Abstract
Background
Gut microbiota has been suggested to play a role in stroke patients. Nevertheless, little is known about gut microbiota and the clinical indexes in stroke patients.
Methods
Total of 30 cerebral ischemic stroke (CI) patients and 30 healthy control were enrolled in this study and the fecal gut microbiota was profiled via Illumina sequencing of the 16S rRNA V1-V2. The National Institutes of Health Stroke Scale (NIHSS) were used to quantify stroke severity and modified Rankin scale (mRS) to assess outcome for CI patients. The correlations between the clinical indexes and microbiota were evaluated.
Results
Though the microbial α-diversity and structure is similar between CI patients and healthy controls, the gut microbiota of CI patients had more short chain fatty acids producer including Odoribacter, Akkermansia, Ruminococcaceae_UCG_005 and Victivallis. We also found that the special microbes were correlation with serum index, such as norank_O_ _Mollicutes_RF9, Enterobacter, Ruminococcaceae_UCG-002 were negative correlation with LDL (r = − 0.401, P < 0.01), HDL (r = − 0.425, P < 0.01) and blood glucose (r = − 0.439, P < 0.001), while the HDL was significantly positive correlation with the genus Ruminococcus_1 (r = 0.443, P < 0.001). The Christensenellaceae_R-7_group and norank_f_Ruminococcaceae was significantly positive correlation with NIHSS1M (r = 0.514, P < 0.05; r = 0.449, P < 0.05) and mRS (r = 0.471, P < 0.05, r = 0.503, P < 0.01), respectively. On the other hand, the genus Enterobacter was significantly negative correlation with NIHSS1M (r = 0.449, P < 0.05) and mRS (r = 0.503, P < 0.01).
Conclusions
This study suggests that CI patients showed significant dysbiosis of the gut microbiota with enriched short chain fatty acids producer, including Odoribacter, Akkermansia. This dysbiosis was correlation with the outcomes and deserves further study.
Electronic supplementary material
The online version of this article (10.1186/s12866-019-1552-1) contains supplementary material, which is available to authorized users.
Keywords: Cerebral ischemic stroke, Microbiota, Dysbiosis, Outcomes
Background
Cerebrovascular accidents (stroke) is brain injury caused by disruption the supply of blood to a brain region, which could result in permanent neurological deficits or death [1]. Now, stroke is global health problem and now became to the second leading cause of death as well as the third leading cause of disability [2]. Basing on the underlying pathology, the stroke was classified to ischemic and hemorrhagic stroke [2]. The cerebral Ischemic stroke (CI) is responsible for 85% of all strokes, and the hemorrhagic stroke accounts for about the rest 15% [2]. The most ischemic stroke is caused by the middle cerebral artery occlusion, resulting in the brain tissue damage in the affected territory, which is followed by inflammatory and immune response [1]. Despite the stroke debilitated the neurological deficits, infection is the superior cause of death after stroke [3]. It was found that 90 % of stroke cases may be correlated with behavioral factors including poor diet, smoking, and low physical activity as well as metabolic factors including obesity, hypertension, diabetes mellitus. Previous study also found that gut microbiota might be as a risk factor for stroke [4].
The gastrointestinal tract is thought to be a major immune organ which was equipped with the largest pool of immune cells, accounting for more than 70% of the entire immune system. More and more evidences propose that the gut inflammatory along with immune response plays an essential role in the pathophysiology of stroke and this may become an important therapeutic target for treatment of stroke [5]. It was found that gut microbiota plays a vital role in regulation of the immune system. The gut microbiota was also revealed to be an important influence factor for stroke in a mouse model [6–9]. In addition, stroke usually cause gut dysmotility, gut microbiota dysbiosis, gut hemorrhage, as well as gut-origin sepsis, resulting in poor prognosis. However, there are little about the characteristics of gut microbiota from stroke patient [10].
This study aims to determine the characteristic of gut microbiota of stroke patient. We also evaluated the relations between gut microbiota and serum indexes or outcome by Spearman’s rank correlation.
Results
Subjects
Total of 60 subjects, including 30 CI (21 males, 9 females) and 30 healthy control (18 males, 12 females) were recruited in this study (Table 1). It was found that there were no significant differences in gender (male: 70. % vs. 60.0%, P = 0.42) and age (60.47 ± 10.57 vs. 64.17 ± 12.67, P = 0.31) between the CI group and healthy control group. The LDL, GLU, UA, TG, HCY of CI patients were similar with the healthy control. The HDL of CI patients was lower than that of healthy control group (1.10 ± 0.19 vs. 1.29 ± 0.24, P < 0.01) (Table 1). The NIHSS at 7d (NIHSS 1 W) and 1 month (NIHSS 1 M), modulate RANK score (mRS) and serum index of each subjects were shown in Additional file 1: Table S1 and Additional file 2: Table S2.
Table 1.
Characteristics of the study participants
| CI (n = 30) | HC (n = 30) | P | |
|---|---|---|---|
| Sex (male, %) | 21 (70%) | 18 (60%) | 0.42 |
| Age | 60.47 ± 10.57 | 64.17 ± 12.67 | 0.31 |
| LDL | 2.20 ± 0.68 | 2.30 ± 0.50 | 0.51 |
| HDL | 1.10 ± 0.19 | 1.29 ± 0.24 | < 0.01 |
| UA | 303.8 ± 64.5 | 282.3 ± 62.7 | 0.20 |
| GLU | 4.92 ± 0.67 | 5.20 ± 0.76 | 0.14 |
| HCY | 12.94 ± 3.93 | 12.36 ± 3.33 | 0.54 |
| TG | 1.24 ± 0.47 | 1.33 ± 0.31 | 0.39 |
HDL high-density lipoprotein, LDL low-density lipoprotein, GLU glucose of blood, UA uric acid, TG triglycerides, HCY homocysteine
The microbial α-diversity and structure is similar between CI and HC group
We obtained a total of 2,191,980 high-quality sequences by quality-filtering with a coverage above 99.0%. After removing rare OTUs., 19,869 sequences per sample were obtained which were clustered to 955 OTUs. The results of bacterial community richness and diversity were shown in Table 2. It was found that there was no difference in shannon, simpson, ace or chao index between two group.
Table 2.
The index of α-diversity
| CI (n = 30) | HC (n = 30) | P | |
|---|---|---|---|
| Shannon | 3.69 ± 0.52 | 3.59 ± 0.59 | 0.3515 |
| Simpson | 0.09 ± 0.06 | 0.10 ± 0.07 | 0.4786 |
| Ace | 555.68 ± 51.24 | 552.19 ± 43.47 | 0.4679 |
| Chao | 565.64 ± 55.61 | 558.79 ± 48.05 | 0.7775 |
The similarity of the bacterial community structures between CI group and healthy control group was evaluated by PCoA (Fig. 1). There was no obvious difference of the microbiota structure between the 2 groups. Then, ANOSIM were performed and also found a high similarity in the bacterial community between CI patients and the healthy control based on two algorithms (unweighted unifrac, r = 0.0254, P = 0.132; weighted unifrac r = − 0.012, P = 0.649, respectively).
Fig. 1.
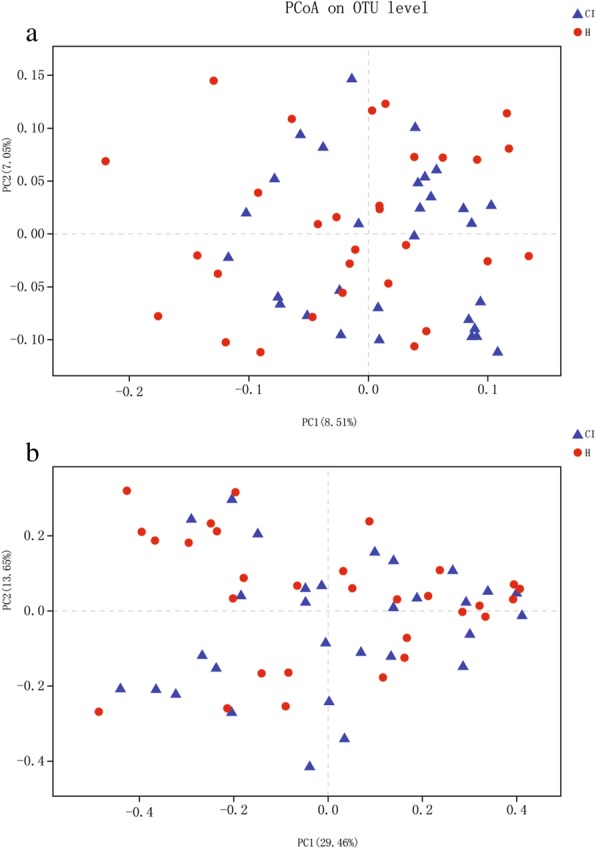
PCoA analysis of the microbiota among CI and healthy control groups. a Weighted unifrac PCoA; b Unweighted unifrac PCoA
Differentially abundant taxon in microbiota of CI patients and healthy control
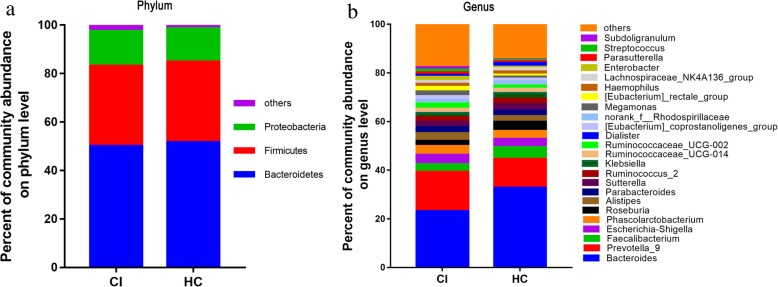
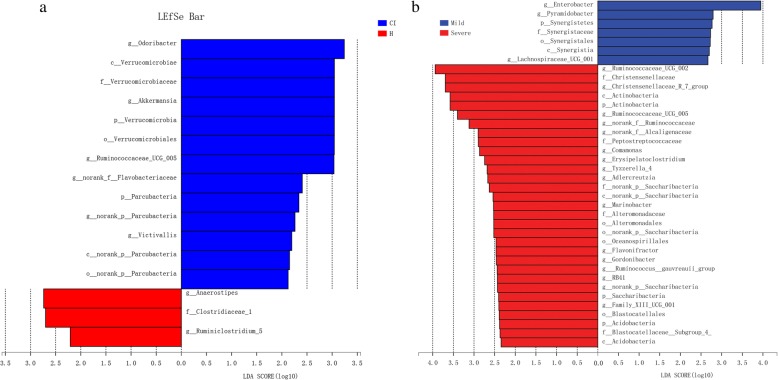
Most of the gut bacteria detected in this study falls into 3 phyla: Bacteroidetes, Firmicutes, and Proteobacteria (Fig. 2a). The main genus of gut micriobiota (the percentages were above 1%) includes 24 genera and they compose up to 80% of the total microbiota, such as Bacteroides, Prevotella, Faecalibacterium, Escherichia/Shigella, Phascolarctobacterium, and Roseburia (Fig. 2b). The taxa that most likely reveal the differences between CI and healthy control group were identified by LEfSe (Fig. 3). At the genus level, Anaerostipes and Ruminiclostridium_5 were significantly enriched in the healthy group, while Odoribacter, Akkermansia, Ruminococcaceae_UCG_005, norank_p_Flavobacteriaceae, norank_p_Parcubacteria, and Victivallis exhibited relatively higher abundance in the CI group (Fig. 3a). We also observed that the genus Enterobacter, Pyramidobacter, and Lachnospiraceae_UCG_001 increased in mild CI patients (NIHSS score ≤ 4) (Fig. 3b). While, genus Ruminococcaceae_UCG-002, Christensenellaceae_R-7_group, Ruminococcaceae_UCG-005 and norank_f_Ruminococcaceae increased in severe stroke patients (NIHSS score > 4) (Fig. 3b).
Fig. 2.
The relative taxa abundance between CI and healthy control groups. a Phylum; b Genus
Fig. 3.
a The most differentially abundant taxa between CI and b healthy control group based on LEfSe analysis
The gut microbiota was related to the clinical indices
The serum indices of each subjects were shown in Additional file 1: Table S1. We try to find relationship between the gut microbiota and the serum indices including TG, LDL, HDL, UA, Glu, and Hcy by Spearman’s rank correlation coefficient. We found strong correlations (r > 0.22 or < − 0.22, P < 0.05) between them as shown in Fig. 4. A significant positive correlation was exhibited between LDL and genus Bacteroides (r = 0.42, P < 0.01) as well as [Eubocterium]_rectole_group (r = 0.336, P < 0.01). While, it was found that norank_O_ _Mollicutes_RF9 was negative correlation with LDL (r = − 0.401, P < 0.01). The HDL was significantly positive correlation with the genus Ruminococcus_1 (r = 0.443, P < 0.001), Ruminococcus_2 (r = 0.381, P < 0.01) and Lachnospiraceae_NK4A136_group (r = 0.338, P < 0.01) and negative correlation with the genus Enterobacter (r = − 0.425, P < 0.01). The UA was found positive correlation with Dialister (r = 0.279, P < 0.05) and the GLU was found significantly negative correlation with Ruminococcaceae_UCG-002 (r = − 0.439, P < 0.001), Alistipes (r = − 0.313, P < 0.05), and Ruminococcus_1 (r = − 0.287, P < 0.05). And the Hcy was found positive correlation with Megamonas (r = 0.291, P < 0.05) and Fusobacterium (r = 0.297, P < 0.05).
Fig. 4.
Heatmap of spearman correlation analysis between the gut microbiota and the serum indices. *p < 0.05, **p < 0.01, ***p < 0.01. HDL, high-density lipoprotein; LDL, low-density lipoprotein; GLU, glucose of blood; UA, uric acid; TG, triglycerides; HCY, homocysteine
The gut microbiota was correlated with the severity and outcome of CI
The severity and outcome of CI patients could be evaluated by NIHSS and mRS (Additional file 2: Table S2). Then, we do the Spearman’s rank correlation coefficient analysis and attempt to explore the relationship between the gut microbiota and severity or outcome including NIHSS 1 W NIHSS 1 M and mRS (Fig. 5). As a result, the genus Christensenellaceae_R-7_group was significantly positive correlation with NIHSS1W (r = 0.488, P < 0.01), NIHSS1M (r = 0.514, P < 0.05) and mRS (r = 0.471, P < 0.05). While, the genus norank_f_Ruminococcaceae was also significantly positive correlation with NIHSS1M (r = 0.449, P < 0.05) and mRS (r = 0.503, P < 0.01). In contrast, the genus Enterobacter was significantly positive correlation with NIHSS1M (r = 0.449, P < 0.05) and mRS (r = 0.503, P < 0.01).
Fig. 5.
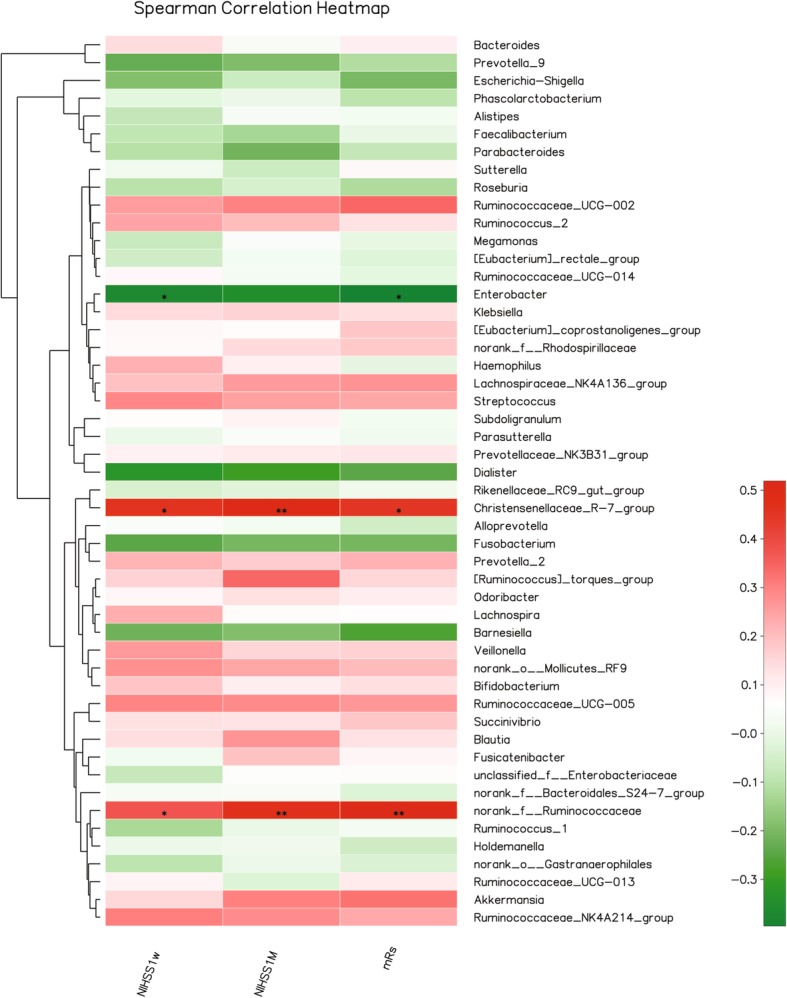
Heatmap of spearman correlation analysis between the gut microbiota and the outcome of CI patients. *p < 0.05, **p < 0.01, ***p < 0.01
Discussion
In this study, the results demonstrate that certain changes in fecal microbiota was happened in ischemic stroke patients, such as the increasement short-chain fatty acids (SCFAs)-producing taxonomies Odoribacter and Akkermansia. Furthermore, the levels the genus Christensenellaceae_R-7_group, norank_f_Ruminococcaceae, and Enterobacter were correlated with the severity, while the level of genus Christensenellaceae_R-7_group was positively correlated with the outcome of CI patients. In addition, we also found that the microbiota was correlated with the human hematological index.
Previous studies have proved that gut microbiota play a vital role in stroke. It was reported that depletion of gut bacteria via antibiotic administration decreased the survival rate of mice following the induction of ischemia [11]. In mice, Houlden et al., found that the composition of caecal microbiota was altered by experimental stroke, such as specific changes of Peptococcaceae and Prevotellaceace [6]. Benakis et al. found that the microbiota dysbiosis would affect the ischemic stroke outcome via suppressing the traffick of effector T cells from the gut to the leptomeninges in middle cerebral artery occlusion model [9]. Though only a few studies have investigated the role of microbiota in stroke patient, dysregulation of the microbiota in patients following stroke have been identified [12–14]. Yin et al., found that ischemic stroke was associated with different microbiota structure and more opportunistic pathogens, such as Megasphaera, Enterobacter, Oscillibacter, along with fewer commensal or beneficial genera such as Prevotella, Bacteroides, and Faecalibacterium [13]. After that, Yamashiro et al., found that ischemic stroke was independently associated with increasement of Atopobium cluster and Lactobacillus ruminis, and decreased numbers of L. sakei subgroup based on qRT-PCR [12]. In another study, gut microbiota dysbiosis was identified with increased abundance of Escherichia, Bacteroides, Megamonas, Parabacteroides, and Ruminococcus in the CI and IS patients compared with the healthy group. In the higher risk of stroke people, there was no significant change in alpha diversity index of the microbiota. However, enrichment of opportunistic pathogens together with low abundance of butyrate-producing bacteria were found in subjects with high risk of stroke [4]. In this study, we also found that the microbial α-diversity and structure was similar between CI patients and healthy control. However, the Anaerostipes and Ruminiclostridium_5 were significantly enriched in the healthy group, while Odoribacter, Akkermansia, Ruminococcaceae_UCG_005, norank_p_Flavobacteriaceae, and Victivallis represented a relatively higher abundance in CI group (Fig. 2a). Of them, Odoribacter, Akkermansia, Ruminococcaceae_UCG_005 and Victivallis are known producer of SCFAs, including acetate, propionate, and butyrate [4, 15–17]. Meanwhile, significant differences within the CI group was also observed that the Enterobacter, Pyramidobacter, and Lachnospiraceae_UCG_001 increased in mild CI patients (NIHSS score ≤ 4). While, genus Ruminococcaceae_UCG-002, Christensenellaceae_R-7_group, Ruminococcaceae_UCG-005 and norank_f_Ruminococcaceae increased in severe stroke patients (NIHSS score > 4) (Fig. 2b). These results suggest that the intestinal flora may have been involved in the human stroke and correlated to the severity of CI.
SCFAs are a major class of bacterial metabolites obtained from fermentation of otherwise indigestible polysaccharides (fibers) in the colon by bacteria [18]. Preview studies indicated that SCFAs have important role in maintaining the intestinal integrity and anti-inflammatory. As a result, beneficial effects of SCFAs have been found in several disease models, such as colitis, metabolic syndrome and so on [19]. In this study, we found that short-chain fatty acids (SCFAs)-producing taxonomies Odoribacter and Akkermansia increased in CI patient. Akkermansia muciniphila is the single species of genus Akkermansia in the human which accounts 1–3% of the total bacterial cells in feces [20]. It was found that A. muciniphila was inversely associated with diabetes, obesity, cardiometabolic diseases and low-grade inflammation [16]. Previous study demonstrated that the shift in mucosal microbial composition induced by the stroke were an increasement of A. muciniphila along with an excessive abundance of Clostridial species in mice [8]. However, it was reported that Akkermansia decreased in CI patient [14]. The Akkermansia was of 20% abundance of the total microbe in one normal control and this might cause statistical bias. In this study, we found that Akkermansia significantly increased in CI patient (P < 0.05) (Fig. 2c). It was reported that A. muciniphila could use mucin to produce high levels acetate which may be consumed by butyrate-producing Ruminococcaceae to promote butyrate production [4]. In this study, genus Odoribacter, a butyrate producer which belongs to phylum Bacteroidetes [21], also increase in CI patient. This indicated that the simultaneous increase of these two bacteria might promote the production of butyrate. Butyrate is a preferential energy source for epithelial cells and it preserves the epithelial heath [22]. Butyrate also could impact the expression level of genes which are aroused by A. muciniphila in epithelial cells [23]. Therefore, Stanley et al. thought that the Akkermansia may play a crucial role in healing of wound damage via promotion the butyrate levels, resulting in consolidate epithelial integrity in the post-stroke mice [8]. In addition, A. muciniphila could induce mucus production and expression of Reg3γ in colon, resulting microbiota remodeling [24]. Thus, further work needs to be done to find out the possible role of Akkermansia and Odoribacter in post-stroke.
Regarding the sequelae of a stroke, predicting the severity and outcome is important to CI patients and their families in guiding the treatment [25]. Many biomarkers offer the potential to predict the outcome of stroke, including TG/HDL-C [26], IL-6, NT-proBNP [27], and serum YKL-40 [28]. In this study, the genus Christensenellaceae_R-7_group and norank_f_Ruminococcaceae was significantly positive correlation with NIHSS1M (r = 0.514, P < 0.05; r = 0.449, P < 0.05) and mRS (r = 0.471, P < 0.05, r = 0.503, P < 0.01), respectively. On the other hand, the genus Enterobacter was significantly negative correlation with NIHSS1M (r = 0.449, P < 0.05) and mRS (r = 0.503, P < 0.01). Yin et al. found that opportunistic pathogens including Enterobacter enriched in stroke patients [13]. However, the relationship between genus Enterobacter and the severity or outcome is unknown. We also found that the genus Enterobacter was negative correlation with the HDL-C (r = − 0.425, P < 0.01), which was considered as a potential protective factor against CVD [29]. Thus, these results suggested that the HDL-C was positive correlation with the NIHSS at 1 month (r = 0.449, P < 0.05) and mRS and this is inconsistent with previous studies [30]. On the other hand, we also found that the genus Enterobacter increased in mild CI patients (NIHSS score ≤ 4). These patients usually have a better outcome than severe CI patients. These might be the reason that the genus Enterobacter was negative correlation with NIHSS 1 M and mRS. Until now, we knew little about the genus of Christensenellaceae_R-7_group and norank_f_Ruminococcaceae. Further study to identify the potent microbial biomarkers for predicting the outcome of stroke patients need to be done.
Conclusion
This study suggests that dysbiosis of the gut microbiota in CI patients with enriched SCFAs producer, including Odoribacter, Akkermansia. This dysbiosis was correlation with the outcomes and might be used as biomarkers for the long-term outcome which clearly deserves further study.
Methods
Subjects
Total of 30 CI patients were selected. They were recruited from Qilu Hospital (Jinan, China) between May 2017 and January 2018, and had been diagnosed by skull computed tomography examination. The subjects enrolled in this study received and written the informed consent. The National Institutes of Health Stroke Scale (NIHSS) > 4 was defined as severe stroke. In addition, 30 healthy volunteers who were examined to ensure that they had no metabolic, cardiovascular or cerebrovascular diseases or cancer were selected as the normal group. All these individuals did not receive antibiotics or probiotic before 4 weeks prior to the specimen collection. This study was approved by the Ethical Committee of the Qilu Hospital of Shandong University (Jinan, China).
Sample collection and 16S rRNA sequencing
The first available fecal samples of CI patients after admission (within 48 h) were collected. The samples and its genomic DNA were immediately frozen in liquid nitrogen and then stored at − 80 °C. Genomic DNA of microbes in fecal samples was extracted by CTAB assay. About 1 mg of each sample was used for DNA quantification by a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
For analyzing the microbial populations, amplification of the V1-V2 region of the 16S rRNA gene was performed via barcode-indexed primers 27F: 5′ AGAGTTTGATCMTGGCTC G, 338R-I 5′ GCWGCCTCCCGTAGGAGT, and 338R-II 5′ GCWGCCACCCGTAGGTGT). Then, we used the QIAquick PCR Purification Kit (Qiagen, Barcelona, Spain) to purified PCR amplicons and the products were quantified by a spectrophotometer as before. After that, it was pooled in equal concentration and 2 nM of pooled amplicons were then sequenced by Illumina MiSeq sequencer following the standard protocols.
Analysis of the microbiota
Trimmomatic was used to demultiplexed and quality-filtered the Raw data. The obtained data was merged by FLASH compliance with rules: (i) The reads attaining an average quality score less than 20 in a sliding window of 50 bp were deleted. (ii) Primers were only allowing 2 nucleotides mismatching, meanwhile reads containing ambiguous bases were deleted. (iii) Sequences containing overlap over 10 bp were merged through the overlap sequence.
Majorbio I-Sanger Cloud Platform was used to analyze the microbiota (https://www.i-sanger.com). We used the UPARSE to clustered the operational taxonomic units (OTUs) with a cutoff of 97% similarity (version 7.1) as well as UCHIME were used to identified and removed chimeric sequences. RDP Classifier was used to analyzed the taxonomy of each sequence through algorithm against the Silva (SSU128) 16S rRNA database by a confidence threshold of 70% (http://rdp.cme.msu.edu/).
We normalized the OTUs abundance of each sample based on the least sequences number of all samples and removed the rare OTUs. Then, it was used to analyze the alpha diversity and beta diversity by QIIME tool. Meanwhile, linear discriminant analysis (LDA) effect size (LEfSe) analyses were carried out through the LEfSe tool with an LDA of 2.0. The analysis of similarity (ANOSIM) test were used to evaluate the differences of microbial structure between CI patients and healthy control.
Biochemical assays
Blood samples were gathered from patients with stroke at admission and from healthy subjects at physical examination center. Serum levels of low-density lipoprotein (LDL), glucose of blood (GLU), high-density lipoprotein (HDL), uric acid (UA), triglycerides (TG), and homocysteine (HCY) were measured using standard techniques.
Statistic methods
All data are demonstrated as mean ± SD. The normality of the distribution had been demonstrated with Kolmogorov-Smirnoff statistic of the data. The Chi-square-tests was used to assess gender. The T-tests was used for analysis of continuous variables. Spearman’s rank correlation was used to analysis of statistical dependence of continuous variables. P-values < 0.05 were considered as statistically significant. Analyses were carried out through the SPSS statistical package, version 24.0.
Additional files
Table S1. The serum index of CI patient and healthy control. (PDF 140 kb)
Table S2. The severity and outcome of CI patients. (PDF 107 kb)
Acknowledgements
Not applicable.
Abbreviations
- CI
Cerebral ischemic stroke
- GLU
Glucose of blood
- HCY
Homocysteine
- HDL
High-density lipoprotein
- LDL
Low-density lipoprotein
- OTU
Operational taxonomic unit
- TG
Triglycerides
- UA
Uric acid
Authors’ contributions
All authors participated in the conception and design of the study; conceived and drafted the manuscript: NL, XCW; performed the experiments: NL, XCW, CCS, XWW, XJZ, NW and XBW; collected the basic patient information, clinical indicators, and imaging data: ML, XY, TW, and XLY; analyzed the data: CCS and XBW; revised the paper: CCS, XWW and XBW. All authors read and approved the final manuscript.
Funding
This research was supported by Shandong Provincial Natural Science Foundation, China (Grant NO. ZR2016HB37), the Shandong Medical Science and Technology Development Plan Project (NO. 2017WS433 and 2017WSA01044), Science and technology projects of Jinan health and family planning commission (NO. 2016-1-50).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Ethics approval and consent to participate
The protocol of this study was approved by the local Ethical Committee of the Qilu Hospital of Shandong University (KYLL-2017(KS)-088). All volunteers received information concerning their participation in the study and gave written informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Na Li, Xingcui Wang, Congcong Sun and Xinwei Wu contributed equally to this work.
References
- 1.Hossmann KA. Pathophysiology and therapy of experimental stroke. Cell Mol Neurobiol. 2006;26(7–8):1057–1083. doi: 10.1007/s10571-006-9008-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parr Emma, Ferdinand Phillip, Roffe Christine. Management of Acute Stroke in the Older Person. Geriatrics. 2017;2(3):27. doi: 10.3390/geriatrics2030027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chamorro Ángel, Urra Xabier, Planas Anna M. Infection After Acute Ischemic Stroke. Stroke. 2007;38(3):1097–1103. doi: 10.1161/01.STR.0000258346.68966.9d. [DOI] [PubMed] [Google Scholar]
- 4.Zeng X, et al. Higher risk of stroke is correlated with increased opportunistic pathogen load and reduced levels of butyrate-producing Bacteria in the gut. Front Cell Infect Microbiol. 2019;9:4. [DOI] [PMC free article] [PubMed]
- 5.Iadecola Costantino, Anrather Josef. The immunology of stroke: from mechanisms to translation. Nature Medicine. 2011;17(7):796–808. doi: 10.1038/nm.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Houlden A., Goldrick M., Brough D., Vizi E.S., Lénárt N., Martinecz B., Roberts I.S., Denes A. Brain injury induces specific changes in the caecal microbiota of mice via altered autonomic activity and mucoprotein production. Brain, Behavior, and Immunity. 2016;57:10–20. doi: 10.1016/j.bbi.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wen Shu Wen, Wong Connie H. Y. An unexplored brain-gut microbiota axis in stroke. Gut Microbes. 2017;8(6):601–606. doi: 10.1080/19490976.2017.1344809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stanley D, Moore RJ, Wong CHY. An insight into intestinal mucosal microbiota disruption after stroke. Sci Rep. 2018;8(1):568. [DOI] [PMC free article] [PubMed]
- 9.Benakis Corinne, Brea David, Caballero Silvia, Faraco Giuseppe, Moore Jamie, Murphy Michelle, Sita Giulia, Racchumi Gianfranco, Ling Lilan, Pamer Eric G, Iadecola Costantino, Anrather Josef. Commensal microbiota affects ischemic stroke outcome by regulating intestinal γδ T cells. Nature Medicine. 2016;22(5):516–523. doi: 10.1038/nm.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sherwin E, Dinan TG, Cryan JF. Recent developments in understanding the role of the gut microbiota in brain health and disease. Ann N Y Acad Sci. 2018;1420(1):5–25. doi: 10.1111/nyas.13416. [DOI] [PubMed] [Google Scholar]
- 11.Winek K, et al. Depletion of cultivatable gut microbiota by broad-Spectrum antibiotic pretreatment worsens outcome after murine stroke. Stroke. 2016;47(5):1354–1363. doi: 10.1161/STROKEAHA.115.011800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamashiro K, et al. Gut dysbiosis is associated with metabolism and systemic inflammation in patients with ischemic stroke. PLoS One. 2017;12(2):e0171521. doi: 10.1371/journal.pone.0171521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin J, et al. Dysbiosis of gut microbiota with reduced trimethylamine-N-oxide level in patients with large-artery atherosclerotic stroke or transient ischemic attack. J Am Heart Assoc. 2015;4(11):e002699. doi: 10.1161/JAHA.115.002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ji W, et al. Analysis of intestinal microbial communities of cerebral infarction and ischemia patients based on high throughput sequencing technology and glucose and lipid metabolism. Mol Med Rep. 2017;16(4):5413–5417. doi: 10.3892/mmr.2017.7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta) genomic data. MBio. 2014;5(2):e00889. doi: 10.1128/mBio.00889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cani PD, de Vos WM. Next-generation beneficial microbes: the case of Akkermansia muciniphila. Front Microbiol. 2017;8:1765. doi: 10.3389/fmicb.2017.01765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zoetendal EG, et al. Victivallis vadensis gen. Nov., sp. nov., a sugar-fermenting anaerobe from human faeces. Int J Syst Evol Microbiol. 2003;53(Pt 1):211–215. doi: 10.1099/ijs.0.02362-0. [DOI] [PubMed] [Google Scholar]
- 18.Cummings JH, et al. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28(10):1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koh A, et al. From dietary Fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165(6):1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 20.Zhai Q, et al. A next generation probiotic, Akkermansia muciniphila. Crit Rev Food Sci Nutr. 2018; 10.1080/10408398.2018.1517725. [Epub ahead of print]. [DOI] [PubMed]
- 21.Fang D, et al. Bifidobacterium pseudocatenulatum LI09 and Bifidobacterium catenulatum LI10 attenuate D-galactosamine-induced liver injury by modifying the gut microbiota. Sci Rep. 2017;7(1):8770. doi: 10.1038/s41598-017-09395-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng L, et al. Microbial-derived butyrate promotes epithelial barrier function through IL-10 receptor-dependent repression of Claudin-2. J Immunol. 2017;199(8):2976–2984. doi: 10.4049/jimmunol.1700105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukovac S, et al. Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. MBio. 2014;5(4). [DOI] [PMC free article] [PubMed]
- 24.Hanninen A, et al. Akkermansia muciniphila induces gut microbiota remodelling and controls islet autoimmunity in NOD mice. Gut. 2018;67(8):1445–1453. doi: 10.1136/gutjnl-2017-314508. [DOI] [PubMed] [Google Scholar]
- 25.Kamtchum-Tatuene J, Jickling GC. Blood biomarkers for stroke diagnosis and management. NeuroMolecular Med. 2019. [DOI] [PMC free article] [PubMed]
- 26.Deng QW, et al. Triglyceride to high-density lipoprotein cholesterol ratio predicts worse outcomes after acute ischaemic stroke. Eur J Neurol. 2017;24(2):283–291. doi: 10.1111/ene.13198. [DOI] [PubMed] [Google Scholar]
- 27.Dieplinger B, et al. Prognostic value of inflammatory and cardiovascular biomarkers for prediction of 90-day all-cause mortality after acute ischemic stroke-results from the Linz stroke unit study. Clin Chem. 2017;63(6):1101–1109. doi: 10.1373/clinchem.2016.269969. [DOI] [PubMed] [Google Scholar]
- 28.Chen XL, et al. Serum YKL-40, a prognostic marker in patients with large-artery atherosclerotic stroke. Acta Neurol Scand. 2017;136(2):97–102. doi: 10.1111/ane.12688. [DOI] [PubMed] [Google Scholar]
- 29.Tall AR. Plasma high density lipoproteins: therapeutic targeting and links to atherogenic inflammation. Atherosclerosis. 2018;276:39–43. doi: 10.1016/j.atherosclerosis.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 30.Prospective Studies C, et al. Blood cholesterol and vascular mortality by age, sex, and blood pressure: a meta-analysis of individual data from 61 prospective studies with 55,000 vascular deaths. Lancet. 2007;370(9602):1829–1839. doi: 10.1016/S0140-6736(07)61778-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The serum index of CI patient and healthy control. (PDF 140 kb)
Table S2. The severity and outcome of CI patients. (PDF 107 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this published article.