Abstract
The broad application of RNA interference for disease prevention is dependent upon the production of dsRNA in an economically feasible, scalable, and sustainable fashion, as well as the identification of safe and effective methods for RNA delivery. Current research has sparked interest in the use of Saccharomyces cerevisiae for these applications. This review examines the potential for commercial development of yeast interfering RNA expression and delivery systems. S. cerevisiae is a genetic model organism that lacks a functional RNA interference system, which may make it an ideal system for expression and accumulation of high levels of recombinant interfering RNA. Moreover, recent studies in a variety of eukaryotic species suggest that this microbe may be an excellent and safe system for interfering RNA delivery. Key areas for further research and development include optimization of interfering RNA expression in S. cerevisiae, industrial-sized scaling of recombinant yeast cultures in which interfering RNA molecules are expressed, the development of methods for large-scale drying of yeast that preserve interfering RNA integrity, and identification of encapsulating agents that promote yeast stability in various environmental conditions. The genetic tractability of S. cerevisiae and a long history of using this microbe in both the food and pharmaceutical industry will facilitate further development of this promising new technology, which has many potential applications of medical importance.
Keywords: RNAi, shRNA, gene therapy, Aedes aegypti, Anopheles gambiae, mosquito, biopharmaceutical, bioengineering
1. INTRODUCTION
Saccharomyces cerevisiae, also known as baker’s or budding yeast, is a non-infectious microbe and genetic model organism. Research in S. cerevisiae has provided key insights into the regulation of many fundamental eukaryotic cellular processes, including the cell cycle, signal transduction, chromatin structure, transcription, and genetic inheritance [1, 2]. Despite the many attributes of this model system, RNA interference (RNAi), a regulatory pathway in eukaryotic cells that silences gene expression through the production of small interfering RNAs (siRNAs), has not traditionally been studied in S. cerevisiae [1], which lacks components of the RNAi pathway [3]. RNAi is an ancient mechanism that protects a diverse number of eukaryotic species, including some species of yeast [1, 3], from nucleic acids such as viruses [4]. The presence of double stranded RNA (dsRNA) in a cell triggers processing of the dsRNA by Dicer, a ribonuclease III (RNAse III) family endonuclease. Dicer cleaves dsRNA molecules into siRNAs, which are then assembled onto the Argonaute effector protein [5]. This results in complementary base pairing interactions that target and inactivate endogenous cellular RNAs or repress transcription [6, 7].
The potential applications of RNAi for the treatment and prevention of various diseases are seemingly endless. As discussed by Tam et al. [8], siRNAs can suppress viral infections, tumorigenesis, inflammatory disorders, and cardiovascular disease. Furthermore, RNAi could potentially be used for the control of human disease vector insects [9, 10]. The broad application of RNAi for applications of medical and commercial importance is dependent upon production of dsRNA in an economically feasible, scalable, and sustainable fashion. dsRNA manufacture has traditionally relied on expensive, carbon-intensive chemical synthesis, resulting in high costs that have made the broad and large-scale use of dsRNA prohibitively expensive [11]. In addition to advancements in the cost-effective scalable manufacture of interfering RNA, broad application of RNAi requires not only the discovery of molecules to be used in various applications but also safe and effective mechanisms for their delivery to humans or other intended organisms [12]. Several studies in which S. cerevisiae was successfully used as an expression and delivery system for dsRNA to other organisms [9, 10, 13, 14] have sparked interest in the use of S. cerevisiae for RNAi-based applications (Table 1).
Table 1. Bioengineering S. cerevisiae for recombinant interfering RNA expression.
| Recombinant Yeast Strains Generated | Resulting Advancements |
|---|---|
| Restoration of Dicer and Argonaut activity in S. cerevisiaea | Functional RNAi activity in yeast. |
| Generation of recombinant S. cerevisiae for delivery of shRNA targeting murine CD40b | Successful delivery of an shRNA expression system to murine intestinal DC cells, leading to successful gene silencing in mice. |
| S. cerevisiae expressing shRNA corresponding to mosquito genesc | Successful delivery of dried inactivated yeast interfering RNA pesticides to mosquitoes; shRNA expression cassette successfully integrated into yeast genome. |
| S. cerevisiae that express long dsRNA molecules targeting Drosophila crop pestsd | Development of S. cerevisiae system for expressing and delivering long dsRNA insecticides to crop pests |
| S. cerevisiae shRNA expression system for improvement of itaconic acid production straine | Optimization of a yeast strain as a metabolic engineering tool; evaluation of parameters that improve shRNA expression |
| RNAi screening libraries generatedf | Facilitated development of tunable yeast biomolecule production systems; evaluation of parameters that improve expression of long dsRNA molecules in S. cerevisiae |
2. ADVANTAGES OF THE S. CEREVISIAE EXPRESSION SYSTEM
A deficiency of functional RNAi machinery in S. cerevisiae, which lacks both Dicer and Argonaute [3], may have initially caused it to be overlooked as a system for producing interfering RNA molecules. However, this lack of RNAi machinery may, in fact, promote the accumulation of bioengineered dsRNAs in yeasts, making it an ideal system for RNA production. In support of this concept, recent studies in plants suggest that dsRNAs can be stably produced in chloroplasts, a cellular component that, like yeast, lacks functional RNAi machinery [18]. When insecticidal dsRNA targeting potato beetles was expressed in the chloroplasts of potato plants, the plants were better protected from this crop pest than plants in which dsRNA had been expressed from the potato plant nuclear genome. Expression of dsRNA in chloroplasts led to accumulation of dsRNA at high levels, up to 0.4% of total cellular RNA. These findings, which have generated interest in using the chloroplast genome to express dsRNAs targeting crop pests [18], may be applicable to yeast dsRNA expression systems.
A number of key advantages make S. cerevisiae an excellent system for expression of recombinant nucleic acids and proteins [2]. First, S. cerevisiae grows rapidly and is both straightforward and inexpensive to culture. As discussed by Roohvan et al. [2], sophisticated genetic manipulations are possible in S. cerevisiae, the first eukaryotic genome to be sequenced [19]. A wide variety of vectors, including episomal, integrative, and copy number controlled vectors are available, as are a wide range of auxotrophic strains that can be rescued through transformation with vectors bearing wild-type copies of the mutated genes. Moreover, a number of different promoters, including both constitutive and inducible promoters, are available, permitting flexibility in the construction of gene expression systems. Although genetic manipulations in S. cerevisiae are as straightforward as prokaryotic systems, the advanced cellular features of eukaryotes, including post-translational modifications and glycosylation patterns that are more comparable to mammals, exist in yeast [2]. S. cerevisiae has also been genetically engineered such that it can generate proteins with more human-like glycosylation patterns, thereby allowing for the production of recombinant proteins that are properly processed in humans [20]. Furthermore, yeasts can be engineered to secrete recombinant proteins into the cell media, which greatly facilitates subsequent purification of these proteins, another key advantage of using yeast expression systems [21]. Relatively high levels of recombinant protein expression, more than 1 g/L [2], have been obtained for several products. Finally, fermentation and downstream processing systems have been established for S. cerevisiae [2]. The many attributes of S. cerevisiae suggest that it could be an excellent system for scaled expression of recombinant interfering RNA molecules.
Regardless of whether the end goal is gene therapy, mosquito control, or crop protection, the successful application of RNAi requires the development of safe and effective delivery systems. The use of bacterial and viral dsRNA delivery systems presents safety risks, as non-infectious agents and means of ensuring that the recombinant DNA does not integrate into the genome of the target organism are preferred [13]. The safety profile of S. cerevisiae makes it a winner in these regards, as well [2]. In three recent studies [9, 10, 13], S. cerevisiae was genetically engineered to express short hairpin RNA (shRNA), an artificial RNA molecule with a hairpin turn that can be used for RNAi. In these studies, the yeast strains were engineered for expression of shRNA corresponding to murine [13] and mosquito [9, 10] genes. The subsequent feeding of these engineered yeast strains to mice and mosquito larvae resulted in effective gene silencing in these organisms. It was further demonstrated that shRNA expression cassettes can be stably integrated into the yeast genome [9], and that the yeast can be heat-inactivated prior to use in oral feeding assays [9, 10], thereby alleviating additional safety concerns. The results of these studies, which will be described in more detail below, suggest that use of this technology has a wide range of potential medical applications, from therapeutics in humans to the control of human disease vector mosquitoes (Table 1). In addition to discussing potential applications, this review summarizes requisite research and development that will facilitate the commercialization of this technology.
3. POTENTIAL APPLICATIONS
3.1. A Yeast System for Delivery of shRNA to Mammals
S. cerevisiase has been investigated as a potential vaccine carrier, as its complex cell wall components and appropriate size facilitate the specific uptake of this microbe by Dendritic Cells (DCs) [22], professional antigen-presenting cells that function in innate and adaptive immune responses [23-25]. Importantly, S. cerevisiae appears to withstand simulated human digestive environment conditions, suggesting that it may be a useful oral delivery system for biological molecules to intestinal DC cells [26]. Recognition of S. cerevisiae through dectin, mannose-fucose, and toll-like receptors on DC cells permits the phagocytosis of yeast by these cells, resulting in DC maturation [27-29]. DNA and mRNA have been successfully delivered to mammalian phagocytic cells using a recombinant yeast delivery system [30]. S. cerevisiae has also been utilized for the successful delivery of DNA and protein to DCs [27, 31, 32]. Based on these observations, Zhang et al. [13] hypothesized that S. cerevisiae might serve as a potential oral delivery system for shRNA to mouse intestinal DCs (Table 1).
To test their hypothesis, Zhang et al. [13] chose to silence CD40, an activation receptor [33] that functions in DC maturation [34], the inhibition of which was expected to impact the immune response. shRNA corresponding to the sequence of CD40 was expressed in recombinant S. cerevisiae. For the construction of the expression vector, the shRNA expression cassette was placed under control of a mammalian U6 promoter followed by a small nuclear RNA leader sequence. This shRNA targeting sequence was inserted into an endogenous mouse micro RNA (miRNA), miR30 [35-37]. The authors anticipated that DC cells would take up the recombinant yeast through phagocytosis, after which the shRNA expression vector would be taken into the murine nucleus where shRNA would be transcribed. Following shRNA processing, mature siRNA was expected to promote CD40 mRNA degradation [13].
To verify the function of the shRNA expression vector, it was transfected into a cultured mammalian cell line with a CD40-GFP reporter. GFP expression was reduced, confirming that shRNA had been successfully produced and had effectively silenced the reporter transcript. shRNA expression was also confirmed through reverse transcription of total RNA from the cells and PCR amplification of DNA corresponding to the shRNA. Mice were then fed with either recombinant yeast expressing the shRNA or control yeast bearing a vector that lacked the shRNA expression cassette. Animals fed with yeast expressing shRNA displayed reduced CD40 levels in intestinal dendritic cells. Analysis of mouse serum demonstrated that CD40 silencing resulted in altered cytokine levels. The results of this investigation suggested that a recombinant yeast delivery system could be used for efficient delivery of an shRNA expression system to murine dendritic cells for the purpose of gene silencing and immunomodulation. Zhang et al. [13] commented that the effective silencing of CD40 in humans could have therapeutic applications for the treatment of various autoimmune disorders, vascular disease, and rejection of transplanted tissues. This group also published a protocol with detailed instructions for the construction of shRNA expression vectors and delivery of recombinant yeast through oral administration to mice [38]. Their publication of detailed methodology will hopefully permit other researchers to explore the use of this delivery system for additional applications.
3.2. Targeting of Disease Vector Mosquitoes with Yeast Interfering RNA Pesticides
Although it is beginning to attract attention in agricultural biotechnology communities [18], the use of RNA interference (RNAi) is an innovative yet still largely unexplored approach for control of disease vector mosquitoes. Over 3 billion people worldwide are at risk for contracting malaria, which results from infection with Plasmodium spp. parasites that are transmitted to humans through the bites of infected Anopheles mosquitoes [39]. In 2016, there were >200 million cases of malaria world-wide, and 445,000 deaths resulted from malaria infection [39]. Dengue, a leading cause of morbidity in the tropics, Zika, which was designated a public health emergency of international concern in 2016, as well as yellow fever and chikungunya, result from infections with arboviruses transmitted through the bites of Aedes mosquitoes [40]. Cases of Zika, which has been linked to severe birth defects and neurological disorders, are currently occurring in many countries in the Americas, and Zika has rapidly spread to previously unaffected geographic areas [41]. Dengue has an annual incidence of approximately 400 million cases resulting in ~50,000 deaths annually worldwide [42]. These statistics highlight the vital need to combat these pathogens and the mosquitoes that transmit them to human hosts. Due to the lack of progress in vaccine development, production and distribution, mosquito control is the primary means of controlling mosquito-borne illnesses. However, increased emergence of insecticide resistance and a general lack of funding for and support of mosquito control programs threaten current strategies for managing mosquitoes [40]. Larviciding, the application of microbial or chemical agents to kill mosquito larvae in aquatic habitats, is a key component of integrated mosquito control and disease prevention strategies. Aedes mosquitoes lay eggs in natural and artificial water-filled containers within or close to human dwellings in urban areas, and are therefore susceptible to larviciding [43]. When used as a supplement to insecticide treated nets and indoor residual spraying, larviciding is cost-effective for control of Anopheles (malaria vector) mosquitoes in urban settings where vector breeding sites are few, fixed, and findable [44]. Given the increase of reported insecticide resistance to existing larvicides and the rising concern for negative effects of pesticides on non-target organisms, new larvicidal agents are vitally needed to address current and emerging arthropod-borne infectious diseases. To this end, we have pursued RNAi approaches for the control of mosquito larvae [9, 10, 45-48].
Though most mosquito researchers use longer (300-400 bp) dsRNA molecules for RNAi, the short length (25 bp) of custom siRNAs and their shRNA counterparts facilitates the design of interfering RNA that recognizes conserved target sites in multiple mosquito species, but not in non-target organisms. We initiated high-throughput screens designed to enrich for the selection of siRNA larvicides that generate high levels of mosquito mortality and that can be used against multiple disease vector mosquito species, but which lack sequence homology in non-targeted species. Our goal is to create an siRNA arsenal that can combat pesticide resistance that arises from point mutations in any one mosquito gene target sequence. These screens led to the discovery of hundreds of siRNAs that induce mosquito larval lethality [9, 10]. To facilitate the cost-effective production of interfering RNA and delivery of RNA pesticides to mosquitoes in the field, S. cerevisiae was engineered to produce shRNA corresponding to select genes/target sequences identified in the siRNA larvicide screens [9, 10] (Table 1).
shRNA expression cassettes corresponding to several siRNA targets sequences in Aedes [9] and Anopheles [10] genes were cloned into the non-integrating pRS426 GPD yeast shuttle vector [49]. This multi-copy shuttle plasmid was transformed into S. cerevisiae, facilitating expression of shRNA downstream of a constitutive GPD promoter [49]. These yeast strains, as well as a control yeast strain expressing shRNA with no known target in mosquitoes, were initially fed to larvae in an agarose gel-covered formulation containing live yeast, and this formulation generated high levels of larval death. It was determined that heat-killed yeast prepared in a dry pellet formulation killed larvae as effectively as the live yeast formulation. Dried and inactivated yeast interfering RNA pellets generated >90% silencing of target genes and up to 100% larval lethality in laboratory trials [9, 10]. In our hands, the yeast interfering RNA delivery strategy is the most effective method for gene silencing in mosquito larvae that we have tested to date and generates higher levels of larval mortality than other delivery methods examined in our laboratory.
Upon verification that these yeast strains induced significant larval lethality, stable yeast transformants were generated for several strains of interest [9]. In these strains, the shRNA expression cassette was placed under control of an inducible Gal1 promoter, which facilitates induction of shRNA expression with galactose just prior to harvesting and heat killing of the yeast for generation of inactivated yeast tablets. Moreover, for these strains, two copies of shRNA expression cassettes were integrated into the yeast genome, dispensing of the use of plasmids with antibiotic resistance markers and eliminating the risk of horizontal transfer to other species, two modifications that will facilitate the testing of these yeast strains in field trials [9]. Semi-field trials with the yeast larvicides are underway. The development of dried inactivated ready-to-use yeast interfering RNA pellets will facilitate the seamless integration of this technology into existing mosquito control programs. Importantly, interfering RNA is generated through yeast culturing, significantly reducing RNA production costs. Moreover, yeasts have been cultivated worldwide for thousands of years, and this technology can be adapted to resource-limited countries with constrained infrastructures. Dried yeast can be packaged and shipped, which can facilitate regional distribution to communities in need. Furthermore, S. cerevisiae, a natural product that is often used in food and alcoholic beverage preparation and sold (in its inactivated form) as a dietary supplement, is non-toxic, suggesting that it could be much safer for humans than existing chemical pesticides. Finally, this technology is scalable, as yeast can be produced in industrial-sized cultures [9, 10].
Our characterization and development of mosquito yeast interfering RNA larvicides revealed several findings that may be of importance to those working to develop yeast as a delivery system for human therapeutics. First, as discussed above, laboratory trials demonstrate that the larvicidal capacity of this yeast is maintained even when the microbes are heat-killed and inactivated [9, 10]. Given that consumption of large quantities of live yeast is typically not advised, it would be preferable to heat-inactivate yeast used in human therapies. Indeed, most studies with Whole Yeast Vaccines (WYVs) use heat-killed vaccines as opposed to live yeast [28]. Moreover, while Zhang et al. [13] prepared shRNA expression cassettes that would be delivered to and expressed in murine cells, we instead induced shRNA expression in yeast from transgenes integrated into the yeast genome, heat-killed the yeast, and then delivered this shRNA to insects in yeast cells. This shRNA, which was only produced in yeast rather than in insect cells, effectively suppressed insect gene expression. A similar procedure may be a viable option in mice or humans, and it would be interesting to directly compare the levels of CD40 silencing that result from use of the two different systems in the mouse system. Furthermore, we succeeded in the development of dried inactivated ready to use yeast interfering RNA pellets. These pellets were modeled after nutritional yeast, a human dietary supplement. This dried pellet formulation could therefore be suitable for human consumption.
3.3. Mass Production and Purification of Interfering RNA
While it may be beneficial to use yeast as a delivery system, the development of yeast systems for large-scale production of interfering RNA that could be subsequently purified would also be of interest. To date, microbial dsRNA expression systems have focused on the development of bacterial systems for dsRNA expression. In fact, shortly after the discovery of RNA silencing in Caenorhabditis elegans [50], an Escherichia coli dsRNA expression and delivery system was developed [51]. In the bacterial system, fragments of the gene to be silenced are flanked by opposing T7 RNA polymerase promoters, allowing for the production of dsRNA molecules. dsRNA expression is improved by transformation of E. coli strains lacking RNAse III [51], which permits higher levels of dsRNA accumulation. Even larger quantities of dsRNA can be produced in RNAse III deficient E. coli that overexpress T7 polymerase [52].
Huang et al. [53] later developed a method for mass production of siRNA in E. coli that capitalizes on expression of the siRNA binding protein p19. Expression of recombinant His-tagged p19 protein stabilized siRNA that was produced in the E. coli while also facilitating subsequent nickel-column purification of the siRNA [54]. This system was recently further optimized, and production was successfully scaled using a high-cell density fed-batch fermentation method in a bioreactor [55]. This system generated 10 mg purified RNA yields, yields that were significantly higher than those achieved in other bacterial systems. Interestingly, the authors found that production of hairpin RNA constructs resulted in higher yields, higher percentages of the target sequence, and better silencing efficiencies than siRNAs that were generated with convergent overlapping sense and antisense transcripts. Two convergent T7 promoters flanking hairpin RNA could further increase siRNA production. The authors concluded that this method could be adapted for production of siRNAs in a renewable and cost-effective manner, though they expect that this will require further process development. In particular, they noted that there would be a need for further purification to reduce levels of contaminating dsRNAs present in the bacteria [55].
Surprisingly, large-scale interfering RNA production systems in S. cerevisiae have yet to be described in the literature, perhaps once again as a result of the lack of functional RNAi machinery in S. cerevisiae. However, as discussed above, it is possible that this lack of RNAi machinery may allow interfering RNA molecules to accumulate at higher levels in the yeast. It therefore seems logical to consider the development of such systems. Although S. cerevisiae lacks Dicer, it does have one RNAse III gene, Rntp1 [56, 57]. While we were able to produce shRNA in yeast in the presence of a functional copy of this gene [9, 10], the hairpin loop used in our studies is not recognized by Rntp1 [58-61]. Depending on the interfering RNA molecule produced, it may be useful in some cases to modify the levels of this gene. It would be interesting to recapitulate the Huang et al. [53] p19 system in S. cerevisiae, and doing so should be fairly straightforward. Recombinant p19 could be expressed in yeast, and larger-scale fermentations in a bioreactor would need to be optimized, but are certainly possible. Given the lack of RNAi machinery in S. cerevisiae, it is possible that fewer endogenous dsRNAs would be available to bind to p19, which might result in the isolation of purer recombinant dsRNAs that can be produced in E. coli.
4. PROSPECTS AND CHALLENGES FOR COMMERCIAL DEVELOPMENT
4.1. Overview
Due to the many attributes of S. cerevisiae, it is considered to be an exceptional workhorse for biopharmaceutical production [2]. Roughly 20% of today’s biopharmaceuticals are produced in yeasts, with S. cerevisiae, the most genetically developed yeast species, most often selected for production purposes [62]. Biopharmaceutical production in yeast initiated in the 1980s when ZymoGenetics, the first company to succeed in the industrialization of S. cerevisiae for production of pharmaceuticals, engineered yeast that produced recombinant human insulin. Today, nearly half the global supply of insulin is produced in S. cerevisiae [21]. In the 1990s, S. cerevisiae-based production of a hepatitis B subunit vaccine, recombinant human granulocyte macrophage-colony stimulating factor, and human platelet derived growth factor followed, as did the production of the human papillomavirus subunit vaccine and a kallikrein inhibitor, all of which were produced after the turn of the century [21]. The uptake of yeast by immune cells combined with the ease of producing pathogens and tumor antigens in yeast cells has resulted in the use of yeasts as a new vaccine platform [2]. In particular, the potential for using whole yeast vaccines for oral immunization strategies is gaining momentum [2, 63]. Thus, a strong history of using yeast for production of biopharmaceuticals suggests that this platform could be used for the production of shRNA-based biopharmaceuticals and biopesticides.
Following development of the initial concept and selection of target molecules in microbes, the process of bioengineering strains for production of commodity chemicals involves the characterization of target molecule expression and optimization of expression levels, overcoming any toxicity that may develop in the host strain, optimizing host metabolism to enhance biomolecule production, and scale-up of the production to commercial reactors [64] (Fig. 1). The CD40 silencing and mosquito pesticide studies discussed above illustrate that target molecules selection and expression can be achieved in S. cerevisiae. However, the path from target shRNA molecule selection and initial engineering of shRNA expression strains to scaled production of commercial volumes is less clear. As noted above, S. cerevisiae has been used to produce a number of commercially manufactured biopharmaceuticals [20]. Unfortunately, none of these products were RNA products, and so optimized and scaled production of interfering RNA molecules in S. cerevisiae will likely require considerable research and development.
Fig. (1).
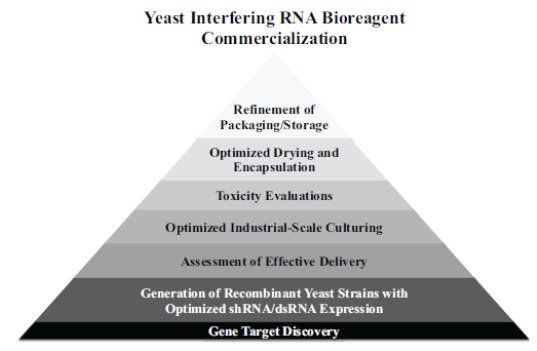
Pathway to commercialization of yeast interfering RNA bioreagent technology.
Chubukov et al. [64] discuss the challenges of using synthetic and systems biology for producing commodity chemicals in microbes, and many of their findings may be applicable to the development of recombinant yeast-interfering RNA expression systems. The authors explain that to date, the production of biomolecules by microbes has largely focused on pharmaceutical production, as these molecules will enter high-price low-volume markets where they have a higher likelihood of making a more immediate and tangible impact [65]. However, as more of these molecules are successfully developed, both the need and the potential for producing high-volume and low-cost molecules, such as biopesticides, will likely grow. In either case, the commercial potential of these molecules must be assessed, typically during the development phase prior to scale-up, but this is not an easy task. The development of reasonable estimates requires knowledge of the chemical properties and efficacy of the product, as well as assessment of the price and feasibility of producing the biomolecules by microbes at scale. Likewise, it relies on the ability to predict additional developmental costs required for scale-up, as well as down-stream purification costs. Such predictions are difficult at best but will likely become more straightforward as the volume of successful biomolecule production schemes developed in S. cerevisiae increases [64]. Given the many potential applications of yeast-interfering RNA technology, optimization of the scaled production of interfering RNA molecules in yeast could yield high dividends, but it is very difficult at this juncture to estimate the costs of successfully scaling and commercializing this technology. Moreover, although some parameters will undoubtedly vary depending on the interfering RNA molecules to be produced for each given application, investment into the optimization of this process for several recombinant RNA-producing yeast strains is likely to be broadly applicable to many different RNA-based biomolecule production schemes in yeast. Given the potential benefits, some of the key areas that will need to be developed will now be discussed.
4.2. Optimizing shRNA Production in S. cerevisiae
Although S. cerevisiae lacks recognizable orthologs of both Dicer and Argonaute, introduction of these proteins into S. cerevisiae generates RNAi activity [3] (Table 1). Using S. cerevisiae in which RNAi machinery had been reconstituted through heterologous expression of Argonaute and Dicer, Crook et al. [15] worked to optimize yeast as a metabolic engineering tool (Table 1). Their overall goal was to use RNAi to identify genetic targets that can be modulated to improve yeast strains used for synthesis of various biological molecules, but their findings may also be applicable more generally to the commercialization of dsRNA molecule production in yeasts. To begin to optimize their yeast shRNA expression system, the authors evaluated how hairpin expression levels, hairpin length, and copy number of the hairpin expression plasmid influenced target gene silencing. A yellow fluorescent protein (YFP) gene that had been artificially expressed in yeast cells served as the target gene in these studies, as detection of YFP fluorescence readout permitted straightforward monitoring of gene silencing. Increased downregulation of YFP fluorescence levels was observed when the shRNA was expressed from a strong pTDH3 promoter as opposed to a weak pCYC1 promoter, with 80% downregulation of YFP expression observed when the strong promoter drove shRNA expression. They also observed an increase in gene silencing when the hairpin length was increased from 100 to 200 bp. Expression of a 200 bp shRNA with the strong promoter resulted in 94% downregulation of YFP fluorescence, though the authors reported difficulty in construction and propagation of strains with an inverted repeat of this size. They suspected that the problem might be the result of interference with DNA replication machinery [66], which was resolved through inclusion of an intron-containing spacer that increased plasmid stability [67].
Next, Crook et al. [15] examined the impact of shRNA expression plasmid copy number on YFP silencing. Yeast 2 μ-based plasmids, which are maintained at copy numbers of 5-30 per cell in in S. cerevisiae, can facilitate high transgene expression levels, but these plasmids can be difficult to maintain and may be less stable [68]. Centromeric plasmids are another option; these plasmids bear an autonomously replicating origin and a yeast centromere, but only a single copy is present in each yeast cell [69], making it difficult to promote high expression levels [68]. After exploring the use of several different vectors, Crook et al. [15] determined that silencing was improved through the use of a low copy auxotrophic TRP1 marker vector that enabled 93% downregulation of YFP fluorescence, though they ultimately concluded that a mechanism other than copy number was responsible for the improved silencing observed when using low copy auxotrophic vectors in their assays. Another option that was not explored by Crook et al. [15], but that was recently used in our own yeast interfering RNA system [9], is to stably integrate expression cassettes into the yeast genome by coupling integration with selection of a deficient marker. Although several multiple integration approaches have been developed, there is not always a linear correlation between copy number and yield [68]. Indeed, we have not observed higher levels of interfering RNA larvicide activity in strains in which the copy number was increased from two to four copies of the integrated RNA expression cassette (MDS, unpublished).
After optimizing their system, Crook et al. [15] were able to quickly use RNAi for improvement of itaconic acid production in S. cerevisiae. More recently, the development of S. cerevisiae dsRNA expression libraries has facilitated tunable RNAi screens aimed at optimizing yeast biomolecule production systems through RNAi-assisted genome engineering [16, 17] (Table 1). These systems will make it possible to use RNAi to optimize expression of a wide variety of therapeutics, such as insulin and vaccines, in yeast. The results of this study may also have implications for the use of yeast-shRNA expression systems for the silencing of genes in humans or insects, though it is difficult to know how well the results will translate to improved silencing efficiency in non-yeast cells. It should also be noted that silencing efficacy must be optimized for each separate target gene, and so the results of YFP gene silencing studies may not be applicable to every target gene in yeast or non-yeast cells.
Rather than cloning shRNA expression cassettes, when their RNAi screening libraries were produced, both Si et al. [16] and Crook et al. [17] used a different approach for production of dsRNA in yeast. Both groups opted to clone larger pieces of yeast DNA (i.e. 200-400 bp fragments) between converging reporters that were used for production of dsRNA corresponding to the sequence cloned between the two promoters. A similar approach was recently used by Muprhy et al. [14] to produce yeast-based dsRNA insecticides for control of Drosophila pest species (Table 1). These longer pieces of dsRNA induced efficient gene silencing both in yeast and in Drosophila. Crook et al. [17] made an effort to optimize RNAi efficiency when they constructed their libraries. Using their YFP expression system to evaluate RNAi efficacy, they determined that the addition of introns improved downregulation of YFP. They again found that use of a low-copy vector actually significantly improved YFP downregulation as it had in their previous shRNA study [15]. Although comparisons of silencing rates induced by shRNA vs. dsRNA have yet to be performed, cloning longer dsRNA molecules is undoubtedly more straightforward that cloning shRNA expression cassettes and therefore much more suitable for construction of whole yeast genome RNAi screening libraries. Moreover, examining the expression levels of shRNAs that are very short in length, such as those used in the Hapairai et al. [9] and Mysore et al. [10] studies, is extremely difficult. Due to the secondary structure of these molecules, it is difficult to PCR-amplify cDNA prepared from shRNA or to detect it with Northern blots. In these cases [9, 10], measure of RNAi efficacy required examination of target gene RNA transcript levels, which were reduced by ~90%. This issue could be resolved by the use of longer shRNA molecules, such as those used by Zhang et al. [13], the levels of which could be directly monitored by qRT-PCR.
The use of inducible promoters for expression of RNA should also be explored further. In our study [9], although shRNA expression was initially under control of a constitutive promoter, expression of shRNA was placed under an inducible Gal1 promoter when the shRNA expression cassettes were integrated into the yeast genome. We reasoned that use of this promoter would be necessary when the expression cassettes were stably integrated, as it could potentially make up for the lower copy number (initially one, and then increased to two) of the integrated expression cassettes. This did appear to be the case, as the copy number was reduced ~10 fold, but comparable larvicide activities resulted. However, it has been argued that the use of galactose for induction of gene expression is not realistic for large-scale industrial fermentations, as production is complicated by the requirement to use galactose as the carbon source, and galactose is more expensive than glucose [70]. Also, a lack of Gal1 promoter induction has been observed when the carbon source is switched from glucose to galactose under anaerobic conditions [71, 72]. It may therefore be advisable to choose other promoters, and assessment of promoters that may be more readily used in industrial-sized cultures will be important.
Clearly, although much work has been undertaken and significant strides were made, much more work could be pursued to define the best parameters for production of interfering RNA in yeast. For example, future studies should directly compare the efficacy of shRNA vs. longer dsRNA yeast expression systems. Parameters such as the duration of target gene silencing, dsRNA half-life, and propensity for off-site targeting should also be included in these comparative studies. These parameters are quite likely to change depending on the gene being studied and the organism in which the yeast will ultimately be used, and so it will be useful to collect data for multiple transgenes and in different tissues.
4.3. Scaling Yeast Production
Recent advances in molecular genetics, systems and synthetic biology have revolutionized the potential for advancing yeast biosynthetic systems. Further advancement of this potential into commercial production requires scaling production from lab cultures sizes (i.e. one ml to one L culture volumes) to industrial scaled commercial biorecators (i.e. one hundred to one million L volumes). Unfortunately, promising strategies that work well in the laboratory do not always directly scale well under industrial fermentation conditions or are not robust enough to withstand changes in system parameters [64]. Chubukov et al. [64] comment that implementation of ideas in biological engineering requires significantly more effort than creating them and can require 5-10 years for further development and optimization of the process. A difficulty in predicting the behavior of large-scale fermentations is at the root of this problem [73], underscoring the utility of piloting scaled production during early phases of a project.
As discussed by Chubukov et al. [64], a variety of factors can contribute to difficulties in scale-up, resulting in reduced yields of the desired product, undesired side products, and problems with batch consistency. In particular, production schemes that exhibit susceptibility to minor variations in cultivation conditions in the laboratory or which cannot be easily reproduced within or between laboratories are not likely to fare well when scaled [64]. Physiological and metabolic states can be different in large versus small scale cultures, leading to altered growth rates. Challenges in mixing associated with large bioreactors can result in non-homogenous conditions, with gradients of glucose and oxygen leading to poor yields as a result. Re-optimizing culturing conditions, including pH, aeration, and carbon source feeding rates may therefore be necessary when production size is increased [68]. Chubukov et al. [64] also emphasize that it is important to identify optimal flux distributions by manipulating host metabolism to direct as much flux as possible to production of the desired product. As discussed above, dsRNA screening libraries have led to the identification of genetic factors that could be manipulated to improve production of desired products in S. cerevisiae [16, 17]. Now that these screening libraries have been constructed, such strategies, which result directly from the genetic tractability of this species, are likely to be invoked more frequently and promise to have a profound impact on the yeast biomolecule production industry.
While toxicity of the product to the host strain can be a problem in yeast cultures of any size, toxicity is often a problem at the scale-up stage [64]. Toxicity, which has primarily been studied in the context of recombinant protein production, could also be problematic when dsRNA molecules are expressed at high levels in yeast. The lack of native RNA machinery in S. cerevisiae could offer some protection against dsRNA toxicity, though non-processed dsRNA could potentially have cytotoxic effects as well. However, though we have constructed and characterized many different shRNA expression strains in our laboratory [9, 10] (MDS unpublished), we have not yet noticed any significant toxicity in these strains. However, it should be noted that we have not yet scaled culture sizes to more than 1 L in volume. Interestingly, the LD50 and LD90 values for most of the yeast interfering RNA larvicide strains produced by our group to date are fairly comparable (MDS, unpublished). It is therefore possible that some sort of negative feedback on shRNA production in yeast, shRNA stability in yeast, processing of shRNA, or the silencing of target genes in insect cells, may exist. When toxicity results from protein production in S. cerevisiae, problems with strain toxicity are often addressed through manipulation of transcriptional control, which is highly modular and well characterized in this species [Chubukov, 2016 #70]. Given that the goal will be to produce RNA rather than protein as a product, the wealth of knowledge regarding transcriptional regulation in S. cerevisiae will be an asset for overcoming toxicity issues that might arise when RNA is produced in yeast. Finally, in addition to considering toxicity of the RNA to the yeast strain, when developing yeast interfering RNA strains for commercialization, it is of course also critical to evaluate the potential toxicity of these strains to humans or other non-target organisms. This review has touched upon several efforts to improve the safety of this technology (i.e. heat-inactivation of yeast, generation of stable transformants that eliminate the need for use of plasmids with antibiotic resistance markers). While this subject is much broader than the scope of this review, it is nevertheless a critical aspect of evaluating the commercial potential of yeast interfering RNA technology.
4.4. Considerations for Drying and Encapsulation
Beyond scaling yeast production, if recombinant yeast is to be produced in formulations that can be consumed by humans or used to treat containers in which mosquitoes are breeding, the yeast will need to be formulated, packaged, shipped, and stored, all the while maintaining the integrity of the recombinant interfering RNA molecules (Fig. 1). Based on our success with dried and inactivated yeast interfering RNA larvicides (Fig. 2A), we anticipate that the desired end product is an inactivated, dried, ready-to use tablet. Given that the tablets we produced [9, 10] were modeled after yeast nutritional tablets, a human dietary supplement, it is quite possible that a similar preparation could be used for human gene therapy applications. Likewise, one could envision that a similar formulation could be used for whole yeast vaccine applications [2, 63] or for the delivery of mRNA, DNA, or proteins to intestinal DC cells [27, 30-32], and so further development of this technology could have multiple applications (Fig. 2B). Unfortunately, our current procedure for generating dried inactivated tablets is a laborious process that produces only several tablets at a time (Fig. 2A) [9, 10]. Clearly, the process of producing dried inactivated yeast tablets will need to be scaled for commercial production.
Fig. (2).
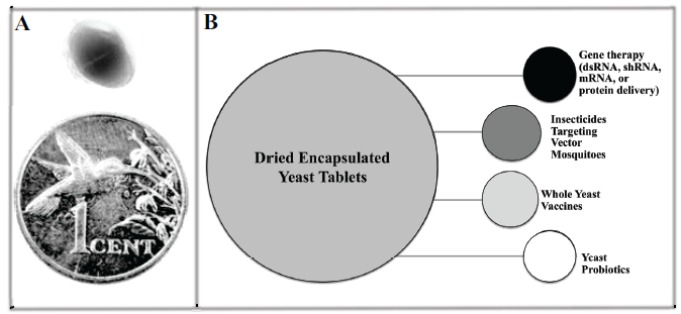
Potential applications for dried encapsulated yeast tablets. (A) A dried inactivated yeast tablet prepared as described [9, 10] is shown at top; a penny is shown for scale below it. (B) Delivery of interfering RNA, mRNA, or proteins through yeast requires the development of dried encapsulated tablet formulations. The development of whole yeast vaccines and yeast probiotic therapeutics would also benefit from research and development on yeast encapsulation and drying.
Modern food drying technologies for processing natural compounds aim to effectively dry the materials while maintaining high concentrations of bioactive compounds [74]. Freeze drying is a popular method of preserving microbial cultures and is often used for the preservation of live yeast strains that are heat-labile [75]. However, it is a somewhat expensive and laborious process, and the hygroscopic product can pick up moisture from the atmosphere [76]. Fluidized bed drying is cost effective and has been successfully used to produce products in the food, pharmaceutical, and agricultural industries [74]. This system has many advantages, including high drying rates and high thermal efficiency, and it has been used to dry yeast without significant loss of vitamins [74]. Spray drying, which may increase product shelf life, has also been explored in yeast [74, 76]. The spray drying method is widely used because it is cost-effective, enhances evaporation of liquids, and is readily scaled [74]. Spray drying at lower temperatures (i.e. 40 – 60o C) has led to better preservation of a number of biological compounds [74, 77], and this is likely to be the case for RNA molecules, as well. It is not presently known whether dsRNA or shRNA expressed in yeast withstands any of these drying processes, and so it will be critical to explore this concept further. Following drying, methods for large-scale tableting of the dried yeast will also need to be employed.
Although we have recently succeeded in the development of yeast interfering RNA tablets (Fig. 2A) that are stable at room temperature for at least two weeks (MDS, unpublished), shelf life stabilities of 1-2 years are more typical for commercial products. Moreover, exposure to extreme temperatures and variable humidity conditions is common when materials are shipped, and the final formulation must be stable under such conditions. It is therefore critical that packaging and storage conditions are optimized in preparation for commercialization (Fig. 1). Encapsulation of yeast may offer some protection against environmental conditions [76]. Furthermore, encapsulation can also help facilitate controlled release of microorganisms [78], which is likely useful given that time-release yeast interfering RNA formulations will likely be preferred for both medical and mosquito control applications. A wide range of food grade materials have been developed for microencapsulation and controlled release technologies applied to tableted preparation [76, 78, 79]. The development of methods for microencapsulation and time-release formulation of yeast has grown in recent years, as interest in the use of yeast probiotics has increased [79]. Although yeast used in probiotic applications must be viable, which may not be necessary for yeast RNA delivery systems (see above), these studies may nevertheless be of interest for RNA delivery applications and would likely benefit from further research in this area (Fig. 2). Alginate-chitosan microencapsulation of Saccharomyces has been explored in recent studies [80, 81]. The use of chitosan is of interest, as chitosan itself can also be used for delivery of dsRNA/ siRNA molecules [45, 82, 83], and the potential to combine yeast and chitosan delivery systems is intriguing. Jantzen et al. [84] explored the use of whey as a bacterial substrate and encapsulation matrix within a coupled fermentation and spray drying process, concluding that this technology is an efficient method for industrial production of probiotics. Thus, many potential agents for microbial encapsulations are available, and this will be a critical area of exploration for improvement of yeast RNA delivery systems.
CONCLUSION
In conclusion, S. cerevisiase may be an excellent system for production and delivery of interfering RNA molecules, and the development of yeast in this capacity clearly has many potential applications of medical importance. This review has highlighted the many advantages of the S. cerevisiae system, while also outlining key areas that will require further research and development. We have only begun to explore how to optimize shRNA and dsRNA expression levels in yeast, and more work must be pursued, particularly in industrial-scaled cultures. It will be critical to pilot larger-scaled culturing of yeast interfering RNA production strains in the near future. This will help us examine the potential for culturing these strains at industrial-sized scale and to understand how the strains and their growth conditions can be further optimized in preparation for commercial-scale production. Moreover, it will be critical to develop scaled methods for drying yeast that preserve the integrity of the interfering RNA molecules. The identification of encapsulating agents that promote yeast stability in various environmental conditions, both prior to and during its use, as well as to facilitate the controlled release of yeast will also be key. As discussed by Chubukov et al. [64], the implementation of ideas in biological engineering can sometimes require significantly more effort than the creation of these ideas. Thus, while this technology shows a great deal of promise in the laboratory, a good deal more work will be required for the successful commercialization of this technology. The genetic tractability of S. cerevisiae and the long history of using this microbe in both the food and pharmaceutical industry will undoubtedly benefit the development of this microbe as a production and delivery system for interfering RNA molecules.
ACKNOWLEDGEMENTS
Thank you to the Innovative Vector Control Consortium, David W. Severson, Na Wei, Nicole Achee, John Grieco, and to the Scheel laboratory for useful discussions.
LIST OF ABBREVIATIONS
- DC
Dendritic cell
- dsRNA
Double stranded RNA
- miRNA
micro RNA
- RNAse III
Ribonuclease III
- RNAi
RNA interference
- siRNAs
Small interfering RNAs
- RNAse III
Ribonuclease III
- shRNA
Short hairpin RNA
- WYV
Whole yeast vaccine
- YFP
Yellow fluorescent protein
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
Funding for work on yeast interfering RNA larvicides in the lab of MDS is funded by the United States Agency for International Development (Award AID-OAA-F-16-00097 to MDS), the NIH/NIAID (Award 1R21 AI128116-02 to MDS, David W. Severson, and Na Wei), the Department of Defense (Award W81XWH-17-1-0294 to MDS and David W. Severson), and the Showalter Trust. The funders of this research program did not impact the content or opinions presented in this review article nor the decision to publish it. Indiana University has submitted a patent application on which MDS is named as an inventor of technologies related to the work described herein. This application did not influence the content of this review article, the opinions presented herein, nor the decision to publish this article.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Moazed D. Molecular biology. Rejoice--RNAi for yeast. Sci. 2009;326(5952):533–534. doi: 10.1126/science.1182102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roohvand F., Shokri M., Abdollahpour-Alitappeh M., Ehsani P. Biomedical applications of yeast- a patent view, part one: yeasts as workhorses for the production of therapeutics and vaccines. Expert Opin. Ther. Pat. 2017;27(8):929–951. doi: 10.1080/13543776.2017.1339789. [DOI] [PubMed] [Google Scholar]
- 3.Drinnenberg I.A., Weinberg D.E., Xie K.T., et al. RNAi in budding yeast. Science. 2009;326(5952):544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malone C.D., Hannon G.J. Small RNAs as guardians of the genome. Cell. 2009;136(4):656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomari Y., Zamore P.D. Perspective: machines for RNAi. Genes Dev. 2005;19(5):517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 6.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Ghildiyal M., Zamore P.D. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009;10(2):94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tam C., Wong J.H., Cheung R.C.F., Zuo T., Ng T.B. Therapeutic potentials of short interfering RNAs. Appl. Microbiol. Biotechnol. 2017;101(19):7091–7111. doi: 10.1007/s00253-017-8433-z. [DOI] [PubMed] [Google Scholar]
- 9.Hapairai L.K., Mysore K., Chen Y., et al. Lure-and-kill yeast interfering RNA larvicides targeting neural genes in the human disease vector mosquito Aedes aegypti. Sci. Rep. 2017;7(1):13223. doi: 10.1038/s41598-017-13566-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mysore K., Hapairai L.K., Sun L., et al. Yeast interfering RNA larvicides targeting neural genes induce high rates of Anopheles larval mortality. Malar. J. 2017;16(1):461. doi: 10.1186/s12936-017-2112-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LaMattina J. Big pharma's turn on rnai shows that new technologies don't guarantee R & D Success. Forbes. 2014 [Google Scholar]
- 12.Tiemann K., Rossi J.J. RNAi-based therapeutics-current status, challenges and prospects. EMBO Mol. Med. 2009;1(3):142–151. doi: 10.1002/emmm.200900023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang L., Zhang T., Wang L., et al. In vivo targeted delivery of CD40 shRNA to mouse intestinal dendritic cells by oral administration of recombinant Sacchromyces cerevisiae. Gene Ther. 2014;21(7):709–714. doi: 10.1038/gt.2014.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy K.A., Tabuloc C.A., Cervantes K.R., Chiu J.C. Ingestion of genetically modified yeast symbiont reduces fitness of an insect pest via RNA interference. Sci. Rep. 2016;6:22587. doi: 10.1038/srep22587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crook N.C., Schmitz A.C., Alper H.S. Optimization of a yeast RNA interference system for controlling gene expression and enabling rapid metabolic engineering. ACS Synth. Biol. 2014;3(5):307–313. doi: 10.1021/sb4001432. [DOI] [PubMed] [Google Scholar]
- 16.Si T., Luo Y., Bao Z., Zhao H. RNAi-assisted genome evolution in Saccharomyces cerevisiae for complex phenotype engineering. ACS Synth. Biol. 2015;4(3):283–291. doi: 10.1021/sb500074a. [DOI] [PubMed] [Google Scholar]
- 17.Crook N., Sun J., Morse N., Schmitz A., Alper H.S. Identification of gene knockdown targets conferring enhanced isobutanol and 1-butanol tolerance to Saccharomyces cerevisiae using a tunable RNAi screening approach. Appl. Microbiol. Biotechnol. 2016;100(23):10005–10018. doi: 10.1007/s00253-016-7791-2. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J., Khan S.A., Hasse C., et al. Pest control. Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids. Sci. 2015;347(6225):991–994. doi: 10.1126/science.1261680. [DOI] [PubMed] [Google Scholar]
- 19.Mewes H.W., Albermann K., Bahr M., et al. Overview of the yeast genome. Nat. 1997;387(6632) Suppl.:7–65. doi: 10.1038/42755. [DOI] [PubMed] [Google Scholar]
- 20.Wang G., Huang M., Nielsen J. Exploring the potential of Saccharomyces cerevisiae for biopharmaceutical protein production. Curr. Opin. Biotechnol. 2017;48:77–84. doi: 10.1016/j.copbio.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 21.Meehl M.A., Stadheim T.A. Biopharmaceutical discovery and production in yeast. Curr. Opin. Biotechnol. 2014;30:120–127. doi: 10.1016/j.copbio.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Roeder A., Kirschning C.J., Rupec R.A., Schaller M., Korting H.C. Toll-like receptors and innate antifungal responses. Trends Microbiol. 2004;12(1):44–49. doi: 10.1016/j.tim.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 23.Banchereau J., Steinman R.M. Dendritic cells and the control of immunity. Nat. 1998;392(6673):245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 24.Palucka K., Banchereau J. Dendritic cells: A link between innate and adaptive immunity. J. Clin. Immunol. 1999;19(1):12–25. doi: 10.1023/a:1020558317162. [DOI] [PubMed] [Google Scholar]
- 25.Qian C., Qian L., Yu Y., et al. Fas signal promotes the immunosuppressive function of regulatory dendritic cells via the ERK/beta-catenin pathway. J. Biol. Chem. 2013;288(39):27825–27835. doi: 10.1074/jbc.M112.425751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blanquet S., Meunier J.P., Minekus M., Marol-Bonnin S., Alric M. Recombinant Saccharomyces cerevisiae expressing P450 in artificial digestive systems: a model for biodetoxication in the human digestive environment. Appl. Environ. Microbiol. 2003;69(5):2884–2892. doi: 10.1128/AEM.69.5.2884-2892.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stubbs A.C., Martin K.S., Coeshott C., et al. Whole recombinant yeast vaccine activates dendritic cells and elicits protective cell-mediated immunity. Nat. Med. 2001;7(5):625–629. doi: 10.1038/87974. [DOI] [PubMed] [Google Scholar]
- 28.Franzusoff A., Duke R.C., King T.H., Lu Y., Rodell T.C. Yeasts encoding tumour antigens in cancer immunotherapy. Expert Opin. Biol. Ther. 2005;5(4):565–575. doi: 10.1517/14712598.5.4.565. [DOI] [PubMed] [Google Scholar]
- 29.Howland SW, Tsuji T, Gnjatic S, et al. Inducing efficient crosspriming using antigen-coated yeast particles. J Immunother (Hagerstown, Md : 1997) 2008;31(7):607-19. doi: 10.1097/CJI.0b013e318181c87f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walch B., Breinig T., Schmitt M.J., Breinig F. Delivery of functional DNA and messenger RNA to mammalian phagocytic cells by recombinant yeast. Gene Ther. 2012;19(3):237–245. doi: 10.1038/gt.2011.121. [DOI] [PubMed] [Google Scholar]
- 31.Haller A.A., Lauer G.M., King T.H., et al. Whole recombinant yeast-based immunotherapy induces potent T cell responses targeting HCV NS3 and core proteins. Vaccine. 2007;25(8):1452–1463. doi: 10.1016/j.vaccine.2006.10.035. [DOI] [PubMed] [Google Scholar]
- 32.Wansley E.K., Chakraborty M., Hance K.W., et al. Vaccination with a recombinant Saccharomyces cerevisiae expressing a tumor antigen breaks immune tolerance and elicits therapeutic antitumor responses. Clin. Cancer Res. 2008;14(13):4316–4325. doi: 10.1158/1078-0432.CCR-08-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lutgens E., Daemen M.J. CD40-CD40L interactions in atherosclerosis. Trends Cardiovasc. Med. 2002;12(1):27–32. doi: 10.1016/s1050-1738(01)00142-6. [DOI] [PubMed] [Google Scholar]
- 34.Ma D.Y., Clark E.A. The role of CD40 and CD154/CD40L in dendritic cells. Semin. Immunol. 2009;21(5):265–272. doi: 10.1016/j.smim.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silva J.M., Li M.Z., Chang K., et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat. Genet. 2005;37(11):1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 36.Paddison P.J., Silva J.M., Conklin D.S., et al. A resource for large-scale RNA-interference-based screens in mammals. Nat. 2004;428(6981):427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- 37.Zeng Y., Wagner E.J., Cullen B.R. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell. 2002;9(6):1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 38.Xu K., Liu Z., Zhang L., Zhang T., Zhang Z. siRNA in vivo-targeted delivery to murine dendritic cells by oral administration of recombinant yeast. Methods Mol. Biol. 2016;1364:165–181. doi: 10.1007/978-1-4939-3112-5_14. [DOI] [PubMed] [Google Scholar]
- 39.WHO. World Malaria Report 2016. Geneva: World Health Organization; 2016. [Google Scholar]
- 40. CDC. Surveillance and control of Aedes aegypti and Aedes albopictus. 2016: Available from: http://www.cdc.gov/chikungunya/ resources/vector-control.html [Accessed January, 2016].
- 41. CDC. Zika virus. 2018: Available from: https://www.cdc.gov/zika/ index.html [Accessed January 2018].
- 42. CDC. Dengue. 2018: Available from: https://www.cdc.gov/dengue/ index.html [Accessed January 2018].
- 43.WHO. Dengue guidelines for diagnosis, treatment, prevention and control: new edition. Geneva: World Health Organization; 2009. [PubMed] [Google Scholar]
- 44.WHO. Larval source management: a supplementary measure for malaria vector control: an operational manual. Geneva: World Health Organization; 2013. [Google Scholar]
- 45.Mysore K., Flannery E.M., Tomchaney M., Severson D.W., Duman-Scheel M. Disruption of Aedes aegypti olfactory system development through chitosan/siRNA nanoparticle targeting of semaphorin-1a. PLoS Negl. Trop. Dis. 2013;7(5):e2215. doi: 10.1371/journal.pntd.0002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mysore K., Andrews E., Li P., Duman-Scheel M. Chitosan/siRNA nanoparticle targeting demonstrates a requirement for single-minded during larval and pupal olfactory system development of the vector mosquito Aedes aegypti. BMC Dev. Biol. 2014;14:9. doi: 10.1186/1471-213X-14-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mysore K., Flannery E., Leming M.T., et al. Role of semaphorin-1a in the developing visual system of the disease vector mosquito Aedes aegypti. Dev. Dyn. 2014;243(11):1457–1469. doi: 10.1002/dvdy.24168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mysore K., Sun L., Tomchaney M., et al. siRNA-mediated silencing of doublesex during female development of the dengue vector mosquito Aedes aegypti. PLoS Negl. Trop. Dis. 2015;9(11):e0004213. doi: 10.1371/journal.pntd.0004213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mumberg D., Muller R., Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156(1):119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 50.Fire A., Xu S., Montgomery M.K., et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nat. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 51.Timmons L., Court D.L., Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263(1-2):103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 52.Tenllado F., Martinez-Garcia B., Vargas M., Diaz-Ruiz J.R. Crude extracts of bacterially expressed dsRNA can be used to protect plants against virus infections. BMC Biotechnol. 2003;3:3. doi: 10.1186/1472-6750-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang L., Jin J., Deighan P., et al. Efficient and specific gene knockdown by small interfering RNAs produced in bacteria. Nat. Biotechnol. 2013;31(4):350–356. doi: 10.1038/nbt.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang L., Lieberman J. Production of highly potent recombinant siRNAs in Escherichia coli. Nat. Protoc. 2013;8(12):2325–2336. doi: 10.1038/nprot.2013.149. [DOI] [PubMed] [Google Scholar]
- 55.Kaur G., Cheung H.C., Xu W., et al. Milligram scale production of potent recombinant small interfering RNAs in Escherichia coli. Biotechnol. Bioeng. 2018;115(9):2280–2291. doi: 10.1002/bit.26740. [DOI] [PubMed] [Google Scholar]
- 56.Chanfreau G., Legrain P., Jacquier A. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J. Mol. Biol. 1998;284(4):975–988. doi: 10.1006/jmbi.1998.2237. [DOI] [PubMed] [Google Scholar]
- 57.Lamontagne B., Tremblay A., Abou Elela S. The N-terminal domain that distinguishes yeast from bacterial RNase III contains a dimerization signal required for efficient double-stranded RNA cleavage. Mol. Cell. Biol. 2000;20(4):1104–1115. doi: 10.1128/mcb.20.4.1104-1115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gaudin C., Ghazal G., Yoshizawa S., Elela S.A., Fourmy D. Structure of an AAGU tetraloop and its contribution to substrate selection by yeast RNase III. J. Mol. Biol. 2006;363(2):322–331. doi: 10.1016/j.jmb.2006.08.029. [DOI] [PubMed] [Google Scholar]
- 59.Lamontagne B., Hannoush R.N., Damha M.J., Abou Elela S. Molecular requirements for duplex recognition and cleavage by eukaryotic RNase III: discovery of an RNA-dependent DNA cleavage activity of yeast Rnt1p. J. Mol. Biol. 2004;338(2):401–418. doi: 10.1016/j.jmb.2004.02.059. [DOI] [PubMed] [Google Scholar]
- 60.Sam M., Henras A.K., Chanfreau G. A conserved major groove antideterminant for Saccharomyces cerevisiae RNase III recognition. Biochem. 2005;44(11):4181–4187. doi: 10.1021/bi047483u. [DOI] [PubMed] [Google Scholar]
- 61.Wu H., Henras A., Chanfreau G., Feigon J. Structural basis for recognition of the AGNN tetraloop RNA fold by the double-stranded RNA-binding domain of Rnt1p RNase III. Proc. Natl. Acad. Sci. USA. 2004;101(22):8307–8312. doi: 10.1073/pnas.0402627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nielsen J. Production of biopharmaceutical proteins by yeast: advances through metabolic engineering. Bioengin. 2013;4(4):207–211. doi: 10.4161/bioe.22856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ardiani A., Higgins J.P., Hodge J.W. Vaccines based on whole recombinant Saccharomyces cerevisiae cells. FEMS Yeast Res. 2010;10(8):1060–1069. doi: 10.1111/j.1567-1364.2010.00665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chubukov V., Mukhopadhyay A., Petzold C.J., Keasling J.D., Martin H.G. Synthetic and systems biology for microbial production of commodity chemicals. NPJ Syst. Biol. Appl. 2016;2:16009. doi: 10.1038/npjsba.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baeshen N.A., Baeshen M.N., Sheikh A., et al. Cell factories for insulin production. Microb. Cell Fact. 2014;13:141. doi: 10.1186/s12934-014-0141-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Voineagu I., Narayanan V., Lobachev K.S., Mirkin S.M. Replication stalling at unstable inverted repeats: interplay between DNA hairpins and fork stabilizing proteins. Proc. Natl. Acad. Sci. USA. 2008;105(29):9936–9941. doi: 10.1073/pnas.0804510105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoshimatsu T., Nagawa F. Control of gene expression by artificial introns in Saccharomyces cerevisiae. Sci. 1989;244(4910):1346–1348. doi: 10.1126/science.2544026. [DOI] [PubMed] [Google Scholar]
- 68.Kim H., Yoo S.J., Kang H.A. Yeast synthetic biology for the production of recombinant therapeutic proteins. FEMS Yeast Res. 2015;15(1):1–16. doi: 10.1111/1567-1364.12195. [DOI] [PubMed] [Google Scholar]
- 69.Fang F., Salmon K., Shen M.W., et al. A vector set for systematic metabolic engineering in Saccharomyces cerevisiae. Yeast. 2011;28(2):123–136. doi: 10.1002/yea.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Papapetridis I., Goudriaan M., Vazquez Vitali M., et al. Optimizing anaerobic growth rate and fermentation kinetics in Saccharomyces cerevisiae strains expressing Calvin-cycle enzymes for improved ethanol yield. Biotechnol. Biofuels. 2018;11:17. doi: 10.1186/s13068-017-1001-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van den Brink J., Akeroyd M., van der Hoeven R., et al. Energetic limits to metabolic flexibility: responses of Saccharomyces cerevisiae to glucose-galactose transitions. Microbiology. 2009;155(Pt 4):1340–1350. doi: 10.1099/mic.0.025775-0. [DOI] [PubMed] [Google Scholar]
- 72.Guadalupe-Medina V., Wisselink H.W., Luttik M.A., et al. Carbon dioxide fixation by Calvin-Cycle enzymes improves ethanol yield in yeast. Biotechnol. Biofuels. 2013;6(1):125. doi: 10.1186/1754-6834-6-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Takors R. Scale-up of microbial processes: impacts, tools and open questions. J. Biotechnol. 2012;160(1-2):3–9. doi: 10.1016/j.jbiotec.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 74.Nicula A.N., Nicula A.T., Socaciu C., Du Breucq P. Application of advanced drying technologies for obtaining bioactive beer. Bulletin UASVM Agriculture. 2009;66(2):581–590. [Google Scholar]
- 75.Bond C. Freeze drying of yeast cultures. Methods Mol. Biol. 2007;368:99–107. doi: 10.1007/978-1-59745-362-2_6. [DOI] [PubMed] [Google Scholar]
- 76.Chandralekha A., Tavanandi A.H., Amrutha N., et al. Encapsulation of yeast (Saccharomyces cereviciae) by spray drying for extension of shelf life. Dry. Technol. 2016;34(11):1307–1318. [Google Scholar]
- 77.Sultana A., Miyamoto A., Hy Q.L., et al. Microencapsulation of flavors by spray drying using Saccharomyces cerevisiae. J. Food Eng. 2017;199:36–41. [Google Scholar]
- 78.Lakkis J.M. Encapsulation and controlled release technologies in food systems. Oxford, U.K.: Blackwell Publishing; 2007. [Google Scholar]
- 79.Ghorbani-Choboghlo H., Zahraei-Salehi T., Ashrafi-Helan J., et al. Microencapsulation of Saccharomyces cerevisiae and its evaluation to protect in simulated gastric conditions. Iran. J. Microbiol. 2015;7(6):338–342. [PMC free article] [PubMed] [Google Scholar]
- 80.Graff S., Hussain S., Chaumeil J.C., Charrueau C. Increased intestinal delivery of viable Saccharomyces boulardii by encapsulation in microspheres. Pharm. Res. 2008;25(6):1290–1296. doi: 10.1007/s11095-007-9528-5. [DOI] [PubMed] [Google Scholar]
- 81.Song H., Yu W., Liu X., Ma X. Improved probiotic viability in stress environments with post-culture of alginate-chitosan microencapsulated low density cells. Carbohydr. Polym. 2014;108:10–16. doi: 10.1016/j.carbpol.2014.02.084. [DOI] [PubMed] [Google Scholar]
- 82.Zhang X., Zhang J., Zhu K.Y. Chitosan/double-stranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae). Insect Mol. Biol. 2010;19(5):683–693. doi: 10.1111/j.1365-2583.2010.01029.x. [DOI] [PubMed] [Google Scholar]
- 83.Zhang X, Mysore K, Flannery E, et al. Chitosan/interfering RNA nanoparticle mediated gene silencing in disease vector mosquito larvae. J Vis Exp. 2015;97 doi: 10.3791/52523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jantzen M., Gopel A., Beermann C. Direct spray drying and microencapsulation of probiotic Lactobacillus reuteri from slurry fermentation with whey. J. Appl. Microbiol. 2013;115(4):1029–1036. doi: 10.1111/jam.12293. [DOI] [PubMed] [Google Scholar]


