agents Chemother. 2015;60:507–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Institute CaLS. Performance standards for antimicrobial susceptibility testing Twenty-First Informational Supplement CLSI document M100-S24. Wayne, Pennsylvania, USA: Clinical and Laboratory Standards Institute; 2014. [Google Scholar]
- [17].Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Weber N, Liou D, Dommer J, MacMenamin P, Quinones M, Misner I, et al. Nephele: a cloud platform for simplified, standardized and reproducible microbiome data analysis. Bioinformatics. 2018;34:1411–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. [DOI] [PubMed] [Google Scholar]
- [20].Rideout JR, He Y, Navas-Molina JA, Walters WA, Ursell LK, Gibbons SM, et al. Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ. 2014;2:e545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73:1576–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lozupone C, Hamady M, Knight R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Fierer N, Lauber CL, Zhou N, McDonald D, Costello EK, Knight R. Forensic identification using skin bacterial communities. Proc Natl Acad Sci U S A. 2010;107:6477–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zakrzewski M, Proietti C, Ellis JJ, Hasan S, Brion MJ, Berger B, et al. Calypso: a user-friendly web-server for mining and visualizing microbiome-environment interactions. Bioinformatics. 2017;33:782–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ruengsomwong S, La-Ongkham O, Jiang J, Wannissorn B, Nakayama J, Nitisinprasert S. Microbial Community of Healthy Thai Vegetarians and Non-Vegetarians, Their Core Gut Microbiota, and Pathogen Risk. J Microbiol Biotechnol. 2016;26:1723–35. [DOI] [PubMed] [Google Scholar]
- [29].Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Malinen E, Krogius-Kurikka L, Lyra A, Nikkila J, Jaaskelainen A, Rinttila T, et al. Association of symptoms with gastrointestinal microbiota in irritable bowel syndrome. World J Gastroenterol. 2010;16:4532–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Koch C, Amati AL, Hecker A, Hoxter M, Hirschburger M, Matejec R, et al. Microbiomic Analysis of Intra-Abdominal Infections by Using Denaturing High-Performance Liquid Chromatography: A Prospective Observational Study. Surg Infect (Larchmt). 2017;18:596–602. [DOI] [PubMed] [Google Scholar]
- [32].Ganesh BP, Klopfleisch R, Loh G, Blaut M. Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella Typhimurium-infected gnotobiotic mice. PLoS One. 2013;8:e74963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gibson MK, Pesesky MW, Dantas G. The yin and yang of bacterial resilience in the human gut microbiota. J Mol Biol. 2014;426:3866–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:204. [DOI] [PubMed] [Google Scholar]
- [35].Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Curtis MM, Hu Z, Klimko C, Narayanan S, Deberardinis R, Sperandio V. The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe. 2014;16:759–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ferreira RB, Gill N, Willing BP, Antunes LC, Russell SL, Croxen MA, et al. The intestinal microbiota plays a role in Salmonella-induced colitis independent of pathogen colonization. PLoS One. 2011;6:e20338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–5. [DOI] [PubMed] [Google Scholar]
- [39].Jernberg C, Lofmark S, Edlund C, Jansson JK. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology. 2010;156:3216–23. [DOI] [PubMed] [Google Scholar]
- [40].Cho I, Yamanishi S, Cox L, Methe BA, Zavadil J, Li K, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Langdon A, Crook N, Dantas G. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med. 2016;8:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Shukla SK, Ye Z, Sandberg S, Reyes I, Fritsche TR, Keifer M. The nasal microbiota of dairy farmers is more complex than oral microbiota, reflects occupational exposure, and provides competition for staphylococci. PLoS One. 2017;12:e0183898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Deng H, Li Z, Tan Y, Guo Z, Liu Y, Wang Y, et al. A novel strain of Bacteroides fragilis enhances phagocytosis and polarises M1 macrophages. Sci Rep. 2016;6:29401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wang Y, Deng H, Li Z, Tan Y, Han Y, Wang X, et al. Safety Evaluation of a Novel Strain of Bacteroides fragilis. Front Microbiol. 2017;8:435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ouwehand AC, Forssten S, Hibberd AA, Lyra A, Stahl B. Probiotic approach to prevent antibiotic resistance. Ann Med. 2016;48:246–55. [DOI] [PubMed] [Google Scholar]